Abstract
DNA synthesis and postreplication mismatch repair were measuredin vitro using cell-free extracts from cultured human SY5Y neuroblastoma and WI38 fibroblast cells in different growth states. All extracts, including differentiated SY5Y and quiescent WI38 fibroblasts, catalyzed SV40 origin-dependent DNA synthesis, totally dependent on SV40 T-antigen. Thus, although differentiated neuroblastoma and quiescent fibroblasts cells were essentially nondividing, their extracts were competent for DNA replication using DNA polymerases δ, α, and possibly ε, with proliferating cell nuclear antigen. Nonreplicative DNA synthesis and lesion bypass by either α- or β-polymerases were detected independently in extracts using primed or gapped single-stranded DNA templates. Long-patch postreplication mismatch repair was measured for the first time in neuroblastoma cell-free extracts. Extracts from subconfluent and high-density SY5Y cells catalyzed postreplication mismatch repair with efficiencies comparable to those of HeLa cell extracts. No significant differences were observed in repair between SY5Y differentiated and undifferentiated cell extracts. Mismatch repair efficiencies were threefold lower in extracts from subconfluent WI38 cells, and repair in WI38 quiescent cells was fourfold less than in subconfluent cells, suggesting that mismatch repair may be regulated. The spectrum of mismatch repair in SY5Y extracts closely resembled the mismatch removal specificities of HeLa extracts: T · G and G · G mismatches were repaired most efficiently; C · A, A · A, A · G and a five-base loop were repaired with intermediate efficiency; repair of G · A, C · C, and T · T mismatches was extremely inefficient.
Keywords: DNA replication, mismatch repair, abasic lesion bypass, SY5Y neuroblastoma, WI38 fibroblasts, human cell-free extracts, differentiation, Pol α, Pol β
DNA constantly undergoes structural modifications in vivo, caused by exogenous and endogenous sources triggering DNA repair. Base excision repair (BER) of single nucleotide (nt) gaps eliminates apurinic and apyrimidinic (abasic) sites resulting from spontaneous loss of bases and corrects T · G mispairs caused by deamination of methylated cytosine (Bestor and Coxon, 1993; Lindahl, 1993). Nucleotide excision repair (NER) acts to remove UV damage, generating patches of 12 nt in prokaryotes and 29 nt in eukaryotes (Sancar, 1996). Postreplication mismatch repair (MMR), a long-patch nucleotide excision repair pathway, is required to maintain genetic integrity in actively dividing cells. In contrast to BER, MMR can correct a wide variety of mismatched base pairs and small unpaired loops requiring excision and resynthesis of hundreds to perhaps thousands of nucleotides and thus requires coordinated action of mismatch binding proteins, endonucleases and exonucleases, ligases, and DNA polymerases (Kolodner, 1996; Modrich and Lahue, 1996). MMR deficiencies in prokaryotes produce strong mutator phenotypes (Lu et al., 1983), and defective MMR may be the primary cause of pathogenicity in Escherichia coli and Salmonella strains (LeClerc et al., 1997). In eukaryotes, defective MMR is responsible for a wide spectrum of cancers, including a hereditary form of colon cancer (Leach et al., 1993; Bronner et al., 1994).
Mutations occurring in nondividing cells may also have important biological consequences. Frameshifts were found in nondividing E. coli (Foster, 1993) and recently in mRNA from Brattleboro rat neuronal cells (Evans et al., 1994) and in proteins in human brain neurons (Evans et al., 1996). The appearance of frameshifts and other mutations in nondividing cells implies a capacity for DNA synthesis, perhaps requiring MMR to reduce errors. MMR proteins can bind to natural base mispairs as well as to UV and chemically induced DNA lesions (Duckett et al., 1996), suggesting that MMR may not be restricted to repairing DNA polymerase-catalyzed errors.
Studies on DNA replication and repair activities in nondividing cell populations such as brain are complicated by issues of quantity and heterogeneity of cell types in a tissue. In this paper, we investigate DNA synthesis and MMR in vitro using two different cultured cell systems, SY5Y neuroblastoma cells and WI38 fibroblasts, in which essentially homogeneous cell populations can be uniformly induced to slow their growth rates or even to stop dividing. SY5Y, a subclone of the human neuroblastoma cell line SK-N-SH, is a nearly diploid cell population that undergoes terminal differentiation when treated with agents such as retinoic acid (Sidell et al., 1983), nerve growth factor (Perez-Polo et al., 1979; LoPresti et al., 1992), 12-O-tetradecanoylphorbol-13-acetate (TPA) (Pahlman et al., 1990), or staurosporine (Shea and Beermann, 1991; Jalava et al., 1993). WI38 cells are normal diploid human fibroblasts with a defined replicative lifespan in culture (Hayflick and Moorhead, 1961; Hayflick, 1965). After SV40 transformation, WI38 human fibroblasts cells become immortal. Each of these cell systems allows us to analyze origin-dependent DNA replication, abasic lesion bypass, and mismatch repair relative to cellular proliferative state.
MATERIALS AND METHODS
Nucleotides. Nonradioactive nucleotides were purchased from Pharmacia (Piscataway, NJ). [α-32P]dATP was purchased from ICN Radiochemicals (Costa Mesa, CA).
DNA substrates. The plasmid DNA used for replication reactions was pSV011, which contains the SV40 origin of replication (200 bp fragment) from HindIII to SphI (nucleotides 5171–128) in a pUC18 plasmid and is 2.9 kb long (Tsurimoto et al., 1989)
For studies of insertion opposite an abasic site, 42 mer DNA templates with and without a tetrahydrofuran abasic lesion (Eritja et al., 1987) and various primers were synthesized on an Applied Biosystems (Foster City, CA) 392 DNA/RNA synthesizer.
For mismatch repair studies M13mp2 mutants were a gift from Dr. T. Kunkel (National Institute of Environmental Health Sciences, Research Triangle Park, NC). M13mp2 single-stranded (+) DNA and RFI DNA were propagated in E. coli CSH50 and isolated. A double-stranded M13mp2 molecule containing a defined mispair, and a nick on the (−) strand at a unique AvaII site was prepared according to the methods of Kunkel and Soni (1988) and Thomas et al., (1991) .
Bacterial strains. Strains CSH 50 and NR 9162 were kindly provided by Dr. T. Kunkel (National Institute of Environmental Health Sciences).
