Abstract
The eag family of K+ channels contains three known subtypes: eag, elk, anderg. Genes representing the first two subtypes have been identified in flies and mammals, whereas the third subtype has been defined only by the human HERG gene, which encodes an inwardly rectifying channel that is mutated in some cardiac arrhythmias. To establish the predicted existence of a Drosophila gene in the erg subfamily and to learn more about the structure and biological function of channels within this subfamily, we undertook a search for the Drosophila counterpart of HERG. Here we report the isolation and characterization of theDrosophila erg gene. We show that it corresponds with the previously identified seizure (sei) locus, mutations of which cause a temperature-sensitive paralytic phenotype associated with hyperactivity in the flight motor pathway. These results yield new insights into the structure and evolution of theeag family of channels, provide a molecular explanation for the sei mutant phenotype, and demonstrate the important physiological roles of erg-type channels from invertebrates to mammals.
Keywords: K+ channels, eag family, seizure mutation, neurogenetics, LQT syndrome, HERGchannels, hyperexcitability
Analysis of Drosophila mutants enabled the initial molecular isolation and characterization of several distinct types of K+ channels. Shaker (Sh) encodes a voltage-activated K+ channel (Kamb et al., 1987;Temple et al., 1987; Pongs et al., 1988). Subsequently, a family of at least four Sh-related K+ channel genes (Sh, Shab, Shal, and Shaw) was identified in Drosophila and mammals (Salkoff et al., 1992). The slowpoke (slo) gene encodes a Ca2+-activated K+ channel. Mouse and humanslo homologs were isolated (Atkinson et al., 1991; Butler et al., 1993; Pallanck and Ganetzky, 1994), but no additionalslo subtypes have been reported. A third type of K+ channel is encoded by ether a go-go (eag) (Drysdale et al., 1991; Warmke et al., 1991).
Mutations of eag cause spontaneous repetitive firing in motor axons and enhanced transmitter release at neuromuscular junctions (Ganetzky and Wu, 1983, 1985). The eag polypeptide is distantly related to the Sh family of voltage-activated K+ channels, but it also contains novel features, including a segment in the C-terminal cytoplasmic region that is homologous to cyclic nucleotide-binding domains (cNBD) (Guy et al., 1991; Warmke et al., 1991). In Xenopus oocytes, eag channels conduct a voltage-activated, K+-selective outward current (Bruggeman et al., 1993; Robertson et al., 1993). These results indicate that eag encodes a new type of voltage-activated K+ channel.
Moreover, eag is the progenitor of a conserved family of K+ channel genes that parallels the Sh family (Warmke and Ganetzky, 1994). Three subfamilies were identified:eag, elk (eag-like K+ channel), anderg (eag-related gene). Although eagand elk genes were identified in both Drosophilaand mammals, only a single representative of the third subfamily, humanerg (HERG), was found. Recently, HERG mutations were shown to cause long-QT (LQT) syndrome, a type of cardiac arrhythmia (Curran et al., 1995). In Xenopus oocytes, HERG forms voltage-sensitive K+ channels with a distinctive inactivation mechanism that attenuates efflux during depolarization (Sanguinetti et al., 1995; Trudeau et al., 1995; Smith et al., 1996). These channels most closely resemble those of native cardiac IKr channels, although their exact identity remains to be established.
To confirm the existence of a Drosophila erg gene and to learn more about the structure and biological function oferg channels in an organism more amenable to experimental manipulation than humans, we undertook the isolation and characterization of Drosophila erg. We show thaterg is encoded by the seizure (sei) locus, originally defined by temperature-sensitive paralytic mutations. The underlying defect in sei has been uncertain because behavioral and electrophysiological phenotypes suggested a hyperexcitability defect, whereas other results indicated thatsei causes a reduction in Na+ channels (Jackson et al., 1984, 1985; Kasbekar et al., 1987; O’Dowd and Aldrich, 1988;Elkins and Ganetzky, 1990). Our results demonstrate that the primary defect in sei is in a K+ channel polypeptide. The phenotypes of erg mutations in flies and humans emphasize the important physiological roles of erg channels from invertebrates to mammals.
MATERIALS AND METHODS
Mutants.seits1 andseits2 were isolated in a screen performed >15 years ago for ethylmethane sulfonate (EMS)-induced temperature-sensitive paralytic mutations on the second chromosome (B. Ganetzky and C.-F. Wu, unpublished observations). Flies homozygous for mutagenized second chromosomes were placed in glass vials preheated to 37.5°C in a water bath, and those that were paralyzed, unable to right themselves, or unable to climb the sides of the vial within 5 min were saved for further analysis. Wild-type flies remain mobile for at least 30 min under the same conditions. A characteristic feature of both seits1and seits2 is the bout of uncontrolled flight motor activity exhibited by the mutants 10–15 sec after initial exposure to the elevated temperature, causing the flies to bounce erratically around the vial. After longer exposure the mutants fall to the bottom of the vial but continue to show uncoordinated motor activity. More detailed descriptions of the behavioral and electrophysiological phenotypes of seimutants have been reported elsewhere (Jackson et al., 1984, 1985;Kasbekar et al., 1987; O’Dowd and Aldrich, 1988; Elkins and Ganetzky, 1990).
Two additional sei alleles,seiRK3 andseiRK4, were newly generated as part of the studies reported here. Wild-type (Canton-S) males were irradiated with γ-rays (4000 R) and mated toseits2bw females.bw is an eye color marker closely linked to sei. Progeny from the mating were placed in preheated glass vials and exposed to 37.5°C in a water bath. Under these conditionsseits2/sei+flies behave normally. Among ∼10,000 offspring scored, two flies were recovered that displayed the usual sei paralytic phenotype. The mutagenized second chromosomes from these two flies were recovered, and each was made homozygous in subsequent matings. To recover the newly mutagenized chromosomes instead of theseits2-bearing chromosome, we identified the chromosomes carrying bw+ in these matings. Retests confirmed that the newly isolated mutations failed to complement both seits1 andseits2 and therefore represent bona fide new sei alleles, which we namedseiRK3 andseiRK4.
