Abstract
Swiss cheese (sws) mutant flies develop normally during larval life but show age-dependent neurodegeneration in the pupa and adult and have reduced life span. In late pupae, glial processes form abnormal, multilayered wrappings around neurons and axons. Degeneration first becomes evident in young flies as apoptosis in single scattered cells in the CNS, but later it becomes severe and widespread. In the adult, the number of glial wrappings increases with age. The sws gene is expressed in neurons in the brain cortex. The conceptual 1425 amino acid protein shows two domains with homology to the regulatory subunits of protein kinase A and to conceptual proteins of yet unknown function in yeast, worm, and human. Sequencing of two sws alleles shows amino acid substitutions in these two conserved domains. It is suggested that the novel SWS protein plays a role in a signaling mechanism between neurons and glia that regulates glial wrapping during development of the adult brain.
Keywords: Drosophila, adult central nervous system, apoptosis, glia–neuron interaction, neurodegeneration, reduced life span
Glial cells are well known to have a critical role in development and maintenance of the nervous system (Raff et al., 1993, Travis, 1994). Glia–neuron interactions in the peripheral nervous system, between Schwann cells and motoneurons, have been particularly well studied. Defects in the wrapping of peripheral nerves by Schwann cells cause various human diseases (Chance and Fischbeck, 1994; Chance and Reilly, 1994). Charcot-Marie-Tooth neuropathy includes a heterogeneous group of diseases, all involving demyelination; some are caused by mutations in the myelin genes myelin protein 0 (Hayasaka et al., 1993) and peripheral myelin protein 22 (PMP-22) (Valentijn et al., 1992). Another type is caused by a mutation in the gene for connexin 32, a major component of gap junctions in Schwann cells (Bergoffen et al., 1993). Not only absence of myelin, but also hypermyelination, has been observed in peripheral neuropathies. Patients with hereditary neuropathy with liability to pressure palsies (HNPP) develop so-called tomaculae, formations caused by swelling of the myelin sheath (Verhagen et al., 1993). Interestingly, HNPP is also caused by mutations in the PMP-22 gene (Mariman et al., 1993); whereas HNPP is caused by a deletion, Charcot-Marie-Tooth cases are caused by duplication of the gene (Le Guern et al., 1994). Mutation of the PMP-22 gene in the mouse is also responsible for a demyelination disorder; the mouse mutant trembler was identified as a point mutation in the so-called growth arrest-specific gene gas-3 (Suter et al., 1992, 1993). Another mouse neuropathy is associated with thejimpy mutation, which causes a drastic reduction of all myelin components in the brain (Hogan and Greenfield, 1984). The defect was shown to be attributable to abnormal development and subsequent degeneration of oligodendrocytes, the brain equivalents of Schwann cells in the peripheral nervous system (Knapp et al., 1986).
A genetically and molecularly accessible system for the study of basic mechanisms of neurodegeneration is provided by Drosophila. Some mutations in Drosophila that cause late-onset neurodegeneration show glial defects. In the drop-deadmutant (Buchanan and Benzer, 1993), glial cells have shortened processes; therefore neurons lack the complete glial sheath provided by the glia. In reversed polarity (repo) flies, glial cells of the visual system degenerate in adulthood, leading to subsequent degeneration of lamina neurons and photoreceptors (Xiong and Montell, 1995). Molecular data show that the repo gene encodes a glial-specific homeodomain protein required for the correct development of glial cells in both embryo and adult (Xiong and Montell, 1994; Halter et al., 1995).
Here we describe the cloning of a gene, which when mutated causes degeneration in all brain regions of adult Drosophila. Swiss cheese (sws) mutant flies first show hyperwrapping of neurons by glial sheaths, followed by vacuolization and apoptosis of neurons. The gene encodes a novel protein with homology to the regulatory subunit of protein kinase A (PKA), a protein complex suggested to be involved in glia–neuron interaction in vertebrates (Mews and Meyer, 1993). SWS could be the first identified member of a new family of conserved proteins involved in glia–neuron interaction.
MATERIALS AND METHODS
Drosophila stocks. All stocks were maintained and raised under standard conditions. The five extant mutant alleles ofsws were induced by ethylmethane sulfonate (EMS) (Lewis and Bacher, 1968) in the Berlin wild type and were isolated in screens for structural brain defects using the mass histology procedure ofHeisenberg and Böhl (1979). Thesws3 allele, isolated by Merriam (Lindsley and Zimm, 1992), has been lost. The olfE mutant and the olfE genomic transformant have been described byHasan (1990). The duplicationDp(1;3)sn13a1 and deficiencyDf(1)snC128 were described by Lindsley and Zimm (1992).
Mosaic flies. Flies mosaic forsws4 were obtained by crossing females carrying the ring-X chromosomeR(1)2,In(1)wvC to males of the genotypey w sws4. InR(1)2,In(1)wvC/y w sws4 progeny, tissue in which the ring-X is lost at an early stage of development is genotypically male and the recessive y w, and sws phenotypes are uncovered. Heterozygous female y w sws4/ring-X tissue is wild type for these phenotypes. The external phenotype marked by eye and cuticle color was used as an indicator of underlying internal tissue (Hotta and Benzer, 1972); in the great majority of mosaic flies, external eye and cuticle color correlates with the genotype of the underlying brain (Kankel and Hall, 1976). Animals were killed after ≥8 d at 25°C.
Tissue sections for light and electron microscopy. Larval, pupal, and adult tissues were prepared for light and electron microscopy as described by Renfranz and Benzer (1989). For light microscopy, 1 μm serial sections were cut and stained with 1% toluidine blue and 1% borax. LacZ staining was performed on dissected whole-mount brains as described previously (Kretzschmar et al., 1992); these were post-fixed in 6% glutaraldehyde, embedded in Epon plastic, sectioned, and counterstained with 1% pyronin Y. Ultrathin Epon plastic sections were post-stained with 2% uranyl acetate, followed by Reynolds’ lead citrate (Reynolds, 1963), and stabilized for transmission electron microscopy by carbon coating. Examination was done with a Philips 201C electron microscope at 40–80 kV. Glial cell material was clearly identified by its characteristically higher electron density (Saint Marie and Carlson, 1983a,b, 1990).
