Abstract
Appicans are secreted or cell-associated brain chondroitin sulfate proteoglycans produced by glia cells and containing Alzheimer amyloid precursor protein (APP) as a core protein. Here, we report that rat C6 glioma cells transfected with appican displayed a dramatic change in their phenotypic appearance compared with untransfected cells or cells transfected with APP. Appican-transfected cells lost the round appearance of the untransfected control C6 cells, acquired a flat morphology, and elaborated more processes than control cells. Untransfected, or APP-transfected C6, cells were completely dissociated from their substrate after 40 min of treatment with cell dissociation solution. Under the same conditions, however, <20% of the appican-transfected C6 cells were dissociated from their substrate, suggesting that the appican-transfected glia cells attach more avidly to their substrate than do untransfected or APP transfected control cells. In contrast, appican-transfected fibroblast cells showed no morphological changes and dissociated from their substrate similarly to untransfected fibroblast cells. Extracellular matrix (ECM) prepared from appican-transfected C6 cell cultures contained high levels of appican and was a significantly better substrate for the attachment of C6 cells than ECM from either untransfected or APP-transfected cultures. Furthermore, cell adhesion to ECM was independent of the level of appican expression of the plated cells. ECM from appican-transfected C6 cultures stimulated adhesion of other neural cells including primary astrocytes, Neuro2a neuroblastoma, and PC12 pheochromocytoma, but not fibroblast cells. Conditioned media from appican-transfected C6 cultures failed to promote cell adhesion. Together, these data suggest that secreted appican incorporates into ECM and promotes adhesion of neural cells. Furthermore, our data suggest that the chondroitin sulfate chain engenders APP with novel biological functions.
Keywords: Alzheimer disease; appican; APP; cell adhesion; extracellular matrix, proteoglycan
The amyloid precursor proteins (APPs) are a family of type I transmembrane glycoproteins involved in the development of Alzheimer disease (AD). They are the precursors of Aβ peptide, which aggregates to form the amyloid deposition characteristic of AD pathology (for review, see Robakis, 1994). In addition, certain APP mutations seem to be sufficient for the induction of the AD phenotype (for review, see St. George-Hyslop et al., 1994). The APP gene contains at least 19 exons, and several distinct APP species have been detected as a result of alternative exon splicing. APPs are expressed ubiquitously and have been shown to regulate cell growth (Saitoh et al., 1989), promote cell adhesion (Schubert et al., 1989; Breen et al., 1991; Chen and Yankner, 1991), and enhance neurite extension (Robakis et al., 1990; Milward et al., 1992; Qiu et al., 1995).
Appicans are brain chondroitin sulfate proteoglycans (CSPGs) containing APP as a core protein and are mainly expressed by glia cells (Shioi et al., 1992, 1995; Pangalos et al., 1995a). Recent reports showed that appicans contain one chondroitin sulfate (CS) chain attached to L-APP, a splicing variant of APP without exon 15. Absence of this exon is essential for the formation of the appropriate consensus sequence that allows the attachment of the CS glycosaminoglycan chain to the core L-APP protein (Pangalos et al., 1995b). Therefore, APP isoforms containing exon 15 are not expected to be present in the proteoglycan form. Appican core proteins containing the Kunitz protease inhibitor (KPI) insert of APP are called KL-APP. In KL-APP733protein, which contains 733 amino acid residues, the CS chain of appican is attached to residue Ser619 located 16 amino acids N terminus to the Aβ sequence. Construction of a KL-APP733 mutant containing alanine instead of serine at position 619, called KL-APP733S/A, failed to produce any appican (Pangalos et al., 1995b). It was reported recently that other members of the APP family of proteins including APLP2 may also occur as CSPGs (Thinakaran and Sisodia, 1994; Pangalos et al., 1995a).
Cell surface appican contains full-length L-APP, whereas secreted appican contains a truncated APP without the transmembrane and cytoplasmic regions produced after the core protein of cellular appican is cleaved by α-secretase (Shioi et al., 1992, 1993). Although the biological function(s) of appicans is not known, CSPGs have been postulated to be involved in a number of key cellular functions (Jackson et al., 1991; Oohira et al., 1994; Small et al., 1996). In the brain, CSPGs have been suggested to have neuroprotective properties and to modulate cell adhesion, axonal growth, and neural patterning (Snow et al., 1990; Brittis et al., 1992; Margolis and Margolis, 1993). Proteoglycans, including heparan, dermatan, and CSPGs, have been found in and around senile plaques and neurofibrillary tangles, suggesting that they may play a role in the pathogenesis of AD (Snow et al., 1992;Su et al., 1992; Dewitt et al., 1993). Appicans were found in human and rat brains, and they are produced by glial cells rather than by neurons in rat brain primary cultures (Shioi et al., 1995). Expression of appican in transformed cell lines is also cell-type specific and depends on the growth conditions of the culture (Pangalos et al., 1995a). In the present study, we show that transfection of glial cells with appican changes dramatically both their morphology and cell adhesion properties. Moreover, extracellular matrix (ECM) prepared from appican-transfected glia cultures stimulated adhesion of both glia and neuronal cells.