Isolation of Drosophila erg genomic and cDNA clones.A partial cDNA for the rat erg gene containing the segment from the pore region to the 3′ end (S. Titus and B. Ganetzky, unpublished observations) was used to screen a wild-type (Canton-S) genomic library (Maniatis et al., 1978) at medium stringency [5× SSCP (1× SSCP = 120 mm sodium chloride/15 mm sodium citrate/20 mm sodium phosphate), 0.1% SDS, 10× Denhardt’s solution, salmon sperm DNA (250 μg/ml), and 30% formamide at 42°C; wash, 0.2× SSC and 0.1% SDS at 42°C]. To distinguish erg clones from other known family members, we screened a second lift from the library at high stringency [2× SSCP, 0.1% SDS, 10× Denhardt’s solution, and salmon sperm DNA (250 μg/ml) at 65°C; wash, 0.2× SSC and 0.1% SDS at 65°C] withDrosophila eag and elk probes. Positive clones from both screens were subjected to PCR using a set of degenerate primers (Ransom Hill Biosciences) corresponding to segments of the S5 (5′ TGGHTNGCNTGYATHTGGTA 3′) and pore (5′ AAGCTTNCCRAANCCCAC 3′) regions of known members of the eag family. The predicted size of the PCR fragment was ∼200 bp of translated sequence plus the size of any introns present in the segment spanned by the primers. The various genomic clones tested fell into three separate classes on the basis of the size of the PCR product they produced: 310 bp, 450 bp, or 1.0 kb. The PCR products were examined by Southern blot hybridization at high stringency [2× SSCP, 0.1% SDS, 10× Denhardt’s solution, and salmon sperm DNA (250 μg/ml) at 65°C; wash, 0.2× SSC and 0.1% SDS at 65°C] with mixed eag andelk probes. The 450 bp and 1.0 kb bands hybridized with the probe, but the 310 bp fragment did not, indicating that it probably represented a new family member. Accordingly, the 310 bp fragment was subcloned into pBluescript II (KS+) (Stratagene, La Jolla, CA) and sequenced. Two small introns of 65 and 61 bp were embedded in an open reading frame (ORF) encoding an amino acid sequence distinct from, but closely related to, the eag and elkpolypeptides in the S5–pore region.
The 310 bp PCR product was used to screen an adult head cDNA library (provided by T. Schwarz, Stanford University, Stanford, CA), and a 1.0 kb cDNA (SFW16) encoding a segment extending from the pore region to the cNBD was recovered. An additional 14 cDNAs falling into six different classes were isolated by rescreening the same cDNA library with SFW16. Two of these classes were fusions of ergsequences with unrelated cDNAs, and all of them contained some unprocessed introns. The cDNA that extended farthest in the 5′ direction included 1 kb of sequence beyond the S1 segment. Because the deduced amino acid sequence of this cDNA upstream of the S1 segment had essentially no similarity with other members of the eagfamily, it was important to prove that this sequence did not represent another spurious cDNA fusion (see below). The cDNA that extended farthest in the 3′ direction contained 200 bp beyond the cNBD, but it was incomplete because it ended in frame before reaching a termination codon. This cDNA was used to rescreen a different head cDNA library (provided by P. Salvaterra, City of Hope, Duarte, CA) that was primed with oligo dT. A single positive clone was recovered that contained 310 bp of translated sequence distal to the cNBD before reaching a termination codon. A poly(A+)-addition signal and string of 13 A residues beyond the termination codon indicated that this cDNA was complete at the 3′ end. However, the encoded amino acid segment distal to the cNBD was considerably shorter than that in all other knowneag family members. Consequently, it was also necessary to confirm the sequence at the 3′ end (see below).
Verification of 5′ and 3′ ends of erg sequence.The sequence at the 5′ end of erg was confirmed by two independent methods. First, we determined an EcoRI map of the erg genomic clones by Southern blot analysis and subcloned the 5′-most EcoRI fragment (6 kb) for sequence analysis. Second, adult poly(A+)-selected RNA was used for Marathon 5′ RACE. After synthesis of double-stranded cDNA, this cDNA was used as a PCR template for 5′ RACE according to the manufacturer’s instructions (Clontech, Cambridge, UK), using an oligonucleotide (Ransom Hill Biosciences) corresponding to a sequence in the S1-encoding segment of erg as primer (5′ GCGGATCCTTAAAGGGCGAGTAGTGC 3′). From two independent reactions two 5′ RACE clones, both ∼1.3 kb, were recovered and subcloned into pBlueScript II (KS+). Sequence analysis indicated that both RACE products contained a contiguous ORF of ∼1 kb, including an initiating Met. The two RACE products shared identical sequence with each other, with the corresponding segment from the genomic clone, and with the 5′-most cDNA isolated from library screens.
To confirm the sequence at the 3′ end of erg, we identified and subcloned the 3′-most genomic EcoRI fragment (4.3 kb). A segment that extended from the cNBD to the termination codon was sequenced and found to be identical to the sequence of the 3′-most cDNA obtained from library screens.
In situ hybridization. A digoxygenin-labeled DNA probe from erg was synthesized by random priming according to the manufacturer’s instructions (Genius kit, Boehringer Mannheim, Indianapolis, IN). The template was a 1.25 kb BamHI fragment, isolated from a partially processed cDNA, that encoded a segment from S1 through the cNBD. Before this fragment was used as template, it was digested with EcoRV to yield pieces ≤300 bp. The labeled probe was used for both chromosomal and tissue in situ hybridization according to previously published protocols (Drysdale et al., 1991; Hong and Ganetzky, 1994).
