Abstract
The circadian timekeeping system of Drosophilafunctions from the first larval instar (L1) onward but is not known to require the expression of clock genes in larvae. We show that period (per) andtimeless (tim) are rhythmically expressed in several groups of neurons in the larval CNS both in light/dark cycles and in constant dark conditions. Among the clock gene-expressing cells there is a subset of the putative pacemaker neurons, the “lateral neurons” (LNs), that have been analyzed mainly in adult flies. Like the adult LNs, the larval ones are also immunoreactive to a peptide called pigment-dispersing hormone. Their putative dendritic trees were found to be in close proximity to the terminals of the larval optic nerve Bolwig’s nerve, possibly receiving photic input from the larval eyes. The LNs are the only larval cells that maintain a strong cycling in PER from L1 onward, throughout metamorphosis and into adulthood. Therefore, they are the best candidates for being pacemaker neurons responsible for the larval “time memory” (inferred from previous experiments). In addition to the LNs, a subset of the larval dorsal neurons (DNLs) expresses per andtim. Intriguingly, two neurons of this DNL group cycle in PER and TIM immunoreactivity almost in antiphase to the other DNLs and to the LNs. Thus, the temporal expression of per andtim are regulated differentially in different cells. Furthermore, the light sensitivity associated with levels of the TIM protein is different from that in the heads of adultDrosophila.
Keywords: larval CNS, period, timeless, pigment-dispersing hormone, pacemaker, circadian, Bolwig’s nerve
The period(per) and timeless (tim) genes specify important components of the circadian clock inDrosophila. This is supported by the isolation of arrhythmic as well as period-altered mutants at both the per andtim loci (Konopka and Benzer, 1971; Sehgal et al., 1994;Rutila et al., 1996), cycling of the abundance of their RNA and protein products, and the fact that these molecular oscillations are dependent on the normal functions of both genes (Hardin et al., 1990; Zerr et al., 1990; Sehgal et al., 1994, 1995; Zeng et al., 1994, 1996; Price et al., 1995; Hunter-Ensor et al., 1996; Myers et al., 1996).
Regulation of these molecular oscillations is thought to involve transcriptional (Hardin et al., 1992) as well as post-transcriptional mechanisms (Lee et al., 1996; Zeng et al., 1996; Dembinska et al., 1997). Temporally varying transcriptional control also may be responsible for the cycling of putative clock output genes (Van Gelder et al., 1995; Van Gelder and Krasnow, 1996). These are some of the reasons for the intense focus of recent studies on molecular features of circadian rhythmicity in Drosophila.
The adult head has been used as the tissue source for these studies because it controls the best-studied circadian phenotype, the fly’s locomotor activity rhythm (Konopka et al., 1983; Ewer et al., 1992; Frisch et al., 1994). In the adult head, immunocytochemical studies showed that per is rhythmically expressed in a variety of cells, including the putative pacemaker cells—the lateral neurons (LNs) located between the central brain and the optic lobes (Siwicki et al., 1988; Zerr et al., 1990; Ewer et al., 1992; Frisch et al., 1994). Two groups of LNs can be distinguished, a more dorsal group (LNd) and a ventral one (LNv) (Ewer et al., 1992). Most of the LNvs are also immunoreactive to antibodies against pigment-dispersing hormone (PDH,Helfrich-Förster, 1995). Preliminary immunocytochemical studies using an antiserum against TIM indicate that tim also is rhythmically expressed at least in the LNvs (Hunter-Ensor et al., 1996).
Whereas PER and TIM oscillations are well documented in the adult brains and per expression in the CNS (including the brain hemispheres) of mid to late embryos and pupae has been reported (Liu et al., 1988; Siwicki et al., 1988; Konopka et al., 1995), previous attempts to detect a significant level of per expression in larvae have been unsuccessful, in large part (Bargiello and Young, 1984; James et al., 1986; Bargiello et al., 1987; Liu et al., 1988;Siwicki et al., 1988). These negative, descriptive results are intriguingly problematical given the following observation: the phase of eclosion (Brett, 1955) and of adult activity (Sehgal et al., 1992) rhythms can be set by light pulses delivered to animals entering or progressing through the first larval stage (L1). These results strongly suggest the presence of a clock and therefore imply that some clock genes are expressed during postembryonic development.
Here we demonstrate that the products of per andtim are detectable in a limited number of neurons in the larval CNS; the expression patterns in several such cells is cyclical. Among these neurons five laterally located ones (which seem to be precursors of the aforementioned LNvs) expressper from early larval stage through metamorphosis, suggesting that they may be responsible for the larval time memory implied above. In another cluster of larval cells, oscillations of PER and TIM were found to occur almost in antiphase from that in all of the other cells that express per in a cyclical manner—either elsewhere in the larval brain or in the various tissues of the adult. Application of further cell-marking reagents leads us to propose a possible light entrainment pathway that may connect larval photoreceptors to the clock neurons.
MATERIALS AND METHODS
Strains
Flies were grown in a light/dark (LD) cycle of 12 hr L and 12 hr D at 25 or 22°C. A wild-type Canton-S strain, or awhite-mutant strain previously made isogenic with Canton-S, were the sources of “clock normal” (per+tim+) in the anti-PER, anti-TIM, and anti-PDH immunostaining and in the control for endogenous galactosidase activity (described below, used in conjunction with certain transgenic strains).per01 was used as a control for anti-PER immunohistochemistry as well as for the study of TIM cycling in this genetic background.tim01 flies (also carrying therosy506 eye marker) were used as a control for anti-TIM immunohistochemistry [see Sehgal et al. (1994) for the first report of this tim null mutant].
Larvae from transgenic strains involving two differentper-lacZ fusion genes, SG and BG, were stained for β-galactosidase (β-gal) activity (encoded by lacZ) to assess per-reported expression in the larval CNS. The fusion genes share the same 4.2 kb 5′ upstream region (5′-flanking), but they differ in the length of per coding sequences (Liu et al., 1988; Stanewsky et al., 1997a). SG, which encodes the N-terminal 637 amino acids of the PER polypeptide, is expressed in a noncyclical manner in all of the classes of theper-expressing cells identified so far in the adult head (Liu et al., 1988; Ewer et al., 1992; Stanewsky et al., 1997a). The staining intensity of the SG-encoded protein in most of these cells is stronger than signals of anti-PER staining (Ewer et al., 1992). Therefore, detection of β-gal expression in this line was considered to be the most sensitive way of studying the spatial expression pattern of this gene. Hence, application of the SG lines was emphasized to maximize the chance of revealing per expression pattern inL1 larvae.
In addition to the SG strains, we applied anotherper-β-gal fusion gene construct called BG. It contains an additional 230 amino acids’ worth of per-coding sequence extending C-terminal to those encoded by SG (Dembinska et al., 1997;Stanewsky et al., 1997a). In adult heads the level of β-gal in BG lines cycles with a similar phase and amplitude, as does endogenous PER in wild-type (Dembinska et al., 1997; Stanewsky et al., 1997a). It also was found that the β-gal expression pattern in BG more closely reflects endogenous PER expression (better than in the case of SG). For instance, the (inferred) quantitative levels of both BG and endogenous PER are higher in LNs than in CNS glia, but the reverse is true for SG (Ewer et al., 1992; Stanewsky et al., 1997a). However, no qualitative difference in the types of cells stained has been detected between BG and SG lines in adults. Therefore, the BG fusion gene transformant also was used to study per expression in larvae.
To check for chromosomal position effects, we used separate transgenic lines with different chromosomal insertion sites for the SG and BG transgenes: SG3 (chromosome 2) and SG10 (X-chromosome), BG6a (chromosome 2) and BG/TM2 (chromosome3); the TM2 abbreviation refers to a third chromosomal balancer chromosome, applied here because this particular insert of BG transgene is homozygous lethal. With the exception of BG/TM2, all of the transformants were studied in developing animals homozygous for the transgenes.
Staging of larvae and pupae
To obtain L1 individuals of defined age, we collected freshly emerged larvae during a 10 min period in 1 hr intervals. They were allowed to grow until they reached the desired age, and then they were dissected (after 0, 1, 2, 3, 4, 5, 8, 12, 15, and 20 hr). With the use of SG strains, this could be done at any time during the light phase of the LD cycle, because the per-lacZfusion product is present at a high level throughout the daily cycle (Stanewsky et al., 1997a). In BG, the per-lacZfusion product cycles with a peak at ∼Zeitgeber Time (ZT) 21 (Stanewsky et al., 1997a). Therefore, dissections were done at ZT 0 (by convention, the beginning of the light phase) to minimize the effect of light during dissection. Dissections also were done at ZT 12 (the beginning of the dark phase). To obtain 1- to 8-hr-old L1animals at ZT 0, we collected freshly hatched larvae at night under red light. Animals in the second (L2) and third (L3) larval instar stages were determined on the basis of the morphology of the anterior spiracles and the mandibular hooks (Bodenstein, 1950). Because the L3 stage lasts ∼2 d at 25°C, it was possible to investigate the cycling of PER and the per-lacZ fusion product in BG without further staging of the larvae. With regard to pupae, the white prepupa was considered as 0-hr-old pupa (0%-pupa). White prepupae of BG were collected at ZT 0, ZT 6, and ZT 12 and then dissected in 12–24 hr intervals at ZT 0, ZT 6, or ZT 12. The ages of the pupae investigated for per-lacZ expression were 12 hr (10% = the first 10% of the pupal period), 24 hr (20%), 48 hr (40%), 60 hr (50%), 72 hr (60%), and 96 hr (80%).
β-Galactosidase histochemistry
To stain for β-gal activity of the SG and BG fusion proteins, we performed “X-gal” histochemistry by two slightly different methods, described below. The first method was more sensitive than the second one. The main advantage of the first method was that the incubation time could be reduced to 5 hr, and this shortened incubation time yielded better results in subsequent immunohistochemistry. Therefore, this method was applied predominantly for X-gal and anti-PDH double-labeling on animals expressing the BG transgene. For staining pupal brains of this transformant type, the incubation time was 12 hr, unless otherwise stated. The second method was used mainly for scoring of β-gal signal intensities in the L3 brains of BG transformant, because its lower sensitivity enabled us to detect differences in β-gal expression level better than the first method.
In the first method the CNS was fixed for 30 min in 1% glutaraldehyde, subsequently rinsed in phosphate buffer (PB), pH 7.2, for 3 × 5 min, and incubated at 37°C for 5–12 hr in an X-gal staining solution (Simon et al., 1985) containing 0.2% 5-bromo-4-chloro-3-indolyl-b-d-galactopyranoside (X-gal); this β-gal substrate first was dissolved in 1:50 volume of dimethyl sulfoxide before being mixed with the buffer. After incubation, the specimens were washed again in PB for 3 × 5 min and fixed for 3 hr, either in 6.25% glutaraldehyde in PB or in Zamboni’s fixative (4% paraformaldehyde/7.5% picric acid in PB), if PDH immunostaining subsequently was performed (see below).
In the second method the larval CNS was fixed in 0.2% glutaraldehyde and 0 or 0.3% Triton X-100 (TX-100) in 100 mm sodium cacodylate, pH 7.3, for 5 min, rinsed in PBS, pH 7.2, plus 0 or 0.1% TX-100 for 3 × 5 min, and incubated for 24 hr at 37°C in X-gal staining solution (0.2% substrate in a buffer containing 0.01m PB, pH 7.2, 0.15 m NaCl, 1 mmMgCl2, 5 mm potassium ferrocyanide, 5 mm potassium ferrocyanide, and 0.1 or 0.3% TX-100). In this case, X-gal was dissolved in 1:10 volume of dimethyl sulfoxide before being mixed in the staining buffer. After incubation, specimens were washed in a series of concentrations of glycerol (w/v 30, 60, and 90%) in PBS and mounted.
Scoring for staining intensities
Two methods were used to score the staining intensities. For each, a Zeiss Axiophot microscope (Oberkochen, Germany) equipped with both an epifluorescent and a halogen light source was used. The initial scoring of staining intensity for X-gal histochemistry in L3was done as follows.
Brains stained at each time point were scored for staining versus nonstaining for each cell cluster in each hemisphere. The percentage of brain hemispheres stained at each cell cluster was plotted against ZT.
Because this scoring method was not done blindly, the scoring method ofStanewsky et al. (1997a) also was used—for samples stained for β-gal, TIM and PER—with the following modifications: after each sample was mounted individually, slides were shuffled and coded by an unbiased party (not one of the authors). Then staining intensities subjectively were scored “blind” by one of us, using an intensity scale of 0–4 (in which 0 is no staining) in increments of 1 (cf. Zerr et al., 1990). The mean and SEM associated with the score for a given genotype, cell cluster, and time point were calculated. In cases for which the staining intensity varied among cells within a cluster, values were taken from the most strongly staining cell in that cluster.
