Abstract
Zone-to-zone projection of olfactory and vomeronasal sensory axons underlies the topographic and functional mapping of chemoreceptor expression zones of the sensory epithelia onto zonally arranged glomeruli in the main and accessory olfactory bulbs. Here we identified OCAM (R4B12 antigen), an axonal surface glycoprotein expressed by subsets of both olfactory and vomeronasal axons in a zone-specific manner. OCAM is a novel homophilic adhesion molecule belonging to the immunoglobulin superfamily with striking structural homology to neural cell adhesion molecule. In both the main and accessory olfactory systems, OCAM mRNA is expressed by sensory neurons in restricted chemoreceptor expression zones, and OCAM protein-expressing axons project to the glomeruli in the corresponding zones of the main and accessory bulbs. OCAM protein is expressed on subsets of growing sensory axons in explant cultures even in the absence of the target bulb. These results demonstrate a precisely coordinated zonal expression of chemoreceptors and OCAM and suggest that OCAM may play important roles in selective fasciculation and zone-to-zone projection of the primary olfactory axons.
Keywords: OCAM, cell adhesion molecule, immunoglobulin superfamily, NCAM, olfactory system, odorant receptors, primary olfactory axons, zone-to-zone projection
The mammalian olfactory system possesses sophisticated molecular and cellular mechanisms underlying discrimination among odor molecules and perception of olfactory images of objects (Shepherd, 1994; Mori and Yoshihara, 1995; Buck, 1996). Odor ligands are recognized by odorant receptors on the cilia of sensory neurons in the olfactory epithelium (OE). To discriminate among odors, the central olfactory nervous system needs to determine which of the numerous odorant receptors are activated by the odor molecules. One of the cellular bases for this task is that individual olfactory sensory neurons express a single or only a few odorant receptor genes among as many as 1000 repertoires (Buck and Axel, 1991; Nef et al., 1992;Strotmann et al., 1992; Chess et al., 1994). Accordingly, to distinguish which receptors are activated by the odor molecules, the central olfactory system only needs to know which sensory neurons have been activated. Another important cellular basis lies in the pattern of olfactory axon connectivity with the main olfactory bulb (MOB); all of the sensory neurons expressing a given odorant receptor project and converge their axons onto two glomeruli among the 1500–3000 within the MOB (Ressler et al., 1994; Vassar et al., 1994; Mombaerts et al., 1996). Thus, individual glomeruli seem to constitute functional units, each representing one type or a few types of odorant receptors. This axonal connection pattern underlies the response specificity of the second-order neurons in the MOB to odor molecules. Individual mitral/tufted cells are activated only by a range of odor molecules with a similar chemical structure, which can bind to a given odorant receptor (Imamura et al., 1992; Mori et al., 1992; Katoh et al., 1993;Mori, 1995; Mori and Yoshihara, 1995).
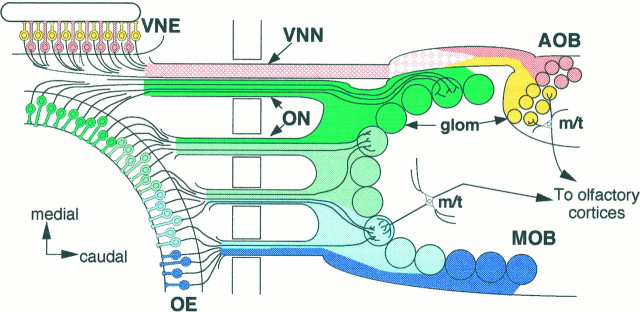
In situ hybridization studies reveal yet another principle of odorant receptor expression. Sensory neurons expressing a given odorant receptor gene are distributed sparsely within one specific circumscribed zone out of the four zones in the OE (Ressler et al., 1993; Vassar et al., 1993; Strotmann et al., 1994). In addition, it has been hypothesized that olfactory sensory neurons within a given OE zone project their axons to glomeruli in a corresponding zone of the MOB: a zone-to-zone projection (Fig. 1) (Mori and Yoshihara, 1995).
Fig. 1.
Zonal organization of the main and accessory olfactory systems. Schematic dorsal view of the olfactory systems. Olfactory sensory neurons in the four zones (zone I, dark green; zone II, light green; zone III,light blue; zone IV, dark blue) of the olfactory epithelium (OE) project their axons to four corresponding zones of the main olfactory bulb (MOB). Similarly, vomeronasal sensory neurons in the two zones (apical,yellow; basal, pink) of the vomeronasal epithelium (VNE) project their axons to two zones (rostral, yellow; caudal, pink) of the accessory olfactory bulb (AOB). Note that olfactory axons from different zones already are segregated spatially at the exit points of the OE, whereas the vomeronasal axons from the different zones are intermingled in the vicinity of theVNE and become segregated gradually near theAOB. ON, Olfactory nerves;VNN, vomeronasal nerves; glom, glomeruli;m/t, mitral and tufted cells.
A similar but distinct type of zone-to-zone projection has been suggested in the accessory olfactory system. Studies on the expression of pheromone receptors (Dulac and Axel, 1995) and G-proteins (Halpern et al., 1995; Berghard and Buck, 1996) reveal that two zones (apical and basal) are stacked in the vomeronasal epithelium (VNE). Vomeronasal neurons in the apical zone project their axons to glomeruli in the rostral zone of the accessory olfactory bulb (AOB), whereas sensory neurons in the basal zone send their axons to glomeruli in the caudal zone (Fig. 1).
Monoclonal antibody (mAb) R4B12 recognizes a membrane protein expressed by subsets of rabbit primary olfactory axons in both the main and accessory olfactory systems (Fujita et al., 1985; Imamura et al., 1985;Mori et al., 1985, 1987; Mori, 1993; Yoshihara et al., 1993). mAb R4B12 labels olfactory axons that originate from sensory neurons located in a ventrolateral region, which may correspond to the odorant receptor expression zones II, III, and IV, whereas it does not bind olfactory axons in a dorsomedial region presumably corresponding to the zone I (Mori and Yoshihara, 1995). We report here the isolation of cDNAs encoding R4B12 antigen and its intimate relationship with neural cell adhesion molecule (NCAM). Because of the characteristic expression by olfactory axons and remarkable structural similarity to NCAM, we designate R4B12 antigen as OCAM. We demonstrate here that the OCAM expression region corresponds precisely with odorant receptor expression zones.
MATERIALS AND METHODS
Purification of OCAM and peptide sequencing. mAb R4B12 (Fujita et al., 1985) was prepared from ascites fluid of hybridoma-injected mice (BALB/c), purified with an EZ-Sep kit (Pharmacia, Piscataway, NJ), and coupled to CNBr-activated Sepharose 4B (Pharmacia). The large form of OCAM was purified from CHAPS-solubilized membrane of rabbit brain by affinity chromatography with a mAb R4B12–Sepharose 4B column. The purified material was subjected to reduced carboxymethylation and digested with lysyl-endopeptidase (Boehringer Mannheim, Indianapolis, IN). The peptide fragments were purified by reverse-phase HPLC with a Cosmosil 5C18 column (Nacalai Tesque). Selected peptides were subjected to microsequencing on a gas phase sequencer (Applied Biosystems, Foster City, CA).
cDNA cloning of OCAM. Two degenerate oligonucleotides (5′-ga[ag]at[act]gcicciaci[at][cg]iga[ct]aa[ct]ga[ct]tt[ct]gg-3′ and 5′-tt[ct]tcna[ag]cca[ct]tg[ag]tc[ct]tc-3′) corresponding to the amino acid sequences of OCAM peptide EIAPTSDNDFG and EDQWLEK, respectively, were synthesized and used in a PCR with rabbit, rat, and mouse brain cDNA as templates. The PCR product (545 bp) was cloned directly into pCRII vector (Invitrogen, San Diego, CA) and sequenced by the dideoxy chain termination method (Sanger et al., 1977) with a Sequenase kit (Amersham Japan, Tokyo, Japan). An adult mouse OE λgt10 cDNA library was constructed and screened with the mouse OCAM PCR product labeled with 32P by random priming (Boehringer Mannheim). Six positive clones of 3.2 × 105 recombinants screened were obtained and subcloned into the SalI site of pBluescript II SK+ (Stratagene, La Jolla, CA). Restriction mapping and partial sequencing revealed that they were categorized into two groups with partially identical sequences. The complete nucleotide sequence was determined for both strands of the two clones 102 and 104.
Northern blot analysis. Total RNA was isolated from mouse (ddY) tissue by the acid guanidinium thiocyanate-phenol-chloroform method (Chomczynski and Sacchi, 1987), fractionated in 1% agarose–formaldehyde gels, and transferred to nylon membranes (Hybond N+, Amersham Japan). Three cDNA fragments of OCAM were used as probes: 480 nucleotides in the first and second Ig-like domains to detect both OCAM-glycosylphosphatidylinositol (GPI) and OCAM-transmembrane (TM), 549 nucleotides in the GPI-anchoring domain and 3′-noncoding region to detect OCAM-GPI specifically, and 822 nucleotides in the cytoplasmic region to detect OCAM-TM specifically. The probes were labeled by random priming. Hybridization and washing were performed as described previously (Yoshihara et al., 1994a).