MATERIAL AND METHODS
Cell cultures and transfections. All cell lines were from ATCC cell bank (Shioi et al., 1993). Transformed cell lines rat C6 glioma, mouse Neuro2a (N2a) neuroblastoma, or human embryonic fibroblasts 293 cells were grown either in DMEM (Life Technologies, Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS, JRH Biologicals, Lenexa, KA) or in DMEM supplemented with 1 mg/ml transferrin, 5 μg/ml insulin, 30 nm sodium selenite, 20 nm progesterone, and 100 μm putrescine, and maintained at 37°C in an atmosphere of 5% CO2 and 100% relative humidity. Rat pheochcromocytoma PC12 cells were grown in RPMI-1640 medium plus 10% horse serum (Gemini Bio-Products, Calabasas, CA) and 5% FBS. Chinese hamster ovary (CHO) cells were grown in McCoy 5A medium (Sigma, St. Louis, MO) supplemented with 10% FBS. Primary astrocyte cultures were prepared from rats at postnatal day 2 as described (Shioi et al., 1995). cDNA species encoding K-APP751 (the KPI and exon 15 containing APP), KL-APP733, or KL-APP733S/A, where serine 619 is mutated to alanine, were subcloned into vector pRc-CMV, and the resultant plasmids were used to transfect the appropriate cell lines as described (Pangalos et al., 1995b). Transfectants were maintained in 400 μg/ml Geneticin (Life Technologies). To eliminate clonal effects, experiments were performed initially on pooled colonies of stable appican transfectants and then repeated on individual colonies.
Cell dissociation assay. Untransfected or stably transfected cells were plated at a cell density of ∼5 × 105 cells per 35 mm Falcon tissue culture dish (Falcon Primaria, Lincoln Park, NJ). Cells were grown in the presence of 10% FBS, until they were ∼80% confluent, and then washed with DMEM and cultured for an additional 2–3 d in DMEM medium with supplements in the absence of serum. At the end of the incubation, cells were washed carefully with 1 ml PBS and then treated with 0.8 ml of the nonenzymatic EDTA-containing cell dissociation solution (Sigma) for various times on a shaker. Detached cells were aspirated, and cells that remained attached to the plate were dissociated by trypsinization and counted using a hemocytometer.
Preparation of ECM. ECM was prepared as described (Fairbairn et al., 1985). Briefly, untransfected or transfected C6 cells were trypsinized, resuspended, and counted. Cells were then plated at a density of 3× 105 cells per plate on 35 mm tissue culture dishes in DMEM containing 10% FBS and were grown for 3 d until they were ∼80% confluent. Cells were then fed with DMEM containing 1% FBS and cultured for an additional 2–3 d. At the end of the incubation, cells were washed once with 2 ml of PBS containing 5 mm EDTA (buffer A) and then incubated in the same solution at room temperature with gentle shaking until all cells detached from the plate. Plates were washed twice with PBS and then fixed and sterilized for 15 min in 70% ethanol followed by 20 min of UV irradiation. Plates were stored in sterile PBS at 4°C and kept for a maximum of 10 d before use.
Cell adhesion assay. Untransfected or transfected cells were trypsinized and suspended in DMEM containing 10% FBS (C6, N2a, and 293 cells) or 10% FBS plus 5% horse serum (PC12 cells). Approximately 1× 106 cells were pipetted on to 35 mm ECM plates prepared as described above. Cells were allowed to adhere to the plates for 30 min or 2 hr, depending on the cell type, and unattached cells were removed by aspiration. Plates were then washed once with PBS, and attached cells were trypsinized and counted as above.
Determination of ECM appican. Approximately 2 × 106 C6 cells were plated on a 100 mm dish and grown in DMEM plus 10% FBS. At 70–80% confluency, cells were changed to DMEM medium containing 1% FBS and cultured until they reached confluency. Cells were then incubated for 24 hr in sulfate-free DMEM plus serum-free supplements in the presence of [35SO4] (carrier-free, NEN, 400 μCi/5 ml). At the end of the incubation, cells were dissociated from plates with buffer A and gentle shaking, and the dissociated cells were collected and cellular protein was determined as described (Pangalos et al., 1995a). Plates were then washed several times with buffer A to ensure removal of all cells, and the remaining ECM fraction was extracted as described, with modification (Kahari et al., 1991). Briefly, 500 μl of a buffer containing 0.125 m Tris-Cl, pH 6.8, 2% SDS, 1 mm phenylmethylsulfonyl fluoride (Boehringer Mannheim, Indianapolis, IN), 25 μg/ml pepstatin (Boehringer Mannheim), 25 μg/ml leupeptin (Sigma), 50 μg/ml aprotinin (Sigma), and 1 mm 1,10-phenanthroline (Sigma) were added to each plate, and the ECM was then extracted into the buffer by scraping with a rubber cell scraper. Extracts were collected, and plates were rinsed three times with PBS to completely remove the ECM. More than 95% of the SDS was then removed using the Extrati-Gel D Detergent Removing Gel system according the manufacture’s directions (Pierce, Rockford, IL). After the final centrifugation, the supernatant was collected and ECM appican was then immunoprecipitated using anti-α5 antibody, which is specific for human APP (Pangalos et al., 1995b). Chondroitinase ABC treatment of the sample was performed as described (Shioi et al., 1995).
Statistics. Nonlinear regression was used to fit an exponential increase curve to dissociation data. Means were obtained from at least three independent experiments and were compared with Student’s t test and judged for significance (p < 0.05). Error bars indicate SD.
