Abstract
Mutations in the Drosophila rdgB gene, which encodes a transmembrane phosphatidylinositol transfer protein (PITP), cause a light-enhanced retinal degeneration. Cloning of mammalianrdgB orthologs (mrdgB) reveal predicted proteins that are 39% identical to rdgB, with highest homology in the N-terminal PITP domain (62%) and in a region near the C terminus (65%). The human mrdgB gene spans ∼12 kb and maps to 11q13.1, a locus where several retinal diseases have also been mapped. Murine mrdgB maps to a syntenic region on the proximal region of chromosome 19. MrdgB is specifically expressed in the retina and brain. In the retina, MrdgB protein is localized to photoreceptor inner segments and the outer and inner plexiform layers. Expression of murine mrdgB in mutant flies fully rescues both the rdgB-dependent retinal degeneration and abnormal electroretinogram. These results suggest the existence of similarities between the invertebrate and mammalian retina that were not previously appreciated and also identifymrdgB as a candidate gene for retinal diseases that map to 11q13.1.
Keywords: retina, retinal degeneration, phosphatidylinositol transfer protein, photoreceptors, phototransduction, retinal degeneration B
The basic mechanisms of vertebrate and invertebrate vision share certain similarities, but they also demonstrate dramatic differences. The differences are significant enough that it has been argued as to whether the vertebrate and invertebrate eye arose by convergent or divergent evolution (Nilson, 1966). Regardless of their origin, however, the complementary study of the vertebrate and invertebrate visual systems has provided important insights. In vertebrates, phototransduction is initiated by photon absorption by a visual pigment (rhodopsin or one of the cone opsins), which activates a heterotrimeric G-protein (transducin) (for review, see Koutalos and Yau, 1993). The resultant stimulation of a cGMP phosphodiesterase leads to closure of membrane cation channels and hyperpolarization of the cell surface. In Drosophila, perhaps the best studied invertebrate visual system, phototransduction also begins with photon capture by rhodopsin (NinaE) and subsequent activation of a G-protein, Dgqα (Lee et al., 1994; Scott et al., 1995; Zuker, 1996). However, the DrosophilaG-protein activates phospholipase C (NorpA), which hydrolyzes phosphatidylinositol 4,5-bisphosphate to generate the second messengers inositol 1,4,5-triphosphate (IP3) and diacyl glycerol (DAG). The IP3 is thought to release intracellular calcium, which in turn opens membrane channels and leads to membrane depolarization, whereas the DAG activates a protein kinase C (InaC), which is thought to be involved in light adaptation.
Although many of the important molecules in vertebrate phototransduction are well characterized, it is becoming increasingly clear that there are other less well understood molecules that play significant roles in the visual process. One powerful approach to identify potentially important molecules is to look for homologs of genes that have been identified in invertebrate genetic screens (Pak, 1995). Perhaps most interesting are homologs for which there are noa priori expectations that a functional vertebrate counterpart exists, such as those that act distally in invertebrate phototransduction. In this paper we describe the discovery of one such protein, the mammalian homolog of the Drosophila retinal degeneration B (rdgB) gene.
rdgB was one of the first Drosophila retinal degeneration mutants identified (Hotta and Benzer, 1969; Pak et al., 1970; Heisenberg, 1971). The mutation is characterized by multiple phenotypes. The phototransduction defects are evidenced by abnormal termination of the light response and profound loss of the electroretinogram (ERG) amplitude shortly after initial light exposure. The mutation also causes a light-enhanced retinal degeneration (Harris and Stark, 1977; Stark et al., 1983). In addition, some rdgBalleles demonstrate olfaction defects (Woodard et al., 1992). The ERG changes occur before any morphological evidence of retinal degeneration, which becomes evident several days after eclosion. Changes are first observed at the photoreceptor terminals (Carlson et al., 1985; Stark et al., 1989). When reared in the light, mutant flies demonstrate evidence of photoreceptor somal and axonal degeneration by 3 d after eclosion (Stark and Carlson, 1983). By 7 d, the synaptic terminals lack synaptic vesicles, and the cell bodies demonstrate unusual liposomes and lysosome-like bodies. Ultimately, rhabdomeres are lost, photoreceptors die, and holes appear in the retina.
Genetic, biochemical, and pharmacological studies all suggest that RdgB functions, at least partially, in the phototransduction pathway, probably subsequent to phospholipase C and protein kinase C. Application of the nonhydrolyzable GTP analog guanosine 5′-3-O-(thio)triphosphate or expression of a constitutively active Dgq mutation, both of which mimic light activation, cause rapid photoreceptor degeneration in dark-reared rdgB flies (Rubinstein et al., 1989; Lee et al., 1994). Similarly, phorbol ester, which activates protein kinase C, also causes enhanced photoreceptor degeneration (Minke et al., 1990). Conversely, mutations inninaE and norpA, which inhibit light activation, suppress the light-enhanced degeneration in rdgB mutant flies (Harris and Stark, 1977; Stark et al., 1983).
Cloning of the rdgB gene revealed that it codes for a 1054 amino acid residue polypeptide with six putative transmembrane domains (Vihtelic et al., 1991, 1993). It contains an N-terminal phosphatidylinositol transfer (PITP) domain and an adjacent calcium-binding region. The N-terminal PITP domain (residues 1–276), which was initially defined by homology to the rat brain-soluble PITPα protein, demonstrates PITP activity in vitro. Within the retina, the RdgB protein has been immunolocalized to both photoreceptor axons and subrhabdomeric cisternae (SRC) (Vihtelic et al., 1993; Suzuki and Hirosawa, 1994), an extension of the endoplasmic reticulum that has been implicated in rhodopsin transport and as an intracellular calcium store (Walz, 1982; Matsumoto-Suzuki et al., 1989;Suzuki and Hirosawa, 1991). However, despite the accumulated information about rdgB, its actual function in vivo remains largely a mystery.
We have been screening for conserved mammalian genes that are differentially expressed in the retina and retinal pigment epithelium (RPE) in an attempt to find novel genes involved in retinal development and function as well as to provide new candidate genes for the study of inherited retinal diseases. In this process, we identified a mammalian homolog of rdgB (mrdgB). Based on strong sequence conservation and similarity of the expression pattern at both the RNA and protein levels, we suggest that mrdgB is in fact the ortholog of rdgB. Most importantly, and perhaps surprisingly given the significant differences between mammalian and invertebrate phototransduction, expression of murine mrdgB inrdgB mutant flies fully rescues the mutant phenotypes. These results suggest the existence of novel aspects of vertebrate photoreceptor signal transduction that were not appreciated previously.
MATERIALS AND METHODS
Generation of bovine RPE/retina-subtracted cDNA library. Detailed description of the library will be published elsewhere (J. T. Chang, N. Della, C. Chew, S. Zhang, P. A. Campochiaro, and D. J. Zack, unpublished data). In brief, a library was constructed in Uni-ZAP XR (Strategene, La Jolla, CA) using cDNA that was generated from bovine RPE RNA; the library was in vivo excised and made single-stranded, hybridized in several rounds with an excess of biotinylated heart and liver RNA; the resulting RNA–DNA hybrids and unhybridized RNA were removed by phenol extraction after the addition of streptavidin; and the remaining unhybridized plasmid DNA was electroporated into MC1061 cells.
Fluorescent in situ hybridization. Fluorescentin situ hybridization (FISH) mapping was performed by standard methods (Lichter et al., 1990). Identical results were obtained with two independent but overlapping P1 clones. The clones were identified from high-density filters and were processed according to the supplier’s direction (Genome Systems).
Interspecific mouse backcross mapping. Interspecific backcross progeny were generated by mating (C57BL/6J × Mus spretus) F1 females and C57BL/6J males as described (Copeland and Jenkins, 1991). A total of 205 N2 mice were used to map the Mrdgb locus (see text for details). DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, Southern blot transfer, and hybridization were performed essentially as described (Jenkins et al., 1982). All blots were prepared with a Hybond-N+ nylon membrane (Amersham, Arlington Heights, IL). The probe, a ∼4.2 kb fragment of mouse cDNA, was labeled with [α32P]dCTP using a nick translation labeling kit (Boehringer Mannheim, Indianapolis, IN); washing was done to a final stringency of 0.8× SSCP and 0.1% SDS, 65°C. Fragments of 20.0, 7.4, and 1.9 kb were detected in HindIII-digested C57BL/6J DNA, and fragments of 9.4, 7.4, and 1.9 kb were detected inHindIII-digested M. spretus DNA. The presence or absence of the 9.4 kb HindIII M. spretus-specific fragment was followed in backcross mice.