Western blot analysis. Polyclonal antibodies against mouse OCAM were produced by immunizing a rabbit with the recombinant OCAM/Fc fusion protein. The antiserum was used for Western blot analysis (3000× dilution) and immunohistochemistry (1000× dilution). Monoclonal anti-NCAM antibody (clone H28-123-16, rat IgG2a) was purchased from Boehringer Mannheim. Brains from embryonic days (E) 13 and 18, postnatal days (P) 1, 7, and 21, and adult mice were homogenized and subjected to Western blot analysis as described previously (Yoshihara et al., 1993).
Cell aggregation assay. L929 mouse fibroblast cells were grown in DMEM with 10% fetal bovine serum. A linearized plasmid (pEF–OCAM-TM) containing a full-length OCAM-TM cDNA subcloned into the mammalian expression vector pEF–BOS (Mizushima and Nagata, 1990) was cotransfected with pSTneoB by using Lipofectamine (BRL, Bethesda, MD) according to the manufacturer’s procedure. Stable transfectants were selected in medium containing 400 μg/ml G418. Cells were screened for OCAM-TM expression by Western blotting, and a cell line with the highest expression of OCAM-TM was chosen and used for the aggregation assay. L1-expressing L929 cells (Miura et al., 1992) were kindly provided by Dr. H. Asou (Tokyo Metropolitan Institute of Gerontology, Tokyo, Japan). The cell aggregation assay was performed essentially as described (Miura et al., 1992; Yoshihara et al., 1994b). Briefly, cells in monolayer culture were dispersed into single cells at 1 × 106 cells per milliliter in Ca2+- and Mg2+-free HBSS. The cell suspension was incubated in a polystyrene tube at 37°C without rotation, and aliquots were taken at the indicated times after mixing by several gentle inversions. The number of cell aggregates was counted in a hemocytometer.
Preparation of OCAM/Fc, NCAM/Fc, and OCAM/AP proteins. For preparation of recombinant soluble OCAM/Fc protein, the expression plasmid was constructed with the extracellular region of mouse OCAM cDNA and the Fc region of the human IgG1 gene (Nishimura et al., 1987) in pEF–BOS. The OCAM/Fc chimeric protein was produced in COS7 cells by the standard DEAE–dextran transfection method and purified to near homogeneity from culture supernatants with a protein A-Sepharose 4FF column (Pharmacia). NCAM/Fc was produced in a similar manner. Levels of OCAM/Fc and NCAM/Fc fusion proteins in the supernatants were in the range of 0.5–0.8 μg/ml. A fusion protein OCAM/AP consisting of the extracellular region of mouse OCAM and the human placental alkaline phosphatase was prepared with an AP–tag-1 expression vector as described previously (Cheng and Flanagan, 1994). COS7 supernatant was concentrated with Centriprep 30 (Amicon, Beverly, MA) and used for the protein binding assay.
OCAM-AP binding assay. Xenobind plastic plates (Xenopore) were coated with 1 μg/well goat anti-human Fc IgG (Sigma, St. Louis, MO) in PBS overnight at 4°C, blocked with 0.4% bovine serum albumin in PBS for 2 hr at room temperature, and then coated with OCAM/Fc, NCAM/Fc, or Fc in PBS for at least 2 hr at room temperature. The concentrated COS7 supernatant, which contains OCAM/AP, was added to the Fc chimeric protein-coated wells and incubated for 60 min at 37°C. OCAM/AP that remained bound after three washes was visualized by AP reaction with p-nitrophenyl phosphate as a substrate, and the absorbance at 405 nm was measured after 24 hr.
In situ hybridization. Riboprobes (300–500 nucleotides in length) for OCAM, NCAM, olfactory marker protein (OMP), and odorant receptors R15, R16, R36, and R38 were prepared with 35S-UTP (Amersham Japan) and an RNA transcription kit (Stratagene). All steps of the in situ hybridization reactions were performed essentially according to Simmons et al. (1989). Frontal sections (15 μm) from paraformaldehyde-perfused P14 mouse nose were cut with a cryostat and mounted on poly-l-lysine-coated glass slides (Matsunami). The sections were dried, treated with proteinase K (10 μg/ml at 37°C for 30 min), acetylated, dehydrated, and air-dried. The sections were hybridized overnight at 56°C in a humidified chamber with 150–200 μl of hybridization solution per section (hybridization solution consists of 50% formamide, 20 mmTris-HCl, pH 7.6, 1 mm EDTA, 0.3 m NaCl, 0.1m dithiothreitol, 0.5 mg/ml yeast tRNA, 1× Denhardt’s solution, and 10% dextran sulfate) containing 1 × 106 cpm/ml 35S-labeled cRNA probe. After hybridization, sections were washed in 4× SSC, treated with RNase A (10 μg/ml at 37°C for 30 min), washed in 0.05× SSC, dehydrated with ethanol, and exposed to βmax x-ray film (Amersham Japan). After autoradiography, slides were dipped in NTB-2 emulsion, exposed, developed in Kodak D-19, fixed with Ren Fix, and counterstained with cresyl violet.
Immunohistochemistry. Immunohistochemical analysis was performed essentially as described (Mori et al., 1987). Mice were anesthetized with pentobarbital sodium and perfused with PBS, followed by 3% paraformaldehyde in phosphate buffer. The OB was removed, post-fixed overnight at 4°C in the same fixative, and kept in cold PBS containing 30% sucrose. Parasagittal sections (40 μm) were cut with a sliding microtome. The sections were blocked with PBS containing 10% normal goat serum, incubated with primary antibodies overnight at room temperature, washed, and incubated with FITC-labeled (Cappel, West Chester, PA) or Cy3-labeled (Jackson Laboratories, Bar Harbor, ME) secondary antibodies. The primary antibodies used were as follows: anti-mouse OCAM/Fc (rabbit antiserum, 1000×), anti-NCAM (clone OB11, mouse IgG, 100×; Sigma), and anti-Goα (rabbit IgG, 1 μg/ml; MBL). The sections were examined with a confocal laser scanning microscopy system (Bio-Rad MRC-600, Hercules, CA) equipped with a Zeiss Axiophot Fl microscope (Oberkochen, Germany).
Explant culture of olfactory and vomeronasal epithelia. E10, E11, or E13 BALB/c mouse noses were removed from the head, digested with 1% trypsin in TBS/EDTA for 1 hr on ice, and treated with 10% FBS to stop the enzymatic reaction. Olfactory and vomeronasal epithelia were removed from the surrounding mesenchymal tissue, cut into small fragments (<500 μm in diameter), and cultured for 3–4 d on coverslips coated with 100 μg/ml poly-l-lysine (Sigma) for 1 hr and then 100 μg/ml laminin (Koken) for 1–4 hr. The culture medium was DMEM/F12 supplemented with 5 μg/ml insulin, 100 μg/ml transferrin, 20 nm progesterone, 20 nmhydrocortisone, 100 μm putrescine, and 20 nmsodium selenite. After fixation with 4% paraformaldehyde, cultured fragments were immunostained with both anti-OCAM/Fc and anti-NCAM antibodies and photographed with a confocal laser scanning microscopy.
RESULTS
Purification and cDNA cloning of OCAM
The large form of OCAM was isolated from a CHAPS extract of nine rabbit brains by immunoaffinity chromatography with a mAb R4B12-coupled Sepharose 4B column. Western blot analysis of the purified material revealed that OCAM bears an HNK-1 carbohydrate epitope, which is shared by a number of cell adhesion molecules in the nervous system (data not shown) (Yoshihara et al., 1991). The purified OCAM (∼80 μg) was digested with Lys-endopeptidase, and the resulting peptides were separated by reverse-phase HPLC and subjected to amino acid sequencing. The sequences of nine different peptides were determined.
Several degenerate oligonucleotides were synthesized on the basis of the peptide sequences and used as primers for PCR with cDNA from rabbit, mouse, and rat brains. One pair of the oligonucleotides yielded a specific PCR product of 545 bp in samples of all three species. The deduced amino acid sequences of rabbit, mouse, and rat PCR products were highly homologous (>93% identity in all of the pairs) and probably represent species homologs of the same gene. The PCR product contained a part of the immunoglobulin (Ig)-like domain and one and one-half of the fibronectin type III (FNIII)-like domain. We used mouse OCAM for detailed analyses. To obtain a full-length sequence, we used the mouse PCR product as a probe to screen a λgt10 cDNA library derived from mouse OE. Six positive clones (101, 102, 103, 104, 106, and 107) were identified and classified into two groups with partially different restriction maps (Fig.2A).
Fig. 2.