Apoptosis staining. For the detection of cells undergoing apoptosis, we used an ApopTag kit from Oncor. Frozen head sections were performed for immunohistochemistry as described by Ashburner (1989), fixed in 10% neutral buffered formalin for 10 min at room temperature, and post-fixed in 2:1 ethanol:acetic acid for 5 min at −20°C. Sections were incubated in 2% hydrogen peroxide for 5 min to quench endogenous peroxidase. The DNA fragments were tailed by adding digoxigenin–deoxyuridine triphosphate with terminal transferase for 1 hr at 37°C. Detection was followed by binding of a peroxidase-coupled anti-digoxigenin antibody for 1 hr and staining with DAB for up to 1 hr. All washes were done with PBS/0.5% Triton X-100.
Characterization of the sws/olfE transcription unit. A detailed description of the sws/olfE genomic region, including the localization of the deficiency and duplication break points, the transcripts, and their expression patterns, as determined by Northern analysis, was given by Hasan (1990). The exon–intron map for the 5.4 kb transcription unit was determined by performing out restriction digests and Southern blots of the genomic clones and hybridizing them to specific restriction fragments from the cDNA clones and by sequencing genomic DNA.
cDNA isolation and sequencing. Both 5′ (pBS-E75) and 3′ (λgt10-E71) cDNA fragments were subcloned into the EcoRI sites of the vectors M13-mp18 and M13-mp19. DNA sequencing was performed with the Sequenase kit (United States Biochemicals, Cleveland, OH), using single-stranded phage templates and standard M13 primers. To obtain sequence beyond 300 bp of the primer site, the initial subclones were sequentially shortened using the ExoIII–mung bean nuclease method (Henikoff, 1984).
In situ hybridization. Frozen head sections of adult wild-type and mutant flies were fixed and hybridized as described byPoeck et al. (1993). Digoxigenin-labeled RNA probes were transcribed following the protocol of the Boehringer Mannheim (Mannheim, Germany) RNA-labeling kit, using pBS-E75 and a PCR fragment containing bp 3570–4934. Hybridizations were performed overnight at 50°C using 25 μl/slide hybridization solution consisting of Boehringer Mannheim standard hybridization buffer containing 50% formamide and digoxigenin-labeled antisense RNA at 2.5–5 μg of RNA/ml.
Protein analysis. The conceptual SWS protein was compared with other proteins in the GenBank, Swiss, and Protein Identification Resource databases by using the BLAST (Altschul et al., 1990) and TFASTA (Pearson and Lipman, 1988) algorithms. Potential protein motifs were sought by MOTIFS (Hodgman, 1993), SAPS (Brendel et al., 1992) and TMpred (Hoffmann and Stoffel, 1993) analysis. SAPS and TMpred both found the first transmembrane domain significant, whereas the other two were only found significant by TMpred. TheCaenorhabditis elegans clone (M98552) was characterized by the Nematode Sequencing Project (MRC, Laboratory of Molecular Biology, Cambridge, UK) and Department of Genetics, Washington University (St. Louis, MO). The yeast clone (Z46729) was isolated by the Sanger Centre, Hinxton, Cambridge, UK; the human cDNA fragment T10299 was isolated by Bento Soares (Columbia University, New York, NY); and T09456 was isolated by M.D. Adams (Institute for Genomic Research, Gaithersburg, MD). By further sequencing the clone T09456, we determined its identity with T10299 and the sequence between these two fragments.
Molecular characterization of the mutant alleles. Oligos of 18–22 bp, complementary to the sense or antisense strand from thesws cDNA, were synthesized. Genomic DNA from the different alleles was isolated using standard conditions (Ashburner, 1989). PCR reactions were performed using the Expand PCR system (Boehringer Mannheim); the amplification products ranged from 500 bp to 3.5 kb. The fragments were visualized on a gel, purified with the Qiagen (Hilden, Germany) QIAquick gel extraction kit, and diluted to 25 ng/μl. The sequence reaction was done according to the Applied Biosystems (ABI, Foster City, CA) manual with 250 ng of purified PCR product and 5pmol of the PCR primers, labeled through dye termination, and run on an ABI sequencer. All PCR fragments were sequenced on both strands, and mutations were verified by sequencing an independent PCR reaction.
RESULTS
sws causes neurodegeneration of the CNS
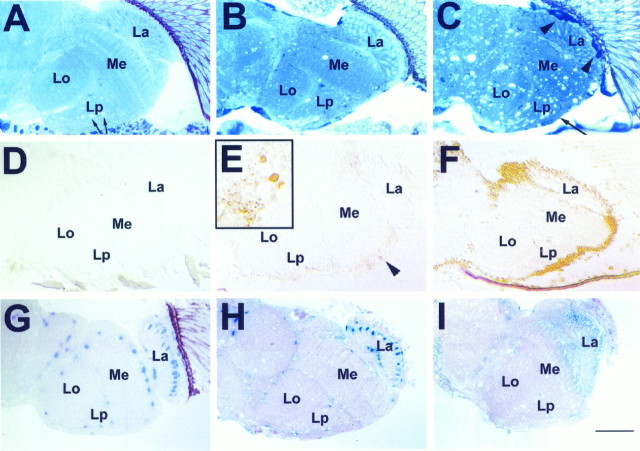
The X-linked swiss cheese (sws) mutation was originally isolated in a screen for mutants showing anatomical brain defects in the adult fly (Heisenberg and Böhl, 1979). Head sections of mutant flies reveal the formation of vacuoles in all brain regions, hence the name swiss cheese. Figure1A–C shows a comparison of head sections from wild type, a young sws fly (5 d at 25°C), and an aged sws fly (20 d at 25°C). The degeneration advances with age, as shown by an increased number of vacuoles and shrinking of the cell cortex (Fig. 1C, arrow). Note that in the wild type the white areas in the lobula plate (Fig.1A, arrows) are not vacuoles but are cross-sections of giant fibers from horizontal system and vertical system neurons. In addition, we detect an increasing number of intensely toluidine blue-stained cells (Fig. 1B) indicating cell death. Degeneration is also enhanced by increased temperature; flies raised for 7 d at 29°C show effects similar to those seen in flies raised for 20 d at 25°C (data not shown).
