Abstract
Native AMPA receptors (AMPARs) were investigated in neocortical fast-spiking (FS) and regular-spiking nonpyramidal (RSNP) cells. The onset of and recovery from desensitization as well as current rectification and single-channel conductance were studied by using fast glutamate application to outside-out patches. The GluR1–4 subunit, flip/flop splicing, and R/G editing expression patterns of functionally characterized cells were determined by single-cell reverse transcription-PCR to correlate the subunit composition of native AMPARs with their functional properties. Our sample, mostly constituted by RSNP neurons, predominantly expressed GluR3 flip and GluR2 flop. In individual cells, flip/flop splicing of each subunit appeared to be regulated independently, whereas for R/G editing all subunits were either almost fully edited or unedited. We confirmed that the relative GluR2 expression controls the permeation properties of native AMPARs, whereas none of the single molecular parameters considered appeared to be a key determinant of the kinetics. FS neurons displayed AMPARs with relatively homogeneous functional properties characterized by fast desensitization, slow recovery from desensitization, marked inward rectification, and large single-channel conductance. In contrast, these parameters varied over a wide range in RSNP neurons, and their combination resulted in various AMPAR functional patterns. Indeed, in different cells, fast or slow desensitization was found to be associated with either slow or fast recovery from desensitization. Similarly, fast or slow kinetics was associated with either strong or weak rectification. Our results suggest that kinetic and permeation properties of native AMPARs can be regulated independently in cortical neurons and probably do not have the same molecular determinants.
Keywords: AMPA receptor, neocortex, interneuron, single-cell RT-PCR, fast glutamate application, subunit, splicing, editing
The fast excitatory synaptic transmission in the CNS is mediated mainly by AMPA receptor channels (AMPARs). These receptors are multimeric assemblies of four different subunits GluR1–4 (for review, see Wisden and Seeburg, 1993; Hollmann and Heinemann, 1994). Further diversity is generated by alternative splicing (Sommer et al., 1990) and mRNA editing (Sommer et al., 1991;Lomeli et al., 1994). In heterologous expression systems recombinant AMPAR permeation properties are controlled by the relative abundance of GluR2 (for review on voltage dependence and calcium permeability, seeHollmann and Heinemann, 1994; Jonas and Burnashev, 1995) [see alsoSwanson et al. (1997) for single channel conductance]. In contrast, their kinetic properties are affected by multiple factors, including subunit composition, flip/flop alternative splicing, and mRNA edition at the R/G site (Sommer et al., 1990; Lomeli et al., 1994; Mosbacher et al., 1994; Partin et al., 1994).
Studies of native AMPARs by patch-clamp recordings combined with single-cell reverse transcription-PCR (single-cell RT-PCR; Lambolez et al., 1992) have confirmed the role played by the GluR2 subunit in their permeation (Bochet et al., 1994; Jonas et al., 1994; Geiger et al., 1995), but the identity of the molecular determinants controlling their kinetics is still controversial. Indeed, slow and fast desensitizations were correlated with either GluR2 flip and GluR4 (Geiger et al., 1995), respectively, or to flip and flop splice variants (Lambolez et al., 1996). The correlation found by Geiger et al. (1995) suggests that permeation and kinetic properties of native AMPARs may not be regulated independently and would range from fast desensitizing–calcium permeable (as found in interneurons) to slow desensitizing–calcium impermeable (as found in pyramidal cells). This is indeed the case for most neural cell types studied so far.
In the present work the kinetic and permeation properties of AMPARs were studied in neocortical nonpyramidal cells and the GluR1–4, flip/flop splicing, and R/G editing expression patterns of electrophysiologically characterized cells were determined by single-cell RT-PCR. Neocortical nonpyramidal neurons comprise the fast-spiking (FS) cells that show relative homogeneity in their morphology, action potential firing, biochemical markers expression, and AMPAR properties and the highly heterogeneous regular-spiking nonpyramidal (RSNP) cells for which the AMPARs have not been studied (McCormick et al., 1985; Hestrin, 1993; Kawaguchi and Kubota, 1993;Jonas et al., 1994; Kawaguchi, 1995; Lambolez et al., 1996; Cauli et al., 1997).
Nonpyramidal cells showed a large diversity of AMPAR functional patterns in which permeation and kinetic properties varied independently. We confirmed that permeation properties of native AMPARs are determined by GluR2. In contrast, none of the single molecular parameters considered appeared to be a key determinant of kinetic properties. Our results suggest that the functional diversity of native AMPARs derives from the combination of independent molecular variables and that specific combinations of these variables vary on a cell type basis.
MATERIALS AND METHODS
Brain slices preparation and patch-clamp recordings.Young Wistar rats (13–17 postnatal days old) were anesthetized with ketamine (65 mg/kg) and xylazine (14 mg/kg) and decapitated. Brains were removed quickly, and 300-μm-thick parasagittal slices were prepared from cerebral frontal cortex with a vibratome (DSK microslicer DTK-1000, Dosaka, Japan), following the procedure ofEdwards et al. (1989). For recordings, slices were transferred to a chamber perfused at 2–3 ml/min with a physiological extracellular saline solution containing (in mm): 121 NaCl, 2.5 KCl, 1.25 NaH2PO4, 2 CaCl2, 1 MgCl2, 5 Na-pyruvate, and 15 glucose, bubbled with a mixture of 95% O2/5% CO2. Patch pipettes were pulled from borosilicate glass tubing and had a resistance of 5–10 MΩ when filled with an internal solution containing (in mm): 144 K-gluconate, 3 MgCl2, 0.2 EGTA, 10 HEPES, 2 ATP, 0.2 GTP, and 100 μm spermine, pH 7.2–7.4, 300 mOsm. Spermine was included because it maintains the voltage dependence of AMPARs in excised patches (Bowie et al., 1995; Kamboj et al., 1995; Koh et al., 1995). All potentials were corrected for junction potential (−12 mV) (Neher, 1992). Whole-cell recordings were performed from layers IV/VI nonpyramidal cells at room temperature. Only neurons with resting membrane potential more negative than −62 mV were considered further.
Cells were identified initially by videomicroscopy with Nomarski optics under infrared illumination (Stuart et al., 1993). Neurons with characteristic pyramidal shapes were excluded from the sample. Furthermore, the kinetics of the action potential firing of recorded cells was analyzed to discard those with electrophysiological patterns of pyramidal cells (see Data Collection and Analysis; McCormick et al., 1985; Connors and Gutnick, 1990; Kawaguchi, 1995; Lambolez et al., 1996).
After intracellular recordings outside-out membrane patches were excised from nonpyramidal cells to study native AMPARs. Fast application experiments on outside-out patches were performed by a method originally developed by Franke et al. (1987). A double-barreled application pipette (100–200 μm tip diameter) was operated by a piezoelectrical device (PZ-150 m, Burleigh Instruments, Fishers, NY). Two solutions were used for rapid perfusion: a control solution containing (in mm) 135 NaCl, 5.4 KCl, 1.8 CaCl2, 1 MgCl, and 10 HEPES (315 mOsm) and a test solution containing, additionally, 10 mm glutamate and 50 mm sucrose. To adjust the osmolarity, we diluted the test solution 1:10 with water before the addition of the glutamate (final concentration 10 mm). Isolated patches were positioned in the control solution near the interface of the agonist-containing stream. The rate of the solution exchange was estimated at the end of each recording by measuring the current resulting from the junction potential between the two solutions with the open patch pipette. The exchange time (20–80%) of considered patches was 0.26 ± 0.1 msec (mean ± SD; n = 57), and the mean rise time (20%–80%) of glutamate-induced currents was 0.35 ± 0.16 msec (n = 66).
Data collection and analysis. Intracellular current-clamp (mode I-Clamp fast) and outside-out voltage-clamp recordings were obtained by a patch-clamp amplifier (Axopatch 200A, Axon Instruments, Foster City, CA) and filtered at 5 and 2 kHz, respectively. Digitized data were acquired and analyzed with Acquis1 software (Gérard Sadoc, Centre National de la Recherche Scientifique, Gif-sur-Yvette, France).
The characteristics of the action potential discharges induced by depolarizing current pulses were analyzed following the procedure described by Cauli et al. (1997). Briefly, the instantaneous discharge frequency was determined all along the discharge and plotted as a function of time at the highest intensity tested (400–600 pA). The instantaneous discharge frequencies between the first two spikes (finitial) and at the end of the stimulation (ffinal) were used to calculate the accommodation of the firing frequency during the discharges according to (finitial −ffinal)/finitial. Two physiological groups of nonpyramidal cells were defined according to their action potential firing behavior, as defined by Cauli et al. (1997). Neurons of the first group corresponded to fast-spiking cells, which fired fast action potentials at a high constant rate. Cells of the second group were unable to sustain high frequencies of repetitive discharges and corresponded to RSNP cells (Kawaguchi, 1995). Uncertain cases were distinguished by measuring the input resistance, the spike amplitude and duration, and the after-hyperpolarization amplitude of the first and second action potentials of the discharges as previously described (for details, see Cauli et al., 1997). Pyramidal cells were discriminated from these two populations of nonpyramidal cells according to their smaller after-hyperpolarization (less than −7 mV), longer spike duration (>1.5 msec), and stronger accommodation of the firing frequency (>80%) (for details, see Cauli et al., 1997).
