Abstract
We have investigated the patterns of postnatal brain expression and DNA binding of Gtx, a homeodomain transcription factor. Gtx mRNA accumulates in parallel with the RNAs encoding the major structural proteins of myelin, myelin basic protein (MBP), and proteolipid protein (PLP) during postnatal brain development; Gtx mRNA decreases in parallel with MBP and PLP mRNAs in the brains of myelin-deficient rats, which have a point mutation in the PLP gene. Gtx mRNA is expressed in differentiated, postmitotic oligodendrocytes but is not found in oligodendrocyte precursors or astrocytes. These data thus demonstrate that Gtx is expressed uniquely in differentiated oligodendrocytes in postnatal rodent brain and that its expression is regulated in parallel with the major myelin protein mRNAs, encoding MBP and PLP, under a variety of physiologically relevant circumstances.
Using a Gtx fusion protein produced in bacteria, we have confirmed that Gtx is a sequence-specific DNA-binding protein, which binds DNA sequences containing a core AT-rich homeodomain binding site. Immunoprecipitation of labeled DNA fragments encoding either the MBP or PLP promoter regions with this fusion protein has identified several Gtx-binding fragments, and we have confirmed these data using an electrophoretic mobility shift assay. In this way we have identified four Gtx binding sites within the first 750 bp of the MBP promoter and four Gtx binding sites within the first 1.3 kb of the PLP promoter. In addition, inspection of the PLP promoter sequence demonstrates the presence of six additional Gtx binding sites. These data, taken together, strongly suggest that Gtx is important for the function of differentiated oligodendrocytes and may be involved in the regulation of myelin-specific gene expression.
Keywords: brain development, DNA binding, Gtx, myelination, oligodendrocytes, gene expression
Myelin is a multilamellar membrane structure ensheathing axons in both the CNS and the peripheral nervous system (PNS) and acts to facilitate nerve conduction. In the CNS, myelin is synthesized by oligodendrocytes (Wood, 1984) and consists of a series of concentrically wrapped extensions of the oligodendrocyte plasma membrane. Myelin is composed mainly of lipid but also contains a number of myelin-specific structural proteins, including proteolipid protein (PLP), myelin basic protein (MBP), cyclic nucleotide phosphodiesterase (CNP), myelin-associated glycoprotein (MAG), and myelin–oligodendrocyte glycoprotein (Campagnoni, 1988; Lemke, 1993).
Synthesis of the proper amounts of the myelin-specific proteins and lipids by oligodendrocytes is critical for both normal myelin sheath formation and normal brain function. Lack of expression of MBP in the mouse mutant shiverer, for example, causes failure of normal myelin compaction and causes a progressive ataxic syndrome in affected animals. Replacement of the missing MBP in affected mice rescues both the clinical and the dysmyelinating phenotypes (Readhead et al., 1987,1994). Lack of PLP expression attributable to a PLP deletion also causes abnormal myelin formation (Raskind et al., 1991; Hodes et al., 1993), producing mental retardation, ataxia, weakness of limbs, and increased muscle tone. In addition, overexpression of the PLP gene in transgenic mice (Kagawa et al., 1994; Readhead et al., 1994) and individuals with a PLP duplication (Hodes et al., 1993; Ellis and Malcom, 1994) causes CNS dysmyelination with a similar neurological syndrome. Thus, too little or too much of the normal myelin constituents can cause dysmyelination in the CNS.
The cellular and molecular processes that regulate the synthesis and assembly of the myelin sheath, although important for proper brain function, are poorly understood. Oligodendrocyte precursors migrate out from regions surrounding the developing ventricles (Yu et al., 1994) to populate developing axon tracts. Before the onset of myelination, these proliferating oligodendrocyte precursors stop dividing and begin to synthesize the major myelin structural proteins in an orderly sequence. CNP appears first, then MBP, MAG, and finally PLP days later (Dubois-Dalcq et al., 1986; Knapp et al., 1987); myelination then commences. Although the myelin proteins appear at different times during oligodendrocyte development, the mRNAs encoding these proteins accumulate with similar temporal profiles both in the developing brain (Scherer et al., 1994) and in oligodendrocytes in culture (Zeller et al., 1985; Collarini et al., 1992; Grinspan et al., 1993; Scherer et al., 1994). Regulation of this process, including the accumulation of myelin-specific mRNAs and proteins, as well as compact myelin, can be accounted for by the transcriptional activation of the set of myelin-specific protein genes during the final stage of oligodendrocyte differentiation (Zeller et al., 1984; Milner, 1985;Zeller et al., 1985; Roach et al., 1983; Kamholz and Wrabetz, 1992). Thus a coordinated activation of myelin-specific gene expression precedes the onset of myelination by oligodendrocytes in the CNS and is critical for normal myelin formation.
The molecular mechanisms underlying the activation of myelin-specific gene expression are not well understood. Several laboratories have demonstrated that relatively small regions of DNA sequence upstream of both the PLP and MBP genes are sufficient for developmental and tissue-specific activation of lacZ expression in oligodendrocytes in transgenic animals (Gow et al., 1992; Goujet-Zalc et al., 1993; Wright et al., 1993; Wrabetz et al., 1995), and transcription factors interacting with these regions have been identified (Berndt et al., 1992; Kim and Hudson, 1992; Haas et al., 1993; Wrabetz et al., 1993). Only one of these factors, MyT1, a member of the zinc finger family of DNA-binding proteins, has been shown to be expressed in oligodendrocytes (Armstrong et al., 1995). The role of MyT1 in the activation of myelin-specific gene expression is uncertain, because its expression declines with the onset of myelination.
The transcription factor Gtx, a novel member of the homeodomain family, has been shown to be expressed in white matter in adult rodents (Komuro et al., 1993). In this paper we have extended this initial observation by investigating in detail the timing of Gtx expression in brain and in cultured oligodendrocytes. We find that Gtx is expressed indifferentiated oligodendrocytes but not in oligodendrocyte progenitors, astrocytes, or other glia. In postnatal rodent brain, Gtx expression is thus confined to oligodendrocytes. Furthermore, Gtx expression parallels that of the myelin-specific mRNAs in a variety of physiologically relevant circumstances, including development and in the brains of myelin-deficient rats, which have a point mutation in the PLP gene. Finally, using a Gtx fusion protein, we have found that Gtx can interact with the MBP and PLP promoters in a sequence-specific manner by way of the core homeodomain binding motif, and we have identified four Gtx binding sites within the first 750 bp of the MBP promoter and four Gtx binding sites within the first 1.3 kb of the PLP promoter. Inspection of the first kilobase of the PLP promoter sequence further demonstrates the presence of six additional Gtx binding sites. Gtx is thus the only transcription factor known that is uniquely expressed in differentiated oligodendrocytes and that also interacts with myelin-specific promoters. These data, taken together, strongly suggest that Gtx is important for the function of differentiated oligodendrocytes and may be involved in the regulation of myelin-specific gene expression.
MATERIALS AND METHODS
Cell culture. Primary mixed glial cultures obtained from cerebral white matter of 6-d-old rats were prepared and grown as described (Grinspan et al., 1990). Complement-mediated cell lysis of oligodendrocytes and their progenitors was performed using anti-GalC and A2B5 antibodies as described (Grinspan et al., 1990). Purified progenitor cultures were prepared as described previously (Collarini et al., 1992).
Northern blot analysis. Northern blot analysis was performed as described (Scherer et al., 1994). Briefly, mouse and rat tissue RNA was isolated by the guanidium thiocyanate–CsCl2 method (Chirgwin et al., 1979). Twenty micrograms of total RNA were electrophoresed on a 1% agarose/2.2 m formaldehyde gel and then transferred overnight to a nylon membrane (Duralon; Stratagene, La Jolla, CA) in 6× SSC. The blots were then UV-cross-linked (0.12 J), prehybridized, hybridized, and washed using standard techniques (Sambrook et al., 1989). The following probes were used: a full-length 1.25 kb Gtx cDNA, a 0.85 kb Pst fragment from a rat CNP cDNA (Bernier et al., 1987), a 1.5 kb EcoRI fragment from a rat MBP cDNA isolated in our laboratory, a 1.4 kb fragment from a rat PLP cDNA (Kamholz et al., 1992), and a 1.3 kb cDNA encoding rat glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Fort et al., 1985).32P-labeled probes were prepared by primer extension with random hexamers using the Prime-a-gene kit (Promega, Madison, WI).
RNase protection assay. RNase protection assays were performed as described by Kamholz et al. (1988). Fragments of Gtx, MBP, and GAPDH cDNAs were cloned into Bluescript SK (Stratagene). Radiolabeled antisense RNA was prepared using either T3 or T7 RNA polymerases (Promega). Fifty thousand to 100,000 cpm of each probe were incubated with 5–20 μg of total RNA in a buffer containing 10 mm PIPES, pH 6.7, 80% formamide, 0.4 m NaCl, and 1 mm EDTA overnight at 47°C in a total volume of 30 μl. Then, 300 μl of RNase digestion buffer (10 mm Tris, pH 7.5, 0.3 m NaCl, 1 mm EDTA, 40 μg/ml RNase A, and 1 μg/ml RNase T1) was added to each hybridization reaction and incubated at 30°C for 1 hr. Next, 20 μl of 10% SDS and 5 μl of Proteinase K (20 mg/ml) were added to each reaction and incubated at 37°C for 30 min. The reactions were then phenol–chloroform-extracted, ethanol-precipitated, and resuspended in 5 μl of 80% formamide/dye-loading buffer. The reactions were analyzed on a 4% acrylamide/7 m urea sequencing gel. The gel was dried and exposed to x-ray film with an intensifying screen.
