Abstract
Neurons have the potential to form thousands of distinct neuronal nicotinic receptors from the eight α and three β subunits that currently are known. In an effort to determine how much of this potential complexity is realized among individual neurons, we examined the nicotinic pharmacological and biophysical properties and receptor subunit mRNA expression patterns in individual neurons cultured from rat epicardial ganglia. Analysis of the whole-cell pharmacology of these neurons showed a diversity of responses to the agonists acetylcholine, nicotine, cytisine, and 1,1-dimethyl-4-phenylpiperazinium, suggesting that a heterogeneous population of nicotinic receptor classes, or subtypes, is expressed by individual neurons. Single-channel analysis demonstrated three distinct conductances (18, 24, and 31 pS), with patches from different neurons containing different combinations of these channel classes. We used single-cell RT-PCR to examine nicotinic acetylcholine receptor (nAChR) subunit mRNA expression by individual neurons. Although mRNAs encoding all eight neuronal nAChR subunits for which we probed (α2–α5, α7, β2–β4) were present in multicellular cultures, we found that individual epicardial neurons express distinct subsets of these nAChR subunit mRNAs. These results suggest that individual epicardial neurons express distinct arrays of nAChR subunits and that these subunits may assemble into functional receptors with distinct and variable subunit composition. This variable receptor subunit expression provides an explanation for the diversity of pharmacological and single-channel responses we have observed in individual neurons.
Keywords: neuronal nicotinic receptors, nicotinic acetylcholine receptors, cardiac ganglia, parasympathetic ganglia, single-cell RT-PCR, nicotinic pharmacology
The neuronal nicotinic acetylcholine receptor (nAChR) subunit gene family currently consists of eleven members: α2–α9 and β2–β4 (Sargent, 1993; Elgoyhen et al., 1994). Neuronal nAChRs are thought to have an architecture similar to that of muscle nAChRs: a pentameric assembly of subunits surrounding an ion pore. The existence of eleven subunits suggests that a neuron could express nAChRs with a variety of subunits and could express multiple distinct nAChR classes that differ in subunit composition. Insight into receptor composition can be attained by examining those subunit combinations that form functional receptors in exogenous expression systems and by identifying which combinations actually are expressed by neurons.
Expression studies using Xenopus oocytes suggest that functional neuronal nAChRs can be formed by pairwise combinations of one kind of α and one kind of β subunit (Boulter et al., 1987; Wada et al., 1988; Duvoisin et al., 1989) in a stoichiometry of two α and three β (Anand et al., 1991; Cooper et al., 1991) or, in some cases, as homomeric assemblies (Schoepfer et al., 1990; Séguéla et al., 1993; Elgoyhen et al., 1994). Complex subunit combinations are also possible, because receptors containing more than one distinct α or β subunit have been shown to form in oocytes (Colquhoun et al., 1993; Ramirez-Latorre et al., 1996). Both α and β subunits make contributions to the pharmacological and biophysical properties of these receptors, giving each distinct subunit combination unique characteristics (Wada et al., 1988; Duvoisin et al., 1989; Papke et al., 1989; Luetje et al., 1990; Luetje and Patrick, 1991; Papke and Heinemann, 1991).
Immunoprecipitation experiments have identified a nAChR in rat brain composed of α4 and β2 subunits, as well as a nAChR in chick ciliary ganglion containing the α3, α5, and β4 subunits (Flores et al., 1991; Vernallis et al., 1993). Pharmacological analysis suggests that the β2 and β4 subunits may coassemble into the same receptor in rat sympathetic neurons (Mandelzys et al., 1995), and a preliminary report suggests formation of neuronal nAChRs of even more complex composition (Forsayeth et al., 1995). Electrophysiological studies have shown that individual neurons can express several distinct nAChR conductance classes (Mathie et al., 1991; Adams and Nutter, 1992; Moss and Role, 1993). This expression of multiple nAChR classes by individual neurons is a complicating factor in attempts to use oocyte expression data to identify nAChR subunit composition in neurons (Covernton et al., 1994).
We now show that the whole-cell nicotinic agonist pharmacology of intracardiac parasympathetic neurons varies dramatically among individual neurons, and we provide evidence of single-channel behavior suggesting a heterogeneous population of nAChRs on these neurons. Then, using RT-PCR analysis, we demonstrate that, although cultures of these neurons express a wide array of neuronal nAChR subunit mRNAs, individual neurons express distinct subsets of these mRNAs. Taken together, these results suggest that individual intracardiac neurons express distinct combinations of several different nAChR subtypes.
MATERIALS AND METHODS
Materials. Collagenase was purchased from Worthington Biomedical (Freehold, NJ). DMEM (containing 10 mm glucose) was obtained from Life Technologies (Grand Island, NY). Fetal calf serum, penicillin/streptomycin, laminin, Na4BAPTA, HEPES-NaOH, acetylcholine (ACh), cytisine, 1,1-dimethyl-4-phenylpiperazinium (DMPP), and nicotine were obtained from Sigma (St. Louis, MO). dNTPs, RNAsin ribonuclease inhibitor, and Maloney murine leukemia virus (MMLV) reverse transcriptase were purchased from Promega (Madison, WI). Random hexamers and S-400 micro spin columns were obtained from Pharmacia Biotech (Piscataway, NJ). All restriction endonucleases were obtained from New England Biolabs (Beverly, MA). PCR primers were synthesized in an on-site facility at the University of Miami.
Cell culture. Neurons from rat atria were isolated and cultured as previously described (Fieber and Adams, 1991). Atria were dissected from neonatal (2–7 d postpartum) rats and incubated in Krebs solution containing 1 mg/ml collagenase for 1 hr at 37°C. Intracardiac ganglia were dissected from the atria and transferred to a sterile culture dish containing culture medium [DMEM with 10 mm glucose, 10% (v/v) fetal calf serum, 100 U/ml penicillin, and 0.1 mg/ml streptomycin], triturated with a fire-polished Pasteur pipette, and then plated onto 18 mm glass coverslips coated with laminin. The dissociated cells were incubated at 37°C under a 95% air/5% CO2 atmosphere. Experiments were performed on cells maintained in tissue culture for 36–72 hr. At the time of experiments, the glass coverslip was transferred to a low volume (0.5 ml) recording chamber and viewed at 400× magnification with an inverted, phase-contrast microscope.
Recording conditions. Agonist-induced responses were measured under voltage clamp in isolated neurons with standard patch-clamp techniques (Hamill et al., 1981). Patch pipettes were pulled from borosilicate glass and had tip resistances of 2–5 MΩ. Membrane currents were recorded with a List EPC-7 patch-clamp amplifier, filtered at 10 kHz (−3 dB) with a low-pass Bessel filter (4-pole, Ithaco 4302, Ithaca, NY), and stored on videotape using an A/D recorder adapter (PCM-1; Medical Systems, NY). Whole-cell currents were monitored with a chart recorder, and individual current traces were recorded on disk with the Axotape program (Axon Instruments, Foster City, CA) for subsequent analysis. Peak current amplitude elicited by each agonist was measured by cursor with the Axotape program.
The extracellular physiological salt solution (PSS) for voltage-clamp experiments consisted of (in mm): 140 NaCl, 1 CaCl2, 7.7 glucose, and 10 histidine, pH 7.2. The intracellular pipette solution for whole-cell and excised outside-out patch recordings contained (in mm): 130 NaCl, 2 Na2ATP, 5 Na4BAPTA, and 10 HEPES-NaOH, pH 7.2. Agonist-mediated whole-cell responses were evoked by brief (10 msec, 15 psi) focal application of ACh, cytisine, DMPP, and nicotine dissolved in PSS at concentrations of 100 μm via a pressure ejection device through an extracellular pipette located 50 μm from the neuronal soma. Each agonist was applied 2–5 times in random order, and responses to ACh were compared at the start and end of each experiment to ensure the stability of the recording conditions. So that receptor desensitization could be minimized, a delay of ∼1 min separated individual pulses of agonist application, and recordings were made during continuous perfusion of the recording chamber (∼2 ml/min) with PSS.