Cell lines. HeLa S-3 cells, human cervical carcinoma cells, were obtained from the Cell Culture Core Facility at the Norris Cancer Hospital and Research Institute (Los Angeles, CA). SY5Y human neuroblastoma cells were a gift from Dr. C. E. Finch (Department of Gerontology, University of Southern California, Los Angeles, CA). WI38 cells (CCl 75; diploid human fetal lung fibroblasts) and WI38-SV40 cells (CCL 75.1; SV40-transformed WI38 fibroblasts) were purchased from the American Type Culture Collection (Rockville, MD).
Cell culture conditions and preparation of cell-free extracts. All cultures were maintained at 37°C in a humidified 5% CO2 atmosphere. HeLa S-3 cells were grown as spinner cultures in S-MEM (Sigma, St. Louis, MO) with 10% fetal calf serum. The cells were harvested by centrifugation in the upper part of their log phase growth. WI38 cells and SV40-transformed WI38 cells were grown in DMEM (Life Technologies, Gaithersburg, MD) containing 10% fetal calf serum (HyClone, Logan, UT), 4 mm glutamine, and antibiotics (50 U/ml penicillin and 50 μg/ml streptomycin). Human fibroblasts were used at passages 20–24 at which >90% were capable of DNA synthesis. “Proliferating” fibroblast cultures were actively dividing and subconfluent. “Quiescent,” nondividing, fibroblasts were harvested 72–96 hr after having been shifted from medium containing 10% serum into medium containing 0.2% serum.
SY5Y cells were cultured in RPMI 1640 supplemented with 10% fetal calf serum (Gemini Bioproducts, Calabassas, CA) and 10 μg/ml penicillin–streptomycin (Sigma). SY5Y cells were counted by trypan blue exclusion and plated at a density of 0.3 × 105 cells/cm2. Undifferentiated cells were harvested at a density of 1 × 105cells/cm2 (“subconfluent” cells) or 2.5 × 105 cells/cm2 (“high density”). To induce differentiation, SY5Y cells were exposed to 10 μm retinoic acid (Sigma) in growth medium, and the medium was changed every other day for 9 d. Differentiated cells had a diminished growth rate as well as morphological changes, notably extended neuritic processes and large growth cones.
HeLa and SY5Y cell-free extracts were prepared according to published protocols (Wold et al., 1989) with minor modifications. For differentiated SY5Y cell extracts, retinoic acid-treated cells were used. WI38 cell-free extracts were prepared as described (Krauss et al., 1997).
In vitro SV40 DNA replication. Replication reactions (50 μl) contained 30 mm HEPES, pH 7.8, 7 mmMgCl2, 200 μm each CTP, GTP, and UTP, 4 mm ATP, 100 μm each dCTP, dGTP, dTTP, and [α-32P]dATP (4000 cpm/pmol), 40 mm creatine phosphate, 100 μg/ml creatine phosphokinase, 600 ng of substrate DNA, 1 μg of SV40 large T-antigen (Chimerx, Milwaukee, WI), and the amount of extract protein indicated. Reactions without T-antigen were used as negative controls. After incubation at 37°C for different periods, reactions were quenched by adding an equal volume of “stop solution” (2% SDS, 2 mg/ml proteinase K, and 50 mm EDTA) and further incubation for 30–60 min at 37°C. Reaction products were purified by extraction with phenol–chloroform–isoamyl alcohol followed by ethanol precipitation. The DNA was resuspended in TE (10 mm Tris-HCl, pH 8.0, and 1 mm EDTA) and separated on a 1% agarose gel. Aliquots from the various samples were treated with DpnI (New England Biolabs, Beverly, MA). Restriction digests (20 μl) containing >200 ng of DNA and 20 units of DpnI were incubated for 30 min at 37°C. Dried gels were analyzed using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Primer extension and bypass of abasic lesions. 5′-End labeling of primers with 32P was performed as described previously by Efrati et al. (1997). Primers were annealed to 42 mer templates with or without a site-directed abasic lesion (1,4-anhydro-2-deoxy-d-ribitol). When assaying for polymerase α (Pol α) activity in the extracts, primer extension reactions were performed with no NaCl present and a 15 mer primer annealed to the template. When assaying polymerase β (Pol β) activity, the reaction buffer contained 100 mm NaCl (see below), and to the primer template used with Pol α conditions another downstream 22 mer 5′-phosphorylated oligonucleotide was annealed forming a 5 base gap. The 5′-phosphate group as well as a gap size of <6 bases are required for processive behavior by Pol β (Singhal and Wilson, 1993). Reactions were performed by mixing equal volumes (5 μl) of annealed primer template and deoxyribonucleotide in Pol α reaction buffer (20 mm Tris-HCl, pH 8.0, 10 mmMgCl2, 0.2 mg/ml bovine serum albumin, and 1 mm β-mercaptoethanol) or Pol β reaction buffer (35 mm Tris-HCl, pH 8.0, 6.7 mmMgCl2, 100 mm NaCl, 0.2 mg/ml bovine serum albumin, 1.5 mm dithiothreitol, and 2% glycerol). The final concentration of primer template was 3.8 nm. All four deoxynucleoside triphosphates (dNTPs) were provided at a final concentration of 100 μm. The indicated amount of protein (extract) was added, and reactions were incubated at 37°C for 10–45 min at which time they were terminated by adding two volumes of 20 mm EDTA and 95% formamide before heating to 100°C for 5 min. Samples were loaded onto 18% polyacrylamide gels, and the extended single-stranded DNA primers were separated according to length by electrophoresis. Gel bands were analyzed using a PhosphorImager (Molecular Dynamics).
Mismatch repair assay. A circular DNA M13mp2 substate contained a mismatch located at a single site in a lacZreporter gene (Thomas et al., 1991) and a site-specific nick, at a unique AvaII restriction site, in the (−) strand at position −264, where position +1 is the first transcribed nucleotide of the LacZ gene. Thus, the continuous strand is the (+) strand viral DNA, whereas the discontinuous strand is derived from the (−) strand of AvaII-linearized RF DNA. Reactions (25 μl) contained 30 mm HEPES, pH 7.8, 7 mmMgCl2, 200 μm each CTP, GTP, and UTP, 4 mm ATP, 100 μm each dCTP, dATP, dGTP, and dTTP, 40 mm creatine phosphate, pH 7.8, 100 μg/ml creatine phosphokinase, 15 mm sodium phosphate, pH 7.5, 1 fmol (5 ng) of substrate DNA, and the amount of extract protein indicated. Reactions were incubated for 30 min at 37°C. The reaction was terminated by the addition of an equal volume of stop solution (2% SDS, 2 mg/ml proteinase K, and 50 mm EDTA) and further incubation at 37°C for 30 min. The substrate DNA was recovered, purified, transfected into MMR-defective E. coli strain NR 9162 (mutS) by electroporation, and plated to score plaques as described (Kunkel and Soni, 1988; Thomas et al., 1991). Taking G · G88 as an example, repair of the (−) strand preserves a TGA codon in the (+) strand, giving rise to a white plaque phenotype; repair of the (+) strand changes the codon to TCA, yielding a blue plaque phenotype (Thomas et al., 1991). Importantly, mixed bursts represent unrepaired molecules. Thus, a decrease in the population of mixed burst plaques is a measure of overall repair efficiency (Thomas et al., 1991). G · G-specific repair of the nicked strand is observed as an increased number of white plaques and decrease in the number of both mixed and blue plaques (Thomas et al., 1991). Total repair efficiency is calculated as 100 × (1 − the ratio of percentages of mixed bursts from extract-treated and untreated samples).
