Abstract
The family Ampullariidae includes both aquatic and amphibious apple snails. They are an emerging model for evolutionary studies due to the high diversity, ancient history, and wide geographical distribution. Insight into drivers of ampullariid evolution is hampered, however, by the lack of genomic resources. Here, we report the genomes of four ampullariids spanning the Old World (Lanistes nyassanus) and New World (Pomacea canaliculata, P. maculata, and Marisa cornuarietis) clades. The ampullariid genomes have conserved ancient bilaterial karyotype features and a novel Hox gene cluster rearrangement, making them valuable in comparative genomic studies. They have expanded gene families related to environmental sensing and cellulose digestion, which may have facilitated some ampullarids to become notorious invasive pests. In the amphibious Pomacea, novel acquisition of an egg neurotoxin and a protein for making the calcareous eggshell may have been key adaptations enabling their transition from underwater to terrestrial egg deposition.
Keywords: mollusc, gastropod, genomics, Hox genes, interchromosome rearrangement, gene duplication
Introduction
Apple snails (i.e., Ampullariidae) represent an excellent model to study speciation, historical biogeography, and adaptation (Hayes, Cowie, Jørgensen, et al. 2009; Van Bocxlaer 2017). As an early diverging family of Caenogastropoda, a superorder comprising over 50,000 species or 60% of gastropod species diversity (Ponder et al. 2008), Ampullariidae holds a unique potential for understanding ancestral genomic features within the ecologically important and biologically diverse caenogastropods. Ampullariidae is widely distributed in tropical and subtropical freshwaters and contains >180 species in nine genera (Cowie 2015; Hayes et al. 2015). The family originated >150 Ma on Gondwana, and later spread and diversified across freshwater habitats in the Old World (Africa and Asia) and New World (Central and South America; only one species in North America) (Cowie 2002; Rawlings et al. 2007; Jørgensen et al. 2008; Hayes, Cowie, and Thiengo 2009; Hayes et al. 2015). Recently, several species indigenous to South America have been introduced into Asia, Europe, North America, and various Pacific islands including Hawaii (Rawlings et al. 2007; Hayes et al. 2015). Some of these invaders, notably Pomacea canaliculata and P. maculata, are among the worst invasive species worldwide because of their severe damage to natural and agricultural (e.g., rice and taro) wetlands (Cowie 2002; Horgan et al. 2014). As intermediate hosts of various parasites, the spread of invasive apple snails also poses a serious threat to human health through causing angiostrongyliasis, cercarial dermatitis, and echinostomiasis (Lv et al. 2009; Hayes et al. 2015).
The successful colonization of new habitats by several Pomacea species (e.g., P. canaliculata and P. maculata) and Marisa cornuarietis has been attributed in part to their broad environmental tolerance (e.g., to desiccation [Sun et al. 2013], low temperature [Matsukura et al. 2016], and hypoxia [Mu et al. 2018]), and to their alimentary versatility (Lach et al. 2000), whereas Lanistes nyassanus is stenotopic and endemic to the Malawi Basin of Africa (Van Bocxlaer 2017). Additionally, whereas Marisa and Lanistes deposit eggs underwater, Pomacea lays eggs on land (Hayes, Cowie, Jørgensen, et al. 2009). The derived condition of terrestrial egg deposition in Pomacea is considered a key adaptation to avoid aquatic predation and/or parasitism, making it the most speciose and widely distributed genus in Ampullariidae (Hayes, Cowie, Jørgensen, et al. 2009; Cowie 2015). However, the transition from aquatic to terrestrial egg-laying required morphological changes, such as elongation of the respiratory siphons and increase in lung size in the adults (Cowie 2002; Hayes, Cowie, Jørgensen, et al. 2009), as well as biochemical changes in the eggs to cope with exposure to heat, desiccation, UV radiation, and terrestrial predators (Yusa 2001; Heras et al. 2008).
Due to the lack of genomic resources in ampullariids and more generally in molluscs, the genetic basis of the behavioral, morphological, and physiological adaptations of Ampullariidae remains largely unknown. The paucity of molluscan genomes has been pointed out by the Global Invertebrate Genomics Alliance, and a species of apple snail (i.e., P. maculata) was specifically proposed as a priority species for whole genome sequencing (Voolstra et al. 2017). The present study thus aimed to sequence and assemble the genomes of the African L. nyassanus and South American P. canaliculata, P. maculata, and M. cornuarietis (fig. 1). By including these Old World and New World species with different reproductive characteristics, we conducted comparative analyses to understand the genomic basis underlying the diversity and invasiveness in Marisa and Pomacea, and the transition to terrestrial egg deposition in the amphibious Pomacea. In a broader taxonomic context, we explored the potential of the ampullariid genomes for use in comparative genomics with other molluscs as well as more distantly related bilaterians.
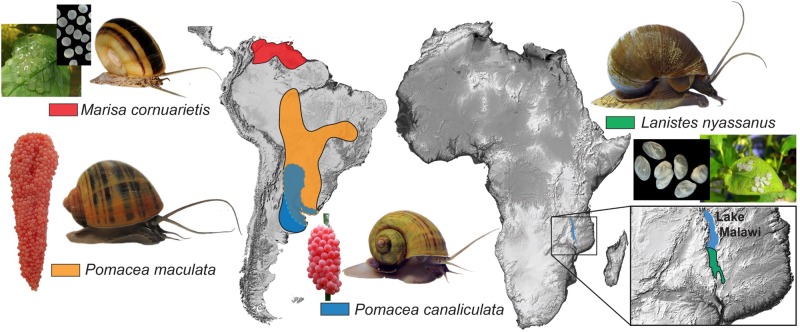
Fig. 1.
Native ranges of the four ampullariid species included in this study, and pictures of their adults and egg masses. Dashed lines demarcate the distribution overlap of Pomacea spp. The pink-reddish calcareous egg masses of the two Pomacea species are deposited terrestrially, whereas the white gelatinous egg masses of Lanistes nyassanus and Marisa cornuarietis are deposited underwater.
Results and Discussion
Genome Assembly and Quality Assessment
The genomes of P. canaliculata, P. maculata, and L. nyassanus were assembled after sequencing short- and long-insert DNA libraries using Illumina sequencing platforms and calculating their heterozygosity (supplementary tables S1 and S2, Supplementary Material online). Among these three species, the largest sequencing effort was placed on P. canaliculata, for which a high-quality reference genome is particularly desirable given the economic damage it causes as a crop pest, and the health risks it poses as an intermediate host of infectious diseases (Hayes et al. 2015). These three genomes range from 432.3 to 510.0 million base pairs (Mb) in total length (table 1), with the two Pomacea genomes being smaller. For P. canaliculata, the assembly produced 3,131 scaffolds, among which 413 scaffolds that accounted for 97.3% of the total genome length were further anchored to 14 chromosomes using Hi-C data produced by another group (Liu et al. 2018) (supplementary fig. S3, Supplementary Material online). The genome of M. cornuarietis, assembled using both Illumina short reads and Nanopore long reads, has a total length of 535.5 Mb (table 1). The contig N50 (the shortest sequence length at 50% of the genome size) of the four genomes ranges between 33.9 and 98.2 kilobase pairs (kb), and scaffold N50 between 316.6 kb and 32.6 Mb. The final scaffold/contig N50 values of the P. canaliculata (32.6 Mb) and M. cornuarietis genome (4.4 Mb) exceed those of most other sequenced molluscan genomes considerably, showing the high continuity of our assemblies.
Table 1.
Key Assembly Statistics of Molluscan Genomes.