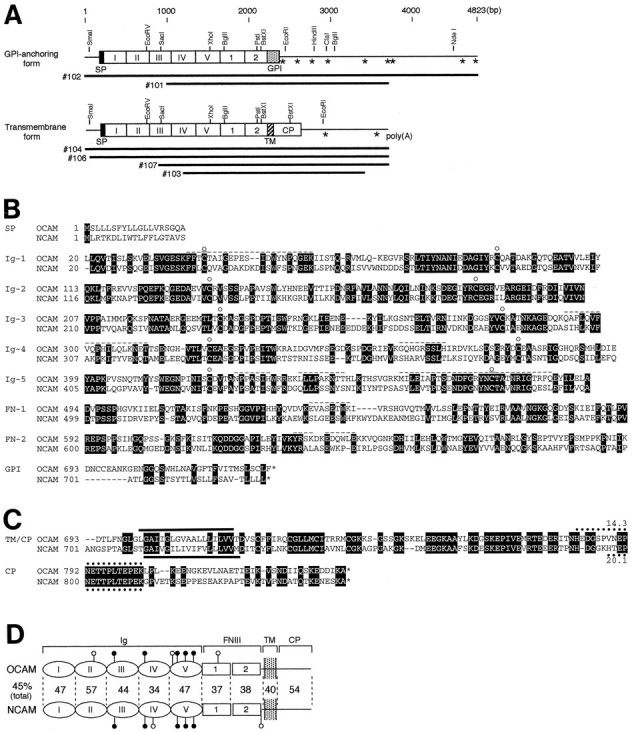
Structure of OCAM in comparison with NCAM.A, Organization of OCAM cDNAs and alignment of clones. OCAM exists in two forms with different membrane attachment modes: a GPI-anchoring form and a transmembrane form. Upper barswith columns show the overall structure deduced from overlapping cDNA clones that are given below. Restriction endonuclease sites are shown above thebars. Columns represent putative protein-coding regions. The five Ig-like and two FNIII-like domains are indicated by Roman and Arabic numerals, respectively. Positions of the putative mRNA-destabilizing signals (ATTTA motifs) in the 3′ noncoding regions are indicated byasterisks. SP, Signal peptide; GPI, GPI-anchoring region; TM, transmembrane region.B, Amino acid sequence alignment of the extracellular and GPI-anchoring regions of OCAM and NCAM. The residues conserved between OCAM and NCAM are highlighted. Theopen circles denote positions of the highly conserved Cys residues in the Ig-like domains. The nine peptide sequences obtained from purified OCAM are shown by dashed overlines. C, Amino acid sequence alignment of the transmembrane and cytoplasmic regions of OCAM and NCAM. The putative transmembrane segments are indicated by solid bars. The PEST sequences are shown by dotted lines with PEST scores (Rogers et al., 1986). The nucleotide sequences of mouse OCAM-GPI and OCAM-TM have been deposited into GenBank with accession numbers AF001286 and AF001287, respectively.D, Proposed model of the domain structure of the transmembrane forms of OCAM and NCAM. The Roman andArabic numerals indicate the Ig-like and FNIII-like domains, respectively. The putative Asn-linked glycosylation sites are shown by lollipop symbols (closed, conserved; open, nonconserved). The sequence conservation (percentage of amino acid identity) between OCAM and NCAM is shown between individual corresponding domains.
OCAM is a new member of the NCAM family
An analysis of the OCAM cDNA clones indicated that they encode two related proteins with different modes of plasma membrane anchorage. Two predicted proteins shared 692 identical amino acids at the N terminus, which contains a putative signal peptide (19 amino acids) and a long extracellular region with five Ig-like domains and two FNIII-like domains (Fig. 2A,B). There were eight potential Asn-linked glycosylation sites in the extracellular region of OCAM (Fig. 2D). All of the peptide sequences determined by microsequencing appeared in the deduced amino acid sequences (Fig.2B, dashed overlines).
The OCAM cDNA clones diverged in sequence downstream of the second FNIII-like domain in two ways. Clones 101 and 102 terminated in a short stretch of hydrophobic amino acids characteristic of proteins attached to the plasma membrane via a GPI linkage (Fig. 2A,B). On the other hand, clones 103, 104, 106, and 107 encoded a protein with a TM domain of a stretch of 20 hydrophobic amino acids and a cytoplasmic region of 119 amino acids at the C terminus (Fig.2A,C). The presence of two forms, OCAM-GPI and OCAM-TM, was in good agreement with the result of Western analysis that mAb R4B12 recognized two protein bands, one of which was released from the membrane by the GPI-specific phospholipase C (Yoshihara et al., 1993).
Within the analyzed 3′ untranslated region (2.5 kb) of OCAM-GPI, there were nine ATTTA motifs (Fig. 2A, asterisks), which are believed to confer instability to the mRNA (Shaw and Kamen, 1986;Malter, 1989). In contrast, only two ATTTA motifs were found in the 3′ untranslated region (0.8 kb) of OCAM-TM. OCAM-TM contained a conserved PEST sequence in the cytoplasmic region (Fig. 2C). The PEST sequence is found in proteins with high turnover rates and is postulated to be involved in protein degradation (Rogers et al., 1986;Rechsteiner, 1988). Thus, turnover of OCAM-GPI and OCAM-TM in cells may be rapid via a regulated degradation at mRNA and protein levels, respectively.
In the Ig superfamily OCAM is classified into the NCAM family, which includes vertebrate NCAM (Cunningham et al., 1987; Santoni et al., 1989), Aplysia apCAM (Mayford et al., 1992), and insect fasciclin II (Fas II) (Harrelson and Goodman, 1988; Grenningloh et al., 1991). All four molecules have a similar domain organization and occur in both GPI and TM forms. In particular, the greatest similarity was found between OCAM and NCAM, with overall amino acid identity of 45% (Fig. 2D). Both OCAM and NCAM showed similar degrees of identity (26–27%) with both apCAM and Fas II (Table1).
Table 1.
Amino acid sequence identity of OCAM and NCAM with apCAM and fasciclin II
| Total | Ig-1 | Ig-2 | Ig-3 | Ig-4 | Ig-5 | FN-1 | FN-2 | CP | |
|---|---|---|---|---|---|---|---|---|---|
| OCAM versus apCAM | 26 | 34 | 32 | 27 | 24 | 26 | 25 | 22 | 24 |
| NCAM versus apCAM | 27 | 25 | 25 | 30 | 30 | 23 | 25 | 23 | 32 |
| OCAM versus Fas II | 26 | 22 | 34 | 28 | 30 | 29 | 28 | 15 | 24 |
| NCAM versus Fas II | 26 | 22 | 33 | 27 | 28 | 29 | 27 | 16 | 24 |
Schwob and Gottlieb (1986) have reported a similar antigen molecule RB8, which is expressed by subsets of rat olfactory axons projecting to a ventrolateral area of the MOB. The reported N-terminal 13 amino acid sequence of purified rat RB8 antigen (Schwob and Gottlieb, 1988) completely matched that of mouse OCAM, indicating that RB8 antigen is a rat homolog of mouse OCAM.
Northern and Western blot analyses of OCAM
Northern blot analysis was performed to survey the distribution of OCAM mRNA in adult mouse tissue (Fig.3A). By using a probe for the Ig-like domains of OCAM (which recognizes both GPI and TM forms), we detected OCAM transcripts not only in the OE but also in the brain and retina. However, no signal was observed in the peripheral tissues such as lung, heart, thymus, liver, spleen, kidney, testis, and skeletal muscle. Distribution of GPI and TM forms of OCAM was compared by Northern analysis, using GPI- and TM-specific probes (Fig.3B). The OCAM-GPI-specific probe recognized a single band of 7.8 kb, which was expressed predominantly by the OE. In contrast, the OCAM-TM-specific probe detected two major bands of 7.8 and 3.6 kb, with highest expression in the brain.
Fig. 3.
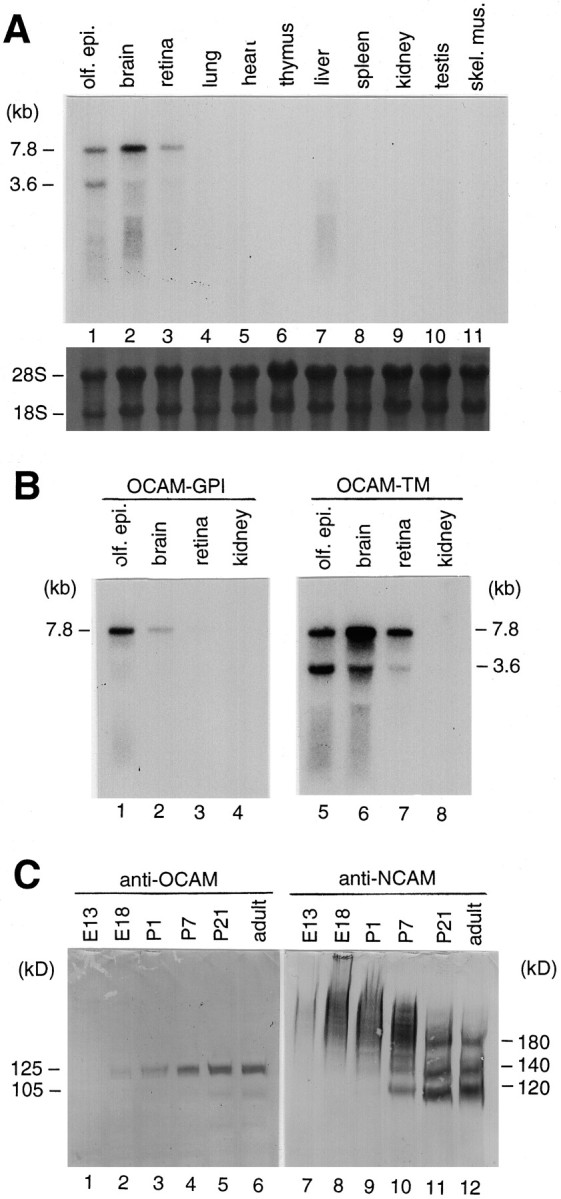
Northern and Western blot analyses of OCAM.A, Tissue distribution of OCAM mRNA expression. Total RNAs from a variety of tissues of adult mouse were analyzed as described in Materials and Methods. The upper panelshows an autoradiogram of the blot probed with OCAM Ig-like domain cDNA, which recognizes both GPI and TM forms. The lower panel shows the same blot stained with methylene blue. The size of OCAM transcripts estimated from the mobility of RNA standards is indicated on the left in kilobases. olf.epi., Olfactory epithelium; skel. mus., skeletal muscle. B, Identification of OCAM-GPI and OCAM-TM transcripts, using specific probes. Northern blot analysis was performed with isoform-specific probes for OCAM-GPI (left panel) and OCAM-TM (right panel). Note that the OCAM-GPI transcript (7.8 kb) was expressed predominantly in the olfactory epithelium, whereas the OCAM-TM transcripts (7.8 and 3.6 kb) were detected in all of the neural tissues tested.C, Ontogeny of OCAM protein expression in comparison with NCAM. Total proteins from mouse brains of different ages (from E13 to adult) were subjected to Western blot analysis with rabbit anti-OCAM/Fc antibody (left panel) and rat anti-NCAM antibody (right panel). NCAM inE18, P1, and P7 samples appeared as slowly migrating diffuse bands because of polysialylation. Note that OCAM was detected as nonpolysialylated sharp bands.