In situ hybridization. In situ hybridization of mixed glial cultures was performed as described by Ghandour and Skoff (1991), and the slides were developed using nitroblue tretrazolium 5-bromo-4-chloro-3-indoyl phosphate.
Production of a maltose-binding protein (mbp)–Gtx fusion protein. An NcoI–SphI fragment containing the Gtx open reading frame was blunt-ended with T4 DNA polymerase (Promega) and cloned in frame with maltose-binding protein (mbp) into the BamHI site of pMal-c2 (New England Biolabs, Beverly, MA), and the fusion site was verified by DNA sequencing. To produce an mbp–Gtx fusion protein, TB1 bacteria were transformed with the above construct, grown to an OD600 of 0.6, and induced with 0.3 mm isopropyl-1-thio-β-d-galactopyranoside for 30 min. The bacteria were harvested by centrifugation; the pellet was resuspended in (in mm) 200 NaCl, 10 Tris, pH 7.4, 1 EDTA, and 1 DTT; and the soluble protein was released by freeze-thawing and sonication. The bacterial extract was then centrifuged for 30 min at 10,000 rpm (Sorvall SS-34 rotor), and the supernatant was passed through an amylose resin column. Fractions were eluted with (in mm) 10 maltose, 200 NaCl, 10 Tris, pH 7.4, 1 EDTA, and 1 DTT and dialyzed overnight at 4°C in a buffer containing 5% glycerol and (in mm) 50 NaCl, 10 Tris, pH 7.4, 1 EDTA, and 1 DTT. The protein concentration of the fractions was determined using a Bio-Rad (Richmond, CA) protein assay kit. The fractions were then analyzed by SDS-PAGE followed by Coomassie blue staining. The identity of the mbp–Gtx fusion was confirmed by immunoblotting with an antibody against maltose-binding protein (New England Biolabs). Maltose-binding protein without the Gtx fusion was produced and isolated in an identical manner as above.
To remove the maltose-binding protein moiety from mbp–Gtx, the fusion protein was subjected to proteolytic cleavage by factor Xa protease, as described by the supplier (New England Biolabs). Briefly, 20 μg of affinity-purified mbp–Gtx fusion protein was incubated with 1 μg of factor Xa protease in a 100 μl reaction in elution buffer for varying amounts of time at room temperature. SDS-PAGE analysis of the cleavage products visualized by Coomassie blue staining demonstrated that complete cleavage occurred in 24 hr and that the cleavage products were of the predicted size for mbp and Gtx.
DNA immunoprecipitation assays. DNA immunoprecipitation assays were performed as described (Bharucha et al., 1994). Plasmid DNAs containing the promoter region of interest, either MBP or PLP, were digested with restriction enzymes as described in the legend to Figure 7. The pool of fragments was end-labeled with [α-32P]dCTP by filling in with Klenow DNA polymerase. Approximately 500,000 cpm of the labeled fragments were mixed with 4 μl (0.5 μm) mbp–Gtx or with 4 μl of a similar fraction of mbp in a volume of 20 μl in a buffer containing (final concentration) 5% glycerol, 50 mm NaCl, 10 mmTris, pH 7.4, 1 mm EDTA, 1 mm DTT, and 1 μg poly(dI-dC) (Sigma, St. Louis, MO), and the mixture was incubated on ice for 1 hr. Two microliters of anti-mbp antisera (New England Biolabs) were then added to the reaction and incubated on ice for an additional 30 min. Twenty-two microliters of 50% protein A–Sepharose beads (Pharmacia, Piscataway, NY) were then added to the reaction, and the incubation continued for 20 min on ice. The reaction was centrifuged for 30 sec in an Eppendorf microcentrifuge at maximum speed to pellet the Sepharose, and the supernatant was removed. The beads were then washed twice with 1 ml of ice-cold wash solution (100 mm NaCl, 50 mm Tris, pH 7.4, 5 mmEDTA, and 0.1% NP-40), briefly air-dried, resuspended in 5 μl of loading buffer (80% formamide, 0.01N NaOH, 1 mm EDTA, and 1 mg/ml bromophenol blue and xylene cyanole), heated at 95°C for 5 min, and then loaded on a 6% acrylamide/7 m urea sequencing gel. After electrophoresis the gel was dried and exposed to x-ray film with an intensifying screen.
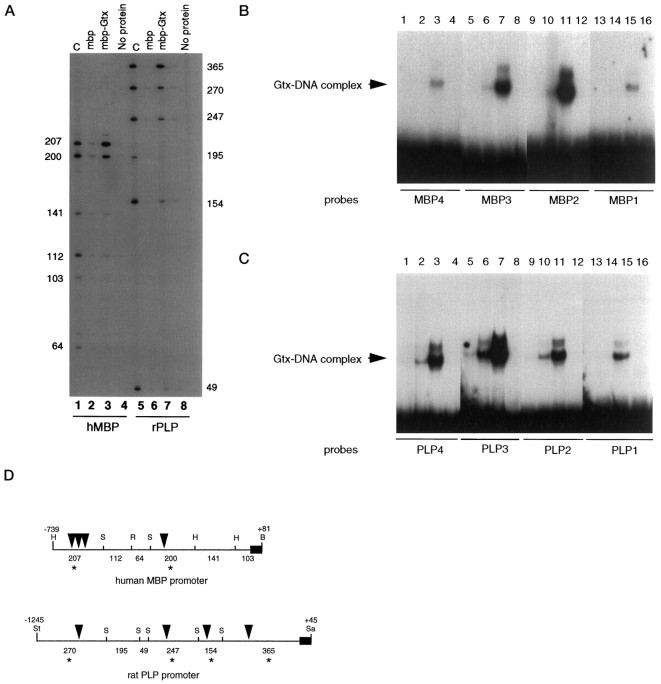
Fig. 7.
Gtx binds to multiple sites on the MBP and PLP promoters. A, Immunoprecipitation analysis of the MBP and PLP promoters. The 750 bp proximal fragment of the human MBP (hMBP) promoter and the 1.3 kb proximal fragment of the rat PLP (rPLP) promoter were digested with various restriction enzymes, and the fragments were end-labeled and incubated with the mbp–Gtx fusion protein, mbp alone, or no protein. Protein–DNA complexes were then immunopreciptated with a polyclonal rabbit anti-mbp antiserum and Staphylococcus protein A-Sepharose beads and separated on a 4%/7 m urea DNA-sequencing gel. Labeled fragments not subjected to immunoprecipitation analysis were run as a control for each promoter (lane C). Sizes (in base pairs) of the labeled fragments from each promoter are indicated on the left andright. B, Electrophoretic mobility shift assay (EMSA) analysis of human MBP promoter–Gtx interactions. Four double-stranded oligonucleotide probes, corresponding to nucleotides −331 to −304 (MBP1), −621 to −598 (MBP2), −636 to −614 (MBP3), and −656 to −630 (MBP4) of the human MBP promoter were each incubated with 1 nm (lanes 1, 5, 9, 13), 10 nm (lanes 2, 6, 10, 14), and 100 nm (lanes 3, 7, 11, 15) Gtx and 1000 nm maltose-binding protein (lanes 4, 8, 12, 16). Protein–DNA complexes were resolved on a nondenaturing polyacrylamide gel. The position of the Gtx–DNA complex is indicated at the left.C, EMSA analysis of rPLP promoter–Gtx interactions. Four double-stranded oligonucleotide probes, corresponding to nucleotides −286 to −259 (PLP1), −434 to −409 (PLP2), −671 to −644 (PLP3), and −1214 to −1188 (PLP4) of the rat PLP promoter were each incubated with 1 nm (lanes 1, 5, 9, 13), 10 nm (lanes 2, 6, 10, 14), and 100 nm (lanes 3, 7, 11, 15) Gtx or 1000 nm maltose-binding protein (lanes 4, 8, 12, 16). Protein–DNA complexes were resolved on a nondenaturing polyacrylamide gel. The position of the Gtx–DNA complex is indicated at the left.D, Summary of human MBP and rat PLP promoter–Gtx interactions. The solid boxed area in each promoter represents the mRNA sequence, beginning at the transcription start site; the thin line represents the upstream promoter sequence. The numbering of the nucleotide sequence for each promoter fragment begins (+1) at the known transcription start site.Positive numbers represent coding sequence;negative numbers represent promoter sequence. The sizes of the labeled fragments from the immunoprecipitation experiment inA are shown below each diagram. Immunoprecipitated fragments are indicated by stars.Arrowheads show the positions of the AT-rich regions corresponding to the probes used in the EMSA in B,C. The arrowheads most proximal to the transcription start sites mark the MBP1 and PLP1 sequences, respectively. Restriction enzymes used were HinfI (H), StyI (S), EcoRI (E), PstI (P), StuI (St), andBamHI (B).
Electrophoretic mobility shift assay. Varying amounts of protease Xa-cleaved Gtx were incubated with 40,000 cpm of end-labeled, double-stranded oligonucleotides at 4°C in 5% glycerol, 10 mm Tris, pH 7.4, 50 mm NaCl, 1 mmEDTA, 1 mm DTT, and 1 μg of poly(dI-dC) at a final volume of 20 μl. After 1 hr, protein–DNA complexes were loaded onto an 8% nondenaturing polyacrylamide gel, and the gel was electrophoresed at 20 mA in a running buffer containing (in mm) 6.7 Tris, pH 7.4, 3.3 sodium acetate, and 1 EDTA cooled to 4°C. Gels were then dried and exposed to x-ray film overnight at −70°C with an intensifying screen.