Fig. 1.
Progression of brain degeneration insws flies. A–C, One micrometer horizontal plastic head sections stained with toluidine blue. Vacuolization and darkly stained structures (arrowheadsin C) increase with advancing age. These are accompanied by a thinning of the cortex (long arrow inC). A, Wild type, age 20 d (arrows indicate white areas in the lobula plate that are not vacuoles but cross-sections of giant fibers). B, sws1 5 d old. C, sws1 20 d old. D–F,Apoptosis staining on 8 μm horizontal cryosections. D,Wild type 20 d old. E, In a 5-d-oldsws1 fly, single cells are intensely stained (arrowhead and inset). Single dying cells are already indicated by the dark toluidine blue staining in B (arrows). F, In a 20-d-old sws1 fly, there is widespread staining, suggesting apoptosis in most cortical cell bodies.G–I, Effect of degeneration on LacZ-expressing cells of the glial-specific enhancer–trap line rC56: 1 μm horizontal plastic brain sections stained with X-galactosidase (X-Gal).G, Heterozygous rC56/wild-type fly 10 d old. H, 8-d-oldsws4/y;rC56/wild-type fly. The number of cells showing LacZ staining is decreased. I, Fourteen-day-oldsws4/y;rC56/wild-type brain. The LacZ staining is almost completely absent, indicating loss of expression or degeneration of many glial cells. Brains were dissected and stained with X-Gal and then embedded in plastic, sectioned, and counterstained with pyronin Y. All flies were raised at 25°C. La, Lamina; Me, medulla;Lo, lobula; Lp, lobula plate. Anterior is at the top. Scale bar: A–I, 50 μm;inset in E, 15 μm.
To examine the cell death in the mutant brain further, we used an immunohistochemical method for visualizing DNA fragmentation, a characteristic step in apoptotic cell death (ApopTag; see Materials and Methods). Brain sections from wild-type flies typically show little or no staining (Fig. 1D). In brain sections from 5-d-old mutant flies, a few intensely stained cells are seen (Fig.1E, arrowhead and inset), indicating apoptosis. By 20 d at 25°C, the entire brain cortex is intensely labeled, suggesting that dying neurons in sws undergo apoptotic cell death (Fig. 1F). This does not seem to be restricted to neurons; glial cells also appear to be dying. This can be seen in the enhancer trap line rC56, in which thelacZ gene is specifically expressed in a subset of glial cells (Fig. 1G) (Brunner et al., 1994; Menne and Klämbt, 1994). In an sws background (sws/Y;rC56/+) the expression of lacZ in very young (1- to 2-d-old) flies is normal, but in 8-d-old flies it has begun to disappear in most of these glial cells, with the exception of the epithelial and marginal glia in the lamina neuropil (Fig.2H). In older flies (14 d), staining of individual cells is almost completely gone (Fig. 1I). The loss of marker expression shows either degeneration or, at least, dysfunction of glial cells with aging in the mutant.
Fig. 2.
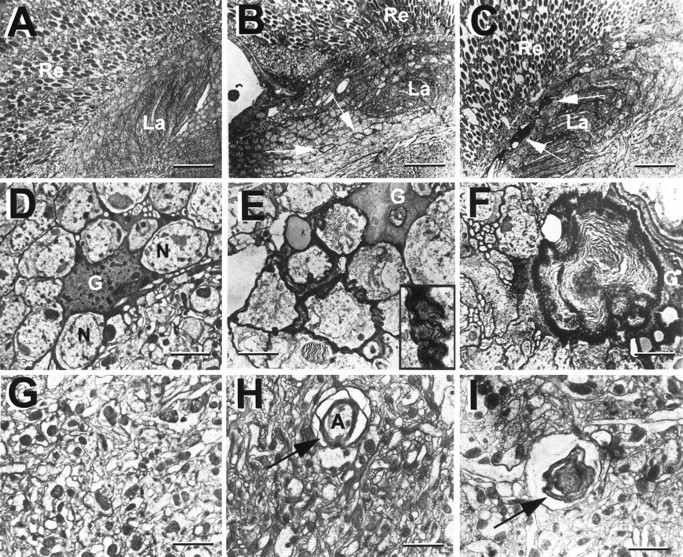
Multilayered glial membranes in swsmutant brains as seen in EM horizontal sections. A, D, G, Wild type. A, Retina and lamina.D, Cell bodies of the lamina cortex, showing a glial cell recognizable by its dark cytoplasm. Extensions of the glial cell surround various neurons with single-layered wrappings.G, Medulla neuropil, a tangle of synapses. B, E, H, In newly eclosed sws1flies, glial processes form multiple sheaths around neurons in the cortex (arrows in B, inset inE) and around axons in the neuropil (arrow inH). C–I, In 7-d-oldsws1 flies, large membrane whorls are visible (F, I). These are especially prominent in the lamina cortex (arrows in C). Flies were kept at 25°C. Re, Retina, La, lamina; G, glial cell; N, neuron;A, axon. Scale bars: A–C, 3 μm;D–I, 0.5 μm.
Staining of plastic-embedded sws brain sections with toluidine blue reveals not only holes and degeneration of cell bodies but also compact, dark-staining bodies in the cortex, which are especially prominent in the lamina cortex (Fig. 1C, arrowheads). These structures first appear in the late pupa, long before vacuoles are formed, and increase in size during aging of the adult. To resolve their morphology, we used electron microscopy (EM).
EM studies on newly eclosed mutant flies showed that ∼10% of the neurons in the brain cortex were enwrapped by multilayered membranes, which originate from nearby glia (Fig. 2B,E). In the wild type, glia typically enwrap neurons with a single or sometimes two processes (Fig. 2D). In the mutant, the occurrence of multiple ensheathing in cortex as well as in neuropil (Fig.2H) is greatly increased; this is already visible in newly eclosed flies. In older sws flies (7 d), we find an increasing frequency of structures consisting of membranous whorls (Fig. 2F,I). In some cases, these are embedded within glial cell bodies, which are identifiable by their electron-dense cytoplasm (Saint Marie and Carlson, 1983a,b, 1990). The EM studies on these older flies also reveal cells with the typical features of apoptotic cells, such as shrinkage or the formation of pyknotic nuclei (data not shown).