RESULTS
SV40 origin-dependent DNA replication in neuroblastoma and fibroblast extracts
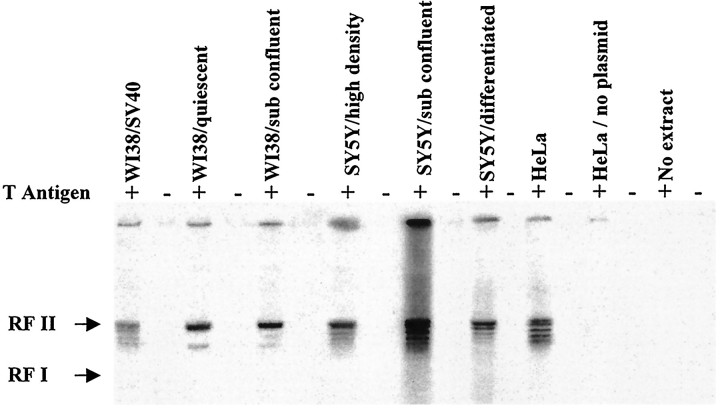
We investigated the capacity of cell-free extracts from SY5Y neuroblastoma cells and WI38 fibroblasts to initiate DNA synthesis from an SV40 origin of replication on a covalently closed, double-stranded plasmid, pSV011 (Fig. 1). In this assay system, DNA replication by HeLa cell-free extracts is T-antigen-dependent and requires the presence of the origin-bearing plasmid. The DNA products are predominantly RF II (nicked, double-stranded circles) along with a series of covalently closed topoisomers running between the RF II and RF I marker DNA, consistent with product DNA patterns reported previously (Wobbe et al., 1985; Wold et al., 1989; Waga et al., 1994). To verify that semiconservative replication was occurring, the product DNA was treated withDpn1, which only cuts fully methylated 5′-GATC sequences. As expected, hemimethylated or nonmethylated product DNAs resulting from semiconservative replication were resistant to cleavage (data not shown).
Fig. 1.
Origin-dependent replication using cell-free extracts from human fibroblasts and SY5Y neuroblastoma cells. DNA synthesis was performed as described in Materials and Methods (4 hr, 37°C) in the presence or absence of T-antigen. DNA was purified and subjected to electrophoresis in a 1% agarose gel, followed by ethidium bromide staining (0.2 mg/ml) and autoradiography. DNA standards were run in adjacent lanes. The positions of the different forms of replicated DNA are indicated at the left. HeLa extract was used as a positive control for replication that was both Tag- and plasmid-dependent. To verify that semiconservative replication was occurring, the product DNA was treated with DpnI, which only cuts fully methylated 5′-GATC sequences. Hemimethylated product DNAs resulting from semiconservative replication, dependent on the presence of T-antigen in the reaction, were refractory to cleavage (data not shown). Extract protein concentration in all the reactions involving WI38 cells was 2 mg/ml, in reactions with SY5Y was 1 mg/ml, and in reactions with HeLa was 3 mg/ml. The bands at thetop are caused by radioactive material remaining in the wells.
Significant DNA synthesis is observed using cell-free extracts prepared from SV40-transformed WI38 cells, from both proliferating and quiescent WI38 cells, and from subconfluent and high-density undifferentiated SY5Y cultures (Fig. 1). In all cases, the replication observed is T-antigen-dependent. An attractive feature of the neuroblastoma SY5Y cell line is that cells can be induced to differentiate on exposure to retinoic acid (Sidell et al., 1983) or other agents. When SY5Y cells are grown in the presence of retinoic acid, they proliferate at a much slower rate than control cultures. In addition, retinoic acid causes SY5Y cells to undergo distinct morphological alterations so that they take on the appearance of differentiated neurons. A comparison of SY5Y cells grown in either the absence or presence of retinoic acid is shown in Figure 2A. The typical morphology for undifferentiated neuroblasts is shown in Figure2A, left. After retinoic acid treatment, the cells extend long neuritic processes and present large growth cones (Fig. 2A, right).
Fig. 2.

Differentiation of SY5Y cells by retinoic acid.A, Phase-contrast micrographs of SY5Y cells with and without retinoic acid treatment. SY5Y cells were left untreated or were treated with 10 μm retinoic acid for 9 d. Retinoic acid induced neurite formation as well as enlargement of cell bodies (right) compared with untreated SY5Y cells (left). Magnification, 100×. B, Effect of retinoic acid on SY5Y cell growth. SY5Y cells were plated at a density of 0.3 × 105cells/cm2 in the presence (open squares) or absence (closed squares) of 10 μm retinoic acid for 9 d. Harvested cells were centrifuged, resuspended, and counted (trypan blue exclusion) each day. For each data point, the SE is ∼10% of the mean.
In retinoic acid-treated cultures, the number of viable cells (measured by trypan blue exclusion) did not increase with time, indicating an exceedingly slow proliferation rate on average (Fig.2B). In contrast, control cell populations grown without retinoic acid increased fivefold in 9 d. After differentiation by exposure to retinoic acid for 9 d, SY5Y cell-free extracts retained a full and active complement of replication enzymes capable of supporting origin-dependent DNA synthesis despite a very low level of proliferation (Fig. 1). Taken together, these data on SV40 origin-dependent synthesis demonstrate that neuroblastoma and fibroblast extracts contain proteins that can form an active replication complex. This complex includes cellular replication DNA polymerases δ and α (and possibly ε), along with the proliferating cell nuclear antigen (PCNA) that acts as a sliding processivity clamp for Pols δ and ε, clamp-loading accessory proteins (RFC), and human single-stranded binding protein (Waga et al., 1994; Podust et al., 1995).
