Abstract
Two new potassium channel genes, erg2 anderg3, that are expressed in the nervous system of the rat were identified. These two genes form a small gene family with the previously described erg1 (HERG) gene. Theerg2 and erg3 genes are expressed exclusively in the nervous system, in marked contrast toerg1, which is expressed in both neural and non-neural tissues. All three genes are expressed in peripheral sympathetic ganglia. The erg3 channel produces a current that has a large transient component at positive potentials, whereas the other two channels are slowly activating delayed rectifiers. Expression of theerg1 gene in the sympathetic nervous system has potential implications for the etiology of the LQT2 form of the human genetic disease long QT syndrome.
Keywords: potassium channel, erg, long QT syndrome, sympathetic neuron, gene, delayed rectifier
A family of three related voltage-gated potassium channel genes (eag, erg, and elk) has been described in either Drosophilaor mammals (Warmke et al., 1991; Ludwig et al., 1994; Warmke and Ganetzky, 1994; Titus et al., 1997; Wang et al., 1997). These channels share the six-membrane-spanning architecture of the Kv class (Shaker-related) of voltage-gated potassium channels but otherwise are distantly related to the Kv class channels (Warmke and Ganetzky, 1994). The channels encoded by the eag-related genes are relatively slowly activating, as compared with Kv class potassium channels (Ludwig et al., 1994; Sanguinetti et al., 1995;Terlau et al., 1996), and have some similarities to slowly activating potassium currents that are important in determining the threshold firing properties of neurons (Brown, 1988; Yamada et al., 1989; Wang and McKinnon, 1995). As might be anticipated from the biophysical properties of these channels, mutations in either the eaggene or the erg gene of Drosophila result in a hyperexcitable phenotype (Ganetzky and Wu, 1983; Titus et al., 1997;Wang et al., 1997).
The erg gene recently has become the center of considerable interest because mutations in this gene have been shown to underlie one form of a human genetic disease known as long QT syndrome, which gives rise to arrhythmias and an increased incidence of sudden death (Curran et al., 1995; Sanguinetti et al., 1996a). It is thought that theerg gene encodes the channel that underlies a previously identified potassium current known as IKr(Sanguinetti et al., 1995, 1996a; Trudeau et al., 1995). This current was described originally in cardiac myocytes (Shibasaki, 1987;Sanguinetti and Jurkiewicz, 1990), and it has been suggested that mutations in the erg gene may increase the susceptibility to arrhythmia, possibly by causing a prolongation of the cardiac action potential (Curran et al., 1995; Sanguinetti et al., 1996a). Alternatively, it had been suggested previously that dysfunction in the control of sympathetic outflow to the heart can induce the life-threatening arrhythmias that are characteristic of long QT syndrome (Schwartz et al., 1994).
We have shown that transcripts from the erg gene are expressed abundantly in the rat and human nervous systems, suggesting that the erg gene product might contribute to nervous system function (Wymore et al., 1997). The observation that mutations in theerg gene of Drosophila produce clear neurological defects adds further weight to this possibility (Titus et al., 1997;Wang et al., 1997). To examine further the role of the erggene in the mammalian nervous system, we have conducted a systematic screen to identify genes that are related to erg. We have concentrated particularly on the sympathetic nervous system because of the potential clinical importance of erg channels in this tissue. In this paper we describe two new members of the erggene family (erg2 and erg3) that are expressed exclusively in the nervous system and are expressed abundantly in sympathetic ganglia.
MATERIALS AND METHODS
Isolation of partial cDNA clones. Two sets of degenerate oligonucleotides directed against conserved regions of theeag, erg, and elk gene family were designed.
Set 1 was directed against the amino acid sequences “APQNTF” and “FK(AT)(ITV)WDW”: forward, GC(ACGT) CC(ACGT) CA(AG) AA(CT) AC(ACGT) TT(CT); and reverse, CCA (AG)TC CCA (ACGT)(AG)(CT) (ACGT)G(CT) (CT)TT (AG)AA.
Set 2 was directed against the amino acid sequences “FK(AT)(ITV)WDW” and “HWLACIWY”: forward, TT(CT) AA(AG) (AG)C(ACGT) (AG)(CT)(ACGT) TGG GA(CT) TGG; and reverse, (AG)TA CCA (AGT)AT (AG)CA (ACGT)GC (ACGT)AG CCA (AG)TG.
Using these two sets of oligonucleotides, we prepared cDNA fragments by reverse transcription and PCR amplification from total cellular RNA isolated from rat superior cervical ganglia (SCG). The amplified cDNA fragments were gel-purified and then subcloned into pBluescript SK II (Stratagene, La Jolla, CA) and analyzed by manual sequencing. Using this procedure, we identified two novel classes oferg-related cDNA clones. The erg3 cDNA initially was isolated by using the first set of oligonucleotides, and theerg2 cDNA was identified by using the second set.
Isolation of full-length ERG2 and ERG3 cDNA clones.Full-length rat erg2 and erg3 cDNA sequences were obtained first by performing a modified 5′ and 3′ rapid amplification of cDNA ends (RACE) protocol (essentially as described byFrohman, 1994), using anchor oligonucleotides complementary to the partial erg2 and erg3 clones. This required several rounds of RACE in both directions to obtain sequences with complete open reading frames. For erg2 the tissue source for synthesizing the cDNA used in the RACE protocol was celiac ganglia, and for erg3 the tissue source was brain.
Once cDNAs were obtained that extended beyond both the 3′ and 5′ ends of the open reading frame, oligonucleotides complementary to noncoding regions at either end of the coding sequence were designed, and full-length cDNA clones were obtained by using the Expand High Fidelity PCR system (Boehringer Mannheim, Indianapolis, IN) for PCR amplification. For erg2, the following oligonucleotides were used to amplify full-length cDNA clones from rat celiac ganglia RNA, giving a 32 and 497 bp 5′ and 3′ untranslated region (UTR), respectively: forward, GAG TAA CTC CCA GCA AGT GC; and reverse, ACT GTT ATG AGA GTC TCA GGG G.
For erg3, the following oligonucleotides were used to amplify full-length cDNA clones from rat brain RNA, giving a 182 and 37 bp 5′ and 3′ UTR, respectively: forward, GAT GGA TTG GAC TTC GGC; and reverse, GCA CTT ACA TTG GAT GTG GAG.
Full-length cDNAs were subcloned into the pBluescript SK II vector. Twoerg2 clones were sequenced in their entirety, using a combination of manual and automatic sequencing. One erg3 clone was sequenced in its entirety, using a combination of manual and automatic sequencing; a second independent sequence was obtained by sequencing the erg3 clones obtained by RACE. Differences between the two sequences were resolved by partial sequencing of a second full-length erg3 cDNA clone, which also was obtained from brain. Sequence alignment was performed with the Clustal W program (Thompson et al., 1994). Sequences were submitted to GenBank, with accession numbers AF016192 and AF016191 for erg2 anderg3, respectively.