The oligonucleotides used were as follows: Hox A5/A6, 5′-AAGAGGTAGTAATTAGATCTGTCAATTT-3′ and 3′-CATCATTAATCTAGACAGTTAAAG-5′; mutant Hox A5/A6, 5′-AAGAGGTAGTGGCCAGATCTGTCAATTT-3′ and 3′-CATCACCGGTCTAGACAGTTAAAG −5′; brain creatine kinase (CK), 5′-GGCTATAAATAGCCGCCA-3′ and 3′-CCGATATTTATCGGCGGTGTGA-5′; PLP1, 5′-CACTTAATTTCCACCCACAATTACATTC-3′ and 3′-TGAATTAAAGGTGGGTGTTAATGTAAG-5′; PLP2, 5′-ATGTTTGGTAATATAGCAAGTAGGGT-3′ and 3′-ACAAACCATTATATCGTTCATCCCA-5′; PLP3, 5′-AATCATTAATACTTCTGGCTCTTCTTGA-3′ and 3′-TTAGTAATTATGAAGACCGAGAAG-5′; PLP4, 5′-GAAGAAAATAATTCCCCAGTAAACTC-3′ and 3′-TTCTTTTATTAAGGGGTCATTTGAG-5′; MBP1, 5′-TGCACATATTCTGTGGGTTTTATAGGAG-3′ and 3′-CGTGTATAAGACACCCAAAATATCCTC-5′; MBP2, 5′-TCCTTGCATATTTAACTTATG-3′ and 3′-AGGAACGTATAAATTGAATACGTG-5′; MBP3, 5′-TTGTCAAATAAATGCTCCTTGCA-3′ and 3′-CAGTTTATTTACGAGGAACGT-5′; and MBP4, 5′-AAGAAAACTAAAAACACCTTTTGTC-3′ and 3′-TTCTTTTGATTTTTGTGGAAAACAGTT-5′.
RESULTS
Gtx mRNA increases in parallel with the major myelin-specific mRNAs during postnatal brain development
Komuro and colleagues (1993) initially identified Gtx as a homeodomain protein expressed in adult white matter. Because white matter contains at least three cell types, oligodendroctes, oligodendrocyte precursors, and astrocytes, this localization is not sufficient to identify which cell is expressing Gtx or to describe the developmental profile of Gtx expression. To evaluate the timing of Gtx expression during brain development, we analyzed the steady state levels of Gtx mRNA in mouse cerebrum from the day of birth [postnatal day 1 (P1)] to P120 (adult) by Northern blotting. As can be seen in Figure 1A, two Gtx mRNAs, a major species of 1.5 kb and a minor species of 2.0 kb, were detected in developing mouse brain. Both Gtx mRNAs can first be detected in the developing mouse cerebrum at P15, reach a peak between P25 and P30, and decline somewhat in the adult. A similar profile of mRNA accumulation was also found for the mRNAs encoding MBP, PLP, and CNP1, the larger of the two CNP transcripts. The smaller CNP transcript, encoding the CNP2 isoform, is expressed much earlier in development, probably by oligodendrocyte precursors (Scherer et al., 1994; Yu et al., 1994).
Fig. 1.
Analysis of Gtx mRNA accumulation in developing mouse cerebrum. A, Twenty micrograms of total mouse cerebrum RNA from P1 to adult were electrophoresed, blotted, and successively hybridized with radiolabeled cDNA probes encoding Gtx, CNP, PLP, MBP, and GAPDH. B, Twenty micrograms of the same total mouse cerebrum RNA as in A were hybridized to a uniformly labeled, singled-stranded cRNA probe encoding mouse Gtx. The resulting hybrids were digested with RNase A and T1, and the protected fragments were separated on a 4% acrylamide/7 murea DNA-sequencing gel. A schematic diagram of the Gtx cDNA is shownbelow the autoradiogram; the cRNA probe used, encoding nucleotides 672–1052, is underlined. The locations of the undigested probe of 447 bp (lane C) and the protected fragments of 377 bp are indicated at theright.
Because the homeodomain family of transcription factors shares significant amino acid homology within the homeodomain, we wished to demonstrate that the signal detected by Northern blotting was specific for Gtx. We thus repeated the experiment shown in Figure1A using the technique of RNase protection to analyze the steady state levels of Gtx mRNA. The results of this experiment and the specific probe used are shown in Figure 1B. The predicted Gtx-specific band of 377 nucleotides can first be detected at postnatal day 15, peaks between postnatal days 15 and 25, and declines in the adult. These data, taken together with those above, demonstrate that Gtx mRNA increases in parallel with the major myelin-specific mRNAs during development and suggest that Gtx is expressed by myelinating oligodendrocytes.
Gtx is expressed predominantly in brain and thymus, but not by myelinating Schwann cells in the PNS
To determine the tissue distribution of Gtx expression in adult animals, we analyzed a number of organs for the presence of Gtx transcripts by RNase protection. The results of this experiment, shown in Figure 2, demonstrate that Gtx expression is limited to a few adult tissues, including brain and thymus. Gtx is not expressed in adult spleen, lung, heart, liver, or kidney. Furthermore, Gtx is not expressed in adult sciatic nerve, which contains Schwann cells, the myelinating cells of the peripheral nervous system, or in purified Schwann cell cultures (data not shown). The demonstration that Gtx is expressed in thymus is important, because several other myelin-related transcripts, including those encoding PLP/DM20 (Pribyl et al., 1996), CNP (Scherer et al., 1994), and GOLLI–MBP (Pribyl et al., 1993), are also expressed in this tissue. These data suggest that Gtx is also coordinately expressed along with myelin transcripts in thymus as well as in brain.
Fig. 2.
RNase protection analysis of Gtx mRNA expression in neural and non-neural adult mouse tissues. Twenty micrograms of total RNA isolated from various adult mouse tissues were hybridized to a uniformly labeled, singled-stranded cRNA probe encoding mouse Gtx and digested with RNase A and T1, and the protected fragments were separated on a 4% acrylamide/7 m urea DNA-sequencing gel. The locations of the undigested probe of 447 bp (lane C) and the protected fragments of 377 bp are indicated at theright.
Gtx is expressed by oligodendrocytes in primary cultures derived from cerebral white matter
Because our developmental Northern blot data suggest that Gtx mRNA accumulates in myelinating oligodendrocytes, we investigated the pattern of Gtx mRNA expression in mixed cerebral white matter cultures by RNase protection and in situ hybridization. Primary cultures of cerebral white matter, which contain both astrocytes and differentiating oligodendrocytes (Grinspan et al., 1990, 1993), were examined for Gtx and MBP mRNA. As can be seen in Figure3A, both Gtx and MBP mRNAs accumulate in parallel as oligodendrocytes differentiate in the presence of PDGF (Grinspan et al., 1993). However, when oligodendrocytes and their precursors are removed from this culture by complement-mediated cell lysis before extracting RNA, no Gtx or MBP transcripts can be detected. These data thus strongly suggest that Gtx is expressed by differentiated oligodendrocytes but not by astrocytes. To confirm this hypothesis directly, mixed cerebral white matter cultures derived from postnatal mouse brain, which also contain both differentiating oligodendrocytes and astrocytes (Knapp et al., 1987), were analyzed for Gtx mRNA expression by in situhybridization. These data, shown in Figure 3B, demonstrate reaction product in cells with the typical branched morphology of postmitotic oligodendrocytes but not in cells from the astrocyte bed layer. The reaction product is localized predominantly to the perinuclear cytoplasm. The faint staining of the oligodendrocyte processes and astrocyte membranes represents nonspecific staining. Taken together, the above data demonstrate that Gtx mRNA is found exclusively in oligodendrocytes but not in astrocytes in primary cultures prepared from cerebral white matter.
Fig. 3.

Gtx mRNA is expressed in oligodendrocytes.A, Ten micrograms of total RNA, prepared from rat cerebral white matter (CWM) cultures treated with PDGF for various times, were hybridized simultaneously with uniformly labeled cRNA probes encoding Gtx, MBP, and GAPDH. The resulting hybrids were digested with RNase A and T1, and the protected fragments were separated on a 4% acrylamide/7 m urea DNA sequencing gel. RNA from P34 rat brain was used as a control. Astrocyte RNA was prepared from CWM cultures, which had been depleted of oligodendrocytes by complement-mediated cell lysis (Astrocytes lane). The appropriately sized protected fragments of 377 bp (Gtx), 101 bp (MBP), and 324 bp (GAPDH) were observed for each probe. B, Mixed glial cultures, composed of both oligodendrocytes and astrocytes, were prepared from neonatal mouse brain, and the cells fixed onto coverslips and hybridized with a digoxigenin-labeled cDNA full-length probe encoding mouse Gtx. After hybridization the coverslips were washed and developed with nitroblue tretrazolium 5-bromo-4-chloro-3-indoyl phosphate. Note that the reaction product, representing hybridization to the labeled Gtx probe, is concentrated around the oligodendrocyte nucleus (large arrows) and in the oligodendrocyte perinuclear cytoplasm (arrowhead) but is not found in the underlying bed layer of astrocytes (*).