Sequence analysis of sei mutations. Genomic DNA isolated from seits1,seits2,seiRK3, andseiRK4 was used as template for DNA sequencing by the dye termination method (Lee et al., 1992;Rosenthal and Charnock-Jones, 1992) using ABI (Foster City, CA) Prism models 373 and 377 automated sequencers according to the manufacturer’s instructions. A series of 12 oligonucleotides spanning the entire 3.8 kb of genomic DNA that includes the complete ORF was used to prime the DNA sequencing reactions. The entire ORF ofseiRK3 andseiRK4 and all but 40 amino acids of the ORF of seits1 andseits2 were determined by automated sequencing. The remaining sequence ofseits1 andseits2 was obtained manually by the dideoxy method (Sambrook et al., 1989) using a Sequenase version 2 kit (United States Biochemicals, Cleveland, OH).
DNA sequence analysis. A sequence for a Caenorhabditis elegans erg gene was found by using the GCG (version 8.0, Devereux et al., 1984) Blast program to search the GenBank database with theDrosophila erg sequence. This search uncovered two 10 kb pieces of genomic DNA from the C. elegans genome project (Accession numbers U02425 and U02453) encoding predicted amino acid sequences that aligned respectively to the N- and C-terminal halves of the Drosophila erg polypeptide. The c-erggene is 5800 bp in length with a 2200 bp ORF interrupted by 14 introns.
GenBank accession number. The accession number for the seizure sequence data reported in this paper is U42204.
RESULTS
Isolation and sequence analysis of erg cDNA
In previous attempts to identify additional members of theeag family in Drosophila by low stringency screens of head cDNA libraries, only one additional gene,elk, was found (Warmke and Ganetzky, 1994). However, because both eag and elk are highly conserved betweenDrosophila and mammals (Warmke and Ganetzky, 1994; Ludwig et al., 1994; S. A. Titus, J. W. Warmke, B. Ganetzky, unpublished observations), it seemed probable that a Drosophilacounterpart of HERG remained to be found. The discovery of an association between one form of LQT syndrome with mutations inHERG (Curran et al., 1995) provided additional impetus to identify and characterize the counterpart gene in an organism more amenable to experimental manipulation than humans.
In subsequent efforts to identify a member of the ergsubfamily in Drosophila, we first screened a genomic library with a rat erg probe to avoid potential problems associated with low representation of desired target sequences in cDNA libraries (see Materials and Methods). Degenerate primers corresponding to segments of the S5 and pore regions of known members of theeag family were used to generate PCR amplification products from the positive clones. The positive clones fell into three separate classes on the basis of the size of the PCR products they generated and the hybridization of these products to eag andelk probes at high stringency (see Materials and Methods). A 310 bp amplification product from one of the positives seemed to represent a new family member. Sequence analysis of this fragment confirmed that it was a member of the eag family distinct from eag and elk, but closely related toHERG. Then this PCR fragment was used to screen aDrosophila head cDNA library at high stringency. Sequence analysis of cDNAs from this screen confirmed the identity of this gene as Drosophila erg. Because the ORF was incomplete at both the 5′ and 3′ ends, it was necessary to obtain several overlapping cDNAs by rescreening cDNA libraries to obtain the complete coding sequence. Sequence at the 5′ and 3′ ends of the ORF was confirmed by analysis of RACE and genomic clones (see Materials and Methods).
The composite erg cDNA encodes a deduced amino acid sequence of 855 amino acids (Fig. 1). The presumptive initiating Met is preceded by an almost perfect match to the Cavener consensus sequence (Cavener, 1991) for translation initiation inDrosophila (AAAA ATG vs CAAA/C ATG). As shown in the sequence alignment in Figure 1, there is a strikingly high degree of amino acid identity among nematode,Drosophila, and human erg polypeptides extending from the region that just precedes the first presumptive membrane-spanning segment (S1) until just after the cNBD-like segment in the C terminus. Amino acid similarity drops off sharply at the N and C termini. Surprisingly, across the first 100 amino acids of their N termini, there is much more similarity between eag andHERG than between erg and eag or between erg and HERG. Another distinctive feature of Drosophila erg, as compared with eag or with other members of the erg subfamily, is the very short sequence at the C terminus.
Fig. 1.

Amino acid sequence of the Drosophila erg polypeptide and its alignment with other members of theeag family of K+ channel polypeptides. Identical residues are shaded in black. The approximate locations of the presumptive membrane-spanning regions (S1–S6), the pore region (P), and the region of homology to a cyclic nucleotide-binding domain (cNBD) are overlined. Gaps in the alignment are indicated by dashes. Vertical bars beneath the alignment mark the positions of introns in theDrosophila erg genomic sequence. The Drosophila eag sequence and the human HERG sequence have been published previously (Warmke et al., 1991; Warmke and Ganetzky, 1994). The nematode erg sequence (C-erg) was obtained by using the GCG Blast program to search the GenBank database (see Materials and Methods).
From sequence analysis of genomic clones, as compared with the cDNA sequence, the genomic organization of the erg transcription unit also was determined. The ORF of erg is interrupted by a total of 13 introns ranging in size from 50 to 220 bp, and another intron is located immediately upstream of the translational start site. Additional introns that we did not detect could interrupt the 5′ or 3′ untranslated sequences. The location of many of these introns has been highly conserved. For example, introns 2, 4, 5, 6, 9, and 12 in theDrosophila erg gene occupy the identical locations as introns 1, 3, 4, 5, 7, and 9, respectively, of the nematodeerg gene. In addition, three introns in the HERGgene that have been identified occupy exactly the same locations as introns 5, 8, and 12 of Drosophila erg. The extent of genomic DNA spanned by the erg transcription unit from 50 bp upstream of the translational start site to the termination codon is only ∼3.8 kb, which is considerably less than that for other known ion channel structural genes in Drosophila, such aspara and eag (Loughney et al., 1989; Drysdale et al., 1991).
