Abstract
Pathogenic microbes can exert extraordinary evolutionary pressure on their hosts. They can spread rapidly and sicken or even kill their host to promote their own proliferation. Because of this strong selective pressure, immune genes are some of the fastest evolving genes across metazoans, as highlighted in mammals and insects. Drosophila melanogaster serves as a powerful model for studying host/pathogen evolution. While Drosophila melanogaster are frequently exposed to various pathogens, little is known about D. melanogaster’s ecology, or if they are representative of other Drosophila species in terms of pathogen pressure. Here, we characterize the genome of Drosophila innubila, a mushroom-feeding species highly diverged from D. melanogaster and investigate the evolution of the immune system. We find substantial differences in the rates of evolution of immune pathways between D. innubila and D. melanogaster. Contrasting what was previously found for D. melanogaster, we find little evidence of rapid evolution of the antiviral RNAi genes and high rates of evolution in the Toll pathway. This suggests that, while immune genes tend to be rapidly evolving in most species, the specific genes that are fastest evolving may depend either on the pathogens faced by the host and/or divergence in the basic architecture of the host’s immune system.
Keywords: Drosophila, viral immunity, Toll, antiviral
Introduction
Pathogens are a substantial burden to nearly every species on the planet, providing a strong selective pressure for individuals experiencing infection to evolve resistance. There is considerable evidence for selection acting on genes involved in resistance to this pathogenic burden, as highlighted by several studies across Metazoans (Kimbrell and Beutler 2001; Enard et al. 2016) including Drosophila (Sackton et al. 2007; Obbard, Welch et al. 2009). Much work concerning the evolution of the invertebrate immune system has focused on the D. melanogaster. While pathogen pressure is ubiquitous, the diversity of pathogens that hosts face and the selection for resistance they trigger vary tremendously (Sackton et al. 2007, 2009; Hetru and Hoffmann 2009; Merkling and van Rij 2013; Martinez et al. 2014; Hanson et al. 2016; Martinson et al. 2017; Palmer, Hadfield, et al. 2018). So, while it is abundantly clear that immune genes are often among the fastest evolving genes in the genome (Obbard et al. 2006, 2009; Sackton et al. 2007; Enard et al. 2016; Shultz and Sackton 2019), it is less clear whether the same genes are fast evolving in species that are genetically, geographically and/or ecologically diverged.
Based on genetic work, a number of pathways have been implicated in immune response based on work in D. melanogaster (Hoffmann 2003). Three of these pathways are Toll, IMD, and JAK-STAT, implicated in the defense response to Gram-positive bacteria and Fungi, defense response to Gram-negative bacteria, and general stress response, respectively (Ekengren and Hultmark 2001; Hoffmann 2003; Hultmark 2003; Hetru and Hoffmann 2009; Obbard, Gordon et al. 2009). In all three pathways, several genes are found to be rapidly evolving, likely due to Host/parasite arms races (Obbard et al. 2006, 2009; Sackton et al. 2007, 2009). In fact, orthologs of some effector molecules in these pathways are difficult to identify due to their rapid evolution (Ekengren and Hultmark 2001; Sackton et al. 2007; West and Silverman 2018).
In D. melanogaster and several members of the Sophophora subgenus, the canonical antiviral RNA interference (RNAi) genes are some of the fastest evolving genes in the genome, and are the focus of many studies of the evolution of antiviral pathways (Obbard et al. 2006, 2009; Palmer, Hadfield, et al. 2018). These studies suggest that viruses, specifically RNA viruses, are a major selective pressure in D. melanogaster, requiring a rapid evolutionary response from the host (Obbard et al. 2006, 2009; Daugherty and Malik 2012; Palmer, Joosten, et al. 2018). The primary antiviral pathway characterized in D. melanogaster is an RNA interference system, which uses small interfering RNAs (siRNA), generated from double stranded viral messenger RNAs (mRNAs) and Argonaute-family proteins (Hutvagner and Simard 2008; Sabin et al. 2009), to bind complimentary sequences and degrade them, preventing their use as a translation template and stopping viral replication (Wang et al. 2006; Obbard et al. 2009; Ding 2010). Importantly, these pathways have mostly been validated only in D. melanogaster, therefore, a broader view of antiviral immune gene evolution across Drosophila is warranted.
One particular group in the Drosophila genus with a rich history of ecological study, and with great potential as a host/pathogen study system, is the quinaria group (Jaenike and Perlman 2002; Perlman et al. 2002; Dyer et al. 2005; Jaenike and Dyer 2008; Unckless 2011; Unckless and Jaenike 2011). This species group is mostly mycophagous, found developing and living on the fruiting bodies of several (sometimes toxic) mushrooms. These mushrooms are commonly inhabited by parasitic nematode worms, trypanosomes, and a host of parasitic microbes (Dyer et al. 2005; Martinson et al. 2017) which are likely significant pathogenic burdens, requiring a strong immune response. One member of the quinaria group of particular interest concerning host/pathogen coevolution is Drosophila innubila (Patterson and Stone 1949). While many species in the quinaria group are broadly dispersed across temperate forests (including the sister species D. falleni and outgroup species D. phalerata) (Patterson and Stone 1949; Markow and O’Grady 2006), Drosophila innubila, is limited to the “Sky Islands,” montane forests and woodlands Southwestern United States and Mexico. It likely colonized the mountains during the previous glaciation period, 10–100 thousand years ago (Patterson and Stone 1949; Jaenike et al. 2003). The flies are restricted to elevations of 900–1,500 m, and are active only during the rainy season (late July to September) (Jaenike et al. 2003). Drosophila innubila is also the only species in the quinaria group frequently (25–46% in females) infected by a male-killing Wolbachia strain (wInn), leading to female biased sex-ratios (Dyer and Jaenike 2004). This Wolbachia is closely related to wMel, which infects D. melanogaster (but does not kill males) (Jaenike et al. 2003). Interestingly, D. innubila is also frequently (35–56%, n > 84) infected by Drosophila innubila Nudivirus (DiNV), thought to have spread to D. innubila during their expansion in the glaciation period (Unckless 2011; Hill and Unckless 2017). In contrast, DiNV is found at lower frequencies in D. innubila’s sister species, D. falleni (0–3%, n = 95) and undetected in the outgroup species, D. phalerata (0%, n = 7) (Unckless 2011). DiNV reduces both lifespan and fecundity of infected hosts (Unckless 2011), with related viruses also causing larval lethality (Payne 1974; Wang and Jehle 2009). When infected with a similar DNA virus (Kallithea virus), Drosophila melanogaster show a standard antiviral immune response, including the induction of the antiviral siRNA pathway along with other identified antiviral pathways (Palmer, Hadfield, et al. 2018; Palmer, Joosten, et al. 2018). Thus, despite lacking the genomic resources of D. melanogaster, D. innubila, and D. falleni are potentially useful model systems to understand the evolution of the immune system to identify if the signatures of selection are conserved across pathways between the highly genetically and ecologically diverged melanogaster and quinaria groups. Like D. melanogaster (Sackton et al. 2007; Obbard, Welch et al. 2009), genes involved in immune defense will be rapidly evolving in this species group. Though the difference in pathogen pressure may affect which pathways are rapidly evolving.
These results suggest species with different pathogenic burdens may have different rates of immune evolution. To this end, we surveyed the evolutionary divergence of genes within a trio of closely related mycophagous Drosophila species (D. innubila, D. falleni, and D. phalerata). As a first step, we sequenced and assembled the genome of D. innubila, resulting in an assembly which is on par with the Drosophila benchmark, D. melanogaster release 6 (Dos Santos et al. 2015). Using short read alignments to D. innubila of two closely related species (D. falleni and D. phalerata), we found evidence of selective constraint on the antiviral RNAi pathway, but rapid evolution of genes in several other immune pathways, including several conserved broad immune pathways such as the Toll and JAK-STAT pathways. These suggest that pathogen pressure differences may lead to drastic differences in immune evolution, or environmental changes may cause differences in general stress response and developmental pathways.
Results
Genome Sequencing and Assembly
Drosophila innubila is a mushroom-feeding species found across the sky islands of the Southwestern United States and Western Mexico (Patterson and Stone 1949). It is in the quinaria group of the Drosophila subgenus, ∼50 My diverged from the research workhorse, D. melanogaster (Dyer and Jaenike 2004; Markow and O’Grady 2006). D innubila has a sister species in northern North America, D. falleni, and the pair share an old-world outgroup species, D. phalerata. These species are highly diverged from all other genome-sequenced Drosophila species and represent a genomically understudied group of the Drosophila subgenus (Jaenike et al. 2003; Markow and O’Grady 2006).