RESULTS
It was reported recently that C6 glioblastoma cells transfected with KL-APP733, the KPI-containing isoform of L-APP, produce and secrete high levels of appican compared with untransfected, K-APP751-transfected, or KL-APP733S/A-transfected C6 cells (Pangalos et al., 1995b; see also introductory remarks). We observed that C6 cells transfected with the appican-producing KL-APP733 displayed a clear change in their morphology compared with either untransfected cells or cells transfected with nonappican-producing APP. At low confluency, untransfected or KL-APP733S/A-transfected C6 cells appeared round with few processes when grown for 3–4 d in the presence of nonserum DMEM plus supplements (Fig.1B,C). At high confluency, these cells appeared small and round and adhered to each other in clumps, which caused large areas of the plate to be free of cells (Fig. 1E,F). In contrast, C6 cells transfected with the appican-producing KL-APP733 displayed a cellular morphology distinctly different from that of the untransfected or KL-APP733S/A-transfected cultures (Fig.1E,D). At low confluency, the appican-transfected cells appeared flattened and were larger than control cells. In addition, appican-transfected cells had substantially more processes than control cells and elaborated fillopodia-like projections that were not observed in control cultures (Fig.1A). At high confluency, these cells also appeared flattened out, did not form clumps, and covered the entire area of the tissue culture plate (Fig. 1D). C6 cells transfected with K-APP751, the KPI isoform of APP, which contains exon 15, showed morphological characteristics similar to those observed in untransfected or KL-APP733S/A-transfected C6 cells (data not shown), suggesting that the morphological changes observed in the KL-APP733-transfected C6 cultures are related to the appican production rather than to the absence of exon 15 from the KL-APP (Pangalos et al., 1995b). Fibroblast cell line 293 transfected with KL-APP733 also produced and secreted significantly higher levels of appican than either untransfected or KL-APP733S/A-transfected 293 fibroblasts (Pangalos et al., 1995b). However, all fibroblast cultures maintained their original round appearance (data not shown), suggesting that the morphological changes observed in the appican-transfected C6 cells may be cell-type specific.
Fig. 1.
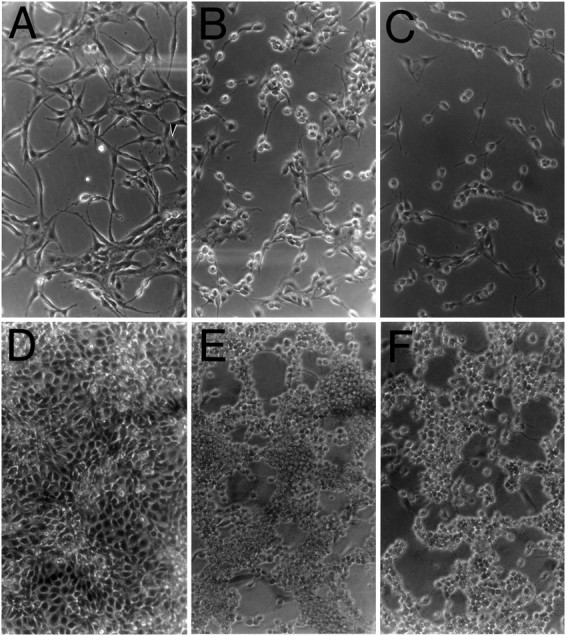
Morphological changes of appican-transfected C6 cells. Untransfected or transfected C6 cells were cultured in DMEM plus 10% FBS for 2 d. After ∼100% confluency, the culture medium was changed to DMEM plus supplements in the absence of serum for an additional 3–4 d. Pictures were then taken under a phase-contrast microscope at 200× magnification.A–C, Cells cultured at low density. D–F, Confluent cell cultures.A, D, C6 cells transfected with the appican-producing KL-APP733. B,E, C6 cells transfected with KLAPP733S/A.C, F, Untransfected C6 cells.Arrow in A indicates a cell elaborating several processes.
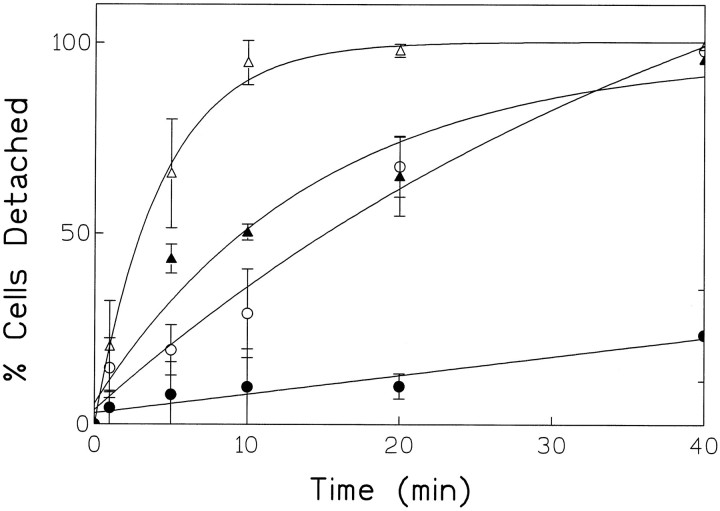
The change in the morphological characteristics of the C6 cells transfected with KL-APP733 suggested that appican may modulate the substrate adhesion of these cells. To examine this hypothesis, we took advantage of a nonenzymatic EDTA-containing cell dissociation solution (see Materials and Methods) used routinely to suspend attached cells. It can be seen in Figure 2 that untransfected C6 cells dissociated rapidly from the tissue culture plates, with ∼70% of the total number of starting cells detached from the plate after 5 min of incubation with the dissociation solution. After 30 min of treatment, practically all cells were detached from the substrate. KL-APP733S/A-transfected C6 cells detached somewhat slower than the untransfected cells. Approximately 30% of the total starting cells dissociated from the plates after 5 min of treatment and >70% of the total cells detached after 30 min. C6 cells transfected with the appican-producing KL-APP733, however, detached from the plates at the slowest rate, with ∼5% or 20% of the total number of starting cells detached from the plates after 5 or 30 min of treatment with dissociation solution, respectively. C6 cells transfected with K-APP751 showed a dissociation rate similar to that of C6 cells transfected with KL-APP733S/A. The slower dissociation rate of cells transfected with either KL-APP733S/A or K-APP751, compared with untransfected C6 cells, is in agreement with data indicating that APP itself may act as a cell adhesion agent (Schubert et al., 1989; Breen et al., 1991; Chen and Yankner, 1991). Determination of the total APP secreted by each one of our cultures showed that all transfectants produced comparable levels of APP (data not shown) (see also Pangalos et al., 1995b), suggesting that the observed differences in dissociation are not attributable to differences in APP expression. The specificity of the effects of appican on cell–substrate dissociation was examined using fibroblast 293 and CHO cells. Although KL-APP733-transfected cell cultures of both cell lines produced high levels of appican (Pangalos et al., 1995b) (M. Pangalos and N. Robakis, unpublished observations), they dissociated from their substrate in a manner similar to that of untransfected control cultures (data not shown). These data suggest that as with the morphological changes (see above), the appican effects on cell dissociation may be cell-type specific.