Preparation of RNA. Tissue samples were quick-frozen in liquid N2 and then homogenized in guanidinium thiocyanate. Total RNA was prepared by pelleting the homogenate over a CsCl step gradient as described previously (Dixon and McKinnon, 1996). Poly(A+) RNA was prepared with paramagnetic poly-dT beads (Dynal, Oslo, Norway) as described previously (Wymore et al., 1997). All RNA samples were quantitated carefully by spectrophotometric analysis.
RNase protection assay. RNA probes were prepared as described previously (Dixon and McKinnon, 1994). In all cases a significant amount of nonhybridizing sequence (∼50–80 bp) was included in the probe to allow for easy distinction between the probe and the specific protected band. The specificity of the assay was such that there was no evidence for unwanted cross-reaction between any probe and another nonspecific potassium channel transcript.
RNase protection assays were performed as described previously (Dixon and McKinnon, 1996). For each sample point 5 μg of total RNA or 1 μg of poly(A+) RNA was used in the assay. A species-specific cyclophilin probe was included in the hybridization as an internal control to confirm that the sample was not lost or degraded during the assay. Five micrograms of yeast tRNA were used as a negative control to test for the presence of probe self-protection bands. RNA expression was quantitated directly from dried RNase protection gels with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Expression of erg channels in Xenopusoocytes. Full-length erg1, erg2, anderg3 cRNA transcripts were synthesized in vitro. The erg1 clone was a human cDNA that has been described previously (Sanguinetti et al., 1995) (a gift from Dr. Michael Sanguinetti, University of Utah).
Oocytes were prepared from mature female Xenopus laevis, using established procedures (Colman, 1984). Defolliculation was performed by incubation for 2 hr in 2 mg/ml collagenase (Type VIII, Sigma, St. Louis, MO) in Ca2+-free OR2 oocyte medium with gentle agitation. Oocytes were stored in OR3 solution [50% L-15 medium (Life Technologies, Gaithersburg, MD), 1 mmglutamine, 15 mm Na-HEPES, pH 7.6, and 0.1 mg/ml gentamicin] at 18°C. Oocytes were injected with 50 nl of cRNA (∼0.3 ng/nl) by using a microdispenser and a micropipette with a tip diameter of 10–20 μm. Injected oocytes were incubated at 18°C for 24–48 hr before analysis.
Oocytes were voltage-clamped with a two-microelectrode voltage clamp. Intracellular electrodes filled with 3 m KCl with resistances of 0.5–3 MΩ were used. The standard extracellular recording solution contained (in mm): 80 NaCl, 5 KCl, 1.8 CaCl2, 1 MgCl2, and 5 Na-HEPES, pH 7.6. Data collection and analysis were performed with pClamp software (Axon Instruments, Foster City, CA). The methanesulfonanilide E4031 was obtained from Eisai Company (Tokodai, Japan).
RESULTS
Isolation and properties of the erg2 anderg3 cDNA clones
Two partial cDNA clones, which identified two novel potassium channel genes, were isolated from rat SCG cDNA. The cDNAs were cloned by PCR, with primers that were directed against conserved regions of the eag, erg, and elk gene families. These partial sequences were closely related to the previously described h-erg (or HERG) gene (Warmke and Ganetzky, 1994), which we will refer to in the following discussions as erg1. Full-length erg2 and erg3 cDNA clones were obtained (see Materials and Methods for details) and sequenced in their entirety (Fig. 1). Identification of the initiator methionine and the start of the open reading frame was facilitated by the high degree of similarity in this region among the three genes. There is a short region of conservation in the three sequences at the C terminus immediately before the presumptive stop codon, which suggests that the derived amino acid sequences correctly identify the entire coding regions of all three genes.
Fig. 1.

Alignment of the erg1,erg2, and erg3 deduced amino acid sequences. There is 63% identity between erg2 anderg1, 57% identity between erg3 anderg1, and 61% identity between erg2 anderg3. Residues that are identical in all three sequences are shown with black shading, residues identical in two sequences are shown with dark gray shading, similar residues are shown with light gray shading, and nonconserved residues are shown without shading. Theerg1 sequence corresponds to the humanerg gene (Warmke and Ganetzky, 1994). Theerg2 and erg3 sequences are from rat. The six hydrophobic domains (S1–S6), the pore (P), and the putative cyclic nucleotide binding domain (cNBD) are overlined.
At the amino acid level the three mammalian erg genes had identity scores of ∼60% over the entire deduced amino acid sequence (Table 1). The erg2 sequence was 63% identical and the erg3 sequence was 57% identical to the erg1 gene, and the erg2 sequence was 61% identical to the erg3 sequence. All three mammalianerg sequences had similar identity scores (∼42%) in comparisons with the Drosophila erg gene, suggesting that they all derive from a common gene. Identity with the other members of the eag gene family (eag and elk) was significantly lower, ∼30% overall. This result suggests that theerg1, erg2, and erg3 genes form a distinct subfamily.
Table 1.
Amino acid identity among the erg family of potassium channels
| h-erg1 | r-erg2 | r-erg3 | d-erg | m-eag | d-elk | |
|---|---|---|---|---|---|---|
| erg1 | 100 | 63 | 57 | 43 | 31 | 27 |
| erg2 | 100 | 61 | 41 | 32 | 32 | |
| erg3 | 100 | 44 | 32 | 25 | ||
| d-erg | 100 | 26 | 24 | |||
| m-eag | 100 | 29 | ||||
| d-elk | 100 |
Multiple sequence alignment of entire deduced amino acid sequence was performed using the ClustalW program (Thompson et al., 1994). h-erg1 is human erg (Warmke and Ganetzky, 1994), r-erg2 and r-erg3 are the rat sequences reported in this paper, d-erg is Drosophila erg (Titus et al., 1997), m-eag is mouse eag (Warmke and Ganetzky, 1994), and d-elk is Drosophila elk(Warmke and Ganetzky, 1994).
There are three highly conserved domains in the three sequences: the initial N-terminal sequence, the hydrophobic core, and the putative cyclic nucleotide binding domain. Curiously, the Drosophila erg sequence lacks this conserved N-terminal domain (Titus et al., 1997; Wang et al., 1997), although it is present in all three mammalianerg sequences as well as in the eag andelk sequences. It is known that the mammalian erggenes can produce alternative spliced transcripts that lack the N-terminal coding region (London et al., 1997), and it is possible that there is also alternative splicing of the Drosophila erggene products and that some Drosophila erg mRNA species may include this domain. A short, completely conserved motif (SDPG) is found in the extreme C terminus of all three mammalian ergsequences.