We sequenced and assembled the genome of D. innubila using a combination of MinION long reads with HiC scaffolding (Oxford Nanopore Technologies Inc., Oxford, UK), and Illumina short reads for error correcting (Illumina, San Diego, CA) (fig. 1A and table 1; NCBI accession: SKCT00000000). The genome is 168 Mb with 50% of the genome represented in scaffolds 29.59 Mb or longer (N50 = 29.59 Mb), eclipsing release 6 of D. melanogaster (N50 = 25.29 Mb, Clark et al. 2007; Dos Santos et al. 2015; Gramates et al. 2017). Additionally, the D. innubila N90 is 25.98 Mb compared with 23.51 Mb in D. melanogaster. At this quality of assembly, the N50 and N90 statistics are approaching full chromosome lengths in Drosophila and therefore likely do not represent how contiguous the assembly is, given the genome of interest. To better discern between the quality of assemblies, we calculated the proportion of the entire genome found in the six largest contigs (for the six Drosophila Muller elements) for each reference genome. In D. innubila, 97.71% of the genome is found in these six largest contigs, compared with 95.99% of the D. melanogaster genome (supplementary tables 1–3, Supplementary Material online).
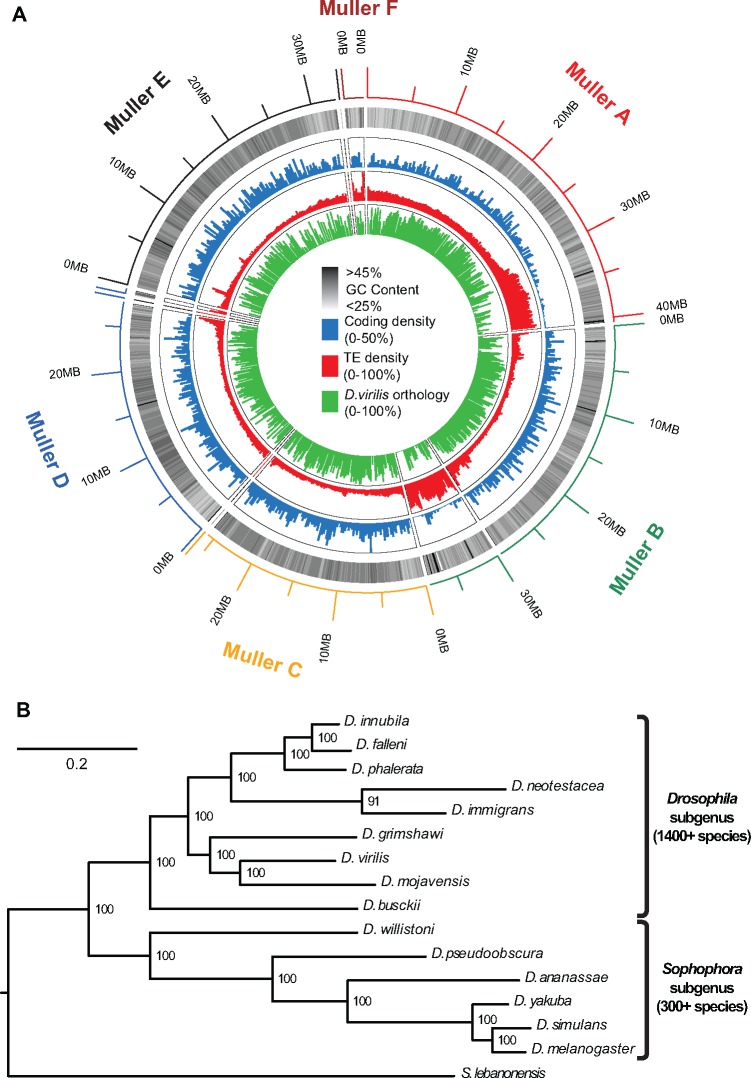
Fig. 1.
The Drosophila innubila genome. (A) A circular summary of the D. innubila genome limited to the assembled acrocentric chromosomes on ten major scaffolds. The rings (from the outside in) are the chromosome identity, the length of each segment, the percentage GC content, the coding density, the transposable element density (250-kb windows, sliding 250 kb), and the percentage of each window that aligns to D. virilis (250 kb windows, sliding 250 kb). (B) Phylogenetic relationships of D. innubila, D. falleni, D. phalerata and the main lineages of Drosophila generated using 100 concatenated genes, with 500 bootstraps (shown as % support on the nodes).
Table 1.
Genome Summary.
| Scaffold (Muller element—D. melanogaster ortholog—ID) | Total Length (kb) | Total Gene Count | Orphan Count | Mean Gene Length (bp) | Mean Repeat Content (%) |
|---|---|---|---|---|---|
| A-X-3 | 40,479 | 2,012 | 73 | 5,090.7 | 31.7 |
| B-2L-4 | 29,570.5 | 2,249 | 99 | 4,394.6 | 13.9 |
| B-2L-5 | 8,037.1 | 160 | 13 | 4,679.2 | 62.9 |
| C-2R-1 | 25,683.3 | 2,518 | 58 | 4,033.3 | 12.2 |
| C-2R-27 | 16.5 | 0 | 0 | NA | 92.4 |
| D-3L-0 | 27,707.2 | 2,345 | 68 | 4,248.1 | 12.6 |
| D-3L-29 | 45.7 | 0 | 0 | NA | 83.7 |
| E-3R-2 | 32,746.5 | 2,863 | 73 | 4,092.4 | 11.4 |
| E-3R-11 | 18.4 | 1 | 1 | 282 | 75.6 |
| F-4-6 | 2,027.1 | 106 | 7 | 6,992 | 35.5 |
| Unassembled (324 contigs) | 1,163.5 | 46 | 1 | 1,429 | 39.1 |
| Mitochondria | 16.1 | 18 | 0 | 1,658.7 | 0 |
| Total | 168,031 | 12,318 | 393 | 4,350.7 | 13.5 |
Note.—Summary of the major assembled scaffolds in the Drosophila innubila genome, including the length in kilobase pairs (kb), number of genes, number of orphan genes, gene lengths in base pairs (bp), and proportion of the scaffold that is repetitive content.
Using a combined transcriptome assembly of all life stages and protein databases from other species, we annotated the genome and found 12, 318 genes (including the mitochondrial genome), with coding sequence making up 11.5% of the genome, at varying gene densities across the Muller elements (fig. 1A blue and table 1). Of the annotated genes, 11, 925 (96.8%) are shared with other Drosophila species (among the 12 genomes available on Flybase.org), 7, 094 (57.6%) have orthologs in the human genome, and the annotation recovered 97.2% of the Dipteran BUSCO protein library (Simão et al. 2015). The D. innubila genome has an average of 36.57% GC content, varying across the Muller elements (fig. 1A black/white). Using dnaPipeTE (Goubert et al. 2015), RepeatModeler (Smit and Hubley 2008) and RepeatMasker (Smit and Hubley 2015), we estimated that 13.53% of the genome consists of transposable elements (TEs, fig. 1A red), which is low for Drosophila (Sessegolo et al. 2016). Using Mauve (Darling et al. 2004) we compared our assembly to the best assembled genome within the Drosophila subgenus, D. virilis, and found that most regions in the D. innubila genome have some orthology to the D. virilis reference genome (fig. 1A green). A summary of the assembled genome including codon bias, TE content, orphan genes, duplications, and expression changes across life stages can be found in the supplementary results (supplementary tables 7–20 and figs. 4–12, Supplementary Material online).
Immune Pathways Evolve Differently in D. innubila and D. melanogaster
We aimed to determine whether evolutionary rates in different functional categories are conserved across D. innubila and D. melanogaster, given the genetic and ecological divergence between the two species (Patterson and Stone 1949; Jaenike et al. 2003; Markow and O’Grady 2006). Using short reads from D. falleni and D. phalerata mapped to the D. innubila genome, we identified DNA sequence divergence and generated consensus gene sequences for each species. We then aligned the DNA sequence from each species for each gene to the D. innubila ortholog (PRANK –codon +F) (Löytynoja 2014). For each ortholog set, we identified the proportion of synonymous (dS) substitutions and amino acid changing, nonsynonymous substitutions (dN) (per possible synonymous or nonsynonymous substitution, respectively) occurring on each branch of the phylogeny (codeML branch-based approach, model 0) (Yang 2007; McKenna et al. 2010; DePristo et al. 2011; Löytynoja 2014). This allowed us to calculate dN/dS to identify genes showing signatures of rapid or unconstrainted evolution specifically on the D. innubila branch of the tree (elevated dN/dS, fig. 2A). Conversely, we can also identify genes under strong purifying selection (reduced dN/dS) (Yang 2007). We also performed this analysis across the D. melanogaster clade using D. melanogaster, D. simulans, and D. yakuba reference genomes but focusing on the D. melanogaster branch (Clark et al. 2007; Gramates et al. 2017). These trios are of similar levels of divergence (fig. 1B), The rate of evolution, as measured by dN/dS, is significantly positively correlated between the species across individual genes (species-specific branch comparison, Spearman rank correlation = 0.6856, P value = 1.52×10−06) and most genes are under selective constraint; only 25.6% of genes have dN/dS >0.5 (fig. 2B).