Different characteristics of AMPARs were studied simultaneously in a single isolated patch. To study the kinetics of desensitization of these receptors, we applied 10 mm glutamate steps of 100 msec at −72 mV (step frequency 0.2 Hz). The desensitization time constant was obtained by fitting the decay of the averaged current (10–50 traces) by a single exponential function. A nonstationary noise analysis was made from glutamate-induced responses (22–50 traces) that did not show any rundown to determine the weighted mean single-channel conductance, as previously described (Sigworth, 1980; Cull-Candy et al., 1988; Hestrin, 1993; Silver et al., 1996). For this purpose the variance–amplitude (ς2–I) relationship was plotted. The shape of the ς2–I relationship was either linear (n = 22) or parabolic (n = 6), and the plot was fit with a linear regression function ( of the points) or with a parabolic function (all points), respectively. The fact that the shape of the ς2–I relationship was often linear suggests that the open probability of AMPARs is low in our experimental conditions (see Silver et al., 1996). The slope of the variance–amplitude relationship was considered as a weighted mean single-channel current and divided by the driving force. We assumed that the driving force was −70 mV, determined from the reversal potential (Erev = −2 mV) calculated from ion concentrations of the external and internal solutions.
Furthermore, a rectification index (RI) was calculated from current–voltage (I–V) curves with the following expression (Ozawa et al., 1991; Bochet et al., 1994):
| Equation 1 |
where I28 andI−72 correspond to the normalized current at 28 and −72 mV, respectively, and Erev corresponds to the experimental reversal potential. Currents were normalized with respect to the I−72 for comparison among cells.
Finally, the recovery from desensitization was studied by applying pairs of 1 msec pulses of glutamate (Hestrin, 1993; Lomeli et al., 1994). The two application pulses were separated by increasing time intervals (from 5 to 350 msec). For each pulse pair we measured the amplitude of the first current (current 1) and the amplitude of second current (current 2); the ratio of current 2/current 1 was plotted against time intervals. The time required for 50% recovery from desensitization (t50%) was estimated by interpolation.
Statistical data are given as mean ± SD. Correlations between functional and molecular parameters (see below) were tested with the nonparametrical Spearman test. We computed the Spearman rank linear correlation coefficient (rs), and we calculated U = ✓(n − 1)rs, which follows the distribution of Laplace–Gauss (Lebart et al., 1982).
Molecular analysis by single-cell RT-PCR. A second whole-cell recording was performed on functionally characterized cells. The patch pipette (3–5 MΩ) used to harvest the cytoplasm was filled with 8 μl of autoclaved RT-PCR internal solution containing (in mm): 144 K-gluconate, 3 MgCl2, 0.2 EGTA, and 10 HEPES, pH 7.4, 300 mOsm. The cell content was aspirated under visual control by application of a gentle negative pressure into the pipette. The harvesting was interrupted as soon as the seal was lost. Then the content of the pipette was expelled into a test tube and RT performed in a final volume of 10 μl, as described (Lambolez et al., 1992).
GluR1–4 cDNAs amplification. The RT products were amplified with the common GluR1–4 primers (sense primer, CCTTTGGCCTATGAGATCTGGATGTG; antisense primer, TCGTACCACCATTTGTTTTTCA) with 40 PCR cycles, as described (Lambolez et al., 1992). In 50% of the harvested cells the RT-PCR yielded amplified GluR1–4 cDNAs, as seen by agarose gel electrophoresis of 10 μl of the reaction. The product of positive samples was purified on a 1.5% low-melting-point agarose gel and saved for further analysis.
Quantification of the GluR1–4 relative proportions. The procedure was performed essentially as described by Lambolez et al. (1996), except that quantification was based on fluorescence instead of radioactivity. Aliquots of the purified product from the first PCR were reamplified with the same set of primers, the antisense primer being carboxyfluorescein (FAM) fluorescently labeled at the 5′ end. In all, 25 cycles (94°C, 30 sec; 49°C, 30 sec; 72°C, 45 sec) were performed. The product of the second PCR was cut by BglI,Bsp1286I, Eco47III, or EcoRI, which selectively cut the GluR1, 2, 3, or 4 PCR fragment, respectively (for details, see Lambolez et al., 1992, 1996). The restriction reactions were performed on 1 μl aliquots of the PCR product in a 5 μl volume and, after the addition of 30 μl of H2O, were run by capillary electrophoresis [2.3% fragment analysis reagent (FAR), nondenaturing conditions] in an ABI 310 Genetic Analyzer (Perkin-Elmer, Foster City, CA) and analyzed with the GeneScan software (Perkin-Elmer). Because only the antisense primer was labeled, each restriction cut generated only one labeled restriction fragment. For each run, corresponding to a subunit-specific cut, the total fluorescence present in both the cut and the uncut peaks was normalized to 100%. The percentage of fluorescence present in the cut peak thus represented the percentage of the corresponding subunit in the GluR1–4 amplified products.
Quantification of the flip/flop proportions of GluR1–4.Aliquots of the purified product of the first amplification were submitted to a second amplification with the following sets of primers: the sense primers were specific for GluR1 (R1, GGACGAGACCAGACAACCAG at position 1717; position 1 is the first nucleotide of the initiation codon), GluR2 (R2, TGAAGATGGAAGAGAAACACAAAG at position 1731), GluR3 (R3, ACCCACAAAGCCCTCCTG at position 1757), or GluR4 (R4, GAAGGACCCAGTGACCAGCC at position 1747), the common antisense primer was that used for the first amplification, but FAM 5′ end-labeled. Then 25 cycles (94°C, 30 sec; 49°C, 30 sec; 72°C, 45 sec) were performed. The products of these second amplifications (632, 639, 628, and 630 bp for GluR1, 2, 3, and 4, respectively) were digested with enzymes specific of either the flip or the flop form, yielding fragments of the following size: 568 and 64 for GluR1 flip cut by BfaI, 571 and 68 for GluR2 flop cut by HpaI, 560 and 68 for GluR3 flop cut by HpaI, and 562 and 68 for GluR4 flop cut byHpaI (Bochet et al., 1994; Lambolez et al., 1996).
The restriction reactions were performed on 1 μl aliquots of the PCR product in a 5 μl volume and, after addition of 30 μl H2O, were run by capillary electrophoresis (2.3% FAR, nondenaturing conditions) in an ABI 310 Genetic Analyzer and analyzed with the GeneScan software. For each run, corresponding to a variant-specific cut, the total fluorescence present in both the cut and the uncut peaks was normalized to 100%. The percentage of fluorescence present in the cut band thus represented the percentage of the corresponding flip or flop variant for each subunit. The percentage of the complementary variant was calculated assuming that flip plus flop variants represent 100%. For each cell these relative values were multiplied by the relative amounts of the GluR1 to four subunits previously determined (see above) to obtain the relative proportions of the GluR1–4 flip or flop expressed.
Quantification of the R/G edited proportions of GluR2–4.Aliquots of the products of the subunit-specific second amplification (see above) were submitted to an additional amplification with the following primers common to GluR2, 3, and 4: the sense primer (TGGATTCCAAAGGCTA, positions 2243, 2258, and 2246 on GluR2, 3, and 4, respectively) was FAM 5′ end-labeled and the antisense primer was that used for the previous amplifications. In all, 18 cycles (94°C, 30 sec; 49°C, 30 sec; 72°C, 15 sec) were performed. The product of this amplification (127 bp) was digested by the editing variant-specific enzyme MseI yielding fragments of the following sizes: 127* bp for GluR2–4 flip edited (G form), 82 and 45* bp for GluR2–4 flip unedited (R form), 69 and 58* bp for GluR2–4 flop edited, and 69, 45*, and 13 bp for GluR2–4 flop unedited (* indicates the fragment bearing the FAM fluorophore).
The restriction reactions were performed on 2 μl aliquots of the PCR product in a 5 μl volume and, after addition of 30 μl of H2O, were run by capillary electrophoresis (2.5% FAR, nondenaturing conditions) in an ABI 310 Genetic Analyzer and analyzed with the GeneScan software. In the cases in which only one of the flip or flop forms was present for a given subunit, the percentage of fluorescence present in the 45 bp band thus represented the percentage of the unedited variant for this subunit. When a mix of flip and flop forms was present for a given subunit, both of their unedited variants contributed to the fluorescence of the 45 bp band. The amount of fluorescence contributed by the unedited flop form (oR) was calculated according to the following equation:
| Equation 2 |
where iG (edited flip) is the fluorescence amount in the 127 bp peak, oG(edited flop) is the fluorescence amount in the 58 bp peak,A is the flip/flop ratio (previously determined), andB is the fluorescence amount in the 45 bp peak. Then the percentage of unedited flop [oR/(oR +oG)] and of unedited flip [(B −oR)/(B − (oR + iG))] were calculated. For each cell these relative values were multiplied by the relative amounts of the GluR2–4 flip or flop subunits previously determined (see above) to obtain the relative proportions of the GluR2–4/flip-flop/R-G expressed.
RESULTS
Native AMPARs were studied from layer IV/VI nonpyramidal cells in the sensory motor cortex of 13- to 17-d-old rats. The functional and molecular properties of these receptors were analyzed by combining fast glutamate application to outside-out patches and single-cell RT-PCR. Outside-out patches from 66 nonpyramidal neurons were recorded, and in 27 cells a successful combined analysis was made.