| Class | Species | GC Content | Assembled Length | No. of Scaffolds | Longest Scaffold | No. of Scaffolds >1 Mb | Unknown Sequences | Scaffold N50 | Contig N50 | No. of Missing BUSCOs | Source |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cephalopoda | Octopus bimaculoides | 0.31 | 2,338.2 Mb | 151,674 | 4.1 Mb | 351 | 0.151 | 475.2 kb | 6.6 kb | 62 (6.3%) | Albertin et al. (2015) |
| Bivalvia | Crassostrea gigas | 0.30 | 557.7 Mb | 7,659 | 1.96 Mb | 60 | 0.118 | 401.7 kb | 32.6 kb | 38 (3.9%) | Zhang et al. (2012) |
| Bivalvia | Patinopecten yessoensis | 0.34 | 987.6 Mb | 82,659 | 7.50 Mb | 248 | 0.081 | 803.6 kb | 66.76 kb | 53 (5.5%) | Wang et al. (2017) |
| Bivalvia | Bathymodiolus platifrons | 0.30 | 1,658.2 Mb | 65,662 | 2.79 Mb | 164 | 0.118 | 343.3 kb | 13.2 kb | 38 (3.9%) | Sun et al. (2017) |
| Bivalvia | Modiolus philippinarum | 0.32 | 2,629.6 Mb | 74,573 | 715.3 kb | 0 | 0.048 | 100.1 kb | 19.7 kb | 55 (5.6%) | Sun et al. 2017 |
| Bivalvia | Pinctada fucata | 0.33 | 815.3 Mb | 29,306 | 1.26 Mb | 2 | 0.067 | 167.0 kb | 21.9 kb | 45 (4.6%) | Takeuchi et al. (2016) |
| Gastropoda | Pomacea canaliculata | 0.41 | 440.2 Mb | 24 | 45.4 Mb | 14 | 0.0002 | 31.5 Mb | 1.1 Mb | 34 (3.5%) | Liu et al. (2018) |
| Gastropoda | P. canaliculata | 0.41 | 447.7 Mb | 3,131 | 45.35 Mb | 14 | 0.035 | 32.6 Mb | 81.4 kb | 28 (2.9%) | This study |
| Gastropoda | P. maculata | 0.40 | 432.3 Mb | 3,914 | 2.52 Mb | 38 | 0.0085 | 375.9 kb | 91.9 kb | 31 (3.2%) | This study |
| Gastropoda | Marisa cornuarietis | 0.41 | 535.5 Mb | 665 | 24.67 Mb | 117 | 0 | 4.4 Mb | 4.4 Mb | 29 (3.0%) | This study |
| Gastropoda | Lanistes nyassanus | 0.42 | 510.0 Mb | 34,401 | 1.79 Mb | 28 | 0.020 | 316.6 kb | 33.9 kb | 37 (3.8%) | This study |
| Gastropoda | Radix auricularia | 0.34 | 909.8 Mb | 4,823 | 2.97 Mb | 153 | 0.064 | 578.7 kb | 26.7 kb | 53 (5.5%) | Schell et al. (2017) |
| Gastropoda | Aplysia californica | 0.32 | 927.3 Mb | 4,332 | 610.3 kb | 258 | 0.204 | 917.5 kb | 9.6 kb | 56(5.8%) | GenBank GCA_000002075 |
| Gastropoda | Biomphalaria glabrata | 0.35 | 916.4 Mb | 331,401 | 2.18 Mb | 23 | 0.019 | 48.1 kb | 16.6 kb | 66 (6.8%) | Adema et al. (2017) |
| Gastropoda | Lottia gigantea | 0.28 | 359.5 Mb | 4,475 | 9.39 Mb | 98 | 0.169 | 1.87 Mb | 96.0 kb | 32 (3.3%) | Simakov et al. (2013) |
| Gastropoda | Haliotis discus hannai | 0.38 | 1,865.4 Mb | 80,032 | 2.21 Mb | 67 | 0.063 | 200.1 kb | 41.0 kb | 36 (3.7%) | Nam et al. (2017) |
| Gastropoda | Lymnaea stagnalis | 0.37 | 833.2 Mb | 328,378 | 95.6 kb | 0 | 0 | 5.8 kb | 5.8 kb | 61 (6.3%) | Davison et al. (2016) |
Note.—Detailed BUSCO analysis results can be found in supplementary table S3, Supplementary Material online.
Genome completeness was accessed using BUSCO analysis (Simão et al. 2015), which revealed 93.5–95.1% of the 978 universal metazoan single-copy orthologous genes in the four ampullariid draft genomes, exceeding the completeness of most other published molluscan genomes (table 1 and supplementary table S3, Supplementary Material online). In addition, paired-end reads were mapped to the ampullariid genomes to evaluate their accuracy using REAPR analysis (Hunt et al. 2013), which showed that 71.0–85.4% of our ampullariid genomes were error-free, which exceeded the values for two recently published gastropod genomes (59.0% in Biomphalaria glabrata and 66.83% in Radix auricularia) (supplementary table S4, Supplementary Material online). Compared with a recently published P. canaliculata genome assembled from hybrid Illumina/PacBio reads (Liu et al. 2018), our P. canaliculata assembly is very similar in GC content, assembled genome size (table 1), and macrosynteny pattern (supplementary figs. S5 and S6, Supplementary Material online), but has a higher percentage of putative error-free bases (85.44% vs. 71.01%), fewer putative fragment coverage distribution errors (2,093 vs. 18,801), more complete single copy BUSCOs (930 vs. 925), and fewer missing BUSCOs (28 vs. 34). Nevertheless, our P. canaliculata assembly has more scaffolds (3,131 vs. 24) and more putative collapsed repeats (44,915 vs. 26,291) (supplementary tables S3 and S4, Supplementary Material online). Annotation combining the evidence from transcripts, homologous proteins, and in silico prediction revealed 18,263, 23,464, 23,827, and 20,938 gene models from P. canaliculata, P. maculata, M. cornuarietis, and L. nyassanus, respectively (supplementary table S5, Supplementary Material online). The fewer gene models in P. canaliculata might be due to more transcriptomic and proteomic data used for refinement of the gene model prediction in this species.
Genome Size and Structural Characteristics
With sizes ranging from 432.3 to 535.4 Mb, the assembled ampullariid genomes are among the most compact molluscan genomes (supplementary fig. S4, Supplementary Material online). The evolution of genome structure in Mollusca was assessed by comparing with ten well-annotated molluscan genomes covering the size range of 0.36–2.63 Gb (supplementary table S5, Supplementary Material online). The total exon length of our ampullariid genomes are comparable to these other genomes (47.9–70.6 Mb vs. 21.0–74.7 Mb; Wilcoxon rank sum test: W = 23; P = 0.733), but their intergenic regions are substantially shorter (153.1–284.8 Mb vs. 234.7–2,193.1 Mb; W = 3; P = 0.014). The intron length of ampullariids is similar to that of other gastropods (151.8–246.6 Mb vs. 58.7–245.6; W = 8; P = 1.000), but it is significantly shorter than that of other molluscs (173.7-597.6 Mb; W = 2; P = 0.003). Our analyses thus indicate that changes in intronic and intergenic regions largely account for genome-size differences in molluscs.
The repeat contents in the apple snail genomes were identified using three de novo predictors and by comparison with a database of known repeats (Supplementary Material online). The ampullariid genomes contain 20.5–30.8% repeat content, which is significantly less than most other molluscan genomes (24.2–61.1%; W = 4; P = 0.024), although the owl limpet (Lottia gigantea, 24.2%) and the Yesso scallop (Patinopecten yessoensis, 27.8%) fall within the range observed for ampullariids (supplementary table S6, Supplementary Material online). Among the four apple snail genomes, the larger Lanistes and Marisa genomes have ∼55 Mb more repeats than the smaller Pomacea genomes. Because transposable elements (TEs), including DNA transposons, long terminal repeats (LTRs), long interspersed nuclear elements (LINEs), and short interspersed nuclear elements (SINEs) account for a substantial proportion of these repeats, we further compared the composition of TEs among the four ampullariid species, which showed that LINEs (4.08–9.64%) and DNA transposons (3.10–4.44%) are the major classes of classified TEs in these genomes. However, a large proportion of the TEs in these ampullariid genomes (8.89–15.11%) remains unclassified.
Phylogenetics and Molecular Dating
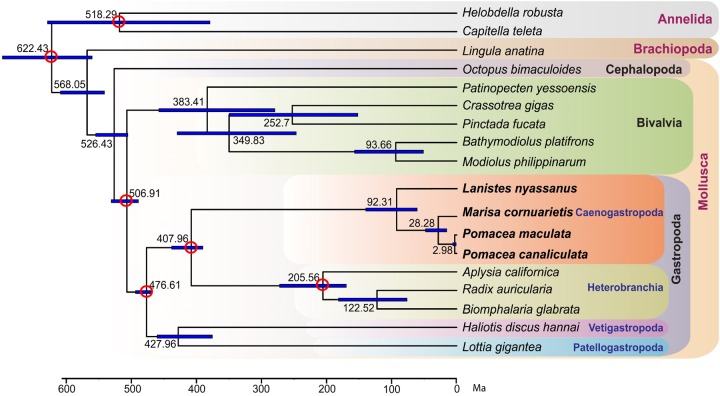
The assembled genomes allow a genome-wide assessment of deep relationships among major molluscan groups, as well as determination of their divergence times (fig. 2). A maximum-likelihood phylogenetic tree, constructed using 1,357 single-copy orthologous genes with a total of 455,177 amino acids, provides strong support (bootstrap value = 100) for all nodes (fig. 2). The arrangement of major molluscan clades within Gastropoda—Archaeogastropoda (Patellogastropoda and Vetigastropoda) as sister group to Apogastropoda (Heterobranchia and Caenogastropoda)—is consistent with one of the phylogenetic hypotheses proposed in previous studies (Smith et al. 2011; Zapata et al. 2014), and the Approximately Unbiased test significantly rejected (P < 0.05; supplementary table S7, Supplementary Material online) alternative hypotheses about Archaeogastropoda–Apogastropoda relationships (Kocot et al. 2011; Zapata et al. 2014). Fossil calibration of six nodes in this phylogenetic tree provides new divergence estimates on nodes within the Ampullariidae: the split between the New World lineage (Pomacea and Marisa) and the Old World lineage (Lanistes) could have occurred 92.3 Ma (95% confidence interval of 61.3–139.0 Ma), which is consistent with the time of Gondwana breakup ∼120 Ma (Jokat et al. 2003). Marisa and Pomacea appear to have split ∼28.3 Ma (95% confidence interval of 16.0–47.3 Ma), and this process might have been initiated by watershed changes related to major orogenic activities during Andean uplift in the Eocene (40–50 Ma) (Hoorn et al. 2010). The two species of Pomacea appear to have diverged ∼3.0 Ma (95% confidence interval of 2.0–5.0 Ma), which is consistent with the fact that these two species can hybridize and produce viable F1 progeny (Matsukura et al. 2013), but this estimate is considerably more recent than a previous estimate (∼15 Ma) based on the mitochondrial COI gene with an assumed rate of divergence of 1.63% Ma−1 (Hayes, Cowie, and Thiengo 2009).