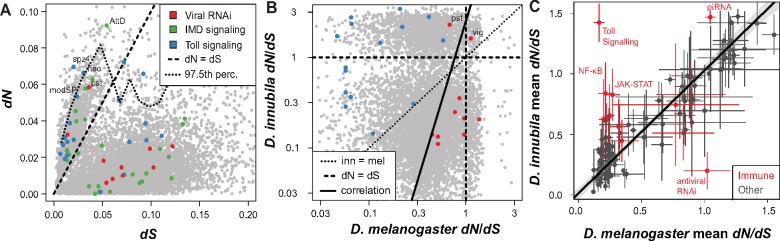
Fig. 2.
Toll and antiviral genes evolve differently between Drosophila melanogaster and D. innubila. (A) The nonsynonymous (potentially adaptive) divergence (dN) for each gene across the D. innubila genome, compared with the gene’s synonymous (neutral) divergence (dS). Genes involved in antiviral and antibacterial (Toll and IMD) immune signaling pathways are colored. The upper 97.5th percentile is shown as a dotted line, dN/dS = 1 is shown as a dashed line. (B) Comparison of dN/dS in D. innubila and D. melanogaster. Toll and antiviral genes are shown for comparison. The dotted line highlights when innubila and melanogaster dN/dS are equal. The dashed line highlights when dN=dS in either species. The solid line highlights the spearman correlation between D. melanogaster and D. innubila dN/dS. (C) Mean dN/dS for genes within core 116 GOslim categories (Consortium et al. 2000; Carbon et al. 2017) in both D. melanogaster and D. innubila, alongside specific immune and RNAi categories of interest. Bars are shown giving the SE for each category in D. melanogaster (X-axis) and D. innubila (Y-axis). Categories are colored by immune categories (red) or background categories (gray), with immune categories of interest highlighted. The fitted line is for all background categories, with immune categories added post hoc. Abbreviations: AttD, attacin D; modSP, modular serine protease; nec, necrotic; pst, pastrel; spz4, spaetzle 4; vig, vasa intronic gene.
To determine whether similar classes of genes were under similar selection pressures in the two species, we grouped genes by gene ontology (GO) categories using GOSlim (Consortium et al. 2000; Carbon et al. 2017). There was a significant linear correlation between mean values of GO categories in each species (fig. 2C black points, Spearman rank correlation = 0.847, GLM t = 17.01, P value = 4.613×10−33), suggesting that, in general, pathways may be under similar selective pressures (either constrained evolution or positive selection, fig. 2C). We identified GO categories enriched in the top 10% of genes for dN/dS to find categories evolving rapidly in each species. Several enriched GO immune categories were common to both D. innubila and melanogaster, including defense response to bacteria and antimicrobial peptide regulation (supplementary table 4, Supplementary Material online). However, several GO terms related to Toll signaling were enriched exclusively in D. innubila, while gene silencing by RNA and RNA splicing were enriched in D. melanogaster but not D. innubila (fig. 2B red points and supplementary table 4, Supplementary Material online).
We fitted a model to contrast D. innubila dN/dS and D. melanogaster dN/dS per gene and extracted the studentized residuals. We examined the upper 10% of residuals, which should contain genes fast evolving in only D. innubila. We found Toll receptor signaling pathways and metabolic processes enriched, among others (supplementary table 4, Supplementary Material online and fig. 2B and C, P value <0.000775, though this is not significant after correcting more multiple tests). Conversely, in the lower 10% (which should be genes fast-evolving only in D. melanogaster) we found RNAi and response to virus genes enriched (supplementary table 4, Supplementary Material online and fig. 2B and C, P value <0.000998, though this is not significant after correcting for multiple tests). These lines of evidence suggested that, while many functional categories are evolving similarly, Toll and antiviral RNAi pathways are evolving quite differently between D. melanogaster and D. innubila and motivated a more thorough examination of the differences in the evolution of genes involved in immune defense.
Immune Evolution Differs between Species Groups, Even after Controlling for Synonymous Divergence
While we found no correlation between dN/dS and gene length in either species (GLM t = 0.34, P value = 0.81), we did find a significant negative correlation between dN/dS and dS in D. innubila (fig. 2A, GLM t =−64.94, P value = 2.2×10−16). Most genes with high dN/dS had lower values of dS, possibly due to the short gene branches (and low neutral divergence) between species inflating the proportion of nonsynonymous substitutions (fig. 2A). We also found slightly different distributions of dN/dS in each species, suggesting which may cause the differences seen (supplementary fig. 1, Supplementary Material online). Because of these effects, we attempted to control for differences by extracting genes that were in the upper 97.5% dN/dS of genes per 0.01 dS window and with dN/dS >1 and labeled these 166 genes as the most rapidly evolving on the D. innubila branch. Contrasting D. melanogaster (Obbard et al. 2006), these genes were not enriched for antiviral RNAi genes and were instead significantly enriched for several metabolic and regulation pathways, as was found previously (table 2). The most common type of gene with elevated dN/dS were those involved in the regulation of the Toll pathway (table 2, figs. 2 and 3, and supplementary table 5, Supplementary Material online, GOrilla FDR P value = 0.00015 after multiple testing correction, enrichment = 14.16) (Eden et al. 2009). Specifically, we found four Toll signaling genes; spatzle4, necrotic, spheroide, and modSP; were the fastest evolving genes in this pathway, and among the fastest in the genome (fig. 2A, above the dotted line, supplementary table 6, Supplementary Material online). Most of these rapidly evolving genes are signaling genes, which were, as a class, not particularly fast evolving in the D. melanogaster clade (Sackton et al. 2007). Additionally, Pp1apha-96A and Attacin D, genes involved in Gram-negative bacterial response, were rapidly evolving (upper 97.5% of genes, dN/dS >1).
Table 2.
Gene Enrichments.
| Gene Ontology Category | No. Genes dN/dS >0.25% | Total No. Genes in GO Category | Enrichment | P value | P Value (after multiple testing correction) |
|---|---|---|---|---|---|
| Regulation of the toll pathway | 4 | 20 | 14.16 | 2.62×10−8 | 0.000153 |
| Metabolic process | 160 | 2,317 | 1.22 | 6.29×10−4 | 1 |
| Cellular response to light stimulus | 5 | 25 | 6.77 | 6.98×10−4 | 1 |
| Vesicle uncoating | 2 | 2 | 33.85 | 8.68×10−4 | 1 |
| Organic hydroxy compound metabolic process | 9 | 87 | 3.5 | 9.85×10−4 | 1 |
Note.—Gene ontology groups enriched for high dN/dS on the Drosophila innubila branch. Table includes the number of genes in each pathway found in the upper 0.25% for dN/dS, the enrichment of each gene category as well as significance of the category before and after multiple testing correction.