Fig. 2.
Dissociation kinetics of appican-transfected C6 cells. Untransfected or transfected C6 cells were cultured in DMEM plus 10% FBS for 3–4 d until the cells became ∼80% confluent. The culture medium was then changed to DMEM plus nonserum supplements for another 2–3 d. Cells were dissociated at different time points using a nonenzymatic cell dissociation solution with gentle shaking and the number (A) (see equation in legend to Fig. 3) of cells remaining attached on the plates were determined (see Materials and Methods). The total number (B) of cells on the plates was obtained by direct trypsinization and counting. The detached cell number was calculated by subtracting A from B. Solid circles, KL-APP733-transfected C6 cells; open circles, KL-APP733S/A-transfected C6 cells;solid triangles, K-APP751-transfected C6 cells; open triangles, untransfected C6 cells.
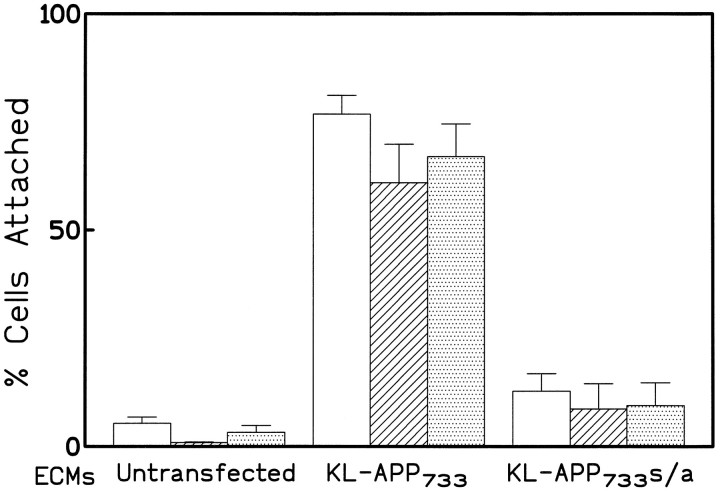
To study further the effects of appican on cell adhesion, we examined the rate of attachment of untransfected and KL-APP733-transfected C6 cells on tissue culture plates. Approximately 50% of the suspended untransfected C6 cells attach to tissue culture dishes within the first 30 min of plating (A. Wu and N. Robakis, unpublished observations). Using this system, no difference in the rate of attachment was observed among untransfected C6 cells or cells transfected with KL-APP733, KL-APP733S/A, or K-APP751 (data not shown), suggesting that the level of intracellular appican does not affect the rate at which C6 cells attach to their substrate. Furthermore, when suspensions of untransfected cells were allowed to attach to tissue culture dishes in the presence of conditioned media obtained either from untransfected or from any of the APP-transfected C6 cultures, no difference was detected in the rate of cell attachment (data not shown), even though the conditioned media from the KL-APP733-transfected C6 cells contained significantly higher levels of appican than conditioned media from the other cell cultures (Pangalos et al., 1995b). These observations suggest that the levels of the soluble appican in the culture media do not affect the adhesion of the C6 cells. All C6 cells, however, untransfected or transfected, attached at a significantly higher rate to ECM plates (see Materials and Methods) prepared from C6 cultures transfected with the appican-producing protein KL-APP733 than to ECM plates prepared from either untransfected or KL-APP733S/A-transfected C6 cultures (Fig.3). Furthermore, all C6 cells attached at a similar rate to each specific ECM, irrespective of their APP transfection, suggesting that ECM, rather than production of cellular appican, was the critical factor in the adhesion of these cells. C6 cells attached slightly better to ECM from the APP-transfected cells than to ECM from wild-type cells (compare far left and far rightsets of bars in Fig. 3). This result is in accord with the mild cell adhesion properties of the APP proteins (see above).
Fig. 3.
Cell attachment on ECM from appican-transfected cultures. All three types of C6 cells, untransfected (open bars), KL-APP733-transfected (hatched bars), and KL-APP733S/A-transfected (dotted bars), were plated on each ECM prepared from untransfected, KL-APP733-transfected, or KL-APP733S/A-transfected C6 cells. The total number (B) of cells plated was determined and after 30 min of incubation at 37°C, media were aspirated and cells were washed once with PBS. The number (A) of cells attached on the ECM plates was determined as described in Figure 2. The percentage (C) of cells attached was calculated using the formula: C = (A/B) · 100%.