The sequence presented in Figure 1 is derived from cDNAs obtained from a library constructed from the Oregon-R wild-type strain. In the course of our sequence analysis of mutations generated on a Canton-S wild-type background (see below), we also obtained the corresponding sequence from Canton-S genomic DNA. Comparison of the ORF from Oregon-R and Canton-S revealed the existence of three polymorphisms, all of which were located in the nonconserved N-terminal domain: amino acids 136–138 of the Oregon-R sequence missing in the Canton-S sequence; a single base change from T to C in the second position of codon 140 of Oregon-R, causing an Ile to Thr replacement; and a single base change from G to A in the first position of codon 166 of Oregon-R, causing a Val to Met replacement. Except for these differences, the rest of the ORF is identical between Oregon-R and Canton-S.
Expression of the erg transcript
On Northern blots of poly(A+)-selected RNA isolated from whole adults, erg cDNA probes detect a single transcript between 2.8 and 3.0 kb in size (Fig. 2). In contrast with other ion channel genes that have been identified inDrosophila, the mRNAs of which are generally 2–3 times larger than the ORF, the erg mRNA is approximately the same size as its ORF and therefore lacks long untranslated sequences at the 5′ and 3′ ends.
Fig. 2.
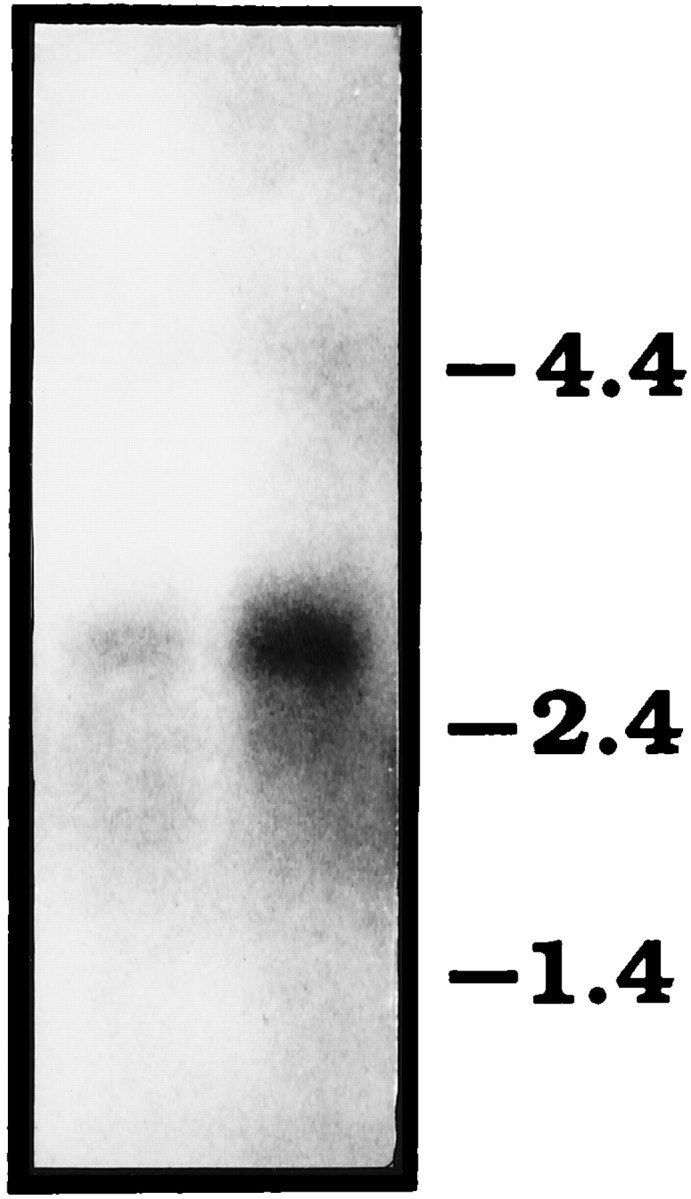
Northern blot analysis of poly(A+)-selected RNA isolated from wild-type (Canton-S) adults and hybridized with an erg cDNA probe.Left lane contains 4 μg of RNA; right lane, 6 μg. The position of size markers is indicated on theright.
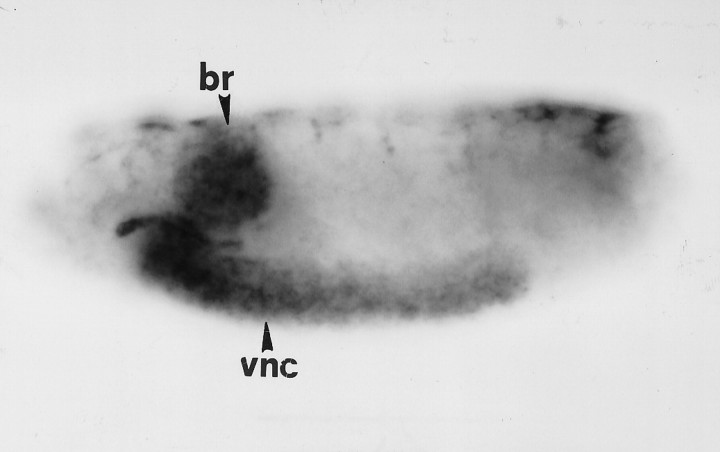
As shown by in situ hybridization, the ergtranscript is expressed throughout the CNS in embryos (Fig.3). No reproducible expression was found elsewhere in the embryo. Compared with para, a Na+ channel gene expressed in most or all CNS neurons (Hong and Ganetzky, 1994), the staining pattern for the erg transcript is fainter and more diffuse, and it seems that erg is expressed only in a subset of neurons.