Given the differences between species, we compared molecular evolution of genes in each of immune gene category for the entire melanogaster trio (D. melanogaster, simulans, and yakuba) with the evolution of those same genes in the entire D. innubila trio (D. innubila, falleni, and phalerata), alongside comparing specifically D. innubila and D. melanogaster. Because nonsynonymous divergence is elevated in genes with low-synonymous divergence on the D. innubila branch but not the D. melanogaster branch (fig. 2A and supplementary fig. 1, Supplementary Material online), we attempted to control for its effect. For each focal immune gene, we extracted genes on the same chromosome with dS within 0.01 of the focal gene. We then calculated the differences in the median dN/dS of these control genes and the focal genes, for each branch on the tree, and categorized these differences by immune category based on Flybase gene ontologies (Gramates et al. 2017). We also separated antiviral genes into those associated with antiviral RNAi and those involved in other pathways (such as NF-κB signaling molecules). Using this method, we found most immune categories had slightly positive differences compared with the controls, suggesting faster evolution than the background (fig. 3A), consistent with results across the entire genus (Sackton et al. 2007). Specifically, the Toll signaling, JAK-STAT, response to Gram-positive infection, response to Gram-negative infection and other genes associated with resistance to viral infection (Magwire et al. 2011, 2012; Palmer, Hadfield, et al. 2018) were significantly higher than the background in the D. innubila trio (fig. 3A and supplementary fig. 3 and table 5, Supplementary Material online, t-test t = 2.39, P value <0.05, all categories are normally distributed, Shapiro–Wilk test P value >0.0521). Toll genes also had significantly higher rates of evolution in D. innubila than D. melanogaster (Wilcoxon Rank Sum Test, W = 226, P value = 0.01051). Again, in contrast to D. melanogaster (Obbard et al. 2006), there was no significant elevation of the rates of evolution in antiviral RNAi genes in D. innubila, suggesting selective constraint (t-test t = 1.0798, P value = 0.3082). In fact, only one antiviral RNAi gene, pastrel, appears to be fast-evolving in D. innubila, with most genes in this category close to the median dN/dS for the innubila genome (figs. 2A, 2B, and 3A). Interestingly, pastrel is among the slowest evolving antiviral gene in D. melanogaster (though still in the upper 25% of all genes). Variation in pastrel has been associated with survival after Drosophila C virus infection (DCV) in D. melanogaster, but is not likely involved with antiviral RNAi (Magwire et al. 2011, 2012; Barbier 2013).
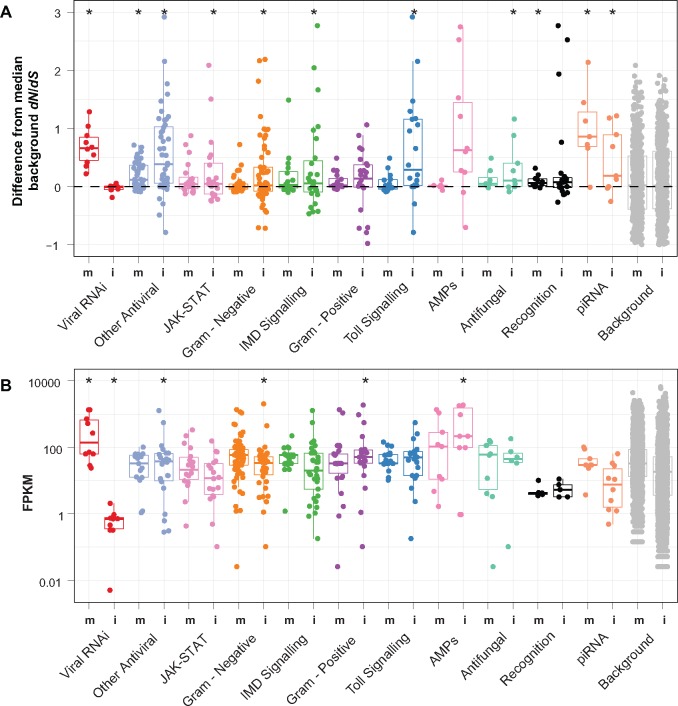
Fig. 3.
Fast evolving immune gene categories differ between species. (A) For each immune gene or RNAi gene, we have calculated the difference in dN/dS between the gene and the mean of background genes of similar dS (+-0.01dS). i refers to the Drosophila innubila branch while m refers to the D. melanogaster branch. A P value (from a single sample, two-sided t-test looking for significant differences from 0) of 0.05 or lower is designated with *. (B) Expression as read counts per 1 kb of exon for each gene (FPKM) by immune gene in each species. For each category, we have overlaid a boxplot showing the median (center line) and interquartile range for each category in both species groups, with whiskers to 97.5% of the next interquartile. i refers to D. innubila while m refers to D. melanogaster. Categories marked with a * are significantly different from the background category with a Mann–Whitney U test (P <0.05).
As dN/dS may give false signals of rapid evolution due to multiple nucleotide substitutions occurring per site (Venkat et al. 2018), we calculated δ (another measurement of the rate of evolution) using a method that controls for multinucleotide substitutions in a single site (Pond et al. 2005; Venkat et al. 2018). δ was broadly positively correlated with dN/dS in both species (Spearman correlation = 0.17, P value = 0.0192). Using this method, we corroborated our previous finding that antiviral genes are rapidly evolving exclusively in the D. melanogaster trio compared with the background (supplementary fig. 2, Supplementary Material online, GLM t = 4.398, P value = 1.1×10−05), while bacterial response genes (both Gram-positive and -negative) are rapidly evolving only in the D. innubila trio compared with the background (supplementary fig. 2, Supplementary Material online, GLM t > 2.682, P value <0.00731).
On some branches of the D. innubila trio phylogeny, we find differing signatures from the total D. innubila trio phylogeny. Specifically, while Gram-positive bacterial response is fast evolving in D. falleni, antifungal and Gram-negative bacterial responses are fast evolving in D. innubila (supplementary table 5, Supplementary Material online, t-test t = 2.11, P value <0.05), whereas none of these three groups are fast evolving in D. phalerata. This potentially highlights differences in the pathogens and environments encountered by the three species. Interestingly, Toll signaling, but not Gram-positive defense response, is fast evolving in D. innubila (fig. 3 and supplementary table 5, Supplementary Material online), suggesting Toll may play a separate role from signaling Gram-positive defense response in D. innubila, possibly directed more acutely toward antiviral or antifungal defense (Takeda and Akira 2005; Zambon et al. 2005; Palmer, Medd, et al. 2018) or directed toward Toll’s role in the regulation of development (Keshishian et al. 1993; Valanne et al. 2011).
Several Alternative Antiviral Immune Genes Are Rapidly Evolving in Both Species
We separated the known antiviral pathways and viral interacting genes into specific categories, and examined their evolution in both D. melanogaster, D. innubila and across each clade to find pathways showing consistent rates of evolution. The JAK-STAT pathway (Janus kinase signal transduction and activation of transcription) is a conserved signaling pathway involved in processes such as immunity, cell death, and general stress response, and is implicated in the DNA virus response (Hultmark 2003; West and Silverman 2018). We find this pathway is significantly faster evolving compared with the background across the D. innubila trio, while NF-κB, Toll, and putative viral-capsid interacting genes are evolving significantly faster than background genes of similar dS in both D. innubila and D. melanogaster trios (supplementary fig. 3, Supplementary Material online, t-test t = 2.90, P value <0.05). Several genes within these categories also showed consistent signatures across the D. innubila and D. melanogaster trios (figs. 2 and 3; supplementary tables 4 and 5, Supplementary Material online). Genes rapidly evolving in both lineages include the JAK-STAT cytokines upd2 and upd3, JAK-STAT regulatory genes CG30423 and asrij, the Toll pathway genes grass and GNBP1, and the NF-κB signaling molecules relish and Aos1. After controlling for multiple nucleotide substitutions per site with δ (Pond et al. 2005; Venkat et al. 2018), we found that JAK-STAT, NF-κB, Toll, and viral capsid associated genes are rapidly evolving in both trios (supplementary fig. 2, Supplementary Material online, GLM t > 3.22, P value >0.00128), however the putatively viral capsid associated genes are evolving most rapidly in the innubila trio (supplementary fig. 2, Supplementary Material online, GLM t = 4.124, P value = 3.73×10−05).