Because of the mutation, sws flies have a reduced life span compared with wild type. The severity of adult lethality varies among the five extant alleles examined [a sixth allele,sws3 (Lindsley and Zimm, 1992), has been lost]; 50% survival time at 29°C ranges from 3 to 10 d (Fig.3), compared with 15 d for the wild-type control (Berlin) and flies heterozygous for the fully recessive sws mutations under the same conditions. Flies hemizygous for any of the sws alleles over a deficiency showed survival times similar to flies homozygous for the same alleles (data not shown), suggesting that they are hypomorphs or null. At 18 and 25°C sws flies live considerably longer than at 29°C (data not shown), but at both temperatures the mutants still show significant reduction in life span compared with wild-type flies.
Fig. 3.
Survival curves for the five swsalleles at 29°C. At least 200 flies were tested per allele.
Mosaic studies suggest a local requirement for the swsgene product
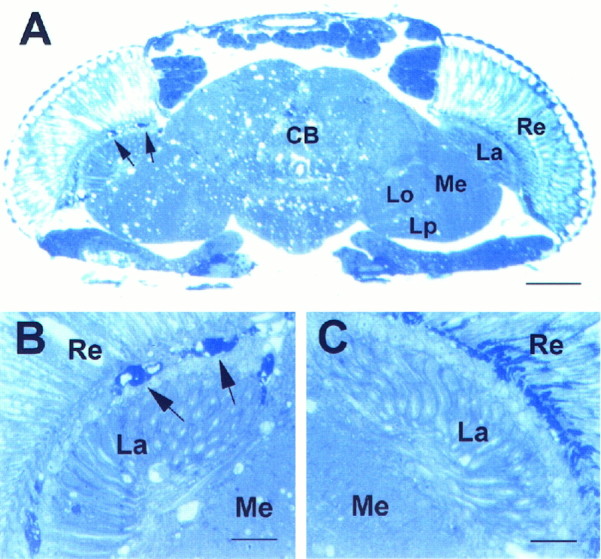
To address the question of whether the sws gene product might be a diffusible factor with a long-range effect, we created mosaics by the use of gynandromorphs (Hotta and Benzer, 1972). In these gynandromorphs, female tissue was heterozygous for the mutation and the cuticle markers yellow and white, whereas male tissue was hemizygous for sws, yellow, andwhite. Flies with half of a wild-type and half of a mutant head (as judged by yellow cuticle and white eyes) were sectioned. In six of seven cases, the tissue underlying the mutant whiteeye showed the sws membrane structures (Fig.4B, arrows), whereas the wild-type side was free of them (Fig. 4C). The occurrence of the dark-stained bodies, together with increased vacuolization, only underneath the genotypically mutant retina makes a long-range effect of the gene product highly unlikely. It suggests a cell-autonomous or short-range function for the sws gene product, especially because vacuoles in the wild-type half may be caused by degeneration of mutant neurons that project into the wild-type side.
Fig. 4.
Head section ofsws4 mosaic fly, 8 d old.A, Left side, B, Darkly stained bodies characteristic of the sws mutation (arrows). These are missing toward the wild-type side at the right (A, C). The few vacuoles on the wild-type side may be caused by mutant neurons projecting from the mutant side to the wild-type side.Re, Retina; La, lamina;Me, medulla; CB, central brain;Lo, lobula; Lp, lobula plate. Scale bars:A, 50 μm; B, C, 10 μm.
sws mapping
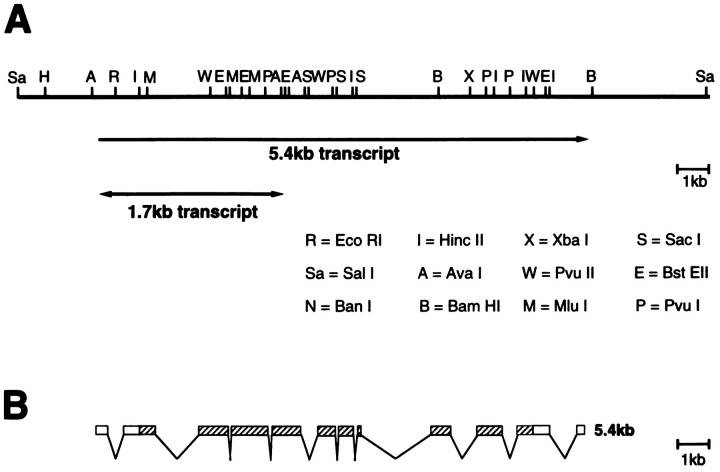
To map the sws gene on the X chromosome, we used a set of deficiencies and duplications kindly provided by the Bloomington and Mid-America stock centers. The deficiencyDf(1)snC128, lacking bands 7D1–D5, uncovers the sws phenotype. This deficiency overlaps at the 7D1 break point, ∼25kb, of the duplicated region inDp(1;3)sn13a1 (Hasan, 1990). Because we were able to rescue the sws phenotype completely by using the Dp(1;3)sn13a1 translocation, we could localize the sws gene to the 25 kb region in 7D1. The region produces two alternatively spliced transcripts (Fig.5A); the larger one of 5.4 kb is expressed in adult heads and bodies, whereas the smaller, less abundant one of 1.7 kb is only detectable in heads (Hasan, 1990). The 14 kb genomic fragment, shown in Figure 5A, which contains this transcribed region but no other detectable transcripts (Hasan, 1990), was used to create a P element-transformed line. Crossing this transformant into different sws mutants (sws1, sws2, and sws4) rescued all their sws phenotypes. In addition, it rescues another mutant, olfE, mapped in this region. Complementation analysis of heterozygous flies, carryingsws1 orsws2 over olfE, exhibited neither the degeneration phenotype of sws nor the mutant olfactory behavior of olfE. The complementation would suggest that olfE and sws are either two independent genes or complementing alleles of a complex gene locus. In the first case, olfE would be encoded by a separate transcription unit, but none have yet been detected, either by Hasan or in an independent transcript analysis done by J. Paterson and K. O’Hare (Hasan, 1990). In the latter, olfE andsws would be similar to the previously described case ofsmellblind and para. Certain allelic combinations of para and smellblind complement both the olfactory and the paralytic phenotype of this gene (Lilly et al., 1994). It is possible that the olfE mutation could specifically affect the smaller transcript (because of the lack of cDNAs for the 1.7 kb transcript, this remains elusive) and its function, whereas sws affects the larger one. As shown below, the identity of the sws gene with this transcription unit was unquestionably confirmed by sequencing sws mutant lines, which identified point mutations within the open reading frame of the larger 5.4 kb transcript.