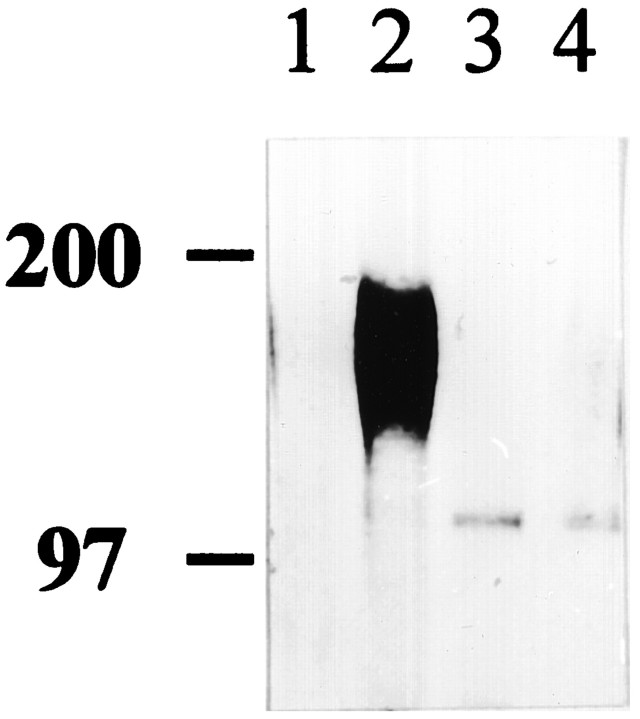
Together, these observations suggested that ECM-incorporated appican, rather than secreted or cellular appican, was the mediator of the observed increased cell adhesion of C6 cells transfected with KL-APP733. To examine whether appican actually incorporates into the ECM and to determine its levels in the ECM of untransfected, KL-APP733-, or KL-APP733S/A-transfected C6 cells, appican was labeled with [35SO4], which incorporates in the CS chain of appicans (Shioi et al., 1993), and ECM plates were prepared from each culture. The ECM levels of appican were then determined by immunoprecipitation using α-5 antibody (see Material and Methods). As shown in Figure 4, the characteristic diffuse banding pattern of appican between 100–200 kDa (Shioi et al., 1992, 1995;Pangalos et al., 1995a,b) was clearly detected in the ECM from the KL-APP733--transfected C6 cultures (lane 2) but not in the ECM from either wild-type or KL-APP733S/A-transfected cells (lanes 1,4). As expected (Shioi et al., 1992, 1993,1995), chondroitinase ABC digestion of the samples resulted in the degradation of the appican and the concomitant appearance of the core APP protein, which is labeled because of a sulfated tyrosine residue (Weidemann et al., 1989). Combined, our data indicated that appican incorporates into ECM and mediates the attachment of C6 cells to their substrate.
Fig. 4.
Detection of appican in ECM. ECM was prepared from untransfected, KL-APP733-transfected, and KL-APP733S/A-transfected C6 cell cultures labeled with [35SO4 ] for 24 hr. Labeled ECM fractions were extracted and immunoprecipitated with α-5 antibody, and immunoprecipitates were analyzed on 6% SDS-PAGE (see Materials and Methods). Lane 1, ECM fraction from untransfected C6 cells; lane 2, ECM fraction from KL-APP733-transfected C6 cells; lane 3, ECM fraction from KL-APP733-transfected C6 cells treated with chondroitinase ABC; lane 4, ECM fraction from KL-APP733S/A-transfected C6 cells.
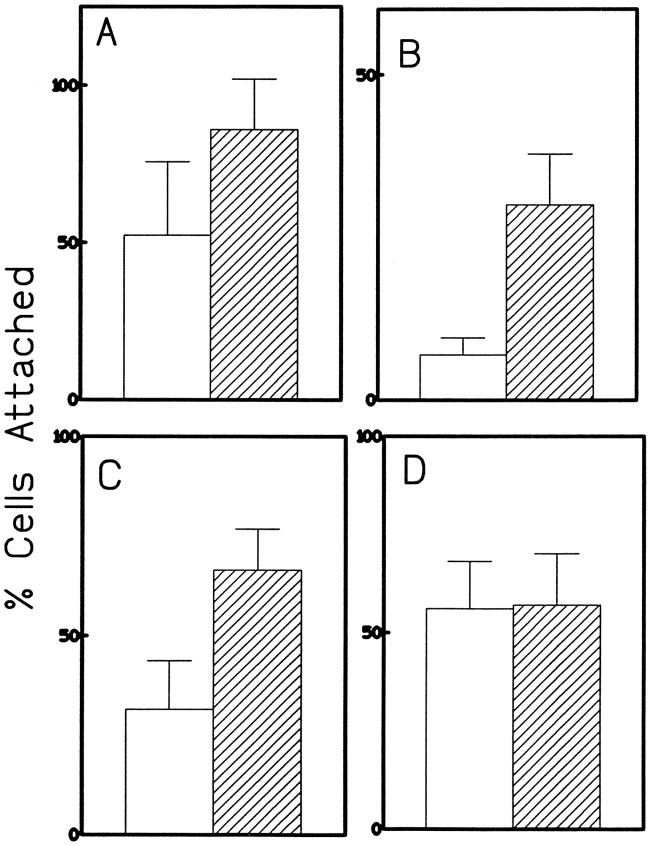
Appicans are found in brain and have been shown to be produced mainly by astrocytes in vitro (Shioi et al., 1995). Therefore, it was of interest to see whether ECM prepared from appican-transfected cells would promote attachment of other neural cells. Figure 5A shows that primary rat brain astrocytes prepared as described (Shioi et al., 1995) attached significantly faster to ECM plates prepared from C6 cultures transfected with KL-APP733 than to ECM plates from KL-APP733 S/A-transfected C6 cultures. Similar results were obtained with PC12 and N2a cells, both of which are of neural origin (Fig. 5B,C). However, 293 fibroblast cells attached equally well to both ECM preparations (Fig.5D), suggesting that the appican-containing ECM specifically promoted the adhesion of neural cells. Furthermore, ECM prepared from appican-producing fibroblast 293 cells transfected with KL-APP733 (Pangalos et al., 1995b) did not promote adhesion of C6 cells, providing additional evidence for the cell specificity of the observed cell-adhesion effects of appican (data not shown). Presently, it is not clear whether the inability of the ECM from appican-transfected fibroblasts to promote adhesion of C6 cells is attributable to a lack of appican incorporation into this ECM or to other differences between the C6 and fibroblast ECM preparations.