DNA Pol β and Pol α in neuroblastoma and fibroblast cel extracts
The cellular repair DNA Pol β can be assayed independently of the other polymerases present in cell-free extracts by measuring primer extension activity under conditions optimal for Pol β. Pol β exhibits maximum activity and processivity for gaps up to six nt long (Singhal and Wilson, 1993), in which a 5′-phosphoryl group on the downstream primer functions as a direct contact point for the repair enzyme (Singhal and Wilson, 1993). We measured extension of a32P-labeled primer to fill in a short, five nt gap in the presence of 100 mm NaCl (Fig.3A). For comparison, the same extracts were used to copy DNA using conditions favoring synthesis by Pol α (Fig. 3B).
Fig. 3.
Abasic lesion bypass by cell-free extracts from human fibroblasts and neuroblastoma under A, Pol β-favoring conditions, and B, Pol α-favoring conditions. Reactions were performed and analyzed as described in Materials and Methods. Cell-free extracts containing 3 μg/reaction of WI38 or 2 μg/reaction of SY5Y or HeLa cells were added to annealed primer template. Left, Primer extension on a normal non-lesion-bearing template; right, primer extension on a template with an abasic site, X. The DNA template sequence is shown on the left of A and B.P, Position of the nonextended primer.
Primer termination bands are observed opposite each of the normal template sites (Fig. 3A, left). The low-intensity bands at central T and 5′-adjacent C sites indicate that the polymerase incorporates nucleotides with relatively high processivity at these sites, consistent with the behavior of purified Pol β (Singhal and Wilson, 1993; Efrati et al., 1997). A transient displacement or melting of the 5′ end of the downstream primer allows the incorporation of as many as seven nt when copying the natural template strand (Fig.3A, left). Similar weak primer displacement synthesis has been demonstrated using purified human Pol β on the same primer template construct shown in Figure 3 (data not shown). DNA synthesis is strongly inhibited by the chain terminator dideoxy-TTP, further documenting that Pol β rather than Pol α is primarily responsible for filling the five nt gap (Kornberg and Baker, 1992) (data not shown).
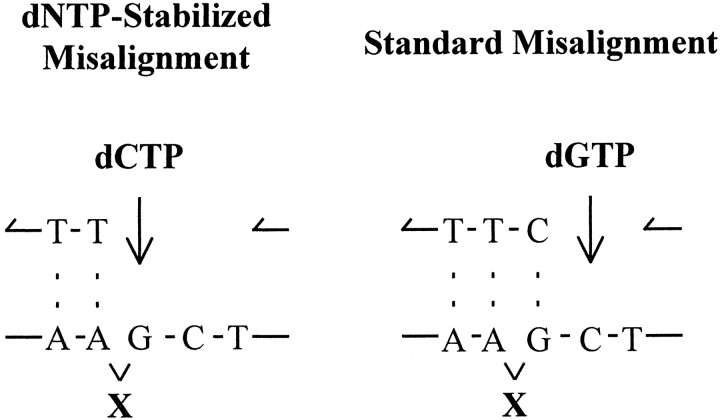
We also used cell-free extracts to assay Pol β activity on a template containing a single, site-directed abasic (tetrahydrofuran) lesion X in the center of the gap (Fig. 3A, right). Purified Pol β has been shown to copy abasic lesions (X) with relatively high efficiency and relaxed nucleotide insertion specificity (Efrati et al., 1997). We measured Pol β activity in cell extracts using a gapped template containing X (Fig. 3A, right) to see whether the distinctive properties of the purified repair polymerase in copying the noncoding lesion are discernible in crude samples containing Pol β in the presence of active replicative polymerases (Fig. 1). Unlike most polymerases, which act in accordance with an “A-rule,” strongly favoring incorporation of dAMP opposite X (Sagher and Strauss, 1983; Schaaper et al., 1983; Strauss, 1991), Pol β favors incorporation of a nucleotide complementary to a template base immediately downstream from the lesion, i.e., incorporation of dGMP opposite C by skipping over the lesion, generating a single-base deletion (Efrati et al., 1997). This skipping mode of synthesis that we have called “dNTP-stabilized misalignment” could occur if the tetrahydrofuran moiety were located out of the helical plane, as diagrammed in Figure 4 (Efrati et al., 1997).
Fig. 4.
Primer template misalignment models for synthesis at an abasic site. In the dNTP-Stabilized Misalignmentmodel (left), the abasic lesion is out of the helical plane with no H-bonds downstream to stabilize it. Instead, the incoming dNTP (dCTP), bound in the Pol β-active site, forms H-bonds with the downstream template base (G). In the Standard Misalignment model (right), the lesion is also out of the helical plane, but the misaligned template strand forms H-bonds with the primer on both sides of the lesion.
A combination of the two modes of synthesis, misalignment and direct incorporation opposite the lesion, produces a doublet band situated opposite X (Efrati et al., 1997) (Fig. 3A,right). The doublet band corresponding to the position of the abasic lesion X is typical of synthesis by purified Pol β on templates containing an abasic lesion. The lower band originates from incorporation occurring directly opposite the lesion, and the upper band corresponds to incorporation of dGMP opposite the downstream template C on a misaligned primer template DNA (Efrati et al., 1997) (Fig. 4); short single-stranded DNA terminated with G migrates more slowly than DNA terminating with any of the other three bases.
The dark band at the template A site immediately before X arises because the lesion provides a strong block to continued DNA synthesis (Fig. 3A, right). Polymerase dissociation at template site A is favored over nucleotide insertion opposite X. A comparison of the band intensities at the two template C sites downstream from the lesion shows that the intensity is generally greater opposite the C site adjacent to X compared with the last C site (Fig. 3A, right). A relatively weak band opposite the last C site implies that the five nt gap is filled by addition of four nt; i.e., the product DNA corresponds to a one nt deletion (Efrati et al., 1997). In contrast, the intense band opposite the last C in the unmodified DNA template demonstrates that the entire five nt gap is copied (Fig. 3A, left). All of the cell-free extracts contain polymerase activity characteristic of purified Pol β.
Pol α synthesis can be measured in primer extension reactions under Pol α-favoring conditions, without a downstream oligonucleotide (i.e., without a gap) and in the absence of NaCl (Fig. 3B). Under these conditions, we analyzed synthesis using normal templates as well as templates containing an abasic lesion. With both templates, significant synthesis occurred in HeLa cell extracts and in both proliferating subconfluent and high-density undifferentiated SY5Y cells. Growing and quiescent WI38 cell extracts, however, appeared to have lower Pol α activity. WI38 cells transformed by SV40 exhibited increased Pol α activity (Fig. 3B). Unlike Pol β, Pol α shows barely detectable synthesis opposite X (Fig. 3B,right). In all cases, DNA synthesis was essentially unaltered when ddNTPs were included in the reactions (data not shown). Because chain-terminating ddNTPs are incorporated into DNA by Pol β, but not by Pol α (Kornberg and Baker, 1992), this observation further confirms that this assay measures Pol α activity. In contrast to Pol α, levels of Pol β activity appears to be roughly similar in each extract (Fig. 3A), in agreement with previous data showing Pol β levels are insensitive to the proliferative state of the cell (Hubscher et al., 1977; Waser et al., 1979; Krauss and Linn, 1982; Wahl et al., 1988; Wong et al., 1988; Verri et al., 1992).