Fig. 2.
Maximum-likelihood phylogenetic tree, constructed using RaxML version 8.2.4 (Stamatakis 2014) with the GTR + Γ model assigned to each partition. The tree shows the relationships and divergence times of 15 molluscan species, with 2 annelids and 1 brachiopod served as outgroups. Blue lines indicate the 95% confidence interval on estimated node depths. The names of four ampullariid species used in this study are written in bold font. Red circles indicate that the nodes are constrained with fossil calibration data (see Supplementary Material for details). All nodes have a bootstrap support of 100.
New Discovery of Hox Cluster Rearrangement and Conserved Karyotype Features
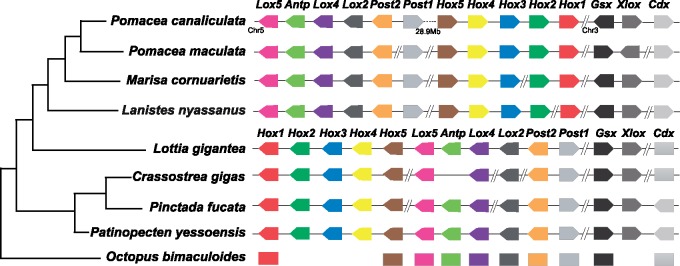
Hox and ParaHox genes are important in the early development of metazoans as they mediate the patterning of the anterior–posterior body axis (Wray 2001; Hui et al. 2009; Holland 2013). Since their numbers are assumed to be conserved across Lophotrochozoa (Simakov et al. 2013; Wang et al. 2017), these genes were investigated in the assembled ampullariid genomes. Like most other well-assembled molluscan genomes, all four ampullariid genomes contained 11 Hox and 3 ParaHox genes (fig. 3), further supporting the completeness of the assemblies. Because the order of the 11 Hox genes is identical in the limpet Lo. gigantea and the scallop Pa. yessoensis, it was assumed that these animals have preserved the ancestral molluscan Hox gene order (Wang et al. 2017). However, our chromosome-level assembly of P. canaliculata shows that, although the anterior subcluster Hox1-Hox5 and the posterior subcluster Lox5-Post1 are located on the same chromosome, Hox1-Hox5 has undergone reverse transposition to become Hox5-Hox1 and is located downstream of Lox5-Post1. Moreover, there is a 28.9-Mb gap between these Lox5-Post1 and Hox5-Hox1, which is much longer than the whole Hox cluster in Lo. gigantea (0.47 Mb) and Pa. yessoensis (1.75 Mb). The same search in the only independently assembled P. canaliculata genome confirmed the reverse transposition of the anterior subcluster of Hox genes on chromosome LG2 as well as a large gap between the two subclusters in this species (Liu et al. 2018). Moreover, there are long sequences both upstream (3.60 Mb) and downstream (4.92 Mb) of Lox5-Post1 in the M. cornuarietis genome (contig Mco_637), as well as upstream (0.20 Mb) and downstream (0.13 Mb) of Lox5-Post1 in the L. nyassanus genome (scaffold Lny_18170), all indicating a disrupted Hox cluster; and the transcription directions of Hox genes indicate reverse transposition of the ancestral Hox1-Hox5 subcluster in Ampullariidae. This is by far the first report of reverse transposition of Hox genes due to intrachromosomal rearrangement in Mollusca. Previous studies of Hox gene expression show that, in polyplacophorans Hox genes are expressed in a staggered fashion along the anterior–posterior body axis, which is in accordance with the proposed ancestral mode of colinear expression in bilaterians (Wollesen et al. 2018). However, in several conchiferans including the gastropod Gibbula varia and the cephalopod Octopus bimaculoides, Hox genes have lost the ancestral function of pattering the anterior–posterior axis. Instead, Hox genes are co-opted to pattern various Mollusca-specific structures such as the larval foot and prototroch (Wanninger and Wollesen 2019). Our finding of the disrupted Hox gene cluster in Ampullariidae may explain why Hox genes do not exhibit temporal colinear expression, and their derived roles of patterning structures off the anterior–posterior axis.
Fig. 3.
Schematic illustration of Hox and ParaHox gene clusters in four ampullariid species and five other selected molluscans. Data for non-ampullariid genomes were adopted from Wang et al. (2017). Gene names are labeled on top of the colored graphs, and species names are labeled at the left of each row. Genes located on the same scaffold or chromosome are connected with a line but the line length is not proportional to their sequence length. The symbol “//” indicates a break between different scaffolds. Transcription direction, when available, is indicated by a bullet head.
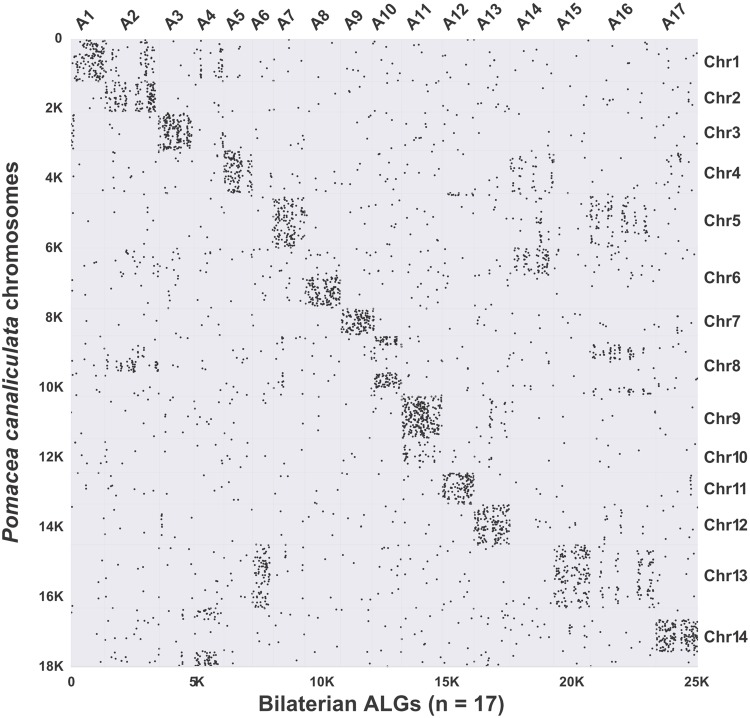
Comparative macrosynteny analysis with the 17 presumed ancestral bilaterian linkage groups (ALGs) (Simakov et al. 2013) indicates that P. canaliculata possesses a conserved ancient bilaterian karyotype (conservation index [CI] = 0.58; fig. 4). From all bilaterians examined before (four deuterostomes and seven protostomes of which three are lophotrochozoans and four ecdysozoans), only the scallop Pa. yessoensis (CI = 0.89) has a more conserved karyotype; all other species have more derived karyotypes (CI = 0.00–0.42) (Wang et al. 2017). Four interchromosome rearrangements (i.e., translocation of ALG6 and fusion of ALG4, ALG14, and ALG16) were found to correspond to a reduction from 17 chromosomes in ancient bilaterians to 14 chromosomes in P. canaliculata. Since all three previously studied bivalves (Pa. yessoensis, Crassostrea gigas, and Pinctada fucata) display two interchromosome rearrangements, that is, the partial translocation of ALG2 and the fusion of ALG5 and ALG16 (Wang et al. 2017), these authors suggested that the rearrangements were present prior to the origin of Bivalvia, which is the sister group of Gastropoda (fig. 2). Because these two chromosome rearrangements do not occur in P. canaliculata, our results indicate that the interchromosome arrangement patterns might have been already different between the common ancestor of gastropods and that of bivalves. Alternatively, the novel interchromosome arrangement pattern in P. canaliculata might be a derived trait during the divergence of ampullariids. These alternative hypotheses can be tested when more chromosome-level gastropod genomes become available. Together, our results show that the P. canaliculata genome can be used as a model for comparative studies of karyotype evolution and interchromosome arrangement.
Fig. 4.
Dotplot showing the conserved macrosynteny between the 14 chromosomes of Pomacea canaliculata and the 17 bilaterian ancient linkage groups (ALGs). Pomacea canaliculata has a chromosome conservation index (CI) of 0.58, which is smaller than that of the scallop Patinopecten yessoensis (0.89), but larger than those of all other bilaterians (0.00–0.42) whose chromosome-level assembly has been examined by Wang et al. (2017), including seven protostomes (three lophotrochozoans and four ecdysozoans) and four deuterostomes. The analysis and the resultant plot were made using MCScanX (Wang et al. 2012).