A description of the probes and restriction fragment length polymorphisms (RFLPs) for the loci linked to mrdgB, including Adrbk1 and Cd5, has been reported previously (Benovic et al., 1991). One locus has not been reported previously for this interspecific backcross. The probe for galanin (Galn) was a ∼750 bp fragment of rat cDNA that was kindly provided by Rob Nickells (University of Wisconsin) and detected a 6.6 kb EcoRI fragment in C57BL/6J DNA and a 9.2 kb fragment ofM. spretus DNA. Recombination distances were calculated using Map Manager, version 2.6.5. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
Northern analysis. Northern analysis was performed essentially as we have described previously (Lee et al., 1996). Ten micrograms of total RNA were electrophoresed through a formaldehyde–agarose gel and transferred onto GeneScreen (NEN Research) using 20× SSC. The membrane was hybridized with a32P-labeled murine mrdgB probe at 42°C in hybridization buffer (50% formamide, 5× SSC, 5× Denhardt’s solution, 0.1% SDS, and 150 μg/ml salmon sperm DNA), washed at room temperature twice for 10 min each in 2× SSC/0.1% SDS, and twice at 65°C for 20 min each in 0.1× SSC/0.1% SDS. The membrane was autoradiographed with an intensifying screen. The membrane was then stripped, rehybridized with an 18 S ribosomal RNA (rRNA) probe, and processed as above.
Generation of rabbit antibody against MrdgB fusion peptide.A DNA fragment containing sequence coding for residues 254–434 of murine MrdgB was PCR amplified, cloned into the glutathioneS-transferase (GST) vector pGEX-4T-2 (Pharmacia, Piscataway, NJ), and sequenced. The fusion peptide was expressed inEscherichia coli SG13009 (pREP4; Qiagen) and purified with glutathione–Sepharose 4B, and its size and purity were confirmed by SDS-PAGE. One rabbit was immunized with the fusion peptide by HRP, Inc. (Denver, PA). The polyclonal antiserum obtained from the immunized rabbit was preabosorbed with GST and affinity purified with fusion peptide bound to Affi-Gel 15 (Bio-Rad, Richmond, CA).
Murine immunoblot analysis. Mouse tissues were homogenized using an Omni 5000 homogenizer in a buffer containing 50 mmsodium phosphate, pH 7.8, and 300 mm NaCl. The amount of total protein was quantified using the Bio-Rad protein assay system. Protease inhibitor mix (10 μg/ml each leupeptin, antipain, chymostatin, and pepstatin; Sigma, St. Louis, MO) was then added to each sample. About 20 μg of total protein/lane was electrophoresed through an 8% SDS-polyacrylamide gel and transferred onto a nitrocellulose membrane. The membrane was blocked for 1 hr (0.5% nonfat dry milk in 1× PBS buffer and 0.1% Tween 20) and incubated for 1 hr at room temperature with affinity-purified rabbit antibodies against murine MrdgB (1:200). After several washes with 1× PBS and 0.1% Tween-20, the membrane was incubated with peroxidase-conjugated anti-rabbit secondary antibody (New England Biolabs, 1:1000), and the signal was detected using an ECL kit as described by the manufacturer (Amersham).
In situ hybridization. In situ hybridization with mouse retinal frozen sections was performed using digoxigenin-labeled antisense and sense RNA probes, as we have described previously (Della et al., 1996; Lee et al., 1996). The antisense and sense probes were generated from a pBluescript SK+ plasmid containing murine mrdgB sequences from nucleotides 1457–2138.
Murine retinal immunohistochemistry. Enucleated mouse eyes were quick frozen in OCT and stored at −80°C. The tissue was cryostat-sectioned (12 μm), fixed with 4% paraformaldehyde for 30 min, and washed three times for 10 min each in 1× PBS. The sections were then blocked with 10% normal goat serum in 1× PBS and 1% BSA. Affinity purified rabbit anti-MrdgB (1:200) was incubated with sections overnight at 4°C. After several washes in 1× PBS and 0.3% Tween 20, sections were incubated for 30 min in either fluorescein- or rhodamine-conjugated anti-rabbit antibody (Sigma). After washing, the slides were mounted in Aqua Poly/Mount (Polysciences, Inc.) and examined by either standard or confocal fluorescence microscopy.
Expression of murine mrdgB in flies. A full-length murine mrdgB cDNA was cloned into the plasmid pTVh1 downstream of the Drosophila ninaE minimal promoter. A fragment containing the promoter and mrdgB cDNA was excised by partial digestion with KpnI and complete digestion withXbaI, gel purified, and ligated into pCaSpeR-4 (Ashburner, 1989a,b). CsCl density gradient-purified DNA was then coinjected with Δ2–3 helper DNA into w1118 embryos (Ashburner, 1989a,b). Analyses were performed after crossing the P[mrdgB] into a w+ orrdgB2 background.
Histology of Drosophila retinal sections. Flies were raised in a 12 hr light/dark cycle for 30 d or until degeneration was observed through deep pseudopupil analysis. Heads from flies lacking a wild-type deep pseudopupil or 30-d-old flies were bisected and fixed at room temperature for 4 hr in sodium cacodylate, pH 7.4, 2% formaldehyde, and 2% glutaraldehyde, followed by incubation in the same solution containing 1% tannic acid overnight at 4°C. The heads were washed three times (10 min each wash) in 0.1m sodium cacodylate and placed for 2 hr in 2% osmium tetraoxide and 0.1 m sodium cacodylate. After three 10 min washes in water, the heads were dehydrated through an ethanol wash series (50, 70, 80, 90, and 100%) for 5 min each. The 100% wash was repeated twice more, and then the heads were placed in xylene/ethanol (1:1), followed by xylene, for 30 min each. Heads were placed in xylene/Polybed 812 at a 3:1 ratio for 30 min and then at a 1:1 ratio for 30 min, followed by overnight incubation in 100% Polybed 812 at room temperature. This was followed by transfer of the heads to fresh 100% Polybed and placement in molds to cure at 35°C overnight. The molds were incubated at 45°C for 8 hr and then shifted to 60°C for the following 3 d. One micrometer sections were cut and stained with methylene blue–axure II.
Electrophysiology. One-day-oldrdgB+, rdgB2 and 30-d-old rdgB2, P[mrdgB+] flies were raised in a 12 hr light/dark cycle. Before ERG analysis (Zars and Hyde, 1996), the flies were subjected to 5 min of saturating light followed by 5 min of dark recovery. The ERG response to a 2 sec pulse of white light (intensity = 1.2 × 10−3W/cm2) was recorded and processed using a MacAdios II analog to digital converter and the SuperScope II software program (GW Instruments).
Drosophila immunoblot analysis. Newly eclosed flies were decapitated in room light. Three heads were homogenized in 30 μl of RdgB extraction buffer (2% SDS, 2 mm KCl, 3% urea, 10 mm Tris, pH 8.0, 2 mm EDTA, 2 mmEGTA, and 5 mm DTT), boiled for 5 min, and then centrifuged at 16,000 × g for 10 min. After pelleting of the debris, 80% of the supernatant volume was added to the appropriate volume of 5× SDS-loading buffer and boiled for 5 min. Before 7.5% PAGE, of the supernatant volume of 10× iodoacetamide (92 mg/ml iodoacetamide in distilled, deionized H20) was added to the homogenate. Fifteen microliters (∼1.5 heads) of homogenate were electrophoresed per lane, and the proteins were transferred from the gel to nitrocellulose using the Bio-Rad semidry transfer apparatus set at 20 V for 1 hr. After transfer, the membrane was blocked with 5% Blotto for 1 hr. The membrane was washed in TTBS (0.05% Tween 20 in TBS) for 20 min and incubated in either undiluted anti-RdgB monoclonal antibody supernatant or anti-MrdgB polyclonal antibody diluted 1:1500 in 2% Blotto (5% nonfat dry milk in TBS) overnight. The blot was washed three times for 10 min each with TTBS and incubated in secondary antibody (horseradish peroxidase-conjugated) for 1–2 hr before a final series of four TTBS washes for 10 min each. Bands were visualized using ECL detection according to the protocol of the manufacturer (Amersham).