These two methods gave essentially the same results for BG cycling inper+ andper01 genetic backgrounds. All of the plots shown in Results reflect our application of the second scoring method, as described above.
For scoring the staining intensity in pupal brains (which were stained by the X-gal method), we used an intensity scale of 1–3, because staining method 1 (the more sensitive of the two) gave a smaller range of staining intensities. Otherwise, scoring was done blindly, as described above.
Anti-PER and anti-TIM immunohistochemistry
Two different polyclonal antisera against PER, one made in rabbit (Stanewsky et al., 1997a) and the other in rat [Edery et al. (1994); provided by Michael Rosbash, Brandeis University, Waltham, MA], were used in this study. For the anti-TIM antibody, rat-derived polyclonal antiserum against TIM was used [reagent 307, Myers et al. (1996); provided by Michael W. Young, Rockefeller University, New York, NY). So that nonspecific binding could be reduced, anti-PER and TIM antisera were preabsorbed overnight by fixed embryos ofper01,tim01, androsy506, respectively (Patel, 1994). All of the procedures were performed at room temperature except for the antibody incubations, which were at 10°C. PER immunoreactivity was visualized by the peroxidase antiperoxidase (PAP) method (Sternberger, 1979), combined with the diaminobenzidine (DAB) color reaction (applied only for the rat-derived anti-PER) as well as by immunofluorescent staining. TIM immunoreactivity was visualized by immunofluorescent staining only. For the PAP method, larvae were dissected in 100 mm PIPES, 1 mmMgCl2, and 1 mm EGTA (PEM), pH 6.9; the CNS subsequently was fixed in 3.7% formaldehyde in PEM (PEM-FA) for 30 min, followed by methanol for 15 min. For immunofluorescent staining, to avoid light-induced degradation of TIM (also see below), we did dissections in PEM-FA. The CNS subsequently was fixed in PEM-FA for 30–35 min. To check whether this procedure minimized TIM degradation, we completed two of six experiments as follows: larvae from dark time points were roughly disrupted under the infrared light and fixed immediately in PEM-FA in the dark for 30 min. After the fixation, the CNS was dissected further out. No apparent difference in staining intensity was found between the “dark” and the “usual condition” methods. Thus, the former seems an unnecessary precaution. Also, the CNS often was deformed by this “dark” method. Therefore, the results from the more standard (and more easily effected) procedure are presented.
After the fixation the CNS was washed in 0.1% bovine serum albumin and 0.1% TX-100 in PBS (PBT) for 3 × 5 min, permeabilized in PBS containing 1% TX-100 for 10 min, blocked in 5% normal donkey serum (NDS, Jackson ImmunoResearch West Grove, PA) in PBT for >30 min, and incubated in the primary antibody diluted in PBT plus 5% NDS for 48–72 hr. Dilutions for rabbit anti-PER, rat anti-PER, and anti-TIM antisera were 1:15000, 1:2000, and 1:800, respectively. For the PAP staining, the secondary and tertiary antibodies were anti-rat IgG (produced in donkey; Jackson ImmunoResearch), diluted at 1:100, and rat PAP at 1:300 (Jackson ImmunoResearch). For immunofluorescent staining, secondary and tertiary reagents were biotin-SP-conjugated anti-rabbit or rat IgG (produced in donkey; Jackson ImmunoResearch) and fluorescein isothiocyanate (FITC)-conjugated streptavidin, respectively (Jackson ImmunoResearch). They were diluted at 1:200. Secondary antibodies and rat PAP were incubated for 24–48 hr, and FITC-streptavidin was incubated for 12–24 hr. Subsequent to each incubation, the CNS was washed extensively in PBT. For peroxidase color reaction, CNSs were stained for 2–3 min with DAB substrate kit (Vector Laboratories, Burlingame, CA). After the color reaction, brains were dehydrated in ethanol concentration series, cleared in xylene, and mounted in Permount (Fisher Scientific, Pittsburgh, PA). A given fluorescently labeled CNS was mounted with 2% n-propyl gallate in 80% glycerol in PB, pH 7.4. To study subcellular localization, we counterstained immunofluorescently stained samples (by FITC) by propidium iodide before they were mounted (Whitfield et al., 1990). Preparations were observed under a Zeiss Axiophot microscope or an MRC600 laser-scanning confocal microscope (Bio-Rad, Richmond CA).
Double-labeling
X-gal and PDH. X-gal and PDH double-labeling was performed on the L3 CNSs from a BG transgenic line (BG6a) at ZT 0. Anti-PDH antiserum was kindly provided by Dr. Heinrich Dircksen (Rheinische Friedrich-Wilhelms-Universität, Institut für Zoophysiologie, Bonn, Germany). First, X-gal histochemistry was performed as described above (the first such method). After fixation in Zamboni’s fixative, PDH immunohistochemistry was performed as described by Helfrich-Förster (1997). Before the DAB reactionL3 CNSs were mounted in PB on a slide, and the X-gal-stained cells were photographed. After the DAB reaction was performed, the same CNSs were photographed again. The stained whole mounts subsequently were dehydrated. Some of the samples were incubated for 1 hr in methylbenzoate to remove the blue X-gal stain. After being mounted in DPX (Fluka, Neu-Ulm, Germany) under glass coverslips, these preparations were photographed once more. This procedure yielded three different images of the stained cells and allowed a reliable judgment about double staining of cells: the first picture showed only the BG-expressing cells in blue, the second showed the BG-expressing cells in blue and the PDH-immunoreactive cells in brown, and the last picture showed only the PDH-immunoreactive cells in brown.
Immunofluorescent double-labeling. Three immunofluorescent double-labeling experiments were performed on the CNS of theL3 animals: anti-β-gal (rabbit-derived polyclonal; Organon Teknika, Durham, NC) and monoclonal antibody (mAb) 22C10 (cf. Zipursky et al., 1984), applied to BG6a transformant strain; anti-β-gal and anti-ELAV (mAb 5D; O’Neill et al., 1994), applied to SG3; and anti-PER (rabbit-derived polyclonal; cf. Stanewsky et al., 1997a) and anti-TIM applied on a wild-type strain. mAb 22C10 was provided by Seymour Benzer (California Institute of Technology, Pasadena, CA), and mAb 5D was provided by Kalpana White (Brandeis University, Waltham, MA). Dissection, fixation, and washes were performed as described for the anti-PER immunohistochemistry, except that methanol fixation sometimes was used here (this occurred for 1–3 min or was omitted), and fixation was done in PEM-FA for double-labeling involving anti-TIM. The dilutions for anti-β-gal, mAb 22C10, and anti-ELAV were 1:1500–2000, 1:20, and 1:10, respectively. Two primary antibodies were incubated simultaneously for 48–72 hr. For anti-PER and TIM double-labeling, lissamine rhodamine sulfonyl chloride (LRSC)-conjugated anti-rabbit IgG and FITC-conjugated anti-rat IgG (polyclonals derived from donkey; Jackson ImmunoResearch) were used as the secondary antibodies. For the other experiments FITC-conjugated anti-rabbit IgG and LRSC-conjugated anti-mouse IgG (Jackson ImmunoResearch) were used. Two secondary antibodies were incubated simultaneously for 24–48 hr at a concentration of 1:100 for each. Little cross-reactivity or background staining was found, except that LRSC-conjugated anti-mouse IgG itself led to some background staining; this was distinguished easily from mAb 22C10- or anti-ELAV-mediated staining. After being mounted in 2%n-propyl gallate plus 80% glycerol plus 20% PB, pH 7.4, stained samples were viewed on an MRC600 laser-scanning confocal microscope, equipped with an argon–krypton laser with dual-channel scanning. Colocalization was verified by merging the two channels in two colors. In the case of the ELAV/β-gal double-labeling experiment, colocalization was observed in at least three samples for each SG-expressing clusters, except for LNs, for which SG expression was too weak for this experiment.
RESULTS
per-Expressing cells in the final larval stage
In CNS specimens from third-instar Drosophila larvae (L3), per expression was determined by X-gal histochemistry and anti-β-gal immunohistochemistry in SG and BGper reporter transgenics. Further assessments of the expression of this clock gene came from anti-PER immunohistochemistry, using wild-type animals. Examples of the staining patterns for SG, BG, and anti-PER are shown in Figures 1, 2, and 3, respectively. The numbers of stained samples and the numbers of cells of each neuronal cluster are summarized in Tables 1 and 2.
Fig. 1.
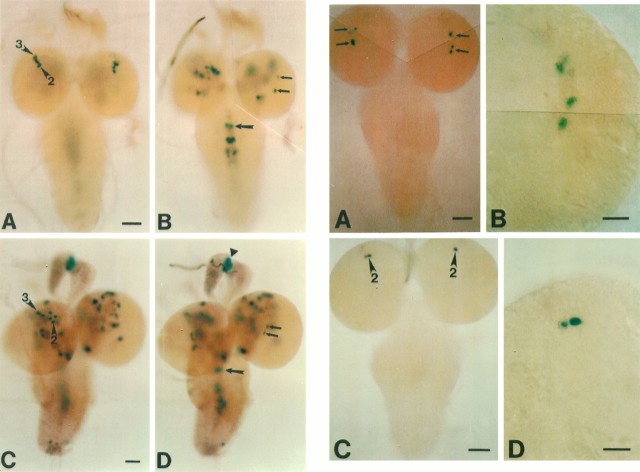
Left. CNS expression pattern in third-instar larvae of per-lacZ transgenics (SG) revealed by X-gal histochemistry. The staining pattern for the SG3 line is shown inA and B and for SG10 in Cand D. A, Dorsoanterior clusters; this dorsal cluster of labeled neurons could be divided into two subclusters corresponding to PER-immunoreactive DN2L(arrowhead marked by 2) and DN3L (arrowhead marked by3). Scale bar, 50 μm. B, Staining in the ventral region of the brain is shown on the left hemisphere and the LNs (arrows) on the right. Cells near the midline of the ventral ganglion are stained (large arrow points to the staining in the first thoracic neuromere). Smaller stained clusters in the abdominal neuromeres are out of focus here. Scale bar, same as inA. C, Dorsal clusters; the clusters corresponding to PER-immunoreactive DN2L and DN3L are indicated witharrowheads (marked with 2 and3, respectively). More cells are stained in this line than in SG3. Cells at the tip of the ventral ganglion are shown as well. Scale bar, 50 μm. D, Focus on the LNs (arrow); stained cells in the ventral region are slightly out of focus in this image; strong staining in the ring gland (filled triangle) and staining in the ventral ganglion (large arrow pointing to the first thoracic neuromere) also are shown. Scale bar, same as inC.
Fig. 2.
Right. CNS expression pattern in third-instar larvae of per-lacZ transgenics (BG) revealed by X-gal histochemistry. The staining pattern (for strain BG6a) at ∼ZT 23 (1 hr before lights on) is shown in Aand B and at ∼ZT 12 (the time of lights off) inC and D. A, Only the LNs (arrows) were stained strongly at ZT 23 (1 hr before lights on). B, Higher magnification of the right hemisphere in A. C, Only DN2Ls (arrowheads marked by2) were stained strongly at ZT 12 (the time of lights off). D, Higher magnification of the left hemisphere inC. Scale bars: 50 μm in A andC; 20 μm in B andD.
Fig. 3.
PER- and TIM-immunoreactive cells in the CNS of third-instar larvae. PER-immunoreactive cells at ∼ZT 0 (the time of lights on) are shown in A, B,D, and E and at ∼ZT 12 inC and F; TIM-immunoreactive cells are shown at ∼ZT 21 in G and at ∼ZT 10 inH. A, Five PER-immunoreactive LNs are shown in both hemispheres; the right-most LN in the right hemisphere is slightly out of focus; scale bar, 50 μm. B, Weakly stained DNL clusters, DN1L (arrowhead marked by1), DN2L(arrowheads marked by 2), and DN3L (arrowhead marked by3); scale bar, same as in A.C, DN2Ls (arrowhead marked by 2) are shown; they are slightly out of focus in the right hemisphere and thus display weaker than in the left hemisphere; scale bar, same as inA. D, Higher magnification of LNs in the right hemisphere in A; scale bar, 20 μm.E, Higher magnification of B; two cells are stained for each of DN1L and DN2L clusters. DN3L seems to be composed of two to three subclusters. Only one of the subclusters of DN3L is in focus here; scale bar, same as inD. F, Higher magnification of the left hemisphere in C; note that DN2Ls are stained more strongly in F than in E; scale bar, same as in D. G, TIM immunoreactivity in DN1L(arrowheads marked by 1), DN2L (arrowheads marked by2), and LNs (arrows); scale bar, 50 μm.H, DN2L(arrowheads marked by 2) is stained more strongly here than in G; scale bar, same as inG.