The FS cells and the RSNP cells were discriminated by the frequency accommodation of the action potential trains induced by depolarizing current injections. The average firing accommodation of FS neurons was 33 ± 11% (n = 16), significantly different from that of RSNP neurons (60 ± 12%; n = 47; Student’s t test, p < 0.001) (see Materials and Methods). The proportion of FS neurons (24%) was smaller than that of RSNP neurons (76%).
Analysis of the AMPAR properties in an RSNP neuron
Four functional parameters were measured on glutamate-induced responses: the desensitization time constant of the current (τ), the recovery from desensitization (t50%), the rectification of the I–V curves (RI), and the weighted mean single-channel conductance (g) (see Materials and Methods).
Figure 1 illustrates an analysis of the AMPAR properties in a neuron identified as an RSNP cell by its firing properties (data not shown). The desensitization time constant was determined from the decay of AMPAR currents elicited by glutamate steps of 100 msec (τ = 5.95 msec; Fig. 1A). Recovery from desensitization was determined by applying two pulses of 1 msec at increasing time intervals (from 5 to 350 msec). For short time intervals the current elicited by the second pulse (current 2) was smaller than the current elicited by the first pulse (current 1) (data not shown). The first pulse thus was sufficient to desensitize the glutamate-induced currents. As shown for the RSNP cell of Figure 1, the ratio of current 2/current 1 was plotted against the time between the two pulses. The recovery from desensitization (t50%), interpolated from the plot, was estimated by the time needed for current 2 to reach 50% of current 1 (t50% = 35 msec; Fig.1B).
Fig. 1.
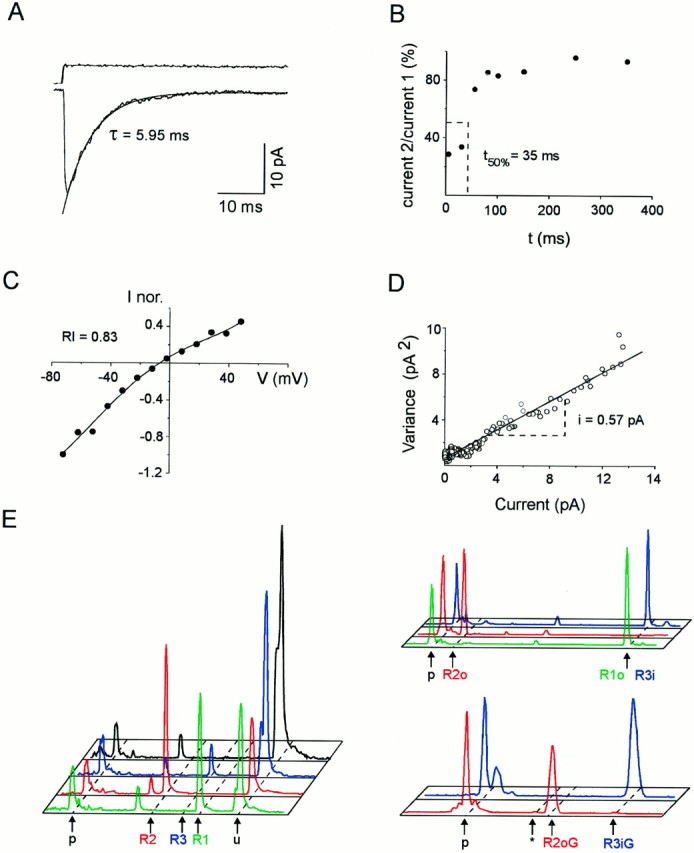
Functional and molecular analysis of AMPARs on a single RSNP neuron. Functional properties were investigated on an excised patch. Subsequently, a second whole-cell recording was obtained on the same cell, and the cellular content was harvested to investigate the GluR1–4 combination expressed. A, Averaged response (bottom trace) to 100 msec step applications (top trace) of 10 mm glutamate (holding potential, −72 mV). The current decay was fit by a single exponential function (τ = 5.95 msec) superimposed to the averaged current; average of 39 responses. B, Recovery from desensitization between currents induced by pairs of 1 msec pulses of glutamate separated by increasing time intervals (from 5 to 350 msec). The ratio of current 2/current 1 was plotted against time intervals. The time required for 50% recovery from desensitization was determined by interpolation (t50% = 35 msec). C,I–V curve of glutamate-induced responses. The averaged amplitudes of peak currents (averages of five responses) were measured at different potentials (from −72 to 48 mV) and normalized with respect to the peak current at −72 mV. Note the linear shape of the curve (RI = 0.83 andErev = −6 mV). The pointswere fit by a fourth-order polynomial function. D, Variance–amplitude plot obtained by nonstationary noise analysis of glutamate-induced responses (holding potential, −72 mV). Two-thirds of the plot was fit with a linear regression. The estimated single-channel current is obtained from the slope of the variance–amplitude relationship (i = 0.57 pA). The weighted mean single-channel conductance for AMPARs on this patch was 8.1 pS.E, Quantification of GluR1–4, flip/flop, and R/G edition proportions. GluR1–4 cDNA fragments were amplified with a fluorescent primer, cut with restriction enzymes, and submitted to capillary electrophoresis. The fluorescence profiles obtained are displayed with subunit-specific colors:green, GluR1; red, GluR2;blue, GluR3; black, GluR4. Time of migration is represented horizontally (fragment size increasing from left to right), and fluorescence intensity is represented vertically. Unincorporated primers peaks (p) are indicated.Left, GluR1–4 proportions. GluR1–4 cDNA fragments were coamplified and cut with subunit-specific enzymes. Positions of uncut (u; GluR1–4 cDNAs resistant to the specific enzyme) and cut cDNA fragments are indicated. No GluR4 was found in this cell. Thesmall peak present on all profiles at the same position (left of R2 peak) is attributable to a PCR artifact uncut by the enzymes. Top right panel, Flip/flop proportions of GluR1–4 subunits expressed. Subunit-specific amplifications were performed, followed by restriction with splice variant-specific enzymes. GluR1 and GluR3 were uncut (flop and flip, respectively), whereas GluR2 was cut (flop). Bottom right panel, R/G edition proportions. After subunit-specific amplification, products were cut with an editing variant-specific enzyme. Positions of edited flip (R3iG), edited flop (R2oG), and unedited (asterisk; not present but found in other analyzed cells) peaks are indicated. In this cell GluR3 flip and GluR2 flop were edited.
The RSNP neuron analyzed in Figure 1 displayed AMPAR responses with an I–V curve almost linear (RI = 0.83; Fig. 1C). Nonstationary noise analyses were performed on glutamate-induced responses to determine the weighted mean single-channel conductance of AMPARs. For the RSNP neuron illustrated in Figure 1, the estimated single-channel current obtained from the slope of the variance–amplitude relationship was i = 0.57 pA (Fig. 1D). Assuming a driving force of −70 mV, the mean single-channel conductance of AMPARs on this patch was 8.1 pS.
After characterizing the functional properties of AMPARs, we obtained a second whole-cell recording on the same cell, and the cellular content was harvested. Then an RT-PCR was performed on the mRNAs harvested to investigate the GluR1–4 combination expressed in the cell (see Materials and Methods).
Figure 1E shows the molecular analysis performed on the RSNP cell for which the glutamate-induced responses are described above (Fig. 1A–D). The relative proportions of GluR1–4 were quantified after a second PCR, using the first PCR product as a template and the primer pair common to all subunits (the antisense primer was fluorescently labeled; see Materials and Methods). The GluR1–4 subunits were cut with subunit-specific restriction enzymes. Then the restriction reactions were run by capillary electrophoresis, and the obtained fluorescence profiles were used to quantify the GluR1–4 proportions (Fig. 1E, left). For this RSNP cell the proportions of the different subunits in the amplified products were 38.9% GluR1, 48.7% GluR2, and 12.4% GluR3.
The flip/flop proportions for subunits expressed in individual cells were analyzed in a second step. After second PCRs that used the first PCR product as a template and subunit-specific primers (the antisense primer was fluorescently labeled), each of the resulting products was cut by a splice variant-specific enzyme and run by capillary electrophoresis. The fluorescence profiles of Figure 1E, upper right panel, showed that the RSNP cell expressed GluR1 flop, GluR2 flop, and GluR3 flip. The flip/flop percentages obtained for each subunit were expressed as percentages of the total of all subunits found in the cell.
The R/G proportions of individual subunits were analyzed in a third step (the GluR1 subunit was not analyzed because it is not edited). A PCR using the subunit-specific PCR products as a template and a nested fluorescently labeled sense primer was performed. Each of the resulting products was cut by an editing variant-specific enzyme and run by capillary electrophoresis. Figure 1E, lower right panel, shows the profiles obtained for the RSNP cell used as an example. The profile corresponding to GluR2 (red) showed that this subunit was edited (the cut peak was at the position corresponding to the G form of flop subunits, 58 bp; see Materials and Methods). The profile corresponding to GluR3 (blue) showed that this subunit was edited (this peak was uncut, at the position corresponding to the G form of flip subunits, 127 bp; see Materials and Methods).