Gtx mRNA is expressed by postmitotic, differentiated oligodendrocytes
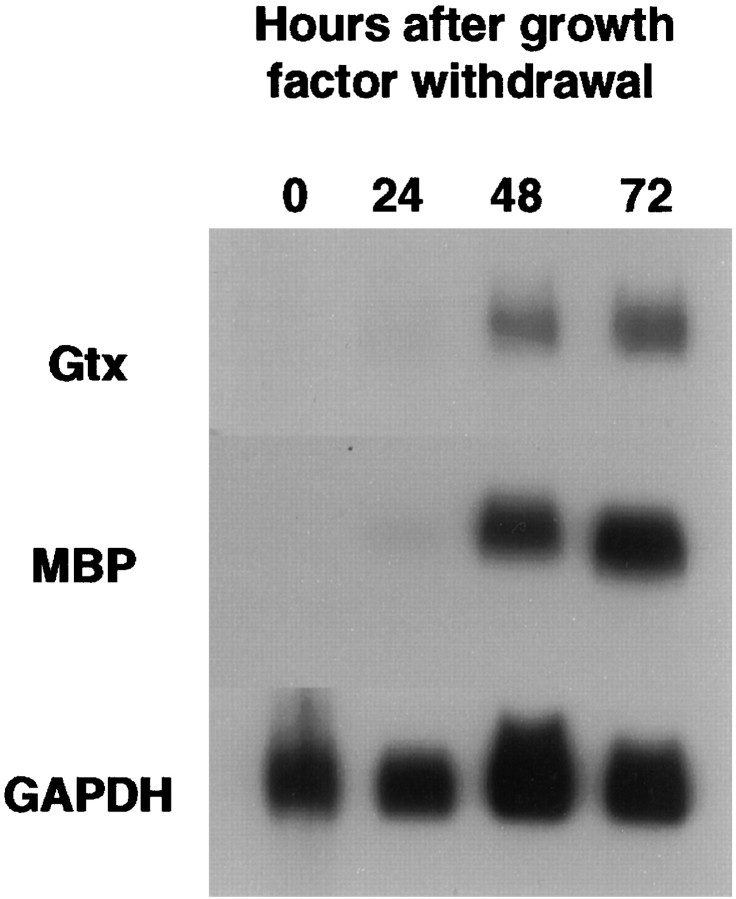
To determine at which stage of oligodendrocyte development Gtx mRNA could first be detected, we analyzed RNA extracted from a differentiating population of growth factor-“synchronized” oligodendrocytes for the presence of both Gtx and MBP mRNAs by Northern blotting. This culture system contains a purified population of oligodendrocyte precursors isolated from neonatal rat brain by immunopanning, which can proliferate indefinitely in the presence of basic FGF (bFGF) and PDGF. When these growth factors are removed, however, the cells cease dividing and differentiate within 24–48 hr (Collarini et al., 1992; Scherer et al., 1994). As can be seen in Figure 4, neither Gtx nor MBP mRNA can be found in dividing oligodendrocyte precursors cultured in the presence of growth factors (time 0). When the growth factors are removed and the cells differentiate, both MBP and Gtx mRNAs can be detected after 48 hr. These data thus demonstrate directly that Gtx mRNA is expressedin vitro in differentiated, postmitotic oligodendrocytes but not their proliferating precursors.
Fig. 4.
Northern blot analysis of Gtx mRNA expression in growth factor-stimulated cultures of developing oligodendrocytes. Total RNA was prepared from purified oligodendrocyte precursors cultured in the presence of bFGF and PDGF and at 24, 48, and 72 hr after growth factor withdrawal. Ten micrograms of RNA were electrophoresed per lane, blotted, and sequentially probed with radiolabeled cDNAs encoding Gtx, MBP, and GAPDH.
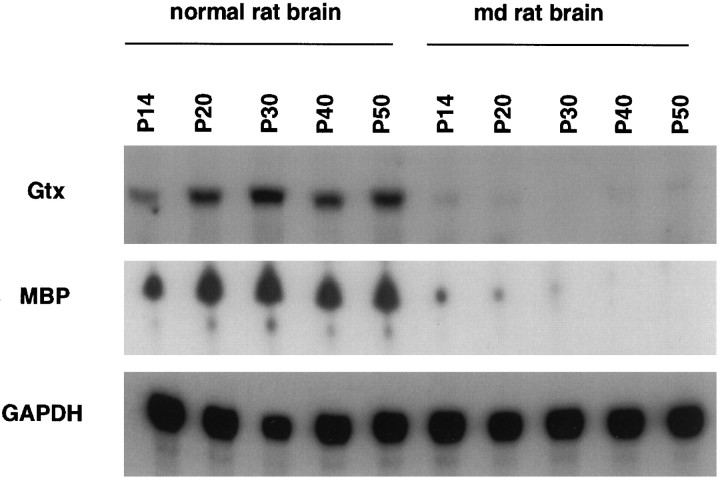
Gtx mRNA is coordinately downregulated with other myelin-specific mRNAs in md rat brain
The data presented above demonstrate that Gtx mRNA is expressed in postnatal brain exclusively in myelinating oligodendrocytes. Because Gtx mRNA accumulates in parallel with the major myelin-specific mRNAs both during brain development in vivo and oligodendrocyte differentiation in vitro, Gtx expression seems to be regulated as part of a coordinated program of myelin-specific gene expression. To evaluate this point further, we analyzed the steady state levels of Gtx mRNA in myelin-deficient (md) rats, which have a point mutation in the major myelin protein, PLP, causing a marked decrease in myelin-specific mRNA expression because of failure of oligodendrocyte maturation (Nadon and Duncan, 1995). RNase protection analysis of md rat brain RNA, shown in Figure5, shows a marked decrease in both MBP and Gtx mRNAs at all time points compared with controls. This experiment thus demonstrates that Gtx mRNA is decreased in parallel with that of MBP mRNA in the brains of md rats. Gtx mRNA expression is thus coordinately regulated with the major myelin protein genes, both during development when oligodendrocytes differentiate and in a pathological situation in which oligodendrocyte gene expression is known to be downregulated.
Fig. 5.
RNase protection analysis of Gtx mRNA inmd rat brain. Ten micrograms of total brain RNA frommd and normal rats of various ages were hybridized simultaneously with uniformly labeled cRNA probes encoding Gtx, MBP, and GAPDH. The appropriately sized protected fragments of 377 bp (Gtx), 101 bp (MBP), and 324 bp (GAPDH) were observed for each probe.
Gtx is a sequence-specific DNA-binding protein that interacts with an AT-rich core homeodomain binding site
Using a glutathione S-transferase (GST) fusion protein containing only the homeodomain portion of Gtx in a PCR-based selection assay, Komuro and colleagues (1993) could not identify a unique Gtx-specific binding sequence. To evaluate Gtx–DNA interactions further, we expressed the full-length Gtx in bacteria fused to the Escherichia coli mbp and then purified the mbp–Gtx fusion by maltose affinity chromatography. A Western blot of whole-cell extracts prepared from bacteria expressing the fusion protein, probed with an anti-maltose binding protein antibody, demonstrates a single band of 72 kDa, the appropriate size of the mbp–Gtx fusion protein (data not shown). We then evaluated affinity-purified mbp–Gtx fusion protein from which the mbp moiety had been cleaved by factor Xa protease for its ability to bind to two known AT-rich homeodomain core binding sites using an electrophoretic mobility shift assay. The first site, containing the core sequence TAATTA, has been shown by Odenwald and co-workers (1989) to interact with both Hox A5 and Hox A6 proteins expressed in baculovirus (J. Garbern, unpublished observations); the second site, TATAAAT, containing the MEF-2 sequence found in the brain CK promoter (Horlick et al., 1990), was shown by Komuro and colleagues (1993) to bind to a GST–Gtx homeodomain fusion protein.
As can be seen in Figure 6, the full-length Gtx protein shifts the oligonucleotide containing the core sequence TAATTA in a dose-dependent manner. A 10 nmconcentration of protein is capable of producing an observable gel shift, and competition with 200-fold excess of this unlabeled oligonucleotide completely inhibits binding in the presence of 50 nm Gtx. When the homeodomain binding site is mutated to TGGCCA, however, no binding is observed with 100 nmprotease-cleaved Gtx, and 200-fold excess of this mutant oligonucleotide does not inhibit binding to the unmutated site in the presence of 50 nm Gtx. In contrast, Gtx binds to the MEF-2 site from the brain CK promoter with less affinity than the TAATTA sequence, because 100 nm protein is required to produce an observable gel shift. These data thus demonstrate that Gtx interacts with DNA in a sequence-specific manner by way of an AT-rich core homeodomain binding site. In addition, they further demonstrate that the strength of specific Gtx–DNA interactions is sequence-dependent. Gtx binds with higher affinity to the TAATTA site, which contains a core TAAT homeodomain binding sequence, than to the MEF-2 site, which does not.
Fig. 6.
Gtx is a sequence-specific DNA-binding protein. Various amounts of mbp–Gtx (0, 1, 10, 50, and 100 nm) cleaved with factor Xa protease to remove the mbp moiety were incubated with an end-labeled, double-stranded oligonucleotide encoding a Hox A5/A6 binding site (H, lanes 1–5); 0, 1, 10, or 100 nm Gtx was also incubated with an end-labeled, double-stranded oligonucleotide encoding brain CK MEF-2 site (lanes 11–15). A 1000 nm concentration of mbp alone was incubated with probe H (lane 6) and probe CK (lane 15); 100 nm Gtx was incubated with a mutant Hox A5/A6 site (Hm; lane 7). A 50 nm concentration of Gtx was incubated with probe H (lane 8), to which 200-fold excess unlabeled probe H (lane 9) or 200-fold excess unlabeled probe Hm (lane 10) was added. Protein–DNA complexes were resolved on a nondenaturing polyacrylamide gel and are indicated at the left.
The MBP and PLP promoters contain multiple Gtx binding sites
The pattern of Gtx expresion suggests that it is involved, either directly or indirectly, in regulating the program of myelin-specific gene expression in oligodendrocytes. If Gtx were directly involved in this process, it should interact with several of the myelin-specific gene regulatory regions. To evaluate this possibility, we adapted an immunoprecipitation assay to identify specific Gtx–DNA interactions within the promoters of the MBP and PLP genes. The regulatory regions evaluated, the proximal 750 bp of the human MBP promoter (Wrabetz et al., 1993) and the proximal 1.3 kb of the rat PLP promoter (Boison et al., 1989), have been shown to drive oligodendrocyte-specific, developmentally regulated gene expression in transgenic mice (Nadon et al., 1994; Wrabetz et al., 1995) and thus contain the DNA sequences sufficient for this purpose. Using this assay, we found that Gtx could interact with four of six fragments within the proximal 1.3 kb of the PLP promoter and two of six fragments within the proximal 739 bp of the MBP promoter (see Fig. 7A). Each of these interactions required the mbp–Gtx fusion, because a control E. coli extract, containing mbp purified in the same manner as mbp–Gtx, did not immunoprecipate significant amounts of any of the promoter fragments.