Using these methods, we identified a total of six to eightper-expressing cell clusters in the nervous system ofL3. All cell clusters consisted of neurons, as shown by their immunoreactivity to ELAV, a neuron-specific nuclear antigen ofDrosophila (see Fig. 4G–I; Bier et al., 1988;Robinow and White, 1988, 1991) or PDH (in the case of PDH-immunoreactive lateral neurons; see Fig. 9).
Fig. 4.
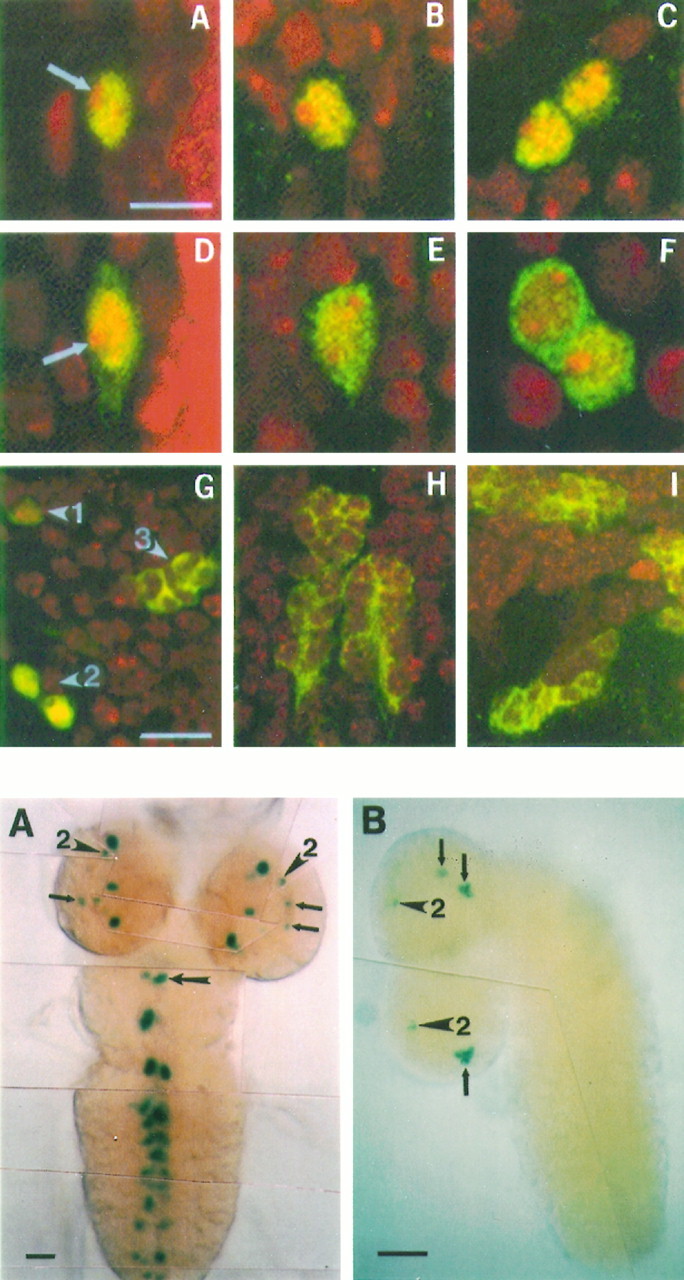
Top. Confocal images showing subcellular localizations of PER, TIM, and SG. A–C, Wild-type L3 brains stained by anti-PER (green) also were stained by propidium iodide (red) to reveal nuclei. A, An LN at ZT 23; B, a DN1L at ZT 23;C, two DN2Ls at ZT 12. Anti-PER signals overlap with red DNA staining except in nucleoli, where there is no PER staining (an arrow in A points to a nucleolus). This indicates that PER is predominantly nuclear in these cells at their peak time points. D–F, Anti-TIM staining (green) on wild-type L3brains counterstained by propidium iodide (red).D, An LN at ZT 21; E, a DN1L at ZT 21; F, two DN2Ls at ZT 10. Unlike PER, TIM is localized both in nuclei and cytoplasm but is excluded from nucleoli (anarrow in D points to a nucleolus). This cytoplasmic (and nuclear as well) staining also was observed in LNs at ZT 23 and DN2Ls at ZT 13 (data not shown).G–I, L3 brains from the SG3 transformant strain were double-labeled by anti-β-gal (green) and anti-ELAV (red). All of the SG-expressing cells were double-labeled, indicating that they are neurons. G, Dorsal region of the brains showing the three dorsal neuronal clusters, DN1L, DN2L, and DN3L(arrowheads marked by 1, 2, and3, respectively). SG is expressed predominantly in nucleus in DN1L and DN2Lclusters but is excluded from nuclei in DN3Ls. The anti-β-gal staining in DN1L is very weak; therefore, it appears almost orange here. Contrastingly, anti-β-gal staining in DN2Ls is so strong that it appears as yellow-green. H, SG signals near the midline of the first segment of the ventral ganglion. They are predominantly cytoplasmic. I, Ventral region of the brain, showing cytoplasmic staining of β-gal. Scale bars: 5 μm inA–F; 10 μm in G–I.
Fig. 9.
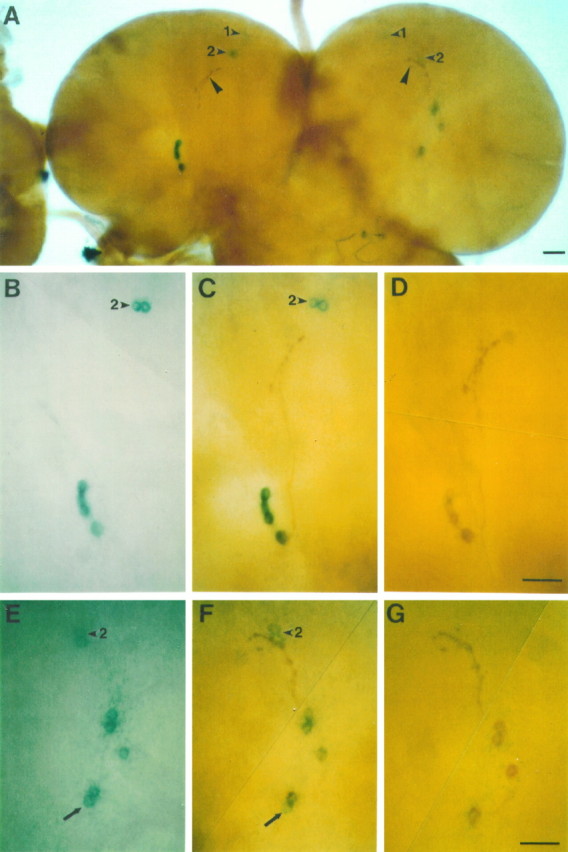
X-gal and anti-PDH double-labeling in the brain of a third-instar larva carrying a per-lacZ fusion gene. The transgenic strain was BG6a. A, Cells in both brain hemispheres labeled by X-gal and by an antibody against pigment-dispersing hormone at relatively low magnification; four and five LNs are prominently stained in the left and right brain hemispheres, respectively; the arrowheads point to the DNLs that are stained in the dorsoanterior brain hemispheres; the DN1Ls (arrowheads marked by 1) are stained only faintly and are slightly out of focus; the DN2Ls (arrowheads marked by 2) are located close to the terminals (large arrowheads) of the LNs, especially in the right brain hemisphere.B–D, LNs and DN2Ls in the left brain hemisphere inA at higher magnification; this brain region was first photographed after X-gal histochemistry had been performed (B), then rephotographed after anti-PDH immunohistochemistry was performed (C), and photographed for a third time after the blue X-gal stain had been dissolved by methylbenzoate (D); all four LNs were marked by X-gal. E–G, LNs and DN2Ls in the right brain hemisphere, marked by X-gal staining only (E), by X-gal and anti-PDH (F), and by anti-PDH only (G); four of five LNs stained by X-gal also were labeled by anti-PDH; the fifth LN (arrows inE and F) and both DN2Ls (marked by 2) were not PDH-immunoreactive; scale bars, 20 μm.
By X-gal histochemistry, some differences in staining intensity and numbers of stained cells were found between the two SG transgenic lines (Fig. 1, Table 1). These differences probably are attributable to chromosomal position effects. However, the similarities of the staining pattern suggest that SG expression is controlled mainly by the regulatory sequences of this construct. The staining patterns for the two BG per reporter lines (BG6a and BG/TM2) were identical and overall very similar to those found by anti-PER immunohistochemistry in the wild type (compare Fig.2 with Fig.3).
Table 1.
β-Gal staining of L3 CNSs expressing the SGper-lacZ fusion gene
| Type of cells and tissues | SG3 strain | SG10 strain | ||
|---|---|---|---|---|
| % stained | No. signals | % stained | No. signals | |
| Dorsoanterior (DNLs) | 100 (1181-a) | 3.8 ± 0.1 | 100 (261-a) | 9.9 ± 0.3 |
| Dorsoposterior | 0 (1181-a) | – | 100 (261-a) | 4.6 ± 0.4 |
| Lateral (LNs) | 27 (1181-a) | 0.3 ± 0.1 | 100 (261-a) | 2.6 ± 0.2 |
| Ventroanterior (ventral cells) | 100 (1181-a) | – | 100 (261-a) | – |
| Ventroposterior | 83 (1181-a) | 1.0 ± 0.1 | 89 (261-a) | 1.5 ± 0.2 |
| Thoracic neuromeres | 100 (59) | – | 100 (13) | – |
| Abdominal neuromeres | 93 (54) | – | 100 (12) | – |
| Mouth parts | 96 (44) | – | 8 (12) | – |
| Ring gland | 26 (50) | – | 100 (11) | – |
The second of the two staining procedures described in Materials and Methods was used. For larvae from the SG3 and the SG10 transgenic lines, percentages of stained samples are indicated in the columns headed “% stained.” The numbers of valid samples are shown in parentheses. In the columns indicating “No. signals,” these means are quoted ± SEM.
Number of brain hemispheres; –, not determined.
In the BG lines and in wild-type larvae, fewer cells were stained than in the SG lines. These differences mainly were quantitative (see below). The most important difference among BG, wild-typeper+, and SG expression was that the SG-encoded fusion protein did not cycle, but BG and endogenous PER exhibited cyclically varying staining intensities at least in some cells. This is consistent with the results recently obtained for SG- and BG-mediated expression of β-gal in the adult head (Dembinska et al., 1997; Stanewsky et al., 1997). The SG-encoded fusion protein obviously has an increased stability (Dembinska et al., 1997), which also might account for the observation that more cells were stained in the SG-bearing larvae than in those bearing the BG construct.
In the following subsections the characteristic expression patterns associated with the different per-expressing cell clusters are described. They have been divided mainly into two categories: the cycling cells, in which cycling of BG and endogenous PER was clearly detected, and the noncycling cells, in which temporal variations of reporter or anti-PER signals were not detected.
Cycling cells
There are three clusters that belong to cycling cells: lateral neurons (LNs) and two dorsoanteriorly located clusters, dorsal neurons-1Larval (DN1L) and dorsal neurons-2Larval(DN2L).
Lateral neurons (LNs). This cluster is located between the optic lobe primordium and the central brain of L3 larvae. SG staining was strong in SG10 but relatively weak in the SG3 transgenic type, for which many of the samples did not show any staining (Fig. 1, Table 1).
In contrast, three to five LNs (mean = 4.0 ± 0.1;n = 80) were stained strongly by X-gal histochemistry applied to the BG lines at ZT 23 (Fig. 2A,B). No such staining was observed at ZT 12, indicating that the BG fusion protein cycles in this cluster (Fig. 2C,D).
Via anti-PER immunohistochemistry on CNSs from wild-type larvae, five LNs were stained relatively strongly at ZT 0–1, but not at ZT 12 (Fig.3A,D, Table 2). No staining was observed in larvae expressing theper01 loss-of-function mutation (data not shown). Via confocal microscopy on fluorescently stained samples, up to four additional cells near the five LNs were found to express PER very weakly (data not shown).
Table 2.
Anti-PER staining of wild-type L3 larvae
| Type of cells and tissues | ZT 0–1 | ZT 11–12 | |||
|---|---|---|---|---|---|
| % stained | No. cells | % stained | No. cells | ||
| Dorsoanterior | DN1Ls | 98 (452-a) | 1.9 ± 0.1 | 0 (382-a) | – |
| DN2Ls | 95 (432-a) | 1.6 ± 0.1 | 100 (382-a) | 1.9 ± 0.0 | |
| DN3Ls | 86 (442-a) | – | 89 (372-a) | – | |
| Lateral | LNs | 100 (462-a) | 5.0 ± 0.0 | 0 (382-a) | – |
| Ventroanterior | ventral cells | 70 (20) | – | 53 (19) | – |
| Thoracic neuromeres | 67 (21) | – | 100 (19) | – | |
Number of brain hemispheres. In the columns indicating “No. cells,” these means are quoted ± SEM; –, not determined. ZT 0–1, the first hour of day light in a 12 hr light/12 hr dark (LD) cycle; ZT 11–12, the first hour of darkness in the same kind of LD cycle.