Functional diversity of AMPARs in nonpyramidal cells
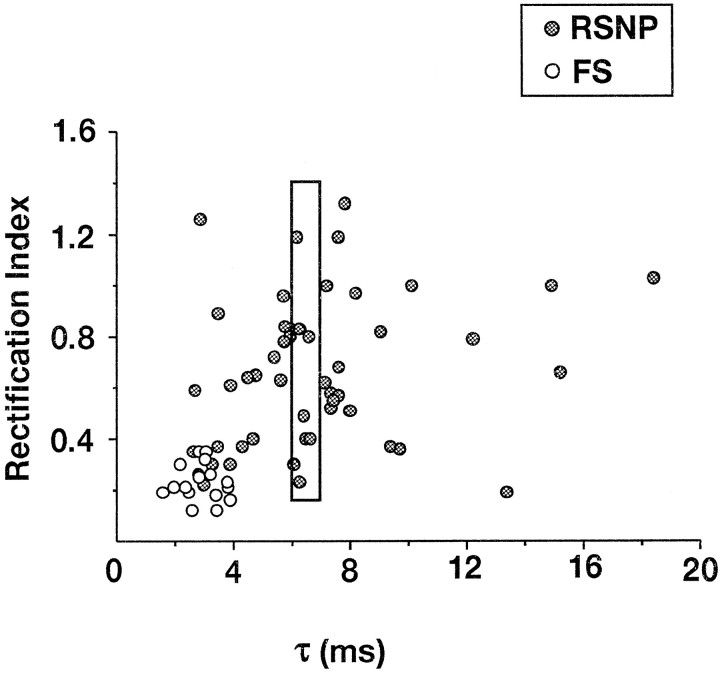
Figure 2 shows the distribution of the desensitization time constant and the recovery from desensitization as well as that of the RI and the mean single-channel conductance of AMPARs in our sample of nonpyramidal cells. Mean values of these functional properties are summarized in Table1. In our sample τ andt50% presented a wide distribution with values ranging from 1.56 to 18.4 msec (n = 66; Fig.2A) and from 5 to 181 msec (n = 32; Fig. 2B), respectively. As observed for the kinetic properties, RI widely varied in our sample (n = 65) with values ranging from 0.12 to 1.32 (Fig.2C). Single-channel conductances ranged from 5.3 to 36.3 pS, but relatively high values were predominant (n = 28; Fig. 2D).
Fig. 2.
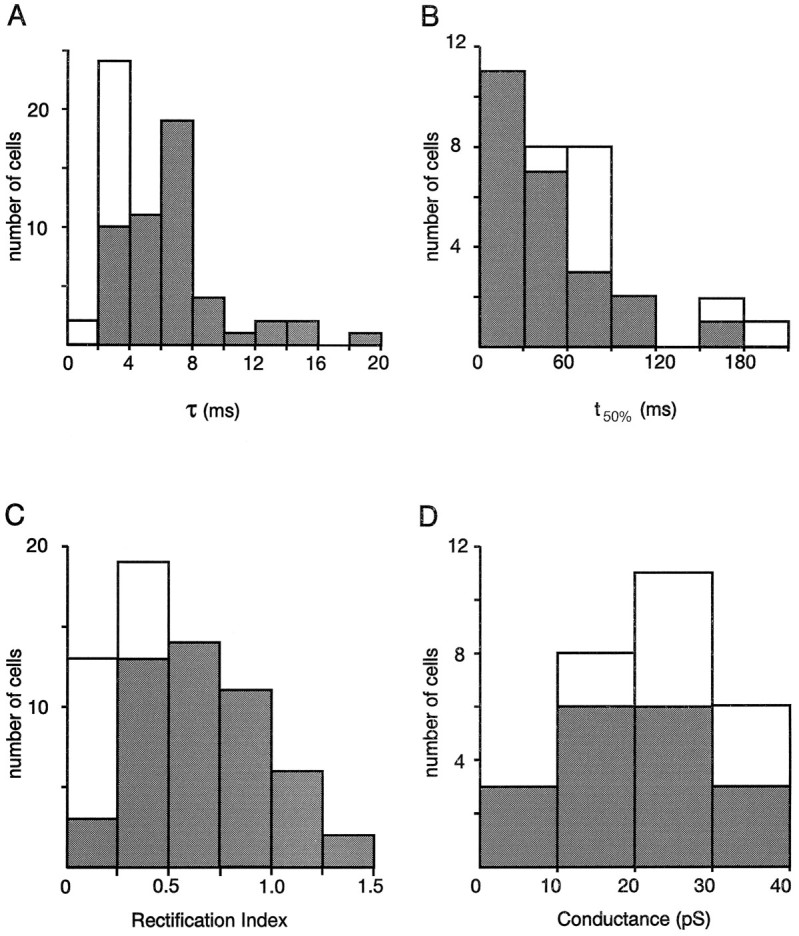
Functional properties of AMPARs in FS and RSNP neurons. Open and filled bars correspond to FS and RSNP cells, respectively. A, Desensitization time constants (τ; n = 66 cells).B, Recovery from desensitization (t50%; n = 32).C, Rectification index values calculated fromI–V curves (RI; n = 65). D, Weighted mean single-channel conductances (g; n = 28). Note that RSNP cells show wide distributions of the four functional parameters in comparison to FS cells.
Table 1.
Mean values of four functional properties of AMPARs in nonpyramidal cells
| FS cells | RSNP cells | Sample | |
|---|---|---|---|
| τ (msec) | 2.88 ± 0.68 | 6.70 ± 3.34 | 5.89 ± 3.38 |
| (n = 16) | (n = 50) | (n = 66) | |
| t50% (msec) | 97.2 ± 44.6 | 44.4 ± 35.6 | 57.6 ± 43.9 |
| (n = 8) | (n = 24) | (n = 32) | |
| RI | 0.23 ± 0.07 | 0.66 ± 0.29 | 0.55 ± 0.31 |
| (n = 16) | (n = 49) | (n = 65) | |
| g(pS) | 26.1 ± 6.6 | 19.6 ± 8.7 | 21.9 ± 8.9 |
| (n = 10) | (n = 18) | (n = 28) |
The desensitization time constant (τ), the recovery from desensitization (t50%), the rectification index (RI), and the single-channel conductance (g) were measured from glutamate-induced responses of FS and RSNP cells. Mean values were calculated from data of individual cells for all of the samples and separately for these two groups of neurons.
The FS and RSNP cell subgroups displayed AMPARs with different functional properties (Table 1). Indeed, the mean τ andt50% of FS were significantly different from those of RSNP cells (Student’s t test; p < 0.01 and p < 0.001 respectively; Table 1). Similarly, the mean RI of FS was significantly different from that of RSNP cells (p < 0.001; Table 1). In contrast, the average of the weighted mean single-channel conductance of FS cells was not significantly different from that of RSNP cells (significance level 0.05; Table 1).
The functional diversity of AMPARs appeared more restricted when only FS cells were considered. For instance, the scattering of the τ andRI values was attributable mostly to the AMPAR responses of RSNP neurons. Indeed, in these cells, AMPAR currents presented desensitization time constants between 2.61 and 18.4 msec andRI between 0.19 and 1.32 (Fig. 2A,C, filled bars). In contrast, FS cells always showed AMPARs with fast kinetics of desensitization (between 1.56 and 3.87 msec) and pronounced inward rectification of the I–V curves (between 0.12 and 0.35) (Fig. 2A,C, open bars). The distribution ofg values also was more restricted for FS cells (Fig.2D), but large variations oft50% values were observed in both the RSNP and FS cell samples (Fig. 2B).
Correlations between permeation properties or between kinetic properties of AMPARs were searched by analyzing data obtained from individual cells (Spearman test; see Materials and Methods).
The two permeation properties of these receptors, RI and the mean single-channel conductance, were negatively correlated (p < 0.01; Table2), single-channel conductances decreasing as a function of increasing RI.
Table 2.
Correlations between functional and molecular parameters of AMPARs
| U | p | n | |
|---|---|---|---|
| Between functional parameters | |||
| τ–RI | 4.53 | <0.001 | 65 |
| τ–RI (only for RSNP cells) | 1.86 | <0.04 | 49 |
| RI–g | −2.81 | <0.01 | 28 |
| RI–g (only for RSNP cells) | −2.16 | <0.02 | 16 |
| τ–t50% | −2.61 | <0.01 | 32 |
| τ–t50% (only for RSNP cells) | 1.34 | <0.11 NC | 24 |
| RI–t50% | −2.37 | <0.01 | 32 |
| RI–t50% (only for RSNP cells) | 0.163 | <0.44 NC | 24 |
| τ–g | −2.21 | <0.02 | 28 |
| τ–g (only for RSNP cells) | −1.31 | <0.10 NC | 18 |
| Between subunitsa | |||
| GluR1–GluR2 | 0.06 | <0.48 NC | |
| GluR3–GluR2 | −3.75 | <0.001 | |
| GluR3–GluR1 | −1.78 | <0.04 | |
| Between functional and molecular parametersa | |||
| τ–GluR1 | 0.47 | <0.32 NC | |
| τ–GluR1 flip | 1.86 | <0.04 | |
| τ–GluR1 flop | −0.43 | <0.34 NC | |
| τ–GluR2 | 1.82 | <0.04 | |
| τ–GluR2 flip | 2.01 | <0.025 | |
| τ–GluR2 flop | 1.08 | <0.14 NC | |
| τ–GluR3 | −1.04 | <0.15 NC | |
| τ–GluR3 flip | −0.32 | <0.38 NC | |
| τ–GluR3 flop | −1.48 | <0.07 NC | |
| τ–total flip | 1.27 | <0.11 NC | |
| τ–total flop | −1.12 | <0.13 NC | |
| t50%–GluR2–4 edition | 0.93 | <0.18 NC | |
| t50%–GluR2–4 flip edition | 0.59 | <0.28 NC | |
| t50%–GluR2–4 flop edition | −0.15 | <0.44 NC | |
| RI–GluR1 | 1.05 | <0.15 NC | |
| RI–GluR1 flip | 0.36 | <0.36 NC | |
| RI–GluR1 flop | 0.64 | <0.26 NC | |
| RI–GluR2 | 2.65 | <0.001 | |
| RI–GluR2 flip | 1.83 | <0.04 | |
| RI–GluR2 flop | 1.59 | <0.06 NC | |
| RI–GluR3 | −1.88 | <0.03 | |
| RI–GluR3 flip | −1.22 | <0.12 NC | |
| RI–GluR3 flop | 0.20 | <0.42 NC | |
| RI–total flip | 0.79 | <0.22 NC | |
| RI–total flop | −0.42 | <0.34 NC |
Statistical analyses of data obtained from individual cells.U = √(n − 1)rs, follows the distribution of Laplace–Gauss.rs, Spearman rank linear correlation coefficient; p, significance level. NC, not correlated.
a n = 27.