The outside-out patch configuration was used to record unitary currents to verify the ACh dependence of the single-channel currents. Patch pipettes were pulled from thick-walled borosilicate glass and had tip resistances of 8–15 MΩ. To decrease noise at high bandwidth, we coated patch electrodes with dental wax. ACh-evoked unitary currents were filtered at 3 kHz (4-pole low-pass Bessel filter) and sampled at 15 kHz (Tecmar Labmaster DMA interface) with a threshold detection device (AI2020A event detector, Axon Instruments) for analysis on a PC 80486 computer using pCLAMP programs (Axon Instruments). The bath contained 1 μm AChCl dissolved in PSS. All numerical data, except where noted, are presented as the mean ± SEM, with the number of experiments in parentheses.
Cytoplasm harvest of individual neurons. The cellular contents of individual neurons were obtained by applying suction on a patch pipette in the whole-cell recording configuration. The pipettes were filled with sterile intracellular pipette solution. The contents of the pipette (4–6 μl) were expelled into a microfuge tube, quickly frozen on dry ice, and stored at −80°C for up to 48 hr. Negative controls were obtained by aspirating extracellular solution near the location of the harvested neuron into a separate pipette. This control was carried through all subsequent reactions to exclude the possibility of false positives caused by contamination with cytoplasm from nearby cells or by contamination with nAChR subunit cDNAs used routinely in the laboratory.
Reverse transcription. Total RNA present in the isolated neuronal cytoplasm was reverse-transcribed in a 20 μl reaction (1 mm each dNTP, 100 pmol of random hexamers, 20 U of ribonuclease inhibitor, and 40 U of MMLV reverse transcriptase) for 1 hr at 37°C. Total RNA from rat brain or whole dishes of dissociated cardiac neurons were isolated by using the protocol of Chomczynski and Sacchi (1987). A “template minus” negative control was initiated at the beginning of the RNA isolation.
PCR primers and controls. A set of degenerate primers was designed to amplify each nAChR subunit cDNA (Table 1). One primer pair annealed to α2, α3, and α4; one primer pair annealed to β2 and β4; one primer pair annealed to α5 and β3; the α7 subunit required a specific primer pair. Degenerate primers were used to reduce the number of PCR reactions into which we would divide the reverse-transcribed cytoplasm from a single neuron. The use of PCR as an analysis tool, especially with the use of degenerate primers, required us to address several concerns. First, the possibility of contamination by nAChR subunit cDNAs used routinely in the laboratory was ruled out by inclusion of a template minus negative control in every experiment (as described above). Second, the possibility that we had amplified genomic DNA was obviated by the fact that the primers were designed so that an intron–exon boundary was located either within a primer sequence or between the two primers. Third, the possibility of false positives caused by primers annealing to untargeted cDNA was ruled out by our use of a restriction digestion identification strategy (see below). Fourth, because we were using a degenerate set of primer pairs, the concern of potential “primer bias” had to be addressed and is discussed in Results. Fifth, the potential for variability in our amplification protocol had to be considered. This did not seem to be a problem for identification of α3 mRNA, because every neuron was positive for α3 (Table2). However, for other subunits, such as α7, only a subset of the neurons presented in Table 2 was found to be positive.
Table 1.
PCR primer sequences
| Subunit | Range | Primer sequence | Enzyme | Fragments | |||
|---|---|---|---|---|---|---|---|
| α2 | 665 –1215 | FWD | GACATCGTCCTCTACAACAAYGCDGAT | BspD1 | 348 | 125 | 78 |
| REV | AGCGGGATGACCAGCGAGGTGGAMGG | ||||||
| α3 | 490 –1040 | Ava1 | 279 | 272 | |||
| α4 | 379 –929 | Aat2 | 420 | 131 | |||
| β2 | 534 –1075 | FWD | GACGTCGTGCTATACAACAATGCYGAYGG | Nhe1 | 474 | 49 | |
| REV | AGCGGTACGTCGAGGGAGGTGGGAGG | ||||||
| β4 | 402 –943 | Afl2 | 327 | 214 | |||
| α5 | 336 –1058 | FWD | ACGACGAATGTCTGGYTGAAGCAGG | Sal1 | 383 | 340 | |
| REV | GGCAAAGACAGTCACCATAATGGATAGGG | ||||||
| β3 | 280 –810 | FWD | ACGACGAATGTCTGGYTGAAGCAGG | Nco1 | 236 | 219 | 75 |
| REV | GACCGTGAGAAAAGACAACCCCAGG | ||||||
| α7 | 338 –813 | FWD | GGAGTGAAGAATGTTCGTTTTCCAGATGG | Hae2 | 398 | 78 | |
| REV | AGCAAGAATACCAGCAGAGCCAGGG | ||||||
Shown are the sequences of PCR primers used in this study, the sequence range of the PCR products, the restriction enzymes used, and the size of the resulting restriction fragments. The α2, α3, and α4 products share a degenerate set of primers, as do the β2 and β4 products. The α5 and β3 products share a degenerate forward primer but have specific reverse primers. Digestion fragments used to identify particular products are underlined.
Table 2.
Expression of nAChR subunit mRNAs among individual intracardiac parasympathetic neurons
| Neuron | α2 | α3 | α4 | α5 | α7 | β2 | β3 | β4 |
|---|---|---|---|---|---|---|---|---|
| A | + | + | + | + | + | |||
| B | + | + | + | |||||
| C | + | + | + | + | ||||
| D | + | + | + | + | ||||
| E | + | + | + | |||||
| F | + | + | ||||||
| G | + | + | + | + | ||||
| H | + | + | + | |||||
| I | + | + | + | + |
The possibility that this result might be attributable to variability in our amplification protocol is addressed in the following control experiment. Reverse-transcribed neuronal cytoplasms were each divided into five aliquots. One aliquot was amplified with the α2/3/4 primer set as a positive control to rule out failure caused by RNA degradation. The other four aliquots were each amplified with the α7 primer set. Of the neurons positive for the α2/3/4 product, two neurons were negative for α7 in all four aliquots, whereas one neuron was positive for α7 in all four aliquots. This control experiment suggests that a negative result in the amplification reaction for some subunit mRNAs in some neurons is not attributable to variability in the amplification protocol but is a reflection of the subunit mRNA expression pattern of that neuron.
Polymerase chain reaction. The RT reaction was divided into aliquots, one for each primer pair, and taken to the first-round PCR by addition of 10× PCR buffer, 1 mm dNTPs, 2.5 mmMgCl2, 10 pmol of each primer, 1.25 U Taqpolymerase, and dH2O to a final volume of 50 μl. PCR was performed using a Perkin-Elmer TC-1 (Norwalk, CT). The cycling parameters were five cycles of 94° for 1 min, 55° for 2 min, ramp to 72° in 51 sec, and 72° for 1.5 min, followed by 35 cycles of 94° for 1 min, 65° for 2 min, and 72° for 1.5 min. Excess primers and primer dimers were removed with S-400 micro spin columns. Then a second round of PCR was performed with the same set of primers and reagents at the same concentrations as in the first round. The cycling parameters for the second round were 35 cycles of 94° for 1 min, 65° for 2 min, and 72° for 1.5 min. These second-round products were gel-purified. In some experiments, the α5 and β3 products were amplified separately.
Restriction digestion. To identify which subunit sequences are present in the second-round PCR products, we developed a restriction digestion strategy such that the PCR product for each subunit could be cleaved by a restriction endonuclease with a cleavage site unique to that product and yield at least one uniquely sized fragment (Table 1). Digestion of the α2 (BspD1), α3 (AvaI), and α4 (Aat2) products would yield fragments uniquely identifying α2 (348 bp), α3 (272 and 279 bp), and α4 (420 bp). Digestion of the β2 (NheI) and β4 (Afl2) products would yield fragments uniquely identifying β2 (474 bp) and β4 (327 and 214 bp). The α5 (SalI) and β3 (NcoI) products were digested separately, yielding uniquely identifying fragments for α5 (340 and 383 bp) and β3 (219 and 236 bp). The α7 (HaeII) product digested to yield uniquely identifying fragments of 78 and 398 bp. The following factors—that the primers were designed specifically for neuronal nAChR subunit cDNAs, that enzymes were used that cleave at sites unique to each product, and that uniquely sized fragments were produced—render the possibility of false positives caused by primers annealing to cDNAs other than those encoding neuronal nAChR subunits as extremely unlikely.