Tissue distribution of erg2 and erg3 gene expression
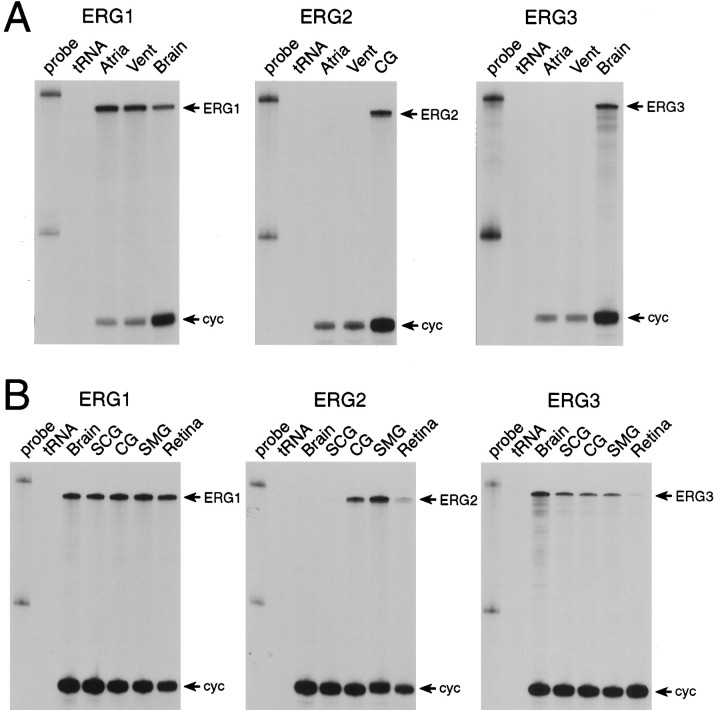
The erg1 gene is expressed in the hearts of all species tested to date (Wymore et al., 1997) and is thought to encode one component of the delayed rectifier potassium current found in heart (Sanguinetti et al., 1995; Trudeau et al., 1995). Mutations in theerg1 gene have been shown to be associated with the LQT2 form of the human genetic disease long QT syndrome, which involves a significantly increased susceptibility to arrhythmias triggered by emotional or physical stress (Curran et al., 1995; Sanguinetti et al., 1996a). For this reason, we first examined erg2 anderg3 mRNA expression in heart. Neither gene is expressed at detectable levels in either atrial or ventricular muscle, in marked contrast to the results obtained with the erg1 gene (Fig.2A).
Fig. 2.
erg potassium channel mRNA expression in heart and neural tissues determined by RNase protection analysis. A, Neither erg2 norerg3 mRNA is expressed at detectable levels in atrial or ventricular (Vent) muscle, in marked contrast toerg1, which is abundant in both tissues (arrows). The positive control samples are celiac ganglia (CG) or brain mRNA. B, All threeerg genes are expressed in neural tissue. Samples tested were brain, superior cervical ganglia (SCG), celiac ganglia (CG), superior mesenteric ganglia (SMG), and retina. The cyclophilin gene (cyc, arrows) was used as an internal positive control; as has been shown previously, cyclophilin expression was always lower in muscle tissues, as compared with other tissues.
As shown previously (Wymore et al., 1997), the erg1 gene is expressed abundantly in brain and in retina. Intriguingly, given the clinical symptoms associated with mutations in the erg1 gene, erg1 mRNA is expressed abundantly in sympathetic ganglia (Fig. 2B). This result suggests that mutations in the erg1 gene could affect sympathetic regulation of cardiac function in addition to having direct effects on myocardial function.
The erg2 gene has a very restricted distribution in neural tissues and is not expressed at detectable levels in brain. Although initially cloned by PCR amplification from SCG mRNA, erg2 expression in SCG is also very low. The peripheral sympathetic nervous system has two anatomically and functionally distinct components: the paravertebral ganglia and the prevertebral ganglia. It has been shown previously that the electrophysiological properties of neurons in the paravertebral ganglia, such as the SCG, are very uniform, whereas there are at least two electrophysiologically distinct types of neurons in the prevertebral ganglia (Cassell et al., 1986; Wang and McKinnon, 1995, 1996). The molecular basis for this differentiation currently is undetermined (Dixon and McKinnon, 1996). The erg2 gene is expressed abundantly in the two prevertebral sympathetic ganglia examined: the celiac ganglia (CG) and the superior mesenteric ganglia (SMG) (Fig. 2B). This result suggests that theerg2 channel could contribute to the electrophysiological differentiation of prevertebral neurons, but identification of the native current corresponding to the erg2 channel is necessary before this conclusion can be established. There is also a low level of erg2 expression in retina.
The erg3 gene is broadly expressed throughout the nervous system, similar to erg1. In addition to brain,erg3 mRNA is expressed in all of the sympathetic ganglia tested and also is expressed at low levels in retina (Fig.2B).
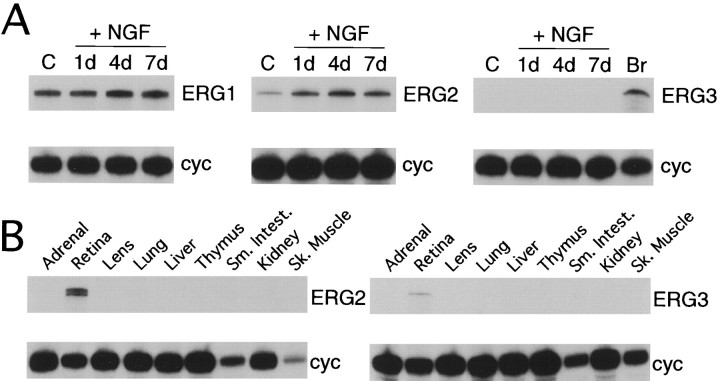
Expression of all three erg genes in sympathetic ganglia prompted us to examine expression in PC12 cells, a cell line that often is used as a cell culture analog of sympathetic neurons. Both theerg1 and erg2 genes are expressed in PC12 cells, whereas the erg3 gene is not expressed (Fig.3A). A further elaboration of the neuronal phenotype of PC12 cells can be induced by treatment with nerve growth factor (NGF). Exposure to NGF resulted in little change inerg1 mRNA expression in PC12 cells. In contrast, upregulation of erg2 mRNA was marked and relatively rapid (∼threefold after 1 d). No induction of erg3 gene expression was observed after NGF treatment.
Fig. 3.
erg potassium channel mRNA expression in PC12 cells and non-neural tissues determined by RNase protection analysis. A, erg mRNA expression in PC12 cells in control media or after 1, 4, or 7 d of treatment with nerve growth factor (NGF). Brain (Br) mRNA was used as a positive control for theerg3 experiment. B, Expression oferg2 and erg3 mRNA in non-neural tissues. Retinal RNA was used as the positive control.
Neither the erg2 nor the erg3 gene was expressed in any of the non-neural tissues that were tested (Fig. 3B), suggesting that expression of these genes is nervous system-specific. This is in marked contrast to the erg1 gene, which is expressed in several non-neuronal tissues, including adrenal gland, thymus, and lung (Wymore et al., 1997).