Thus the LN cluster (in one side of the larval brain) consists of at least five cells in which the BG PER-β-gal fusion protein and the PER protein itself cycle strongly, with a peak near the end of the night or beginning of the day.
Subcellular localization of PER in this cluster was checked near its peak time point in the LD condition by confocal microscopy on immunofluorescently stained samples. It was found to be predominantly nuclear (Fig. 4A).
Dorsal neurons-1Larval(DN1L). The DN1Ls are located at the most medial part of the dorsoanterior region of the brain and consist of one to two cells. They were not detectable by X-gal histochemistry applied to SG larvae, although signals were detectable by the more sensitive method of anti-β-gal combined with confocal microscopy (Fig. 4G).
Via X-gal histochemistry of BG, the DN1Ls were stained weakly in ∼10% (n = 80) of the brain hemispheres at ZT 23, but not at ZT 12, indicating that BG cycles in this dorsal neuronal cluster (data not shown). Using anti-PER immunohistochemistry on CNSs from wild-type larvae, we detected weak staining in almost all of the hemispheres stained at ZT 0–1 (Fig.3B,E, Table 2), but no staining was observed in the samples stained at ZT 12.
Thus the DN1Ls are two cells in the dorsoanterior region of the brain that express PER and BG weakly, but cyclically, with a peak near the end of the night or beginning of the day.
In this cluster subcellular localization of the SG fusion protein and endogenous PER was checked by the same method as that used for LNs. SG was found to be in nuclei at least at ZT 12 (Fig. 4G). PER was predominantly nuclear at ZT 0 (Fig. 4B).
Dorsal neurons-2Larval(DN2L). The DN2Ls consist of one to two cells located in a region lateral and posterior to that of the DN1Ls in the dorsal region of the brain. In SG larvae the staining in this cluster often was difficult to distinguish from that of another dorsal cluster (DN3Ls, see below), and the number of cells in the clusters was difficult to count (Fig. 1A,C). Therefore, the signals for these clusters taken together are compiled in Table 1.
In BG larvae the DN2Ls were stained most strongly at ZT 12 (Fig. 2C,D). One to two cells (mean = 1.6 ± 0.1; n = 78) were counted by this method. Similarly, anti-PER staining was stronger at ZT 12 than at ZT 0 (Fig.3B,C,E,F).
In summary, DN2Ls consist of one to two neurons in which PER and BG cycle with a peak near the end of the day.
Subcellular localization of SG fusion protein and endogenous PER was checked by confocal microscopy. SG (Fig. 4G) and PER (Fig.4C) were nuclear at least at ZT 12.
Noncycling cells
This class of cells were categorized into three to five clusters by position. They are cells on both sides of the midline at ventral ganglion and two to four clusters in the brain hemispheres dorsoanterior (DN3L), dorsoposterior, ventroanterior, and ventroposterior (see Fig. 1, Table 1). All of them seem to consist of multiple cells and could be divided into multiple subclusters.
In all of the noncycling cells, SG is expressed most strongly (Fig. 1, Table 1), with BG and endogenous PER giving much weaker signal (data not shown). Especially in the case of BG, staining in ventral clusters and cells in the ventral ganglion were detected only after anti-β-gal immunohistochemistry combined with confocal microscopy (data not shown). BG staining in DN3Ls was detected only in 5% (n = 311) of the brain hemispheres.
Subcellular localization of SG fusion protein in noncycling cells was checked by confocal microscopy on samples immunofluorescently stained by anti-β-gal. It was found to be cytoplasmic at least at ZT 12 (SG; see Fig. 4G–I).
In summary, SG, BG, and endogenous PER are expressed in at least six classes of cells within the CNS of third-instar larvae: LNs, DN1Ls, DN2Ls, DN3Ls, ventral cells, and cells in the ventral ganglion. The SG-encoded fusion protein does not cycle, but that stemming from BG and the endogenous per+gene cycle, at least in LNs, DN1Ls, and DN2Ls. In the other cells SG is expressed at a high level, but BG and endogenous PER are low.
Outside the CNS
In addition to the CNS, SG staining was observed in tissues around the mouth hook and in the ring gland (Table 1). The staining in the ring gland was mainly in the corpus allatum (Fig.1D). All of these signals are real, because no staining was observed in these regions in wild-type larval controls. Neither BG nor endogenous PER was detected in any of these structures.
Expression pattern of per throughout postembryonic development
Earliest per expression
The SG and BG per-lacZ fusion gene transgenics were used for determining the first stage of per expression in the larval CNS. For the analysis of SG-mediated expression, CNSs from SG larvae were stained for each of early L1 (1–2 hr after hatching), mid L1 (12–14 hr after hatching), and earlyL2 (24–26 hr after hatching; Table3). For the study of BG-reportedper expression, L1 and L2 larvae carrying the BG6a transgene were dissected at ZT 0 and ZT 12—the times of approximate maximal and minimal BG expression in adults (Stanewsky et al., 1997a). The L1 stage was divided into 0–3, 4–8, 12, and 15 hr after hatching (see Table4). Examples of SG and BG staining patterns are shown in Figure 5.
Table 3.
Staining signals in per-lacZ larvae (SG): stages L1 and L2
| Cell cluster: age | No. staining signals | ||
|---|---|---|---|
| L1: 1–2 hr | L1: 12–14 hr | L2: 24–26 hr | |
| Dorsoanterior (DNLs) | – (17) | – (16) | 1.0 ± 0.1 (18) |
| Lateral (LNs) | – (17) | 0.9 ± 0.1 (16) | 2.0 ± 0.1 (18) |
| Ventroanterior | 2.7 ± 0.3 (17) | 2.4 ± 0.2 (16) | 2.2 ± 0.1 (18) |
| Ventroposterior | 1.6 ± 0.1 (17) | 2.4 ± 0.2 (16) | 3.3 ± 0.2 (18) |
| Ventral gangliona | 12.4 ± 0.2 (9) | 12.3 ± 0.2 (9) | 12.9 ± 0.1 (8) |
Larvae carrying the SG fusion gene (strain SG3) were used, as was the second of the two staining procedures indicated in Materials and Methods. L1: 1–2 hr, first larval instar, 1–2 hr after hatching from the embryo; L1 12–14 hr, 12–14 hr after hatching; L2 24–26 hr, second larval instar, 24–26 hr after hatching. The “No. staining signals” are quoted ± SEM, except in a, for which the numbers of segments showing signals ± SEM in the ventral ganglia of the CNSs are indicated. The number of histologically interpretable brain hemispheres or ventral ganglia observed (last row) is indicated in parentheses; –, no staining was observed.
Table 4.
X-gal-stained neurons in per-lacZ transgenic animals (BG) at various postembryonic stages and at the beginning of the day
| Stage | n | No. stained cells | |||||
|---|---|---|---|---|---|---|---|
| Small LNv | Large LNv | LNd | DN1L/DN1 | DN2L/DN2 | DN3L/DN3 | ||
| L1 0–3 hr | 11 (22) | – | – | – | – | – | – |
| L1 4–8 hr | 7 (14) | 2.1 ± 0.4 | – | – | – | – | – |
| L1 12 hr | 7 (14) | 3.4 ± 0.3 | – | – | – | – | – |
| L115 hr | 6 (12) | 3.5 ± 0.4 | – | – | 0.3 ± 0.1 | 1.0 ± 0.3 | – |
| L2 | 7 (14) | 3.8 ± 0.2 | – | – | 1.4 ± 0.1 | 1.7 ± 0.1 | – |
| L3 | 22 (41) | 4.0 ± 0.1 | – | – | 1.4 ± 0.1 | 1.8 ± 0.1 | – |
| P20–40% | 8 (16) | 4.1 ± 0.1 | – | – | – | 0.1 ± 0.1 | – |
| P50% | 10 (18) | 4.1 ± 0.1 | 2.8 ± 0.2 | – | 2.4 ± 0.2 | 1.2 ± 0.2 | – |
| P60% | 8 (16) | 4.1 ± 0.1 | 3.3 ± 0.2 | 2.2 ± 0.4 | 3.7 ± 0.3 | 1.8 ± 0.1 | – |
| P70–90% | 11 (22) | 4.1 ± 0.1 | 4.1 ± 0.2 | 4.5 ± 0.2 | 6.8 ± 0.3 | 1.7 ± 0.1 | 16.4 ± 1.3 |
| Adult | 6 (12) | 3.8 ± 0.3 | 4.2 ± 0.2 | 5.3 ± 0.4 | 8.4 ± 1.1 | 1.8 ± 0.2 | 27.0 ± 2.9 |
Developing or adult animals carrying the BG (strain BG6a)per reporter protein fusion were used, as was first X-gal staining procedure described in Materials and Methods. The animals were killed at ZT 0, the beginning of the light period in a 12 hr/12 hr LD cycle. The “No. stained cells” in each entry are quoted ± SEM. The different life cycle stages are indicated by L10–3 hr, first instar larvae (0–3 hr after embryonic hatching); other intra-L1 times as indicated; L2 (24–26 hr after embryonic hatching); L3 (6–7 d old); P20–40%, 20–40% of the way through pupal (P) development; other intra P times as indicated. n, Numbers of samples. Numbers of histologically interpretable brain hemispheres are shown in parentheses; –, no staining was observed.
Fig. 5.
Bottom. Spatial expression patterns of aper-promoted reporter enzyme in early larval stages revealed by X-gal histochemistry. The staining pattern in the CNS of anL2 larva (24–26 hr after hatching) from the transgenic strain SG3 is shown in A and of an L1larva (∼15 hr, ZT 0) from the BG6a strain in B.A, The arrowheads (marked by2) point to the dorsal clusters that seem to correspond to the PER-immunoreactive DN2L neurons; thearrows point to the stained cells in the lateral region, corresponding to the PER-immunoreactive LNs; all of the unmarked cells in the brain hemispheres are in the ventral region. In the ventral ganglion stained clusters near the midline of every neuromere are shown (large arrow pointing to the first thoracic neuromere).B, As in A, dorsal and lateral clusters—corresponding to the PER-immunoreactive DN2L (arrowheads marked by2) and LNs (arrows)—are shown. Scale bars, 20 μm.
In LNs, BG was found first in 4- to 5-hr-old L1s dissected at ZT 0 (Table 4). SG was found in mid-L1 (12–14 hr after hatching) and L2, but not in early L1. In SG transformants only three or fewer cells were stained, whereas in BG transformants up to five LNs were stained. Importantly, no BG-mediated LN signals were detected in animals dissected at ZT 12, indicating that the amount of BG fusion protein in LNs of these earlier stages cycles with a similar phase, as in L3 (data not shown).
Two DNL clusters, DN1Ls and DN2Ls, were found in L1 larvae of BG older than 15 hr after hatching (Table 4). Up to two cells were stained for each cluster. DN1Ls were detectable more readily at ZT 0 than at ZT 12 (data not shown). DN2Ls were stained more prominently at ZT 12 than at ZT 0, indicating that antiphase cycling of PER begins during this early stage of development (data not shown).
In the SG transformant only DN2Ls (but not DN1Ls) were detected by X-gal histochemistry inL2 larvae (Fig. 5, Table 3). As in L3, the ventral region of the brain lobes and the ventral ganglion were stained prominently in SG, but not in BG. In contrast to the cycling cells, they were observed already in early L1. UnlikeL3, in which larvae strong SG staining was observed only in the three thoracic segments (Fig. 1B,D), signals were seen in 12–13 segments both in L1 and L2 (Fig.5A, Table 3). The difference between L1–L2versus L3 may be attributable to the enlargement of the thoracic nervous system and the shortening of the abdominal nervous system during development.
BG expression patterns at different pupal stages
To follow the fate of the larval LNs and DNs, we stained pupal brains of the BG6a transformant for X-gal at different developmental stages, as well as at ZT 0, ZT 6, and ZT 12. For ZT 0, 8–11 brains (16–22 hemispheres) were stained for each age category, 20–40, 50, 60, and 70–90% of pupal development (Table 4). For ZT 6 and ZT 12, 2–11 brains (4–22 hemispheres) for each of 20–40, 50, and 70–90% of pupal development were observed.