Involvement of Expanded Gene Families in Environmental Sensing and Plant Digestion
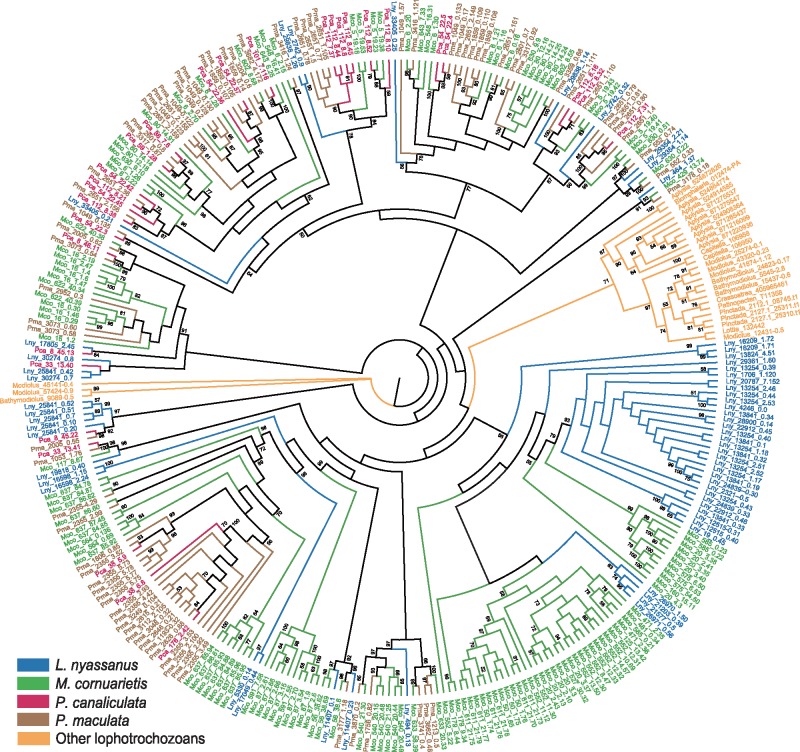
Because changes in gene family size are recognized as a primary driver of evolution (Demuth and Hahn 2009), we compared the number of genes in the gene families shared by the four ampullariids and other lophotrochozoans (supplementary table S8, Supplementary Material online). We found that 28 gene families are substantially expanded in Ampullariidae, and that some of them are functionally annotated as environmental sensing, a key trait for survival in ephemeral habitats and for successful colonization. One example is the massive expansion of the GRL101 family of G-protein coupled receptors (GPCRs, de Mendoza et al. 2014), which is involved in chemoreception in aquatic snails (Adema et al. 2017). A total of 29 to 132 GRL101 genes were found in the four ampullariids, but only zero to nine genes in other lophotrochozoans (supplementary table S8, Supplementary Material online). Phylogenetic analysis shows that all ampullariid GRL101 sequences are clustered in several big clades sister to two small clades of other molluscans and annelids (fig. 5), which indicates at least two major bursts of gene duplication events in the GRL101 family after the divergence of ampullariids from other molluscs. Examining the tissue-specific transcriptome data showed that many of these GRL101 genes are highly expressed in the cephalic tentacles and labial palps of P. canaliculata, indicating their active role in environmental sensing. Many of the ampullariid GRL101 genes are located very closely in identical scaffolds, strongly supporting the hypothesis that they were produced by tandem duplication events. For instance, scaffold 112 on chromosome 14 contains 11 GRL101 genes, among which 9 are tightly grouped in a cluster, and 7 of them (Pca_112_8.10, Pca_112_8.16, Pca_112_7.37, Pca_112_8.52, Pca_112_8.32, Pca_112_8.45, Pca_112_8.8) are highly expressed in the cephalic tentacles and labial palps (supplementary fig. S7, Supplementary Material online).
Fig. 5.
Unrooted maximum-likelihood tree showing the massive expansion of the G-protein coupled receptor (GPCR) GRL101 gene family in Ampullariidae. Among the 18 lophotrochozoan genomes examined, there are 299 sequences from the 4 ampullariids. Only 11 sequences are available for other gastropods (1 for Biomphalaria glabrata, 9 for Aplysia californica, 1 for Lottia gigantea), 15 for bivalves (1 from each of Lo. gigantea, Patinopecten yessoensis, and Crassostrea gigas, 3 from each of Pinctada fucata and Bathymodiolus platifrons, 6 from Modiolus philippinarum), and 2 for the annelid Capitella teleta. Each gene is labeled with a prefix of either the genus name for non-ampullariids or the first letter of the genus name and the first two letters of the species name for ampullariids (e.g., Pca for Pomacea canaliculata). Numbers on the nodes are bootstrap values (>50%).
Many ampullariids are omnivorous, with a diet that includes algae, plants, detritus, zooplankton, surface films, carrion, and other invertebrates (Hayes et al. 2015). This trophic versatility has probably contributed to the evolutionary success of many species in their native range, as well as some species of Pomacea and Marisa to become notorious invasive pests (Qiu and Kwong 2009; Saveanu and Martín 2014). Part of the trophic versatility of ampullariids may have been facilitated by their expanded cellulase gene family (7–12 genes), which is less common in other lophotrochozoans (0–6 genes) (supplementary table S8, Supplementary Material online), and other animal groups including arthropods (Watanabe and Tokuda 2001; Gutekunst et al. 2018). In P. canaliculata, the cellulases are located on chromosomes 8 and 14, but substantial distances exist between these genes (3.89–17.65 Mb). Likewise, in M. cornuarietis, the cellulases are located on three scaffolds, and in those scaffolds with multiple cellulases there are substantial intergenic distances (e.g., 6.13 Mb between two cellulases on Mco_46). These results thus indicate that the cellulases have not undergone recent tandem duplication in ampullariids. Protein domain analysis shows that these genes belong to the glycosyl hydrolase family 10 (GHF10), which are also present in two other herbivorous molluscs with fewer copies (i.e., six in the owl limpet Lo. gigantea and two in the abalone Haliotis discus hannai; supplementary fig. S8A, Supplementary Material online), indicating that the ampullariid cellulases originated from ancestral molluscan cellulases via substantial duplications in the ancestors of ampullariids. In P. canaliculata, these genes are similar in having 9–13 exons and a total predicted length of 230–597 amino acids. Analysis of GHF10 genes in the Pomacea assemblies allowed the correct identification of four previously reported cellulase genes. For instance, Pca_48_7.91A from P. canaliculata and Pma_952_3.84B from P. maculata are almost identical to two cellulase genes previously cloned from Ampullaria crosseana (99% sequence similarity) (Wang et al. 2003). This result indicates that P. canaliculata or P. maculata was misidentified as A. crosseana in that study. In addition, Pca_48_7.91A is highly similar to cellulase EGX1 and EGX3 (99% similarity), both cloned from P. canaliculata in another study (Imjongjirak et al. 2008). Consistent with their putative function of digesting macrophytes, a primary food source of Pomacea species, all these cellulases are highly expressed in the digestive gland but no other studied tissues of P. canaliculata (supplementary fig. S8B, Supplementary Material online). For the first time, our study shows that endogenous cellulases are diverse in ampullariids, and their tissue-specific expression pattern indicates that they are actively engaged in food digestion.
Calcareous Eggshell as a Key Adaptation to Enable Water-to-Land Transition
Since new genes provide an important source for evolutionary innovation (Hilgers et al. 2018), we searched the four ampullariid genomes for genes that can only be found in the two species of amphibious Pomacea. In total, 635 such lineage-specific genes were uncovered for Pomacea (supplementary fig. S9 and table S1, Supplementary Material online). Subsequently, we examined these genes in the context of the transition from aquatic to terrestrial egg deposition which as mentioned earlier must have required substantial changes in biochemical composition of the eggs (Cadierno et al. 2017; Pasquevich et al. 2017). Here, we examined the phylogenetics of some of these genes, and validated their functions by comparing gene expression patterns between aquatic (Lanistes and Marisa) and terrestrial egg depositors (Pomacea).