Fig. 5.
Attachment assay for primary astrocytes, PC12, N2A, and 293 cells on appican-containing ECM. Attachment experiments were performed as described in Figure 3. Primary astrocytes (A), PC12 (B), N2a (C), and 293 cells (D) were plated on ECM plates prepared from C6 cells transfected with KL-APP733 (hatched bars) or KL-APP733S/A (open bars). The incubation time for primary astrocytes, N2a cells, and 293 cells was 30 min, whereas the time for PC12 cells was 2 hr, attributable to the slow attachment displayed by these cells. Percentage of cells attached was calculated as described in Figure 3.
DISCUSSION
CSPGs participate in a wide range of cellular functions. They have been found both on the cell surface, where they react with ECM molecules, thus promoting cell adhesion (Conget and Minguell, 1994;Henke et al., 1996), and as components of the ECM in several tissues including brain. Although it has been suggested that in the brain, depositions of extracellular CSPGs may provide inhibitory cues for growing neurites (Snow et al., 1990), ECM CSPGs seem essential for the adhesion between neural retina and retinal pigmented epithelium (Lazarus and Hageman, 1992). The experiments reported here show that increased appican expression in glial cells changes profoundly both their morphology and their process elaboration. Specifically, appican-transfected C6 glioblastoma cells appeared larger, had a flattened morphology, and elaborated more processes than either untransfected C6 cells or cells transfected with nonappican-producing APP. Furthermore, in response to a standard cell dissociation solution, appican-transfected glial cells dissociated from their substrate at a significantly slower rate than either untransfected or APP-transfected control cells, suggesting that appican increases the avidity of substrate attachment of glial cells. On the contrary, increased appican production in transfected fibroblasts or CHO cells affected neither their morphological appearance nor their substrate dissociation, indicating that the appican effects are cell-type specific. Taken together, these data show that attachment of the CS chain on the core APP protein changes profoundly the effects of APP on cell adhesion and morphology.
Examination of the rate of substrate attachment of medium-suspended glial cells showed that increased levels of either cellular or medium appican had no effect on this rate. However, glial cells attached significantly faster to ECM substrates prepared from appican-transfected C6 cell cultures than to ECM prepared from either untransfected or APP-transfected cultures. Combined, these observations suggested that secreted appican may incorporate into the ECM and promote adhesion of glial cells. This suggestion was strongly supported by the high levels of appican detected in the ECM prepared from appican-transfected C6 cultures. Importantly, ECM from appican-transfected glial cultures promoted adhesion of other cell lines of neural origin, including N2a neuroblastoma cells, whereas it failed to promote adhesion of fibroblasts, providing additional support for the conclusion that the appican effects on cell adhesion are cell-type specific. The specificity of the appican effects is consistent with the endogenous expression of this molecule in cells of neural origin (Shioi et al., 1992, 1995; Pangalos et al., 1995a), as well as with the absence of endogenous appican expression in fibroblast. Because expression of appican was detected in the brain (Shioi et al., 1995), this proteoglycan may also modulate the adhesion and morphogenesis of glial cells in the CNS.
ECM, a structure that plays important roles in cellular morphogenesis and differentiation, is a dynamic assemplance of interacting molecules, and often a change in the expression of one ECM component may alter the molecular composition and function of the ECM (Reing et al., 1992). Our data show that appican incorporates into the ECM of glial cells and changes both their morphology and their adhesion. However, it is not clear whether appican functions directly as a cell adhesion molecule or whether it regulates the expression of other ECM components, which, ultimately, may be responsible for the observed appican effects on cell adhesion and morphology. We obtained evidence that appican expression also enhanced adhesion of C6 cells to laminin-coated plates (Pangalos and Robakis, unpublished observations). Additional work should clarify whether the ECM appican interacts directly with specific integrins on the surface of glia cells, thus promoting cell–substrate adhesion, or whether its effects are mediated through its interaction with other integrin ligands of the ECM.
Proteoglycans have been shown to bind the Aβ peptide and modulate its aggregation to amyloid fibers (Brunden et al., 1993; Buee et al., 1993). In addition, CSPGs have been found in both senile plaques and neurofibrillary tangles (DeWitt et al., 1993), suggesting that these molecules, and possibly appicans, are involved in the pathology of AD. Strong evidence in support of a direct role of APP in the development of AD derives from genetic studies, indicating that certain APP mutations are closely linked to AD in a small number of families (FAD). However, it is not clear whether the FAD mutations cause AD by increasing Aβ production or by altering the biological properties of APP (Regland and Gottfries, 1992; Robakis and Pangalos, 1994; Saitoh and Brugge, 1994). Our results provide evidence that appicans may mediate cell adhesion in the brain and may affect the morphology of glia cells. It remains an interesting question whether the FAD mutations of APP affect the cellular function of appican and its relation to the development of AD.
Footnotes
This work was supported by National Institutes of Health Grants AG08200 and AG05138.
A.W. and M.N.P. contributed equally to this work.