Using an antibody against recombinant OCAM/Fc fusion protein (see below), we examined the ontogeny of OCAM expression in mouse brain by Western blot analysis in comparison with NCAM (Fig.3C). OCAM-TM in the mouse brain was expressed more highly than OCAM-GPI, as previously reported for the rabbit brain (Yoshihara et al., 1993). OCAM protein was detectable at E18, increased thereafter, and reached maximal levels at P21. OCAM was detected as two sharp protein bands. In contrast, NCAM was detected as diffuse protein bands representing a highly polysialylated form in the embryonic and perinatal brains and gradually shifted to sharp bands representing three isoforms of 180, 140, and 120 kDa (Rutishauser, 1996). This result suggests that polysialic acid is absent on the carbohydrate moieties of OCAM.
OCAM is a homophilic adhesion molecule
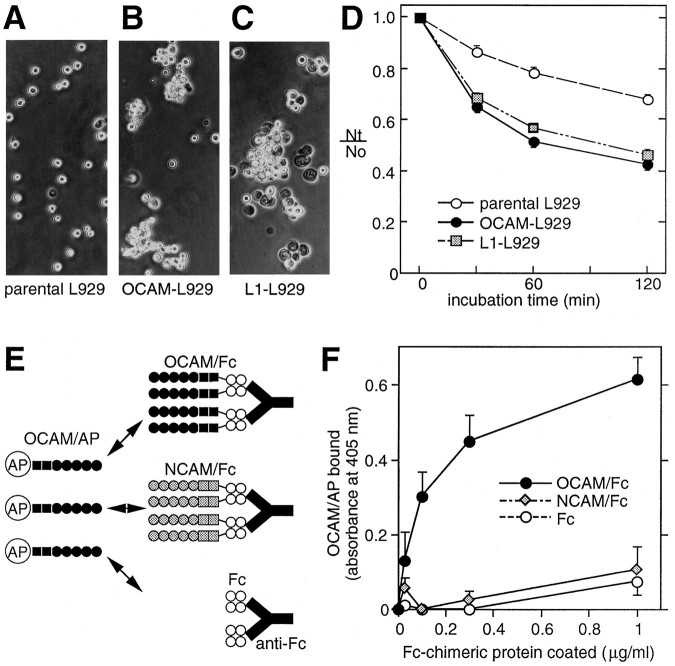
On the basis of the striking structural similarity with NCAM, we speculated that OCAM would function as an adhesion molecule in the nervous system. To clarify whether OCAM can bind in a homophilic manner, as several Ig superfamily molecules do (Brümmendorf and Rathjen, 1994), we performed two experiments. First, we stably transfected mouse OCAM-TM cDNA into mouse fibroblast L929 cells, which normally have little adhesive activity and do not express OCAM. Adhesive properties of OCAM-expressing cells were examined by a static cell aggregation assay, with parental L929 cells as a negative control and L1-expressing L929 cells as a positive control. L1 is another member of the Ig superfamily, which mediates homophilic adhesion in this assay system (Miura et al., 1992). As shown in Figure4A–D, both the OCAM- and L1-expressing L929 cells showed a marked aggregation in contrast to the parental L929 cells, implicating a homophilic adhesive activity of OCAM.
Fig. 4.
Homophilic binding activity of OCAM assessed by cell aggregation assay (A–D) and protein binding assay (E, F). A–C, Phase-contrast micrographs. Parental (A), OCAM-expressing (B), or L1-expressing (C) L929 cells were dissociated in EDTA medium, incubated at 37°C for 30 min, and then photographed. D, Kinetics of cell aggregation. Values are the mean ± SEM for three independent experiments. Nt andN0 are the total number of particles at incubation times t and 0, respectively. The extent of cell aggregation is represented by the indexNt/N0.E, Schematic diagram of protein binding assay. OCAM/AP was added to wells precoated with OCAM/Fc, NCAM/Fc, or Fc via anti- human Fc antibody and incubated for 60 min at 37°C. Bound OCAM/AP was visualized by a chromogenic reaction. F, Specific binding of OCAM/AP to OCAM/Fc. OCAM/AP was added to wells precoated with the indicated concentration of recombinant chimeric proteins. Values are the mean ± SEM for three independent experiments. Note that OCAM/AP (filled circles) specifically bound to OCAM/Fc, but not to NCAM/Fc (shaded diamonds) and Fc (open circles).
In the second experiment we directly measured the homophilic binding of OCAM by using recombinant soluble proteins. OCAM/AP is a chimeric protein, consisting of the extracellular region of OCAM fused to human placental alkaline phosphatase having an enzyme activity that can be traced quantitatively by a simple chromogenic assay (Cheng and Flanagan, 1994). OCAM/Fc comprises the extracellular region of OCAM fused to the Fc region of human IgG1. NCAM/Fc also was prepared to examine the possibility of heterophilic interaction between OCAM and NCAM. OCAM/AP was added to wells precoated with Fc chimeric proteins via anti-human IgG polyclonal antibod- ies (Fig.4E). As shown in Figure 4F, OCAM/AP specifically bound to OCAM/Fc, but not to NCAM/Fc or Fc. The binding of OCAM/AP was dependent on the amount of precoated OCAM/Fc. This result strengthens the idea that OCAM is a homophilic adhesion molecule and also suggests the absence of heterophilic binding between OCAM and NCAM.
OCAM expression in the main olfactory system
To determine whether the OCAM expression region corresponds to the odorant receptor expression zones in the OE, we investigated localization of OCAM mRNA and protein in the main olfactory system byin situ hybridization and immunohistochemistry. The mammalian OE consists of at least four spatially segregated zones that are defined by the expression of odorant receptor mRNAs (Ressler et al., 1993; Vassar et al., 1993). Individual types of odorant receptor mRNA were expressed by olfactory sensory neurons that were distributed selectively within one zone among the four circumscribed zones (Fig.5F–I). All four zones expressed NCAM mRNA (Fig. 5B,E) as well as olfactory marker protein mRNA (Fig. 5C). In contrast, OCAM mRNA was expressed by sensory neurons within the three ventrolateral zones [zone II, III, and IV inRessler et al. (1993); zone 3, 2, and 1 in Mombaerts et al. (1996)], but not in the most dorsomedial zone [zone I in Ressler et al. (1993); zone 4 in Mombaerts et al. (1996)] (Fig. 5A). [In this paper we adopt the zone numbering scheme of Ressler et al. (1993) in Roman numerals I, II, III, and IV from dorsomedial to ventrolateral.] At higher magnification OCAM mRNA expression revealed a clear boundary between zone II (OCAM-positive) and zone I (OCAM-negative) neurons (Fig. 5D, arrowheads). The precise correspondence of the OCAM expression region to zones II, III, and IV was observed in all sections examined along the rostrocaudal axis of the OE (data not shown).
Fig. 5.