Fig. 3.
Embryonic expression pattern of ergdetermined by tissue whole-mount in situ hybridization. Lateral view of a stage 16 embryo oriented with anterior to theleft and dorsal up. Expression of theerg transcript can be detected throughout the CNS, including the ventral nerve cord (vnc) and the brain hemispheres (br). Staining in salivary glands, as seen in this embryo, occurred only sporadically.
The erg gene corresponds to thesei locus
To initiate genetic analysis of erg function inDrosophila, we mapped the gene by chromosomal in situ hybridization to position 60B1–2 on the polytene map (Fig.4). This cytological location corresponds closely with that reported for sei (Jackson et al., 1985). This gene was defined by the isolation of two EMS-induced temperature-sensitive paralytic mutations (seits1 andseits2) >15 years ago (Ganetzky and Wu, unpublished observations). Several different studies have suggested that sei mutations reduce Na+ channel expression or activity (Jackson et al., 1984, 1985; O’Dowd and Aldrich, 1988). However, electrophysiological studies in the adult flight motor pathway showed that the most pronounced phenotype associated withseits1 andseits2 is a substantial enhancement of spontaneous neural activity (Kasbekar et al., 1987; Elkins and Ganetzky, 1990). The bursts of spontaneous firing in the motor pathway parallel the behavioral phenotype of the mutants, which includes a distinctive bout of uncontrolled flight activity on exposure to the restrictive temperature. These hyperexcitable phenotypes are consistent with a possible defect in K+ channels.
Fig. 4.
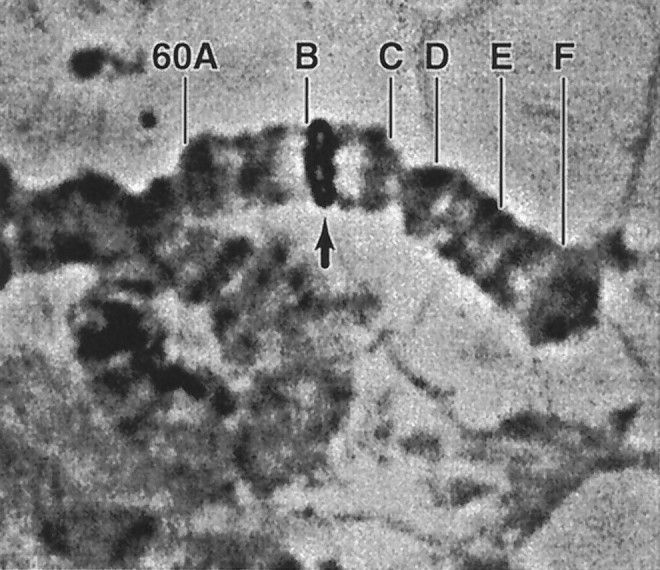
Cytological mapping of the erglocus by chromosomal in situ hybridization. The site of hybridization (arrow) relative to the cytological landmarks in numbered region 60 is shown. The hybridization signal lies directly on top of salivary bands 60B1–2.
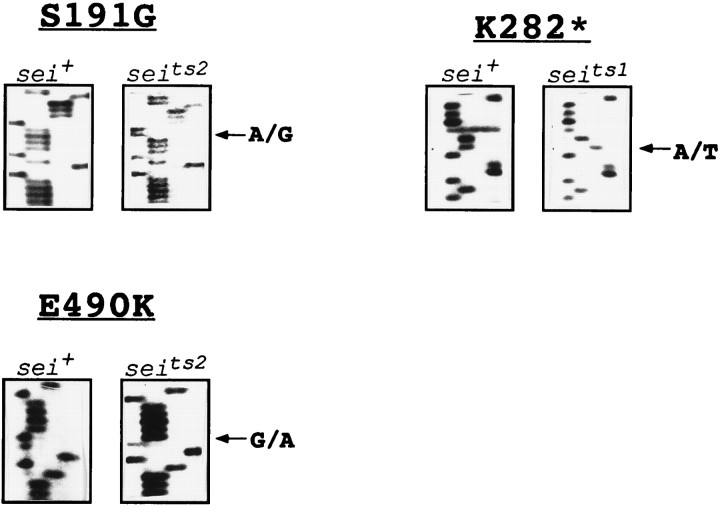
Consequently, we sequenced bothseits1 andseits2 alleles to determine whether they contained lesions in the erg polypeptide (Fig.5). Two amino acid differences from wild-type are present in the seits2 sequence. The first results from a substitution of Gly for Ser at position 191 in the cytoplasmic segment of the N terminus. The second results from a substitution of Lys for Glu at position 490 in the extracellular loop that just precedes the pore domain. A mutation in the ergsequence also was found in seits1. An A to T transversion at the first position of codon 282 results in a change from Lys (AAG) to a stop (TAG) codon. This premature stop codon before the first membrane-spanning segment of the ergpolypeptide must certainly result in complete loss of ergchannel activity. The presence of different mutational lesions in theerg polypeptide in two independently isolated alleles ofsei strongly suggested that the erg polypeptide is the sei gene product.
Fig. 5.
Sequence analysis of mutational changes caused byseits1 andseits2 in theerg polypeptide. In all gels, the sequencing reactions are loaded (from the left) in the order G, A, T, C. Segments of sequence from the sense strand are shown. Two lesions are present in seits2, an A to G substitution at nucleotide position 571 of the ORF, resulting in a replacement of Ser by Gly at amino acid position 191, and a G to A substitution at nucleotide position 1468, resulting in a replacement of Glu by Lys at amino acid position 490. Inseits1, there is an A to T substitution at nucleotide position 844, resulting in the change of a Lys codon to a stop codon at amino acid position 282.