Kinetic properties of the erg2 and erg3 potassium channels
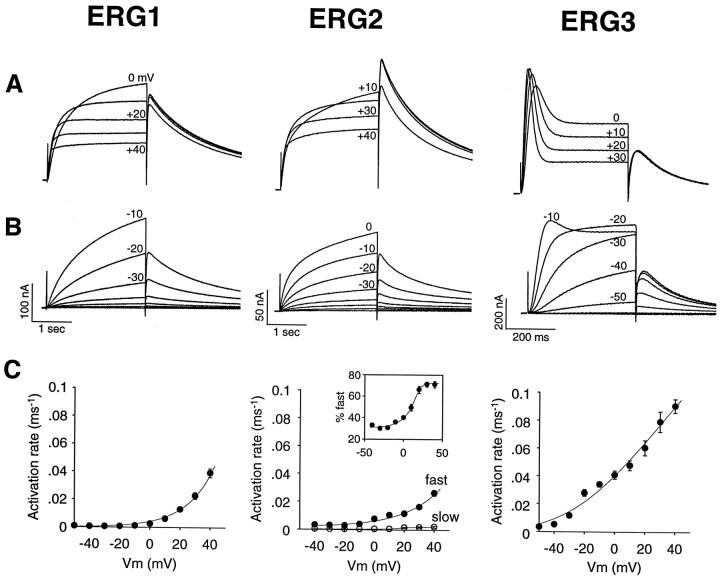
The kinetic properties of the erg1, erg2, and erg3 channels were compared by expressing the channels in Xenopus oocytes. The waveform of the current elicited in response to a depolarizing voltage step to +20 mV was markedly different for the erg3 channel as compared with the other two channels (Fig. 4). Theerg1 and erg2 channels were relatively slowly activating delayed rectifiers, whereas the erg3 current had a predominant transient component that decayed to a sustained plateau. The kinetic properties of the erg1 channel were very similar to those described previously (Sanguinetti et al., 1995; Spector et al., 1996).
Fig. 4.
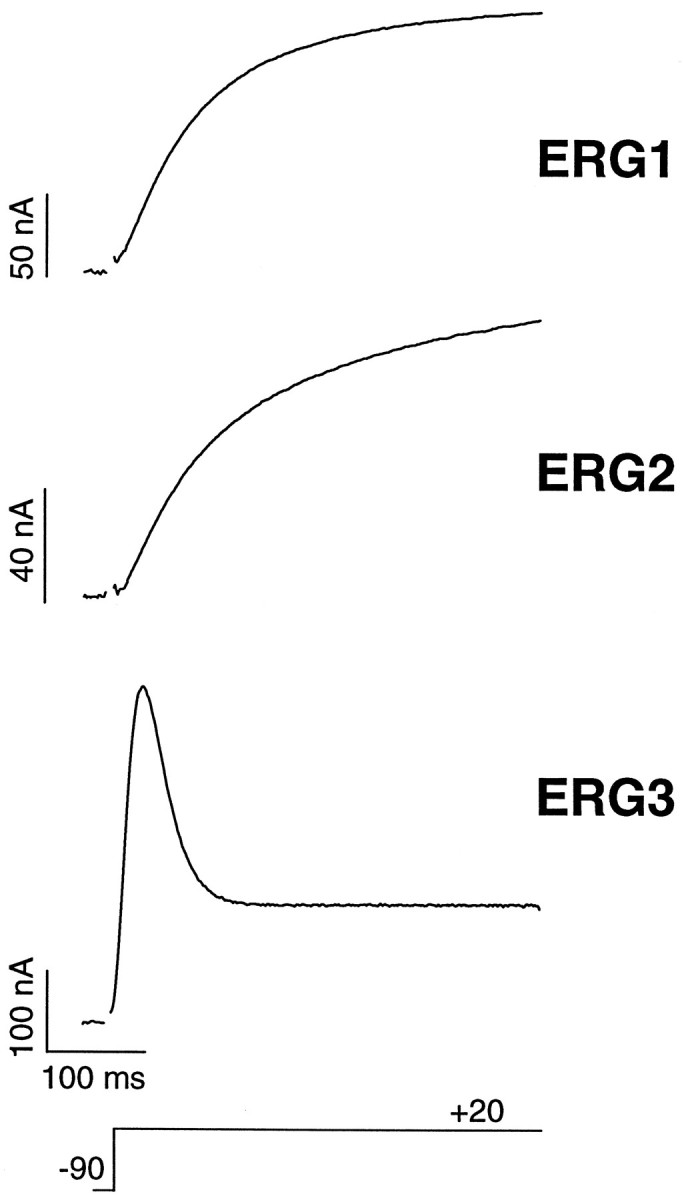
Current responses of the erg1,erg2, and erg3 channels to a depolarizing voltage step. The holding potential was −90 mV, and the step was to +20 mV. Recordings were from Xenopus oocytes and were performed with two-electrode voltage clamp. Current records were leakage-subtracted, and the capacitance artifact at the beginning of the voltage step was blanked.
The kinetic basis for the different waveforms was determined by examining the activation and inactivation kinetics of the three channels. The erg3 channel activated at significantly faster rates than did either the erg1 or erg2 channels over the entire voltage range tested (Fig.5: note that the time scale for theerg3 records is fivefold faster than for the other two channels). The erg1 channel had generally faster activation rates than did the erg2 channel. Over the time course studied, activation of the erg1 channel was approximated reasonably well by a single exponential. Activation of theerg2 channel clearly required two exponentials, with the faster component becoming predominant at more positive potentials. Activation of the erg3 channel was clearly sigmoidal at negative potentials. The transient component of the erg3 current became prominent at step potentials positive to −10 mV. All three channels display a characteristic reduction in steady-state current at positive potentials, producing a negative slopeI–V relationship. The large size of the tail currents relative to the currents elicited by the depolarizing voltage steps is attributable to the rapid relief of steady-state inactivation after the step back to −70 mV (see Fig. 7B).
Fig. 5.
Activation rates of the erg1,erg2, and erg3 channels.A, B, Current traces showing channel activation and deactivation in response to voltage steps to various potentials from a holding potential of −90 mV. Tail currents were recorded at −70 mV. Note the much faster time scale forerg3 as compared with the erg1 anderg2 channels. Current records were leakage-subtracted.C, Comparison of the activation rates of theerg1, erg2, and erg3 channels. Activation rates were measured as the inverse of the time constant of single or double exponentials fit to the current traces. A single exponential gave a good fit for erg1. Forerg2, two exponentials were required, and the fraction of the fast component is shown in the inset. Forerg3, activation was clearly sigmoidal at negative potentials. In these cases the activation time course was fit with a single exponential after a delay, to allow for direct comparison with the other two channels. Data are averages from seven or eight cells; error bars are SEM.