RESULTS
Parasympathetic intracardiac neurons display diverse whole-cell agonist pharmacologies
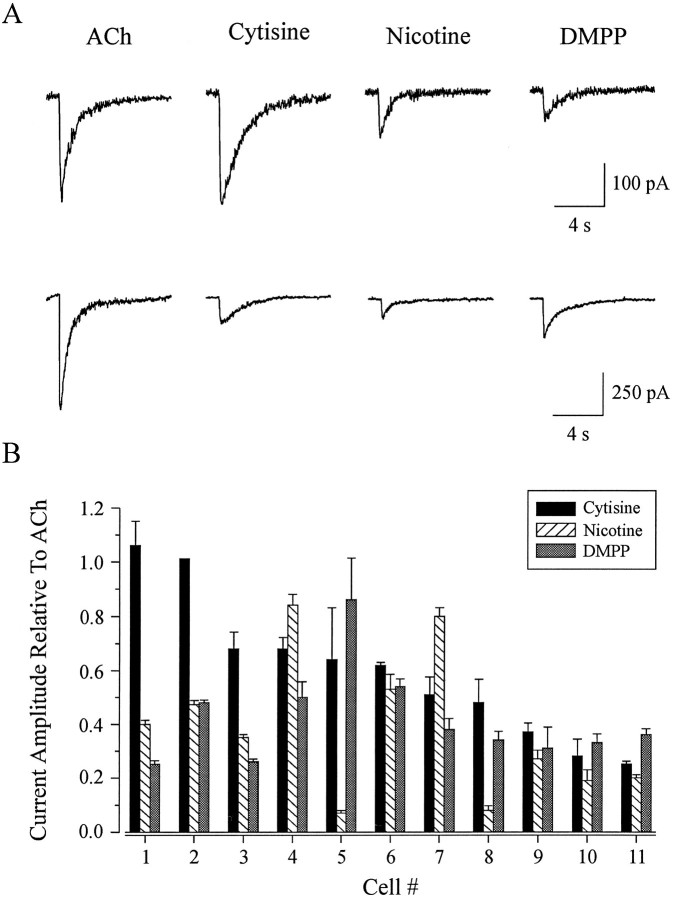
To determine whether these neurons express a heterogeneous population of nicotinic receptors, we used the four agonists that were used in exogenous expression experiments (Luetje and Patrick, 1991). The rank order of potency, determined from the peak current amplitude in response to application of 100 μm concentrations of ACh, cytisine, nicotine, and DMPP, varied from cell to cell. Whole-cell current responses of two different neurons to agonist applications are shown in Figure 1A. The cell represented in the top traces responded in the order cytisine ≥ ACh > nicotine > DMPP, whereas the cell in the bottom traces had a rank order potency of ACh > DMPP > cytisine > nicotine. The whole-cell current amplitude evoked by cytisine, nicotine, and DMPP relative to that elicited by ACh is shown for 11 cells in Figure 1B. The neurons in Figure1B are plotted in the order of decreasing responsiveness to cytisine, with cell 1 having the largest response and cell 11 the smallest response, relative to ACh. As we show, some of the cells shared the same pharmacological profile (i.e., cells 4 and 7 and cells 10 and 11). Each of the other neurons displayed a unique profile.
Fig. 1.
Whole-cell currents in rat parasympathetic neurons evoked by nicotinic agonists. A, Whole-cell current responses evoked by 10 msec focal applications of 100 μmconcentrations of ACh, cytisine, nicotine, and DMPP in two different cells. Rank order of potency was determined by the peak current amplitude evoked by each agonist. One cell responded with a rank order of agonist potency of cytisine ≥ ACh > nicotine > DMPP (top current records), whereas the second cell responded in the order ACh > DMPP > cytisine > nicotine (bottom current records). Holding potential, −90 mV. B, Rank order of potency to nicotinic agonists in 11 parasympathetic intracardiac neurons. Current amplitude evoked by cytisine (filled bars), nicotine (hatched bars), and DMPP (shaded bars) is plotted relative to ACh. Current records shown in A correspond to cell 1 (top records) and cell 11 (bottom records). Cells are plotted in the order of decreasing response to cytisine.
Our results in parasympathetic neurons differ from those obtained in oocyte expression experiments. The pharmacological profiles (agonist rank order of potency) of these neurons do not match the profiles of nicotinic receptors expressed in oocytes. In addition, these neurons display a greater number of rank orders of potency than have been observed in oocyte expression studies. There are several possible explanations for these differences. First, each individual neuron may express a distinct nAChR composed of a combination of subunits not previously characterized in exogenous expression studies. If this were true, then we would expect to see one receptor class in each neuron, with subunit expression varying from neuron to neuron. Second, each individual neuron may express the same combination of several different nAChRs, with differences in whole-cell pharmacology being attributable to individual neurons expressing different proportions of the same set of receptors. If this were true, then we would expect to see the same several receptor classes in each neuron, with subunit expression being the same in each neuron. Third, each individual neuron may express a different subset of a number of different nAChR classes, with differences in whole-cell pharmacology being attributable to the presence of different receptors. If this were true, we would expect to see different combinations of receptor classes in each neuron, with subunit expression varying from neuron to neuron. Fourth, these neurons may be expressing receptors containing subunits that have not yet been cloned. Investigation of this possibility is beyond the scope of this work. These possibilities are, of course, not mutually exclusive. However, by using biophysical techniques in excised outside-out patches and by examining the subunit mRNA expression patterns in individual neurons with RT-PCR, we have attempted to determine which of the first three possibilities most accurately describes the situation in these neurons.
Multiple nAChR channel classes are expressed by parasympathetic intracardiac neurons
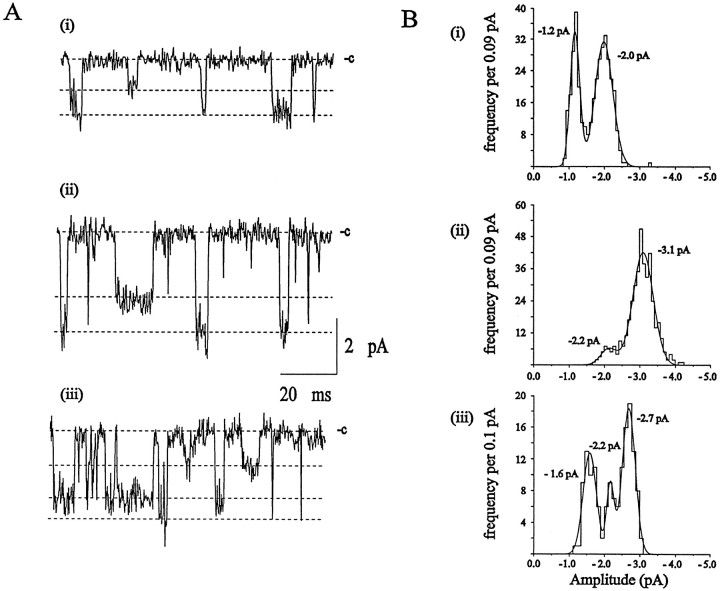
We observed three distinct single-channel conductances in excised patches from rat intrinsic cardiac neurons. ACh-activated unitary currents were recorded from outside-out patches in response to application of 1 μm ACh. Current amplitudes obtained at a holding potential of −90 mV ranged from 0.9–4.2 pA, corresponding to single-channel conductances of 10.0–46.6 pS. This range of conductances is much larger than would be expected for a single-channel class, consistent with the nicotinic receptors in these neurons being a population of heterogeneous channels.
ACh-activated single-channel currents exhibited more than one discrete amplitude level in all patches examined (n = 24). Single-channel currents recorded from three excised membrane patches (−90 mV) and the amplitude histograms obtained from the corresponding experimental records are shown in Figure2A,B. Channel openings of at least two distinct amplitude levels are evident in the patch shown in Figure2Ai. These amplitude components correspond to mean single-channel conductances of 13.3 ± 1.1 pS for the small and 22.2 ± 3.3 pS for the large openings (Fig.2Bi). The patch represented in Figure2Aii also contained two distinct conductance levels, with means of 24.4 ± 2.2 and 34.4 ± 3.3 pS (Fig.2Bii). Three distinct amplitude classes are observed in the patch shown in Figure 2Aiii, with conductances of 17.7 ± 2.2 pS for the small amplitude events, 24.4 ± 1.1 pS for the medium-sized events, and 30.0 ± 2.2 pS for the large amplitude events (Fig. 2Biii).
Fig. 2.