To confirm the relationship between sei and erg, we generated two new sei mutations,seiRK3 andseiRK4, after γ-ray mutagenesis on a defined wild-type (Canton-S) background (see Materials and Methods). Genomic DNA from these two alleles was sequenced and compared with the corresponding sequence from Canton-S. Both of these alleles were found to contain newly induced lesions, resulting in coding changes in theerg polypeptide. InseiRK3, a 6 bp insertion occurred after the second base of codon 669, resulting in the insertion of two amino acids (Gln and Ala) between amino acids 669 (Ala) and 670 (Phe) of the wild-type sequence (data not shown). InseiRK4, a 3 bp deletion was found that removed codon 417 (data not shown). This lesion deletes a conserved Ala residue from the S3 domain. The occurrence of identifiable lesions in the erg sequence in two newly generated mutations isolated on the basis of their failure to complement known sei alleles provides conclusive evidence that the erg polypeptide is the sei gene product.
DISCUSSION
We have identified the Drosophila erg gene and demonstrated that it corresponds to the sei locus. These results offer important new insights about the eag family of K+ channels and provide a molecular explanation of thesei mutant phenotype.
Beginning with eag, which has been shown to encode a distinct type of voltage-activated K+ channel (Bruggeman et al., 1993; Robertson et al., 1993), we previously recovered cDNAs from three additional genes, elk, m-eag, andHerg, in screens of Drosophila, mouse, and human libraries. Sequence comparisons indicated that these genes defined three subfamilies (Warmke and Ganetzky, 1994). Extrapolating from these genes, we predicted that members of these subfamilies would be present from Drosophila to mammals. Identification of additional related genes in library screens and database searches supports this prediction. Two different members of the eag subfamily have been identified in rat and human libraries (Ludwig et al., 1994; Titus and Ganetzky, unpublished observations). Rat and human counterparts ofelk also have been found (Titus and Ganetzky, unpublished observations). The isolation of Drosophila erg now completes the prediction. In general, two members of the same eagsubfamily from different species share ∼60-70% amino acid identities in the region spanning S1 through the cNBD segment. In contrast, two different subfamily members within the same species share only ∼40-50% amino acid identities across the same region. This situation closely parallels the relationships among the four subtypes of K+ channel polypeptides in the Sh family (Salkoff et al., 1992).
There has been considerable interest in HERG since the discovery that it is mutated in the chromosome 7 form of LQT syndrome (Curran et al., 1995). In addition, HERG channels expressed in Xenopus oocytes have properties that distinguish them from other eag family members (Sanguinetti et al., 1995;Trudeau et al., 1995; Smith et al., 1996). Most notably,HERG channels display inward rectification resulting from a rapid inactivation mechanism that attenuates K+ efflux during depolarization but is relieved on hyperpolarization. It will be of interest to determine whether erg shares these properties.
The distinctive sequence of the erg N terminus is surprising, because all other members of the eag family share characteristic sequences within this region. The fact thateag, elk, and HERG share many amino acid identities in their N termini suggests that this sequence predates the evolutionary expansion of the eag family into distinct subfamilies and that this sequence has been preserved inHERG, but not in erg. The rapid evolution of this region also is indicated by the divergence of c-erg from all of its counterparts in this segment.
Identification of lesions in the erg sequence in four independently isolated sei mutations demonstrates that thesei locus encodes the erg polypeptide. Because toxin-binding assays (Jackson et al., 1984, 1985) and whole-cell recordings of Na+ currents in embryonic neurons (O’Dowd and Aldrich, 1988) indicated that sei mutations reduce Na+ channel expression, the finding that seiencodes a K+ channel polypeptide was unexpected. Nonetheless, a K+ channel defect is consistent with and accounts for other characteristic phenotypes of sei mutants. In particular, sei mutants display greatly elevated spontaneous neural activity in the flight motor pathway that apparently underlies the convulsive seizures and paralysis ofseits1 andseits2 at elevated temperatures (Kasbekar et al., 1987; Elkins et al., 1990). These hyperexcitable phenotypes are consistent with a defect in K+ channels. One possible explanation for the apparent pleiotropic effect ofsei mutations is that Na+ channel expression is altered via some regulatory mechanism as a secondary consequence of a perturbation in erg K+ channels.
Two amino acid substitutions were found inseits2, a Ser-to-Gly substitution at position 191 and a Glu-to-Lys substitution at position 490. However, we believe that the Glu-to-Lys change is primarily responsible for the mutant phenotype. The presence of an acidic residue at the position corresponding to 490 in the erg polypeptide is conserved completely in all known members of the eagfamily and is likely to be a functionally important site. The Ser-to-Gly substitution is a more conservative change occurring in a region that may be less critical for channel function. Furthermore, this substitution exists as a normal polymorphism in varioussei+ strains that we have examined (E. Massa and B. Ganetzky, unpublished observations).
In seits1, a severely truncated polypeptide lacking all membrane-spanning segments results from a mutation to a premature stop codon. In contrast withseits2,seits1 is fully recessive tosei+ and acts phenotypically like a complete loss-of-function mutation. This suggests that, even if a stable truncated N-terminal segment is produced byseits1, it does not interfere with the assembly or function of wild-type subunits. These results imply that it is the loss of erg activity, rather than the production of a temperature-sensitive polypeptide per se, that renders the seits1 flies more sensitive than normal to elevated temperatures. Because the temperature-sensitive behavioral and electrophysiological phenotypes ofseits2 andseits1 are very similar, it is likely that the temperature sensitivity inseits2 also involves a severe decrement in erg function, the physiological consequences of which become exacerbated at elevated temperatures. Several other examples are known in Drosophila, in which it is the complete or nearly complete loss of function of a protein involved in neural signaling that results in a temperature-sensitive paralytic phenotype (Atkinson et al., 1991; Zinsmaier et al., 1994; Feng et al., 1995).