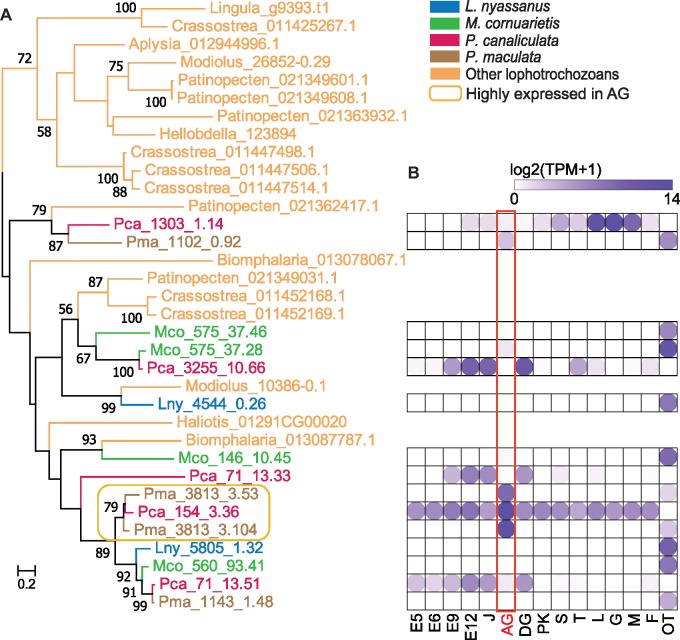
Several proteins in the egg perivitelline fluid (PVF) have previously been associated with the physiological changes related to the environment of egg deposition, but their evolutionary history remains unclear (Heras et al. 2008; Sun et al. 2012; Dreon et al. 2013; Mu et al. 2017). Among the taxon-specific genes in Pomacea, we found a calcium binding protein (CaBP), which may play an important role in forming the hard eggshell that physically protects the egg and prevents desiccation (Heras et al. 2007). Reanalysis of previously published proteomic data shows that CaBP is highly abundant in the PVF of the two Pomacea species that lay terrestrial eggs with a calcareous shell (supplementary table S9, Supplementary Material online). However, CaBP is absent from the PVF of M. cornuarietis that lays gelatinous eggs underwater (Ip et al. 2019). The Pomacea CaBP contains an EF-hand domain that has been known to be functionally involved in binding calcium ions during eggshell calcification in birds (Du et al. 2015). Our new proteomic analysis of the eggshell of P. canaliculata revealed a total of 15 proteins, including a single highly abundant CaBP. This CaBP is encoded by a gene (Pca_154_3.36) that is highly expressed in the albumen gland, the organ that secrets the PVF and packs the egg with a shell (supplementary table S9, Supplementary Material online). Compared with its abundance in the PVF (0.09%, supplementary table S10, Supplementary Material online), CaBP exhibited enrichment in the eggshell (7.32%), indicating selective incorporation of this protein into the organic shell matrix during shell formation.
To understand the origin of Pomacea CaBP, we searched the genomes of 18 other lophotrochozoans (fig. 2), and found 15 homologs in the four ampullariids and 19 homologs in 8 other lophotrochozoans. Maximum-likelihood phylogenetic analysis using RAxML ver. 8 (Stamatakis 2014) showed that 8 of the 15 ampullariid sequences are clustered together with other 19 lophotrochozoan sequences, indicating widespread presence of CaBP in Mollusca (fig. 6). Importantly, there is a terminal clade of eight sequences from ampullariids only, indicating expansion of the CaBP gene family after the divergence of ampullariids from other molluscs. Within this 8-gene clade there are three Pomacea genes (Pca_154_3.36, Pma_3813_3.53, Pma_3813_3.104) that are highly expressed in the albumen gland (TPM > 100). None of the other 12 ampullariid CaBP-like genes are highly expressed in the albumen gland. As mentioned earlier, our proteomic analysis showed that Pca_154_3.36 is abundant in the P. canaliculata eggshell. Together, our genomic, transcriptomic, and proteomic analyses revealed lineage-specific expansion and acquisition of a new function (i.e., secretion of CaBP from the albumen gland for forming the unique calcareous eggshell) were responsible for the origin of the CaBP protein in the eggshell of Pomacea, suggesting an important role of this gene in the transition of aquatic to terrestrial egg deposition. We will have to wait until genomic resources become available for Cyclophoridae, a group of fully terrestrial Architaenioglossa that is sister to Ampulariidae (Hayes, Cowie, Jørgensen, et al. 2009), to provide further insight into the evolution of calcareous eggshell in Mollusca.
Fig. 6.
Maximum-likelihood phylogenetic tree of calcium binding proteins (CaBP) in 18 lophotrochozoans, with numbers on nodes showing bootstrap values (>50%) (A), and their expression in different early stages and adult tissues of ampullariids (B). Note that in Pomacea canaliculata, many different development stages and different tissues are available, but in the other species, only AG and OT are available. Abbreviations of developmental stages/tissues: E5-12, day 5–12 embryo; J, juvenile; AG, albumen gland; DG, digestive gland; PK, posterior kidney; S, stomach; T, testis; L, lung; G, gill, M, mantle; F, Foot; OT, Other tissues (pooled tissues of DG, F, M, and T for P. maculata, Marisa cornuarietis, and Lanistes nyassanus, except the latter without testis data). Names of non-ampullariids: Aplysia californica, Biomphalaria glabrata, Haliotis discus hannai, Crassostrea gigas, Modiolus philippinarum, Patinopecten yessoensis, Lingula anatina, and Hellobdella robusta.
Novel Defense Protein as a Key Adaptation to Enable Water-to-Land Transition
Among the acquisitions that may have enabled terrestrial egg-laying are toxic perivitellins. PV2, which constitutes ∼10-20% of the total protein in the egg PVF of both P. canaliculata and P. maculata, has been shown to be highly neurotoxic to mice, and therefore it was considered to be a defensive protein (Dreon et al. 2013). Previous studies indicate that PV2 is a complex of two proteins: a membrane attack complex/perforin (MACPF) subunit (termed PV2-67 of 67 kDa, probably the toxic moiety), and a tachylectin-like subunit (termed PV2-31 of 31 kDa, probably the delivery moiety). Our newly generated ampullariid genomic resources allow for the first time to examine the evolution of this protein complex that is presumably critical for protecting the nutrient-rich eggs from terrestrial predators (Dreon et al. 2013).
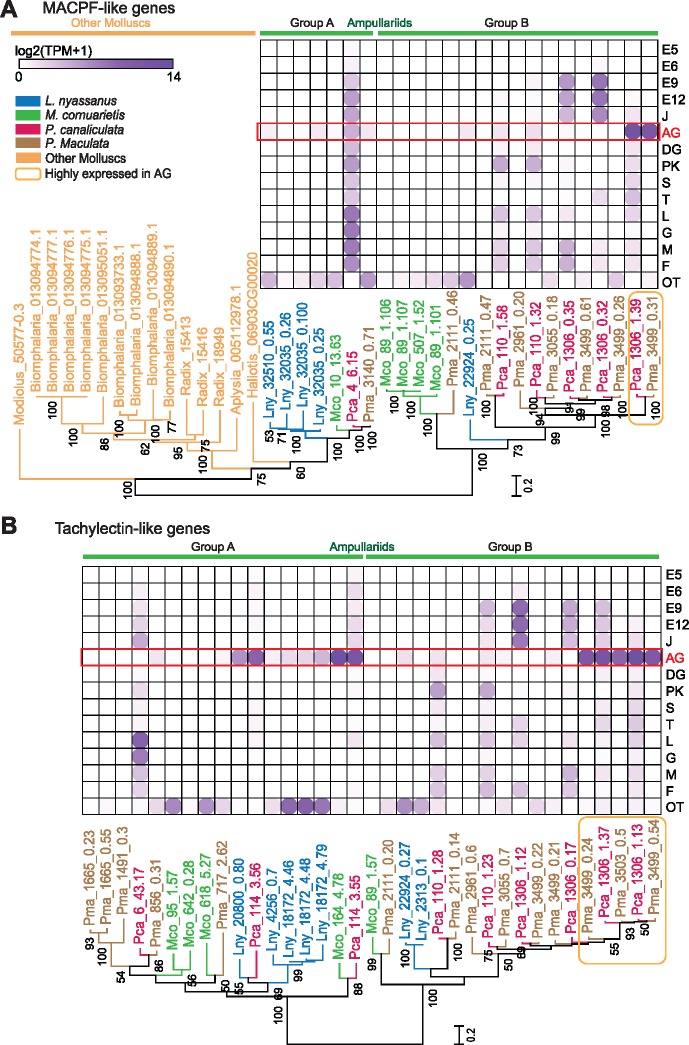
We searched 18 lophotrochozoan genomes (fig. 2), and found 24 MACPF homologues in the four ampullariids species and 15 homologues in five other molluscan species (fig. 7A). There were no MACPF homologues in the more distantly related lophotrochozoans including the Octopus, Lingua, Capitella, and Helobdella. Maximum-likelihood phylogenetic analysis showed that ampullariid MACPF-like genes are clustered into two groups (fig. 7A). Group A comprises seven ampullariid sequences nested among non-ampullariid sequences, indicating a possible ancient origin of these genes dating back to the common ancestor of all molluscs. Group B, however, is specific to the four ampullariids and comprises 17 sequences, which indicates that extensive gene duplication events have occurred after the divergence of Ampullariidae from other molluscs. Within Group B, there is a sequence that encodes the MACPF subunit of P. canaliculata (Pca_1306_1.39) and another encodes the MACPF subunit of P. maculata (Pma_3499_0.31) (supplementary table S10 and figs. S10 and S11, Supplementary Material online). Transcriptomic data reveal that these two genes are the only highly expressed MACPF-like genes in the albumen gland of P. canaliculata and P. maculata (fig. 7A). Other MACPF-like genes show different tissue- or life stage-specific expression patterns in ampullariids, including Pomacea spp., but none of them exhibits high expression in the albumen gland (fig. 7A). These results indicate that acquisition of a new function (i.e., secretion of MACPF by the albumen gland, which was not detected in underwater egg laying Marisa and Lanistes) after gene duplication was probably responsible for the origin of the PV2 toxin where the MACPF subunit is linked to a tachylectin for embryo protection in the terrestrial egg laying Pomacea.