Interpatch variability of single-channel conductance levels activated by ACh. A, Unitary currents evoked by ACh in three separate outside-out membrane patches held at −90 mV. Patches exhibited either the small and medium conductance (i, 9 of 24 patches), the medium and large conductance (ii, 6 of 24 patches), or all three conductance levels (iii, 9 of 24 patches). Conductance levels are designated by the dashed lines, and the closed states of the channels are indicated. Currents were filtered at 3 kHz.B, Amplitude histograms of the experimental records from which the data shown in A were taken. The current amplitude distributions are fit by Gaussian curves with mean ± SD (relative area) of (i) −1.2 ± 0.1 pA (37 ± 3%) and −2.0 ± 0.3 pA (63 ± 4%), (ii) −2.2 ± 0.2 pA (13 ± 4%) and −3.1 ± 0.3 pA (87 ± 4%), and (iii) −1.6 ± 0.2 pA (36 ± 3%), −2.2 ± 0.1 pA (14 ± 3%), and −2.7 ± 0.2 pA (50 ± 4%).
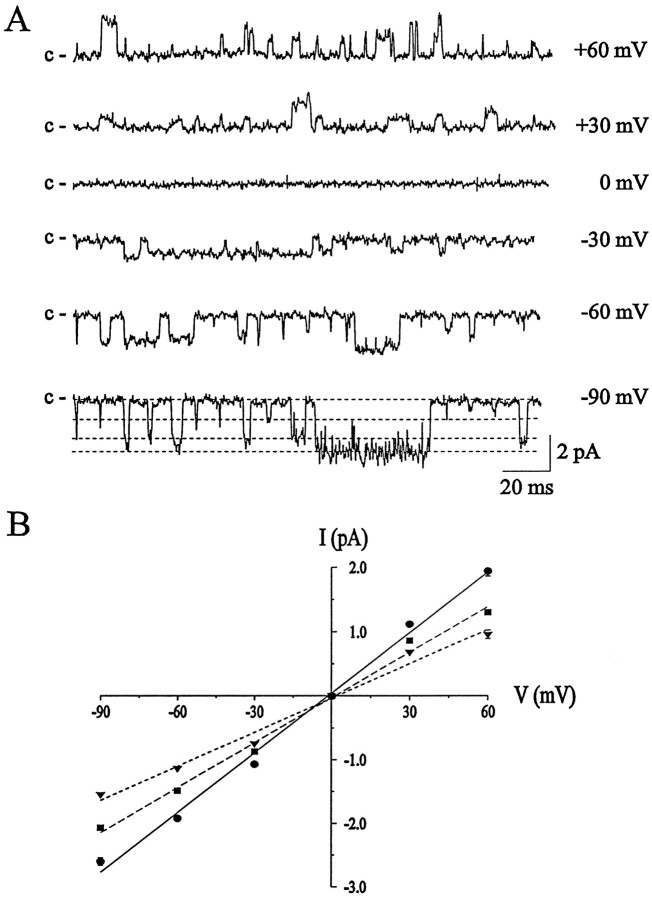
ACh-activated single-channel currents recorded from an excised outside-out patch at membrane potentials ranging from −90 mV to +60 mV are shown in Figure 3A. In this patch, three distinct current amplitude levels were observed and were most evident at the most negative or positive holding potentials. Channel openings were classified according to unitary current amplitude, and the single-channel current–voltage (i–V) relation obtained from 16 different excised patches is shown in Figure3B. Three distinct levels of single-channel current amplitudes were observed, with linear i–V relations and reversal potentials near 0 mV in symmetric Na+ solutions. The i–V relations for the three current levels were fit by linear regression and gave slope conductances of 18 pS for the small current amplitude, 24 pS for the medium-sized current, and 31 pS for the large current amplitude. Linear single-channel i–Vrelationships have been observed previously for the nAChRs in these neurons (Fieber and Adams, 1991; Adams and Nutter, 1992), as well as in rat sympathetic neurons (Mathie et al., 1991), whereas whole-cell nAChRi–V curves can exhibit marked rectification (Mathie et al., 1990). We observed the small and medium conductances in 37.5% of patches (9 of 24), the medium and large in 25% of patches (6 of 24), and the small, medium, and large conductances in 37.5% of patches (9 of 24). Occasionally, larger amplitude openings were seen. The mean conductance of this channel, determined from 89 events in 10 different patches, was 39 ± 2.6 pS. These results suggest that these neurons express multiple nAChR classes and that individual neurons may express different combinations of receptors.
Fig. 3.
Single-channel currents evoked by exogenous ACh in rat cultured parasympathetic neurons. A, Unitary ACh-activated currents obtained in an excised outside-out membrane patch at the membrane potentials indicated. The closed states of the channels (c) are indicated. Three distinct levels are evident in this patch (indicated by the dashed lines in the bottom trace), corresponding to slope conductances of 27.4, 21.8, and 16.6 pS. B, Current–voltage (i–V) relations for single-channel currents activated by ACh. Each point represents the mean ± SEM from 16 excised outside-out patches obtained from different cells, with 65–628 openings per patch at each membrane potential. The data are fit by linear regression and give slope conductances of 18 pS (triangles), 24 pS (squares), and 31 pS (circles). Current records were filtered at 3 kHz and digitized at 67 μsec/point (15 kHz).
Cultures of parasympathetic intracardiac neurons express a wide array of neuronal nAChR subunit mRNAs
Data presented in Figures 1, 2, 3 suggest that these neurons express several distinct classes of nAChR. To generate this pharmacological and biophysical diversity, these neurons may be expressing a large number of distinct nAChR subunits. To examine this possibility, we probed for nAChR subunit mRNA expression in cultures of parasympathetic intracardiac neurons by using RT-PCR, followed by restriction digestion (Lambolez et al., 1992). PCR primers were designed to amplify products from neuronal nAChR subunit mRNAs encoding α2, α3, α4, α5, α7, β2, β3, and β4 (see Table 1). The homology between subunit cDNA sequences allowed design of three degenerate primer sets: one each for the α2/3/4 subunits, the β2/4 subunits, and the α5/β3 subunits. The α7 primers were designed as a specific pair without degeneracy because of the minimal homology between this subunit and the others. The forward primers were placed at the 5′ end of the coding region, near the sequence encoding the cysteine loop. The reverse primers were placed within sequence-encoding transmembrane domains I and II. Use of PCR in these experiments raises several concerns that must be addressed: (1) the potential for false positives caused by contamination with cytoplasm from nearby cells or with nAChR subunit-encoding cDNAs used routinely in the laboratory, (2) genomic contamination, (3) primers annealing to untargeted cDNAs, (4) the potential that the degenerate primer pairs used to detect α2/3/4, β2/4, or α5/β3 might be biased in favor of one subunit cDNA over others, or (5) potential variability in our amplification protocol. These concerns were controlled for or ruled out, as described in Materials and Methods.
The primer bias issue is an important one and deserves attention here. We designed the degenerate primers by manipulating the primer sequence so that amplification of one subunit would not out-compete amplification of other subunits. Potential bias by degenerate primer pairs was tested directly by subjecting mixtures of cDNA templates encoding the target subunits to our PCR and restriction digestion protocol, followed by densitometric determination of the ratio of digested PCR products. A β2/β4 template ratio of 1.0 (100 fg of each) yielded a PCR product ratio of 0.99. β2/β4 template ratios of 0.1 and 10.0 yielded PCR product ratios of 0.27 and 4.12, respectively. The PCR product ratios are the mean of duplicate determinations, with individual values varying <15% from the mean. These results demonstrate that the β2/β4 primer set amplifies the β2 and β4 products without significant bias, although large differences between β2 and β4 products in our experiments may be underestimated. We performed a similar experiment for the α2/α3/α4 primer set. A template ratio of 1:1:1 (100 fg each) yielded a product ratio of 1.0:0.96:1.10. It should be noted that these bias control experiments are not directly comparable to the single-cell amplifications, which contain several orders of magnitude less starting material. Our control experiments do, however, allow us to rule out any gross bias in our primer sets.