RESULTS
Cloning of mammalian orthologs of Drosophila rdgB
A bovine RPE/retinal cDNA library enriched for genes preferentially expressed in the RPE and retina was generated by subtracting an RPE library with bovine heart and liver mRNA (J. T. Chang, N. Della, C. Chew, S. Zhang, P. A. Campochiaro, and D. J. Zack, unpublished data). Northern blot analysis indicated that ∼30% of the clones represent genes that are strongly and preferentially expressed in the RPE. Approximately 11.6% correspond to genes preferentially expressed in the retina, presumably because the original RPE RNA preparation contained some retinal RNA. Database analysis (BLAST, National Center for Biotechnology Information) of a partial sequence from one of the clones revealed homology with rdgB as well as with several sequences in the expressed sequence tag (EST) database (dbest). This bovine cDNA was used as a probe to isolate full-lengthmrdgB clones from a mouse retinal cDNA library. Partial-length EST clones corresponding to the human mrdgBwere procured (Research Genetics) and sequenced. These EST clones were used to obtain human P1 genomic clones, which were sequenced to derive the full open reading frame and to determine the genomic structure ofmrdgB.
The predicted protein sequences of human and murine mrdgBwere compared with Drosophila rdgB (Fig.1A). Also shown is the sequence of the homologous soluble rat brain PITPα (Dickeson et al., 1989). Human and murine mrdgB are 88% identical at the nucleotide level, 92% at the amino acid level. Both share 39% amino acid identity with RdgB. The highest degrees of homology involve the N- and C-terminal regions. The N-terminal PITP domain has 62% amino acid identity. Within the C terminus (human residues 909–1006), the identity is 65%. For comparison, the overall amino acid identity between mammalian andDrosophila rhodopsins is ∼36% (Nathans and Hogness, 1983;O’Tousa et al., 1985).
Fig. 1.
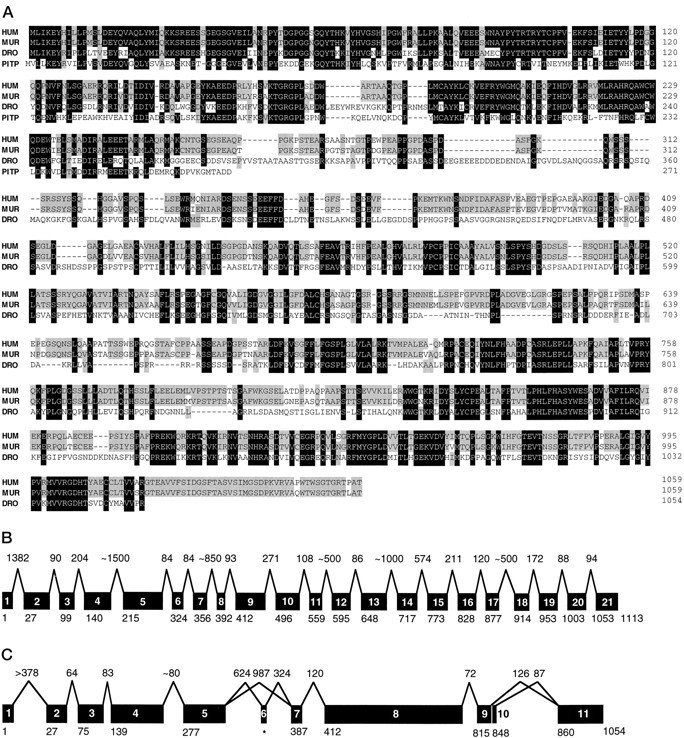
Sequence and gene structure of mrdgB.A, Alignment of the amino acid sequences of human MrdgB (HUM), murine MrdgB (MUR),Drosophila RdgB (DRO), and rat brain soluble PITPα (PITP). Alignment was generated using Geneworks 2.3 (IntelliGenetics, Mountain View, CA) and modified manually. Spaces (dashes) were introduced to maximize homology. Among MrdgB, RdgB, and PITPα residues, at least three residues that are identical are indicated in black, whereas residues that are identical in two of the three species and PITPα are shaded. Complete DNA sequences for murine and human mrdgB will be available from GenBank (pending). B, Schematic structure of the humanmrdgB gene. The individual coding region exons arenumbered. The number above each intron represents the corresponding intron size (in base pairs). The numberbelow each exon represents the number of the first amino acid residue.C, Schematic structure of the Drosophila rdgB gene labeled as in B, except that inC the intron sizes are drawn approximately to scale. Exons 6 and 10 are differentially expressed. Amino acid residues for exon 6 (asterisk) are not numbered, because its sequence does not appear in the published rdgB sequence (Vitelic et al., 1991).
The coding region of the human mrdgB gene contains 21 exons (Fig. 1B), which contrasts with the 10 exons in theDrosophila rdgB gene (Fig. 1C). The introns tend to be small; nine of them are smaller than 125 bp, and the boundaries do not correlate with those of the Drosophila gene. Intron–exon borders are shown in Table 1. Fuller sequences are available from GenBank.
Table 1.
Nucleotide sequences at intron–exon junctions of the coding region of the human mrdgB gene
| Exon | 5′-Intron | Exon sequence | 3′-Intron | Exon size (bp) |
|---|---|---|---|---|
| 1 | ggcccgccgagcgccttcag1-a | GATGCTCATC-----CATGATCCAG | gtgagggcggcggggagagg | 79 |
| 2 | tgctctcccttgggccacag | AAAAAGAGCC-----CCCGAACCCG | gtgagtgggtgagctgggca | 215 |
| 3 | cacccgtgccttctgcccag | GTACACCTGC-----CGCATCCTGG | gtgaggcctggagctatggg | 122 |
| 4 | tgcttgccgaactccctcag | ACACCATCGA-----CATGATGTAG | gtgagcacccagctgcggga | 225 |
| 5 | tgacgcctgggcacctgcag | GTCTGCGTCG-----CAACATGGAG | gtgaggctggggcacaggga1-b | 327 |
| CAACATGGAGG | tgaggctggggcacaggga1-b | |||
| 6 | gaggctgagctggcccggca1-b | GGGCTGTGTC-----GATGCCCACG | gtcagcactgaggccctttt | 95 |
| aggctgagctggcccggcag1-b | GGCTGTGTC | |||
| 7 | acagccccctgttccctcag | AAGGCTTCTC-----GGAACGCCAG | gtaaagataccgcctaggag | 108 |
| 8 | ctggcttcgttgccctgcag | AGCCTGGAGC-----GGACTCAGAG | gcgagtatggtcagccaccc | 62 |
| 9 | caccccattgttccccccag | GGCCTGGATG-----TTGTCTCCAA | gtactagccacgggtggaag | 251 |
| 10 | cacatttccccctcctacag | CCTGAGCCCT-----CTGTGGGCAG | gtcagcggttagggacagtc | 190 |
| 11 | gtgcccctgctcccacccag | GTCGCACTGA-----TGGGAGCATG | gtcagtatagccaaacccgg | 108 |
| 12 | ctgtctcctgttccatccag | AACAATGAGC-----CTCAGAACAG | gtgacgtccctgccccatca | 158 |
| 13 | gcacccctcctcccccacag | CCTTCAGGCA-----GCCCTGGAGG | gtgagtcctaggggctgcgg | 206 |
| 14 | tgtcttctgggtaccagcag | CCCAGATGCG-----CTGCTGCTGG | gtacgcccccaaacaagata | 168 |
| 15 | accttccctccttcccccag | CCGACACTCT-----GTGGTTAAGA | gtaagtcagccagccatggc | 165 |
| 16 | tcgctgccatggccgcgcag | TCCTGGAGCG-----CCTGCGCCAG | gtgggcctgggaatatggaa | 149 |
| 17 | tcgccccctgtcccgtgcag | GTGATCGAGA-----CAAGATCCGG | gtaggtgcccggccccgggc | 111 |
| 18 | tacgcctctcctccctctag | AACGTCACTT-----TGGAGAGAAG | gtcaggacccagaggccctg | 117 |
| 19 | ccgccccttctcccctccag | GTGGATGTCT-----TGGTGGTCAG | gtgagcgcggctggccgggc | 149 |
| 20 | ccccgccccgtgggccgcag | GGGCGACCAC-----ACGTGGTCAG | gtaggagttgccactgccac | 149 |
| 21 | ccgctgcccccgccccacag | GCACTGGCAG-----GGTGCAGGAG | gtgcggcggctggggggagg | 184 |
It has not determined whether this sequence represents 5′-untranslated or intron sequence.