Low-Antiviral RNAi Expression in Drosophila innubila
We examined immunity and RNAi genes in the context of baseline (e.g., constitutive) gene expression in D. innubila compared with D. melanogaster using ModEncode (Consortium et al. 2000). It should be noted that this is not a well-controlled comparison, whereby differences in expression could be due to different laboratory conditions or other experimental variables as opposed to true baseline expression differences. Nevertheless, some of the observed differences are consistent with rates of molecular evolution. We found no effect for life stage, sex, or tissue on immune expression outside of an increase in immune expression when transitioning from embryo to larval stages (GLM P > 0.05, supplementary tables 12–19, Supplementary Material online). Specifically, the Toll pathway genes Toll, GNBP1, and grass had higher expression in larvae than embryos, and this was maintained throughout the rest of the life stages. Because the main shift in gene expression appears to occur as embryos develop into larvae, we focused on adults, as they represent a more stable period of gene expression. We focused on adult whole-body expression differences between D. innubila and D. melanogaster. The viral RNAi pathway is mostly shut off in D. innubila (fig. 3B, only two of seven genes showed expression >1 read per million counts per kb of gene in larvae and adults) and had significantly lower expression than the rest of the genome across all life stages (both before and after adjusting for gene length, Wilcoxon rank sum P value <0.02). In contrast, the piRNA pathway showed appreciable levels of expression and no difference from the background genome at any stage (fig. 3B, Wilcoxon rank sum P value >0.05). This difference in expression between the antiviral RNAi and piRNA pathways may be due to antiviral genes only being expressed upon infection, though other antiviral genes show no significant difference in expression from the background (Wilcoxon Rank Sum test: W = 27,464, P value = 0.1089). Additionally, AMPs, which are highly induced upon infection in insects, here also showed high levels of constitutive expression compared with the background (fig. 3B, Wilcoxon Rank Sum test: W = 11,642, P value <0.05). Furthermore, the antiviral RNAi genes are significantly more highly expressed compared with background genes in the D. melanogaster expression data, even without a known infection, and are further induced upon infection with a DNA virus (Wilcoxon Rank Sum test: W = 80,672, P value <0.005) (Obbard et al. 2006, 2009; Ding 2010; Palmer, Hadfield, et al. 2018; Palmer, Medd, et al. 2018). Antiviral RNAi genes are significantly more highly expressed in D. melanogaster than in D. innubila (Wilcoxon Rank Sum test: W = 2, P value = 0.0003), while immune signaling and immune recognition genes are more highly expressed in D. innubila compared with D. melanogaster (Wilcoxon Rank Sum test: W = 178 P value = 0.0002). Thus, high rates of expression seem to be associated with high rates of evolution in Drosophila immune genes, irrespective of species.
Discussion
Host/parasite coevolution is ubiquitous across the tree of life (Dawkins and Krebs 1979; Lively 1996). It is expected to result in rapid evolution of protein sequence in both the host and the parasite, as both organisms are adapting to escape the selective pressure exerted by the other. In support of this, immune genes in Drosophila, and other organisms, evolve more rapidly than most other gene categories (Kimbrell and Beutler 2001; Sackton et al. 2007; Obbard, Welch et al. 2009; Enard et al. 2016), the fastest among these being the antiviral genes (Obbard et al. 2006, 2009; Enard et al. 2016; Palmer, Hadfield, et al. 2018). Here, we have shown that while immune genes are fast evolving in D. innubila, the categories of genes most rapidly evolving is strikingly different from those most rapidly evolving in D. melanogaster.
There are several explanations for the observed differences in immune evolution between the D. melanogaster and D. innubila trios. First, and most obvious explanation is that different pathogen pressures result in different rates of evolution between species. The most tempting difference to highlight is the high-frequency DNA virus infection in D. innubila but not D. melanogaster (Unckless 2011). DNA virus response in Drosophila involves a larger set of pathways than RNA virus response, which is largely mediated via the siRNA pathway (Coccia et al. 2004; Bronkhorst et al. 2012; Palmer, Joosten, et al. 2018). Many previous studies of RNA and DNA virus immune response in D. melanogaster have implicated the IMD, JAK-STAT, NF-κB, and Toll pathways as vital components of viral defense, all of which are rapidly evolving in D. innubila (Dostert et al. 2005; Zambon et al. 2005; Hetru and Hoffmann 2009; Palmer et al. 2018; West and Silverman 2018). Due to overlapping viral transcripts, infection can still induce the siRNA pathway irrespective of the viral class, but may differ in effectiveness between species and viral class (Webster et al. 2015; Palmer, Medd, et al. 2018). It is currently believed that D. melanogaster is mostly exposed to RNA viruses in nature whereas D. innubila is mostly exposed to DNA viruses (Unckless 2011; Webster et al. 2015; Lewis et al. 2018). Therefore, it makes sense that various immune response pathways are evolving at different rates in the two species groups. In keeping with this, D. falleni and D. phalerata have different rates of immune gene evolution and are not frequently exposed to the DNA virus (Unckless 2011) (supplementary tables 5 and 6, Supplementary Material online).
DNA virus exposure is not the only difference in pathogens seen between D. innubila and D. melanogaster. In fact, most pathways identified as rapidly evolving in D. innubila are involved in the response to infection by multiple pathogens (Toll, for viruses, fungi and Gram-positive bacteria) or are general stress response pathways (JAK-STAT). Given the similar ecologies (including regular exposure to rotting mushrooms) (Perlman et al. 2002; Jaenike et al. 2003) and consistent signatures of rapid evolution in D. innubila and D. falleni, similar pathogen pressures may drive rapid evolution of these pathways in quinaria group flies (Shoemaker et al. 1999; Perlman et al. 2002). For example, the rapid evolution of Toll signaling but not Gram-positive defense response (fig. 3), might suggest that Toll is evolving in response to something other than Gram-positive bacteria such as fungal pathogens, viruses, or even extracellular parasites that uniquely infect the quinaria group (Jaenike and Perlman 2002; Perlman et al. 2002; Hoffmann 2003; Zambon et al. 2005; Hamilton et al. 2014; Palmer, Joosten, et al. 2018).
Previous work in D. melanogaster has highlighted the role that Gram-negative commensal bacteria play in priming the antiviral immune system (Sansone et al. 2015). As both the Toll and IMD signaling pathways are rapidly evolving across the innubila trio, it is conceivable that this priming may play a role in immune defense in D. innubila.
Finally, Toll signaling is also involved in dorso/ventral development and motorneuron development in Drosophila (Keshishian et al. 1993; Hoffmann 2003; Valanne et al. 2011), as eye and neuronal development are almost always enriched in D. innubila but not D. melanogaster (supplementary table 4, Supplementary Material online), this rapid evolution of Toll may have little to do with immune response and instead is involved in changes in the body pattern to adapt to changes in the environment of D. innubila. Though this explanation does not explain the rapid evolution of other immune pathways.
A second hypothesis for the lack of evolution in the antiviral RNAi system, is that the immune response to DNA virus infection has diverged in the approximately 50 My since the quinaria and melanogaster groups last shared a common ancestor. They may fundamentally differ in their immune response to viral infection, and this may be due to the divergence of the siRNA pathways across the Drosophila groups (Lewis et al. 2018). This divergence could also be nonadaptive, in fact, given D. innubila has undergone repeated bottlenecking during habitat invasion, it is possible that changes in effective population size may have led to genetic drift steering the evolution of the immune system in this species, resulting in relaxed constraint on immunity genes. An ineffective antiviral immune system may even explain the high frequency of DiNV infection in D. innubila (Unckless 2011). However, rates of evolution are mostly consistent across the D. innubila trio, and, as broadly dispersed temperate species, D. phalerata and falleni should not have been affected by the same demographic patterns (Markow and O’Grady 2006). Drosophila innubila’s invasion of the “sky islands” is also estimated to be more ancient than D. melanogaster’s global invasion (occurring during the last glaciation period, 10–100 Ka), with current estimates of diversity at similar levels as D. melanogaster (Dyer and Jaenike 2005). If D. innubila’s bottleneck was more severe than D. melanogaster’s, drift may still explain the lack of antiviral evolution in D. innubila. The lack of adaptation of the antiviral RNAi could also be due to the more recent infection by DiNV, estimated to have infected D. innubila 10–30 thousand years ago, however this is more than enough time for adaptation to occur in Drosophila (Aminetzach et al. 2005; Karasov et al. 2010).
There are several other aspects of the host biology that may explain the constrained evolution of the siRNA in D. innubila (figs. 2 and 3). siRNA, alongside piRNA have been implicated in transposon regulation as well as viral suppression (Biryukova and Ye 2015). It is possible siRNA has an alternate, TE-related role in D. innubila, which may contribute to their low TE content (fig. 1 and supplementary fig. 6, Supplementary Material online).
Studies have also identified an interaction between Wolbachia infection and resistance or susceptibility to viral infection (Teixeira et al. 2008; Martinez et al. 2014). The high frequency of Wolbachia infection in D. innubila (Dyer and Jaenike 2005), may therefore provide some resistance to viral infection or may be involved in immune system priming (Sansone et al. 2015). However, Wolbachia has only been shown to protect against RNA viruses (Teixeira et al. 2008), and this effect of Wolbachia was found to be absent in an earlier assessment of DiNV infections in D. innubila (Unckless 2011; Martinez et al. 2014), suggesting Wolbachia may not play a role in viral resistance.