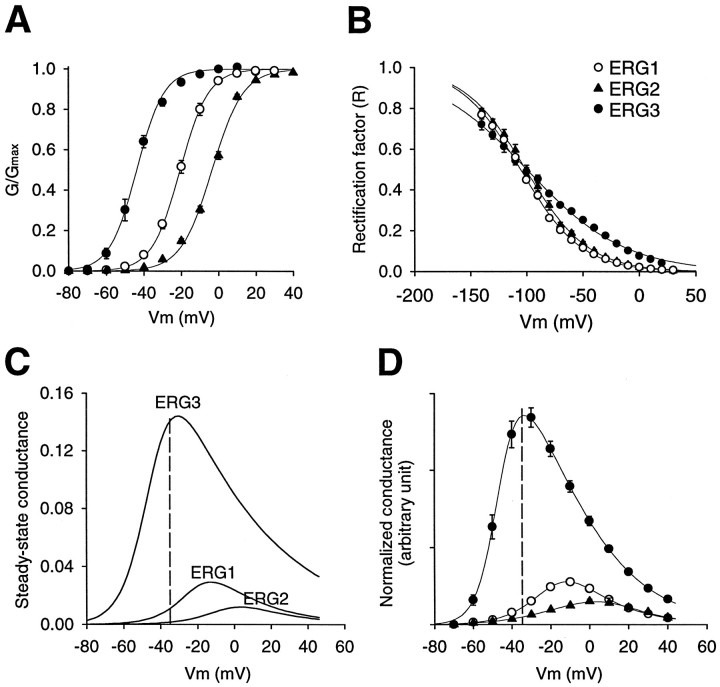
Fig. 7.
Steady-state kinetic properties of theerg1, erg2, and erg3 channels. A, Peak conductance–voltage curves were measured by stepping to the test potential from a holding potential of −90 mV, followed by a step back to the holding potential. The sizes of the tail currents after recovery from inactivation were used as a measure of channel activation during the test step. The step duration was 5 sec for erg1 and erg2 and 1 sec forerg3. Data points are the average of seven or eight cells and were fit with the Boltzmann equation:G/Gmax = 1/(1 + exp ((V −Vh)/kh)), where Vh = −21 ± 1.0, −3.5 ± 0.6, and −44 ± 1.4 mV and kh = −7.6 ± 0.4, −8.3 ± 0.3, and −7.2 ± 0.2 mV forerg1, erg2, and erg3, respectively. The open circle representserg1, the filled triangle representserg2, and the filled circle representserg3. B, Rectification factor or steady-state inactivation curve. This was measured by using a protocol similar to that described previously (Sanguinetti et al., 1995). Channels were fully activated by stepping to +40 mV for 1 sec. Then the fully activated I–V relationship was determined by stepping back to various test potentials. Tail currents were extrapolated back to t = 0 to correct for deactivation where necessary. Slope conductance was determined from theI–V plot between −140 and −120 mV, and then the rectification factor was calculated with the following formula:R =I/(Gslope(Vm −EK)), where R is the rectification factor. Data points are the average of three or four cells and were fit with the Boltzmann equation.Vh = −101 ± 2.4, −105 ± 0.3, and −100 ± 3.0 mV and kh = 28 ± 0.7, 27 ± 0.2, and 43 ± 1.2 mV for erg1,erg2, and erg3, respectively.C, Calculated steady-state conductance–voltage curve. This was calculated by multiplying the fit conductance–voltage and rectification factor curves together for each channel. Thedashed line corresponds to Vm= −35 mV, which is the threshold for spike initiation in a typical sympathetic neuron. D, Normalized steady-state conductance–voltage curve. This was measured by calculating the steady-state conductance–voltage curve and then normalizing to the tail current at −90 mV after complete activation of the current by a step to +40 mV. The tail current was extrapolated back tot = 0 to correct for deactivation. This procedure normalized for different levels of expression between different oocytes and different channels. Although this procedure did not give absolute values for the fractional conductance, it did allow direct comparison of the relative heights and shapes of the G–V curve for the three channels. Symbols have the same representations as inA and B. Data points are averages from six or eight cells; error bars are SEM.
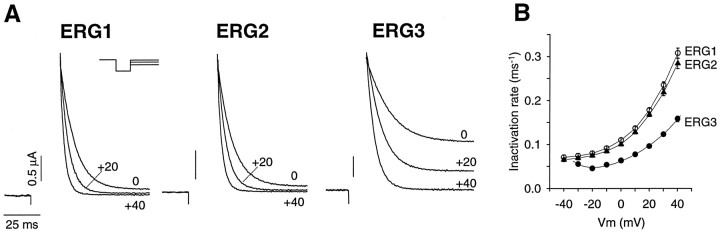
The erg1 channel and the corresponding nativeIKr current are known to have very unusual inactivation kinetics, with the inactivation rate being significantly faster than the activation rate at most membrane potentials (Shibasaki, 1987; Smith et al., 1996; Spector et al., 1996). For this reason, we compared the inactivation rates of the three channels, using the triple pulse protocol described previously for erg1 (Smith et al., 1996; Spector et al., 1996). The inactivation process could be well described by a single exponential for all three channels. Inactivation of the erg1 and erg2 channels had virtually identical time courses over the entire range of potentials tested (Fig.6). In marked contrast, theerg3 channel inactivated significantly more slowly at membrane potentials positive to −40 mV.
Fig. 6.
Inactivation rates of the erg1,erg2, and erg3 channels.A, Current traces showing channel inactivation at 0, +20, and +40 mV. Membrane potential was depolarized to +40 mV for 1000 msec to activate the channels fully. For erg1 anderg2, a brief (15–20 msec) hyperpolarizing step to −95 mV was used to allow for recovery from inactivation before the depolarizing voltage step shown in the recordings. Forerg3, because the rate of deactivation was significantly faster than for the other two channels, a slightly modified protocol was used. The hyperpolarizing step was shorter (7 msec), and the step potential was more positive (−70 mV). With the use of this protocol minimal deactivation (<6%) occurred so that during the subsequent depolarization step the kinetics of inactivation were not significantly contaminated by reactivation. B, Comparison of the inactivation rates of the erg1, erg2, anderg3 channels. Inactivation rates were measured as the inverse of the time constant of a single exponential fit to the current traces. Data are averages from seven cells; error bars are SEM.
It can be seen from these data that the appearance of a transient waveform at positive potentials in the erg3 currents, but not the erg1 and erg2 currents, is attributable to differences in the relative rates of activation and inactivation. The ratio between the inactivation and activation rates of theerg3 channel was close to two at positive potentials, whereas inactivation rates were at least 10-fold faster than activation rates for both the erg1 and erg2 channels in this potential range. For the erg1 and erg2 channels, the inactivation process is in quasi-steady-state relative to the much slower activation process. The transient waveform of theerg3 current is produced by much the same mechanism as for a typical “A current,” which has an inactivation rate that is similar or somewhat slower than the activation rate. The primary difference between the erg3 current and a typical A current is the persistence of a maintained plateau current, which is attributable to the very shallow steady-state inactivation curve of the erg3 channel.
There were other differences among the three erg channels that are of potential relevance to the physiological function of these channels. The peak conductance–voltage relationship was significantly different for each of the three channels (Fig.7A). The midpoint for activation shifted ∼20 mV among the three channels, from −44 ± 1.4 mV for erg3 to −20 ± 1.0 mV for erg1 to −3.5 ± 0.6 mV for erg2 (n = 7 or 8). The erg1 and erg2 channels had generally similar steady-state inactivation, or rectification properties, whereas the slope of the rectification curve for the erg3 channel was significantly shallower, although the midpoint was similar to the other channels (Fig. 7B). This difference in slope meant that the erg3 channel had less steady-state inactivation at potentials positive to −100 mV than did the other two channels.