The exact location of the donor and acceptor sites is ambiguous.
Human mrdgB maps to the site of several known retinal diseases; murine mrdgB maps to a syntenic region
We mapped the human mrdgB gene to 11q13.1 by FISH using two independent but overlapping P1 genomic clones as probes (data not shown). This result is in agreement with both the cDNA-based FISH results of Banfi et al. (1996), who mapped a partial-lengthmrdgB cDNA EST clone (R56391) to 11q13.1, and our finding that mrdgB contains a sequence-tagged site (WI-13814) that maps 381.2 centirays from the terminus of chromosome 11. The 11q13.1 locus is in the immediate vicinity of four previously mapped human retinal diseases: recessive Bardet–Biedl syndrome 1 (11q13) (Leppert et al., 1994), dominant vitelliform macular dystrophy (Best’s Disease; 11q13) (Nichols et al., 1994; Hou et al., 1996), dominant Criswick–Schepens syndrome (dominant familial exudative vitreoretinopathy; 11q13–q23) (Fuhrmann et al., 1995), and dominant neovascular inflammatory vitreoretinopathy (11q13) (Stone et al., 1992).
We determined the mouse mrdgB chromosomal location by interspecific backcross analysis using progeny derived from matings of (C57BL/6J × M. spretus) F1 × C57BL/6J mice (Copeland and Jenkins, 1991). The 9.4 kb HindIIIM. spretus RFLP (see Materials and Methods) was used to follow the segregation of the mrdgB locus in backcross mice. The mapping results indicate that mrdgB is located in the proximal region of mouse chromosome 19 linked to Galn,Adrbk1, and Cd5 (Fig. 2). In total, 116 mice were analyzed for every marker and are shown in the segregation analysis (Fig. 2), and up to 189 mice were typed for some pairs of markers. Each locus was analyzed in pairwise combinations for recombination frequencies using the additional data. The ratios of the total number of mice exhibiting recombinant chromosomes to the total number of mice analyzed for each pair of loci and the most likely gene order are centromere–Galn–0/189–mrdgB– 2/188–Adrbk1–3/119–Cd5. The recombination frequencies [expressed as genetic distances in centimorgans (cM) ± SE] are (Galn, mrdgB), 1.1 ± 0.8–Adrbk1, 2.5 ± 1.4–Cd5. No recombinants were detected between Galn and mrdgBin 189 animals typed in common, suggesting that the two loci are within 1.6 cM of each other (upper 95% confidence limit). The proximal region of mouse chromosome 19 shares homology with human chromosome 11q (summarized in Fig. 2), and placement of mrdgB in this mouse interval is consistent with the human mrdgB localization at 11q13.1.
Fig. 2.
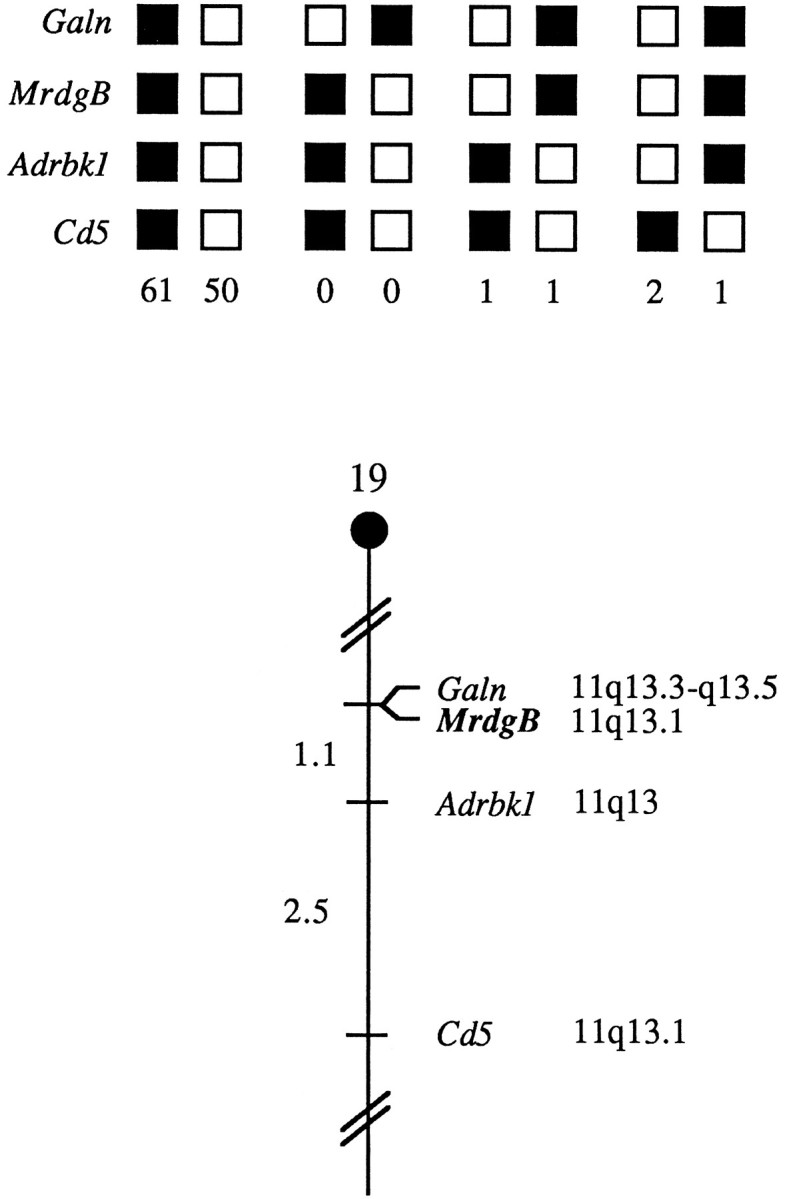
Murine mrdgB maps in the proximal region of mouse chromosome 19. Mrdgb was mapped to mouse chromosome 19 by interspecific backcross analysis. The segregation patterns of mrdgB and flanking genes in 116 backcross animals that were typed for all loci are shown at thetop. For individual pairs of loci, more than 116 animals were typed (see text). Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J ×M. spretus) F1 parent. The shaded boxes represent the presence of a C57BL/6J allele, andwhite boxes represent the presence of an M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial chromosome 19 linkage map showing the location ofmrdgB in relation to linked genes is shown at thebottom. Recombination distances between loci in centimorgans are shown to the left of the chromosome, and the positions of loci in human chromosomes, where known, are shown to the right. References for the human map positions of loci cited in this study can be obtained from the Genome Data Base, a computerized database of human linkage information maintained by The William H. Welch Medical Library (Johns Hopkins University, Baltimore, MD).
To determine whether murine mrdgB is a reasonable candidate gene for any known mouse mutations, we compared our interspecific map of chromosome 19 with a composite mouse linkage map that reports the map location of many uncloned mouse mutations (Mouse Genome Database, The Jackson Laboratory, Bar Harbor, ME). However, the region of the composite map to which mrdgB maps does not contain any mutations with a retinal phenotype (data not shown).
mrdgB is specifically expressed in the retina and brain
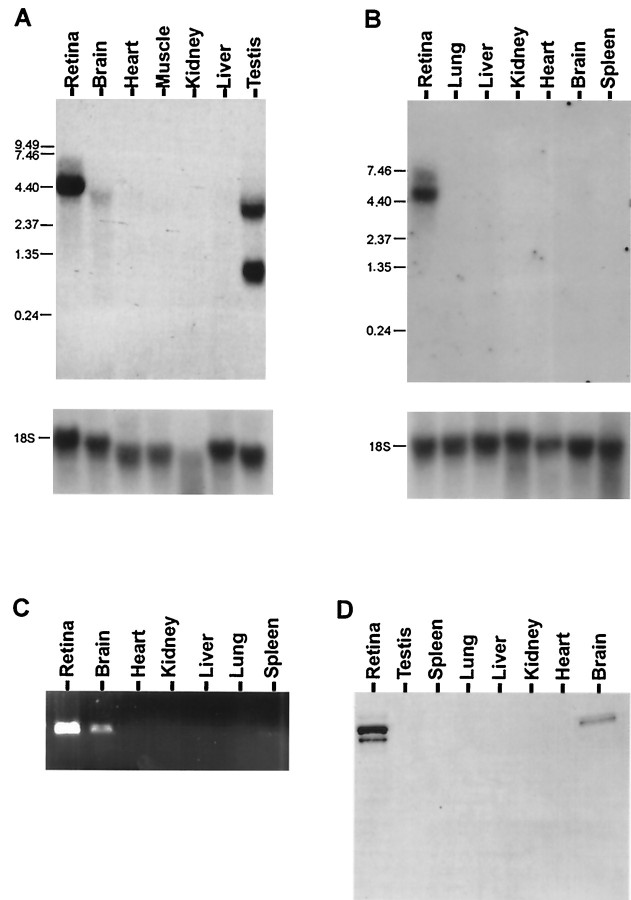
Northern analysis indicates that bovine mrdgB is strongly expressed in the retina and weakly in the brain, with a single major transcript of 4.4 kb (Fig. 3A). In mouse retina there is a major transcript of 4.9 kb and a minor 6.4 kb RNA (Fig. 3B). Additionally, there is a faint 4.9 kb band in the brain. Reverse transcription (RT)-PCR with mouse RNA samples also indicates strong expression in the retina and weak expression in the brain (Fig. 3C). For comparison, Drosophila shows multiple transcripts ranging from 3.9 to 9.5 kb; the mRNA is expressed in the head, but it is not retina-specific (Vihtelic et al., 1991).