We have worked to bring mycophagous Drosophila to the table as a modern genomic model for the study of immune system evolution. Here, we have highlighted that the evolution of the immune system among closely related trios of species may differ drastically across Drosophila genera. Specifically, we found that across the D. innubila genome, even though the immune system is, in general, evolving rapidly; the canonical antiviral RNAi pathways do not appear to be evolving as if in an arms race with viruses. Instead, several alternative immune pathways may be evolving in response to the different pathogen pressures seen in this species. Together these results suggest that the evolution of genes involved in the immune system can be quite specific to the suite of pathogens faced by hosts.
Materials and Methods
DNA/RNA Isolation, Library Preparation, Genome Sequencing, and Assembly
We extracted DNA following the protocol described in (Chakraborty et al. 2017) for D. innubila, D. falleni, and D. phalerata as further described in the Supplementary Material online. We prepared the D. innubila DNA as a sequencing library using the Oxford Nanopore Technologies Rapid 48-h (SQK-RAD002) protocol, which was then sequenced using a MinION (Oxford Nanopore Technologies, Oxford, UK; NCBI SRA: SAMN11037163) (Jain et al. 2016). The same DNA was also used to construct a fragment library with insert sizes of ∼180, ∼3,000, and ∼7,000 bp, we sequenced this library on a MiSeq (300 bp paired-end, Illumina, San Diego, CA; NCBI SRA: SAMN11037164). We prepared the D. falleni and D. phalerata samples as Illumina libraries like D. innubila but with a 300-bp insert size. We sequenced the D. falleni fragment library on one half of a MiSeq (300 bp paired-end, Illumina, San Diego, CA; NCBI SRA: SAMN11037165) by the KU CMADP Genome Sequencing Core. We sequenced the D. phalerata fragment library on a fraction of an Illumina HiSeq 4000 run (150 bp paired end, Illumina, San Diego, CA; NCBI SRA: SAMN11037166).
For gene expression analyses, we obtained two replicate samples of female and male heads and whole bodies (including heads), embryos, larvae (pooled across all three instar stages), and pupae (all nonadults were unsexed). RNA was extracted using a standard Trizol procedure (Simms et al. 1993) with a DNAse step. RNA-sequencing libraries were constructed using the standard TruSeq protocol (McCoy et al. 2014) with ½ volume reactions to conserve reagents. Individually indexed RNA libraries (two replicates from each tissue/sex) were sequenced on one lane of an Illumina “Rapid” run with 100-bp single-end reads (NCBI SRA: SAMN11037167-78). All data used in the assembly and annotation of the D. innubila genome are available in the NCBI BioProject PRJNA524688.
Bases were called post hoc using the built in read_fast5_basecaller.exe program with options: –f FLO-MIN106 –k SQK-RAD002 –r–t 4. Raw reads were assembled using CANU version 1.6 (Koren et al. 2016) with an estimated genome size of 150 million bases and the “nanopore-raw” flag. We then used Pilon to polish the genome with our Illumina fragment library (Walker et al. 2014). The resulting assembly was submitted to PhaseGenomics (Seattle, WA) for scaffolding using Hi-C and further polished with Pilon for seven iterations. With each iteration, we evaluated the quality of the genome and the extent of improvement in quality, by calculating the N50 and using BUSCO to identify the presence of conserved genes in the genome, from a database of 2,799 single copy Dipteran genes (Simão et al. 2015).
Repetitive regions were identified de novo using RepeatModeler (engine = NCBI) (Smit and Hubley 2008) and RepeatMasker (-gff –gcalc –s) (Smit and Hubley 2015).
These sequencing and assembly steps are further described in the supplementary methods, Supplementary Material online, alongside additional steps taken to verify genes, identify additional contigs and genes, and find genes retained across all species. The final version of the genome and annotation is available on the NCBI (accession: SKCT00000000).
Genome Annotation
As further described in the supplementary methods, Supplementary Material online, we assembled the transcriptome, using all Illumina RNA reads following quality filtering, using Trinity (version 2.4.0) (Haas et al. 2013), Oases (velvetg parameters: -exp_cov 100 -max_coverage 500 -min_contig_lgth 50 -read_trkg yes) (Schulz et al. 2012), and SOAPdenovo Trans (127mer with parameters: SOAPdenovo-Trans-127mer -p 28 -e 4 and the following kmers: 95, 85, 75, 65, 55, 45, 35, 29, 25, 21) (Xie et al. 2014) which we combined using EvidentialGene (Gilbert 2013; http://eugenes.org/EvidentialGene/). We used the D. innubila transcriptome as well as protein databases from M. domestica, D. melanogaster, and D. virilis, in MAKER2 (Holt and Yandell 2011) to annotate the D. innubila genome, including using RepeatModeler (Smit and Hubley 2008) to not misassign repetitive regions. This was repeated for three iterations to generate a GFF file containing gene evidence generated by MAKER2 (Holt and Yandell 2011) (NCBI:).
Drosophila Quinaria Group Species on the Drosophila Phylogeny
To build a phylogeny for the Drosophila species including D. innubila, D. falleni, and D. phalerata, we identified genes conserved across all Drosophila and humans and found in the Dipteran BUSCO gene set (Simão et al. 2015). We then randomly sampled 100 of these genes, extracted their coding sequence from our three focal species and 9 of the 12 Drosophila genomes (Limited to nine due to our focus on the Drosophila subgenus and the close relation of several species, rendering them redundant in this instance, Clark et al. 2007). We also searched for genomes in the Drosophila subgenus with easily identifiable copies of these 100 conserved genes, settling on D. busckii (Zhou and Bachtrog 2015), D. neotestactea (Hamilton et al. 2014), D. immigrans, and Scaptodrosophila lebanonensis (Zhou et al. 2012). We aligned these genes using MAFFT (–auto) (Katoh et al. 2002), concatenated all alignments and generated a phylogeny using PhyML with 500 bootstraps (-M GTR, -Gamma 8, -B 500) (Guindon et al. 2010).
Signatures of Adaptive Molecular Evolution among Species
We mapped short read sequencing data of D. innubila, D. falleni, and D. phalerata to the repeat-masked D. innubila reference genome using BWA MEM (Li and Durbin 2009). As similar proportions of reads mapped to the genome (97.6% for D. innubila, 96.1% for D. falleni, and 94.3% for D. phalerata), covering a similar proportion of the reference genome (99.1% for D. innubila, 98.5% for D. falleni, and 97.1% for D. phalerata), we considered the D. innubila genome to be of similar enough to these species to reliably call single nucleotide polymorphisms and indels. We realigned around indels using GATK IndelRealigner then called variants using HaplotypeCaller (default parameters) (McKenna et al. 2010; DePristo et al. 2011). We then used GATK FastaReferenceMaker (default parameters) to generate an alternate, reference genome for each of these species (McKenna et al. 2010; DePristo et al. 2011). We extracted the coding sequence for each gene found in the genomes of D. innubila, D. phalerata, and D. falleni and aligned orthologs using PRANK (-codon +F –f=paml) (Löytynoja 2014). For each PRANK generated gene alignment and gene tree, we used codeML (Yang 2007) for the branches model (M0 model), to identify genes with signatures of rapid evolution on the D. innubila, D. falleni, D. phalerata branches, as well as across the entire clade. Focusing specifically in the D. innubila branch, for genes involved in the immune system pathways, we attempted to rescale for synonymous divergence. For each focal gene, we found genes with dS within 0.01 of the focal gene on the same scaffold. We then found the difference in dN/dS between the focal gene and the median of the control gene group.
For an independent contrast, we downloaded the latest coding sequences for D. melanogaster, D. simulans, and D. yakuba from Flybase.org (Downloaded January 2018, Gramates et al. 2017) and aligned orthologous genes using PRANK (-codon +F –f=paml) (Löytynoja 2014). Following the generation of a gene alignment and gene tree, we used codeML (Yang 2007) to identify genes with adaptive molecular signatures on each branch of the phylogeny (using the branch-based model, M0). Again, we found the difference in dN/dS from background genes of similar dS (with 0.01) on the same scaffold for all immune genes, focusing on the D. melanogaster branch. We compared genes enriched in the top 2.5% for dN/dS (vs. the lower 97.5%) using GOrilla (Eden et al. 2009) in both D. innubila and D. melanogaster. We also performed this analysis using the top 5% and 10% and found no differences in enrichments than the more stringent 2.5% (not shown).
We downloaded genes involved in a core set of gene ontologies from GOslim (Consortium et al. 2000; Carbon et al. 2017) and found the mean and SE of dN/dS for each category in both D. melanogaster and D. innubila. We chose to compare genes in the top 10% for dN/dS in both species in these categories, as no enrichments are found in the top 2.5% or 5% for the GOslim genes alone, instead we chose to broadly examine the fastest evolving genes in each species, even if not significantly enriched.