During the first 40% of pupal development, only the four to five larval LNs were revealed readily by X-gal at ZT 0, except for a single weakly labeled cell near the region of DN2Ls in 1 of 16 hemispheres, dissected out at ZT 0 (Fig.6A, Table 4). This suggests that DNLs stop expressing PER after the stage of pupariation. The BG-expressing LNs seem to correspond to a group of smaller LNs that are located relatively ventrally in adults (small LNvs; see below; also Helfrich-Förster and Homberg, 1993; Helfrich-Förster, 1995).
Fig. 6.
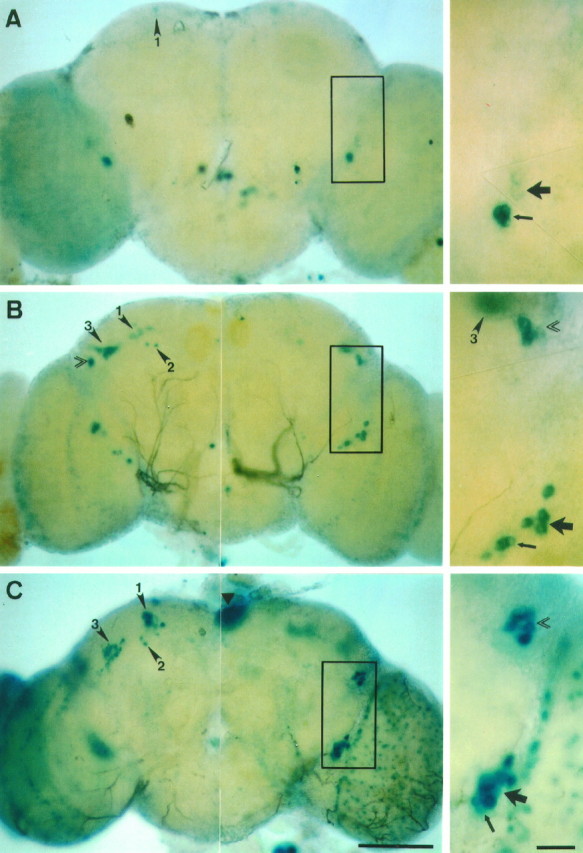
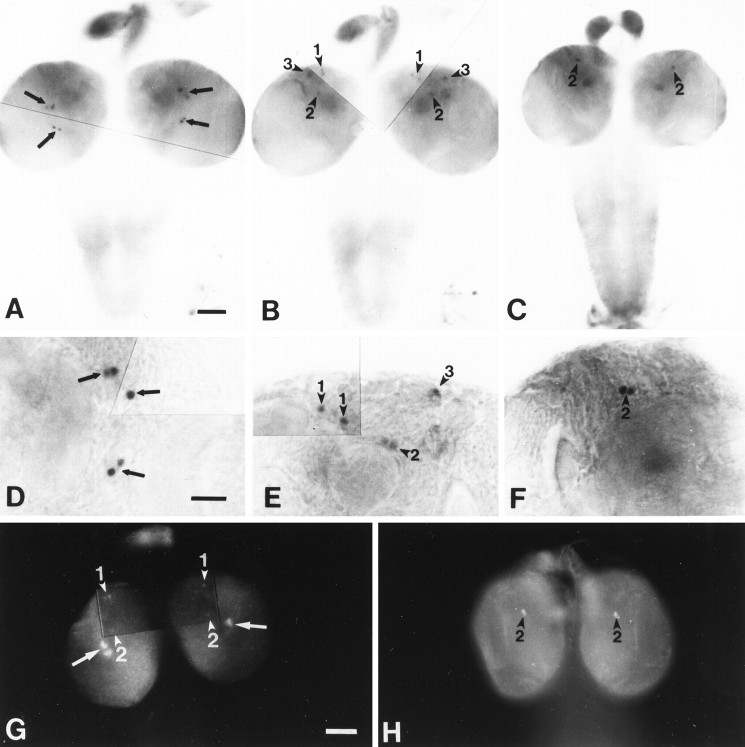
per-lacZ expression pattern in pupal brains revealed by X-gal histochemistry. The transgenic strain used was BG6a. For all of these late developmental stages the left brain hemispheres are shown from a posterior view and reveal the DNs; the right ones are anterior views and reveal the LNs. A, Brain at 50% of pupal development; the arrowhead(marked by 1) in the left brain hemisphere points to two faintly stained DN1s; one of them is slightly out of focus and thus is difficult to see in this image; in the right brain hemisphere four prominently stained small ventral LNs (small LNvs) can be seen, which correspond to the larval LNs; dorsally to them two large LNvs have begun to express the BG-encoded reporter; theboxed section of the brain is shown to theright at higher magnification; the large arrow marks the two faintly stained large LNvs, and the small arrow marks the prominently stained small LNvs. B, Brain at 70% of pupal development; the arrowheads in the left brain hemisphere point to the DN1s, DN2s, and DN3s (marked by 1, 2, and3, respectively); the DN3s are in close vicinity to the dorsolateral neurons (LNds; double arrowhead); in the right hemisphere four small and four large LNvs are stained (small and large arrows in the magnification of the boxed section); furthermore, the more dorsally situated LNds (double arrowhead) are now prominently stained; as in the left brain hemisphere, they are in close vicinity to the DN3s (arrowhead marked by 3).C, Brain of a newly emerged adult fly; all three groups of DNs can be seen in the left brain hemisphere, and the LNds as well as the small and large LNvs can be seen in the right one (also note the higher magnification of theboxed brain section); the LNds (double arrowhead) and the DN1s (arrowhead marked by1) are now clearly separated from each other and cannot be seen in the same plane of focus; in addition to the DNs and LNs, the photoreceptor cells of the compound eyes (data not shown) and the ocelli (filled triangle) were stained prominently; furthermore, numerous glial cells on the surface of the optic lobe and central brain are revealed by X-gal. Scale bars: 100 μm for the pictures of the whole brains; 20 μm for the higher magnifications.
By 80% of the pupal development, five other neuronal clusters were recognized: large LNvs (relatively ventrally located and larger LNs; Helfrich-Förster and Homberg, 1993;Helfrich-Förster, 1995), LNds (relatively dorsolateral per neurons; Ewer et al., 1992), DN1s, DN2s, and a previously undescribed dorsal cluster, DN3 (C. Helfrich-Förster and M. Kaneko, unpublished data; see Fig.6B,C, Table 4).
At the approximate half-way point of the pupal stage, DN1s, DN2s, and large LNvs were recognized first (Fig.6A, Table 4). The DN1s continuously increased in number throughout pupal development (Table 4), whereas the DN2s remained at two cells and stayed at their position near the calyces of the mushroom body (Fig. 6C). At ∼60% of development LNds and DN3s were first marked by X-gal (Table 4). It should be pointed out that larval DNLs and pupal DNs could be different cells despite the similarity in their position, because DNLs stop expressing BG after the pupariation.
Except for the DN2s and DN3s, all pupal BG-expressing cells cycled in β-gal activity with a phase similar to the larval LNs. For example, small LNvs were stained strongly in all of the hemispheres dissected at ZT 0 but were not stained at ZT 6 and ZT 12, except for 2 of 22 hemispheres dissected at ZT 6 during 70–90% of pupal development. Similarly, LNds were stained only at ZT 0 in an experiment involving 8–11 brains dissected for each time point.
BG cycling in pupal DN1s and large LNvs seems to be less pronounced than in the above-mentioned two clusters. Both the DN1s and the cluster of large LNvs were stained relatively strongly at ZT 0 in >90% of the hemispheres dissected at 50–90% of pupal development, whereas at ZT 6 or ZT 12 they were stained only weakly and in relatively few hemispheres (12 of 42 hemispheres in the case of DN1, and 21 of 42 for large LNvs). Pupal DN2s and DN3s were stained at comparable levels at all three time points, and therefore cycling of BG was undetectable in these clusters. A similar result was obtained in a separate set of experiments on X-gal histochemistry on pupae with shorter incubation time (8 hr instead of 12 hr).
Anti-TIM staining
To see if the other clock gene cloned from Drosophila,tim, is expressed in the larval CNS and to compare the expression pattern with that of per, we immunostained CNSs from L3 larvae with an antiserum against TIM. Sixty-six larval brains from eight time points—ZT 1.3, 3.9, 10.1, 13, 15.3, 21.4 and 23—were subjected to anti-TIM immunohistochemistry (Table5).
Table 5.
L3 CNS cells immunopositive to anti-TIM
| ZT | No. cells stained | No. samples | ||
|---|---|---|---|---|
| LNs | DN1Ls | DN2Ls | ||
| 1.3 | 0.3 ± 0.1 | – | 1.7 ± 0.1 | 8 (16) |
| 3.9 | – | – | 2.0 ± 0.0 | 10 (20) |
| 10.1 | – | – | 1.7 ± 0.1 | 10 (20) |
| 10.9 | – | – | 1.6 ± 0.2 | 5 (10) |
| 13 | 3.1 ± 0.4 | – | 1.3 ± 0.2 | 8 (15) |
| 15.3 | 4.9 ± 0.3 | 1.3 ± 0.2 | 0.7 ± 0.1 | 9 (18) |
| 21.4 | 4.9 ± 0.1 | 2.0 ± 0.0 | 0.9 ± 0.1 | 10 (20) |
| 23 | 4.6 ± 0.2 | 1.8 ± 0.1 | 1.1 ± 0.1 | 6 (12) |
In addition to the experiment summarized here, five additional ones with two to four timepoints each were performed. Very similar results (to those tabulated above) were obtained in all experiments. The “No. cells” stained (first three data columns) are quoted ± SEM. In the final column the numbers of histologically interpretable brain hemispheres are indicated in parentheses; –, not stained. See Table 2 for definition of ZT.
In the dorsoanterior brain region only two clusters were stained prominently with anti-TIM (Fig. 3G,H). The staining in DN1Ls was weak, even at its peak time point during late night. DN2Ls were stained strongly all through the day but weakly at night. Up to two cells per hemisphere were recognized for each of these DNL clusters (Table 5). In addition, one of the DN3Lsubclusters was stained very weakly. This faint staining was detected only after confocal microscopy was applied (data not shown).
In the lateral brain region LNs were stained by anti-TIM (Fig.3G). They were stained strongly at night but not during the day. Up to five cells were counted in this cluster by epifluorescent microscope (Table 5). Additional weakly stained cells (up to four cells) were detected by confocal microscopy (data not shown).
In LNs, DN1Ls, and DN2Ls, subcellular localization of TIM was found to be both nuclear and cytoplasmic at least at their peak time point by observation on confocal microscopy (Fig. 4D–F). Taking advantage of this cytoplasmic staining, we determined the sizes of the cell bodies for each cluster. Diameters were similar for all three clusters, within the range of 5–9 μm.
In the ventral region of the brain and ventral ganglion, two to four cells were stained at the tip of the ventral ganglion in both wild-type larvae and those expressing thetim01 loss-of-function mutation (data not shown; Sehgal et al., 1994; Myers et al., 1995). We judge this to be nonspecific staining, not the result of normaltim+ expression. No other staining was observed in the CNS of tim01larvae.
Colocalization of PER and TIM in all of the TIM-expressing clusters was confirmed by a double-labeling experiment, using anti-PER and anti-TIM antisera on nine brain hemispheres from five brains (data not shown).
Cycling phases of BG and TIM in LD
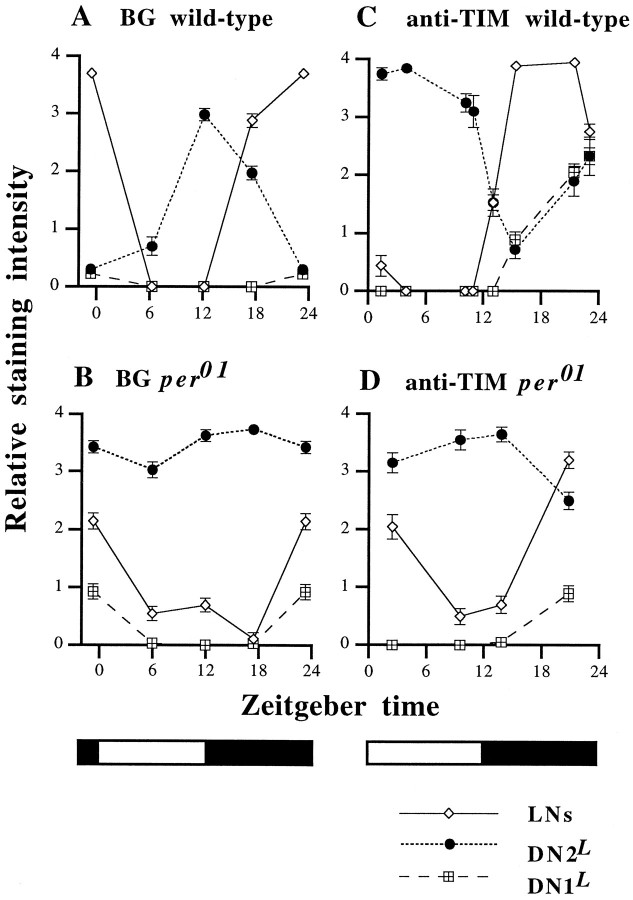
Because the phase of BG cycling seems to be indistinguishable from the endogenous PER cycling (as is also the case in adults; Dembinska et al., 1997; Stanewsky et al., 1997a), the BG line was used for the detailed study of PER cycling in the larval CNS under LD cycle conditions. X-gal histochemistry was performed on the CNS ofL3 from the BG6a strain at four time points: ∼ZT 6, 12, 18, and 23 (Fig. 7). Per time point, 36–40 samples were processed.