Fig. 3.
MrdgB is expressed in the retina and brain. Northern analysis of expression of mrdgB mRNA in bovine (A) and murine (B) tissues, using bovine and murine probes, respectively. After initial hybridization, membranes were stripped and rehybridized with a human 18 S rRNA probe. Size standards (in kilobases) are indicated on theleft margins of the blots. C, RT-PCR using mrdgB-specific primers and first-strand cDNA prepared from the indicated murine tissues. D, Western analysis of MrdgB protein expression in extracts prepared from the indicated murine tissues.
Western analysis also indicates that MrdgB is expressed in both the retina and brain, with an apparent molecular weight of approximately 160 kDa in both mouse (Fig. 3D) and bovine tissue (data not shown). It is interesting to note that the amount of MrdgB in the brain relative to the retina is considerably higher at the protein level compared with the RNA level. Possible explanations for this observation include increased translational efficiency in the brain and increased protein stability in the brain. Perhaps related to degradation, or posttranslational modification, the retinal samples consistently show two similarly sized bands, whereas the brain samples show a single band.
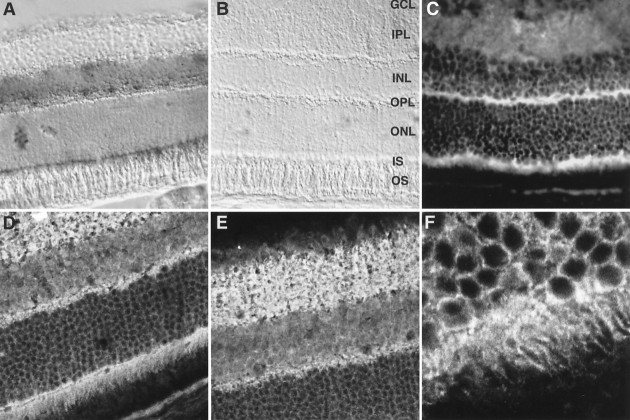
Retinal in situ hybridization indicates mrdgBmRNA expression in photoreceptors and the inner nuclear layer (Fig.4A,B). Retinal immunocytochemistry, using both standard fluorescence and confocal microscopy, demonstrates strong protein expression in photoreceptors, particularly in the inner segments and the outer plexiform layer, in the inner plexiform layer, and possibly in the ganglion cell layer (Fig. 4C–E). These results are consistent with the expression pattern of RdgB inDrosophila, because vertebrate inner segments are analogous to the region of the fly photoreceptor cell that contains the SRC, and the outer plexiform layer is analogous to fly photoreceptor synaptic terminals.
Fig. 4.
Expression pattern of mrdgB in the retina. In situ hybridization of mouse retina with antisense (A) and sense (B) digoxigenin-labeled mrdgB riboprobes.GCL, Ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer;OPL, outer plexiform layer; ONL, outer nuclear layer; IS, inner segments; andOS, outer segments. B, Standard immunofluorescence image of mouse retinal section stained with affinity-purified anti-MrdgB antibody. D–F, Higher-power confocal images of similarly stained sections.F highlights the photoreceptor inner segment staining.
mrdgB fully rescues the rdgB mutant phenotypes in flies
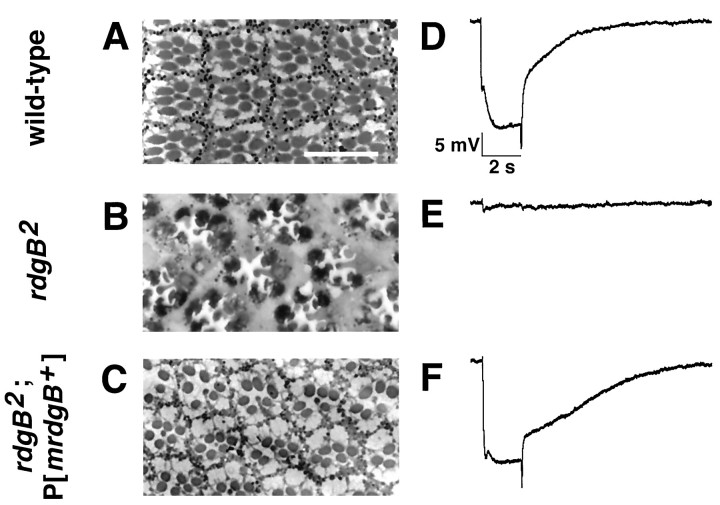
As a first approach to define the function of mrdgB, we expressed the murine mrdgB cDNA in rdgB2null flies to determine whether it could rescue the mutant phenotype. The murine mrdgB cDNA was cloned into a P element transformation vector under the transcriptional control of theninaE promoter and microinjected into embryos. As shown by immunoblot analysis, MrdgB was expressed by the resulting transgenic flies (Fig. 5). Gross examination of the flies expressing the murine mrdgB cDNA revealed that the deep pseudopupil, which was absent in the mutant flies, was restored in the transgenic flies. The deep pseudopupil, a virtual image of the rhabdomeres from ∼20 adjacent ommatidia, reveals the integrity of the rhabdomeres and ommatidial structure (Franceschini, 1972). In the mutant background, under normal light cycling, substantial degeneration and loss of the deep pseudopupil was clearly evident by day 4, whereas flies expressing mrdgB displayed no retinal degeneration even at 30 d. Light microscopic analysis confirmed the prevention of photoreceptor R1–R6 degeneration (Fig.6A–C). Degeneration of the R7 photoreceptor was still evident, presumably because theninaE promoter did not direct mrdgB transgene expression in R7 cells.
Fig. 5.