Finally, to control for possibly multiple nucleotide substitutions in a single site creating false signals of rapid evolution (Venkat et al. 2018), we calculated δ using HyPhy (Pond et al. 2005) based on the method presented in (Venkat et al. 2018). δ was calculated under both the null and alternative models, with the best model selected based on the result of a χ2 test. We then compared δ between each species and across immune gene categories.
RNA Differential Expression Analysis
We used GSNAP (-sam –N 1) (Wu and Nacu 2010) to map each set of D. innubila RNA sequencing short reads to the repeat masked D. innubila genome with the TE sequences concatenated at the end (NCBI SRA: SAMN11037167-78). We then counted the number of reads mapped to each gene per kb of exon using HTSeq (Anders et al. 2015) for all mapped RNA sequencing data and normalized by counts per million per data set. Mapped RNA sequencing information for D. melanogaster across all life stages was downloaded from ModEncode (modencode.org) (Chen et al. 2014). We compared D. melanogaster data to D. innubila data using EdgeR (Robinson et al. 2010) to identify differentially expressed genes, and also compared RNAseq reads per million reads per 1 kb of exon (fragments per kb of exon per million reads, FPKM) between the immune genes of D. innubila and D. melanogaster.
Statistical Analysis
All statistical analyses were performed using R (R Core Team 2018). We used the R packages EdgeR July 2018 version (Robinson et al. 2010), RCircos July 2018 version (Zhang et al. 2013), and ggplot2 (Wickham 2009) for statistical analysis and plot generation/visualization, July 2018 version.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We are thankful to Justin Blumenstiel, Jamie Walters, John Kelly, Carolyn Wessinger, Joanne Chapman and three anonymous reviewers for helpful discussion and advice concerning the manuscript. We thank Brittny Smith and the KU CMADP Genome Sequencing Core (NIH Grant P20 GM103638) for assistance in genome isolation, library preparation and sequencing. Genome annotation was performed in the K-INBRE Bioinformatics Core supported by P20 GM103418. This work was supported by a postdoctoral fellowship from the Max Kade foundation (Austria) and a K-INBRE postdoctoral grant to T.H. (NIH grant P20 GM103418). This work was also funded by NIH grants R00 GM114714 and R01 AI139154 to R.L.U.
References
- Aminetzach YT, Macpherson JM, Petrov D.. 2005. Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science 309(5735):764–767. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W.. 2015. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier V. 2013. Pastrel, a restriction factor for picornalike-viruses in Drosophila melanogaster. Virology. Université de Strasbourg. Thesis: 1–249.
- Biryukova I, Ye T.. 2015. Endogenous siRNAs and piRNAs derived from transposable elements and genes in the malaria vector mosquito Anopheles gambiae. BMC Genomics 16:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronkhorst AW, Cleef KV, Vodovar N, İnce İA, Blanc H, Vlak JM, Saleh M-C, Van RR.. 2012. The DNA virus Invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc Natl Acad Sci U S A. 109(51):E3604–E3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S, Dietze H, Lewis SE, Mungall CJ, Munoz-Torres MC, Basu S, Chisholm RL, Dodson RJ, Fey P, Thomas PD, et al. 2017. Expansion of the gene ontology knowledgebase and resources: the gene ontology consortium. Nucleic Acids Res. 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M, VanKuren NW, Zhao R, Zhang X, Kalsow S, Emerson JJ.. 2018. Hidden genetic variation shapes the structure of functional elements in Drosophila. Nat Genet. 50:20–25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT.. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32(8):1220–1222. [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow T. a, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450(7167):203–218. [DOI] [PubMed] [Google Scholar]
- Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, Cella M, Lande R, Uzé G.. 2004. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol. 34(3):796–805. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball C, Blake J, Botstein D, Butler H, Cherry M, Davis A, Dolinski K, Dwight S, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT.. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. [DOI] [PMC free article] [PubMed]
- Daugherty MD, Malik HS.. 2012. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet. 46:677–700. [DOI] [PubMed] [Google Scholar]
- Dawkins R, Krebs JR.. 1979. Arms races between and within species. Proc R Soc Lond B Biol Sci. 205(1161):489–511. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW. 2010. RNA-based antiviral immunity. Nat Rev Immunol. 10(9):632–644. [DOI] [PubMed] [Google Scholar]
- Dos Santos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM, Brown NH, Kaufman T, et al. 2015. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 43(D1):D690–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler J-L.. 2005. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat Immunol. 6(9):946–953. [DOI] [PubMed] [Google Scholar]
- Dyer KA, Jaenike J.. 2004. Evolutionarily stable infection by a male-killing endosymbiont in Drosophila innubila: molecular evidence from the host and parasite genomes. Genetics 168(3):1443–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer KA, Jaenike J.. 2005. Evolutionary dynamics of a spatially structured host-parasite association: Drosophila innubila and male-killing Wolbachia. Evolution 59(7):1518–1528. [PubMed] [Google Scholar]
- Dyer KA, Minhas MS, Jaenike J.. 2005. Expression and modulation of embryonic male-killing in Drosophila innubila: opportunities for multilevel selection. Evolution 59(4):838–848. [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z.. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekengren S, Hultmark D.. 2001. A family of Turandot-related genes in the humoral stress response of Drosophila. Biochem Biophys Res Commun. 284(4):998–1003. [DOI] [PubMed] [Google Scholar]
- Enard D, Cai L, Gwennap C, Petrov DA.. 2016. Viruses are a dominant driver of protein adaptation in mammals. Elife 5:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubert C, Modolo L, Vieira C, Moro CV, Mavingui P, Boulesteix M.. 2015. De novo assembly and annotation of the Asian tiger mosquito (Aedes albopictus) repeatome with dnaPipeTE from raw genomic reads and comparative analysis with the yellow fever mosquito (Aedes aegypti). Genome Biol Evol. 7(4):1192–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates LS, Marygold SJ, Santos GD, Urbano J-M, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, et al. 2017. FlyBase at 25: looking to the future. Nucleic Acids Res. 45(D1):D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton PT, Leong JS, Koop BF, Perlman SJ.. 2014. Transcriptional responses in a Drosophila defensive symbiosis. Mol Ecol. 23(6):1558–1570. [DOI] [PubMed] [Google Scholar]
- Hanson MA, Hamilton PT, Perlman SJ.. 2016. Immune genes and divergent antimicrobial peptides in flies of the subgenus Drosophila. BMC Evol Biol. 16:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetru C, Hoffmann JA.. 2009. NF-B in the immune response of Drosophila. Cold Spring Harb Perspect Biol. 1(6):a000232.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T, Unckless RL.. 2017. The dynamic evolution of Drosophila innubila Nudivirus. Infect Genet Evol. 57:151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JA. 2003. The immune response of Drosophila. Nature 426(6962):33–38. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultmark D. 2003. Drosophila immunity: paths and patterns. Curr Opin Immunol. 15(1):12–19. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ.. 2008. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 9(1):22–32. [DOI] [PubMed] [Google Scholar]
- Jaenike J, Dyer KA.. 2008. No resistance to male-killing Wolbachia after thousands of years of infection. J Evol Biol. 21(6):1570–1577. [DOI] [PubMed] [Google Scholar]
- Jaenike J, Dyer KA, Reed LK.. 2003. Within-population structure of competition and the dynamics of male-killing Wolbachia. Evol Ecol Res. 5:1023–1036. [Google Scholar]
- Jaenike J, Perlman SJ.. 2002. Ecology and evolution of host‐parasite associations: mycophagous Drosophila and their parasitic nematodes. Am Nat. 160:S23–S39. [DOI] [PubMed] [Google Scholar]
- Jain M, Olsen HE, Paten B, Akeson M.. 2016. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 17:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov T, Messer PW, Petrov D. a.. 2010. Evidence that adaptation in Drosophila is not limited by mutation at single sites. PLoS Genet. 6(6):e1000924.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshishian H, Chiba A, Chang TN, Halfon MS, Harkins EW, Jarecki J, Wang L, Anderson M, Cash S, Halpern ME.. 1993. Cellular mechanisms governing synaptic development in Drosophila melanogaster. J Neurobiol. 24(6):757–787. [DOI] [PubMed] [Google Scholar]
- Kimbrell DA, Beutler B.. 2001. The evolution and genetics of innate immunity. Nat Rev Genet. 2(4):256–267. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM.. 2016. Canu : scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. [DOI] [PMC free article] [PubMed]
- Lewis SH, Quarles KA, Yang Y, Tanguy M, Frézal L, Smith SA, Sharma PP, Cordaux R, Gilbert C, Giraud I, et al. 2018. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat Ecol Evol. 2(1):174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lively C. 1996. Host-parasite coevolution and sex. Bioscience 46(2):107–114. [Google Scholar]
- Löytynoja A. 2014. . Phylogeny-aware alignment with PRANK In: Russell DJ, editor. Multiple sequence alignment methods. Totowa (NJ: ): Humana Press; p. 155–170. [Google Scholar]
- Magwire MM, Bayer F, Webster CL, Cao C, Jiggins FM.. 2011. Successive increases in the resistance of Drosophila to viral infection through a transposon insertion followed by a duplication. PLoS Genet. 7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwire MM, Fabian DK, Schweyen H, Cao C, Longdon B, Bayer F, Jiggins FM.. 2012. Genome-wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster. PLoS Genet. 8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow TA, O’Grady P.. 2006. Drosophila: a guide to species identification.