The γ-ray induced alleles, seiRK3and seiRK4, are associated with the insertion of two amino acids and the deletion of one amino acid, respectively. The two amino acid insertion inseiRK3 falls between the S6 segment and the cNBD-like region. The contribution of this region to the functional properties of channels in the eag family is still unknown, but the isolation of a mutation in this region on the basis of a behavioral phenotype suggests that it has an important role. This conclusion is consistent with the high degree of evolutionary conservation of this region, particularly among members of theerg subfamily, from nematodes to humans. The deletion of a highly conserved Ala residue from the S3 membrane-spanning segment inseiRK4 also would be expected to cause significant perturbations of erg channel functionin vivo. Interestingly, the deleted Ala residue falls within the nine amino acids deleted in one of the HERG mutations (Δ1500–F508) associated with LQT syndrome in humans (Curran et al., 1995).
From studies of HERG channels expressed inXenopus oocytes, it has been proposed that theHERG polypeptide represents a subunit of IKrchannels in cardiac myocytes. A K+ current with properties similar to cardiac IKr has not yet been identified inDrosophila neurons or muscle fibers, so the particular current affected by sei mutants in vivo is not clear. In situ hybridization in Drosophilaembryos shows that erg is expressed primarily throughout the CNS. The increased spontaneous activity in flight motor neurons insei mutants at elevated temperatures indicates thaterg channels are expressed in these neurons and play some role in repolarization of action potentials or in maintaining the resting potential. Thus, there is at least a rough correspondence between the functions of erg in Drosophilaneurons and HERG in human heart. Although HERG is expressed predominantly in the heart, HERG transcripts also are detected in brain, and HERG cDNAS originally were isolated from a human hippocampus library. However, the in vivo function of HERG in human neurons is still unknown. The identified mutations of HERG causing LQT syndrome are all dominant, and none is reported to have associated neurological impairments (Curran et al., 1995). Possibly, cardiac myocytes are more sensitive than neurons to partial loss ofHERG activity, and only individuals homozygous forHERG mutations will show neurological defects. More detailed molecular and electrophysiological studies of sei mutants inDrosophila should facilitate elucidation of the role oferg channels in neural functions.
The striking phenotypes associated with mutations of the ergchannel in both Drosophila and humans further highlight the physiological importance of the eag family of K+channels from insects to mammals. The distinct phenotypes associated with eag and erg mutations indicate that these channel subtypes subserve different and primarily nonoverlapping functions in vivo. Further analysis of the eagfamily should continue to increase our understanding of the expanding diversity of K+ channels and their physiological functions.
Footnotes
This work was supported by National Institutes of Health Grant NS15390. This is paper number 3451 from the Laboratory of Genetics, University of Wisconsin, Madison. We thank Robert Kreber for excellent technical help and for generating theseiRK3 andseiRK4 alleles. We also thank Rob Reenan, Bing Zhang, Gail Robertson, and Enrique Massa for helpful discussion and comments on this manuscript; Bill Feeny for help with figures; and Linda Hall and XinJing Wang for communicating their results before publication.
Correspondence should be addressed to Barry Ganetzky, Laboratory of Genetics, 445 Henry Mall, University of Wisconsin, Madison, WI 53706.
Dr. Warmke’s present address: Department of Genetics and Molecular Biology, Merck Research Laboratories, Rahway, NJ 07065.
REFERENCES
- 1.Atkinson N, Robertson G, Ganetzky B. A structural component of calcium-activated potassium channels encoded by the Drosophila slo locus. Science. 1991;253:551–555. doi: 10.1126/science.1857984. [DOI] [PubMed] [Google Scholar]
- 2.Bruggeman A, Pardo LA, Stuhmer W, Pongs O. Ether a-go-go encodes a voltage-gated channel permeable to K+ and Ca2+ and modulated by cAMP. Nature. 1993;365:445–448. doi: 10.1038/365445a0. [DOI] [PubMed] [Google Scholar]
- 3.Butler AS, Tsunoda S, McCobb DP, Wei A, Salkoff L. mSlo, a complex mouse gene encoding “maxi” calcium-activated potassium channels. Science. 1993;261:221–224. doi: 10.1126/science.7687074. [DOI] [PubMed] [Google Scholar]
- 4.Cavener DC, Ray SC. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curran ME, Splawski I, Timothy KW, Vincent M, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–804. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 6.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drysdale RA, Warmke JW, Kreber R, Ganetzky B. Molecular characterization of eag, a gene affecting potassium channels in Drosophila. Genetics. 1991;127:497–505. doi: 10.1093/genetics/127.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elkins T, Ganetzky B. Conduction in the giant nerve fiber pathway in temperature-sensitive paralytic mutants of Drosophila. J Neurogenet. 1990;6:207–219. doi: 10.3109/01677069009107111. [DOI] [PubMed] [Google Scholar]
- 9.Feng G, Deák P, Hall LM. Cloning and functional analysis of tipE: a novel membrane protein required for Drosophila para sodium channel function. Cell. 1995;82:1001–1011. doi: 10.1016/0092-8674(95)90279-1. [DOI] [PubMed] [Google Scholar]
- 10.Ganetzky B, Wu C-F. Neurogenetic analysis of potassium currents in Drosophila: synergistic effects on neuromuscular transmission in double mutants. J Neurogenet. 1983;1:17–28. doi: 10.3109/01677068309107069. [DOI] [PubMed] [Google Scholar]