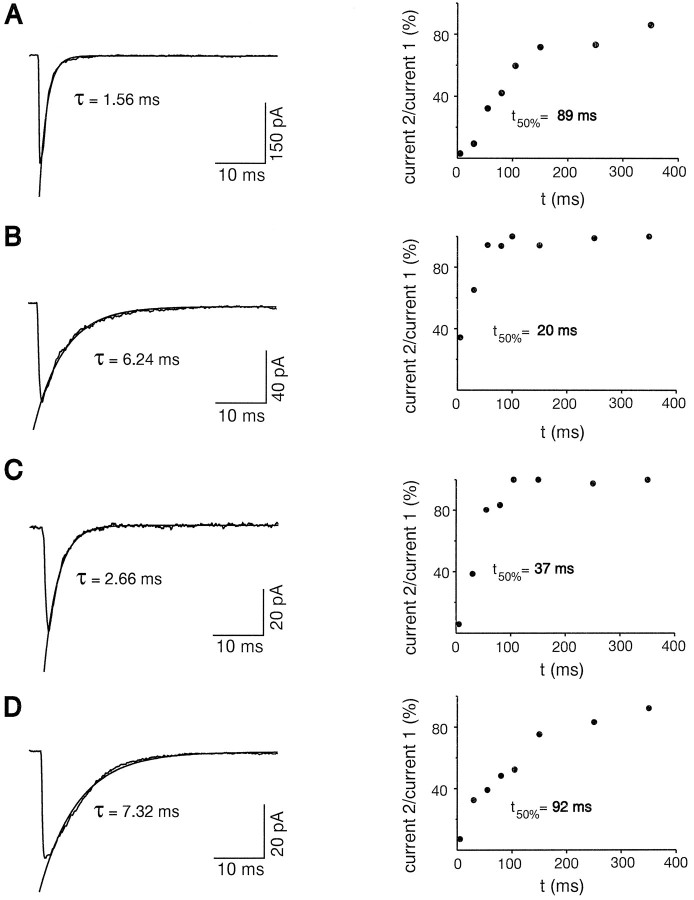
Statistical analyses of kinetic properties of AMPARs showed that the τ and t50% were negatively correlated (p < 0.01; Table 2). This correlation indicated that slow desensitization time constants were associated with fast recoveries from desensitization and vice versa. Typical examples are shown in Figure 3, A andB. The FS cell of Figure 3A had fast desensitizing (τ = 1.56 msec) and slow resensitizing AMPAR currents (t50% = 89 msec). Conversely, the RSNP cell of Figure 3B had slow desensitizing (τ = 6.24 msec) and fast resensitizing AMPAR currents (t50% = 20 msec). However, in 5 of 24 RSNP neurons, we observed the opposite τ–t50% relationships. Figure 3Cshows one of the three RSNP neurons with fast kinetics of desensitization (τ = 2.66 msec) and fast kinetics of recovery from desensitization (t50% = 37 msec). In contrast, Figure 3D illustrates one of the two RSNP neurons that presented a slow desensitization (τ = 7.32 msec) and a slow recovery from desensitization (t50% = 92 msec). Indeed, when data obtained from FS cells, which constitute a relatively homogeneous population of cells as compared with RSNP cells, were removed from the statistical analysis, the τ–t50% correlation disappeared (Spearman test; p < 0.11). In contrast, theRI–g (two permeation properties) correlation was little affected by the same operation (from U = −2.81,p < 0.01 to U = −2.16,p < 0.02; Table 2). These data indicate that desensitization and recovery from desensitization of native AMPARs can be controlled independently.
Fig. 3.
Desensitization and recovery from desensitization of AMPARs in individual cells. Left, Averaged responses of excised patches elicited by step glutamate application. The current decay was fit by a single exponential function shown superimposed to the averaged current. Right, Plot of the ratio of current 2/current 1 against time, obtained by applying pairs of 1 msec pulses of glutamate separated by increasing time intervals (from 5 to 350 msec). A, Patch excised from an FS neuron. Note the fast desensitization (τ = 1.56 msec; average of six responses) and the slow recovery from desensitization (t50% = 89 msec). B, Patch excised from an RSNP neuron with a slow desensitization (τ = 6.24 msec; average of 11 responses) and a fast recovery from desensitization (t50% = 20 msec). C, Patch excised from an RSNP neuron with fast desensitization (τ = 2.66 msec; average of 10 responses) and fast recovery from desensitization (t50% = 37 msec). D, Patch excised from an RSNP neuron. Note the slow desensitization (τ = 7.32 msec; average of 25 responses) and the slow recovery from desensitization (t50% = 92 msec).
Molecular diversity of AMPARs in nonpyramidal cells
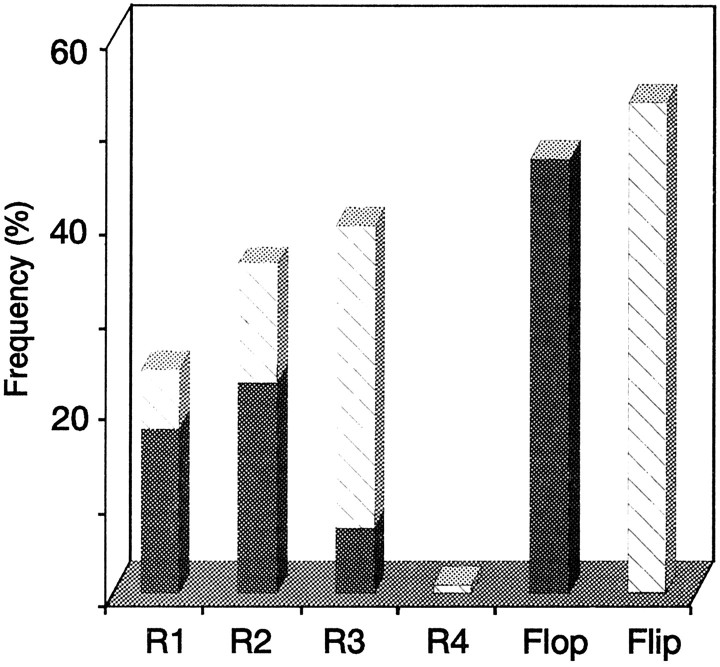
Because in our sample of nonpyramidal cells 23 of 27 molecularly analyzed cells corresponded to RSNP neurons, we did not differentiate the data of FS and RSNP cells. Quantification of the different GluR subunits mRNAs indicated that GluR3 and GluR2 were expressed predominantly, whereas GluR4 was observed rarely (GluR1, 24%; GluR2, 36%; GluR3, 39%; GluR4, 1%; see Fig.4). In our sample the overall abundance of flop variants (47%) was equivalent to that of flip variants (53%;right bars in Fig. 4). However, GluR1 and GluR2 were predominantly flop (73 and 63%, respectively; filled barsin Fig. 4), whereas GluR3 was mostly flip (82%; hatched barin Fig. 4). Moreover, the flip/flop proportion widely varied from one cell to another: among the 27 cells analyzed, 11 expressed >80% of the flop form and 12 expressed >80% of the flip form. In addition, splicing for each subunit appeared to be regulated independently in single cells (for instance, in the cell of Fig. 1, GluR1 and GluR2 were flop, whereas GluR3 was flip; in another cell, GluR1 was flop, whereas GluR2 and GluR3 were flip). In our sample the mean number of GluR1–4 flip/flop subunits expressed per cell was 2.7 ± 1.4.
Fig. 4.
Relative abundance of GluR1–4 flip/flop in nonpyramidal neurons. Shown are mean proportions of GluR1–4 subunits found in 27 nonpyramidal neurons (23 RSNP cells and 4 FS cells). GluR2 and GluR3 mRNAs were expressed predominantly (GluR1, 24 ± 28%; GluR2, 36 ± 36%; GluR3, 39 ± 39%; GluR4, 1 ± 4%). Average percentages of flip (53 ± 43%) and flop (47 ± 43%) variants are indicated by hatched andfilled bars, respectively. The two barson the right represent the total percentages of flip and flop variants calculated from flip and flop proportions of each subunit.
Because the edition at the R/G site predominantly affects the recovery from desensitization of AMPARs (Lomeli et al., 1994), the R/G quantification was performed only on cells for which we could measure this electrophysiological parameter (n = 14). We observed that 11 of 14 neurons presented GluR2–4 subunits mRNAs edited at 100%. In two other cells the proportion of the GluR2–4 edited subunits was also high, with values of 91 and 83%. The remaining cell showed a low proportion of edition (7%; data not shown).