MMR
MMR proteins present in eukaryotic and prokaryotic cells can correct residual errors incurred after DNA replication. It is an open question whether MMR activities differ in dividing versus nondividing cells or are regulated during the cell cycle. To measure MMR in extracts from subconfluent, high-density, and differentiated neuroblastoma cells, we used an assay that detects repair of a single-base mismatch or deletion loop on M13 bacteriophage DNA (Thomas et al., 1991). This assay requires that extracts contain all necessary activities (i.e., mismatch binding proteins, 3′- and 5′-exonucleases, and DNA polymerases and helicases) to correct single-base mismatches and deletion loops (Modrich and Lahue, 1996).
An analysis of G · G88, T · G89, and C · C88 repair is shown in Table 1. The decrease in the number of mixed and nicked (−) strand plaques for G · G and T · G and concomitant increase in the number of (+) strand plaques demonstrate that strand-specific repair is occurring for both mismatches, and that G · G and T · G appear to be corrected with similar efficiencies. In the case of C · C, however, the number of mixed plaques remains constant after treatment with HeLa or SY5Y, indicating that the C · C mispair is not corrected.
Table 1.
Repair efficiencies of G·G, T·G and C·C mispairs by HeLa and SY5Y cell extracts
| Cell extract | Mispair [(−):(+)] | Total number of Plaques | % of plaques | Repair efficiency (%) | ||
|---|---|---|---|---|---|---|
| Mixed | (−) Strand | (+) Strand | ||||
| None | G·G88 | 380 | 28.5 | 55.3 | 16.3 | |
| HeLa | G·G88 | 252 | 8.3 | 33.7 | 58 | 70.9 |
| SY5Y | G·G88 | 310 | 9.9 | 35.9 | 54.2 | 65.3 |
| None | T·G89 | 470 | 27.7 | 40.7 | 31.6 | |
| HeLa | T·G89 | 311 | 6.1 | 25.4 | 68.5 | 78 |
| SY5Y | T·G89 | 280 | 8.2 | 27.5 | 64.3 | 70.4 |
| None | C·C88 | 202 | 30.1 | 43.3 | 26.6 | |
| HeLa | C·C88 | 353 | 30 | 47.2 | 22.8 | 0.3 |
| SY5Y | C·C88 | 397 | 29.2 | 47.1 | 23.7 | 3 |
Cell-free extracts (50 μg of protein) from HeLa and SY5Y cells were assayed for their ability to repair a single mismatch located in alacZ reporter gene on a nicked M13mp2 strand (see Materials and Methods) designated as the (−) strand. For repair efficiency calculations, see Materials and Methods.
To obtain a mismatch repair spectrum for SY5Y, we investigated 10 sets of mismatched heteroduplexes, comprising nine single-base mismatches and a five-base deletion loop (Fig. 5). HeLa cell extracts are used as a positive control for each substrate. The data show similar trends in repair efficiencies for the two cell types. T · G and G · G mismatches are repaired with highest efficiency. A · A, C · A, A · G, and a five-base deletion loop are repaired moderately well, but correction of G · A, C · C, and T · T mispairs is either not detectable or at most slightly above background (Fig. 5). The neuoroblastoma MMR spectrum (Fig. 5) is basically consistent with measurements using extracts from other eukaryotic organisms (Holmes et al., 1990; Thomas et al., 1991). Notably, repair of loops greater than four bases has been observed to occur in eukaryotes but not in E. coli (Umar et al., 1994;Modrich and Lahue, 1996).
Fig. 5.
Mismatch repair of different heteroduplexes with HeLa (filled bars) and SY5Y (open bars) cell-free extracts. Mismatch repair efficiencies of 10 different heteroduplexes (indicated at the bottom) were determined in reactions performed at 37°C for 30 min in the presence of 50 μg of protein. Repair of A · G mismatches was examined at two adjacent DNA positions. The calculation of repair efficiency is described in Materials and Methods. The data are plotted as the mean ± SE. n.d., Not detected.
A comparison of repair of the G · G mismatch was made for different preparations of SY5Y and WI38 cells. We find no significant difference in G · G repair for neuroblastoma extracts prepared from subconfluent and high-density undifferentiated SY5Y cultures. However, repair efficiencies for SY5Y cells appear to be measurably greater than those for cultured fibroblasts (Fig. 6). In comparison to SY5Y cells, extracts from normal and SV40-transformed fibroblasts exhibit an approximate twofold reduction in MMR on a protein basis, and MMR activity in quiescent fibroblasts is lower by approximately a factor of 8 (Fig. 6).
Fig. 6.
Repair efficiencies of G · G mismatches by HeLa, SY5Y, and WI38 cell-free extracts. Mismatch repair assays for a G · G mispair were carried out at 37°C for 30 min in the presence of 50 μg of protein for the extracts indicated belowthe histogram. The data are plotted as the mean ± SE. Percent MMR was calculated as described in Materials and Methods.
To address the question of whether there are significant differences in the MMR activities in undifferentiated versus differentiated cells, we used the SY5Y system to approximate a homogeneous population of human neuron-derived cells, i.e., uncontaminated with glial cells. Extracts were prepared from SY5Y cells cultured without or with retinoic acid to compare activities from proliferating undifferentiated with differentiated cell populations (Fig. 2). We found no significant difference in MMR efficiencies comparing proliferating subconfluent, high-density, or differentiated SY5Y cells (Figs. 6, 7).
Fig. 7.
. Mismatch repair of G · G, T · G, and C · C mismatches by cell-free extracts of undifferentiated SY5Y cells and SY5Y cells differentiated by retinoic acid treatment. Mismatch repair assays using three different heteroduplexes were performed at 37°C for 30 min in the presence of 50 μg of protein from extracts of SY5Y cells with or without retinoic acid treatment. The data are plotted as the mean ± SE. Percent MMR was calculated as described in Materials and Methods.