- 11.Ganetzky B, Wu C-F. Genes and membrane excitability in Drosophila. Trends Neurosci. 1985;8:322–326. [Google Scholar]
- 12.Guy HR, Durell SR, Warmke JW, Drysdale R, Ganetzky B. Similarities in amino acid sequences of Drosophila eag and cyclic nucleotide-gated channels. Science. 1991;254:730. doi: 10.1126/science.1658932. [DOI] [PubMed] [Google Scholar]
- 13.Hong C-S, Ganetzky B. Spatial and temporal expression patterns of two sodium channel genes in Drosophila. J Neurosci. 1994;14:5160–5169. doi: 10.1523/JNEUROSCI.14-09-05160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson FR, Wilson SD, Strichartz GR, Hall LM. Two types of mutants affecting voltage-sensitive sodium channels in Drosophila melanogaster. Nature. 1984;308:189–191. doi: 10.1038/308189a0. [DOI] [PubMed] [Google Scholar]
- 15.Jackson FR, Gitschier J, Strichartz GR, Hall LM. Genetic modifications of voltage-sensitive sodium channels in Drosophila: gene dosage studies of the seizure locus. J Neurosci. 1985;5:1144–1151. doi: 10.1523/JNEUROSCI.05-05-01144.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamb A, Iverson LE, Tanouye MA. Molecular characterization of Shaker, a Drosophila gene that encodes a potassium channel. Cell. 1987;50:405–413. doi: 10.1016/0092-8674(87)90494-6. [DOI] [PubMed] [Google Scholar]
- 17.Kasbekar DP, Nelson JC, Hall LM. Enhancer of seizure: a new genetic locus in Drosophila melanogaster defined by interactions with temperature-sensitive paralytic mutations. Genetics. 1987;116:423–431. doi: 10.1093/genetics/116.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee LG, Connell CR, Woo SL, Cheng RD, McArdle BF, Fuller CW, Halloran ND, Wilson RK. DNA sequencing with dye-labeled terminators and T7 DNA polymerase: effect of dyes and dNTPs on incorporation of dye terminators and probability analysis of termination fragments. Nucleic Acids Res. 1992;20:2471–2483. doi: 10.1093/nar/20.10.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loughney K, Kreber R, Ganetzky B. Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell. 1989;58:1143–1154. doi: 10.1016/0092-8674(89)90512-6. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig J, Terlau H, Wunder F, Bruggeman A, Pardo LA, Marquardt A, Stuhmer W, Pongs O. Functional expression of a rat homologue of the voltage-gated ether a go-go potassium channel reveals differences in selectivity and activation kinetics between the Drosophila channel and its mammalian counterpart. EMBO J. 1994;13:4451–4458. doi: 10.1002/j.1460-2075.1994.tb06767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis T, Hardison R, Lacy E, Lauer J, O’Connell C, Quon D, Sim GK, Efstadiadis A. The isolation of structural genes from libraries of eucaryotic DNAs. Cell. 1978;15:687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- 22.O’Dowd DK, Aldrich RW. Voltage-clamp analysis of sodium channels in wild-type and mutant Drosophila neurons. J Neurosci. 1988;8:3633–3643. doi: 10.1523/JNEUROSCI.08-10-03633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pallanck L, Ganetzky B. Cloning and characterization of human and mouse homologs of the Drosophila calcium-activated potassium channel gene. Hum Mol Genet. 1994;3:1239–1243. doi: 10.1093/hmg/3.8.1239. [DOI] [PubMed] [Google Scholar]
- 24.Pongs O, Kecskemethy N, Muller R, Krah-Jentgens I, Baumann A, Kiltz HH, Canal I, Llamazares S, Ferrus A. Shaker encodes a family of putative potassium channel proteins in the nervous system of Drosophila. EMBO J. 1988;7:1087–1096. doi: 10.1002/j.1460-2075.1988.tb02917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robertson GA, Warmke JW, Ganetzky B. Functional expression of the Drosophila eag K+ channel gene. Biophys J. 1993;64:A340. [Google Scholar]
- 26.Rosenthal A, Charnock-Jones DS. New protocols for DNA sequencing with dye terminators. DNA Seq. 1992;3:61–64. doi: 10.3109/10425179209039697. [DOI] [PubMed] [Google Scholar]
- 27.Salkoff L, Baker K, Butler A, Coarrubias M, Pak MD, Wei A. An essential “set” of K+ channels conserved in flies, mice, and humans. Trends Neurosci. 1992;15:161–166. doi: 10.1016/0166-2236(92)90165-5. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, 2nd Ed. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1989. [Google Scholar]
- 29.Sanguinetti MG, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81:299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 30.Smith PL, Baukrowitz T, Yellen G. The inward rectification mechanism of the HERG cardiac potassium channel. Nature. 1996;379:833–836. doi: 10.1038/379833a0. [DOI] [PubMed] [Google Scholar]
- 31.Temple BL, Papazian DM, Schwarz TL, Jan YN, Jan LY. Sequence of a probable potassium channel component encoded at the Shaker locus of Drosophila. Science. 1987;237:770–775. doi: 10.1126/science.2441471. [DOI] [PubMed] [Google Scholar]
- 32.Trudeau MC, Warmke JW, Ganetzky B, Robertson GA. HERG, a human inward rectifier in the voltage-gated potassium channel family. Science. 1995;269:92–95. doi: 10.1126/science.7604285. [DOI] [PubMed] [Google Scholar]
- 33.Warmke JW, Ganetzky B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci USA. 1994;91:3438–3442. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warmke JW, Drysdale R, Ganetzky B. A distinct potassium channel polypeptide encoded by the Drosophila eag locus. Science. 1991;252:1560–1562. doi: 10.1126/science.1840699. [DOI] [PubMed] [Google Scholar]
- 35.Zinsmaier KE, Eberle KK, Buchner E, Walter N, Benzer S. Paralysis and early death in cysteine string protein mutants of Drosophila. Science. 1994;263:977–980. doi: 10.1126/science.8310297. [DOI] [PubMed] [Google Scholar]