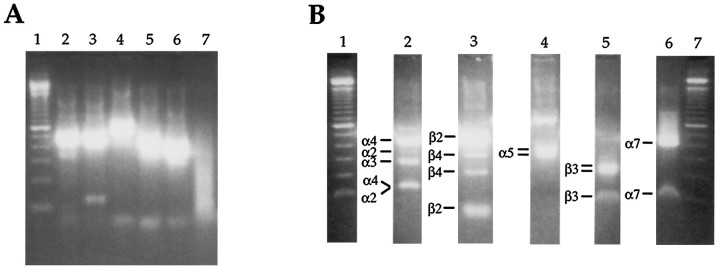
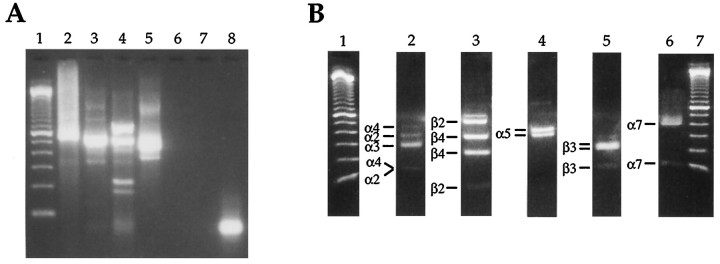
As a test of our primers and the restriction digest strategy, we reverse-transcribed total RNA isolated from adult rat brain. We then subjected this cDNA to our PCR and digestion protocol (Fig.4). The α2/3/4 product (A, lane 2) digested to fragments diagnostic for α2 (348 bp), α3 (272 and 279 bp), and α4 (420 bp) (B, lane 2). The β2/4 product (A, lane 3) digested to fragments diagnostic for β2 (474 bp) and β4 (214 and 327 bp) (B, lane 3). In this experiment, the α5 and β3 products were amplified separately, using the degenerate forward primer and specific reverse primer for each. The α5 product (A, lane 4) digested to fragments diagnostic for α5 (340 and 383 bp) (B, lane 4). The β3 product (A, lane 5) digested to fragments diagnostic for β3 (219 and 236 bp) (B, lane 5). The α7 product (A, lane 6) digested to a fragment diagnostic for α7 (398 bp) (B, lane 6). No products were amplified in the negative control (A, lane 7).
Fig. 4.
RT-PCR identification of mRNA encoding α2, α3, α4, α5, α7, β2, β3, and β4 in rat brain RNA.A, Second-round PCR products. Lane 1, 100 bp standards; lanes 2–6 are the PCR products amplified with the α2/3/4, β2/4, α5, β3, and α7 primers, respectively. In this experiment, α5 and β3 were amplified separately.Lane 7 is the negative control. B, Restriction digestion of PCR products. Lanes 1, 7, 100 bp standards; lanes 2–6 contain digested second-round PCR products for α2/3/4, β2/4, α5, β3, and α7 primers, respectively. Fragments identifying each subunit are labeled to theleft of each lane. For enzymes used and exact fragment sizes, see Table 2. For clarity in presentation, we have removed the undigested sample routinely run next to each digested product.
Next we examined the expression of neuronal nAChR subunit mRNAs by a dish of cultured parasympathetic intracardiac neurons. At 3 d after dissociation, this culture contained ∼100 neurons as well as a number of other cell types (e.g., cardiac myocytes, Schwann cells, and fibroblasts). Total RNA extracted from this sample was reverse-transcribed and subjected to our PCR and digestion protocol (Fig. 5). We found that RNA encoding each of the subunits for which we probed (α2, α3, α4, α5, α7, β2, β3, and β4) was expressed by this dish of cultured neurons. There are several possible explanations for this result. First, each neuron may be expressing the same wide array of subunit mRNAs, a result that would be consistent with the idea that each neuron expresses the same receptor classes, but the proportions of each class vary from neuron to neuron. Second, different neurons may be expressing different combinations of subunit mRNAs, which would be consistent with the hypothesis that individual neurons may assemble different combinations of receptor classes. Third, the neurons may be expressing only a subset of the observed subunit mRNAs, with the rest being expressed by other cell types on the dish. To distinguish among these possibilities, we used the RT-PCR and digestion strategy to examine the expression of subunit mRNAs by individual neurons.
Fig. 5.
Neuronal nicotinic receptor subunit mRNAs encoding α2, α3, α4, α5, α7, β2, β3, and β4 are expressed by cultures of intracardiac parasympathetic neurons. A, Second-round PCR products. Lane 1, 100 bp standards;lanes 2–5 are the PCR products obtained by using the α2/3/4, β2/4, α5/β3, and α7 primers, respectively. In this experiment, α5 and β3 products were amplified in the same reaction (lane 4). Lanes 6 and7 are blank. Lane 8 is the negative control. B, Restriction digestion of PCR products.Lanes 1, 7, 100 bp standards; lanes 2–6contain the digested fragments for the α2/3/4, β2/4, α5, β3, and α7 products, respectively. Note that the α5 and β3 products, although amplified in the same reaction, are digested separately because of different buffer requirements. Fragments identifying each subunit are labeled to the left of each lane. For enzymes used and exact fragment sizes, see Table 1. For clarity in presentation, we have removed the undigested sample routinely run next to each digested product.
Individual parasympathetic intracardiac neurons differ in their neuronal nAChR subunit mRNA expression patterns
To examine the subunit mRNA expression patterns of individual neurons, we isolated the cytosolic contents of each neuron by aspiration with a patch pipette in the whole-cell configuration. Then the contents of each neuron were subjected to a reverse transcription reaction, followed by our PCR and restriction digest protocol. The results for several neurons are shown in Figure 6. Results obtained for these and additional neurons are presented in Table 2.
Fig. 6.
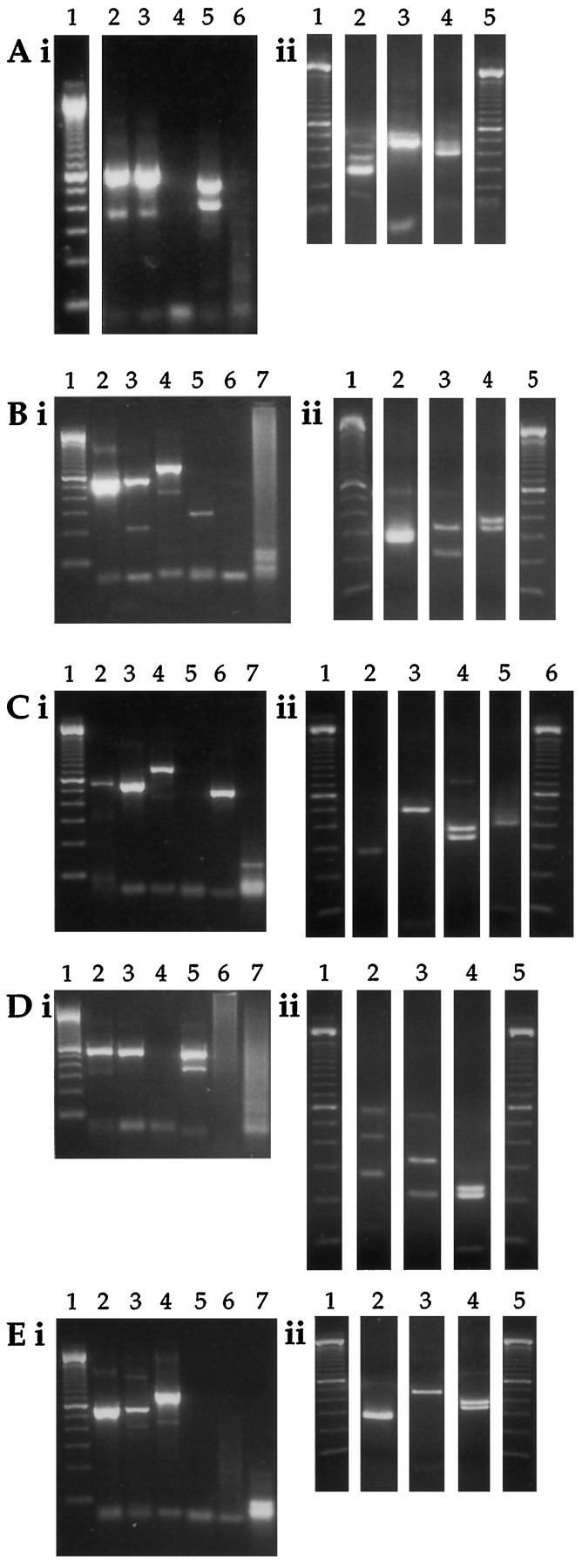
Individual intracardiac parasympathetic neurons express diverse arrays of nAChR subunit mRNAs. Second-round PCR (i) and restriction digestion (ii) results for neurons A–E are shown. A, Neuron expressing α2, α3, and α4 (lanes 2i and 2ii), α7 (lanes 5i and 4ii), and β2 (lanes 3i and 3ii). B, Neuron expressing α3 (lanes 2i and2ii), α5 (lanes 4i and4ii), and β4 (lanes 3i and3ii). C, Neuron expressing α3 (lanes 2i and 2ii), α5 (lanes 4i and 4ii), α7 (lanes 6i and5ii), and β2 (lanes 3i and3ii). D, Neuron expressing α3 and α4 (lanes 2i and 2ii), β3 (lanes 5i and 4ii), and β4 (lanes 3iand 3ii). E, Neuron expressing α3 (lanes 2i and 2ii), α5 (lanes 4i and 4ii), and β2 (lanes 3iand 3ii). Neurons C and D were harvested from the same culture dish. Negative controls are run in lane 6 for neuron A and lane 7 for neurons B–E. 100 bp size standards are run in lane 1 of each second-round PCR gel (Ai–Ei); lanes 1 and 5 inAii, Bii, Dii, and Eii; and lanes 1 and 5 in Cii. For clarity in presentation, we have removed the lanes containing undigested samples routinely run next to each digested product.