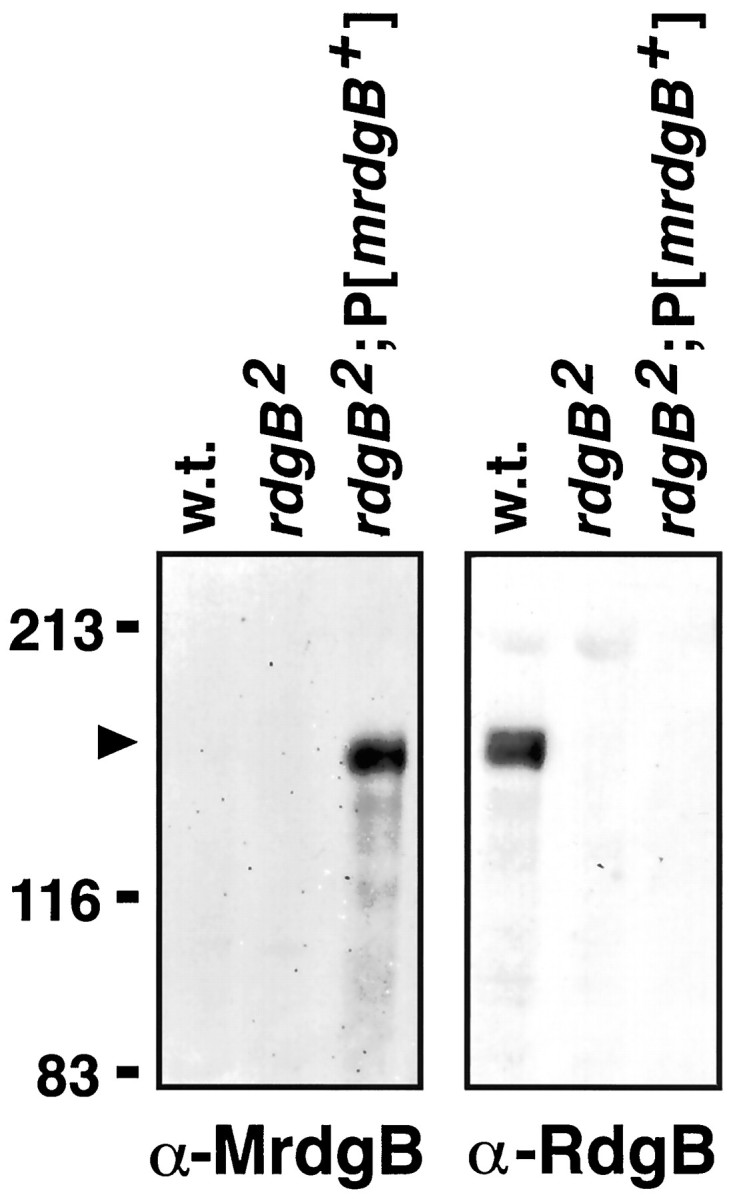
Murine MrdgB is expressed in the heads of transgenic flies. Immunoblot was performed using head extracts from Oregon-R (wild-type), rdgB2 andrdgB2P[mrdgB+] flies. The left panel was probed with anti-MrdgB antiserum, and theright panel was probed with anti-RdgB antiserum. The antibodies used are specific for their respective proteins, because no cross-reacting signals were detected. The position of molecular weight standards (in kilodaltons) is indicated on theleft.
Fig. 6.
Expression of murinemrdgB in rdgB mutant flies prevents retinal degeneration and restores the ERG. One-micrometer-thick plastic sections were cut through the retinas of 30-d-old Oregon-R (wild-type) (A), 7-d-old rdgB2(B), and 30-d-old rdgB2, P [mrdgB+] (C) flies. At a minimum, three flies from each genotype from several time points were examined and are consistent with the time course of deep pseudopupil loss or maintenance (data not shown). TherdgB2 flies (B) undergo retinal degeneration characterized by dark-staining cell bodies and shrinkage and loss of rhabdomeres. The R1–R6 photoreceptor cells are more sensitive in the rdgB2 mutant and degenerate before R7. The R1–R6 cells in both wild-type (A) and rdgB2P[mrdgB+] (C) flies are normal at 30 d. However, the R7 cell is either missing or degenerating in rdgB2, P[mrdgB+] flies, consistent with expression of the mrdgB+ cDNA being limited to the R1–R6 cells. Electroretinogram responses were recorded from 1-d-old wild-type (D), 1-d-oldrdgB2 (E), and 30-d-oldrdgB2, P[mrdgB+] (F) flies stimulated with a 2 sec pulse of white light. Before recording, the flies were exposed to 5 min of intense constant light followed by 5 min of dark. The ERGs shown in D andE are representative of recordings from at least six different rdgB+ orrgdB2 flies. SixrdgB2 flies containing two different P[mrdgB+] lines were examined, and a representative ERG is shown in F. TherdgB+ andrdgB2, P[mrdgB+] flies displayed ERG traces that were not significantly different from each other in any respect.
ERG analysis similarly demonstrated restoration of wild-type function. After a saturating light stimulus, flies were dark-adapted for 5 min and then given a 2 sec light pulse. Wild-type flies produced an ERG light response roughly equivalent to that elicited before light saturation (Fig. 6D). With rdgB flies, however, the saturating light treatment effectively eliminated the subsequent light response (Fig. 6E). The expression of MrdgB in rdgB2 null flies restored the normal recovery after the saturating light stimulation (Fig.6F). In both wild-type and “rescued” flies the recovery time is <30 sec, whereas for mutant flies it is >30 min.
DISCUSSION
Based on the hypothesis that proteins that are both differentially expressed and evolutionarily conserved often play fundamental roles in biological processes, we screened for conserved mammalian genes that are differentially expressed in the neural retina and RPE. Because of the power of Drosophila genetics and the extensive body of knowledge relating to the identification and characterization of fly visual system mutations, we were particularly interested in mammalian genes that show homology to Drosophila genes. In this paper we have described one such gene, mrdgB, and demonstrated it to be the mammalian ortholog of Drosophila rdgB. Our data implicate mrdgB as a potentially important candidate gene for the study of human retinal disease and suggest the existence of important aspects of the vertebrate visual process that were not appreciated previously.
MrdgB as a candidate gene for retinal diseases that map to 11q13.1
A large number of hereditary forms of human retinal degeneration are known to exist, ranging from childhood Leber’s congenital amaurosis, to retinitis pigmentosa, to adult onset forms of macular degeneration (Goldberg and Penie, 1986). Although mutations in several different genes have been identified as the causes of several of these diseases, the known mutations account for only a small minority of the cases (Rosenfeld et al., 1994; Dryja and Berson, 1995). The genes corresponding to the remaining cases must be isolated by positional cloning and/or candidate gene approaches. Drosophila has proven to be a good model for genetically characterizing retinal degeneration because this phenotype is easy to identify and a large number of mutants are known. Additionally, Drosophila can provide candidate genes for analysis in vertebrate systems. For example, the observation that rhodopsin (NinaE) mutations cause retinal degeneration was first made in flies (O’Tousa et al., 1989). More recent NinaE studies have shown that, just as in humans (Dryja et al., 1990), both dominant and recessive NinaE mutations can cause photoreceptor degeneration (Kurada and O’Tousa, 1995).
The rdgB phenotype together with the finding that humanmrdgB maps at or near the site of four retinal diseases [Bardet–Biedl syndrome 1 (Leppert et al., 1994), vitelliform macular dystrophy (Best’s disease) (Stone et al., 1992; Graff et al., 1994;Nichols et al., 1994; Hou et al., 1996), Criswick–Schepens syndrome (dominant famial exudative vitreoretinopathy) (Criswick and Schepens, 1969; Fuhrmann et al., 1995), and dominant neovascular inflammatory vitreoretinopathy (Stone et al., 1992)] make mrdgB a strong candidate gene for these diseases. Ongoing studies will determine whether mrdgB mutations are responsible for any of these or other currently unmapped retinal diseases. If such mutations are found,rdgB null flies will provide a useful assay system to assess the mechanism by which the mutant proteins act.
Functional domains within RdgB and MrdgB
Analysis of the sequence conservation between RdgB and MrdgB is both consistent with the importance of the N-terminal PITP domain and suggestive of an as yet unknown role for the C-terminal region. Unlike all other known PITP proteins, which are soluble,rdgB and mrdgB both encode putative transmembrane proteins. Nonetheless, expression of the soluble PITP domain fromrdgB (residues 1–276) is sufficient to rescue the knownrdgB mutant phenotypes fully (S. Milligan, J. Alb, V. Bankaitis, and D. Hyde, unpublished results). Supportive of the key role of this domain is its unusually high conservation between RdgB and MrdgB (62% amino acid identity). In fact, the conservation in this domain is higher between RdgB and MrdgB than between either of these proteins and any of the other known PITP proteins (maximum homology, 42%). Consistent with this pattern of homology is the finding that PI transferase activity itself is not sufficient to provide rescue. Expression of a mutant form of RdgB (T59E), which demonstrates wild-type PI transfer activity in vitro, only partially rescues the ERG abnormalities and fails to rescue the retinal degeneration (S. Milligan, J. Alb, V. Bankaitis, and D. Hyde, unpublished results). Furthermore, neither the soluble rat brain PITPα protein alone nor the protein in combination with the C terminus of RdgB as a fusion protein provides any detectable degree of rescue. These results, taken together, suggest that the N termini of both RdgB and MrdgB provide important functions in addition to PITP activity.
The results with residues 1–276 raise questions concerning the role of the other 75% of the molecule. Comparison of the RdgB and MrdgB sequences demonstrate that there is a region in the C-terminal part of the molecule (residues 909–1006 in MrdgB and 936–1043 in RdgB) with homology that is as high (65%) as that in the PITP domain. This degree of homology suggests that this region carries out some conserved biological function. Also suggestive of an important function for the C-terminal region are preliminary results indicating that anrdgB mutant allele has a missense mutation in the C-terminal domain and the observation that engineered C-terminal deletions do not fully rescue the rdgB mutant phenotypes (S. Milligan and D. R. Hyde, unpublished results). Although it remains to be determined, the C-terminal domain may be involved in intracellular trafficking or interactions with other proteins. Use of the yeast two-hybrid assay may help to identify some of these putative interacting proteins.