Correspondence should be addressed to Dr. Nikolaos K. Robakis, Mount Sinai School of Medicine, Department of Psychiatry and Fishberg Research Center for Neurobiology, One Gustave L. Levy Place, New York, NY 10029.
Dr. Pangalos’s present address: Janssen Pharmaceutica, Department of Applied Molecular Biology, Turnhoutseweg 30, B-2340 Beerse, Belgium.
REFERENCES
- 1.Breen KC, Bruce M, Anderton BH. β-Amyloid precursor protein mediates neuronal cell-cell and cell-surface adhesion. J Neurosci Res. 1991;28:90–100. doi: 10.1002/jnr.490280109. [DOI] [PubMed] [Google Scholar]
- 2.Brittis PA, Canning DR, Silver J. Chondroitin sulfate as a regulator of neuronal patterning in the retina. Nature. 1992;255:733–736. doi: 10.1126/science.1738848. [DOI] [PubMed] [Google Scholar]
- 3.Brunden KR, Richter CNJ, Chaturvedi N, Frederickson RC. pH-Dependent binding of synthetic beta-amyloid peptides to glycosaminoglycans. J Neurochem. 1993;61:2147–2154. doi: 10.1111/j.1471-4159.1993.tb07453.x. [DOI] [PubMed] [Google Scholar]
- 4.Buee L, Ding W, Delacourte A, Fillit H. Binding of secreted human neuroblastoma proteoglycans to the Alzheimer’s amyloid A4 peptide. Brain Res. 1993;601:154–163. doi: 10.1016/0006-8993(93)91706-x. [DOI] [PubMed] [Google Scholar]
- 5.Chen M, Yankner BA. An antibody to beta amyloid and amyloid precursor protein inhibits cell-substratum adhesion in many cell types. Neurosci Lett. 1991;125:223–226. doi: 10.1016/0304-3940(91)90034-q. [DOI] [PubMed] [Google Scholar]
- 6.Conget P, Minguell JJ. Modifications in the synthesis of membrane-associated chondroitin sulfate proteoglycans in hemoietic progenitor cells are accompanied by alterations in their adhesive properties. J Cell Physiol. 1994;159:142–150. doi: 10.1002/jcp.1041590118. [DOI] [PubMed] [Google Scholar]
- 7.DeWitt DA, Silver J, Canning DR, Perry G. Chondroitin sulfate proteoglycans are associated with the lesions of Alzheimer’s disease. Exp Neurol. 1993;121:149–152. doi: 10.1006/exnr.1993.1081. [DOI] [PubMed] [Google Scholar]
- 8.Fairbairn S, Gilbert R, Ojakian G, Schwimmer R, Quigley JP. The extracellular matrix of normal chick embryo fibroblasts: its effect on transformed chick fibroblasts and its proteolytic degradation by the transformants. J Cell Biol. 1985;101:1790–1798. doi: 10.1083/jcb.101.5.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henke CA, Roongta U, Mickelson DU, Knutson JR, McCarthy JB. CD44-related chondroitin sulfate proteoglycan, a cell surface receptor implicated with tumor cell invasion, mediates endothelial cell migration on fibrinogen and invasion into a fibrin matrix. J Clin Invest. 1996;97:2541–2552. doi: 10.1172/JCI118702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson RL, Busch SJ, Cardin AD. Glycosaminoglycans: molecular properties, protein interactions and role in physiological processes. Physiol Rev. 1991;71:481–539. doi: 10.1152/physrev.1991.71.2.481. [DOI] [PubMed] [Google Scholar]
- 11.Kahari V-M, Larjava H, Uitto J. Differential regulation of extracellular matrix proteoglycan (PG) gene expression. J Biol Chem. 1991;266:10608–10615. [PubMed] [Google Scholar]
- 12.Lazarus HS, Hageman GS. Xyloside-induced disruption of interphotoreceptor matrix proteoglycans results in retinal detachment. Invest Ophthalmol Vis Sci. 1992;33:364–376. [PubMed] [Google Scholar]
- 13.Margolis RK, Margolis RU. Nervous tissue proteoglycans. Experientia. 1993;49:429–446. doi: 10.1007/BF01923587. [DOI] [PubMed] [Google Scholar]
- 14.Milward EA, Papadopoulos R, Fuller SJ, Small D, Beyreuther K, Masters CL. The amyloid protein precursor of Alzheimer’s disease is a mediator of the effects of nerve growth factor on neurite outgrowth. Neuron. 1992;9:129–137. doi: 10.1016/0896-6273(92)90228-6. [DOI] [PubMed] [Google Scholar]
- 15.Oohira A, Katoh-Semba R, Watanabe E, Matsui F. Brain development and multiple molecular species of proteoglycan. Neurosci Res. 1994;20:195–207. doi: 10.1016/0168-0102(94)90088-4. [DOI] [PubMed] [Google Scholar]
- 16.Pangalos MN, Shioi J, Robakis NK. Expression of the chondroitin sulfate proteoglycans of amyloid precursor (appican) and amyloid-like protein 2. J Neurochem. 1995a;65:762–769. doi: 10.1046/j.1471-4159.1995.65020762.x. [DOI] [PubMed] [Google Scholar]
- 17.Pangalos MN, Efthimiopoulos S, Shioi J, Robakis NK. The chondroitin sulfate attachment site of appican is formed by splicing out exon 15 of the amyloid precursor gene. J Biol Chem. 1995b;270:10388–10391. doi: 10.1074/jbc.270.18.10388. [DOI] [PubMed] [Google Scholar]