Fig. 5.
Genomic map of the sws/olfE region and structure of the cDNA. A, Top line, Restriction map of the genomic fragment used for the rescue experiment. The two transcripts are indicated below. B, Exon–intron structure of the 5.4 kb transcript. The open reading frame isstriped.
Analysis of the sws mRNA and its expression
As shown by Northern blot analysis, the larger transcript of 5.4 kb is expressed in all stages, more prominently in young embryos and adult heads and bodies, whereas the smaller 1.7kb transcript was only detectable in adult heads (Hasan, 1990). The 1.7 kb transcript is recognized in Northern blots by the first four exons of the 5.4 kb transcript and is an alternatively spliced transcript from the same transcription unit. A cDNA clone encoding 2994 bp of the 3′ end of the 5.4 kb transcript was isolated from an embryonic λgt10 library. In addition, we obtained a 5′ cDNA clone of 2505 bp, which overlapped with the 3′ cDNA clone by 134 bp. Both cDNA clones were sequenced, yielding an open reading frame (ORF) of 4274 bp. Partial genomic sequencing revealed that the 5.4 kb transcript is spliced together from at least 12 exons; the intervening introns range in size from only 66 bp to ∼2 kb (Fig. 5B).
To assess the expression pattern of the sws mRNA, we performed in situ hybridization on frozen head sections, using a 2.5 kb 5′ cDNA fragment and a 1.4 kb 3′ cDNA fragment, specific for the large transcript. Both probes revealed the same staining in most or all cell bodies of the brain cortex, showing expression in neurons (Fig. 6A). We could not detect expression in glial cells localized in the neuropil, despite the fact that some of those glial cells do degenerate in the mutant, but we cannot exclude expression in some cortical glial cells.
Fig. 6.

In situ hybridization ofsws RNA on wild-type cryosections. A, The antisense probe derived from pBS-E75 detects sws mRNA in the entire brain cortex, suggesting widespread expression in cortical cell bodies. B, Control, using a sense RNA probe.Re, Retina; La, lamina;Me, medulla; Lo, lobula;Lp, lobula plate. Scale bar, 50 μm.
sws encodes a novel protein
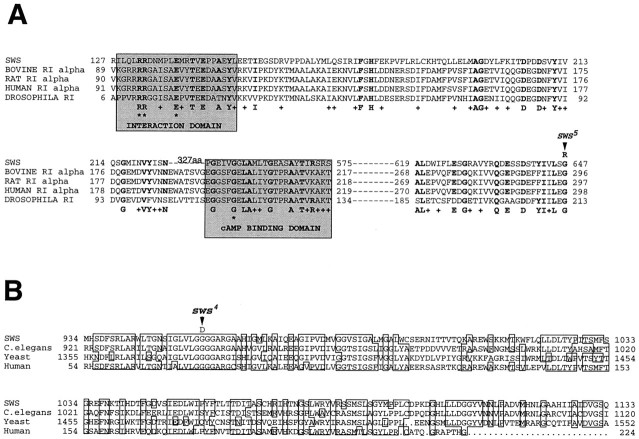
The ORF of the sws cDNA encodes a protein of 1425 amino acids (Fig. 7). A hydrophobicity analysis of the conceptual protein suggests the formation of one (SAPS analysis) or three (TMpred analysis) transmembrane helices; amino acids 35–54 show a highly significant score, whereas 943–964 and 973–994 show lower scores and no signal sequence. These sequences areunderlined in Figure 7. The SWS protein contains a region homologous (21 and 39% identity over two stretches of 127 and 28 amino acids, respectively) to the regulatory subunit of cAMP-dependent protein kinase type Iα (RIα) from various species, ranging from slime mold to human (Fig.8A). This region contains the domain interacting with the catalytic subunit and one of the two cAMP-binding sites that are normally present in the regulatory subunit (Taylor et al., 1990). In addition, homology searches in the available databases revealed highly significant similarity (BLASTp = 1 × 10−231) to a conceptual protein of 1475 amino acids obtained from the C. elegans sequencing project and to a 1679 amino acid conceptual yeast protein (BLAST p = 2 × 10−139). We also found ∼50% identity to a human protein fragment encoded by two cDNAs from a 3-month-old human infant brain (BLAST p = 3 × 10−41). Our further sequencing of one of these cDNAs (clone HIBBU51, kindly provided by M. D. Adams) showed both to be derived from the same transcript. No functions are presently known for any of these conceptual proteins.
Fig. 7.
Conceptual sequence of the SWS protein. Possible transmembrane domains are underlined. The locations of the sws1, sws4, andsws5 mutations are identified byarrows.
Fig. 8.
Comparison of the SWS conceptual protein with other, known sequences. A, Similarity of SWS to regulatory subunit type Iα of cAMP-dependent protein kinase from various species. The domains interacting with the catalytic subunit and the cAMP-binding site are shaded. The two arginines and the glutamic acid in the interaction domain (asterisks) are the only amino acids shared by all the types of regulatory subunits (RIα, RIβ, RIIα, and RIIβ). One of the important amino acids in the cAMP-binding domain is the conserved glycine (asterisk), which, when mutated in mouse lymphoma cells, inactivates the binding site (Taylor et al., 1990). Amino acids conserved between SWS and at least one of the regulatory subunits are shown in boldface; similar ones are indicated by aplus. The amino acid substitution insws5 is indicated (arrow). B, Similarity of a second segment of SWS to conceptual proteins from sequencing projects onSaccharomyces cerevisiae, C. elegans, and human. Amino acids conserved in at least three of the sequences areboxed. The amino acid substitution insws4 is indicated (arrow).