Fig. 7.
BG cycling revealed by X-gal histochemistry and TIM cycling revealed by immunofluorescent staining in LD cycles. LN signals, solid lines and diamonds; DN2Ls, dotted lines andcircles; DN1Ls, broken lines and squares. Error bars represent SEM of 0.1 or larger. (SEMs smaller than 0.1 do not appear in the graphs because of the artifact in the plotting program, but these are quite small errors in any case.) All of the data points are an average of multiple samples. Open and black bars at the bottom of graphs B andD represent the 12 hr light phase and the 12 hr dark phase, respectively. The method for scoring staining intensities reported by Stanewsky et al. (1997a) was used to generate these graphs (see Materials and Methods). A, BG in aper+ genetic background at four time points (∼ZT 6, 12, 17, and 23); four such experiments were performed, in each of which at least 34 brain hemispheres per time point were scored. In one of these experiments a more qualitative scoring procedure (scoring method 1) was used (see Materials and Methods); because all four experiments gave similar results, the one with largest sample sizes (34–42 brain hemispheres per time point) is presented here. B, BG in aper01 background at four time points (∼ZT 6, 12, 17, and 23); similar to BG-mediated staining in per+, two experiments with similar results were performed, in one of which the aforementioned scoring method 1 was applied. The second experiment, in which 38–40 brain hemispheres were scored for each time point, is presented here.C, TIM immunoreactivity in a clock gene normal genetic background; six experiments were performed in which two to eight time points were taken. Because similar results were obtained in each experiment, the one with largest number of time points is presented here; 16, 20, 20, 10, 15, 18, 20, and 12 brain hemispheres were scored at times ZT 1.3, 3.9, 10.1, 10.9, 13, 15.3, 21.4, and 23.D, Anti-TIM immunoreactivity inper01 background; one experiment was done in which 20 brain hemispheres were scored for each of four time points, ∼ZT 2, 10, 14, and 21.
The phases of BG cycling in LNs and DN1Ls were similar to those that have been found in the adult, with a peak at ∼ZT 23 and a trough at ∼ZT 6–ZT 12 (Fig. 7A). The cycling amplitude was low in DN1Ls because of the overall low expression level. In contrast, BG-encoded fusion protein was found to cycle in almost opposite phase in the DN2Ls, with a peak at ∼ZT 12 and a trough at ∼ZT 0. The exact peak time point in DN2Ls may be a few hours later than ZT 12, because the level of BG staining was found to be higher at ZT 17 than at ZT 12 in one of four such experiments.
The peak level in the DN2Ls was lower than that in the LNs, and some staining in the DN2Ls could be detected in ∼40% of the brain hemispheres even at their trough time point. Therefore, the BG-encoded fusion protein in the DN2Ls seems to cycle with lower amplitude than in the LNs.
TIM cycling in wild-type larvae was assessed by anti-TIM immunohistochemistry at eight time points (Fig. 7C). As forper-reported expression in the BG transgenics, TIM was found to cycle with different phases in different cell types. Especially in DN2Ls, where BG cycling was found to be almost antiphase of that in LNs and DN1Ls, TIM cycling was also antiphase (Fig. 7C). Furthermore, TIM was high all through the day in this cluster. In addition, there is a difference in the phase relationships of the TIM and BG curves, when the LN and DN2L clusters are considered separately (compare Fig. 7A with 7C). In DN2Ls the rising phase of BG curve in large part lagged behind that of TIM curve, yet in LNs the rising phases of BG and TIM curves were different by only a few hours.
In adults, BG protein level cycles in LD cycle conditions even in aper01 genetic background, yet with lower amplitude than in per+(Dembinska et al., 1997; Stanewsky et al., 1997a). This cycling of BG protein level in per01 seems to be dependent on fluctuations of TIM in response to LD cycles (Dembinska et al., 1997; Stanewsky et al., 1997b). Note that in adult heads light causes TIM levels to “crash,” and this leads to lowered PER levels (Hunter-Ensor et al., 1996; Myers et al., 1996; Zeng et al., 1996). Therefore, BG and TIM cycling in theper01 background was tested in the larval CNS (Fig. 7B,D). In LNs, the amplitude of BG and TIM cycling was reduced in theper01 background. This was confirmed in experiments in which staining intensity forper+ andper01 larvae were scored simultaneously (data not shown).
Furthermore, the cycling phases of TIM and BG are slightly different from those in per+. For example, TIM did not decrease as quickly in response to light inper01 as inper+. This could be explained by a higher level of tim mRNA available inper01, as compared withper+, at the dark/light transition—which corresponds to the trough time point of tim RNA in genetically normal adults (Sehgal et al., 1995). In contrast to the case in LNs, high levels of BG and TIM were observed in DN2Ls at all four time points (Fig.7B,D). BG and TIM cycling in this group of cells was undetectable. The important point is that, in all of the cycling cells in larvae, TIM and BG fluctuate with a similar profile in theper01 genetic background (Fig.7D), indicating the dependency of BG cycling on the level of TIM in per01 background, as has been shown in adults (Dembinska et al., 1997; Stanewsky et al., 1997b).
Cycling of PER and TIM in constant dark
The above results indicate that PER and TIM cycle at least in the three neuronal clusters in the larval CNS under LD cycle conditions. If these rhythms are true circadian ones and not driven by the external LD cycle, they should persist under free-running conditions. To test this, we performed PER and TIM immunohistochemistry on L3 brains of wild-type larvae under constant dark (DD) conditions for 2 d, starting at the fifth day after egg laying. Fluctuations of PER and TIM in a preceding LD cycle also were assessed.
Cycling of PER and TIM immunoreactivity was detectable in LNs, DN1Ls, and DN2Ls for 2 d in DD (Fig. 8). In LNs and DN1Ls, the peak times in DD were in phase with those in LD, indicating free-running periods of ∼24 hr. In DN2Ls, the cycling phases for both PER and TIM seem to be advanced by a few hours on the second day in DD. For instance, the trough time point of PER was at ∼circadian time 18 in DD but at ∼ZT 23 in LD. Similarly, the peak time point for TIM was at ∼circadian time 23 in DD, but at ∼ZT 6 in LD. The cycling amplitudes of both PER and TIM seemed to decrease in both DNL clusters. In DN1Ls, this was attributable mainly to the decrease in the peak levels, whereas in DN2Ls the amplitude decrement occurred because of the increase in the trough levels. It should be pointed out that this amplitude-dampening effect could be attributable to the age difference as well as to the environmental condition (DD), because 2 d elapsed between the first day (LD) and the last day (the second one in DD) in this experiment. The amplitude dampening was not apparent in LNs. In all three of the clusters, TIM rose a few hours earlier than PER did in both LD and DD conditions (cf. Marrus et al., 1996). In DN2Ls the large phase difference between PER and TIM, as was observed under LD conditions for BG and TIM, was not obvious in DD.
Fig. 8.
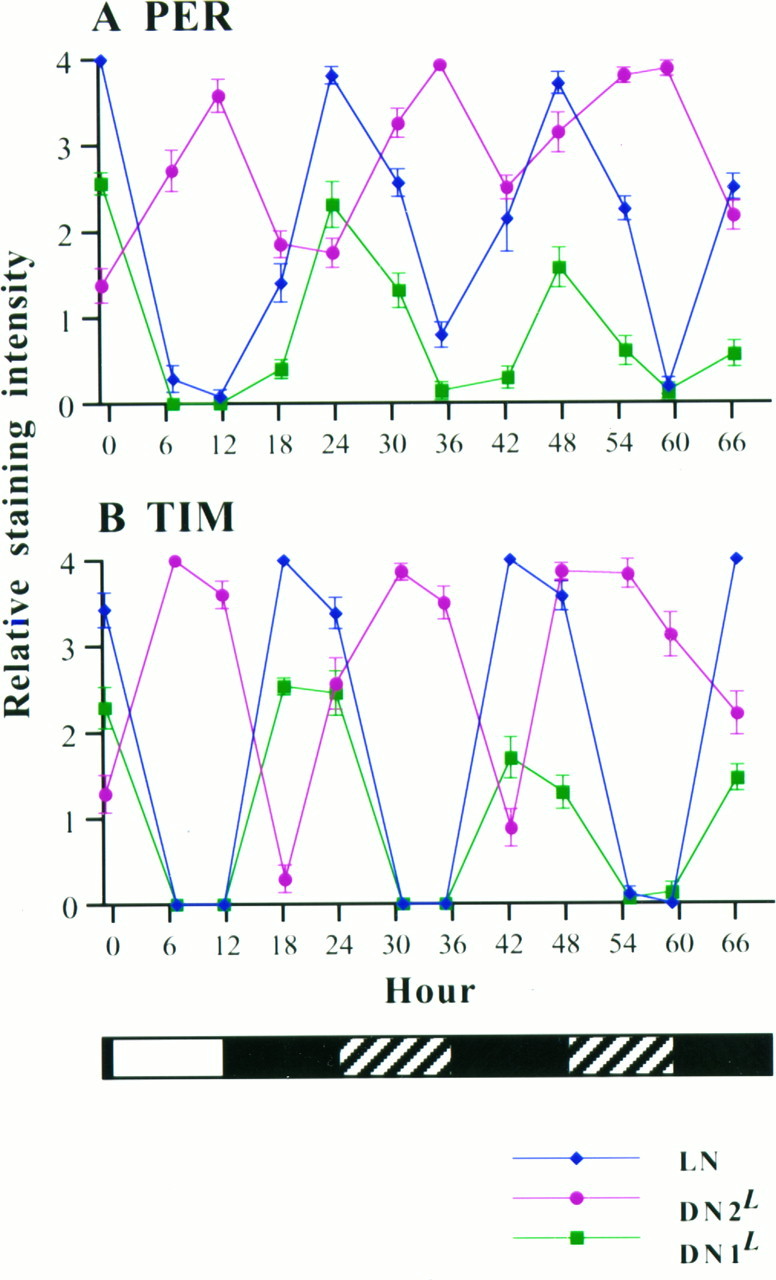
PER and TIM cycling revealed by immunohistochemistry in an LD cycle, followed by 2 d of DD. Fluctuation of PER immunoreactivity is shown in A, and TIM immunoreactivity is shown in B. LN signals,blue lines and diamonds; DN2Ls, pink lines andcircles; DN1Ls, green lines and squares. Error bars represent SEM of 0.09 or larger. The light regime is shown at the bottomof B, where the white bar indicates the last period when lights were on, the black bars indicate when the lights were off, and the hatched bars indicate when the lights would have been on had the LD cycle been continued. For all of the data points, 5–10 (mean = 7.8) samples (10–20 brain hemispheres) were scored. Larvae were raised in LD cycles for 4 d after 24 hr of egg laying. Starting on the fourth day, 10–20 larvae were dissected at ∼ZT 23, 6, 12, 18, and 23 in an LD cycle (first day in the graphs, as indicated by the black and open bars at the bottom). Therefore, 0 hr on theabscissa corresponds to ZT 0 on the fourth day. Then the remaining larvae were maintained in constant darkness (DD) for 2 further days (in particular, 42 hr of DD subsequent to the end of the “D” portion of the LD cycle), and 10–20 larvae were dissected every ∼6 hr. By the seventh day many of the larvae had completed pupariation. One-half of the brains dissected at each time point were stained for PER immunoreactivity, and the other half were stained for TIM immunoreactivity. After brains from all of the time points were stained, PER and TIM immunoreactivities were blindly scored simultaneously by the second such method (see Materials and Methods). The resulting mean and SEM values for each time point were plotted: inA, PER immunoreactivity; in B, TIM immunoreactivity.