Fig. 7.
Phylogeny and expression of homologues of PV2 subunits in lophotrochozoans. (A) MACPF-like genes and (B) tachylectin-like genes. Ampullariid genes are colored by species, whereas albumen gland overexpressed genes are highlighted with an orange box. Numbers on nodes are bootstrap values (>50%). Gene expression levels are in logarithmic scale. E5-12, 5- to 12-day embryo; J, juvenile; AG, albumen gland; DG, digestive gland; F, Foot; PK, posterior kidney; L, lung; M, mantle; S, stomach; T, testis; OT, other tissues. Of the 14 non-ampullariid examined, only the following have MACPF-like genes, but none of them have a tachylectin-like gene: Aplysia californica; Biomphalaria glabrata; Radix auricularia; Haliotis discus hannai; Modiolus philippinarum.
For tachylectin-like genes, we found 35 homologues in the four species of ampullariids but no such homologues in the other 14 lophotrochozoans examined (fig. 2). Maximum-likelihood phylogenetic analysis showed that ampullariid tachylectin-like genes are also divided into two groups (fig. 7B). Group A comprises 17 ampullariid sequences that include three exhibiting high expression in the albumen gland. However, none of them has a corresponding egg PVF protein, indicating that posttranscriptional mechanism may have prevented the translation of mRNAs into proteins, or that the proteins produced do not have in vivo half-lives long enough for them to be detected (Greenbaum et al. 2003). Group B comprises 18 ampullariid sequences, with 16 Pomacea sequences forming a terminal clade, indicating that extensive duplication events may have occurred after the divergence of Pomacea from other ampullariids. Among them, two from each of the two species of Pomacea are highly expressed in the albumen gland, and they are the exact genes that encode the tachylectins found in the egg PVF (fig. 7B). The other gene of this group (Pma_3503_0.5) that is also highly expressed in the albumen gland does not have a signal peptide (supplementary fig. S11, Supplementary Material online). As such, it is not secreted and has no corresponding protein in the PVF (supplementary table S10, Supplementary Material online). Other Group B tachylectin-like genes exhibit different tissue-specific expression patterns, but none of them is highly expressed in the albumen gland (fig. 7B). Together, these phylogenetic, gene expression, and proteomics data indicate that neofunctionalization after gene duplication may be responsible for the origin of the novel tachylectin subunit present in Pomacea eggs.
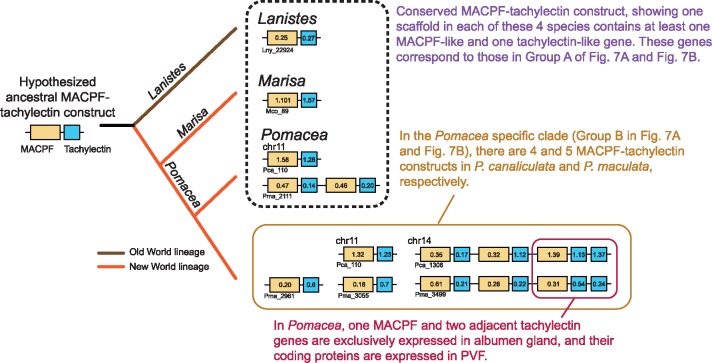
Further, interrogating the ampullariid genomes reveals at least one scaffold with both MACPF-like and tachylectin-like genes in all four species (fig. 8). Such a two-gene configuration, with a gap as small as 420 bp between them, is present only once in L. nyassanus and M. cornuarietis, suggesting that it was probably present as a single copy in the common ancestor of all ampullariids. However, it occurs five times in P. canaliculata and seven times in P. maculata, indicating multiple duplications after the divergence of Pomacea from its common ancestor with Marisa. For instance, chromosome 14 of P. canaliculata contains three tandem repeats of this 2-gene configuration, with an additional duplication of the tachylectin gene (fig. 8). This gene configuration indicates that some of the duplication events for the two genes could have occurred simultaneously. In both Pomacea species, the functional PV2 subunits are encoded by the most downstream MACPF and two adjacent tachylectin genes within the same scaffold with the above-mentioned tandem repeat sequences. As reported earlier, the MACPF and the two tachylectins are among the most abundant PVF proteins in Pomacea eggs (Sun et al. 2012; Mu et al. 2017).
Fig. 8.
Schematic illustration of the evolution of the MACPF–tachylectin complex in ampullariids. Based on the genomic arrangements of the MACPF and tachylectin genes, the model proposes that a single copy of MACPF–tachylectin complex was present in the common ancestor of ampullariids. Only in the two species of Pomacea has it become highly diversified, with both MACPF-tachylectin genes that are conserved across Ampullariidae, and multiple Pomacea specific MACPF-tachylectin genes that were generated by tandem duplication. The final 1-MACPF and 2-tachylectin configuration is exclusively expressed in the albumen gland of Pomacea and the proteins detected in their egg PVF. Numbers below and inside gene diagram boxes are scaffold numbers and gene numbers in the scaffold, respectively. For instance, Lanistes nyassanus contains a MACPF gene (Lny_22924_0.25) and a tachylectin gene (Lny_22924_0.27) in scaffold Lny_22924. For P. canaliculata, the chromosome numbers are shown above the gene diagram boxes.
Overall, our analyses clearly illustrate the involvement of gene duplication and neofunctionalization as some of the key mechanisms for the acquisition of the PV2 protein complex. This combination of MACPF-like moiety (likely the membrane attack component) and tachylectin-like moiety (likely the delivery component) has only been found in plant and bacterial AB toxins before (Dreon et al. 2013). The occurrence in PV2 in Pomacea eggs and its toxicity to mice indicate its defensive role against terrestrial predators. The co-option of PV2 into the eggs might be an important innovation that enabled the transition of aquatic to terrestrial oviposition in ampullariids.
Conclusions
Here, we report the comparative genomics of four ampullariid species that display two drastically different ovipositioning strategies in the context of the entire phylum of Mollusca. Our results indicate that these gastropods have conserved ancestral bilaterian karyotype features, and that the pattern of interchromosome rearrangements is different from that of their sister group, the bivalves. Remarkably, different from other molluscs examined, the Hox cluster in ampullariids is discontinuous, with a reverse transposition of the ancestral anterior subcluster. These results, together with the basal position of Ampullariidae within Caenogastropoda that accounts for >60% of all gastropod species (Ponder et al. 2008), indicate that our genomic data represent a valuable resource for comparative studies with other molluscs, for which only few published genomes are available (Halanych and Kocot 2017; Sun et al. 2017; Voolstra et al. 2017), in comparison to their massive species diversity (∼100,000 species) and high body plan disparity (Aguilera et al. 2017; Sigwart 2017).
Comparative genomic analyses of Ampullariidae in a phylogenetic context, together with the study of expression patterns and proteomic validations revealed several genetic predispositions that may have driven diversification in Ampullariidae. Specifically, enhanced environmental sensing and digestive capability for macrophytes may have augmented the potential enabling some species of Pomacea and Marisa to become notorious invaders. Proteomic analyses also provided clues on the transition from aquatic to terrestrial egg deposition. For instance, acquisitions of a novel protein involved in making the hard calcareous eggshell, and a novel protein involved in embryo defense may have been critical evolutionary adaptations. Given that several species of invasive ampullariids are notorious agricultural pests, our genomic resources could be used to develop effective control strategies for these pests, such as synthesizing chemical molluscicides and designing genetic control measures using RNAi and gene drives (Jiang et al. 2006).
Materials and Methods
Supplementary Material online contains details of the Materials and Methods. In brief, for each of the four species, genomic DNA was extracted from a piece of the snail foot, DNA libraries of various sizes were prepared, sequenced on a second and/or third generation sequencing platform, and assembled. To annotate the genomes and study gene expression, total RNA was extracted from various developmental stages (for P. canaliculata) and adult tissues (for the four species) and sequenced using an Illumina platform. Genome analyses mainly involved bench-marking with other assembled molluscan genomes, determination of genomic structure, phylogenetic tree construction, and gene expression profiling. Comparative analyses were conducted to understand the roles of selected expanded gene families in the diversity and invasiveness of ampullariids. Furthermore, to understand the molecular basis of transition from underwater to terrestrial egg deposition, published LC-MS/MS data from the ampullariid PVF proteomes were reanalyzed, and calcareous eggshells of P. canaliculata were collected and analyzed to determine their protein composition in the organic matrix. Possible origins of genes encoding selected PVF proteins were explored through analyzing their genomic location and phylogenetic relationships.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was supported by the Research Grants Council of Hong Kong (261312 to J.W.Q.), Hong Kong Baptist University (SDF15-1012-P04 to J.W.Q.), Scientific and Technical Innovation Council of Shenzhen and Department of Education of Guangdong Province (827000012, 2017KTSCX161, KQJSCX20170330110206042 to Y.Z.), the Howard Hughes Medical Institute (to A.S.A.), the Stowers Institute for Medical Research (to A.S.A.), the Fund for Scientific and Technological Research (FONCYT-Argentina, to A.C.V.), the Society for Developmental Biology (SDB Emerging Models grant to A.A.), the American Association of Anatomists (postdoctoral fellowship to A.A.), FWO Vlaanderen (12N3915N to B.V.B.), the French Agence Nationale de la Recherche (ANR-JCJC-EVOLINK to B.V.B.), and the U.S. National Science Foundation (DEB0949061 to R.H.C.). H.M., J.C.H.I., and T.X. received a postgraduate studentship from Hong Kong Baptist University.