Sequencing of the sws mutations
To verify the relation of the sws gene to the 5.4kb transcript, we sequenced the genomic region containing the large transcript open reading frame in three sws alleles. Insws1, we identified a C to A nucleotide exchange at position 1616, producing an amber stop codon in place of a codon for serine. The SWS protein in the mutant should thus be shortened to about one-fourth of its normal length and would therefore probably be nonfunctional. It would still contain the region similar to the interaction domain of RIα but lack the putative cAMP-binding site. sws5 showed a G to A exchange at nucleotide 2431, leading to an arginine instead of a glycine. This glycine is one of the conserved amino acids in the region homologous to RIα (Fig. 8A). Insws4, nucleotide 3357 is also changed from G to A, substituting asparagine for glycine in the second region, which is conserved among Drosophila, yeast, C. elegans, and human (Fig. 8B). We could not detect a mutation in the open reading frame of the large transcript inolfE, indicating that the olfE mutation may affect only the small transcript. The demonstration that point mutations in three different mutant sws alleles all affect the 5.4 kb transcript proves that this transcript indeed encodes thesws gene product.
DISCUSSION
The sws mutation causes age-dependent neurodegeneration in adult Drosophila, preceded by an excess in glial wrapping of neurons, as seen in the multilayered glial sheaths in late pupae. The extension of glial processes happens during the second half of wild-type pupal development (Buchanan and Benzer, 1993). The first signs of neuronal apoptosis in sws flies are visible after 3–4 d of adulthood. Therefore, the occurence of the glia phenotype already in the late pupal stage suggests that the hyperwrapping is a defect in glia differentiation and not a response of glia to dying neurons. In the case of sws, such a defect in the glial sheaths could negatively influence the survival of neurons, leading to the massive degeneration visible as the mutant flies age. In another neurodegeneration mutant, drop-dead, it was also shown that glial sheaths were defective; in that case glial wrappings were incomplete (Buchanan and Benzer, 1993). The development and maintenance of correct wrapping would therefore seem to be important for the function and survival of neuronal networks in both vertebrates and invertebrates. In vertebrates, it is known that the correct number of glial cells is achieved by neuron–glia interaction; the proliferation of glial precursors and the survival of developing glial cells in rats is correlated with the number and length of axons of motoneurons (Barres and Raff, 1994). Only glial cells that manage to make contact with an unwrapped axon survive and begin myelination. However, how the correct amount of wrapping is regulated is unknown.
The phenotype of sws suggests an involvement of the SWS protein in mechanisms regulating proper ensheathment. Although the expression pattern shows that the mRNA is expressed in neurons, the phenotype is first visible in the hyperwrapping by glia. SWS might be part of a mechanism, generated in neurons, signaling glia that the right amount of wrapping has been established, or switching off a signal that promotes wrapping. If that fails, glial cells continue to extend the hypertrophic processes seen in the mutant flies. The lack of interaction would finally lead to apoptosis and breakdown of the entire nervous sytem.
The protein structural analysis indicates a possible membrane localization, with one to three transmembrane domains. Considering only the highly significant transmembrane domain, the SWS protein could be anchored in the membrane with this N-terminal domain [amino acids (aa) 36–55], which does not score for a signal sequence (Van Heijne, 1986), with the remaining 1370 aa extending into the cytoplasm. If there are three transmembrane domains, as predicted by the TMpred program, SWS could span the membrane with the N-terminal domain, form a long intracellular loop containing the region homologous to protein kinase A, and then span the membrane twice (aa 944–965 and 974–995), ending with a short intracellular stretch at its C-terminal end. The latter two putative transmembrane domains and the C-terminal end are shared by the homologous proteins in yeast, worm, and human. It has been shown in rat brain that cAMP-dependent protein kinases are localized to membranes of cell organelles. Interestingly, the membrane localization is mediated by the regulatory subunits. These are predominantly of the RIIβ type in neurons; nearly equal proportions of the RIβ and RIIβ types occur in myelin-producing oligodendrocytes (Stein et al., 1987).
The homology of the SWS protein to the RIα subunit of PKA could point to an involvement of SWS in a cAMP-dependent signal transduction pathway. In cultured Schwann cells, elevating the cAMP level mimics axonal contact (Mews and Meyer, 1993; Chiu et al., 1994). Also, cAMP can induce the expression of myelin-associated glycoprotein in oligodendrocytes (Jung et al., 1995) and neuron–glia adhesion molecules in neurons (Kallunki et al., 1995), suggesting a functional role of PKA in glia–neuron interactions in vertebrates. PKA has a quarternary structure consisting of two regulatory and two catalytic subunits. Because of the presence of a domain showing similarity with the interaction domain, SWS could bind to the catalytic subunit. However, SWS lacks the characteristic dimerization region. Moreover, SWS has homology to only the first of the two cAMP-binding sites of the regulatory subunit, and we do not know whether this region can bind cAMP. SWS is therefore significantly different from the standard regulatory subunit and might have an inhibitory function. The protein kinase inhibitor also shows major differences, and, in the interaction domain, only the two arginines are conserved. The major importance of this homologous region for the function of SWS is shown by the effect of the point mutation in sws5, which exchanges one of the conserved amino acids in that region. Phenotypically, sws5 is as severe assws1, a putative null allele containing a mutation that should shorten the protein to one-fourth of its normal size. The second mutation involving an amino acid substitution,sws4, also seems to be localized in a functionally important domain, as indicated by the high conservation of this region in different species (Fig. 8B). The point mutation changes a glycine to an arginine in a stretch of four glycines. The latter lie in the middle of a region of 10 amino acids that are identical in all proteins obtained by the homology searches, ranging from yeast to human. These proteins show a very high conservation in approximately the last third of the molecule, suggesting a common function about which, unfortunately, nothing is known. The conservation in the amino-terminal part, containing the domain similar to PKA, is much weaker, and a strech of the yeast protein seems not to be contained at all in the others. Further characterization of sws and functional studies of the protein could therefore be important to our understanding of a group of novel proteins conserved in organisms as distant as yeast and human.