DISCUSSION
Understanding mechanisms underlying aberrant DNA synthesis and long-patch MMR in eukaryotic cells is important for elucidating normal and pathological processes. For example, at least 12 neurodegenerative diseases are characterized by large expansions of triplet repeats attributed to strand slippage during DNA replication (Boyer, 1995). Furthermore, frameshifts have been detected in mRNA and proteins in postmitotic neurons of the Brattleboro rat (Evans et al., 1994) and human brain (Evans et al., 1996). Future DNA sequence analysis of the relevant genes is needed to confirm that these frameshifts are encoded in the DNA. It has been proposed (Bridges, 1997; Finch and Goodman, 1997) that nonreplicative DNA synthetic processes may be responsible for generating such mutations in nondividing prokaryotic (Foster, 1993) and eukaryotic cells.
Adult brain neurons were recently shown to correct T · G mismatches, arising from deamination of 5-methyl cytosine, by the BER pathway (Brooks et al., 1996). Although Brooks et al. (1996) suggest that G · T are the only types of mismatches that can arise in nondividing cells, there are other potential mechanisms such as spontaneous loss of DNA bases (Lindahl, 1982) and oxidative DNA damage (Farr and Kogoma, 1991; Pacifici and Davies, 1991; Michaels and Miller, 1992) that can give rise to mismatches in nondividing cells. Mismatches can also be generated during transcription-coupled repair (Hanawalt, 1994), a process that involves the action of MMR (Mellon and Champe, 1996). It is therefore important to investigate replication and repair pathways using human-derived cell systems as an in vitromodel to investigate the biochemistry of replication and postreplication repair.
Identification of individual DNA polymerases in extracts from cultured neuroblastoma and fibroblast cells
Studies on DNA polymerases purified from mammalian cells show that replicative polymerases, Pol α and Pol δ, appear to be regulated as a function of cell cycle (Wahl et al., 1988; Zeng et al., 1994), as does PCNA, the Pol δ-associated sliding clamp (Morris and Mathews, 1989). However, levels of Pol β, involved in DNA repair (Waser et al., 1979; Frosina et al., 1996; Sobel et al., 1996), remain relatively constant (Verri et al., 1992; Hubscher et al., 1977). We asked whether extracts prepared from neuroblastoma and fibroblast cells in various replicative states can catalyze origin-dependent, semiconservative DNA replication requiring Pols δ and α, PCNA, RFC, and RFA (Waga et al., 1994). Our data definitively demonstrate that all extracts examined, even differentiated neuroblastoma cells, are capable of replication in vitro (Fig. 1).
We analyzed the behavior of Pol β in crude extracts and found that its properties resemble those of the purified enzyme; it carries out processive gap-filling synthesis, catalyzes efficient bypass at an abasic lesion (Figs. 3A, 4) (Efrati et al., 1997), and is inhibited by incorporation of a chain-terminating ddNTP (data not shown). All cell extracts examined contain approximately similar levels of Pol β activity. In contrast, assays under conditions optimal for Pol α activity show variable degrees of synthesis with different cell-free extracts (Fig. 3B). The reduced levels of incorporation found for WI38 quiescent and subconfluent cells may reflect the presence of a majority of nondividing cells. Mixing experiments did not detect inhibitors of replication in extracts from nondividing cells. The presence of similar levels of Pol α-like activities in subconfluent and high-density populations of SY5Y neuroblastoma cells is consistent with the inability of undifferentiated SY5Y cells to become contact-inhibited or to cease dividing.
Postreplication mismatch repair activity in extracts from cultured neuroblastoma and fibroblast cells
In neuroblastoma cell extracts, we observed DNA strand-specific mismatch repair for T · G, G · G, C · A, A · A, and A · G mispairs and a five base loop; G · A, C · C, and T · T mispairs are essentially not repaired (Fig.5). Efficient repair of T · G mismatches in neuroblastoma extracts is noteworthy in light of the observation that adult brain neurons can catalyze T · G → C · G repair (Brooks et al., 1996). The substrates used were short DNA heteroduplexes (34 mer) containing a single mismatched base pair to detect enzymes involved in BER. In contrast, the nicked M13 DNA heteroduplexes in our study measure long-patch MMR. MMR repair tracts can be hundreds of bases, reflecting the distance between a nick and the mismatch (Modrich and Lahue, 1996). Nicking at C · A mismatches was not observed in the study of Brooks et al. (1996), whereas significant C · A mismatch repair is observed in MMR assay (Fig. 5). However, G · A mismatches do not appear to be corrected by either BER (Brooks et al., 1996) or NER (Fig.5).
The mismatch repair profile for neuroblastoma cell extracts (Fig. 5), closely parallels that observed for HeLa as well as other eukaryotic cells (Thomas et al., 1991; Umar et al., 1994). Measurable C · C repair appears to be absent in neuroblastoma cells (Fig. 5), as has been reported for other prokaryotic and eukaryotic systems (Thomas et al., 1991); exceptions include CV1 and Drosophila cell extracts that repair C · C heteroduplexes with high efficiency (Modrich and Lahue, 1996) (A. Kaur, M. F. Goodman, and J. Tower, unpublished observations).
The degree to which MMR proteins may be regulated in eukaryotes is not known. The E. coli Mut S mismatch binding protein decreases 10-fold when bacteria enter late-stationary phase or are starved for a carbon source (Feng et al., 1996). Using G · G mismatches as a representative measure of mismatch repair efficiency, we find no evidence for regulation of mismatch repair in subconfluent versus high-density undifferentiated SY5Y neuroblastoma cells (Fig. 6). We also observe no measurable difference in G · G, T · G, and C · C mismatch repair efficiencies comparing SY5Y differentiated and undifferentiated cells (Fig. 7).
Thus, in contrast to E. coli in which the mismatch recognition protein Mut S is 10-fold lower in stationary compared with exponentially growing cells (Feng et al., 1996), it appears that postreplication mismatch repair may be present at roughly similar levels in dividing and nondividing SY5Y cells. It is well documented that DNA synthesis is occurring in nondividing prokaryotic cells (Foster, 1993) and likely to be occurring in eukaryotic cells (Evans et al., 1994, 1996). Perhaps prokaryotes can “afford” to downregulate MMR, owing to the high fidelity of proofreading-proficient Pols II and III, shown to be involved in DNA synthesis in nondividing E. coli (Foster et al., 1995). However, in nondividing eukaryotic cells, it might be necessary to retain active MMR to counteract potentially high levels of proofreading-deficient Pol β-catalyzed errors.