Author Contributions
J.W.Q., P.Y.Q. Y.Z., and J.S. designed research; K.A.H., R.H.C., H.H., A.C.V., and J.W.Q. conceptualized research; H.M., J.C.H.I., H.H., S.I., A.C.V., I.A.V. B.V.B., A.A., K.N., and Y.Y. collected samples; J.C.H.I., H.M., Y.L., and A.A. extracted DNA and RNA. J.S. and Y.L. performed Nanopore sequencing; J.S. and R.L. assembled genomes; J.S. annotated genomes; J.S., J.C.H.I., and Y.S. performed phylogenetic analysis; J.S. and E.R. assembled and annotated transcriptomes; J.S., A.C.V., and I.A.V. estimated genome sizes; R.L. analyzed HiC data; B.V.B. statistically compared genome assemblies; H.M. and T.X. analyzed gene expression; J.S. and H.M. analyzed gene families; J.C.H.I. and J.S. analyzed repeat contents; J.C.H.I. analyzed CaBP and PV2; J.W.Q., Y.Z., P.Y.Q., A.A., A.S.A., and A.C.V. contributed materials/reagents; J.S., H.M., J.C.H.I., R.L., P.Y.Q., and J.W.Q. drafted the article; K.A.H., R.H.C., A.A., A.S.A., H.H., S.I., A.C.V., and B.V.B. revised the draft substantially.
Data reported in this article have been deposited in the GenBank database: genome assemblies and reads (Bioproject No: PRJNA523959 for P. canaliculata, PRJNA523958 for P. maculata, PRJNA445755 for M. cornuarietis, and PRJNA523095 for L. nyassanus) and transcriptome sequences (Bioproject PRJNA473031 and PRJNA473253). Whole Genome Shotgun project numbers SRJH00000000, SRHC00000000, SMMW00000000 and SMGT00000000. The genome assemblies and annotations are also available from the Dryad Digital Repository at https://doi.org/10.5061/dryad.15nd8v3. Whole Genome Shotgun project numbers SRJH00000000, SRHC00000000, SMMW00000000 and SMGT00000000. The genome assemblies and annotations are also available from the Dryad Digital Repository at https://doi.org/10.5061/dryad.15nd8v3. Some of the raw transcriptome data used in this study can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1311.
References
- Adema CM, Hillier LW, Jones CS, Loker ES, Knight M, Minx P, Oliveira G, Raghavan N, Shedlock A, do Amaral LR, et al. 2017. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat Commun. 8:15451.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera F, McDougall C, Degnan BM.. 2017. Co-option and de novo gene evolution underlie molluscan shell diversity. Mol Biol Evol. 34(4):779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertin CB, Simakov O, Mitros T, Wang ZY, Pungor JR, Edsinger-Gonzales E, Brenner S, Ragsdale CW, Rokhsar DS.. 2015. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524(7564):220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadierno MP, Dreon MS, Heras H.. 2017. Apple snail perivitellin precursor properties help explain predators’ feeding behavior. Physiol Biochem Zool. 90(4):461–470. [DOI] [PubMed] [Google Scholar]
- Cowie RH. 2002. Apple snails (Ampullariidae) as agricultural pests: their biology, impacts and management In: Barker GM, editor. Molluscs as crop pests. Wallingford (United Kingdom: ): CAB International; p. 145–192. [Google Scholar]
- Cowie RH. 2015. The recent apple snails of Africa and Asia (Mollusca: Gastropoda: Ampullariidae: Afropomus, Forbesopomus, Lanistes, Pila, Saulea): a nomenclatural and type catalogue. The apple snails of the Americas: addenda and corrigenda. Zootaxa 3940:1–92. [DOI] [PubMed] [Google Scholar]
- Davison A, McDowell G, Holden JM, Johnson HF, Koutsovoulos GD, Liu MM, Hulpiau P, Van Roy F, Wade CM, Banerjee R.. 2016. Formin is associated with left-right asymmetry in the pond snail and the frog. Curr Biol. 26(5):654–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendoza A, Sebé-Pedrós A, Ruiz-Trillo I.. 2014. The evolution of the GPCR signaling system in eukaryotes: modularity, conservation, and the transition to metazoan multicellularity. Genome Biol Evol. 6(3):606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth JP, Hahn MW.. 2009. The life and death of gene families. Bioessays 31(1):29–39. [DOI] [PubMed] [Google Scholar]
- Dreon MS, Frassa MV, Ceolín M, Ituarte S, Qiu JW, Sun J, Fernández PE, Heras H.. 2013. Novel animal defenses against predation: a snail egg neurotoxin combining lectin and pore-forming chains that resembles plant defense and bacteria attack toxins. PLoS One 8(5):e63782.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Hincke MT, Rose-Martel M, Hennequet-Antier C, Brionne A, Cogburn LA, Nys Y, Gautron J.. 2015. Identifying specific proteins involved in eggshell membrane formation using gene expression analysis and bioinformatics. BMC Genomics 16:792.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D, Colangelo C, Williams K, Gerstein M.. 2003. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4(9):117.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst J, Andriantsoa R, Falckenhayn C, Hanna K, Stein W, Rasamy J, Lyko F.. 2018. Clonal genome evolution and rapid invasive spread of the marbled crayfish. Nat Ecol Evol. 2(3):567–573. [DOI] [PubMed] [Google Scholar]
- Halanych KM, Kocot KM.. 2017. Genome evolution: shellfish genes. Nat Ecol Evol. 1(5):142.. [DOI] [PubMed] [Google Scholar]
- Hayes KA, Burks RL, Castro-Vazquez A, Darby PC, Heras H, Martín PR, Qiu J-W, Thiengo SC, Vega IA, Wada T, et al. 2015. Insights from an integrated view of the biology of apple snails (Caenogastropoda: ampullariidae). Malacologia 58(1-2):245–302. [Google Scholar]
- Hayes KA, Cowie RH, Jørgensen A, Schultheiß R, Albrecht C, Thiengo SC.. 2009. Molluscan models in evolutionary biology: apple snails (Gastropoda: ampullariidae) as a system for addressing fundamental questions. Am Malacol Bull. 27(1-2):47–58. [Google Scholar]
- Hayes KA, Cowie RH, Thiengo SC.. 2009. A global phylogeny of apple snails: Gondwanan origin, generic relationships, and the influence of outgroup choice (Caenogastropoda: ampullariidae). Biol J Linn Soc. 98(1):61–76. [Google Scholar]
- Heras H, Dreon MS, Ituarte S, Pollero RJ.. 2007. Egg carotenoproteins in neotropical Ampullariidae (Gastropoda: arquitaenioglossa). Comp Biochem Physiol C Toxicol Pharmacol. 146(1-2):158–167. [DOI] [PubMed] [Google Scholar]
- Heras H, Frassa MV, Fernandez PE, Galosi CM, Gimeno EJ, Dreon MS.. 2008. First egg protein with a neurotoxic effect on mice. Toxicon 52(3):481–488. [DOI] [PubMed] [Google Scholar]
- Hilgers L, Hartmann S, Hofreiter M, von Rintelen T.. 2018. Novel genes, ancient genes, and gene co-option contributed to the genetic basis of the radula, a molluscan innovation. Mol Biol Evol. 35(7):1638–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PW. 2013. Evolution of homeobox genes. Wiley Interdiscip Rev Dev Biol. 2(1):31–45. [DOI] [PubMed] [Google Scholar]
- Hoorn C, Wesselingh FP, ter Steege H, Bermudez MA, Mora A, Sevink J, Sanmartín I, Sanchez-Meseguer A, Anderson CL, Figueiredo JP, et al. 2010. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science 330(6006):927–931. [DOI] [PubMed] [Google Scholar]
- Horgan FG, Stuart AM, Kudavidanage EP.. 2014. Impact of invasive apple snails on the functioning and services of natural and managed wetlands. Acta Oecol. 54:90–100. [Google Scholar]
- Hui JH, Raible F, Korchagina N, Dray N, Samain S, Magdelenat G, Jubin C, Segurens B, Balavoine G, Arendt D, et al. 2009. Features of the ancestral bilaterian inferred from Platyneries deumerilii ParaHox genes. BMC Biol. 7:43.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M, Kikuchi T, Sanders M, Newbold C, Berriman M, Otto TD.. 2013. REAPR: a universal tool for genome assembly evaluation. Genome Biol. 14(5):R47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imjongjirak C, Amparyup P, Sittipraneed S.. 2008. Cloning, genomic organization and expression of two glycosyl hydrolase family 10 (GHF10) genes from golden apple snail (Pomacea canaliculata). DNA Seq. 19(3):224–236. [DOI] [PubMed] [Google Scholar]