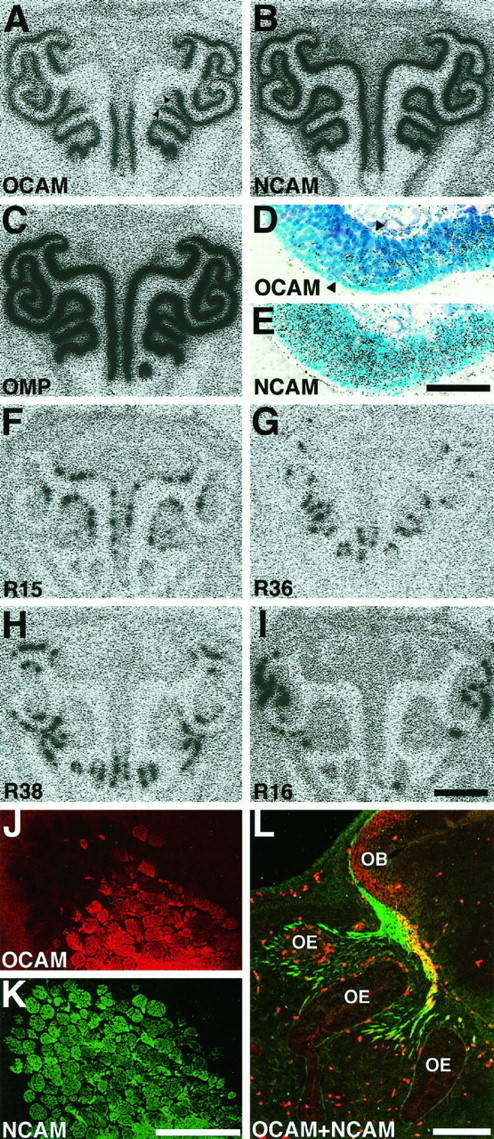
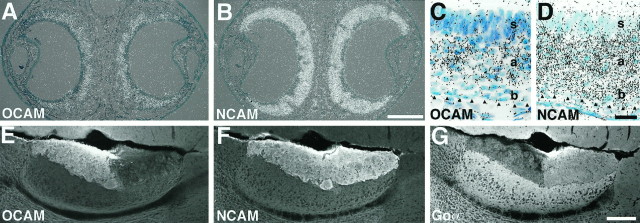
Zone-specific expression of OCAM in the main olfactory system. A–I, Autoradiographs (A–C and F–I) and bright-field micrographs (D, E) of coronal sections of P14 OE hybridized with OCAM (A, D), NCAM (B, E), olfactory marker protein (OMP, C), and four odorant receptor (F–I) cRNA probes. Dorsal is at the top. The four circumscribed zones of the OE are visualized by differential expression of the representative odorant receptor mRNAs [R15 in zone I (F); R36 in zone II (G); R38 in zone III (H); R16 in zone IV (I)]. Note that OCAM mRNA is expressed in the three ventrolateral zones (zone II, III, and IV), but not in the most dorsomedial zone (zone I), whereas NCAM and OMP mRNAs are expressed in all four zones. The region around the boundary of OCAM-positive and OCAM-negative zones (arrowheads in A) is magnified in D and E. Both OCAM (D) and NCAM (E) mRNAs are expressed by olfactory sensory neurons in the middle layer of the OE, but not by supporting cells in the most superficial layer or stem cells in the basal layer. J, K, Micrographs of a parasagittal section at the medial surface of adult MOB double-stained with anti-OCAM (J) and anti-NCAM (K) antibodies. Dorsal is at thetop; rostral is at the left. Although NCAM is expressed by all glomeruli at the terminal of olfactory axons, OCAM is expressed by caudoventral glomeruli, but not by rostrodorsal ones, showing the zone-to-zone projection of olfactory axons to the MOB. L, Micrograph of a parasagittal section of E13 mouse double-stained with anti-OCAM (red) and anti-NCAM (green) antibodies. Dorsal is at thetop; rostral is at the left. Note that expression of OCAM is already restricted to olfactory axons projecting from the ventral region of the OE (shown inyellow), whereas NCAM is expressed by all olfactory axons (shown in green and yellow). Blood cells show nonspecific autofluorescence. Scale bars: 1 mm forA–C and F–I; 50 μm for D, E; 500 μm for J, K; 200 μm forL.
Next, parasagittal sections of adult mouse MOB were double-labeled with anti-OCAM (Fig. 5J) and anti-NCAM (Fig.5K) antibodies. The glomerular layer appeared tessellated in a parasagittal section of the medial part of the MOB. As shown in Figure 5J, strong expression of OCAM protein was observed in the olfactory nerve layer and in the ventrocaudal glomeruli, but not in the dorsorostral glomeruli. A clear boundary between the glomeruli was apparent, reflecting the segregated termination of OCAM-positive and OCAM-negative axons originating from olfactory sensory neurons in zone II (OCAM-positive) and zone I (OCAM-negative). In contrast, NCAM was present in all of the glomeruli (Fig. 5K). Thus, the zonal organization defined in the OE was preserved also in the MOB.
The distribution of OCAM and NCAM proteins in the developing olfactory system was examined in a parasagittal section of E13 mouse when olfactory axons normally just reach the OB at the tip of the telencephalon (Fig. 5L). Axons projecting to the ventral region of the OB expressed both OCAM and NCAM (Fig. 5L, yellow), whereas axons projecting to the rostrodorsal region were OCAM-negative but NCAM-positive (Fig.5L, green). OCAM already was expressed by subsets of olfactory axons in a region-specific manner at this early developmental stage, whereas NCAM was expressed by all of the olfactory axons.
OCAM expression in the accessory olfactory system
We investigated whether OCAM is expressed in a zone-specific manner in the accessory olfactory system also. The VNE consists of two zones, apical and basal, that are defined by complementary expression of two G-protein α subunits, Gi2α and Goα (Halpern et al., 1995;Berghard and Buck, 1996). Vomeronasal neurons in the apical and basal zones projected their axons to two spatially segregated zones (rostral and caudal, respectively) in the AOB. OCAM mRNA was expressed specifically by vomeronasal neurons in the apical zone, but not in the basal zone (Fig. 6A,C), whereas NCAM mRNA was present in all neurons of the VNE (Fig.6B,D). Corresponding to the restricted localization of OCAM mRNA in the vomeronasal apical zone, the OCAM protein product was detected in vomeronasal nerve fibers terminating in glomeruli in the rostral zone of the AOB (Fig. 6E), complementary to the expression of Goα in the caudal zone (Fig.6G). NCAM was detected in all vomeronasal nerve fibers and their terminals (Fig. 6F).
Fig. 6.
Zone-specific expression of OCAM in the accessory olfactory system. In situ hybridization analysis of coronal sections of the VNE from a P14 mouse (A–D) and immunohistochemical analysis of parasagittal sections of adult AOB (E–G).A, B, Superimposed images of bright-field and dark-field micrographs showing expression of OCAM (A) and NCAM (B) mRNAs in the crescent-shaped VNE. Dorsal is at the top. C, D, High-power bright-field micrographs of the center region of the VNE. The lumen is located at the top. The basal lamina is shown byarrowheads. Note that OCAM is expressed in the apical zone (a) of the VNE, but not in the basal zone (b) or supporting cell layer (s) (C), whereas NCAM is expressed in both the apical and basal zones of the VNE (D).E–G, Micrographs showing expression of OCAM (E), NCAM (F), and Goα (G) proteins in the adult mouse AOB. E and F are different images from a double-stained section. G is a section adjacent toE and F. Dorsal is at thetop; rostral is at the left. OCAM is expressed by a subset of vomeronasal axons that project into glomeruli in the rostral zone of the AOB (E), whereas NCAM is expressed by all vomeronasal axons (F). Complementary to OCAM, Goα is expressed in the caudal zone of the AOB (G). Scale bars: 200 μm forA, B; 20 μm for C, D; 200 μm forE–G.
Zonal expression of OCAM by growing olfactory axons cultured without target MOB
Finally, we investigated how the zonal pattern of OCAM expression by olfactory axons is established during development. One possibility is that the zonal organization is determined by some retrograde signals from the target MOB after the axons innervate it. The other possibility is that whether or not an axon expresses OCAM is predetermined without the MOB at the time of axonal outgrowth. To check these possibilities, we dissected out and cultured OE at various embryonic stages for 3–4 d. All of the neurites extending from the OE fragments were NCAM-positive and peripherin-positive (data not shown), indicating that the axons originated from olfactory sensory neurons. Cultured fragments of E13 OE extended both OCAM-positive and OCAM-negative axons even in the absence of the MOB (Fig. 7A–C). Furthermore, most of the OE fragments extended either OCAM-positive axons or OCAM-negative axons exclusively (Fig. 7A–C), showing that the spatial segregation of OCAM-positive and OCAM-negative axons found in vivo (Fig. 5L) also occurredin vitro. In contrast, OCAM-positive and OCAM-negative axons from cultured E13 VNE fragments were intermingled (Fig.7D–F), consistent with the OCAM expression pattern in the vomeronasal nerve in vivo (Imamura et al., 1985; Mori et al., 1987). Similar segregation of OCAM-positive and OCAM-negative olfactory axons also was observed in cultured OE fragments of E11 and E10 mice (data not shown), when no axons reach the primordial MOB. These results suggest that the formation of a zonal expression pattern of OCAM in the main olfactory system does not require any retrograde signals from the target but already has been determined at the time of axonal projection.
Fig. 7.
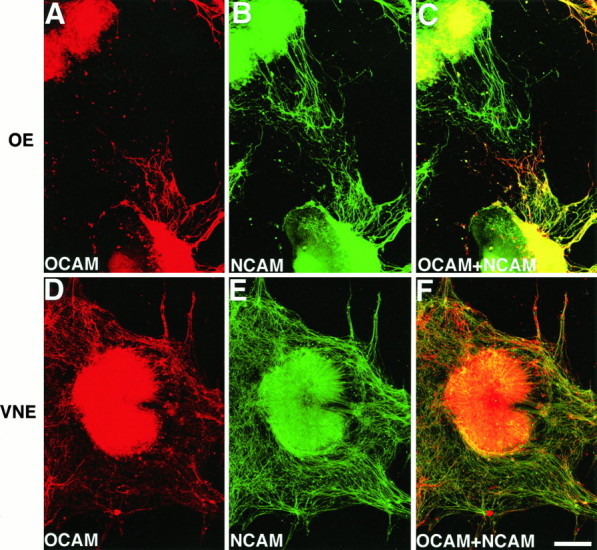
OCAM expression on olfactory and vomeronasal axons in explant cultures. Micrographs of cultured E13 olfactory (A–C) and vomeronasal (D–F) epithelial fragments that were double-labeled with anti-OCAM and anti-NCAM antibodies. A, D, Expression of OCAM. B, E, Expression of NCAM.C, F, Superimposed images of OCAM (red) and NCAM (green) expressions. Note that OCAM is expressed on extending axons from the lower OE fragment, but not from the upper one, whereas NCAM is positive for both OE fragments (A–C). Note also that OCAM-positive and OCAM-negative axons from the VNE fragment are intermingled (D–F). Similar results were obtained consistently in all of the OE explants examined. Of the 40 NCAM-positive OE fragments examined, most of the OE fragments extended either OCAM-positive axons (25 fragments) or OCAM-negative axons (8 fragments) exclusively, whereas both OCAM-positive and OCAM-negative axons were observed in seven explants. Scale bar, 200 μm.