- Martinez J, Longdon B, Bauer S, Chan YS, Miller WJ, Bourtzis K, Teixeira L, Jiggins FM.. 2014. Symbionts commonly provide broad spectrum resistance to viruses in insects: a comparative analysis of Wolbachia strains. PLoS Pathog. 10(9):e1004369.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson VG, Douglas AE, Jaenike J.. 2017. Community structure of the gut microbiota in sympatric species of wild Drosophila. Ecol Lett. 20(5):629–639. [DOI] [PubMed] [Google Scholar]
- McCoy RC, Taylor RW, Blauwkamp TA, Kelley JL, Kertesz M, Pushkarev D, Petrov DA, Fiston-Lavier AS.. 2014. Illumina TruSeq synthetic long-reads empower de novo assembly and resolve complex, highly-repetitive transposable elements. PLoS One 9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky AM, Garimella KV, Altshuler D, Gabriel SB, Daly MJ, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Proc Int Conf Intellect Capital Knowl Manag Organ Learn. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkling SH, van Rij RP.. 2013. Beyond RNAi: antiviral defense strategies in Drosophila and mosquito. J Insect Physiol. 59(2):159–170. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM.. 2009. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci. 364(1513):99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Jiggins FM, Halligan DL, Little TJ.. 2006. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol. 16(6):580–585. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Welch JJ, Kim KW, Jiggins FM.. 2009. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 5(10):e1000698.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer WH, Hadfield JD, Obbard DJ.. 2018. RNA interference pathways display high rates of adaptive protein evolution in multiple invertebrates. Genetics 300567:2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer WH, Joosten J, Overheul GJ, Jansen PW, Vermeulen M, Obbard J, Rij RV.. 2018. Induction and suppression of NF-κB signalling by a DNA virus of Drosophila J. Virol. 93(3):e01443–18. [DOI] [PMC free article] [PubMed]
- Palmer WH, Medd N, Beard PM, Obbard DJ.. 2018. Isolation of a natural DNA virus of Drosophila melanogaster, and characterisation of host resistance and immune responses. PLoS Pathog. 14:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JT, Stone WS.. 1949. Studies in the genetics of Drosophila. Univ Texas Publ. 4920:7–17. [Google Scholar]
- Payne CC. 1974. The isolation and characterization of a virus from Oryctes rhinoceros. J Gen Virol. 25(1):105–116. [DOI] [PubMed] [Google Scholar]
- Perlman SJ, Spicer GS, Dewayne Shoemaker D, Jaenike J.. 2002. Associations between mycophagous Drosophila and their Howardula nematode parasites: a worldwide phylogenetic shuffle. Mol Ecol. 12(1):237–249. [DOI] [PubMed] [Google Scholar]
- Pond S, Frost S, Muse S.. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21(5):676–679. 1–57. [DOI] [PubMed] [Google Scholar]
- R Core Team 2018. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S.. 2009. Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell 138(2):340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton TB, Kulathinal RJ, Bergman CM, Quinlan AR, Dopman EB, Carneiro M, Marth GT, Hartl DL, Clark AG.. 2009. Population genomic inferences from sparse high-throughput sequencing of two populations of Drosophila melanogaster. Genome Biol Evol. 1:449–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG.. 2007. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 39(12):1461–1468. [DOI] [PubMed] [Google Scholar]
- Sansone CL, Cohen J, Yasunaga A, Xu J, Osborn G, Subramanian H, Gold B, Buchon N, Cherry S.. 2015. Microbiota-dependent priming of antiviral intestinal immunity in Drosophila. Cell Host Microbe. 18(5):571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E.. 2012. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28(8):1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessegolo C, Burlet N, Haudry A, Biémont C, Vieira C, Tenaillon M, Hollister J, Gaut B, McClintock B, Mackay T, et al. 2016. Strong phylogenetic inertia on genome size and transposable element content among 26 species of flies. Biol Lett. 12:521–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker DD, Katju V, Jaenike J.. 1999. Wolbachia and the evolution of reproductive isolation between Drosophila recens and Drosophila subquinaria. Evolution 53:1157–1164. [DOI] [PubMed] [Google Scholar]
- Shultz A, Sackton TB.. 2019. Immune genes are hotspots of shared positive selection across birds and mammals. Elife 8:e41815.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Simms D, Cizdziel P, Chomczynski P.. 1993. TRIzol: a new reagent for optimal single-step isolation of RNA. Focus 15(4):99–102.
- Smit AFA, Hubley R.. 2008. RepeatModeler Open-1.0. Available from: www.repeatmasker.org
- Smit AFA, Hubley R.. 2015. RepeatMasker Open-4.0. Available from: http://www.repeatmasker.org/RepeatModeler/.
- Takeda K, Akira S.. 2005. Toll-like receptors in innate immunity. Int Immunol. 17(1):1–14. [DOI] [PubMed] [Google Scholar]
- Teixeira L, Ferreira A, Ashburner M.. 2008. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6(12):e2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unckless RL. 2011. A DNA virus of Drosophila. PLoS One 6(10):e26564.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unckless RL, Jaenike J.. 2011. Maintenance of a male-killing Wolbachia in Drosophila Innubila By male-killing dependent and male-killing independent mechanisms. Evolution 66:678–689. [DOI] [PubMed] [Google Scholar]
- Valanne S, Wang J-H, Ramet M.. 2011. The Drosophila Toll signaling pathway. J Immunol. 186(2):649–656. [DOI] [PubMed] [Google Scholar]
- Venkat A, Hahn MW, Thornton J.. 2018. Multinucleotide mutations cause false inferences of lineage-specific positive selection. Biorxiv 2018:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-HX-H, Aliyari R, Li W-X, Li H-W, Kim K, Carthew R, Atkinson P, Ding S-W.. 2006. RNA interference directs innate immunity against viruses in adult Drosophila. Science 312(5772):452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jehle JA.. 2009. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: new insights on an old topic. J Invertebr Pathol. 101(3):187–193. [DOI] [PubMed] [Google Scholar]
- Webster CL, Waldron FM, Robertson S, Crowson D, Ferrari G, Quintana JF, Brouqui J-M, Bayne EH, Longdon B, Buck AH, et al. 2015. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol. 13(7):e1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West C, Silverman N.. 2018. p38b and JAK-STAT signaling protect against invertebrate iridescent virus 6 infection in Drosophila. Plant Pathol. 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2009. . ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; Available from: http://ggplot2.org [Google Scholar]
- Wu TD, Nacu S.. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26(7):873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Wu G, Tang J, Luo R, Patterson J, Liu S, Huang W, He G, Gu S, Li S, et al. 2014. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30(12):1660–1666. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z.. 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25(21):2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon RA, Nandakumar M, Vakharia VN, Wu LP.. 2005. The Toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci U S A. 102(20):7257–7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Meltzer P, Davis S.. 2013. RCircos : an R package for Circos 2D track plots. BMC Bioinformatics 14(244):1–5. [DOI] [PMC free article] [PubMed]
- Zhou Q, Bachtrog D.. 2015. Ancestral chromatin configuration constrains chromatin evolution on differentiating sex chromosomes in Drosophila. PLoS Genet. 11:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhu H, Huang Q, Zhao L, Zhang G, Roy SW, Vicoso B, Xuan Z, Ruan J, Zhang Y, et al. 2012. Deciphering neo-sex and B chromosome evolution by the draft genome of Drosophila albomicans. BMC Genomics 13(1):109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.