The physiological function of the erg channels in the nervous system is currently obscure, although the Drosophila erg channel clearly acts to reduce neuronal excitability (Titus et al., 1997; Wang et al., 1997). We examined the potential contribution of the erg channels to the steady-state current around the threshold for spike initiation (approximately −35 mV) in two independent ways. Initially, the fraction of channels activated at each potential (Fig. 7A) was multiplied by the fraction of channels inactivated at that potential (Fig. 7B) to give the fraction of channels open at steady-state (Fig. 7C). A second, more direct approach was to measure the conductance at the end of a sustained voltage step; then, to normalize for differences in expression between different oocytes and channels, the conductance at each potential was divided by the maximum conductance to give a normalized steady-state conductance (Fig. 7D). There was good agreement between the two methods used for calculating the shape and relative peaks of the steady-state conductance (compare Fig. 7,C and D).
The steady-state conductance or “window current” was large for theerg3 channel, whereas the erg1 anderg2 channels passed significantly less current. Below −35 mV, the region important for control of spike initiation, the difference was even more striking, with the steady-stateerg3 conductance close to a maximum and the erg1 and erg2 conductances at relatively low values.
Pharmacological properties of the erg2 anderg3 potassium channels
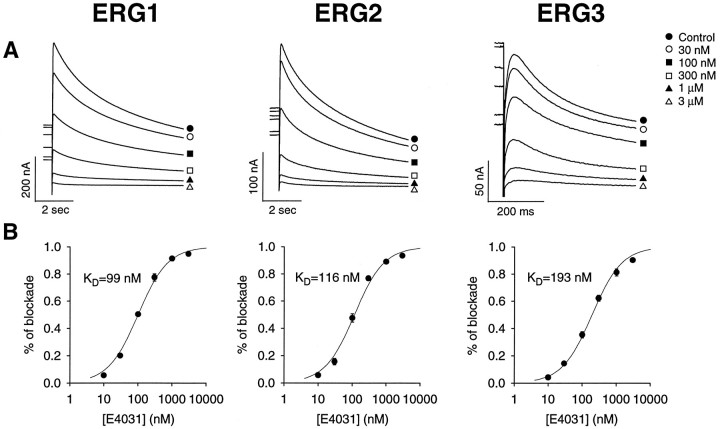
A characteristic pharmacological property of theIKr current, which is thought to be encoded by the erg1 gene, is its sensitivity to methanesulfonanilides (Sanguinetti and Jurkiewicz, 1990). The sensitivity of all three channels to blockade by the methanesulfonanilide E4031 was determined (Fig. 8). The KDfor blockade by E4031 was similar for all three channels (99, 116, and 193 nm for erg1, erg2, anderg3, respectively), which is consistent with the high degree of conservation in the pore region of these channels. As has been described previously for the blockade of the erg1 channel by other methanesulfonanilide drugs (Snyders and Chaudhary, 1996; Spector et al., 1996), E4031 acted as an open channel blocker. The on-rate for binding was apparently very slow, and repeated depolarizing pulses were required to reach quasi-steady-state for drug binding, particularly at low drug concentrations.
Fig. 8.
Inhibition of the erg1,erg2, and erg3 channels by the methanesulfonanilide E4031. A, Tail currents in the presence of increasing concentrations of E4031. The procedure used to measure the degree of blockade at each drug concentration was similar to that described previously (Snyders and Chaudhary, 1996). Because the drug is an open channel blocker with a very slow on-rate of binding, it was necessary to depolarize the cell repetitively to reach equilibrium binding. A 20 sec step to +20 mV was applied at 0.033 Hz until no further reduction in current was seen for that particular drug concentration. At that point a test step to +10 mV for 1 sec was applied, and the tail current at −60 mV was measured.B, Hill plots of E4031 inhibition of theerg1, erg2, and erg3 channels showing the KD for channel blockade. Data points were fit with the Hill equation: percentage of blockade = 1/(1 + (KD/[E4031])), whereKD = 99 ± 10, 116 ± 11, and 193 ± 18 nm for erg1,erg2, and erg3, respectively. Data points are averages from three or four cells; error bars are SEM.
DISCUSSION
In this paper we describe the identification, mRNA distribution, and biophysical properties of two new members of the ergpotassium channel gene family: erg2 and erg3. Both genes are expressed exclusively in neural tissue, in marked contrast to the previously described erg1 gene, which is expressed in a wide range of tissues in addition to the nervous system (Wymore et al., 1997). The erg3 gene is expressed abundantly in brain and sympathetic ganglia. Distribution of erg2 gene expression is restricted considerably more and is located primarily in a subset of sympathetic ganglia known as the prevertebral ganglia.
The physiological role of the erg channels in the mammalian nervous system is currently uncertain. In Drosophila it has been shown that mutations in the erg gene produce a hyperexcitable phenotype (Titus et al., 1997; Wang et al., 1997), suggesting that the erg channel inhibits neuronal excitability. The biophysical properties of the Drosophila erg channel have yet to be characterized, however, and the native current produced by the erg channels has not been described. A current with pharmacological and kinetic properties that are very similar to those of erg channels has been described in a rat dorsal root ganglia cell line (Farvelli et al., 1996). Pharmacological blockade of this current results in a significant decrease in spike frequency adaptation and an increase in excitability in response to maintained depolarizing stimuli (Chiesa et al., 1997), suggesting thaterg channels can contribute to the control of neuronal excitability in some mammalian cells. The observation that all threeerg channels are very sensitive to blockade by methanesulfonanilides such as E4031 should make it possible to determine the function of the erg channels in the nervous system, although the relatively slow onset and voltage dependence of binding means that some caution will be required in interpreting negative results.
Several differences in the functional properties of the threeerg channels were found that could be of physiological importance. Most obvious was the difference in the waveform of the currents produced in response to voltage steps to positive membrane potentials. The current produced by the erg3 channel had a large transient component at positive potentials, whereas the other two channels produced slowly activating currents that resembled classical delayed rectifiers. The kinetic basis for the switch between the two waveforms was somewhat paradoxical. For the erg1 anderg2 channels, the delayed rectifier waveform was produced by an increase in the inactivation rate relative to the inactivation rate of the erg3 channel. This result is attributable to the very unusual kinetic properties of theerg1 and erg2 channels, which have significantly faster inactivation than activation rates (typically at least 10-fold faster). The activation rate was significantly slower for theerg1 and erg2 channels than for theerg3 channel, and this difference also contributed to the production of different waveforms.