Possible novel aspects of vertebrate visual processing involving a phospholipase C/rdgB pathway
The finding that mrdgB fully rescues the majorrdgB phenotypes (retinal degeneration and altered electrophysiological light response) in mutant flies is surprising given that evolution has adapted seemingly divergent paradigms for invertebrate and vertebrate vision. The identification and characterization of mrdgB may provide new insight into vertebrate visual processing. Because RdgB seems to function, at least partially, in the recovery phase after light stimulation, it seems reasonable to hypothesize that MrdgB also functions in light recovery. This aspect of vertebrate vision is less well understood than the “on” pathway of the phototransduction cascade. Recent evidence suggests an important role for calcium, as is also the case with invertebrates (Polans et al., 1996).
The possibility that MrdgB functions in vertebrate phototransduction is supported by the discovery of vertebrate retina-specific homologs of the Drosophila photoreceptor phospholipase C (NorpA) (Ghalayini et al., 1991; Ferreira et al., 1993;Alvarez et al., 1995). Given that RdgB seems to function distal to NinaE and NorpA in the fly, MrdgB may function in a similar cascade in the mammalian retina. Although the specific function of the mammalian retinal phospholipase Cs remains to be clearly defined, experiments in which murine phospholipase Cβ4 was knocked out demonstrate altered visual processing and decreased a- and b-waves in the rod ERG (Jiang et al., 1996). It is hoped that future experiments will help elucidate the biology and biochemistry of this postulated mammalian phospholipase C/MrdgB pathway.
Note added in proof. It has recently been reported that mutation of PITPα causes neurodegeneration in thevibrator mouse [Hamilton et al. (1997) Neuron 18:711–722].
Footnotes
This work was supported by National Eye Institute Grants EY09769, EY05951, and EY08058; a grant from The Foundation Fighting Blindness; funds from the National Cancer Institute, Department of Health and Human Services, under contract with ABL; the Rebecca P. Moon, Charles M. Moon Jr, and Dr. P. Thomas Manchester Research Fund; and Research to Prevent Blindness, Inc. D.J.Z. is a recipient of a Career Development Award from Research to Prevent Blindness. We thank Yan Cheng, Debra J. Gilbert, and Mary Barnstead for their technical expertise.
J.T.C. and S.M. contributed equally to the work described in this manuscript.
Correspondence should be addressed to Donald J. Zack, Johns Hopkins University School of Medicine, 809 Maumenee, 600 North Wolfe Street, Baltimore, MD 21287-9289.
REFERENCES
- 1.Alvarez RA, Ghalayini AJ, Xu P, Hardcastle A, Bhattacharya S, Rao N, Pettenati MJ, Anderson RE, Baehr W (1995) cDNA sequence and gene locus of the human retinal phosphoinositide-specific phospholipase-Cβ4 (PLCB4). [DOI] [PubMed]
- 2.Ashburner M. Drosophila: a laboratory handbook. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989a. [Google Scholar]
- 3.Ashburner M. Drosophila: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989b. [Google Scholar]
- 4.Banfi S, Borsani G, Rossi E, Bernard L, Guffanti A, Rubboli F, Marchitiello A, Giglio S, Coluccia E, Zollo M, Zuffardi O, Ballabio A. Identification and mapping of human cDNAs homologous to Drosophila mutant genes through EST database searching. Nat Genet. 1996;13:167–174. doi: 10.1038/ng0696-167. [DOI] [PubMed] [Google Scholar]
- 5.Benovic JL, Onorato JJ, Arriza JL, Stone WC, Lohse M, Jenkins NA, Gilbert DJ, Copeland NG, Caron MG, Lefkowitz RJ. Cloning, expression, and chromosomal localization of β-adrenergic receptor kinase 2. J Biol Chem. 1991;266:14939–14946. [PubMed] [Google Scholar]
- 6.Carlson SD, Stark WS, Chi C. Rapid light induced degeneration of photoreceptor terminals in rdgB mutant of Drosophila. Invest Ophthalmol Vis Sci [Suppl] 1985;26:131. [Google Scholar]
- 7.Copeland NG, Jenkins NA. Development and applications of a molecular genetic linkage map of the mouse genome. Trends Genet. 1991;7:113–118. doi: 10.1016/0168-9525(91)90455-y. [DOI] [PubMed] [Google Scholar]
- 8.Criswick VG, Schepens CL. Familial exudative vitreoretinopathy. Am J Ophthalmol. 1969;68:578–594. doi: 10.1016/0002-9394(69)91237-9. [DOI] [PubMed] [Google Scholar]
- 9.Della NG, Campochiaro PA, Zack DJ. Localization of timp-3 mRNA expression to the retinal pigment epithelium. Invest Ophthalmol Vis Sci. 1996;37:1921–1924. [PubMed] [Google Scholar]
- 10.Dickeson SK, Lim CN, Schuyler GT, Dalton TP, Helmkamp GMJ, Yarbrough LR. Isolation and sequence of cDNA clones encoding rat phosphatidylinositol transfer protein. J Biol Chem. 1989;264:16557–16564. [PubMed] [Google Scholar]
- 11.Dryja TP, Berson EL. Retinitis pigmentosa and allied diseases. Implicatons of genetic heterogeneity. Invest Ophthalmol Vis Sci. 1995;36:1197–1200. [PubMed] [Google Scholar]
- 12.Dryja TP, McGee TC, Reichel E, Hahn LB, Cowley GS, Yandell DW, Sandberg MA, Berson EL. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–366. doi: 10.1038/343364a0. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira PA, Shortridge RD, Pak WL. Distinctive subtypes of bovine phospholipase C that have preferential expression in the retina and high homology to the norpA gene product of Drosophila. Proc Natl Acad Sci USA. 1993;90:6042–6046. doi: 10.1073/pnas.90.13.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franceschini N. Pupil and pseudopupil in the compound eye of Drosophila. In: Wehner R, editor. Information processing in the visual systems of arthropods. Springer; Berlin: 1972. pp. 75–82. [Google Scholar]
- 15.Fuhrmann C, Duvigneau C, Muller B, Schwinger E, Julier C, Laqua H, Gal A. Autosomal dominant exudative vitreoretinopathy: linkage analysis and its clinical application. Ger J Ophthalmol. 1995;4:43–46. [PubMed] [Google Scholar]
- 16.Ghalayini AJ, Tarver AP, Mackin WM, Koutz CA, Anderson RE. Identification and immunolocalization of phospholipase C in bovine rod outer segments. J Neurochem. 1991;57:1405–1412. doi: 10.1111/j.1471-4159.1991.tb08307.x. [DOI] [PubMed] [Google Scholar]
- 17.Goldberg M, Penie WA. Goldberg’s genetic and metabolic eye disease. Little, Brown; Boston: 1986. [Google Scholar]
- 18.Graff C, Forsman K, Larsson C, Nordstrom S, Lind L, Johansson K, Sandgren O, Weissenbach J, Holmgren G, Gustavson KH, Wadelius C. Fine mapping of Best’s macular dystrophy localizes the gene in close proximity to but distinct from the D11S480/ROM1 loci. Genomics. 1994;24:425–434. doi: 10.1006/geno.1994.1648. [DOI] [PubMed] [Google Scholar]
- 19.Harris WA, Stark WS. Hereditary retinal degeneration in Drosophila melanogaster: a mutant defect associated with the phototransduction process. J Gen Physiol. 1977;69:261–291. doi: 10.1085/jgp.69.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heisenberg M. Separation of receptor and lamina potentials in the electroretinogram of normal and mutant Drosophila. J Exp Biol. 1971;55:85–100. doi: 10.1242/jeb.55.1.85. [DOI] [PubMed] [Google Scholar]
- 21.Hotta Y, Benzer S. Abnormal electroretinogram in visual mutants in Drosophila. Nature. 1969;222:354–356. doi: 10.1038/222354a0. [DOI] [PubMed] [Google Scholar]