- 18.Qiu WQ, Ferreira A, Miller C, Koo EH, Selkoe DJ. Cell-surface beta-amyloid precursor protein stimulates neurite outgrowth of hippocampal neurons in an isoform-dependent manner. J Neurosci. 1995;15:2157–2167. doi: 10.1523/JNEUROSCI.15-03-02157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Regland B, Gottfries CG. The role of amyloid beta-amyloid in Alzheimer’s disease. Lancet. 1992;340:467–469. doi: 10.1016/0140-6736(92)91780-c. [DOI] [PubMed] [Google Scholar]
- 20.Reing J, Durkin ME, Chung AE. Laminin B1 expression is required for laminin deposition into extracellular matrix of PC12 cells. J Biol Chem. 1992;267:23143–23150. [PubMed] [Google Scholar]
- 21.Robakis NK. β-Amyloid and amyloid precursor protein. In: Terry R, Kattman R, Bicke E, editors. Alzheimer’s disease. Raven; New York: 1994. pp. 317–326. [Google Scholar]
- 22.Robakis NK, Pangalos MN. Involvement of amyloid as a central step in the development of Alzheimer’s disease. Neurobiol Aging. 1994;15:S127–S129. doi: 10.1016/0197-4580(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 23.Robakis NK, Altstiel LD, Refolo LM, Anderson JP. Function and metabolism of the protease inhibitor containing Alzheimer amyloid precursors. In: Miyatake T, Selkoe DJ, Ihara Y, editors. Molecular biology and genetic of Alzheimer’s disease. Elsevier; New York: 1990. pp. 179–188. [Google Scholar]
- 24.Saitoh T, Brugge K. Is amyloid casually involved in pathophysiology of Alzheimer’s disease? Neurobiol Aging. 1994;15:461–462. doi: 10.1016/0197-4580(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 25.Saitoh T, Sundsmo H, Roch JM, Kimura N, Cole G, Schubert D, Oltersdorf T, Schenk DB. Secreted forms of amyloid protein precursor is involved in the growth regulation of fibroblasts. Cell. 1989;58:615–622. doi: 10.1016/0092-8674(89)90096-2. [DOI] [PubMed] [Google Scholar]
- 26.Schubert D, LaCorbiere M, Saitoh T, Cole G. The regulation of amyloid b protein precursor and its modulatory role in cell adhesion. Neuron. 1989;3:689–694. doi: 10.1016/0896-6273(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 27.Shioi J, Anderson JP, Ripellino JA, Robakis NK. Chondroitin sulfate proteoglycan form of the Alzheimer’s beta-amyloid precursor. J Biol Chem. 1992;267:13819–13822. [PubMed] [Google Scholar]
- 28.Shioi J, Refolo LM, Efthimiopoulos S, Robakis NK. Chondroitin sulfate proteoglycan form of cellular and cell-surface Alzheimer amyloid precursor. Neurosci Lett. 1993;154:121–124. doi: 10.1016/0304-3940(93)90186-o. [DOI] [PubMed] [Google Scholar]
- 29.Shioi J, Pangalos MN, Ripellino JA, Vassilacopoulou D, Mytilineou C, Margolis RU, Robakis NK. The Alzheimer amyloid precursor proteoglycan (appican) is present in brain and is produced by astrocytes but not by neurons in primary neural cultures. J Biol Chem. 1995;270:11839–11844. doi: 10.1074/jbc.270.20.11839. [DOI] [PubMed] [Google Scholar]
- 30.Small DHS, Mok SS, Williamson TG, Nurcombe V. Role of proteoglycan in neural development, regeneration, and the aging brain. J Neurochem. 1996;67:889–899. doi: 10.1046/j.1471-4159.1996.67030889.x. [DOI] [PubMed] [Google Scholar]
- 31.Snow AD, Mar H, Nochlin D, Kresse H, Wight TN. Peripheral distribution of dermatan sulfate proteoglycans (decorin) in amyloid-containing plaques and their presence in neurofibrillary tangles of Alzheimer’s disease. J Histochem Cytochem. 1992;40:105–113. doi: 10.1177/40.1.1370306. [DOI] [PubMed] [Google Scholar]
- 32.Snow DM, Lemmon V, Carrino DA, Caplan AI, Silver J. Sulfated proteoglycans in astroglial barriers inhibit neurite outgrowth in vitro. Exp Neurol. 1990;94:767–776. doi: 10.1016/s0014-4886(05)80013-5. [DOI] [PubMed] [Google Scholar]
- 33.St. George-Hyslop P, McLachlan DC, Tsuda T, Rogaev E, Lippa CF, Pollen D, Tuda T. Alzheimer’s disease and possible gene interaction. Science. 1994;263:537. doi: 10.1126/science.8290965. [DOI] [PubMed] [Google Scholar]
- 34.Su JH, Cummings BJ, Cotman CW. Localization of heparan sulfate glycosaminoglycan and proteoglycan core protein in aged brain and Alzheimer’s disease. Neuroscience. 1992;51:801–813. doi: 10.1016/0306-4522(92)90521-3. [DOI] [PubMed] [Google Scholar]
- 35.Thinakaran G, Sisodia SS. Amyloid precursor-like protein 2 (APLP2) is modified by the addition of chondroitin sulfate glycosaminoglycan at a single site. J Biol Chem. 1994;269:22099–22104. [PubMed] [Google Scholar]
- 36.Weidemann A, Konig G, Bunke D, Fischer P, Salbaum JM, Master CL, Beyreuther K. Identification, biogenesis and localization of precursors of Alzheimer’s disease A4 amyloid protein. Cell. 1989;57:115–126. doi: 10.1016/0092-8674(89)90177-3. [DOI] [PubMed] [Google Scholar]