Statistical analyses showed that the expression of GluR3 was negatively correlated with those of GluR2 and GluR1 (p < 0.001 and p < 0.04, respectively; Table 2). In other words, high levels of GluR3 were found in cells with low GluR2 (or low GluR1) and vice versa. No correlation was found between GluR2 and GluR1 (Table 2).
Correlation between molecular and functional properties of AMPARs in nonpyramidal cells
We found a positive correlation between the RI and the relative abundance of GluR2 (p < 0.01; Table2). The RI was negatively correlated with the relative abundance of GluR3 mRNAs (p < 0.03; Table 2). Thus, high levels of GluR2 (or low GluR3) were correlated with linearI–V curves and low levels of GluR2 (or high GluR3) with inwardly rectifying I–V curves. It must be noted that, in our sample, the expression of GluR3 was negatively correlated with that of GluR2 (see above). No correlation was found between RIand GluR1 (the correlation with GluR4 was not examined because this subunit was found only in two cells).
We also found a positive correlation between τ and the relative abundance of GluR2 (p < 0.04; Table 2), slower kinetics of desensitization usually being found in cells with high GluR2. This correlation was even better with GluR2 flip (p < 0.025; Table 2). In contrast withRI, no correlation was found between τ and GluR3 (Table2).
No other significant correlation could be found between functional properties and mRNA subunit expression. In particular, the desensitization time constants and the flip/flop ratio or the R/G editing were not correlated (Table 2). For instance, two cells (data not shown) having desensitization time constants in the same range (τ, 5.95 and 7.81 msec) and similar GluR1–4 composition (R1, 38.9 and 49.1%; R2, 48.7 and 38.6%; R3, 12.4 and 12.3%) had opposite flop percentages (87.6 and 0%). On the other hand, two other cells (data not shown) having similar flop percentages (96.1 and 84.5%) and similar GluR1–4 composition (R1, 27.1 and 20.7%; R2, 68.2 and 69.4%; R3, 4.7 and 9.9%) had very different desensitization time constants (τ, 2.85 and 12.2 msec). These two examples suggest that, in addition to the subunit composition, other factors such as protein phosphorylation may regulate the AMPAR kinetics.
Similarly, recovery from desensitization and AMPAR subunits edition was not correlated (Table 2).
We did not estimate the correlation between the single-channel conductance of AMPARs and subunit composition, because this electrophysiological parameter was determined in only 7 of the 27 cells for which the AMPAR mRNAs were studied.
Permeation and kinetics are independent properties in native AMPARs
The correlation of one molecular parameter (GluR2) to bothRI and τ in our sample of neurons suggested that permeation and kinetic properties of AMPARs could be correlated. Indeed, when we statistically analyzed τ and RI from individual patches, we observed a positive correlation between these two parameters (p < 0.001; Table 2). Similarly, we found that τ and g on one hand and RI andt50% on the other hand were negatively correlated (p < 0.02 and p < 0.01, respectively; Table 2). These results indicated that permeation and kinetic properties of AMPARs usually are coupled in our cell sample in a range going from fast desensitization–strong rectification to slow desensitization–linear I–V curve. It further suggested that they might be controlled by the same molecular determinant, GluR2.
However, the plot of τ versus RI (Fig.5) showed a large diversity of relations between the permeation and the kinetic properties of native AMPARs. For example, the data included in the rectangle show the variations of theRI (between 0.23 and 1.19) for desensitization time constants between 6 and 7 msec. Similarly, a large variability of τ (between 3.46 and 18.4 msec) is found for RI between 0.85 and 1.05. Furthermore, when data obtained from FS cells, which constitute a relatively homogeneous population of cells as compared with RSNP cells (open and filled circles, respectively, in Fig. 5), were removed from the statistical analysis, the τ–RI correlation dramatically decreased (fromU = 4.53, p < 0.001 to U = 1.86,p < 0.04; Table 2). A similar decrease in the coefficient of correlation was exhibited by the τ–g andRI–t50% correlations (Table 2). In contrast, as observed above, the RI–g (two permeation properties) correlation was affected little by the same operation. The statistical correlation between permeation and kinetic AMPAR properties obtained in our sample therefore seemed to reflect a cell type-dependent rather than a structure–function-dependent coupling.
Fig. 5.
Desensitization and rectification properties of AMPARs in FS and RSNP neurons. Desensitization time constants andRI were plotted for each cell. Open circles, FS neurons; filled circles, RSNP neurons. The points located inside therectangle show the large variability ofRI found for desensitization time constants of 6–7 msec.
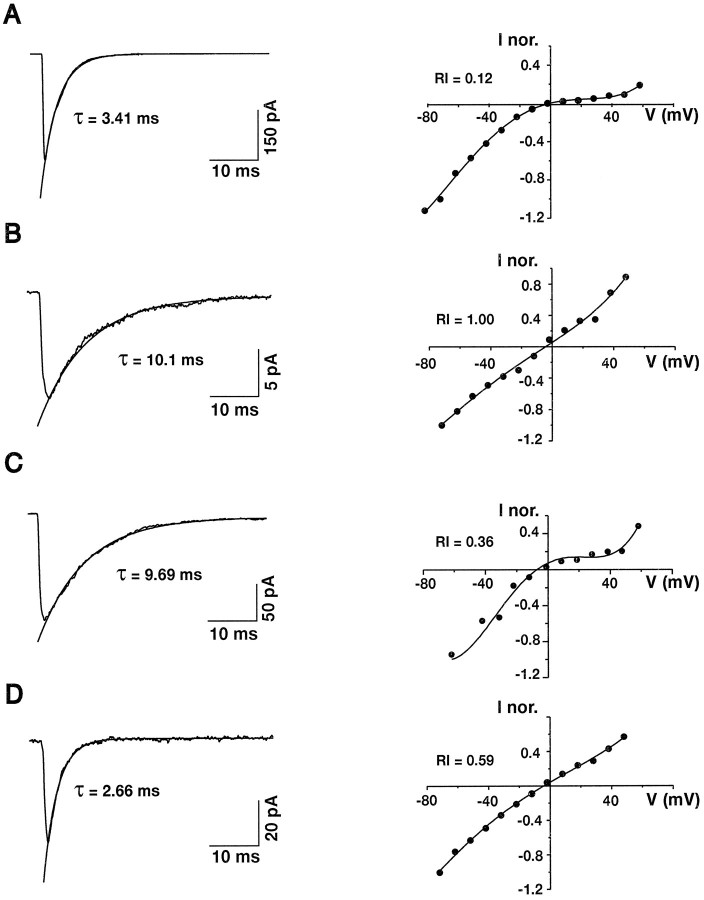
In support of this view, analysis of individual cases revealed that permeation and kinetic properties can be controlled independently. The τ–RI correlation found in our sample predicts functional behaviors exemplified in Figure 6,A and B, showing an FS cell that displayed fast desensitizing (τ = 3.41 msec) and strongly rectifying (RI = 0.12) currents and an RSNP cell that displayed slowly desensitizing (τ = 10.1 msec) and nonrectifying (RI = 1.00) currents, respectively. However, AMPAR responses from 11 of 49 RSNP neurons showed the opposite behaviors. The cell of Figure 6C displayed slowly desensitizing (τ = 9.69 msec) and strongly rectifying (RI = 0.36) currents. This type of AMPAR response was found in seven cells (mean τ = 8.26 ± 2.71 msec; mean RI = 0.32 ± 0.08). In contrast, the cell of Figure 6D displayed fast desensitizing (τ = 2.66 msec) and weakly rectifying (RI = 0.59) currents. This type of AMPAR response was found in four cells (mean τ = 3.21 ± 0.56 msec; meanRI = 0.84 ± 0.31). These results indicate that desensitization and rectification of AMPARs can be controlled independently and thus do not have the same molecular determinants.
Fig. 6.
Desensitization and rectification properties of AMPARs in individual cells. Left, Averaged responses of excised patches elicited by step glutamate application. The current decay was fit by a single exponential function shown superimposed to the averaged current. Right, I–V curves of glutamate-induced responses. The points were fit by third- or fourth-order polynomials. A, Patch excised from an FS neuron. Note the fast desensitization (τ = 3.41 msec; average of 35 responses) and the inward rectification of theI–V curve (RI = 0.12 andErev = −2 mV). B, Patch excised from an RSNP neuron. Note the slow desensitization (τ = 10.1 msec; average of 25 responses) and the linear I–V curve (RI = 1.00 and Erev = −6 mV). C, Patch excised from an RSNP neuron. Note the slow desensitization (τ = 9.69 msec; average of 11 responses) and the inward rectification of the I–V curve (RI = 0.36 and Erev = −5 mV). D, Patch excised from an RSNP neuron. Note the fast desensitization (τ = 2.66 msec; average of 10 responses) and the approximately linear I–V curve (RI= 0.59 and Erev = −5 mV).
DISCUSSION
The present study of neocortical nonpyramidal cells confirms that the permeation properties of native AMPARs (i.e., voltage dependence and single-channel conductance) are regulated by the relative expression of the GluR2 subunit. In contrast, the regulation of kinetic properties could not be attributed to any of the single molecular determinants considered, suggesting more complex structure–function relationships. Native AMPARs showed functional behaviors in which kinetic properties or kinetic and permeation properties varied independently. The diversity of native AMPARs supports the view that their permeation and kinetic properties are coupled in a cell type-dependent manner but do not have the same molecular determinant.