MMR in WI38 cells is lower than in neuroblastoma, with an approximate fourfold decrease in G · G mismatch repair in quiescent versus actively growing fibroblasts (Fig. 6). This observation, although clearly not definitive, is consistent with the observation that some MMR proteins are regulated during the cell cycle. Pol δ and PCNA are examples of regulated replication proteins that are also required for HeLa cell mismatch repair in vitro (Umar et al., 1996;Longley et al., 1997). It is somewhat surprising that we find no change in mismatch repair efficiency in differentiated and nondifferentiated SY5Y cells. A possible explanation is that although the levels of both PCNA and Pol δ decrease after treatment with retinoic acid (Verri et al., 1992), these transformed cells may retain sufficient levels of both to provide ample MMR activity.
Our data demonstrate that both origin-dependent replication and DNA repair synthesis can be analyzed in cell-free extracts from cultured, transformed dividing and differentiated neuronal cells and from normal human diploid fibroblasts in various growth states. We show for the first time that postreplication mismatch repair is taking place via a long-patch NER pathway in cells of neuronal origin; our data complement an earlier demonstration of the occurrence of BER in adult neurons (Brooks et al., 1996). Although SY5Y cells are neuronally derived and display subsets of neuronal phenotypes (Perez-Polo et al., 1979;LoPresti et al., 1992), measurements in similarly prepared cell-free extracts from highly purified primary cultures of neurons are required to establish whether MMR activities in SY5Y cells are indeed comparable to those of neurons. The involvement of replication enzymes in postreplication mismatch repair was discovered using E. coliand HeLa as model systems (Modrich and Lahue, 1996; Umar et al., 1996;Longley et al., 1997). Because we have shown that analogous activities can be measured in neuronal and fibroblast cell systems, it may now be possible to investigate, at a biochemical level, mutational events occurring in the brain and in an aging model system.
Footnotes
This work was supported by National Institutes of Health Grants AG11398 and DK32094. We express our gratitude to Dr. Caleb Finch and Dr. Irina Rozovsky for their generous advice regarding cell culture and for numerous enlightening discussions.
Correspondence should be addressed to Dr. Myron F. Goodman, Department of Biological Sciences, University of Southern California, SHS Room 172, University Park, Los Angeles, CA 90089-1340.
REFERENCES
- 1.Bestor T, Coxon A. The pros and cons of DNA methylation. Curr Biol. 1993;3:384–386. doi: 10.1016/0960-9822(93)90209-7. [DOI] [PubMed] [Google Scholar]
- 2.Boyer JC, Umar A, Risinger JI, Lipford JR, Kane M, Yin S, Barrett JC, Kolodner RD, Kunkle TA. Microsatellite instability, mismatch repair deficiency, and genetic defects in human cancer cell lines. Cancer Res. 1995;55:6063–6070. [PubMed] [Google Scholar]
- 3.Bridges BA. DNA turnover and mutation in resting cells. BioEssays. 1997;19:347–352. doi: 10.1002/bies.950190412. [DOI] [PubMed] [Google Scholar]
- 4.Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, Tannergard P, Bollag RJ, Godwin AR, Ward DC, Nordenskjold M, Fishel R, Kolodner R, Liskay RM. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 5.Brooks PJ, Marietta C, Goldman D. DNA Mismatch repair and DNA methylation in adult brain neurons. J Neurosci. 1996;16:939–945. doi: 10.1523/JNEUROSCI.16-03-00939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duckett DR, Drummond JT, Murchie AIH, Reardon JT, Sancar A, Lilley DMJ, Modrich P. Human MutS-alpha recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc Natl Acad Sci USA. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Efrati E, Tocco G, Eritja R, Wilson SH, Goodman MF. Abasic translesion synthesis by DNA polymerase β violates the “A-rule.”. J Biol Chem. 1997;272:2559–2569. doi: 10.1074/jbc.272.4.2559. [DOI] [PubMed] [Google Scholar]
- 8.Eritja R, Walker PA, Randall SK, Goodman MF, Kaplan BE. Synthesis of oligonucleotides containing the abasic site model compound 1,4-anhydro-2-deoxy-d-ribitol. Nucleosides & Nucleotides. 1987;6:803–814. [Google Scholar]
- 9.Evans DAP, Vanderkleij AAM, Sonnemans MAF, Burbach JPH, Van Leeuwen FW. Frameshift mutations at two hotspots in vasopressin transcripts in post-mitotic neurons. Proc Natl Acad Sci USA. 1994;91:6059–6063. doi: 10.1073/pnas.91.13.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans DAP, Burbach JPH, Swaab DF, Van Leeuwen FW. Mutant vasopressin precursors in the human hypothalamus - evidence for neuronal somatic mutations in man. Neuroscience. 1996;71:1025–1030. doi: 10.1016/0306-4522(95)00501-3. [DOI] [PubMed] [Google Scholar]
- 11.Farr SB, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng G, Ho-Ching TT, Winkler ME. Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. J Bacteriol. 1996;178:2388–2396. doi: 10.1128/jb.178.8.2388-2396.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finch CE, Goodman MF. Relevance of “adaptive” mutations arising in non-dividing cells of microorganisms to age-related changes in mutant phenotypes of neurons. Trends Neurosci. 1997;20:501–507. doi: 10.1016/s0166-2236(97)01143-0. [DOI] [PubMed] [Google Scholar]
- 14.Foster PL. Adaptive mutation-the uses of adversity. Annu Rev Microbiol. 1993;47:467–504. doi: 10.1146/annurev.mi.47.100193.002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster PL, Gudmundsson G, Trimarchi JM, Cai H, Goodman MF. Proofreading-defective DNA polymerase II increases adaptive mutation in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:7951–7955. doi: 10.1073/pnas.92.17.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 17.Hanawalt PC. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 18.Hayflick L. The limited in vitro life span of human diploid strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 19.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 20.Holmes J, Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubscher U, Kuenzle CC, Spadari S. Variations of DNA polymerases α, β and γ during perinatal tissue growth and differentiation. Nucleic Acids Res. 1977;4:2917–2929. doi: 10.1093/nar/4.8.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jalava A, Lintunen M, Heikkila J. Protein kinase C-alpha but not protein kinase C-epsilon is differentially downregulated by bryostatin 1 and tetradecanoyl phorbol 13-acetate in SH-SY5Y human neuroblastoma. Biochem Biophys Res Commun. 1993;191:472–478. doi: 10.1006/bbrc.1993.1242. [DOI] [PubMed] [Google Scholar]
- 23.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 24.Kornberg A, Baker TA. DNA replication, Ed 2. Freeman; New York: 1992. [Google Scholar]