To confirm both the above immunoprecipitation data and to localize specific Gtx binding sequences within the MBP and PLP promoter regions, we scanned the two promoters for the presence of putative AT-rich Gtx binding sites, similar to the Hox A5/A6 site previously shown to bind Gtx with high affinity. In this way we identified four putative Gtx binding sites within the initial 750 bp of MBP promoter and 11 putative Gtx binding sites within the first 1.3 kb of the PLP promoter. Each of the restriction fragments immunoprecipitated above contained two or more of these putative Gtx binding sites, whereas fragments not immunoprecipitated did not contain these sequences.
We then analyzed labeled, double-stranded oligonucleotides encoding putative Gtx binding sites located within each of the immunoprecipitated fragments using an electrophoretic mobility shift assay. The results of these experiments are shown in Figure 7,B and C. Gtx binds three sites within the MBP promoter between nucleotides −598 and −656, located in one of the immunoprecipitated fragments, and another site between nucleotides −304 and −331, located in the second immunoprecipitated fragment. The site between nucleotides −598 and −621 binds Gtx with the highest affinity of the four MBP sites but not as high as the Hox A5/A6 site (data not shown). Binding to each of these four sites is specific, because it can be competed out with 200-fold molar excess unlabeled oligonucleotide encoding a Hox A5/A6 binding site (H), but not with 200-fold molar excess unlabeled oligonucleotide encoding a mutant Hox A5/A6 binding site (Hm) (data not shown). None of the four MBP sites, however, encode a sequence identical to the Hox A5/A6 core sequence.
Gtx also binds at least four sites within the PLP promoter, each of which is also located in one of the four immunoprecipitated fragments. The site located between nucleotides −644 and −671 binds Gtx with the highest affinity, similar to that of the Hox A5/A6 sequence. The sites located between −409 and −434 and −1188 and −1214 bind with less affinity, and the site between −259 and −286 binds with the least affinity. Each of these four sites, however, contains a TAAT core, so that the presence of this core sequence alone cannot explain the differences in binding affinity. Binding to each of these four sites is specific, because it can be competed out with 200-fold molar excess unlabeled oligonucleotide encoding H but not with 200-fold molar excess unlabeled oligonucleotide encoding Hm (data not shown). In addition to these four sites, there are six other putative Gtx binding sites within the PLP promoter, three of which also contain a TAAT core sequence. The location and DNA sequence of the Gtx binding sites within the MBP and PLP promoters are displayed in Table1. These data thus demonstrate that the regulatory regions of the MBP and PLP genes, which are coordinately regulated in oligodendrocytes during myelination, each contain multiple sites capable of binding Gtx.
Table 1.
Gtx binding sites within the MBP and PLP promoters
| Probe | Binding site | Site location | Binding | Immunoprecipitated fragments (bp) |
|---|---|---|---|---|
| MBP1 | CATATTCT | −327 to −320 | + | −356 to −157 (200) |
| MBP2 | GTTAAATA | −606 to −613 | +++ | −739 to −532 (207) |
| MBP3 | AATAAATG | −630 to −623 | ++ | −739 to −532 (207) |
| MBP4 | ACTAAAAA | −650 to −643 | + | −739 to −532 (207) |
| PLP1a | TGTAATTG | −262 to −269 | + | −320 to +45 (365) |
| PLP1b | CTTAATTT | −284 to −277 | + | −320 to +45 (365) |
| PLP2 | GGTAATAT | −428 to −421 | +++ | −474 to −320 (154) |
| PLP3 | ATTAATGA | −662 to −669 | ++++ | −721 to −474 (247) |
| PLP4 | AATAATTC | −1194 to −1201 | +++ | −1245 to −975 (270) |
| PLP1-a | TGTAATCT | −10 to −17 | ND | −320 to +45 (365) |
| PLP1-a | TATAAATG | −245 to −252 | ND | −320 to +45 (365) |
| PLP1-a | CTTAAATC | −296 to −303 | ND | −320 to +45 (365) |
| PLP1-a | TTTAAATG | −331 to −324 | ND | −474 to −320 (154) |
| PLP1-a | AATAATTT | −559 to −566 | ND | −721 to −474 (247) |
| PLP1-a | GATAATCC | −1020 to −1013 | ND | −1245 to −975 (270) |
| CK | TATAAATA | ++ | ||
| H | AGTAATTA | ++++ | ||
| Hm | AGTGGCCA | − |
ND, Not done. Relative binding affinity is indicated by +, with ++++ the affinity of binding to the Hox A5/A6 sequence. PLP1a and PLP1b are located within the region −262 to −284 of the PLP promoter and have been evaluated by EMSA using a single oligonucleotide encoding both sites. The sites are displayed individually for clarity. Numbering of the nucleotide sequence for each promoter begins (+1) at the known transcription start site. Positive numbers represent transcribed sequence; negative numbers represent promoter sequences. Bold areas of sequence are the TA-rich core of the homeodomain binding site.
A putative Gtx binding site, identified by homology to a known binding site.
DISCUSSION
In this study we have investigated the patterns of postnatal brain expression and DNA binding of Gtx, a homeodomain transcription factor. We have shown that Gtx mRNA accumulates in parallel with RNAs encoding two of the major structural proteins of myelin, MBP and PLP, during postnatal brain development, as well as in the brains of myelin-deficient rats. Gtx mRNA is expressed in differentiated oligodendrocytes but not in oligodendrocyte precursors or astrocytes. In addition, we have demonstrated that Gtx can bind DNA sequences containing a core AT-rich homeodomain binding site and have identified four Gtx binding sites in the MBP promoter and an additional four Gtx binding sites in the PLP promoter. These data, taken together, strongly suggest that Gtx is important for the function of differentiated oligodendrocytes and may directly regulate myelin-specific gene expression.
Gtx is expressed by oligodendrocytes
The conclusion that Gtx mRNA is expressed by oligodendrocytes is based on the following observations. First, Gtx mRNA levels decrease inmd rat brain and increase during postnatal brain development similar to the myelin-specific mRNAs expressed by differentiated oligodendrocytes but quite distinct from the pattern of astrocyte-specific gene expression (Brenner et al., 1994). Second, Gtx mRNA can be detected by RNase protection analysis of RNA prepared from mixed cerebral white matter cultures containing both oligodendrocytes and astrocytes, and removal of oligodendrocytes by complement-mediated cell lysis eliminates the Gtx signal. Third, Gtx mRNA can be detected only in oligodendrocytes but not in astroctyes by in situhybridization of mixed cerebral white matter cultures. Komuro and co-workers (1993) found Gtx mRNA predominantly in white matter in the brains of adult rodents by in situ hybridization. Our results thus confirm and extend this study and provide strong evidence that Gtx mRNA is expressed exclusively by differentiated oligodendrocytes but not by oligodendrocyte progenitors, astrocytes, or neurons in postnatal rodent brain.
In contrast to the above data, Komuro and co-workers (1993) found immunoreactive Gtx protein in both cultured astrocytes and oligodendrocytes using an antibody raised in chicken to a GST–Gtx homeodomain fusion protein. Because we have shown that Gtx mRNA cannot be detected in astrocytes, by either in situ hybridization or RNase protection, this result is somewhat surprising. Further analysis of this anti-Gtx antibody thus will be necessary to resolve the discrepancy in these data.
Transcription factors and the regulation of oligodendrocyte development
Only two transcription factors have been implicated by their pattern of expression in the regulation of oligodendrocyte differentiation. The first of these suppressed cAMP-inducible POU (SCIP), a member of the POU homeodomain family, is expressed in proliferating oligodendrocyte progenitors in culture but is markedly downregulated when the cells differentiate and cease dividing (Collarini et al., 1992). An SCIP binding site identified within the neuronal nicotinic acetylcholine α3 receptor promoter (Fyodorov and Deneris, 1996) is identical to one of the highest affinity Gtx binding sites, ATTAATG, located between nucleotides −662 and −669 within the PLP promoter, suggesting that both SCIP and Gtx can interact with this site. Inactivation of SCIP by homologous recombination, however, has little or no effect on CNS myelination (Bermingham et al., 1996) although SCIP-positive oligodendrocyte progenitors have been identifiedin vivo (Arroyo et al., 1994). The second factor, MyT1, a member of the zinc finger family, was initially identified by its ability to bind to a specific DNA sequence in the promoter of the PLP gene (Kim and Hudson, 1992). MyT1 is expressed in oligodendrocyte progenitors, both in vitro and in vivo, although its expression declines soon after the cells differentiate and begin to myelinate (Armstrong et al., 1995). These data suggest that neither MyT1 nor SCIP is necessary for maintaining the phenotype of differentiated oligodendrocytes, although they may play a role in promoting oligodendrocyte differentiation. Gtx is thus unique, because it is the only transcription factor known to be expressed in differentiated oligodendrocytes, but not in other stages of the oligodendrocyte lineage.