Molecular properties of AMPARs in nonpyramidal cells
All of the GluR1–4 subunits, except GluR4 flop, were found to be expressed in our sample of molecularly analyzed nonpyramidal cells, mostly constituted by RSNP neurons. The mean number of subunits expressed per cell was 2.7, intermediate between those reported for FS and pyramidal cells in the neocortex (3.3 and 2.3, respectively;Lambolez et al., 1996). Similarly, the proportion of GluR2 was intermediate between those reported for FS and pyramidal cells (Jonas et al., 1994; Lambolez et al., 1996). However, the predominant expression of GluR3 indicates that RSNP cells expressed AMPARs different from those of FS or pyramidal cells. Indeed, FS neurons are characterized by a high relative abundance of GluR1 mRNAs (Jonas et al., 1994; Lambolez et al., 1996) [see also Kondo et al. (1997), where GluR1 is found colocalized with parvalbumin, an FS cell marker], whereas pyramidal neurons express a high proportion of GluR2 (Jonas et al., 1994; Lambolez et al., 1996).
Although in our sample of neurons we found equivalent amounts of flip and flop forms, predominant expression of one form was observed in most individual cells. Because at the developmental stage used in the present work (13- to 17-d-old rats) the flip/flop variants ratio has reached its adult level (Monyer et al., 1991), our data confirm that the expression of flip/flop subunit proportions is cell type-dependent (Lambolez et al., 1992, 1996; Bochet et al., 1994; Jonas et al., 1994;Geiger et al., 1995). Moreover, the comparison between the flip/flop expression of the different subunits at the single-cell level confirms that splicing is regulated independently for each subunit (Lambolez et al., 1992).
Consistent with developmental studies of the GluR2–4 subunits R/G edition (Lomeli et al., 1994), we found that, in our sample, GluR2–4 subunits were predominantly edited. In individual cells all subunits were either almost fully edited or unedited. This may indicate that, in contrast with the flip/flop splicing, the R/G editing is not regulated independently for each subunit.
Permeation properties of AMPARs
The wide range of RI values observed is reminiscent of similar results obtained on hippocampal nonpyramidal neurons (Isa et al., 1996). The mean RI value obtained in our sample (0.55) was intermediate between that of our FS cells (0.23) and theRI values reported for hippocampal pyramidal neurons (mostly above 1; see Isa et al., 1996). Similarly, g values varied from those reported for FS cells to those reported for pyramidal cells (Hestrin, 1993). The average of g in our population of FS neurons (26 pS) is similar to that previously reported for this cell type (27 pS, Hestrin, 1993). For both permeation properties the FS cells appeared to form a homogeneous population, as compared with RSNP cells.
A negative correlation was found between RI andg, suggesting that these permeation properties are both controlled by the same molecular determinant. Consistent with numerous reports, the positive correlation we found between RI and the relative abundance of GluR2 indicates that this subunit regulates both of these permeation properties. Indeed, the role played by the GluR2 subunit in the rectification of AMPAR currents is well established in heterologous expression systems (for review, seeHollmann and Heinemann, 1994; Jonas and Burnashev, 1995) and for native receptors (Bochet et al., 1994). Similarly, the mean single-channel conductance of recombinant AMPARs is determined by the relative abundance of the GluR2 subunit (Swanson et al., 1997). The negative correlation between RI and the predominantly expressed GluR3 subunit observed in our sample of cells also confirms that permeation properties are regulated by the ratio GluR2/GluR1, 3, 4.
The relative abundance of GluR2 also determines another permeation property of AMPARs, their calcium permeability (for review, seeHollmann and Heinemann, 1994; Jonas and Burnashev, 1995) [see alsoJonas et al. (1994) and Geiger et al. (1995) for native receptors]. From the present study and previous reports it can be inferred that the relative abundance of GluR2 in the neural cell types analyzed so far determines AMPARs for which the permeation properties vary from highg–high calcium permeability–low RI (neocortical FS cell type) to low g–low calcium permeability–highRI (pyramidal cell type).
Kinetic properties of AMPARs
In our sample desensitization values ranged from the fastest reported (medial nucleus of the trapezoid body cells) to the slowest reported (Hilar mossy cells), using 1 mm glutamate applications (1.7 and 16.3 msec, respectively; Geiger et al., 1995). Most of this variability was attributable to RSNP neurons, FS neurons always showing fast desensitization. In a previous study neocortical pyramidal and FS neurons displayed AMPARs with slow and fast recovery from desensitization, respectively (Hestrin, 1993). Here we observed that, in both FS and RSNP cells, t50% values widely varied between these two extremes.
The negative correlation observed between τ andt50% (fast desensitization generally being associated with slow recovery and vice versa) dramatically decreased when data of FS neurons were removed from statistical analyses. Indeed, we found individual RSNP cells with either fast desensitization–fast recovery or slow desensitization–slow recovery, as previously reported for other cell types (Colquhoun et al., 1992; Raman and Trussell, 1995). Together, these observations suggest that these two kinetic properties of AMPARs can be controlled independently and probably do not have the same molecular determinants.
In heterologous expression systems the flip/flop splicing affects the kinetic properties of recombinant AMPARs (Sommer et al., 1990; Lomeli et al., 1994; Mosbacher et al., 1994; Partin et al., 1994). Comparison between neocortical FS and pyramidal cells also suggested that native AMPARs kinetics similarly are affected by the flip/flop splicing (Lambolez et al., 1996). However, as observed in a similar study comparing native AMPARs of a broad cell types sample (Geiger et al., 1995), we did not find any correlation between these molecular and functional properties. In agreement with Geiger et al. (1995), we therefore propose that the flip/flop splicing is not the principal molecular determinant controlling the kinetic properties of native AMPARs.
Recombinant studies have suggested that recovery from desensitization of AMPARs is controlled by the R/G site edition (Lomeli et al., 1994). Although the number of cells analyzed for that molecular property in the present study (n = 14) does not allow us to reject this hypothesis, the absence of correlation betweent50% and editing suggests that recovery from desensitization of native AMPARs is not controlled primarily at that site.
As also found in a previous work (Geiger et al., 1995), we observed a positive correlation between desensitization time constants and the relative abundance of GluR2, suggesting a correlation between kinetic and permeation properties. Consistent with this hypothesis we also found a positive correlation between τ and RI. From this correlation native AMPAR properties are expected to range from fast desensitization–strong rectification to slow desensitization–linearI–V curve. However, when data from the homogeneous population of FS neurons were removed from statistical analyses, the significance of this correlation dramatically decreased. Moreover, analyses of individual RSNP cells showed that, in 11 of 49 neurons, the τ–RI relationship was opposite to that expected from the correlation found on our whole nonpyramidal cell sample. Indeed, seven cells displayed AMPAR currents with slow desensitization and marked inward rectification, and four cells displayed AMPAR currents with fast desensitization and weak rectification, as also reported for nigral GABAergic neurons (Götz et al., 1997). Overall, these data indicate that the permeation and the kinetic properties of native AMPARs can be controlled independently and thus do not have the same molecular determinant. GluR2 therefore does not appear to determine dominantly the native AMPAR kinetics.
In heterologous expression systems many molecular determinants have been shown to affect AMPAR kinetics: the subunit composition, the alternative flip/flop variants, and the mRNA edited at the R/G site of subunits GluR2–4 (Sommer et al., 1990; Lomeli et al., 1994; Mosbacher et al., 1994; Partin et al., 1994). Our data, together with these studies, are consistent with the view that multiple determinants regulate the desensitization of AMPARs. Comparisons between individual cells even suggest that additional molecular determinants, such as protein phosphorylation, also might regulate AMPAR kinetics. This, indeed, would explain the absence of correlation between kinetics and individual molecular parameters in native AMPARs.
Cell type-dependent regulations of AMPAR kinetic and permeation properties
Heterologous expression studies have demonstrated that the fine tuning of AMPAR properties can result in a large variability of functional patterns. Despite cell to cell variations in their GluR1–4 expression, cell types express distinct mean GluR1–4 combinations and display AMPARs with relatively homogeneous functional properties. Among neocortical neurons FS cells express AMPARs with fast desensitization, inwardly rectifying I–V curves, high calcium permeability, and large mean single-channel conductances, whereas pyramidal cells exhibit AMPARs with opposite properties. In contrast, RSNP cells form a highly heterogeneous population (Kawaguchi and Kubota, 1993; Kawaguchi, 1995; Cauli et al., 1997) and display wide variations of AMPAR kinetic and permeation properties, resulting in various functional patterns. In most of neuronal populations investigated in previous studies, kinetic and permeation properties of native AMPARs were found to vary in a range going from fast desensitization–inward rectification to slow desensitization–no rectification. In RSNP neurons we observed two other types of native AMPARs characterized by fast desensitization–no rectification or slow desensitization–inward rectification. Overall, the studies of native AMPARs suggest that their properties are regulated independently according to cell type-specific functions.
Footnotes
This study was supported by Centre National de Recherche Scientifique (France) and by European Union Biotec Grants 960382 and 960589. S.H. was supported by National Eye Institute Grant EY09120. M.C.A. was supported by a fellowship from Instituto Colombiano de Ciencia y Tecnología (Colciencias; Colombia). We thank Nathalie Gibelin, Bruno Cauli, and Gérard Sadoc for help.