- Ip JCH, Mu H, Zhang Y, Sun J, Heras H, Chu KH, Qiu JW.. 2019. Understanding the transition from water to land: insights from multi-omic analyses of the perivitelline fluid of apple snail eggs. J Proteomics. 194:79–88. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Loker ES, Zhang SM.. 2006. In vivo and in vitro knockdown of FREP2 gene expression in the snail Biomphalaria glabrata using RNA interference. Dev Comp Immunol. 30(10):855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokat W, Boebel T, König M, Meyer U.. 2003. Timing and geometry of early Gondwana breakup. J Geophys Res. 108:2428. [Google Scholar]
- Jørgensen A, Kristensen TK, Madsen H.. 2008. A molecular phylogeny of apple snails (Gastropoda, Caenogastropoda, Ampullariidae) with an emphasis on African species. Zool Scripta. 37(3):245–252. [Google Scholar]
- Kocot KM, Cannon JT, Todt C, Citarella MR, Kohn AB, Meyer A, Santos SR, Schander C, Moroz LL, Lieb B, et al. 2011. Phylogenomics reveals deep molluscan relationships. Nature 477(7365):452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lach L, Britton DK, Rundell RJ, Cowie RH.. 2000. Food preference and reproductive plasticity in an invasive freshwater snail. Biol Invasions. 2(4):279–288. [Google Scholar]
- Liu C, Zhang Y, Ren Y, Wang H, Li S, Jiang F, Yin L, Qiao X, Zhang G, Qian W, et al. 2018. The genome of the golden apple snail Pomacea canaliculata provides insight into stress tolerance and invasive adaptation. GigaScience 7:giy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S, Zhang Y, Liu H-X, Hu L, Yang K, Steinmann P, Chen Z, Wang L-Y, Utzinger J, Zhou X-N.. 2009. Invasive snails and an emerging infectious disease: results from the first national survey on Angiostrongylus cantonensis in China. PLoS Negl Trop Dis. 3(2):e368.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura K, Izumi Y, Yoshida K, Wada T.. 2016. Cold tolerance of invasive freshwater snails, Pomacea canaliculata, P. maculata, and their hybrids helps explain their different distributions. Freshw Biol. 61(1):80–87. [Google Scholar]
- Matsukura K, Okuda M, Cazzaniga NJ, Wada T.. 2013. Genetic exchange between two freshwater apple snails, Pomacea canaliculata and Pomacea maculata invading East and Southeast Asia. Biol Invasions. 15(9):2039–2048. [Google Scholar]
- Mu H, Sun J, Cheung SG, Fang L, Zhou H, Luan T, Zhang H, Wong CK, Qiu JW.. 2018. Comparative proteomics and codon substitution analysis reveal mechanisms of differential resistance to hypoxia in congeneric snails. J Proteomics. 172:36–48. [DOI] [PubMed] [Google Scholar]
- Mu H, Sun J, Heras H, Chu KH, Qiu JW.. 2017. An integrated proteomic and transcriptomic analysis of perivitelline fluid proteins in a freshwater gastropod laying aerial eggs. J Proteomics. 155:22–30. [DOI] [PubMed] [Google Scholar]
- Nam BH, Kwak W, Kim YO, Kim DG, Kong HJ, Kim WJ, Kang JH, Park JY, An CM, Moon JY, et al. 2017. Genome sequence of Pacific abalone (Haliotis discus hannai): the first draft genome in family Haliotidae. GigaScience 6(5):1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquevich MY, Dreon MS, Qiu JW, Mu H, Heras H.. 2017. Convergent evolution of plant and animal embryo defences by hyperstable non-digestible storage proteins. Sci Rep. 7:15848.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder WF, Colgan DJ, Healy JM, Alexander N, Simone LRL, Mielke EE.. 2008. Caenogastropoda In: WF Ponder, DR Lindberg, editors. Phylogeny and evolution of the Mollusca. Berkeley (CA: ): University of California Press; p. 331–383. [Google Scholar]
- Qiu JW, Kwong KL.. 2009. Effects of macrophytes on feeding and life-history traits of the invasive apple snail Pomacea canaliculata. Freshwater Biol. 54(8):1720–1730. [Google Scholar]
- Rawlings TA, Hayes KA, Cowie RH, Collins TM.. 2007. The identity, distribution, and impacts of non-native apple snails in the continental United States. BMC Evol Biol. 7:97.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu L, Martín PR.. 2014. Egg cannibalism in Pomacea canaliculata (Caenogastropoda: ampullariidae) from the Southern Pampas: an alternative trophic strategy? Malacologia 57(2):341–351. [Google Scholar]
- Schell T, Feldmeyer B, Schmidt H, Greshake B, Tills O, Truebano M, Rundle SD, Paule J, Ebersberger I, Pfenninger M.. 2017. An annotated draft genome for Radix auricularia (Gastropoda, Mollusca). Genome Biol Evol. 9(3):585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigwart JD. 2017. Zoology: molluscs all beneath the Sun, one shell, two shells, more, or none. Curr Biol. 27(14):R708–R710. [DOI] [PubMed] [Google Scholar]
- Simakov O, Marletaz F, Cho SJ, Edsinger-Gonzales E, Havlak P, Hellsten U, Kuo DH, Larsson T, Lv J, Arendt D, et al. 2013. Insights into bilaterian evolution from three spiralian genomes. Nature 493(7433):526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Smith SA, Wilson NG, Goetz FE, Feehery C, Andrade SC, Rouse GW, Giribet G, Dunn CW.. 2011. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 480(7377):364–367. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Mu H, Zhang H, Chandramouli KH, Qian PY, Wong CKC, Qiu JW.. 2013. Understanding the regulation of estivation in a freshwater snail through iTRAQ-based comparative proteomics. J Proteome Res. 12(11):5271–5280. [DOI] [PubMed] [Google Scholar]
- Sun J, Zhang H, Wang H, Heras H, Dreon MS, Ituarte S, Ravasi T, Qian PY, Qiu JW.. 2012. First proteome of the egg perivitelline fluid of a freshwater gastropod with aerial oviposition. J Proteome Res. 11(8):4240–4248. [DOI] [PubMed] [Google Scholar]
- Sun J, Zhang Y, Xu T, Zhang Y, Mu H, Zhang Y, Lan Y, Fields CJ, Hui JHL, Zhang W, et al. 2017. Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nat Ecol Evol. 1(5):121. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Koyanagi R, Gyoja F, Kanda M, Hisata K, Fujie M, Goto H, Yamasaki S, Nagai K, Morino Y, et al. 2016. Bivalve-specific gene expansion in the pearl oyster genome: implications of adaptation to a sessile lifestyle. Zool Lett. 2:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bocxlaer B. 2017. Hierarchical structure of ecological and non-ecological processes of differentiation shaped ongoing gastropod radiation in the Malawi Basin. Proc Biol Sci. 84:1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voolstra CR GIGA Community of Scientists (COS,) Wörheide G, Lopez JV.. 2017. Advancing genomics through the Global Invertebrate Genomics Alliance (GIGA). Invertebr Syst. 31(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ding M, Li YH, Chen QX, Xu GJ, Zhao FK.. 2003. A monovalent anion affected multi-functional cellulase EGX from the mollusca, Ampullaria crossean. Protein Expr Purif. 31(1):108–114. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhang J, Jiao W, Li J, Xun X, Sun Y, Guo X, Huan P, Dong B, Zhang L, et al. 2017. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat Ecol Evol. 1(5):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T-H, Jin H, Marler B, Guo H, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40(7):e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanninger A, Wollesen T.. 2019. The evolution of molluscs. Biol Rev. 94(1):102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Tokuda G.. 2001. Animal cellulases. Cell Mol Life Sci. 58(9):1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollesen T, Rodríguez Monje SV, Luiz de Oliveira A, Wanninger A.. 2018. Staggered Hox expression is more widespread among molluscs than previously appreciated. Proc R Soc B. 285(1888):20181513.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray G. 2001. Resolving the Hox paradox. Science 292(5525):2256–2257. [Google Scholar]
- Yusa Y. 2001. Predation on eggs of the apple snail Pomacea canaliculata (Gastropoda: ampullariidae) by the fire ant Solenopsis geminata. J Molluscan Stud. 67(3):275–279. [Google Scholar]
- Zapata F, Wilson NG, Howison M, Andrade SC, Jörger KM, Schrödl M, Goetz FE, Giribet G, Dunn CW.. 2014. Phylogenomic analyses of deep gastropod relationships reject Orthogastropoda. Proc Biol Sci B. 281(1794):20141739.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490(7418):49–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.