The temporal correlation of Gtx and MBP mRNA expression in oligodendrocytes and the results of the Gtx–promoter binding studies suggest that the PLP and MBP genes may be downstream targets for Gtx. Consistent with this idea, transgenic mice containing the proximal 750 bp of the human MBP promoter fused to lacZ, with two sets of Gtx binding sites localized between nucleotides −608 and −636 and −301 and −331, express β-galactosidase in an oligodendrocyte-specific, developmentally regulated manner (Wrabetz et al., 1995). Transgenic mice containing shorter human MBP promoter–lacZ fusion constructs containing either 150 or 420 bp of MBP promoter sequence did not express lacZ in brain at postnatal day 18, the peak of myelination (L. Wrabetz, unpublished results), suggesting that the more distal Gtx binding sites are necessary for MBP expression. In addition, 1.3 kb of the PLP promoter, which is sufficient to drive developmentally regulated PLP expression in transgenic mice (Nadon et al., 1994), contains 11 putative Gtx binding sites, several of which bind Gtx with high affinity. Gtx is thus expressed in the right cells at the right time and interacts with the appropriate promoters to regulate myelin-specific gene expression. Further experiments are currently in progress in our laboratory to determine whether Gtx is necessary and/or sufficient for the direct activation of myelin-specific gene expression.
Transcription factors and the regulation of Schwann cell development
Although Gtx is an excellent candidate regulator of oligodendrocyte development and myelin-specific gene expression, it is not expressed in Schwann cells, the myelinating cells of the peripheral nervous system. This is, at least superficially, somewhat suprising, because both MBP and PLP, the major CNS myelin proteins, are also expressed in myelinating Schwann cells (Griffiths et al., 1995; Garbern et al., 1997). Comparison of the known mechanisms of development and regulation of myelin-specific gene expression in Schwann cells and oligodendrocytes, however, demonstrate a number of significant differences. First, the transcription factors known to be necessary for normal Schwann cell maturation and PNS myelination, SCIP (Bermingham et al., 1996; Jaegle et al., 1996) and Krox-20 (Topilko et al., 1994), are not required for normal oligodendrocyte maturation or myelination. In addition, the developmental profile of expression of these proteins is different in the two cell types. SCIP is expressed in promyelinating Schwann cells, which are committed to myelinate, but is found in oligodendrocyte precursors, not differentiated, myelinating cells (Collarini et al., 1992). Furthermore, Krox-20 is not expressed in oligodendrocytes (Topilko et al., 1994; Sock et al., 1997). Second, different regions or modules within the MBP promoter have been shown to be required for oligodendrocyte and Schwann cell gene expression. MBP promoter sequences up to 3.2 kb are sufficient to drive developmentally regulated, oligodendrocyte-specific transgene expression but do not support transgene expression in Schwann cells (Foran and Peterson, 1992; Gow et al., 1992) (L. Wrabetz, unpublished data). Recent data from Dr. Alan Peterson’s laboratory demonstrate the presence of a small sequence farther upstream in the MBP promoter with properties of an enhancer that is necessary for Schwann cell but not oligodendrocyte expression of a lacZ transgene (Garafalo et al., 1995) (A. Peterson, personal communication). Finally, comparison of the set of oligodendrocyte and Schwann cell nuclear proteins that bind to the proximal MBP promoter (Wrabetz et al., 1993; Li et al., 1994) (L. Wrabetz, unpublished data) demonstrates a number of significant differences, suggesting that there is a unique set of transcriptional activators present in the two cell types. The above data strongly suggest that the molecular mechanisms underlying the regulation of Schwann cell and oligodendrocyte development are different. The presence of Gtx in oligodendrocytes but not Schwann cells is further evidence of this notion.
Homeodomain proteins, Gtx, and development
The homeodomain, a 60 amino acid motif, found in a large number of proteins both in vertebrates and invertebrates, has structural similarities to the helix-turn-helix DNA-binding domain of some regulatory proteins in yeast and prokaryotes (Laughon and Scott, 1984). Homeodomain-containing proteins bind DNA and have been shown to act as sequence-specific transcription factors. The Antennapedia class of homeodomain-containing proteins, which share homology with theDrosophila antennapedia protein, are clustered within four chromosomal regions in rodents and humans and have been implicated in the regulation of pattern formation during early embryogenesis (Krumlauf, 1994). Homeodomain proteins not found within these four chromosomal clusters, including Gtx, constitute a diverse set of transcription factors, many of which are expressed in a tissue- or organ-specific manner (Blochlinger et al., 1988; Ingraham et al., 1988;Staudt et al., 1988; Suh et al., 1994). Although downstream or target genes have not been identified for many of these orphan homeodomain proteins, several of them have been implicated in organogenesis and cell differentiation (Roberts et al., 1994; Slack, 1995).
The homeodomain protein most closely related to Gtx is Nkx6.1, a hamster protein of the NK-class of homeodomain proteins, isolated from a pancreatic β-cell cDNA library (Rudnick et al., 1994). The homeodomains of Nkx6.1 and Gtx are 95% identical, including a histidine residue at position 3, which is uniquely found in these two proteins. In addition, the overall amino acid identity between the two proteins is 52%, so there is also significant homology outside the homeodomains. Furthermore, both proteins contain a conserved decapeptide sequence, found in the N-terminal region of the NK family of homeodomain proteins. No other homeodomain protein, including other members of the NK class, shares more than 60% homology with the homeodomain of either Gtx or Nkx6.1 (Komuro et al., 1993). German and colleagues have recently identified a mouse protein with a homeodomain that is identical to Nkx6.1, suggesting that it is the mouse Nkx6.1 homolog (M. S. German, personal communication). Studies of the pattern of Nkx6.1 expression have demonstrated expression in pancreatic β-cell lines and pancreatic β-cells but not in postnatal brain (Rudnick et al., 1994) (M. S. German, personal communication; R. Awatramani, unpublished results). These data, taken together, suggest that Gtx and Nkx6.1 are unique but related proteins that define a new subclass of homeodomain proteins.
Several homeodomain-containing proteins other than Gtx, including Pdx1 in the pancreas (Ohlsson et al., 1993; Ahlgren et al., 1996), TTF-1 in lung and thyroid (Lazzaro et al., 1991; Bruno et al., 1995), and Pit-1 in the pituitary (Nelson et al., 1988; Mangalam et al., 1989; Simmons et al., 1990), have been shown both to be expressed in terminally differentiated cells and to bind to the promoters of tissue specific genes, implicating them in their regulation. Both Pit-1 and Pdx1, however, are also expressed early in development of the pituitary (Li et al., 1990) and pancreas (Jonsson et al., 1994; Ahlgren et al., 1996), respectively, and inactivation of either their expression or their function has demonstrated that they also are necessary for normal organogenesis. TTF-1 is also expressed early in thyroid and lung development (Lazzaro et al., 1991) but has not been evaluated by homologous recombination. In addition, all three proteins have been shown to trans-activate tissue-specific promoters, eitherin vitro or in transgenic mice (Civitareale et al., 1989;German et al., 1992; Ohlsson et al., 1993; Ray et al., 1996). The tissue- and/or cell-specific pattern of expression of these three homeodomain transcription factors coupled with their ability to bind to tissue-specific promoters suggests that they are involved in the regulation of differentiation in the pancreas, thyroid, lung, and pituitary, which has been borne out by further experimentation. The pattern of lineage- and stage-specific expression of Gtx coupled with its ability to bind to myelin-specific promoters, such as Pdx1, TTF-1, and Pit-1, strongly suggests that it has an important function in the differentiation of oligodendrocytes.
Footnotes
This work was supported by grants from the National Multiple Sclerosis Society (J.K., R.S., J.G.), the National Institutes of Health (S.S., J.G.), and the Medical Research Council of Great Britain (E.C.). We thank Dr. Seigo Izumo for the generous gift of the mouse Gtx cDNA clone and chicken anti-Gtx antibodies.
Correspondence should be addressed to Dr. John Kamholz, Department of Neurology, Wayne State University School of Medicine, 3105 Elliman Building, 421 East Canfield, Detroit, MI 48201.