DISCUSSION
OCAM is a new member of the NCAM family
OCAM is a novel protein belonging to the Ig superfamily (Williams and Barclay, 1988), the structure of which is related most closely to that of NCAM (Edelman, 1986; Cunningham et al., 1987; Goridis and Brunet, 1992). OCAM and NCAM display the same domain organization with overall amino acid identity of 45%, indicating that they are derived from a common ancestral gene and constitute a subgroup in the Ig superfamily. However, OCAM lacks several structural properties that are well documented in NCAM, such as an alternatively spliced TM form with a long cytoplasmic region, polysialylation in the fifth Ig-like domain, and a unique alternatively spliced exon in the fourth Ig-like domain (data not shown).
OCAM shows significant structural homology also with apCAM and Fas II, two invertebrate members of the NCAM family. When the amino acid sequences of OCAM and NCAM are compared pairwise with those of apCAM and Fas II, no significant difference is observed (26–27% identity in all pairs), indicating that OCAM and NCAM are related equally to apCAM and Fas II. In fact, OCAM shares many properties with apCAM and Fas II. First, OCAM is expressed by restricted subsets of neurons, as are apCAM (Keller and Schacher, 1990) and Fas II (Harrelson and Goodman, 1988;Grenningloh et al., 1991). In contrast, NCAM shows a more widespread expression pattern throughout the nervous system.
Second, OCAM, apCAM, and Fas II share another property: a neuronal activity-dependent downregulation at synaptic sites. We previously reported that OCAM immunoreactivity in glomeruli of the rabbit MOB disappears after strong and prolonged odor stimulation of the OEin situ and speculated that the disappearance may be attributable to the internalization or conformational alteration of OCAM (Yoshihara et al., 1993). In Aplysia, a similar phenomenon has been described in the serotonin- or cAMP-induced long-term facilitation of the gill withdrawal reflex (Bailey et al., 1992; Mayford et al., 1992). Stimulation of Aplysia sensory neurons causes downregulation (internalization) of apCAM, followed by an increase in the number of synaptic connections between sensory neurons and motoneurons, leading to the long-term facilitation of synaptic efficacy. Fas II also is involved in stabilization, growth, and plasticity of synapses at the Drosophila neuromuscular junction (Schuster et al., 1996a,b). A change in the level of Fas II expression leads to either an increase or decrease in sprouting of motoneuron axon terminals, resulting in alteration of synaptic connection strength. These similarities suggest that the disappearance of OCAM after odor stimulation may relate to the reorganization of synaptic connections within the glomeruli. Downregulation of specific cell adhesion molecules may be a common mechanism for the development of synapses and the plastic changes that follow neuronal activation in both vertebrates and invertebrates.
Role of OCAM in zone-to-zone projection of olfactory axons
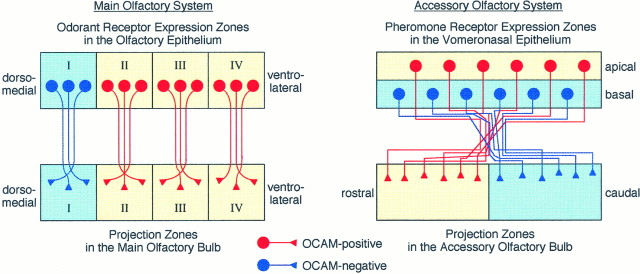
In both the main and accessory olfactory system zone-to-zone projection is one of the basic principles in the formation and maintenance of axonal connections from the sensory epithelia to the brain. OCAM mRNA is expressed by olfactory sensory neurons in the ventrolateral three zones (zones II, III, and IV), but not at all in the most dorsomedial zone (zone I). Accordingly, OCAM protein is present on the surface membrane of olfactory axons projecting from zones II, III, and IV of the OE to corresponding zones of the MOB (Fig.8). Thus, odorant receptors expressed in zone I of the OE are mapped to glomeruli in zone I (OCAM-negative zone) of the MOB, while receptors in zones II, III, and IV are mapped to zones II, III, and IV (OCAM-positive zones).
Fig. 8.
Schematic diagram of zone-to-zone projection of olfactory and vomeronasal axons in relation to zone-specific expression of OCAM. Left, In the main olfactory system four circumscribed zones (I–IV) in the dorsomedial–ventrolateral axis of the OE are defined by expression of odorant receptor genes. OCAM-positive neurons (red) are localized in the three ventrolateral zones (II–IV,yellow) of the OE and project their axons to the corresponding zones (yellow) of the MOB. OCAM-negative neurons (blue) in the most dorsomedial zone (I, light blue) send their axons to the corresponding zone (light blue) of the MOB.Right, In the accessory olfactory system the two zones,apical and basal, are stacked in the VNE as defined by the expression of G-proteins and pheromone receptor genes. OCAM-positive (red) and OCAM-negative (blue) vomeronasal neurons are localized to the apical (yellow) and basal (light blue) zones, respectively. OCAM-positive and OCAM-negative vomeronasal axons are intermingled in the vicinity of the VNE and gradually sort out to make synapses in glomeruli in the rostral (yellow) and caudal (light blue) zones of the AOB, respectively.
In the accessory olfactory system OCAM is expressed by vomeronasal sensory neurons in the apical zone that express Gi2α and several hitherto identified pheromone receptors, whereas OCAM-negative neurons in the basal zone correspond to those expressing Goα protein (Dulac and Axel, 1995; Halpern et al., 1995;Berghard and Buck, 1996). In the AOB, OCAM-positive and OCAM-negative vomeronasal axons terminate within glomeruli in the rostral and caudal zones, respectively. A zone-specific expression is one of the most characteristic properties of OCAM in both the main and accessory olfactory system, as opposed to NCAM, which is expressed by all olfactory and vomeronasal neurons (Fig. 8).
OCAM appears at very early stages in the development of the olfactory system, when pioneering olfactory axons are growing toward the primordial OB (Fig. 5L). The explant culture experiment suggested that at the time of axonal outgrowth it already has been determined whether or not individual olfactory axons express OCAM. Thus, the zone-specific expression of OCAM is established at early stages of development without any retrograde instruction from the OB. This is supported by our preliminary study that OCAM is expressed in a zone-specific manner in the OE of Pdn/Pdn mutant mice in which olfactory axons fail to connect to the brain (Naruse and Keino, 1995; K. Mori and I. Naruse, unpublished observation). Target-independent pattern specification in the OE also was demonstrated by the zonal expression of odorant receptors (Sullivan et al., 1995).
What are the functions of OCAM expressed by olfactory and vomeronasal axons in a zone-specific manner? Immunoelectron microscopical analysis demonstrated that OCAM is localized to closely apposed surface membranes of neighboring olfactory axons in a fascicle (Yoshihara et al., 1993). Because OCAM has a homophilic binding activity, it may mediate fasciculation of subsets of olfactory and vomeronasal axons. A zonal expression of OCAM may serve to sort OCAM-positive and OCAM-negative axons into distinct fascicles in nerve bundles. However, because OCAM commonly is expressed by olfactory axons in zones II, III, and IV, additional cell adhesion molecules with different expression in these zones may be necessary for the establishment of complete zone-to-zone projection. In the accessory olfactory system the sorting of OCAM-positive and OCAM-negative axons in the course of vomeronasal nerve projection seems to be especially critical in the development of zone-to-zone projections, because the two types of axons are intermingled in the vicinity of the VNE and completely segregated at the surface of the AOB (Imamura et al., 1985; Mori et al., 1987).
In addition, OCAM might function as a zone-specific target recognition molecule that mediates specific synapse formation between presynaptic terminals of primary olfactory axons and postsynaptic dendrites of mitral/tufted cells. A search for an OCAM counter-receptor that is expressed on dendrites of mitral/tufted cells in a zone-specific manner and mediates specific heterophilic binding with OCAM might provide insight into the zone-specific connection between the first- and second-order olfactory neurons.
Then, what is the functional meaning of the zone-to-zone projection of olfactory and vomeronasal axons? Odorant receptors can be classified into at least four groups, each being expressed in a specific zone of the OE. They also are categorized into several subfamilies from the primary structure (Ben-Arie et al., 1994). Subsets of receptors expressed in the same spatial zone tend to belong to the same subfamilies with a high degree of sequence homology (Sullivan et al., 1996). Because homologous receptors are assumed to bind distinct but overlapping ranges of odor ligands, the zonal grouping of receptors might relate to olfactory qualities or submodalities. Electro-olfactogram (Ezeh et al., 1995) and Ca2+imaging techniques (Sato et al., 1994) have shown that olfactory sensory neurons with similar response specificity to odor molecules tend to be localized in a certain region (zone) of the OE. The presumptive zone-specific olfactory qualities may be represented in the four projection zones of the MOB. In accord with this idea, electrophysiological recording of mitral/tufted cells in rabbit MOB has demonstrated that the neighboring mitral/tufted cells in a same zone of the MOB are tuned to similar chemical structures of odor ligands, whereas the cells in different zones have systematically different response specificity to odor molecules (Imamura et al., 1992; Mori et al., 1992; Katoh et al., 1993; Mori, 1995; Mori and Yoshihara, 1995).