Correspondence should be addressed to Dr. Bertrand Lambolez, Neurobiologie et Diversité Cellulaire, Centre National de la Recherche Scientifique Unité de Recherche Associée 2054, Ecole Supérieure de Physique et de Chimie Industrielles de la ville de Paris, 10 Rue Vauquelin, 75231 Paris Cedex 5, France.
REFERENCES
- 1.Bochet P, Audinat E, Lambolez B, Crépel F, Rossier J, Lino M, Tsuzuki K, Ozawa S. Subunit composition at the single-cell level explains functional properties of a glutamate-gated channel. Neuron. 1994;12:383–388. doi: 10.1016/0896-6273(94)90279-8. [DOI] [PubMed] [Google Scholar]
- 2.Bowie D, Mayer ML. Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron. 1995;15:453–462. doi: 10.1016/0896-6273(95)90049-7. [DOI] [PubMed] [Google Scholar]
- 3.Cauli B, Audinat E, Lambolez B, Angulo MC, Ropert N, Tsuzuki K, Hestrin S, Rossier J. Molecular and physiological diversity of cortical nonpyramidal cells. J Neurosci. 1997;17:3894–3906. doi: 10.1523/JNEUROSCI.17-10-03894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colquhoun D, Jonas P, Sakmann B. Action of brief pulses of glutamate on AMPA/kainate receptors in patches from different neurones of rat hippocampal slices. J Physiol (Lond) 1992;458:261–287. doi: 10.1113/jphysiol.1992.sp019417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connors BW, Gutnick MJ. Intrinsic firing patterns of diverse neocortical neurons. Trends Neurosci. 1990;13:99–104. doi: 10.1016/0166-2236(90)90185-d. [DOI] [PubMed] [Google Scholar]
- 6.Cull-Candy SG, Howe JR, Ogden DC. Noise and single channels activated by excitatory amino acids in rat cerebellar granule neurones. J Physiol (Lond) 1988;400:189–222. doi: 10.1113/jphysiol.1988.sp017117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards FA, Konnerth A, Sakmann B, Takahashi T. A thin slice preparation for patch-clamp recordings from neurons of the mammalian central nervous system. Pflügers Arch. 1989;414:600–612. doi: 10.1007/BF00580998. [DOI] [PubMed] [Google Scholar]
- 8.Franke C, Hatt H, Dudel J. Liquid filament switch for ultra-fast exchanges of solutions at excised patches of synaptic membrane of crayfish muscle. Neurosci Lett. 1987;77:199–204. doi: 10.1016/0304-3940(87)90586-6. [DOI] [PubMed] [Google Scholar]
- 9.Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 10.Götz T, Kraushaar U, Geiger J, Lübke J, Berger T, Jonas P. Functional properties of AMPA and NMDA receptors expressed in identified types of basal ganglia neurons. J Neurosci. 1997;17:204–215. doi: 10.1523/JNEUROSCI.17-01-00204.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hestrin S. Different glutamate receptor channels mediate fast excitatory synaptic currents in inhibitory and excitatory cortical neurons. Neuron. 1993;11:1083–1091. doi: 10.1016/0896-6273(93)90221-c. [DOI] [PubMed] [Google Scholar]
- 12.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 13.Isa T, Itazawa S, Iino M, Tsuzuki K, Ozawa S. Distribution of neurons expressing inwardly rectifying and Ca2+-permeable AMPA receptors in rat hippocampal slices. J Physiol (Lond) 1996;491:719–733. doi: 10.1113/jphysiol.1996.sp021252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonas P, Burnashev N. Molecular mechanisms controlling calcium entry through AMPA-type glutamate receptor channels. Neuron. 1995;15:987–990. doi: 10.1016/0896-6273(95)90087-x. [DOI] [PubMed] [Google Scholar]
- 15.Jonas P, Racca C, Sakmann B, Seeburg PH, Monyer H. Differences in Ca2+ permeability of AMPA-type glutamate receptor channels in neocortical neurons caused by differential GluR-B subunit expression. Neuron. 1994;12:1281–1289. doi: 10.1016/0896-6273(94)90444-8. [DOI] [PubMed] [Google Scholar]
- 16.Kamboj SK, Swanson GT, Cull-Candy SG. Intracellular spermine confers rectification on rat calcium-permeable AMPA and kainate receptors. J Physiol (Lond) 1995;486:297–303. doi: 10.1113/jphysiol.1995.sp020812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawaguchi Y. Physiological subgroups of nonpyramidal cells with specific morphological characteristics in layer II/III of rat frontal cortex. J Neurosci. 1995;15:2638–2655. doi: 10.1523/JNEUROSCI.15-04-02638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi Y, Kubota Y. Correlation of physiological subgroupings of nonpyramidal cells with parvalbumin- and calbindin D28k-immunoreactive neurons in layer V of rat frontal cortex. J Neurophysiol. 1993;70:387–396. doi: 10.1152/jn.1993.70.1.387. [DOI] [PubMed] [Google Scholar]
- 19.Koh DS, Burnashev N, Jonas P. Block of native Ca2+-permeable AMPA receptors in rat brain by intracellular polyamines generates double rectification. J Physiol (Lond) 1995;486:305–312. doi: 10.1113/jphysiol.1995.sp020813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo M, Sumino R, Okado H. Combinations of AMPA receptor subunit expression in individual cortical neurons correlate with expression of specific calcium-binding proteins. J Neurosci. 1997;17:1570–1581. doi: 10.1523/JNEUROSCI.17-05-01570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambolez B, Audinat E, Bochet P, Crépel F, Rossier J. AMPA receptor subunits expressed by single Purkinje cells. Neuron. 1992;9:247–258. doi: 10.1016/0896-6273(92)90164-9. [DOI] [PubMed] [Google Scholar]
- 22.Lambolez B, Ropert N, Perrais D, Rossier J, Hestrin S. Correlation between kinetics and RNA splicing of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptors in neocortical neurons. Proc Natl Acad Sci USA. 1996;93:1797–1802. doi: 10.1073/pnas.93.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lebart L, Morineau A, Fénelon J-P. Traitement des données statistiques: méthodes et programmes, pp 109–183. Bordas Dunod; Paris: 1982. Méthodes non paramétriques. [Google Scholar]
- 24.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 25.McCormick DA, Connors BW, Lighthall JW, Prince DA. Comparative electrophysiology of pyramidal and sparsely spiny stellate neurons of the neocortex. J Neurophysiol. 1985;54:782–806. doi: 10.1152/jn.1985.54.4.782. [DOI] [PubMed] [Google Scholar]
- 26.Monyer H, Seeburg PH, Wisden W. Glutamate-operated channels: developmentally early and mature forms arise by alternative splicing. Neuron. 1991;6:799–810. doi: 10.1016/0896-6273(91)90176-z. [DOI] [PubMed] [Google Scholar]
- 27.Mosbacher J, Schoepfer R, Monyer H, Burnashev N, Seeburg PH, Ruppersberg JP. A molecular determinant for submillisecond desensitization in glutamate receptors. Science. 1994;266:1059–1062. doi: 10.1126/science.7973663. [DOI] [PubMed] [Google Scholar]
- 28.Neher E. Correction for liquid junction potentials in patch-clamp experiments. Methods Enzymol. 1992;207:123–131. doi: 10.1016/0076-6879(92)07008-c. [DOI] [PubMed] [Google Scholar]
- 29.Ozawa S, Lino M, Tsuzuki K. Two types of kainate response in cultured rat hippocampal neurons. J Neurophysiol. 1991;66:2–11. doi: 10.1152/jn.1991.66.1.2. [DOI] [PubMed] [Google Scholar]
- 30.Partin KM, Patneau DK, Mayer ML. Cyclothiazide differentially modulates desensitization of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor splice variants. Mol Pharmacol. 1994;46:129–138. [PubMed] [Google Scholar]
- 31.Raman IM, Trussell LO. The mechanism of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate receptor desensitization after removal of glutamate. Biophys J. 1995;68:137–146. doi: 10.1016/S0006-3495(95)80168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sigworth FJ. The variance of sodium current fluctuations at the node of Ranvier. J Physiol (Lond) 1980;307:97–129. doi: 10.1113/jphysiol.1980.sp013426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silver RA, Cull-Candy SG, Takahashi T. Non-NMDA glutamate receptor occupancy and open probability at a rat cerebellar synapse with single and multiple release sites. J Physiol (Lond) 1996;494:231–250. doi: 10.1113/jphysiol.1996.sp021487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sommer B, Keinanen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Kohler M, Takagi T, Sakmann B, Seeburg PH. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- 35.Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 36.Stuart GJ, Dodt HU, Sakmann B. Patch-clamp recordings from the soma and dendrites of neurons in brain slices using infrared video microscopy. Pflügers Arch. 1993;423:511–518. doi: 10.1007/BF00374949. [DOI] [PubMed] [Google Scholar]
- 37.Swanson GT, Kamboj SK, Cull-Candy SG. Single-channel properties of recombinant AMPA receptors depend on RNA editing, splice variation, and subunit composition. J Neurosci. 1997;17:58–69. doi: 10.1523/JNEUROSCI.17-01-00058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wisden W, Seeburg PH. Mammalian ionotropic glutamate receptors. Curr Opin Neurobiol. 1993;3:291–298. doi: 10.1016/0959-4388(93)90120-n. [DOI] [PubMed] [Google Scholar]