REFERENCES
- 1.Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in PFP1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong RC, Kim JG, Hudson LD. Expression of myelin transcription factor I (MyTI), a “zinc-finger” DNA-Binding protein, in developing oligodendrocytes. Glia. 1995;14:303–321. doi: 10.1002/glia.440140407. [DOI] [PubMed] [Google Scholar]
- 3.Arroyo EJ, Wegener M, Rosenfeld MG, Kamholz J, Scherer SS. Oligodendrocyte precursors express Tst-1 (SCIP/Oct-6) in vivo. Soc Neurosci Abstr. 1994;20:456. [Google Scholar]
- 4.Berndt JA, Kim JG, Hudson LD. Identification of cis-regulatory elements in the myelin proteolipid protein (PLP) gene. J Biol Chem. 1992;267:14730–14737. [PubMed] [Google Scholar]
- 5.Bernier L, Alvarez F, Norgard EM, Raible DW, Mentaberry A, Schembri JG, Sabatini DD, Colman DR. Molecular cloning of a 2′,3′-cyclic nucleotide 3′-phosphodiesterase: mRNAs with different 5′ ends encode the same set of proteins in nervous and lymphoid tissues. J Neurosci. 1987;7:2703–2710. doi: 10.1523/JNEUROSCI.07-09-02703.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bermingham JR, Scherer SS, O’Connell S, Arroyo E, Kalla KK, Powell FL, Rosenfeld MG. Tst-1/Oct-6/SCIP reglates a unique step in peripheral myelination and is required for normal respiration. Genes Dev. 1996;10:1751–1762. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- 7.Bharucha VA, Peden KWC, Tennekoon GI. SV40 large T antigen with c-Jun downregulates myelin Po gene expression: a mechanism for papovaviral T antigen-mediated demyelination. Neuron. 1994;12:627–637. doi: 10.1016/0896-6273(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 8.Blochlinger K, Bodmer R, Jack J, Jan LY, Jan YN. Primary structure and expression of a product from cut, a locus involved in specifying sensory organ identity in Drosophila. Nature. 1988;333:629–635. doi: 10.1038/333629a0. [DOI] [PubMed] [Google Scholar]
- 9.Boison D, Stoffel W. Myelin-deficient rat: a point mutation in exon III (A—C, Thr75—Pro) of the myelin proteolipid protein causes dysmyelination and oligodendrocyte death. EMBO J. 1989;8:3295–3302. doi: 10.1002/j.1460-2075.1989.tb08490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner M, Kisseberth WC, Su Y, Besnard F, Messing A. GFAP promoter directs astrocyte-specific expression in transgenic mice. J Neurosci. 1994;14:1030–1037. doi: 10.1523/JNEUROSCI.14-03-01030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruno MD, Bohinski RJ, Muelsman KM, Whitsett JA, Korfhagen TR. Lung cell-specific expression of the murine surfactant protein A (SP-A) gene is mediated by interactions between the SP-A promoter and thyroid transcription factor-1. J Biol Chem. 1995;270:6531–6536. doi: 10.1074/jbc.270.12.6531. [DOI] [PubMed] [Google Scholar]
- 12.Campagnoni AT. Molecular biology of myelin proteins from the central nervous system. J Neurochem. 1988;51:1–14. doi: 10.1111/j.1471-4159.1988.tb04827.x. [DOI] [PubMed] [Google Scholar]
- 13.Chirgwin JM, Przbyla AE, MacDonald RJ, Rutter W. Isoation of biologoically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 14.Civitareale R, Lonigro R, Sinclair MJ, DiLauro R. A thyroid-nuclear protein essential for tissue-specific expression of the thyroglobulin promoter. EMBO J. 1989;8:2537–2542. doi: 10.1002/j.1460-2075.1989.tb08391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collarini EJ, Kuhn R, Marshall CJ, Monuki ES, Lemke G, Richardson WD. Down-regulation of the POU transcription factor SCIP is an early event in oligodendrocyte differentiation in vitro. Development. 1992;116:193–200. doi: 10.1242/dev.116.1.193. [DOI] [PubMed] [Google Scholar]
- 16.Dubois-Dalcq M, Behar T, Hudson L, Lazzarini RA. Emergence of three myelin proteins in oligodendrocytes cultured without neurons. J Cell Biol. 1986;102:384–392. doi: 10.1083/jcb.102.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis D, Malcom S. Proteolipid protein gene dosage effect in Pelizaeus-Merzbacher disease. Nat Genet. 1994;6:333–334. doi: 10.1038/ng0494-333. [DOI] [PubMed] [Google Scholar]
- 18.Foran DR, Peterson AC. Myelin acquisition in the central nervous system of the mousee revealed by an MBP-Lac Z transgene. J Neurosci. 1992;12:4890–4897. doi: 10.1523/JNEUROSCI.12-12-04890.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fort P, Marty L, Piechaczyk M, Sabrouty SE, Dani C, Jeanteur P, Blanchard JM. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 1985;13:1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fyodorov D, Deneris E. The POU domain of SCIP/Tst-1/Oct-6 is sufficient for activation of any acetylcholine receptor promoter. Mol Cell Biol. 1996;16:5004–5014. doi: 10.1128/mcb.16.9.5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garafalo L, Foran D, Peterson A. Myelin basic protein promoter elements resulting in high level sustained expression in oligodendrocytes and Schwann cells in transgenic mice. Soc Neurosci Abstr. 1995;21:320. [Google Scholar]
- 22.Garbern JY, Cambi F, Sohi J, Tang X-M, Sima AAF, Bosch EP, Lewis R, Shy M, Kraft G, Vallat JM, Chen KL, Joshi I, Leonard D, Johnson W, Raskind W, Dlouhy SR, Pratt V, Hodes ME, Bird T, Kamholz J (1997) Proteolipid protein is necessary in peripheral as well as central myelin. Neuron, in press. [DOI] [PubMed]
- 23.German MS, Wang J, Chadwick RB, Rutter WJ. Synergistic activation of the insulin gene by a LIM-homeodomain protein and a basic helix-loop-helix protein: building a functional insulin minienhancer complex. Genes Dev. 1992;6:2165–2176. doi: 10.1101/gad.6.11.2165. [DOI] [PubMed] [Google Scholar]
- 24.Ghandour MS, Skoff R. Double labeling in situ hybridization analysis of mRNAs for carbonic anhydrase II and myelin basic protein: expression in developing cultured glial cells. Glia. 1991;4:1–10. doi: 10.1002/glia.440040102. [DOI] [PubMed] [Google Scholar]
- 25.Goujet-Zalc C, Babinet C, Monge M, Timsit S, Cabon F, Gansmuller A, Miura M, Sanchez M, Pournin S, Mikoshiba K, Zalc B. The proximal region of the MBP gene promoter is sufficient to induce oligodendroglial-specific expression in transgenic mice. Eur J Neurosci. 1993;5:624–632. doi: 10.1111/j.1460-9568.1993.tb00528.x. [DOI] [PubMed] [Google Scholar]
- 26.Gow A, Friedrich V, Lazzarini RA. Myelin basic protein gene contains separate enhancers for oligodendrocyte and Schwann cell expression. J Cell Biol. 1992;119:605–615. doi: 10.1083/jcb.119.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths IR, Dickinson P, Montague P. Expression of the proteolipid protein gene in glial cells of the post-natal peripheral nervous system of rodents. Neuropathol Appl Neurobiol. 1995;21:97–110. doi: 10.1111/j.1365-2990.1995.tb01035.x. [DOI] [PubMed] [Google Scholar]
- 28.Grinspan JB, Stern JL, Pustilnik SM, Pleasure D. Cerebral white matter contains PDGF-responsive precursors to O2A cells. J Neurosci. 1990;10:1866–1873. doi: 10.1523/JNEUROSCI.10-06-01866.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grinspan J, Wrabetz L, Kamholz J. Oligodendrocyte maturation and myelin gene expression in PDGF-treated cultures from rat cerebral white matter. J Neurocytol. 1993;22:322–333. doi: 10.1007/BF01195556. [DOI] [PubMed] [Google Scholar]
- 30.Haas S, Gordon J, Khalili K. A developmentally regulated DNA-binding protein from mouse brain stimulates myelin basic protein gene expression. Mol Cell Biol. 1993;13:3103–3112. doi: 10.1128/mcb.13.5.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodes ME, Pratt VM, Dlouhy SR. Genetics of Pelizaeus-Merzbacher disease. Dev Neurosci. 1993;15:383–394. doi: 10.1159/000111361. [DOI] [PubMed] [Google Scholar]
- 32.Horlick RA, Hobson GM, Patterson JH, Mitchell MT, Benfield PA. Brain and muscle creatine kinase genes contain common TA-rich recognition protein-binding regulatory elements. Mol Cell Biol. 1990;10:4826–4836. doi: 10.1128/mcb.10.9.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingraham HA, Chen RP, Mangalam HJ, Elsholtz HP, Flynn SE, Lin CR, Simmons DM, Swanson L, Rosenfeld MG. A tissue-specific transcription factor containing a homeodomain specifies a pituitary phenotype. Cell. 1988;55:519–529. doi: 10.1016/0092-8674(88)90038-4. [DOI] [PubMed] [Google Scholar]
- 34.Jaegle M, Mandemakers W, Broos L, Zwart R, Karis A, Visser P, Grosveld F, Meijer D. The POU factor Oct-6 and Schwann cell differentiation. Science. 1996;273:507–510. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]
- 35.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 36.Kagawa T, Ikenaka K, Inoue Y, Kuriyama S, Tsujii T, Nakao J, Nakajima K, Aruga J, Okano H, Mikoshiba K. Glial cell degeneration and hypomyelination caused by overexpression of myelin proteolipid protein gene. Neuron. 1994;13:427–442. doi: 10.1016/0896-6273(94)90358-1. [DOI] [PubMed] [Google Scholar]
- 37.Kamholz J, Wrabetz L. Molecular genetics of myelin basic protein. In: Martenson RE, editor. Myelin: A treatise. CRC; Boca Raton, FL: 1992. pp. 367–385. [Google Scholar]
- 38.Kamholz J, Toffenetti J, Lazzarini RA. Organization and expression of the human myelin basic protein gene. J Neurosci Res. 1988;21:62–70. doi: 10.1002/jnr.490210110. [DOI] [PubMed] [Google Scholar]
- 39.Kamholz J, Sessa M, Scherer S, Vogelbacker H, Mokuno K, Baron P, Wrabetz L, Shy M, Pleasure D. Structure and expression of proteolipid protein in the peripheral nervous system. J Neurosci Res. 1992;31:231–244. doi: 10.1002/jnr.490310204. [DOI] [PubMed] [Google Scholar]