Footnotes
This work was supported by fellowships to D.K. from the French Foundation for Alzheimer Research and the Deutsche Forschungsgemeinschaft (Kr 1507/1-1), and Research Grants to S.B. from the National Institute of Aging (AG12289), the National Eye Institute (EY09287), the National Science Foundation (MCB-9408718), the McKnight Foundation, and the James G. Boswell Foundation. We thank Rosalind Young and Lynette Dowling for excellent technical assistance and the members of Prof. Benzer’s research group for valuable discussions. Special thanks are due to the sequencing and oligosynthesizing facilities at Caltech.
Correspondence should be addressed to Doris Kretzschmar, Lehrstuhl für Entwicklungsbiologie, Universität Regensburg, 93040 Regensburg, Germany.
REFERENCES
- 1.Altschul S, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner M. Drosophila: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 3.Barres BA, Raff MC. Control of oligodendrocyte number in the developing rat optic nerve. Neuron. 1994;12:935–942. doi: 10.1016/0896-6273(94)90305-0. [DOI] [PubMed] [Google Scholar]
- 4.Bergoffen J, Scherer SS, Wang S, Scott MO, Bone LJ, Paul DL, Chen K, Lensch MW, Chance PF, Fischbeck KH. Connexin mutations in X-linked Charcot-Marie-Tooth disease. Science. 1993;262:2039–2042. doi: 10.1126/science.8266101. [DOI] [PubMed] [Google Scholar]
- 5.Brendel V, Bucher P, Nourbakhsh I, Blaisdell BE, Karlin S. Methods and algorithms for statistical analysis of protein sequences. Proc Natl Acad Sci USA. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunner A, Twardzik T, Schneuwly S. The Drosophila giant lens gene plays a dual role in eye and optic lobe development: inhibition of differentiation of ommatidial cells and interference in photoreceptor axon guidance. Mech Dev. 1994;48:175–185. doi: 10.1016/0925-4773(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan RL, Benzer S. Defective glia in the Drosophila brain degeneration mutant drop-dead. Neuron. 1993;10:839–850. doi: 10.1016/0896-6273(93)90200-b. [DOI] [PubMed] [Google Scholar]
- 8.Chance PF, Fischbeck KH. Molecular genetics of Charcot-Marie-Tooth disease and related neuropathies. Hum Mol Genet. 1994;3:1503–1507. doi: 10.1093/hmg/3.suppl_1.1503. [DOI] [PubMed] [Google Scholar]
- 9.Chance PF, Reilly M. Inherited neuropathies. Curr Opin Neurol. 1994;7:372–380. doi: 10.1097/00019052-199410000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Chiu SY, Scherer SS, Blonski M, Kang SS, Messing A. Axons regulate the expression of Shaker-like potassium channel genes in Schwann cells in peripheral nerve. Glia. 1994;12:1–11. doi: 10.1002/glia.440120102. [DOI] [PubMed] [Google Scholar]
- 11.Halter DA, Urban Y, Rickert C, Ner SS, Ito K, Travers AA, Technau GM. The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system. Development. 1995;121:317–332. doi: 10.1242/dev.121.2.317. [DOI] [PubMed] [Google Scholar]
- 12.Hasan G. Molecular cloning of an olfactory gene from Drosophila melanogaster. Proc Natl Acad Sci USA. 1990;87:9037–9041. doi: 10.1073/pnas.87.22.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayasaka K, Himoro M, Sato W, Takada G, Uyemura K, Shimizu N, Bird TD, Conneally PM, Chance PF. Charcot-Marie-Tooth neuropathy type-1b is associated with mutations of the myelin-P(0) gene. Nat Genet. 1993;5:31–34. doi: 10.1038/ng0993-31. [DOI] [PubMed] [Google Scholar]
- 14.Heisenberg M, Böhl K. Isolation of anatomical brain mutants of Drosophila by histological means. Z Naturforsch [C] 1979;34:143–147. [Google Scholar]
- 15.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 16.Hodgman TC. The elucidation of protein function by sequence motif analysis. Comput Appl Biosci. 1989;5:1–13. doi: 10.1093/bioinformatics/5.1.1. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann K, Stoffel W. TMbase—A database of membrane spanning protein segments. Biol Chem Hoppe Seyler. 1993;34:166. [Google Scholar]
- 18.Hogan EL, Greenfield S. Animal models of genetic disorders of myelin. Plenum; New York: 1984. [Google Scholar]
- 19.Hotta Y, Benzer S. Mapping of behaviour in Drosophila mosaics. Nature. 1972;240:527–535. doi: 10.1038/240527a0. [DOI] [PubMed] [Google Scholar]
- 20.Jung M, Kramer E, Grzenkowski M, Tang K, Blakemore W, Aguzzi A, Khazaie K, Chlichlia K, von Blankenfeld G, Kettenmann H, Trotter J. Lines of murine oligodendroglial precursor cells immortalized by an activated neu tyrosine kinase show distinct degrees of interaction with axons in vitro and in vivo. Eur J Neurosci. 1995;7:1245–1265. doi: 10.1111/j.1460-9568.1995.tb01115.x. [DOI] [PubMed] [Google Scholar]
- 21.Kallunki P, Jenkinson S, Edelman GM, Jones FS. Silencer elements modulate the expression of the gene for the neuron-glia cell adhesion molecule, Ng-CAM. J Biol Chem. 1995;270:21291–21298. doi: 10.1074/jbc.270.36.21291. [DOI] [PubMed] [Google Scholar]
- 22.Kankel DR, Hall JC. Fate mapping of nervous system and other internal tissue in genetic mosaics or Drosophila melanogaster. Dev Biol. 1976;48:1–24. doi: 10.1016/0012-1606(76)90041-5. [DOI] [PubMed] [Google Scholar]
- 23.Knapp PE, Skoff RP, Redstone DW. Oligodendroglial cell-death in jimpy mice—an explanation for the myelin deficit. J Neurosci. 1986;6:2813–2822. doi: 10.1523/JNEUROSCI.06-10-02813.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kretzschmar D, Brunner A, Wiersdorff V, Pflugfelder GO, Heisenberg M, Schneuwly S. giant lens, a gene involved in cell determination and axon guidance in the visual system of Drosophila melanogaster. EMBO J. 1992;11:2531–2539. doi: 10.1002/j.1460-2075.1992.tb05318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Guern E, Sturtz F, Gugenheim M, Gouider R, Bonnebouche C, Ravise N, Gonnaud P-M, Tardieu S, Bouche P, Chazot G, Agid Y, Vandenberghe A, Brice A. Detection of deletion within 17p112 in 7 French families with hereditary neuropathy with liability to pressure palsies (HNPP). Cytogenet Cell Genet. 1994;65:261–264. doi: 10.1159/000133643. [DOI] [PubMed] [Google Scholar]
- 26.Lewis EB, Bacher F (1968) Methods of feeding ethyl methane sulfonate (EMS) to Drosophila males. DrosophilaInf Service 43:193.