Olfactory axon guidance and cell adhesion molecules
Because of the continuous turnover of sensory neurons throughout the animal’s life (Graziadei and Monti-Graziadei, 1979), the olfactory and vomeronasal axon projection provides an excellent model to study molecular mechanisms for the formation and maintenance of functional neuronal connections. What molecules are responsible for the guidance of these axons to their target zones and target glomeruli? One possibility is that the odorant and pheromone receptors themselves play a role in axon guidance (Singer et al., 1995). Mombaerts et al. (1996)developed a genetic approach to visualize axons from olfactory sensory neurons expressing a given odorant receptor, demonstrating that the neurons expressing a particular receptor converge their axons to only two topographically fixed glomeruli of the MOB. They also analyzed whether odorant receptors are involved in axonal guidance by a receptor swap experiment. The coding region of an odorant receptor gene P2 was replaced by that of another receptor gene M12. This experiment yielded the unexpected result that axons of M12-expressing P2 neurons converged onto glomeruli distinct from either the normal P2 or M12 glomeruli, but very close to the normal P2 glomeruli, suggesting that odorant receptors play an instructive role but cannot be the sole determinant in the axon guidance mechanism. We speculate that the function of zone-specific axonal adhesion molecules is a prerequisite for olfactory axon guidance. Because neurons expressing P2 and M12 receptors are distributed in different zones of the OE in the normal animal (P2 in zone II; M12 in zone I), the M12-expressing P2 neurons may have obeyed the instruction by a zone-specific guidance molecule, possibly OCAM, present on normal P2 axons but absent from normal M12 axons. Thus, coordinated expression of cell adhesion molecules with odorant receptors seems to be required for the establishment of functional projections of olfactory axons.
Besides OCAM and NCAM, olfactory and vomeronasal axons express a variety of other Ig superfamily molecules, including L1 (Miragall et al., 1989), TAG-1 (Yamamoto and Schwarting, 1991), Thy-1 (Terkelson et al., 1989), and BIG-2 (Yoshihara et al., 1995; Y. Yoshihara and K. Mori, unpublished observation). It is tempting to speculate that coordinated expression of chemoreceptors and cell adhesion molecules is necessary not only for zone-to-zone projection but also for glomerular convergence of axons from sensory neurons expressing the same type of chemoreceptor. If this is the case, we can expect a clear relationship between chemoreceptor selection by individual sensory neurons and combinatorial expression of cell adhesion molecules on their axons. A search for known and as yet unknown cell adhesion molecules expressed by specific functional subsets of primary olfactory axons will aid our further understanding of molecular mechanisms underlying olfactory and vomeronasal axon guidance.
Footnotes
This work was supported in part by the Special Coordination Funds for Promoting Science and Technology from the Science and Technology Agency of Japan and by grants-in-aid from the Ministry of Education, Science, and Culture of Japan. We thank T. K. Hensch for comments on this manuscript; Y. Watanabe for encouragement; S. Nagata, H. Sugino, H. Nagao, and H. von Campenhausen for discussions; S. Ishii for help in peptide sequencing; H. Asou for L1-expressing L929 cells; C. Goridis and H. Cremer for mouse NCAM cDNAs; and J. G. Flanagan for AP–tag-1 vector.
Correspondence should be addressed to Dr. Yoshihiro Yoshihara, Department of Neuroscience, Osaka Bioscience Institute, 6-2-4 Furuedai, Suita, Osaka 565, Japan, or to Dr. Kensaku Mori, Laboratory for Neuronal Recognition Molecules, Frontier Research Program, The Institute of Physical and Chemical Research (RIKEN), Wako, Saitama 351-01, Japan.
REFERENCES
- 1.Bailey CH, Chen M, Keller F, Kandel ER. Serotonin-mediated endocytosis of apCAM: an early step of learning-related synaptic growth in Aplysia. Science. 1992;256:645–649. doi: 10.1126/science.1585177. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Arie N, Lancet D, Taylor C, Khen M, Walker N, Ledbetter DH, Carrozzo R, Patel K, Sheer D, Lehrach H, North MA. Olfactory receptor gene cluster on human chromosome 17: possible duplication of an ancestral receptor repertoire. Hum Mol Genet. 1994;3:229–235. doi: 10.1093/hmg/3.2.229. [DOI] [PubMed] [Google Scholar]
- 3.Berghard A, Buck LB. Sensory transduction in vomeronasal neurons: evidence for Gαo, Gαi2, and adenylyl cyclase II as major components of a pheromone signaling cascade. J Neurosci. 1996;16:909–918. doi: 10.1523/JNEUROSCI.16-03-00909.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brümmendorf T, Rathjen FG. Cell adhesion molecules 1: immunoglobulin superfamily. In: Sheterline P, editor. Protein profile 1. Academic; London: 1994. pp. 951–1058. [PubMed] [Google Scholar]
- 5.Buck L. Information coding in the vertebrate olfactory system. Annu Rev Neurosci. 1996;19:517–544. doi: 10.1146/annurev.ne.19.030196.002505. [DOI] [PubMed] [Google Scholar]
- 6.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 7.Cheng HJ, Flanagan JG. Identification and cloning of ELF-1, a developmentally expressed ligand for the Mek4 and Sek receptor tyrosine kinases. Cell. 1994;79:157–168. doi: 10.1016/0092-8674(94)90408-1. [DOI] [PubMed] [Google Scholar]
- 8.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham BA, Hemperly JJ, Murray BA, Prediger EA, Brackenbury R, Edelman GM. Neural cell adhesion molecule: structure, immunoglobulin-like domains, cell surface modulation, and alternative splicing. Science. 1987;236:799–806. doi: 10.1126/science.3576199. [DOI] [PubMed] [Google Scholar]
- 11.Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 12.Edelman GM. Cell adhesion molecules in the regulation of animal form and tissue pattern. Annu Rev Cell Biol. 1986;2:81–116. doi: 10.1146/annurev.cb.02.110186.000501. [DOI] [PubMed] [Google Scholar]
- 13.Ezeh PI, Davis LM, Scott JW. Regional distribution of rat electro-olfactogram. J Neurophysiol. 1995;73:2207–2220. doi: 10.1152/jn.1995.73.6.2207. [DOI] [PubMed] [Google Scholar]
- 14.Fujita SC, Mori K, Imamura K, Obata K. Subclasses of olfactory receptor cells and their segregated central projections demonstrated by a monoclonal antibody. Brain Res. 1985;326:192–196. doi: 10.1016/0006-8993(85)91403-9. [DOI] [PubMed] [Google Scholar]
- 15.Goridis C, Brunet JF. NCAM: structural diversity, function, and regulation of expression. Semin Cell Biol. 1992;3:189–197. doi: 10.1016/s1043-4682(10)80015-7. [DOI] [PubMed] [Google Scholar]
- 16.Graziadei PPC, Monti-Graziadei GA. Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J Neurocytol. 1979;8:1–18. doi: 10.1007/BF01206454. [DOI] [PubMed] [Google Scholar]
- 17.Grenningloh G, Rehm EJ, Goodman CS. Genetic analysis of growth cone guidance in Drosophila: fasciclin II functions as a neuronal recognition molecule. Cell. 1991;67:45–57. doi: 10.1016/0092-8674(91)90571-f. [DOI] [PubMed] [Google Scholar]
- 18.Halpern M, Shapiro LS, Jia CP. Differential localization of G-proteins in the opossum vomeronasal system. Brain Res. 1995;677:157–161. doi: 10.1016/0006-8993(95)00159-n. [DOI] [PubMed] [Google Scholar]
- 19.Harrelson AL, Goodman CS. Growth cone guidance in insects: fasciclin II is a member of the immunoglobulin superfamily. Science. 1988;242:700–708. doi: 10.1126/science.3187519. [DOI] [PubMed] [Google Scholar]
- 20.Imamura K, Mori K, Fujita SC, Obata K. Immunochemical identification of subgroups of vomeronasal nerve fibers and their segregated terminations in the accessory olfactory bulb. Brain Res. 1985;328:362–366. doi: 10.1016/0006-8993(85)91050-9. [DOI] [PubMed] [Google Scholar]
- 21.Imamura K, Mataga N, Mori K. Coding of odor molecules by mitral/tufted cells in rabbit olfactory bulb. I. Aliphatic compounds. J Neurophysiol. 1992;68:1986–2002. doi: 10.1152/jn.1992.68.6.1986. [DOI] [PubMed] [Google Scholar]
- 22.Katoh K, Koshimoto H, Tani A, Mori K. Coding of odor molecules by mitral/tufted cells in rabbit olfactory bulb. II. Aromatic compounds. J Neurophysiol. 1993;70:2161–2175. doi: 10.1152/jn.1993.70.5.2161. [DOI] [PubMed] [Google Scholar]
- 23.Keller F, Schacher S. Neuron-specific membrane glycoproteins promoting neurite fasciculation in Aplysia californica. J Cell Biol. 1990;111:2637–2650. doi: 10.1083/jcb.111.6.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malter JS. Identification of an AUUUA-specific messenger RNA binding protein. Science. 1989;246:664–666. doi: 10.1126/science.2814487. [DOI] [PubMed] [Google Scholar]