- 40.Kim JG, Hudson LH. Novel member of the zinc finger superfamily: a C2-HC finger that recognizes a glial-specific gene. Mol Cell Biol. 1992;12:5632–5639. doi: 10.1128/mcb.12.12.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knapp PE, Bartlett WP, Skoff RP. Cultured oligodendrocytes mimic in vivo phenotypic characteristics: cell shape, expression of myelin-specific antigens, and membrane production. Dev Biol. 1987;120:356–365. doi: 10.1016/0012-1606(87)90238-7. [DOI] [PubMed] [Google Scholar]
- 42.Komuro I, Schalling M, Jahn L, Bodmer R, Jenkins NA, Copeland NG, Izumo S. Gtx: a novel murine homeobox-containing gene, expressed specifically in glial cells of the brain and gern cells of testis, has a transcriptional repressor activity in vitro for a serum-inducible promoter. EMBO J. 1993;12:1387–1401. doi: 10.1002/j.1460-2075.1993.tb05783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krumlauf R. Hox genes in vertebrate development. [review]. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 44.Laughon A, Scott MP. Sequence of a Drosophila segmentation gene: protein structure homology with DNA-binding proteins. Nature. 1984;310:25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- 45.Lazzaro D, Price M, De Felice M, Di Lauro R. The transcription factor TTF-1 is expression at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 1991;113:1093–1104. doi: 10.1242/dev.113.4.1093. [DOI] [PubMed] [Google Scholar]
- 46.Lemke G. The molecular genetics of myelination: an update. [review]. Glia. 1993;7:263–271. doi: 10.1002/glia.440070402. [DOI] [PubMed] [Google Scholar]
- 47.Li S, Crenshaw ED, Rawson EJ, Simmons DM, Swanson LW, Rosenfeld MG. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature. 1990;347:528–533. doi: 10.1038/347528a0. [DOI] [PubMed] [Google Scholar]
- 48.Li X, Wrabetz L, Cheng Y, Kamholz J. A novel cyclic AMP response element, CACTTGATC, mediates forskolin induction of the myelin basic protein promoter in the rat Schwannoma line, D6P2T. J Neurochem. 1994;63:28–40. doi: 10.1046/j.1471-4159.1994.63010028.x. [DOI] [PubMed] [Google Scholar]
- 49.Mangalam H, Albert H, Ingraham H, Kapiloff M, Wilson L, Nelson C, Elsholtz H, Rosenfeld M. A pituitary POU domain protein, Pit-1, activates both growth hormone and prolactin promoters transcriptionally. Genes Dev. 1989;3:946–958. doi: 10.1101/gad.3.7.946. [DOI] [PubMed] [Google Scholar]
- 50.Milner RJ, Lai C, Nave KA, Lenoir D, Ogata J, Sutcliffe JG. Nucleotide sequences of two mRNAs for rat brain myelin proteolipid protein. Cell. 1985;42:931–939. doi: 10.1016/0092-8674(85)90289-2. [DOI] [PubMed] [Google Scholar]
- 51.Nadon NL, Duncan ID. Gene expression and oligodendrocyte development in the myelin deficient rat. J Neurosci Res. 1995;41:96–104. doi: 10.1002/jnr.490410111. [DOI] [PubMed] [Google Scholar]
- 52.Nadon NL, Arnheiter H, Hudson LD. A combination of PLP and DM20 transgenes promotes partial myelination in the jimpy mouse. J Neurochem. 1994;63:822–833. doi: 10.1046/j.1471-4159.1994.63030822.x. [DOI] [PubMed] [Google Scholar]
- 53.Nelson C, Albert VR, Elsholtz HP, Lu LI, Rosenfeld MG. Activation of cell-specific expression of rat growth hormone and prolactin genes by a common transcription factor. Science. 1988;239:1400–1405. doi: 10.1126/science.2831625. [DOI] [PubMed] [Google Scholar]
- 54.Odenwald WF, Garbern J, Arnheiter H, Tournier-Lasserve E, Lazzarini RA. The Hox-1.3 homeo box protein is a sequence-specific DNA-binding phosphoprotein. Genes Dev. 1989;3:158–172. doi: 10.1101/gad.3.2.158. [DOI] [PubMed] [Google Scholar]
- 55.Ohlsson H, Karlson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pribyl TM, Campagnoni CW, Kampf K, Kashima T, Handley VW, McMahon J, Campagnoni AT. The human myelin basic protein gene is included within a 179-kilobase transcription unit: expression in the immune and central nervous systems. Proc Natl Acad Sci USA. 1993;90:10695–10699. doi: 10.1073/pnas.90.22.10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pribyl TM, Campagnoni CW, Kampf K, Kashima T, Handley VW, McMahon J, Campagnoni AT. Expression of the myelin proteolipid protein gene in the human fetal thymus. J Neuroimmunol. 1996;69:125–130. doi: 10.1016/0165-5728(96)00058-6. [DOI] [PubMed] [Google Scholar]
- 58.Raskind WH, Williams CA, Hudson LD, Bird TD. Complete deletion of the proteolipid protein gene (PLP) in a family with X-linked Pelizaeus-Merzbacher disease. Am J Hum Genet. 1991;49:1355–1360. [PMC free article] [PubMed] [Google Scholar]
- 59.Ray MK, Chen C-Y, Schwartz RJ, DeMayo FJ. Transcriptional regulation of a mouse Clara cell-specific protein (mCC10) gene by the NKx transcription factor family members thyroid transcription factor 1 and cardiac muscle-specific homeobox protein (CSX). Mol Cell Biol. 1996;16:2056–2064. doi: 10.1128/mcb.16.5.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Readhead C, Popko B, Takahashi N, Shine HD, Sidman RL, Hood L. Expression of a myelin basic protein gene in transgenic shiverer mice: correction of the dysmyelinating phenotype. Cell. 1987;48:703–712. doi: 10.1016/0092-8674(87)90248-0. [DOI] [PubMed] [Google Scholar]
- 61.Readhead C, Schneider A, Griffiths I, Nave K-A. Premature arrest of myelin formation in transgenic mice with increased proteolipid protein gene dosage. Neuron. 1994;12:583–595. doi: 10.1016/0896-6273(94)90214-3. [DOI] [PubMed] [Google Scholar]
- 62.Roach A, Boylan K, Horvath S, Prusiner SB, Hood LE. Characterization of cloned cDNA representing rat myelin basic protein: absence of expression in brain of shiverer mutant mice. Cell. 1983;34:799–806. doi: 10.1016/0092-8674(83)90536-6. [DOI] [PubMed] [Google Scholar]
- 63.Roberts CW, Shutter JR, Korsmyer ST. HoxII controls the genesis of the spleen. Nature. 1994;368:747–749. doi: 10.1038/368747a0. [DOI] [PubMed] [Google Scholar]
- 64.Rudnick A, Ling TY, Odagiri H, Rutter WJ, German MS. Pancreatic beta cell express a diverse set of homeobox genes. Proc Natl Acad Sci USA. 1994;91:12203–12207. doi: 10.1073/pnas.91.25.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 66.Scherer SS, Braun PE, Grinspan J, Collarini E, Wang D-y, Kamholz J. Differential regulation of the 2′, 3′-cyclic nucleotide 3′-phosphodiesterase gene during oligodendrocyte development. Neuron. 1994;12:1363–1375. doi: 10.1016/0896-6273(94)90451-0. [DOI] [PubMed] [Google Scholar]
- 67.Simmons DM, Voss JW, Ingraham HA, Holloway JM, Broide RS, Rosenfeld MG, Swanson LW. Pituitary cell phenotypes involve cell-specific Pit-1 mRNA translation and synergistic interactions with other classes of transcription factors. Genes Dev. 1990;4:695–711. doi: 10.1101/gad.4.5.695. [DOI] [PubMed] [Google Scholar]
- 68.Slack JM. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- 69.Sock E, Leger H, Kuhlbrodt K, Schrieber J, Enderich J, Richter-Landsberg C, Wegner M. Expression of Krox proteins during differentiation of the O-2A progenitor cell line CG-4. J Neurochem. 1997;68:1911–1919. doi: 10.1046/j.1471-4159.1997.68051911.x. [DOI] [PubMed] [Google Scholar]
- 70.Staudt LM, Clerc RG, Singh H, LeBowitz JH, Sharp PA, Baltimore D. Cloning of a lymphoid-specific cDNA encoding a protein binding the regulatory octamer DNA motif. Science. 1988;241:577–580. doi: 10.1126/science.3399892. [DOI] [PubMed] [Google Scholar]
- 71.Suh E, Chen L, Taylor J, Traber PG. A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol Cell Biol. 1994;14:7340–7351. doi: 10.1128/mcb.14.11.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Topilko P, Schneider-Maunoury S, Levi G, Baron-Van Evercooren A, Ben Younes Chennoufi A, Seitanidou T, Baninet C, Charnay P. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- 73.Wood RP. In: Oligodendroglia: advances in neurochemistry (Norton, WT ed), pp 1–46. Plenum; New York: 1984. [Google Scholar]
- 74.Wrabetz L, Shumas S, Grinspan J, Feltri ML, Bozyczko D, McMorris FA, Pleasure D, Kamholz J. Analysis of the human MBP promoter in primary cultures of oligodendrocytes: positive and negative cis-acting elements in the proximal MBP promoter mediate oligodendrocyte-specific expression of MBP. J Neurosci Res. 1993;36:455–471. doi: 10.1002/jnr.490360412. [DOI] [PubMed] [Google Scholar]
- 75.Wrabetz L, Tabeggia C, Ferraresi S, Quattrini A, Feltri M, Messing A, Kamholz J. Expression of a 750 nucleotide MBP promoter-LacZ transgene is reduced in adult shiverer mice. Soc Neurosci Abstr. 1995;21:321. [Google Scholar]
- 76.Wright PA, Duchala CS, Readhead C, Macklin WB. A myelin proteolipid protein-LacZ fusion protein is developmentally regulated and targeted to the myelin membrane in transgenic mice. J Cell Biol. 1993;123:443–454. doi: 10.1083/jcb.123.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu WP, Collarini EJ, Pringle NP, Richardson WD. Embryonic expression of myelin genes: evidence for a focal source of oligodendrocyte precursors in the ventricular zone of the neural tube. Neuron. 1994;12:1353–1362. doi: 10.1016/0896-6273(94)90450-2. [DOI] [PubMed] [Google Scholar]
- 78.Zeller NK, Hunkeler MJ, Campagnoni AT, Sprague J, Lazzarini RA. Characterization of mouse myelin basic protein messenger RNAs with a myelin basic protein cDNA clone. Proc Natl Acad Sci USA. 1984;81:18–21. doi: 10.1073/pnas.81.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zeller NK, Behar TN, Dubois-Dalcg ME, Lazzarini RA. The timely expression of myelin basic protein gene in cultured rat brain oligodendrocytes is independent of continuous neuronal influences. J Neurosci. 1985;5:2955–2962. doi: 10.1523/JNEUROSCI.05-11-02955.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]