The nAChR subunit mRNA expression patterns among individual neurons are diverse. Neuron A (Fig. 6A) expresses mRNA encoding α2, α3, α4, α7, and β2. Neuron B (Fig. 6B) expresses mRNA encoding α3, α5, and β4. Neuron C (Fig.6C) expresses mRNA encoding α3, α5, α7, and β2. Neuron D (Fig. 6D) expresses α3, α4, β3, and β4. Neuron E (Fig. 6E) expresses α3, α5, and β2. Neurons C and D were dissected from the same rat and cultured on the same dish, demonstrating that the differences in mRNA expression are not attributable to differences in culture conditions or “inter-rat” variability. Similarly, three neurons isolated from another heart, cultured together in the same dish and probed for β2 and β4 mRNA expression, also were found to differ (data not shown). Although the nAChR subunit mRNA expression patterns of individual neurons are quite diverse, all neurons were similar in two respects. First, all of the neurons express mRNA encoding the α3 subunit, consistent with numerous observations of the expression of this subunit in peripheral neurons (Boyd et al., 1988; Listerud et al., 1991; Rust et al., 1994). Second, all of the neurons express mRNA encoding either or both the β2 or β4 subunits. Thus, expression of α3 and β2 and/or β4 may be ubiquitous in intrinsic cardiac neurons. The expression patterns for the other subunit mRNAs were diverse, varying from neuron to neuron. Each of the subunit mRNAs shown to be present in RNA isolated from an entire dish of neurons (Fig. 5) is expressed by at least one of the neurons we have examined individually (Fig. 6 and Table 2). Thus, the subunit mRNA expression by neurons can account for all of the subunit mRNA expression seen at the dish level and supports the hypothesis of heterogeneous expression of nAChRs among individual neurons.
DISCUSSION
Our results show that individual parasympathetic neurons of neonatal rat intracardiac ganglia express a heterogeneous population of nAChR subunits. At the whole-cell level, the rank order of potency of several nicotinic agonists varied among individual neurons. Single-channel recordings demonstrate the presence of at least three distinct conductance classes (18, 24, and 31 pS). When the expression of nAChR subunit mRNAs was examined by RT-PCR, these cultures of neurons were found to express a wide array of subunit mRNAs (α2, α3, α4, α5, α7, β2, β3, and β4). At the single-cell level, RT-PCR demonstrated that individual neurons expressed distinctly different arrays of nAChR subunit mRNAs. These results are consistent with the hypothesis that individual neurons in this preparation express distinct subsets of several different nAChR classes.
When we examined the whole-cell agonist pharmacology of rat intracardiac neurons, we observed a greater number of rank orders of potency than seen in exogenous expression of nAChR subunit mRNAs, and none of the profiles in neurons matched those in oocytes (Fig.1B). The dissimilarities of the profiles between neurons and oocytes suggest a greater degree of heterogeneity of subunit expression in neurons than the pairwise or homomeric expression of subunits in oocytes. However, exogenous expression data provide useful observations. For example, in oocyte expression experiments both the α and β subunits contribute to the pharmacological profile of neuronal nAChRs (Luetje and Patrick, 1991). Responsiveness to cytisine seems to be influenced primarily by the identity of the β subunit, because nAChRs containing β4 are several-fold more sensitive to cytisine than to ACh, whereas those containing β2 are many-fold less sensitive to cytisine than to ACh. The cell-to-cell variation in the cytisine-evoked responses in the present study (Fig. 1) could, therefore, reflect differing ratios of β2 and β4 subunits, with cell 1 containing a higher proportion of nAChR channels composed of β4 subunits and cell 11 having channels containing primarily β2 subunits. Similarly, intracardiac parasympathetic neurons expressing varying ratios of nAChR receptors composed of different combinations of α and β subunits could account for the diversity observed for the other agonists.
Our single-channel experiments, conducted on outside-out membrane patches, yielded results consistent with the presence of at least three different conductance levels for nAChR channels in rat intracardiac neurons. The consistent expression of α3 mRNA in every neuron examined and the presence of the medium conductance in every patch suggest that the nAChR responsible for the medium conductance contains α3. The expression of multiple neuronal nicotinic ACh receptors also has been studied in thin slices of rat medial habenula via patch-clamp techniques (Connolly et al., 1995). The heterogeneity of nAChRs in cells of this preparation was suggested by the presence of at least two single-channel conductances (51 and 41 pS) with distinct open times. Detailed analysis concluded that these conductance classes were produced by two molecularly distinct nAChR subtypes. In contrast to our findings, although rat superior cervical ganglion (SCG) neurons express five nAChR transcripts (α3, α5, α7, β2, and β4), Mandelzys et al. (1995) used pharmacological analysis to suggest that the nAChRs on these neurons are a uniform population.
To begin to understand the molecular basis for the pharmacological and biophysical diversity that we have observed, we adapted the RT-PCR strategy of Lambolez et al. (1992)for neuronal nAChR subunits. We found that individual intracardiac parasympathetic neurons express distinct subsets of nAChR subunit mRNAs. The subunit mRNA expression pattern of individual neurons ranged from simple to complex (e.g., in Table 2, compare neuron F, expressing α3 and β4, with neuron A, expressing α2, α3, α4, α7, and β2). Differences in nAChR subunit expression resulting from differential mRNA expression, then, may account for the observed variability in both whole-cell pharmacology and single-channel conductance. It is important to note that protein levels do not necessarily parallel mRNA levels. Thus differential translation, protein stability, or receptor assembly could provide additional levels of regulation. Although it is possible that the heterogeneity we observe might be generated artifactually as a result of studying these neurons in culture, the results of Connolly et al. (1995), as noted above, suggest that this diversity also occurs in vivo. Also, Ullian and Sargent (1995) used subunit-specific monoclonal antibodies to identify at least five distinct classes of neurons in the chick lateral spiriform nucleus on the basis of differential expression of nAChR subunits. These reports, together with our findings, indicate that the classification of nAChRs in neurons will not be a simple task.
Our analysis of nAChR subunit mRNA expression patterns in individual neurons relies on the use of the PCR technique, raising several concerns. We describe our controls in detail in Materials and Methods, but two issues deserve discussion here. One potential problem is that there may be an intrinsic bias within our degenerate primer sets that favors one subunit mRNA over another. The evidence suggesting a lack of bias in the β2/4 primer pair is discussed at length in Results. For the α5/β3 primer set, one or the other of these PCR products does not predominate consistently in our results, and in many of our experiments we probed for the two subunits in separate reactions. Bias conceivably could be a problem with the α2/3/4 primer set, because the α3 product does predominate consistently in our experiments. We can argue against this possibility in two ways. First, although α3 is predominant in all of the cells tested individually (Fig. 6 and Table2) as well as in dishes of neurons (Fig. 5), the α4 PCR product predominates over the α2 and α3 products when we probe rat brain RNA (Fig. 4), consistent with the known predominance of α4 mRNA in rat brain. Second, similar to the β2/4 pair, in control experiments we have demonstrated a lack of bias in the α2/3/4 primer set. Thus, when one degenerate primer set is used in a single reaction, differences in levels of products are likely to be an approximate reflection of differences in levels of mRNA transcripts in the cells. A second concern is the sensitivity of our methods when we are working at the single-cell level. Failure to detect transcripts encoding a particular subunit simply may reflect a level of subunit mRNA expression below our threshold level of detection. Because the reverse-transcribed cytoplasm is split into four or five PCR reactions, we are attempting to detect subunit mRNAs in less than a single cytoplasm. We have been unable to devise an appropriate and feasible control for this, and so expression of subunit mRNAs below our level of detection remains a possibility.