- 25.Krauss SW, Linn S. Changes in DNA polymerases α, β, and γ during the replicative life span of cultured human fibroblasts. Biochemistry. 1982;21:1002–1009. doi: 10.1021/bi00534a027. [DOI] [PubMed] [Google Scholar]
- 26.Krauss SW, Larabell CA, Lockett S, Gascard P, Penman S, Mohandas N, Chasis JA. Structural protein-4.1 in the nucleus of human cells—dynamic rearrangements during cell division. J Cell Biol. 1997;137:275–289. doi: 10.1083/jcb.137.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunkel TA, Soni A. Mutagenesis by transient misalignment. J Biol Chem. 1988;263:14784–14789. [PubMed] [Google Scholar]
- 28.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen LA, Nystrom-Lahti M, Guan XY, Zhang J, Meltzer PS, Yu JW, Kao FT, Chen DJ, Cerosaletti KM, Fournier REK, Todd S, Lewis T, Leach RJ, Naylor SL, Weissenbach J, Mecklin JP, Jarvinen H, Petersen GM, Hamilton SR, Green J, Jass J, Watson P, Lynch HT, Trent JM, de la Chapelle A, Kinsler KW, Vogelstein B. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 29.LeClerc EJ, Li B, Payne WL, Cebula TA. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1997;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 30.Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- 31.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:702–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 32.Longley MJ, Pierce AJ, Modrich P. DNA polymerase δ is required for human mismatch repair in vitro. J Biol Chem. 1997;272:10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 33.LoPresti P, Poluha W, Poluha DK, Drinkwater E, Ross AH. Neuronal differentiation triggered by blocking cell proliferation. Cell Growth Differ. 1992;3:627–635. [PubMed] [Google Scholar]
- 34.Lu A-L, Clark S, Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci USA. 1983;80:4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mellon I, Champe GN. Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide excision repair of the lactose operon in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:1292–1297. doi: 10.1073/pnas.93.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 38.Morris GF, Mathews MB. Regulation of proliferating cell nuclear antigen during the cell cycle. J Biol Chem. 1989;264:13856–13864. [PubMed] [Google Scholar]
- 39.Pacifici RE, Davies KJ. Protein, lipid and DNA repair systems in oxidative stress: the free-radical theory of aging revisited. Gerontology. 1991;37:166–180. doi: 10.1159/000213257. [DOI] [PubMed] [Google Scholar]
- 40.Pahlman S, Mamaeva S, Meyerson G, Mattsson ME, Bjelfman C, Ortoft E, Hammerling U. Human neuroblastoma cells in culture: a model for neuronal cell differentiation and function. Acta Physiol Scand. 1990;552:25–37. [PubMed] [Google Scholar]
- 41.Perez-Polo JR, Werrbach-Perez K, Tiffany-Castiglioni E. A human clonal cell line model of differentiating neurons. Dev Biol. 1979;71:341–355. doi: 10.1016/0012-1606(79)90174-x. [DOI] [PubMed] [Google Scholar]
- 42.Podust LM, Podust VN, Sogo JM, Hubscher U. Mammalian DNA polymerase auxiliary proteins: analysis of replication factor C-catalyzed proliferating cell nuclear antigen loading onto circular double-stranded DNA. Mol Cell Biol. 1995;15:3072–3081. doi: 10.1128/mcb.15.6.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sagher D, Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in DNA during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983;22:4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- 44.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 45.Schaaper RM, Kunkel TA, Loeb LA. Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc Natl Acad Sci USA. 1983;80:487–491. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shea TB, Beermann ML. Staurosporine-induced morphological differentiation of human neuroblastoma cells. Cell Biol Int Rep. 1991;15:161–168. doi: 10.1016/0309-1651(91)90107-t. [DOI] [PubMed] [Google Scholar]
- 47.Sidell N, Altman A, Haussler MR, Seeger RC. Effects of retinoic acid (RA) on the growth and phenotypic expression of several human neuroblastoma cell lines. Exp Cell Res. 1983;148:21–30. doi: 10.1016/0014-4827(83)90184-2. [DOI] [PubMed] [Google Scholar]
- 48.Singhal RK, Wilson SH. Short gap-filling synthesis by DNA polymerase β is processive. J Biol Chem. 1993;268:15906–15911. [PubMed] [Google Scholar]
- 49.Sobel RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-β in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 50.Strauss BS. The ’A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of noninstructional lesions? BioEssays. 1991;13:79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- 51.Thomas DC, Roberts JD, Kunkel TA. Heteroduplex repair in extracts of human HeLa cells. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- 52.Tsurimoto T, Fairman MP, Stillman B. Simian virus 40 DNA replication in vitro: identification of multiple stages of initiation. Mol Cell Biol. 1989;9:3839–3849. doi: 10.1128/mcb.9.9.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Umar A, Boyer JC, Kunkel TA. DNA loop repair by human cell extracts. Science. 1994;266:814–816. doi: 10.1126/science.7973637. [DOI] [PubMed] [Google Scholar]
- 54.Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 55.Verri A, Verzeletti S, Mazzarello P, Spadari S, Negri M, Bunone G, Della Valle G, Hubscher U, Focher F. DNA synthesis enzymes and proliferating cell nuclear antigen in normal and neoplastic nerve cells. Anticancer Res. 1992;12:1099–1106. [PubMed] [Google Scholar]
- 56.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 57.Wahl AF, Geis AM, Spain BH, Wong SW, Korn D, Wang TS-F. Gene expression of human DNA polymerase α during cell proliferation and the cell cycle. Mol Cell Biol. 1988;8:5016–5025. doi: 10.1128/mcb.8.11.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waser J, Hubscher U, Kuenzle CC, Spadari S. DNA polymerase β from brain neurons is a repair enzyme. Eur J Biochem. 1979;97:361–368. doi: 10.1111/j.1432-1033.1979.tb13122.x. [DOI] [PubMed] [Google Scholar]
- 59.Wobbe CR, Dean F, Weissbach L, Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci USA. 1985;82:5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wold MS, Weinberg DH, Virshup DM, Li JJ, Kelly TJ. Identification of cellular proteins required for simian virus 40 DNA replication. J Biol Chem. 1989;264:2801–2809. [PubMed] [Google Scholar]
- 61.Wong SW, Wahl AF, Yuan P-M, Arai N, Pearson BE, Arai K-I, Korn D, Hunkapiller MW, Wang TS-F. Human DNA polymerase α gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988;7:37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng X-R, Hao H, Jiang Y, Lee MYWT. Regulation of human DNA polymerase δ during the cell cycle. J Biol Chem. 1994;269:24027–24033. [PubMed] [Google Scholar]