Anti-PDH signals and coexpression of per
In adults, most of the ventral group of LNs (LNvs) are immunoreactive to the peptide hormone PDH (Helfrich-Förster, 1995). This PDH immunoreactivity in the group of smaller LNvs (small LNvs) was shown to persist from early larval stage through the metamorphosis (Helfrich-Förster, 1997). In the current study BG expression was found to persist in a similar set of LNs (see above). If these BG-expressing LNs in larvae are PDH-immunoreactive, it would support the idea that LNs in larvae are indeed the precursors of a subset of LNvs in the adults. Thus, L3 brains of BG were double-labeled for X-gal and anti-PDH at ZT 0. The double-labeling procedure was performed on 22 brains (44 brain hemispheres) and gave reliable results on 41 brain hemispheres. One example is shown in Figure9.
The numbers of LNs labeled by either BG expression or PDH presence, or both, are shown in Table 6. In ∼80% of the valid brain hemispheres, four or five LNs were revealed for either BG or PDH, or both. In most of the samples in which five LNs were revealed, all of the LNs were stained for BG, but the number of PDH-stained LNs never exceeded four. These PDH-stained LNs also were stained for BG in all 12 such cases. In most of the specimens for which four LNs were revealed, all four of the neurons were indeed double-labeled for PDH and BG.
Table 6.
Lateral neurons of BG transgenic larvae stained for β-gal, by anti-PDH, or both
| No. LNs revealed | 5 LNs | 4 LNs | 3 LNs | 2 LNs |
|---|---|---|---|---|
| No. total hemispheres | 13 | 19 | 8 | 1 |
| No. hemispheres with 4 LNs double-labeled | 126-a | 17 | – | – |
| No. hemispheres with 3 LNs double-labeled | – | 26-c | 7 | – |
| No. hemispheres with 2 LNs double-labeled | 16-b | – | 16-a | – |
| No. hemispheres with 1 LN double-labeled | – | – | – | 16-c |
The number of β-gal-labeled LNs exceeds the number of PDH-labeled LNs by one.
Two other cells were stained only for PDH and one only for BG-encoded β-gal.
The number of PDH-labeled LNs exceeds the number of β-gal-labeled LNs by one.
These results indicate that there are five larval LNs, of which four contain PER and PDH, suggesting that these four doubly expressing neurons correspond to the precursor of small LNvs in adults, whereas one LN contains only PER and had not been identified in adults previously (Helfrich-Förster, 1995).
One further feature of the PDH-related results was that the immunohistochemical signals revealed the projections of the four small LNvs in the double-labeled preparations. Interestingly, their terminals were always located in close proximity to the cell bodies of DN2Ls (Fig.9A,C,F).
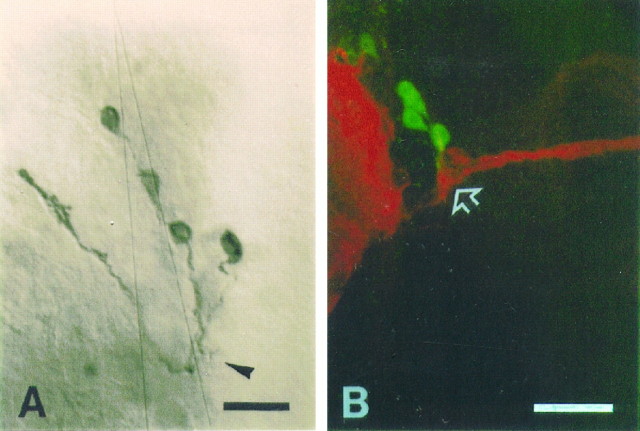
Larval photoreceptors terminate close to LNs
In larvae, LNs are located near the larval optic neuropile, where larval photoreceptor axons called Bolwig’s nerve terminate (for review, see Meinertzhagen and Hanson, 1993), although some of the axons may extend further into the central brain, following the axons of larval optic lobe pioneer cells (OLPs) (Steller et al., 1987; Tix et al., 1989; Green et al., 1993; Campos et al., 1995). Anti-PDH immunohistochemistry showed that the larval LNs send faint processes (putative dendritic trees) into the larval optic neuropile (Fig.10A;Helfrich-Förster, 1997). Therefore, it was of interest to determine whether the LNs are associated with larval photoreceptor axons. Accordingly, L3 larvae carrying the BG transgene were dissected at ∼ZT 0 and then double-labeled with anti-β-gal and the mAb 22C10 to visualize both of these structures. [mAb 22C10 recognizes a cytoplasmic antigen found in all peripheral neurons and in a subset of neurons in CNS (Zipursky et al., 1984).] In particular, this antibody recognizes Bolwig’s nerve and OLPs that fasciculate together (Campos et al., 1995).
Fig. 10.
Spatial relationship between LNs and Bolwig’s nerve. A, Anti-PDH immunohistochemistry on anL3 CNS of a wild-type larva revealed four LNs sending fine dendritic processes (arrowhead) as well as axons.B, Confocal image of an L3 CNS of a BG transgenic larva, double-labeled with anti-β-gal (green) and mAb 22C10 (red). mAb 22C10 revealed Bolwig’s nerve terminating near the border between the central brain and the optic lobe primordium (arrow). The neuropile of the central brain also is strongly labeled by mAb 22C10 on the left side. Five LNs and their short processes were revealed by anti-β-gal. One of the LNs, located anterior to the others, was stained weakly. The processes of the LNs run toward the terminal of the Bolwig’s nerve. Scale bars, 20 μm.
In all, 36 brain hemispheres from larvae of the BG6a strain were observed. In 21 of these hemispheres, five LNs were stained with anti-β-gal. In nine brain hemispheres at least four of five cells are located just next to the terminal of Bolwig’s nerve. In those samples in which they are not close to the terminal of Bolwig’s nerve, short processes projecting from the five cell bodies could be recognized by anti-β-gal staining, and the processes extended in the direction of the terminal of Bolwig’s nerve. In approximately one-half of the brain hemispheres observed, the processes of some of the neurons could be seen very close to the terminal of Bolwig’s nerve (Fig.10B). These results suggest that the five neurons are closely associated with the terminal of Bolwig’s nerve.
DISCUSSION
We have shown that the Drosophila clock genesper and tim are expressed in the CNS. Previous studies on per failed to detect appreciable levels of its expression at this developmental stage, except that a low level ofper expression was found in whole-larval extracts by Northern blotting (Bargiello et al., 1987). The probable reasons for the negative results include the following. (1) perexpression during development seems to be less widespread than in adults. This is true at least for the late embryonic stage, when noper products were detectable outside the nervous system (James et al., 1986; Liu et al., 1988; Siwicki et al., 1988), which would make it difficult to detect the expression of this gene by Northern blotting (Bargiello and Young, 1984; James et al., 1986). (2) The anti-PER antibody used in the current study is more sensitive than one that was used previously (Siwicki et al., 1988). (3) The previous histological study on SG per-lacZ fusion gene expression in larvae was performed on sectioned tissues (Liu et al., 1988). Although some staining was found in CNSs from L1, detailed observation of the staining pattern was difficult on these sections possibly because of weak staining.
Relevance of larval expression to later events
Among the per-expressing cells in such developing animals, larval LNs—which we believe are identical to the small LNvs of adults (cf. Fig. 9 in Helfrich-Förster, 1997)—expressed per from early L1 onward. This coincides well with the time at which the pacemaking system ofDrosophila seems first able to be timed by light pulses (Brett, 1955; Sehgal et al., 1992). Because all of the other larval neurons that express per cyclically begin to be stained later, it seems as if only the LNvs could transferper-related time memory from this early stage into pupae and adults. Furthermore, these LNs are the only larvalper-expressing cells that maintain strong percycling throughout metamorphosis.
A contribution of other per-expressing neurons to the actual “gating” of a developmental event such as eclosion rhythm nevertheless is possible. Such non-LNs, however, would seem not to be suited to transfer larval time memory into pupae and adults, because those other cells (such as the DNLs) either develop too late or do not express per during early metamorphosis.
per-Expressing cells andtim-expressing cells
Among the more intriguing of our developmentally related findings is that spatial expressions of PER and TIM (and temporal ones as well; see below) were not as one might expect from the adult patterns. In larvae, cycling of the clock gene products was detected only in LNs, DN1Ls, and DN2Ls. In the other groups of per neurons, SG fusion protein was expressed strongly in cytoplasm, but BG and endogenous PER were expressed weakly, and TIM was undetectable (at least in terms of epifluorescence microscopy). Our findings suggest that PER and BG (but not SG) stay low in the cells in which TIM is low. This is consistent with biochemical results obtained in adults, which have shown that both TIM and PER are necessary for nuclear translocation and normal molecular cycling of both gene products and that the level of PER is low in a genetic background possessing no tim+ function (Price et al., 1995; Myers et al., 1996; Zeng et al., 1996). The SG fusion protein, on the other hand, may not require TIM for its stability (Dembinska et al., 1997).
Cycling phases of PER and TIM in dorsal neurons of larvae
The most unexpected result in this study was the presence of antiphase cycling of clock gene products in certain dorsally located neurons: the DN2Ls of larvae. In particular, high level of TIM in the DN2Ls during the day was intriguing, considering one of the most salient findings from studying the tim gene product: TIM in the adult head decreases sharply in the face of chronic or pulsatile exposure of the flies to light (for review, see Sehgal et al., 1996). It was suggested that this involves light-induced degradation of TIM, without knowing whether the effect of that stimulus is direct or indirect. One element of the current findings (TIM cycling in the DN2Ls) is to indicate that TIM is not a light-sensitive protein by itself. Factors that may be necessary to degrade TIM quickly in response to light would seem to be lacking from the larval neurons in question.
Previous biochemical studies of adults have revealed similar cycling phases for per in eyes and brains (Zeng et al., 1994), justifying the use of whole adult head as a tissue source for biochemical assays. However, slight but clear differences in PER cycling phases between photoreceptors and CNS cells have been reported (Siwicki et al., 1988; Zerr et al., 1990). The larval pattern indicates more striking intercellular phase differences, in particular, between DN2Ls and LNs.
The mechanisms responsible for these phase differences are unknown. One possibility is that DN2Ls may not be entrained directly by light or the photoreceptors but, rather, are entrained—after a delay—by the LNs; the latter would be entrained directly by the light or by light input from the larval photoreceptors. This hypothesis is supported by the fact that processes from LNs terminate close to DN2Ls, and larval photoreceptors terminate close to LNs.
Furthermore, the relatively high and constant level of the BG fusion protein, as well as that of native TIM, within the DN2Ls of larvae suffering the effects of a loss of per function could be explained by this hypothesis. Aper null genotype, in particular within the larval LNs, would leave such cells always at the trough level time point of PER (despite the presence of BG protein, which is not functional in adults;Stanewsky et al., 1997a). When the amount of PER is at the trough level in the LNs, it is near its peak time point in the DN2Ls. Therefore, under the hypothesis that LNs are entraining DN2Ls,per01 causes the level of BG fusion protein and TIM to stay chronically high in DN2Ls.
However, these findings and interpretations are not in accord with the results of monitoring PER and TIM cycling in DD. Amplitudes for both proteins seem to dampen faster in DN2Ls than in LNs, suggesting that DN2Ls may be influenced more strongly by LD cycles than LNs are. Because TIM is relatively high during most of the daytime, light may induce the production of TIM, stabilize it in DN2Ls, or both. This hypothesis would require a different molecular mechanism related to the effects of light on, as well as the basic oscillations of PER and TIM in the DN2Ls.
Light entrainment pathway from larval photoreceptors to the clock neurons
Eclosion rhythmicity (Brett, 1955) and even the locomotor activity rhythms of adults (Sehgal et al., 1992) can be synchronized by light pulses given to L1 larvae. The photoreceptive system that transfers this environmental information to LNs must, therefore, be fully developed by the onset of the L1. A larval photoreceptor structure called Bolwig’s nerve is present at the beginning of L1 (Steller et al., 1987). Here we show that inL3 the Bolwig’s nerve terminates in close vicinity to the LNs, making it likely that putative circadian pacemakers receive photic information via Bolwig’s nerve. However, extraretinal photoreception seems to be involved in entraining Drosophila’s eclosion rhythm (for review, see Zimmerman and Ives, 1971).
Correspondence of larval clock neurons with adult clock neurons
Four PER-expressing small LNvs are immunoreactive with respect to the PDH neuropeptide and are most likely the precursors of the PDH-immunoreactive small LNvs in adults. However, the fifth LN present in larvae expressed only the clock gene. In principle, this fifth LN could correspond to part of either the LNd or the LNv cluster in pupae and adults. However, BG expression in five LNs was revealed during the early pupal stage before the LNd clusters were first recognized at ∼60% of the way through metamorphosis. Therefore, it seems more likely that the fifth, small PDH-negative LN belongs to the group of small LNvs.