Pharmacological analysis and single-cell RT-PCR performed on the same neuron proved problematic in this study. We believe this is attributable to the extended length of time required to obtain a full pharmacological profile for each neuron. These experiments typically took 30–45 min to complete, during which time the RNA in the cell had ample opportunity for degradation. In any event, it seems that the length of time required by the electrophysiological recordings ruined the chance of success for the PCR. The use of a noninvasive technology (e.g., Ca2+ fluorescence imaging) is being explored for pharmacological analysis in future experiments.
An intriguing issue is the potential importance that heterogeneous expression of neuronal nAChRs may have in the control of cardiac function. Mammalian epicardial ganglionated plexi are composed of different types of neurons, including both cholinergic and adrenergic postganglionic efferents, interneurons, and afferent neurons (Moravec and Moravec, 1987; Armour, 1991; Ardell, 1994). Although ACh is the principal neurotransmitter found in the vagal efferent fibers and the epicardial parasympathetic ganglia (Jacobowitz, 1967; Ehinger et al., 1968; Seabrook et al., 1990), numerous putative neurotransmitters and neuromodulators have been detected within intracardiac ganglia by the use of immunohistochemical techniques (Allen et al., 1994; Steele et al., 1994, 1996). In addition to this molecular complexity, the functional properties of intrinsic cardiac ganglia differ in different regions of the mammalian heart. For example, neurons in the right atrial ganglionated plexus control the sinoatrial node (Butler et al., 1990). The inferior vena cava–inferior atrial plexus modulates the atrioventricular node (Ardell and Randall, 1986). The dorsal atrial and the cranial medial ventricular ganglia modulate contractile tissue (Yuan et al., 1994). Our results demonstrate that individual intracardiac neurons differ in their nAChR expression. Calcium permeability differs among neuronal nAChRs, depending on subunit composition, and thus intracellular calcium levels and neuronal excitability may be affected to differing extents. Neuronal nAChRs also can be differentially localized on neuronal surfaces (Wilson Horch and Sargent, 1996). This differential localization may be determined by subunit composition and could be involved in reception of multiple inputs. Thus, heterogeneous expression of nAChRs by intracardiac neurons is likely to have a profound influence on neural control of cardiac function.
Footnotes
This work was supported by grants to C.W.L. from the American Heart Association Florida affiliate, the Pharmaceutical Research and Manufacturers of America Foundation, and the National Institute on Drug Abuse (DA08102), and to D.J.A. from the National Heart, Lung, and Blood Institute (HL35422). C.W.L. was an Initial Investigator of the American Heart Association Florida affiliate. We thank Drs. Richard Bookman, Richard Kramer, and Jim Patrick for critical reading of this manuscript.
Correspondence should be addressed to Dr. Charles W. Luetje, Department of Molecular and Cellular Pharmacology (R-189), University of Miami School of Medicine, P.O. Box 016189, Miami, FL 33101.
Dr. Cuevas’s present address: Department of Biology, University of California, San Diego, CA 92093.
Dr. Adams’s present address: Department of Physiology and Pharmacology, University of Queensland, Brisbane QLD 4072, Australia.
REFERENCES
- 1.Adams DJ, Nutter TJ. Calcium permeability and modulation of nicotinic acetylcholine receptor channels in rat parasympathetic neurons. J Physiol (Paris) 1992;86:67–76. doi: 10.1016/s0928-4257(05)80009-9. [DOI] [PubMed] [Google Scholar]
- 2.Allen TGJ, Hassall CJS, Burnstock G. Mammalian intrinsic cardiac neurons in cell culture. In: Armour JA, Ardell JL, editors. Neurocardiology. Oxford UP; New York: 1994. pp. 115–138. [Google Scholar]
- 3.Anand R, Conroy WG, Schoepfer R, Whiting P, Lindstrom J. Neuronal nicotinic acetylcholine receptors expressed in Xenopus oocytes have a pentameric quarternary structure. J Biol Chem. 1991;266:11192–11198. [PubMed] [Google Scholar]
- 4.Ardell JL. Structure and function of mammalian intrinsic cardiac neurons. In: Armour JA, Ardell JL, editors. Neurocardiology. Oxford UP; New York: 1994. pp. 95–114. [Google Scholar]
- 5.Ardell JL, Randall WC. Selective vagal innervation of sinoatrial and atrioventricular nodes in canine hearts. Am J Physiol. 1986;251:H764–H773. doi: 10.1152/ajpheart.1986.251.4.H764. [DOI] [PubMed] [Google Scholar]
- 6.Armour JA. Intrinsic cardiac neurons. J Cardiovasc Electrophysiol. 1991;2:331–341. [Google Scholar]
- 7.Boulter J, Connolly J, Deneris E, Goldman D, Heinemann S, Patrick J. Functional expression of two neuronal nicotinic acetylcholine receptors from cDNA clones identifies a gene family. Proc Natl Acad Sci USA. 1987;84:7763–7767. doi: 10.1073/pnas.84.21.7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd RT, Jacob MH, Couturier S, Ballivet M, Berg DK. Expression and regulation of neuronal acetylcholine receptor mRNA in chick ciliary ganglia. Neuron. 1988;1:495–502. doi: 10.1016/0896-6273(88)90180-8. [DOI] [PubMed] [Google Scholar]
- 9.Butler CK, Smith FM, Cardinal R, Murphy DA, Hopkins DA, Armour JA (1990) Cardiac responses to electrical stimulation of discrete loci in canine atrial and ventricular ganglionated plexi. Am J Physiol (Heart Circ Physiol 28) 259:H1108–H1117. [DOI] [PubMed]
- 10.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid-guanidinium-thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Colquhoun L, Dineley K, Patrick J. A hetero-beta neuronal nicotinic acetylcholine receptor expressed in Xenopus oocytes. Soc Neurosci Abstr. 1993;19:1533. [Google Scholar]
- 12.Connolly JG, Gibb AJ, Colquhoun D. Heterogeneity of neuronal nicotinic acetylcholine receptors in thin slices of rat medial habenula. J Physiol (Lond) 1995;484:87–105. doi: 10.1113/jphysiol.1995.sp020650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper E, Couturier S, Ballivet M. Pentameric structure and subunit stoichiometry of a neuronal nicotinic acetylcholine receptor. Nature. 1991;350:235–238. doi: 10.1038/350235a0. [DOI] [PubMed] [Google Scholar]
- 14.Covernton PJO, Kojima H, Sivilotti LG, Gibb AJ, Colquhoun D. Comparison of neuronal nicotinic receptors in rat sympathetic neurones with subunit pairs expressed in Xenopus oocytes. J Physiol (Lond) 1994;481:27–34. doi: 10.1113/jphysiol.1994.sp020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duvoisin RM, Deneris ES, Boulter J, Patrick J, Heinemann S. The functional diversity of the neuronal nicotinic acetylcholine receptors is increased by a novel subunit: β4. Neuron. 1989;3:487–496. doi: 10.1016/0896-6273(89)90207-9. [DOI] [PubMed] [Google Scholar]
- 16.Ehinger B, Falck B, Persson H, Sporrono B. Adrenergic and cholinesterase-containing neurons of the heart. Histochimie. 1968;16:197–205. doi: 10.1007/BF00307848. [DOI] [PubMed] [Google Scholar]
- 17.Elgoyhen AB, Johnson DS, Boulter J, Vetter DE, Heinemann S. α9: an acetylcholine receptor with novel pharmacological properties expressed in rat cochlear hair cells. Cell. 1994;79:705–715. doi: 10.1016/0092-8674(94)90555-x. [DOI] [PubMed] [Google Scholar]