- 27.Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. Academic; San Diego: 1992. [Google Scholar]
- 28.Lilly M, Kreber R, Ganetzky B, Carlson JR. Evidence that the Drosophila olfactory mutant smellblind defines a novel class of sodium channel mutation. Genetics. 1994;136:1087–1096. doi: 10.1093/genetics/136.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mariman E, Gabreels-Festen A, van Beersum SEC, Jongen P, Gabreels F, Ropers HH. Genetic and molecular analysis of families with hereditary neuropathy with liability to pressure palsies. Am J Hum Genet. 1993;53:1041–1041. [Google Scholar]
- 30.Menne TV, Klämbt C. The formation of commisures in the Drosophila CNS depends on the midline cells and on the Notch gene. Development. 1994;120:123–133. doi: 10.1242/dev.120.1.123. [DOI] [PubMed] [Google Scholar]
- 31.Mews M, Meyer M. Modulation of Schwann cell phenotype by TGF-beta 1: inhibition of P0 mRNA expression and downregulation of the low affinity NGF receptor. Glia. 1993;8:208–217. doi: 10.1002/glia.440080308. [DOI] [PubMed] [Google Scholar]
- 32.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poeck B, Hofbauer A, Pflugfelder GO. Expression of the Drosophila optomotor-blind gene transcript in neuronal and glial cells of the developing nervous system. Development. 1993;117:1017–1029. doi: 10.1242/dev.117.3.1017. [DOI] [PubMed] [Google Scholar]
- 34.Raff MC, Barres BA, Burne JF, Coles HS, Ishizaki Y, Jacobsen MD. Programmed cell death and the control of survival: lessons from the nervous system. Science. 1993;262:695–699. doi: 10.1126/science.8235590. [DOI] [PubMed] [Google Scholar]
- 35.Renfranz PJ, Benzer S. Monoclonal antibody probes discriminate early and late mutant defects in development of the Drosophila retina. Dev Biol. 1989;136:411–429. doi: 10.1016/0012-1606(89)90267-4. [DOI] [PubMed] [Google Scholar]
- 36.Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saint Marie RL, Carlson SD. The fine structure of glia in the lamina ganglionaris of the housefly, Musca domestica. J Neurocytol. 1983a;12:213–241. doi: 10.1007/BF01148463. [DOI] [PubMed] [Google Scholar]
- 38.Saint Marie RL, Carlson SD. Glial membrane specializations and the compartmentalization of the lamina ganglionaris of the housefly. J Neurocytol. 1983b;12:243–275. doi: 10.1007/BF01148464. [DOI] [PubMed] [Google Scholar]
- 39.Saint Marie RL, Carlson SD. Structure and function of insect glia. Annu Rev Entomol. 1990;35:597–621. [Google Scholar]
- 40.Stein JC, Farooq M, Norton WT, Rubin CS. Differential expression of isoforms of the regulatory subunit of type II cAMP-dependent protein kinase in rat neurons, astrocytes and oligodendrocytes. J Biol Chem. 1987;262:3002–3006. [PubMed] [Google Scholar]
- 41.Suter U, Moskow JJ, Welcher AA, Snipes GJ, Kosaras B, Sidman RL, Buchberg AM, Shooter EM. A leucine-to-proline mutation in the putative 1st transmembrane domain of the 22 kDa peripheral myelin protein in the trembler-J mouse. Proc Natl Acad Sci USA. 1992;89:4382–4386. doi: 10.1073/pnas.89.10.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suter U, Welcher AA, Snipes GJ. Progress in the molecular understanding of hereditary peripheral neuropathies reveals new insight into the biology of the peripheral nervous system. Trends Neurosci. 1993;16:50–55. doi: 10.1016/0166-2236(93)90015-e. [DOI] [PubMed] [Google Scholar]
- 43.Taylor SS, Buechler JA, Yonemoto W. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 44.Travis J. Glia: the brain’s other cells. Science. 1994;266:970–972. doi: 10.1126/science.7973679. [DOI] [PubMed] [Google Scholar]
- 45.Valentijn LJ, Bolhuis PA, Zorn I, Hoogendijk JE, van den Bosch N, Hensels GW, Stanton VP, Housman DE, Fischbeck KH, Ross DA, Nicholson GA, Meershoek EJ, Dauwerse HG, van Ommen GJB, Baas F. The peripheral myelin gene PMP-22/gas-3 is duplicated in Charcot-Marie-Tooth disease type-1a. Nat Genet. 1992;1:166–170. doi: 10.1038/ng0692-166. [DOI] [PubMed] [Google Scholar]
- 46.Van Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verhagen WIM, Gabreels-Festen AAWM, van Wensen PJM, Joosten EMG, Vingerhoets HM, Gabreels FJM, de Graaf R. Hereditary neuropathy with liability to pressure palsies—a clinical electroneurophysiological and morphological study. J Neurol Sci. 1993;116:176–184. doi: 10.1016/0022-510x(93)90323-q. [DOI] [PubMed] [Google Scholar]
- 48.Xiong WC, Montell C. Defective glia induce neuronal apoptosis in the repo visual system of Drosophila. Neuron. 1995;14:581–590. doi: 10.1016/0896-6273(95)90314-3. [DOI] [PubMed] [Google Scholar]
- 49.Xiong WC, Okano H, Patel NH, Blendy YA, Montell C. repo encodes a glial-specific homeodomain protein required in the Drosophila nervous system. Gen Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]