A more subtle difference, which is of potential importance for the physiology of the cells that express erg channels, was the difference in the position and peak of the steady-state conductance–voltage curve of the three channels. The erg3 channel activated at relatively negative membrane potentials and produced a large window current, which peaked around the threshold for spike initiation in a typical sympathetic neuron (Wang and McKinnon, 1995). The erg3 channels could, therefore, produce a significant inhibitory influence on subthreshold electrical excitability. The conductance–voltage curve for the erg1 and erg2 channels was shifted to the right, and the peak steady-state conductance was significantly lower relative to theerg3 channel. This makes it less likely that these channels could influence the threshold firing properties of neurons, at least as homomultimers. In cardiac myocytes the action potential duration is relatively long, giving the erg1 channels sufficient time to activate fully and contribute to action potential repolarization. The relatively brief duration of the typical neuronal action potential means that there would be relatively little activation of theerg1 or erg2 channels during a single action potential. It is possible that the very slow deactivation kinetics of these channels could result in cumulative activation of the channels during a prolonged burst of action potentials. Even in this case, however, the relatively positive activation threshold of these channels would limit their influence on neuronal excitability. It is possible, or even likely, that the functional properties of the erg1 and erg2 channels are modified by heteromultimer formation. The broad expression of the erg3 channel in the nervous system makes it an obvious candidate, but other, yet to be identified, subunits also could modify the kinetic properties of the channels significantly, as has been shown recently for the KvLQT1 channel (Barhanin et al., 1996; Sanguinetti et al., 1996b).
The discovery that in mammals there is a small family of erggenes may have implications for the etiology of one form of the human genetic disease known as long QT syndrome (LQT). It has been shown that three forms of the autosomal dominant LQT syndrome (Romano–Ward syndrome) are produced by mutations in ion channel genes expressed in the heart. LQT1 syndrome involves mutations in the KvLQT1 gene, which encodes a slowly activating K+ channel that probably underlies IKs in cardiac myocytes (Barhanin et al., 1996; Sanguinetti et al., 1996b). LQT2 syndrome involves mutations in the erg1 (or HERG) gene, which encodes theIKr channel (Sanguinetti et al., 1995; Trudeau et al., 1995), and LQT3 syndrome involves mutations in the cardiac sodium channel gene SCN5 (or hH1) (Wang et al., 1995). Autosomal dominant LQT syndrome characteristically displays no overt neurological symptoms (Schwartz et al., 1994). For the KvLQT1 and SCN5 genes this can be explained by the fact that these two genes are not expressed in the nervous system (Gellens et al., 1992; Wang et al., 1996). For LQT2, however, the situation is more complex, because the erg1 gene is expressed widely in the nervous system (Wymore et al., 1997). Given this distribution pattern, the apparent lack of neurological symptoms in LQT2 patients is somewhat surprising. Identification of theerg2 and erg3 genes provides a potential explanation for this paradox. The cardiac-specific nature of LQT2 syndrome may be explained by the fact that the erg1 gene is the only member of the erg gene family expressed in heart, and, for this reason, cardiac function may be unusually dependent on the presence of functional erg1 channels. In contrast, in neural tissues, two other erg genes are expressed. Although the channels encoded by these genes are not identical in function to the erg1 channel, they are sufficiently similar to suggest that they could compensate for the reduction in functionalerg1 channels.
The observation that the erg1 gene is expressed in sympathetic ganglia does, however, raise an interesting question regarding the genesis of the LQT2 form of LQT syndrome. There have been two hypotheses proposed to account for the underlying etiology of LQT syndrome (Schwartz et al., 1994; Roden et al., 1996). One hypothesis relies on the observation that the life-threatening arrhythmias characteristic of this disease are induced by increased sympathetic outflow stimulated by intense emotional or physical stress. This hypothesis has received support from the finding that the arrhythmias, syncope, and sudden death that are characteristic of the disease can be prevented in large part by β-blockade and/or cardiac sympathetic denervation. The most specific form of this hypothesis invoked a sympathetic imbalance, positing that there was excessive left sympathetic outflow to the heart (Schwartz et al., 1994). The second hypothesis holds that the defect is intrinsic to the heart, affecting the intrinsic electrophysiological function of cardiac myocytes. In the case of LQT1 and LQT3 the intrinsic defect hypothesis is likely to be a sufficient explanation for the syndrome, because the genes underlying these two forms of the disease are not expressed in the nervous system. For LQT2, however, because the erg1 gene is expressed abundantly in sympathetic ganglia and adrenal glands in addition to heart, the situation is more complex. Because erg channels can contribute to the control of neuronal excitability (Chiesa et al., 1997; Titus et al., 1997; Wang et al., 1997), disruption oferg1 gene function conceivably could result in hyperexcitability in sympathetic neurons, thereby affecting sympathetic outflow to the heart, particularly during periods of physiological stress. Alternatively, the adrenal glands might be unusually dependent on normal erg1 channel function because, like the heart, they express erg1, but not the other two erggenes. Mutations in the erg1 gene could result in an increase in circulating catecholamines, at least under some physiological conditions. Either of these effects potentially could contribute to the initiation of arrhythmias in LQT2 syndrome. These possibilities are not exclusive of an intrinsic cardiac myocyte defect in LQT2 and might act synergistically with such a defect. Although the balance of evidence currently favors the intrinsic hypothesis for LQT2 syndrome, it will be of considerable importance to determine the function of the erg channels in the sympathetic/adrenal system to gain further insight into the etiology of the LQT2 form of long QT syndrome.
Footnotes
This work was supported by National Institutes of Health Grants NS-29755, NS-01718, HL-20558, and DK07521. We thank Dr. Michael Sanguinetti (University of Utah) for the gift of theerg1 (HERG) cDNA clone and Drs. Paul Adams and Michael Rosen for helpful comments on this manuscript.
Correspondence should be addressed to Dr. Jane E. Dixon, Department of Neurobiology and Behavior, State University of New York at Stony Brook, Stony Brook, NY 11794-5230.