- 18.Fieber LA, Adams DJ. Acetylcholine-evoked currents in cultured neurones dissociated from rat parasympathetic cardiac ganglia. J Physiol (Lond) 1991;434:215–237. doi: 10.1113/jphysiol.1991.sp018466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flores CM, Rogers SW, Pabreza LA, Wolfe BB, Kellar KJ. A subtype of nicotinic cholinergic receptor in rat brain is composed of α4 and β2 subunits and is up-regulated by chronic nicotine treatment. Mol Pharmacol. 1991;41:31–37. [PubMed] [Google Scholar]
- 20.Forsayeth JR, Kobrin E, Winegar BD, Fitzgerald DJ. The nicotinic acetylcholine receptor in cerebellum is composed of four different subunits. Soc Neurosci Abstr. 1995;21:1583. [Google Scholar]
- 21.Hamill OP, Marty A, Neher E, Sakmann B, Sigworth F. Improved patch-clamp techniques for high resolution current recordings from cells and cell-free membrane patches. Pflügers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 22.Jacobowitz D. Histochemical studies of the relationship of chromaffin cell and adrenergic nerve fibers to the cardiac ganglia of several species. J Pharmacol Exp Ther. 1967;158:227–240. [PubMed] [Google Scholar]
- 23.Lambolez B, Audinat A, Bochet P, Crépel F, Rossier J. AMPA receptor subunits expressed by single Purkinje cells. Neuron. 1992;9:247–258. doi: 10.1016/0896-6273(92)90164-9. [DOI] [PubMed] [Google Scholar]
- 24.Listerud M, Brussaard AB, Devay P, Colman DR, Role LW. Functional contribution of neuronal AChR subunits revealed by antisense oligonucleotides. Science. 1991;254:1518–1521. doi: 10.1126/science.1720573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luetje CW, Wada K, Rogers S, Abramson SN, Tsuji K, Heinemann S, Patrick J. Neurotoxins distinguish between different neuronal nicotinic acetylcholine receptor subunit combinations. J Neurochem. 1990;55:632–640. doi: 10.1111/j.1471-4159.1990.tb04180.x. [DOI] [PubMed] [Google Scholar]
- 26.Luetje CW, Patrick J. Both α- and β-subunits contribute to the agonist sensitivity of neuronal nicotinic acetylcholine receptors. J Neurosci. 1991;11:837–845. doi: 10.1523/JNEUROSCI.11-03-00837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandelzys A, de Koninck P, Cooper E. Agonist and toxin sensitivities of ACh-evoked currents on neurons expressing multiple nicotinic ACh receptor subunits. J Neurophysiol. 1995;74:1212–1221. doi: 10.1152/jn.1995.74.3.1212. [DOI] [PubMed] [Google Scholar]
- 28.Mathie A, Colquhoun D, Cull-Candy SG. Rectification of currents activated by nicotinic acetylcholine receptors in rat sympathetic ganglion neurones. J Physiol (Lond) 1990;427:625–655. doi: 10.1113/jphysiol.1990.sp018191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathie A, Cull-Candy SG, Colquhoun D. Conductance and kinetic properties of single nicotinic acetylcholine receptors channels in rat sympathetic neurones. J Physiol (Lond) 1991;439:717–750. doi: 10.1113/jphysiol.1991.sp018690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moravec J, Moravec M. Intrinsic nerve plexus of mammalian heart: morphological basis of cardiac rhythmical activity? Int Rev Cytol. 1987;106:89–147. doi: 10.1016/s0074-7696(08)61711-8. [DOI] [PubMed] [Google Scholar]
- 31.Moss BL, Role LW. Enhanced ACh sensitivity is accompanied by changes in ACh receptor channel properties and segregation of ACh receptor subtypes on sympathetic neurons during innervation in vivo. J Neurosci. 1993;13:13–28. doi: 10.1523/JNEUROSCI.13-01-00013.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papke RL, Heinemann SF. The role of the beta 4 subunit in determining the kinetic properties of rat neuronal nicotinic acetylcholine alpha 3 receptors. J Physiol (Lond) 1991;440:95–112. doi: 10.1113/jphysiol.1991.sp018698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papke RL, Boulter J, Patrick J, Heinemann SF. Single-channel currents of rat neuronal nicotinic acetylcholine receptors expressed in Xenopus oocytes. Neuron. 1989;3:589–596. doi: 10.1016/0896-6273(89)90269-9. [DOI] [PubMed] [Google Scholar]
- 34.Ramirez-Latorre J, Yu CR, Qu X, Perin F, Karlin A, Role L. Functional contributions to α5 subunit to neuronal acetylcholine receptor channels. Nature. 1996;380:347–351. doi: 10.1038/380347a0. [DOI] [PubMed] [Google Scholar]
- 35.Rust G, Burgunder J-M, Lauterberg TE, Cachelin AB. Expression of neuronal nicotinic acetylcholine receptor subunit genes in the rat autonomic nervous system. Eur J Neurosci. 1994;6:478–485. doi: 10.1111/j.1460-9568.1994.tb00290.x. [DOI] [PubMed] [Google Scholar]
- 36.Sargent PB. The diversity of neuronal nicotinic acetylcholine receptors. Annu Rev Neurosci. 1993;16:403–443. doi: 10.1146/annurev.ne.16.030193.002155. [DOI] [PubMed] [Google Scholar]
- 37.Schoepfer R, Conroy WG, Whiting P, Gore M, Lindstrom J. Brain α-bungarotoxin binding protein cDNAs and MAbs reveal subtypes of this branch of the ligand-gated ion channel gene superfamily. Neuron. 1990;5:35–48. doi: 10.1016/0896-6273(90)90031-a. [DOI] [PubMed] [Google Scholar]
- 38.Seabrook GR, Fieber LA, Adams DJ (1990) Neurotransmission in neonatal rat cardiac ganglion in situ. Am J Physiol (Heart Circ Physiol 28) 259:H997–H1005. [DOI] [PubMed]
- 39.Séguéla P, Wadiche J, Dineley-Miller K, Dani JA, Patrick JW. Molecular cloning, functional properties, and distribution of rat brain α7: a nicotinic cation channel highly permeable to calcium. J Neurosci. 1993;13:596–604. doi: 10.1523/JNEUROSCI.13-02-00596.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steele PA, Gibbins IL, Morris JL, Mayer B. Multiple populations of neuropeptide-containing intrinsic neurons in the guinea-pig heart. Neuroscience. 1994;62:241–250. doi: 10.1016/0306-4522(94)90327-1. [DOI] [PubMed] [Google Scholar]
- 41.Steele PA, Gibbins IL, Morris JL. Projections of intrinsic cardiac neurons to different targets in the guinea-pig heart. J Auton Nerv Syst. 1996;56:191–200. doi: 10.1016/0165-1838(95)00066-6. [DOI] [PubMed] [Google Scholar]
- 42.Ullian EM, Sargent PB. Pronounced cellular diversity and extrasynaptic location of nicotinic acetylcholine receptor subunit immunoreactivities in the chicken pretectum. J Neurosci. 1995;15:7012–7023. doi: 10.1523/JNEUROSCI.15-11-07012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vernallis AB, Conroy WG, Berg DK. Neurons assemble acetylcholine receptors with as many as three kinds of subunits while maintaining subunit segregation among receptor subtypes. Neuron. 1993;10:451–464. doi: 10.1016/0896-6273(93)90333-m. [DOI] [PubMed] [Google Scholar]
- 44.Wada K, Ballivet M, Boulter J, Connolly J, Wada E, Deneris ES, Swanson LW, Heinemann S, Patrick J. Functional expression of a new pharmacological subtype of brain nicotinic acetylcholine receptor. Science. 1988;240:330–334. doi: 10.1126/science.2832952. [DOI] [PubMed] [Google Scholar]
- 45.Wilson Horch HL, Sargent PB. Perisynaptic surface distribution of multiple classes of nicotinic acetylcholine receptors on neurons in the chicken ciliary ganglion. J Neurosci. 1996;15:7778–7795. doi: 10.1523/JNEUROSCI.15-12-07778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan B-Y, Ardell JL, Hopkins DA, Losier AM, Armour JA. Gross and microscopic anatomy of the canine intrinsic cardiac nervous system. Anat Rec. 1994;239:75–87. doi: 10.1002/ar.1092390109. [DOI] [PubMed] [Google Scholar]