- 22.Hou YC, Richards JE, Bingham EL, Pawar H, Scott K, Segal M, Lunetta KL, Boehnke M, Sieving PA. Linkage study of Best’s vitelliform macular dystrophy (VMD2) in a large North American family. Hum Hered. 1996;46:211–220. doi: 10.1159/000154356. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins NA, Copeland NG, Taylor BA, Lee BK. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang HP, Lyubarsky A, Dodd R, Vardi N, Pugh E, Baylor D, Simon MI, Wu DQ. Phospholipase c beta-4 is involved in modulating the visual response in mice. Proc Natl Acad Sci USA. 1996;93:14598–14601. doi: 10.1073/pnas.93.25.14598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koutalos Y, Yau KW. A rich complexity emerges in phototransduction. Curr Opin Neurobiol. 1993;3:513–519. doi: 10.1016/0959-4388(93)90049-5. [DOI] [PubMed] [Google Scholar]
- 26.Kurada P, O’Tousa JE. Retinal degeneration caused by dominant rhodopsin mutations in Drosophila. Neuron. 1995;14:571–579. doi: 10.1016/0896-6273(95)90313-5. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lee J, Della N, Chew CE, Zack DJ. Rin, a neuron-specific and calmodulin-binding small-G protein, and Rit define a novel subfamily of Ras proteins. J Neurosci. 1996;16:6784–6794. doi: 10.1523/JNEUROSCI.16-21-06784.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YJ, Shah S, Suzuki E, Zars T, O’Day PM, Hyde DR. The Drosophila dgq gene encodes a G alpha protein that mediates phototransduction. Neuron. 1994;13:1143–1157. doi: 10.1016/0896-6273(94)90052-3. [DOI] [PubMed] [Google Scholar]
- 30.Leppert M, Baird L, Anderson KL, Otterud B, Lupski JR, Lewis RA. Bardet-Biedl syndrome is linked to DNA markers on chromosome 11q and is genetically heterogeneous. Nat Genet. 1994;7:108–112. doi: 10.1038/ng0594-108. [DOI] [PubMed] [Google Scholar]
- 31.Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto-Suzuki E, Hirosawa K, Hotta Y. Structure of the subrhabdomeric cisternae in the photoreceptor cells of D. melanogaster. J Neurocytol. 1989;18:87–93. doi: 10.1007/BF01188427. [DOI] [PubMed] [Google Scholar]
- 33.Minke B, Rubinstein CT, Sahly I, Bar-Nachum S, Timberg R, Selinger Z. Phorbol ester induces photoreceptor-specific degeneration in a Drosophila mutant. Proc Natl Acad Sci USA. 1990;87:113–117. doi: 10.1073/pnas.87.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nathans J, Hogness DS. Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell. 1983;34:807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- 35.Nichols BE, Bascom R, Litt M, McInnes R, Sheffield VC, Stone EM. Refining the locus for Best vitelliform macular dystrophy and mutation analysis of the candidate gene ROM1. Am J Hum Genet. 1994;54:95–103. [PMC free article] [PubMed] [Google Scholar]
- 36.Nilson D. Eye ancestry: old genes for new eyes. Curr Biol. 1966;6:39–42. doi: 10.1016/s0960-9822(02)00417-7. [DOI] [PubMed] [Google Scholar]
- 37.O’Tousa JE, Baehr W, Martin RL, Hirsh J, Pak WL, Applebury ML. The Drosophila ninaE gene encodes an opsin. Cell. 1985;40:839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- 38.O’Tousa JE, Leonard DS, Pak WL. Morphological defects in oraJK84 photoreceptors cause by mutation in R1–6 opsin gene of Drosophila. J Neurogenet. 1989;6:41–52. doi: 10.3109/01677068909107099. [DOI] [PubMed] [Google Scholar]
- 39.Pak WL. Drosophila in vision research—the Friedenwald lecture. Invest Ophthalmol Vis Sci. 1995;36:2340–2357. [PubMed] [Google Scholar]
- 40.Pak WL, Grossfield J, Arnold K. Mutants in the visual pathway of Drosophila melanogaster. Nature. 1970;227:518–520. doi: 10.1038/227518b0. [DOI] [PubMed] [Google Scholar]
- 41.Polans A, Baehr W, Palczewski K. Turned on by Ca2+—the physiology and pathology of Ca2+ binding proteins in the retina. Trends Neurosci. 1996;19:547–554. doi: 10.1016/s0166-2236(96)10059-x. [DOI] [PubMed] [Google Scholar]
- 42.Rosenfeld PJ, McKusick VA, Amberger JS, Dryja TP. Recent advances in the gene map of inherited eye disorders: primary hereditary diseases of the retina, choroid, and vitreous. J Med Genet. 1994;31:903–915. doi: 10.1136/jmg.31.12.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubinstein CT, Bar-Nachum S, Selinger Z, Minke B. Chemically induced retinal degeneration in the rdgB (retinal degeneration B) mutant of Drosophila. Vis Neurosci. 1989;2:541–551. doi: 10.1017/s0952523800003485. [DOI] [PubMed] [Google Scholar]
- 44.Scott K, Becker A, Sun Y, Hardy R, Zuker CS. Gqα protein function in vivo: genetic dissection of its role in photoreceptor cell physiology. Neuron. 1995;15:919–927. doi: 10.1016/0896-6273(95)90182-5. [DOI] [PubMed] [Google Scholar]
- 45.Stark WS, Carlson SD. Ultrastructure of the compound eye and the first optic neuropile of the photoreceptor mutant oraJK84 of Drosophila. Cell Tissue Res. 1983;233:305–317. doi: 10.1007/BF00238298. [DOI] [PubMed] [Google Scholar]
- 46.Stark WS, Chen D-M, Johnson MA, Frayer KL. The rdgB gene of Drosophila: retinal degeneration in different alleles and inhibition by norpA. J Insect Physiol. 1983;29:123–131. [Google Scholar]
- 47.Stark WS, Sapp R, Carlson SD. Photoreceptor maintenance and degeneration in the norpA (no receptor potential-A) mutant of Drosophila melanogaster. J Neurogenet. 1989;5:49–59. doi: 10.3109/01677068909167264. [DOI] [PubMed] [Google Scholar]
- 48.Stone EM, Kimura AE, Folk JC, Bennett SR, Nichols BE, Streb LM, Sheffield VC. Genetic linkage of autosomal dominant neovascular inflammatory vitreoretinopathy to chromosome 11q13. Hum Mol Genet. 1992;1:685–689. doi: 10.1093/hmg/1.9.685. [DOI] [PubMed] [Google Scholar]
- 49.Stone EM, Nichols BE, Streb LM, Kimura AE, Sheffield VC. Genetic linkage of vitelliform macular degeneration (Best’s disease) to chromosome 11q13. Nat Genet. 1992;1:246–250. doi: 10.1038/ng0792-246. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki E, Hirosawa K. Immunoelectron microscopic study of the opsin distribution in the photoreceptor cell of Drosophila melanogaster. J Electron Microsc (Tokyo) 1991;40:187–192. [PubMed] [Google Scholar]
- 51.Suzuki E, Hirosawa K. Immunolocalization of a Drosophila phosphatidylinositol transfer protein (rdgB) in normal and rdgA mutant photoreceptor cells with special reference to the subrhabdomeric cisternae. J Electron Microsc (Tokyo) 1994;43:183–189. [PubMed] [Google Scholar]
- 52.Vihtelic TS, Hyde DR, O’Tousa JE. Isolation and characterization of the Drosophila retinal degeneration B (rdgB) gene. Genetics. 1991;127:761–768. doi: 10.1093/genetics/127.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vihtelic TS, Goebl M, Milligan S, O’Tousa JE, Hyde DR. Localization of Drosophila retinal degeneration B, a membrane-associated phosphatidylinositol transfer protein. J Cell Biol. 1993;122:1013–1022. doi: 10.1083/jcb.122.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walz B. Calcium-sequestering smooth endoplasmic reticulum in retinula-cells of the blowfly. J Ultrastruct Res. 1982;81:240–248. doi: 10.1016/s0022-5320(82)90079-x. [DOI] [PubMed] [Google Scholar]
- 55.Woodard C, Alcorta E, Carlson J. The rdgB gene of Drosophila: a link between vision and olfaction. J Neurogenet. 1992;8:17–31. doi: 10.3109/01677069209167269. [DOI] [PubMed] [Google Scholar]
- 56.Zuker CS. The biology of vision in Drosophila. Proc Natl Acad Sci USA. 1996;93:571–576. doi: 10.1073/pnas.93.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]