REFERENCES
- 1.Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. KVLQT1 and IsK (minK) proteins associate to form the IKs cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 2.Brown DA. M currents. In: Narahashi T, editor. Ion channels. Plenum; New York: 1988. pp. 55–94. [DOI] [PubMed] [Google Scholar]
- 3.Cassell JF, Clark AL, McLachlan EM. Characteristics of phasic and tonic sympathetic ganglion cells of the guinea-pig. J Physiol (Lond) 1986;372:457–483. doi: 10.1113/jphysiol.1986.sp016020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiesa N, Rosati B, Arcangeli A, Olivotto M, Wanke E. A novel role for HERG K+ channels: spike frequency adaptation. J Physiol (Lond) 1997;501:313–318. doi: 10.1111/j.1469-7793.1997.313bn.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colman A. Translation of eukaryotic messenger RNA in Xenopus oocytes. In: Hames BD, Higgins SJ, editors. Transcription and translation. IRL; Oxford: 1984. pp. 271–302. [Google Scholar]
- 6.Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 7.Dixon JE, McKinnon D. Quantitative analysis of potassium channel mRNA expression in atrial and ventricular muscle of rats. Circ Res. 1994;75:252–260. doi: 10.1161/01.res.75.2.252. [DOI] [PubMed] [Google Scholar]
- 8.Dixon JE, McKinnon D. Potassium channel mRNA and protein expression in prevertebral and paravertebral sympathetic neurons. Eur J Neurosci. 1996;8:183–191. doi: 10.1111/j.1460-9568.1996.tb01179.x. [DOI] [PubMed] [Google Scholar]
- 9.Farvelli L, Arcangeli A, Olivotto M, Wanke E. A HERG-like K+ channel in rat F-11 DRG cell line: pharmacological identification and biophysical characterization. J Physiol (Lond) 1996;496:13–23. doi: 10.1113/jphysiol.1996.sp021661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frohman MA. On beyond classic RACE (rapid amplification of cDNA ends). PCR Methods Appl. 1994;4:S40–S58. doi: 10.1101/gr.4.1.s40. [DOI] [PubMed] [Google Scholar]
- 11.Ganetzky B, Wu CF. Neurogenetic analysis of potassium currents in Drosophila: synergistic effects on neuromuscular transmission in double mutants. J Neurogenet. 1983;1:17–28. doi: 10.3109/01677068309107069. [DOI] [PubMed] [Google Scholar]
- 12.Gellens ME, George AL, Chen LQ, Chahine M, Horn R, Barchi RL, Kallen RG. Primary structure and functional expression of the human cardiac tetrodotoxin-insensitive voltage-dependent sodium channel. Proc Natl Acad Sci USA. 1992;89:554–558. doi: 10.1073/pnas.89.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.London B, Beyer AK, Newton KP, Trudeau MC, Robertson GA. Cloning a cardiac-specific isoform of HERG from the mouse. Biophys J. 1997;72:A224. [Google Scholar]
- 14.Ludwig J, Terlau H, Wunder F, Bruggemann A, Pardo LA, Marquardt A, Stuhmer W, Pongs O. Functional expression of a rat homologue of the voltage-gated ether a go-go potassium channel reveals differences in selectivity and activation kinetics between the Drosophila channel and its mammalian counterpart. EMBO J. 1994;13:4451–4458. doi: 10.1002/j.1460-2075.1994.tb06767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roden DM, Lazzara R, Rosen D, Schwartz PJ, Towbin J, Vincent GM. Multiple mechanisms in the long-QT syndrome. Circulation. 1996;94:1996–2012. doi: 10.1161/01.cir.94.8.1996. [DOI] [PubMed] [Google Scholar]
- 16.Sanguinetti MC, Jurkiewicz NK. Two components of cardiac delayed rectifier K+ current: differential sensitivity to block by class III antiarrhythmic agents. J Gen Physiol. 1990;96:195–215. doi: 10.1085/jgp.96.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanguinetti MC, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81:299–301. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 18.Sanguinetti MC, Curran ME, Spector PS, Keating MT. Spectrum of HERG K+-channel dysfunction in an inherited cardiac arrhythmia. Proc Natl Acad Sci USA. 1996a;93:2208–2212. doi: 10.1073/pnas.93.5.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Keating MT. Coassembly of KVLQT1 and minK (IsK) proteins to form cardiac IKs potassium channel. Nature. 1996b;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz PJ, Locati EH, Napolitano C, Priori SG. The long QT syndrome. In: Zipes DP, Jalife J, editors. Cardiac electrophysiology: from cell to bedside, 2nd Ed. Saunders; Philadelphia: 1994. pp. 788–811. [Google Scholar]
- 21.Shibasaki T. Conductance and kinetics of delayed rectifier potassium channels in nodal cells of the rabbit heart. J Physiol (Lond) 1987;387:227–250. doi: 10.1113/jphysiol.1987.sp016571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith PL, Baukrowitz T, Yellen G. The inward rectification of the HERG cardiac potassium channel. Nature. 1996;379:833–836. doi: 10.1038/379833a0. [DOI] [PubMed] [Google Scholar]
- 23.Snyders DJ, Chaudhary A. High affinity open channel block by dofetilide of HERG expressed in a human cell line. Mol Pharmacol. 1996;49:949–955. [PubMed] [Google Scholar]
- 24.Spector PS, Curran ME, Keating MT, Sanguinetti MC. Class III antiarrhythmic drugs block HERG, a human cardiac delayed rectifier K+ channel. Circ Res. 1996;78:499–503. doi: 10.1161/01.res.78.3.499. [DOI] [PubMed] [Google Scholar]
- 25.Terlau H, Ludwig J, Steffan R, Pongs O, Stuhmer W, Heinemann SH. Extracellular Mg2+ regulates activation of rat eag potassium channel. Pflügers Arch. 1996;432:301–312. doi: 10.1007/s004240050137. [DOI] [PubMed] [Google Scholar]
- 26.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Titus SA, Warmke JW, Ganetzky B. The Drosophila erg K+ channel polypeptide is encoded by the seizure locus. J Neurosci. 1997;17:875–881. doi: 10.1523/JNEUROSCI.17-03-00875.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trudeau MC, Warmke JW, Ganetzky B, Robertson GA. HERG, a human inward rectifier in the voltage-gated potassium channel family. Science. 1995;269:92–95. doi: 10.1126/science.7604285. [DOI] [PubMed] [Google Scholar]
- 29.Wang HS, McKinnon D. Potassium channel expression in prevertebral and paravertebral sympathetic neurones: control of firing properties. J Physiol (Lond) 1995;485:319–335. doi: 10.1113/jphysiol.1995.sp020732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang HS, McKinnon D. Modulation of inward rectifier currents in rat sympathetic neurones by muscarinic receptors. J Physiol (Lond) 1996;492:467–478. doi: 10.1113/jphysiol.1996.sp021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, Shen J, Timothy KW, Vincent GM, de Jager T, Schwartz PJ, Towbin JA, Moss AJ, Atkinson DL, Landes GM, Connors TD, Keating MT. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Reynolds ER, Deak P, Hall LM. The seizure locus encodes the Drosophila homolog of the HERG potassium channel. J Neurosci. 1997;17:882–890. doi: 10.1523/JNEUROSCI.17-03-00882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warmke J, Ganetzky B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci USA. 1994;91:3438–3442. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warmke J, Drysdale R, Ganetzky B. A distinct potassium channel polypeptide encoded by the Drosophila eag locus. Science. 1991;252:1560–1562. doi: 10.1126/science.1840699. [DOI] [PubMed] [Google Scholar]
- 36.Wymore R, Gintant GA, Wymore RT, Dixon JE, McKinnon D, Cohen IS. Tissue and species distribution of mRNA for the IKr-like K+ channel, erg. Circ Res. 1997;80:261–268. doi: 10.1161/01.res.80.2.261. [DOI] [PubMed] [Google Scholar]
- 37.Yamada WM, Koch C, Adams PR. Multiple channels and calcium dynamics. In: Koch C, Segev I, editors. Methods in neuronal modeling. Bradford; Cambridge, MA: 1989. pp. 97–133. [Google Scholar]