- 25.Mayford M, Barzilai A, Keller F, Schacher S, Kandel ER. Modulation of an NCAM-related adhesion molecule with long-term synaptic plasticity in Aplysia. Science. 1992;256:638–644. doi: 10.1126/science.1585176. [DOI] [PubMed] [Google Scholar]
- 26.Miragall F, Kadmon G, Schachner M. Expression of L1 and NCAM cell adhesion molecules during development of mouse olfactory system. Dev Biol. 1989;135:516–531. doi: 10.1016/0012-1606(89)90179-6. [DOI] [PubMed] [Google Scholar]
- 27.Miura M, Asou H, Kobayashi M, Uyemura K. Functional expression of a full-length cDNA coding for rat neural cell adhesion molecule L1 mediates homophilic intercellular adhesion and migration of cerebellar neurons. J Biol Chem. 1992;267:10752–10758. [PubMed] [Google Scholar]
- 28.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- 30.Mori K. Molecular and cellular properties of mammalian primary olfactory axons. Microsc Res Tech. 1993;24:131–141. doi: 10.1002/jemt.1070240205. [DOI] [PubMed] [Google Scholar]
- 31.Mori K. Relation of chemical structure to specificity of response in olfactory glomeruli. Curr Opin Neurobiol. 1995;5:467–474. doi: 10.1016/0959-4388(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 32.Mori K, Yoshihara Y. Molecular recognition and olfactory processing in the mammalian olfactory system. Prog Neurobiol. 1995;45:585–620. doi: 10.1016/0301-0082(94)00058-p. [DOI] [PubMed] [Google Scholar]
- 33.Mori K, Fujita SC, Imamura K, Obata K. Immunohistochemical study of subclasses of olfactory nerve fibers and their projections to the olfactory bulb in the rabbit. J Comp Neurol. 1985;242:214–229. doi: 10.1002/cne.902420205. [DOI] [PubMed] [Google Scholar]
- 34.Mori K, Imamura K, Fujita SC, Obata K. Projections of two subclasses of vomeronasal nerve fibers to the accessory olfactory bulb in the rabbit. Neuroscience. 1987;20:259–278. doi: 10.1016/0306-4522(87)90018-2. [DOI] [PubMed] [Google Scholar]
- 35.Mori K, Mataga N, Imamura K. Differential specificities of single mitral cells in rabbit olfactory bulb for a homologous series of fatty acid odor molecules. J Neurophysiol. 1992;67:786–789. doi: 10.1152/jn.1992.67.3.786. [DOI] [PubMed] [Google Scholar]
- 36.Naruse I, Keino H. Apoptosis in the developing CNS. Prog Neurobiol. 1995;47:135–155. doi: 10.1016/0301-0082(95)00024-p. [DOI] [PubMed] [Google Scholar]
- 37.Nef P, Hermans-Borgmeyer I, Artieres-Pin H, Beasley L, Dionnie VE, Heinemann SF. Spatial pattern of receptor expression in the olfactory epithelium. Proc Natl Acad Sci USA. 1992;89:8948–8952. doi: 10.1073/pnas.89.19.8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimura Y, Yokoyama M, Araki K, Ueda R, Kudo A, Watanabe T. Recombinant human–mouse chimeric monoclonal antibody specific for common acute lymphocytic leukemia antigen. Cancer Res. 1987;47:999–1005. [PubMed] [Google Scholar]
- 39.Rechsteiner M. Regulation of enzyme levels by proteolysis: the role of PEST regions. Adv Enzyme Regul. 1988;27:135–151. doi: 10.1016/0065-2571(88)90014-3. [DOI] [PubMed] [Google Scholar]
- 40.Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 41.Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79:1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 42.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 43.Rutishauser U. Polysialic acid and the regulation of cell interactions. Curr Opin Cell Biol. 1996;8:679–684. doi: 10.1016/s0955-0674(96)80109-8. [DOI] [PubMed] [Google Scholar]
- 44.Sanger F, Nicklen S, Coulson A. DNA sequencing with chain-terminating inhibitor. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santoni MJ, Barthels D, Vopper G, Boned A, Goridis C, Wille WE. Differential exon usage involving an unusual splicing mechanism generates at least eight types of NCAM cDNA in mouse brain. EMBO J. 1989;8:385–392. doi: 10.1002/j.1460-2075.1989.tb03389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato T, Hirono J, Tonoike M, Takebayashi M. Tuning specificities to aliphatic odorants in mouse olfactory receptor neurons and their local distribution. J Neurophysiol. 1994;72:2980–2989. doi: 10.1152/jn.1994.72.6.2980. [DOI] [PubMed] [Google Scholar]
- 47.Schuster CM, Davis GW, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron. 1996a;17:641–654. doi: 10.1016/s0896-6273(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 48.Schuster CM, Davis GW, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. II. Fasciclin II controls presynaptic structural plasticity. Neuron. 1996b;17:655–667. doi: 10.1016/s0896-6273(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 49.Schwob JE, Gottlieb DI. The primary olfactory projection has two chemically distinct zones. J Neurosci. 1986;6:3393–3404. doi: 10.1523/JNEUROSCI.06-11-03393.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwob JE, Gottlieb DI. Purification and characterization of an antigen that is spatially segregated in the primary olfactory projection. J Neurosci. 1988;8:3470–3480. doi: 10.1523/JNEUROSCI.08-09-03470.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 52.Shepherd GM. Discrimination of molecular signals by the olfactory receptor neuron. Neuron. 1994;13:771–790. doi: 10.1016/0896-6273(94)90245-3. [DOI] [PubMed] [Google Scholar]
- 53.Simmons DM, Arriza JL, Swanson LW. A complete protocol for in situ hybridization of messenger RNAs in brain and other tissues with radiolabeled single-stranded RNA probes. J Histotechnol. 1989;12:169–181. [Google Scholar]
- 54.Singer MS, Shepherd GM, Greer CA. Olfactory receptors guide axons. Nature. 1995;376:19–20. doi: 10.1038/377019b0. [DOI] [PubMed] [Google Scholar]
- 55.Strotmann J, Wanner I, Krieger J, Raming K, Breer J. Expression of odorant receptors in spatially restricted subsets of chemosensory neurons. NeuroReport. 1992;3:1053–1056. doi: 10.1097/00001756-199212000-00005. [DOI] [PubMed] [Google Scholar]
- 56.Strotmann J, Wanner I, Helfrich T, Beck A, Breer H. Rostro-caudal patterning of receptor-expressing neurones in the rat nasal cavity. Cell Tissue Res. 1994;278:11–20. doi: 10.1007/BF00305773. [DOI] [PubMed] [Google Scholar]
- 57.Sullivan SL, Bohm S, Ressler KJ, Horowitz LF, Buck LB. Target-independent pattern specification in the olfactory epithelium. Neuron. 1995;15:779–789. doi: 10.1016/0896-6273(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 58.Sullivan SL, Adamson MC, Ressler KJ, Kozak CA, Buck LB. The chromosomal distribution of mouse odorant receptor genes. Proc Natl Acad Sci USA. 1996;93:884–888. doi: 10.1073/pnas.93.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terkelson OBF, Bock E, Mollgård K. NCAM and Thy-1 in special sense organs of the developing mouse. Anat Embryol (Berl) 1989;179:311–318. doi: 10.1007/BF00305057. [DOI] [PubMed] [Google Scholar]
- 60.Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- 61.Vassar R, Chao SK, Sitcheran R, Nuñez JM, Vosshall JB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 62.Williams AF, Barclay AN. The immunoglobulin superfamily—domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 63.Yamamoto M, Schwarting G. The formation of axonal pathway in developing cranial nerves. Neurosci Res. 1991;463:21–27. doi: 10.1016/0168-0102(91)90008-m. [DOI] [PubMed] [Google Scholar]
- 64.Yoshihara Y, Oka S, Ikeda J, Mori K. Immunoglobulin superfamily molecules in the nervous system. Neurosci Res. 1991;10:83–105. doi: 10.1016/0168-0102(91)90033-u. [DOI] [PubMed] [Google Scholar]
- 65.Yoshihara Y, Katoh K, Mori K. Odor stimulation causes disappearance of R4B12 epitope on axonal surface molecule of olfactory sensory neurons. Neuroscience. 1993;53:101–110. doi: 10.1016/0306-4522(93)90288-q. [DOI] [PubMed] [Google Scholar]
- 66.Yoshihara Y, Kawasaki M, Tani A, Tamada A, Nagata S, Kagamiyama H, Mori K. BIG-1: a new TAG-1/F3-related member of the immunoglobulin superfamily with neurite outgrowth-promoting activity. Neuron. 1994a;13:415–426. doi: 10.1016/0896-6273(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 67.Yoshihara Y, Oka S, Nemoto Y, Watanabe Y, Nagata S, Kagamiyama H, Mori K. An ICAM-related neuronal glycoprotein, telencephalon, with brain segment-specific expression. Neuron. 1994b;12:541–553. doi: 10.1016/0896-6273(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 68.Yoshihara Y, Kawasaki M, Tamada A, Nagata S, Kagamiyama H, Mori K. Overlapping and differential expression of BIG-2, BIG-1, TAG-1, and F3: four members of an axon-associated cell adhesion molecule subgroup of the immunoglobulin superfamily. J Neurobiol. 1995;28:51–69. doi: 10.1002/neu.480280106. [DOI] [PubMed] [Google Scholar]