The larval DN1Ls, DN2Ls, and DN3Ls could be the same cells as DN1s, DN2s, and DN3s, respectively, in pupae and adults—judging by the positions of all of these cells, although none of them seems to expressper continuously through metamorphosis. Along these lines, antiphase cycling in DN2Ls, found for PER and BG, was not detectable in the pupal DN2s for BG nor in the adult DN2s, where the cycling phase was in phase with the rest of the brain (M. Kaneko, unpublished observations). Therefore, DN2Ls would have to change their PER cycling phase during the metamorphosis if their pupal and mature adult counterparts are in fact the same cells.
Application of the larval brain to in vitro studies of the circadian clock
In the adult head, per and tim are expressed in photoreceptors, glia, and neurons (Siwicki et al., 1988; Zerr et al., 1990; Ewer et al., 1992; Hunter-Ensor et al., 1996). Notwithstanding this heterogeneity of clock cell types, it usually has been assumed that functions of per and tim are the same in all of these cell types. However, the differences in phases of PER cycling in adults (Zerr et al., 1990), and especially in developing Drosophila (of PER and TIM), suggest the need for studies at the single cell level.
In this regard, individual neurons from the larval CNS ofDrosophila are relatively easy to dissociate from other cells (Wu et al., 1983; Ui et al., 1994). Therefore, separation of LNs from DNLs and mass isolation of each cell type should be relatively easy on this tissue, using a reporter gene such as green fluorescent protein (Brand, 1995) and fluorescence-activated cell sorting (Amrein and Axel, 1997). Such cells could be used not only in biochemical and molecular biological assays but also in electrophysiological and pharmacological studies, as well as those that involve monitoring cyclical clock gene expression in real time (cf. Emery et al., 1996).
Footnotes
This work was supported by National Institutes of Health Grant GM-33205 and Deutsche Forschungsgemeinschaft Fo 207/3. We are grateful for antisera from Michael W. Young (anti-TIM), Michael Rosbash (anti-PER), Ralf Stanewsky (anti-PER), and Heinrich Dircksen (anti-PDH). We thank Kalpana White, Michael Rosbash, Stephen F. Goodwin, and Ralf Stanewsky for comments on this manuscript. The mAb 5D was kindly supplied by Kalpana White and the mAb 22C10 by Seymour Benzer. We appreciate the slide-coding efforts of Alexandre A. Peixoto and Ralf Stanewsky and the excellent photographic assistance provided by Ed Dougherty.
Correspondence should be addressed to Dr. Jeffrey C. Hall, Department of Biology, 235 Bassine Building, Brandeis University, 415 South Street, Waltham, MA02254-9110.
REFERENCES
- 1.Amrein H, Axel R. Genes expressed in neurons of adult male Drosophila. Cell. 1997;88:459–469. doi: 10.1016/s0092-8674(00)81886-3. [DOI] [PubMed] [Google Scholar]
- 2.Bargiello TA, Young M. Molecular genetics of a biological clock in Drosophila. Proc Natl Acad Sci USA. 1984;81:2142–2146. doi: 10.1073/pnas.81.7.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bargiello TA, Saez L, Baylies MK, Gasic G, Young MW, Spray DC. The Drosophila clock gene per affects intercellular junctional communication. Nature. 1987;328:686–691. doi: 10.1038/328686a0. [DOI] [PubMed] [Google Scholar]
- 4.Bier E, Ackerman L, Barbel S, Jan L, Jan YN. Identification and characterization of a neuron-specific nuclear antigen in Drosophila. Science. 1988;240:913–916. doi: 10.1126/science.3129785. [DOI] [PubMed] [Google Scholar]
- 5.Bodenstein D. The postembryonic development of Drosophila. In: Demerec M, editor. Biology of Drosophila. Wiley; New York: 1950. pp. 275–367. [Google Scholar]
- 6.Brand A. GFP in Drosophila. Trends Genet. 1995;11:324–325. doi: 10.1016/s0168-9525(00)89091-5. [DOI] [PubMed] [Google Scholar]
- 7.Brett WJ. Persistent diurnal rhythmicity in Drosophila emergence. Ann Entomol Soc Am. 1955;48:119–131. [Google Scholar]
- 8.Campos AR, Lee KJ, Steller H. Establishment of neuronal connectivity during development of the Drosophila larval visual system. J Neurobiol. 1995;28:313–329. doi: 10.1002/neu.480280305. [DOI] [PubMed] [Google Scholar]
- 9.Dembinska ME, Stanewsky R, Hall JC, Rosbash M. Circadian cycling of a period-lacZ fusion protein in Drosophila: evidence for an instability cycling element in PER. J Biol Rhythms. 1997;12:157–172. doi: 10.1177/074873049701200207. [DOI] [PubMed] [Google Scholar]
- 10.Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emery IF, Noveral JM, Jamison CF, Siwicki KK. Rhythms of Drosophila period gene expression in culture. Proc Natl Acad Sci USA. 1997;94:4092–4096. doi: 10.1073/pnas.94.8.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. J Neurosci. 1992;12:3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 14.Green P, Hartenstein AY, Hartenstein V. The embryonic development of Drosophila visual system. Cell Tissue Res. 1993;273:583–598. doi: 10.1007/BF00333712. [DOI] [PubMed] [Google Scholar]
- 15.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 16.Hardin PE, Hall JC, Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci USA. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helfrich-Förster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc Natl Acad Sci USA. 1995;92:612–616. doi: 10.1073/pnas.92.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helfrich-Förster C. Development of pigment-dispersing hormone immunoreactive neurons in the nervous system of Drosophila melanogaster. J Comp Neurol. 1997;380:335–354. doi: 10.1002/(sici)1096-9861(19970414)380:3<335::aid-cne4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Helfrich-Förster C, Homberg U. Pigment-dispersing hormone-immunoreactive neurons in the nervous system of wild-type Drosophila melanogaster and of several mutants with altered circadian rhythmicity. J Comp Neurol. 1993;337:177–190. doi: 10.1002/cne.903370202. [DOI] [PubMed] [Google Scholar]
- 20.Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- 21.James AA, Ewer J, Reddy P, Hall JC, Rosbash M. Embryonic expression of the period clock gene in the central nervous system of Drosophila melanogaster. EMBO J. 1986;5:2313–2320. doi: 10.1002/j.1460-2075.1986.tb04499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konopka R, Wells S, Lee T. Mosaic analysis of a Drosophila clock mutant. Mol Gen Genet. 1983;190:284–288. [Google Scholar]
- 24.Konopka RJ, Hamblen-Coyle MJ, Jamison CF, Hall JC. An ultrashort clock mutation at the period locus of Drosophila melanogaster that reveals some new features of the fly’s circadian system. J Biol Rhythms. 1995;9:189–216. doi: 10.1177/074873049400900303. [DOI] [PubMed] [Google Scholar]
- 25.Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Lorenz L, Yu Q, Hall JC, Rosbash M. Spatial and temporal expression of the period gene in Drosophila melanogaster. Genes Dev. 1988;2:228–238. doi: 10.1101/gad.2.2.228. [DOI] [PubMed] [Google Scholar]
- 27.Marrus SB, Zeng H, Rosbash M. Effect of constant light and circadian entrainment of perS flies: evidence for light-mediated delay of the negative feedback loop in Drosophila. EMBO J. 1996;15:6877–6886. [PMC free article] [PubMed] [Google Scholar]
- 28.Meinertzhagen IA, Hanson TE. The development of the optic lobe. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1993. pp. 1363–1491. [Google Scholar]
- 29.Myers MP, Wager-Smith K, Wesley CS, Young MW, Sehgal A. Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science. 1995;270:805–808. doi: 10.1126/science.270.5237.805. [DOI] [PubMed] [Google Scholar]
- 30.Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 31.O’Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 32.Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. In: Goldstein LSB, Fryberg EA, editors. Methods in cell biology, Vol 44, Drosophila melanogaster: practical uses in cell and molecular biology. Academic; San Diego: 1994. pp. 445–487. [DOI] [PubMed] [Google Scholar]
- 33.Price JL, Dembinska ME, Young MW, Rosbash M. Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. EMBO J. 1995;14:4044–4049. doi: 10.1002/j.1460-2075.1995.tb00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinow S, White K. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev Biol. 1988;126:294–303. doi: 10.1016/0012-1606(88)90139-x. [DOI] [PubMed] [Google Scholar]
- 35.Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- 36.Rutila JE, Zeng H, Le M, Curtin KD, Hall JC, Rosbash M. The timSL mutant of the Drosophila rhythm gene timeless manifests allele-specific interactions with period gene mutants. Neuron. 1996;17:921–929. doi: 10.1016/s0896-6273(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 37.Sehgal A, Price J, Young MW. Ontogeny of a biological clock in Drosophila melanogaster. Proc Natl Acad Sci USA. 1992;89:1423–1427. doi: 10.1073/pnas.89.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sehgal A, Price JL, Man B, Young MW. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- 39.Sehgal A, Rothenfluh-Hilfiker A, Hunter-Ensor M, Chen Y, Myers MP, Young MW. Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- 40.Sehgal A, Ously A, Hunter-Ensor M. Control of circadian rhythms by two-component clock. Mol Cell Neurosci. 1996;7:165–172. doi: 10.1006/mcne.1996.0013. [DOI] [PubMed] [Google Scholar]
- 41.Simon JA, Sutton CA, Lobell RB, Glaser RL, Lis JT. Determinants of heat shock-induced chromosome puffing. Cell. 1985;40:805–817. doi: 10.1016/0092-8674(85)90340-x. [DOI] [PubMed] [Google Scholar]
- 42.Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC. Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron. 1988;1:141–150. doi: 10.1016/0896-6273(88)90198-5. [DOI] [PubMed] [Google Scholar]
- 43.Stanewsky R, Frisch B, Brandes C, Hamblen-Coyle MJ, Rosbash M, Hall JC. Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: mapping elements of the PER protein involved in circadian cycling. J Neurosci. 1997a;17:676–696. doi: 10.1523/JNEUROSCI.17-02-00676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanewsky R, Jamison CF, Plautz JD, Kay SA, Hall JC (1997b) Multiple circadian-regulated elements contribute to cycling period gene expression in Drosophila. EMBO J, in press. [DOI] [PMC free article] [PubMed]
- 45.Steller H, Fischbach K-F, Rubin GM. Disconnected: a locus required for neuronal pathway formation in the visual system of Drosophila. Cell. 1987;50:1139–1153. doi: 10.1016/0092-8674(87)90180-2. [DOI] [PubMed] [Google Scholar]
- 46.Sternberger LA. Immunocytochemistry. Wiley; New York: 1979. [Google Scholar]
- 47.Tix S, Minden JS, Technau GM. Pre-existing neuronal pathways in the developing optic lobes of Drosophila. Development. 1989;105:739–746. doi: 10.1242/dev.105.4.739. [DOI] [PubMed] [Google Scholar]
- 48.Ui K, Nishihara S, Sakuma M, Togashi S, Ueda R, Miyata Y, Miyake T (1994) Newly established cell lines fromDrosophila larval CNS express neural specific characteristics. In Vitro Cell Dev Biol Anim 30[Suppl A]:209–216. [DOI] [PubMed]
- 49.Van Gelder RN, Krasnow MA. A novel circadianly expressed Drosophila melanogaster gene dependent on the period gene for its rhythmic expression. EMBO J. 1996;15:1625–1631. [PMC free article] [PubMed] [Google Scholar]
- 50.Van Gelder RN, Bae H, Palazzolo MJ, Krasnow MA. Extent and character of circadian gene expression in Drosophila melanogaster: identification of twenty oscillating mRNAs in the fly head. Curr Biol. 1995;5:1424–1436. doi: 10.1016/s0960-9822(95)00280-6. [DOI] [PubMed] [Google Scholar]
- 51.Whitfield WGF, Gonzalez C, Maldonado-Codina G, Glover DM. The A- and B-type cyclins of Drosophila are accumulated and destroyed in temporally distinct events that define separable phases of the G2-M transition. EMBO J. 1990;9:2563–2572. doi: 10.1002/j.1460-2075.1990.tb07437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu C-F, Suzuki N, Poo M-M. Dissociated neurons from normal and mutant Drosophila larval central nervous system in cell culture. J Neurosci. 1983;3:1888–1899. doi: 10.1523/JNEUROSCI.03-09-01888.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng H, Hardin PE, Rosbash M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J. 1994;13:3590–3598. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng H, Qian Z, Myers MP, Rosbash M. A light entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 55.Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zimmerman WF, Ives D. Some photophysiological aspects of circadian rhythmicity in Drosophila. In: Menaker M, editor. Proceedings of the symposium on biochronometry. National Academy of Science; Washington, DC: 1971. pp. 381–391. [Google Scholar]
- 57.Zipursky SL, Venkatesh TR, Teplow DB, Benzer S. Neuronal development in the Drosophila retina: monoclonal antibodies as molecular probes. Cell. 1984;36:15–26. doi: 10.1016/0092-8674(84)90069-2. [DOI] [PubMed] [Google Scholar]