Abstract
A sensitive immunohistochemical technique was used, along with highly specific affinity-purified antibodies to brain-derived neurotrophic factor (BDNF), to generate a detailed mapping of BDNF immunoreactivity (BDNF-ir) throughout the adult rat CNS. A parallel analysis of sites of BDNF synthesis was performed with in situ hybridization techniques using a cRNA probe to the exon encoding mature rat BDNF protein. These combined data revealed (1) groups of cell bodies containing diffuse BDNF-ir throughout the CNS that were strongly correlated with fields of cells containing BDNF mRNA; (2) varying degrees of BDNF-ir outside of cell bodies, in what appeared to be fibers and/or terminals; and (3) many regions containing extremely heavy BDNF-immunoreactive fiber/terminal labeling that lacked BDNF mRNA (e.g., medial habenula, central nucleus of the amygdala, bed nucleus of stria terminalis, lateral septum, and spinal cord). The latter observation suggested that in these regions BDNF was derived from anterograde axonal transport by afferent systems. In the two cases in which this hypothesis was tested by the elimination of select afferents, BDNF immunostaining was completely eliminated. These data, along with the observation that BDNF-ir was rarely found within dendrites or fibers en passage, suggest that BDNF protein produced in adult CNS neurons is polarized primarily along axonal processes and is preferentially stored in terminals within the innervation target.
Keywords: neurotrophin, immunohistochemistry, in situhybridization, neurotrophic factor, BDNF, anterograde axonal transport
The neurotrophins are a related family of growth factors presently consisting of nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin-3 (NT-3), NT-4/5, and NT-6 (Berkemeir et al., 1991; Narhi et al., 1993; Gotz et al., 1994; Ibáñez, 1994). Although much evidence suggests that these factors play an important role in regulating the survival, growth, and differentiation of select populations of peripheral neurons, their physiological role in the developing and adult CNS is less well defined. Achieving an understanding of neurotrophin function in the CNS will likely require several experimental approaches, including elucidation of sites of synthesis and storage for each neurotrophin.
To date, many investigators have reported on the localization of mRNAs for the various neurotrophins, although few of these studies have provided a detailed mapping throughout the CNS. NGF expression has been localized primarily to the hippocampal formation, olfactory bulb, and cortex—all target regions of basal forebrain cholinergic neurons (Korsching et al., 1985; Shelton and Reichardt, 1986; Whittemore et al., 1986; Ernfors et al., 1990; Guthrie and Gall, 1991). Additional studies have indicated that high levels of NGF mRNA are found within the basal forebrain (Lauterborn et al., 1991, 1995), hypothalamus (Spillantini et al., 1989, Ceccatelli et al., 1991), and brainstem (Ceccatelli et al., 1991). BDNF mRNA seems to be broadly distributed throughout the CNS (Ernfors et al., 1990; Phillips et al., 1990;Ceccatelli et al., 1991; Friedman et al., 1991; Guthrie and Gall, 1991;Gall et al., 1992; Castrén et al., 1995). In contrast, NT-3 mRNA is the most narrowly distributed, with high levels of expression restricted to hippocampal CA2 stratum pyramidale, the dentate gyrus granule cells, and the cerebellar granule cells (Ernfors et al., 1990;Maisonpierre et al., 1990; Ceccatelli et al., 1991) and lower levels of NT-3 mRNA in substantia nigra, midline and intralaminar thalamus, and posterior amygdala (Seroogy and Gall, 1993; Lauterborn and Gall, 1994). NT-4/5 expression in the CNS seems to be very low (Timmusk et al., 1993), and its in situ distribution has not been determined. The recently discovered NT-6 is restricted to nonmammalian species, and its distribution remains largely unknown (Gotz et al., 1994).
The cellular localization of neurotrophin proteins in the CNS has proceeded much more slowly, probably reflecting the technical limitations inherent in the immunohistochemical approach. Recently, we have generated antibodies and developed fixation and immunohistochemical protocols permitting the visualization of NGF protein in the normal adult rat brain (Conner et al., 1992). This protocol also has been used successfully to localize NGF in a wide variety of circumstances, including (1) the normal distribution of NGF in human and nonhuman primates (Mufson et al., 1994), (2) changes in NGF accumulation by basal forebrain cholinergic neurons in human Alzheimer’s tissue (Mufson et al., 1995), (3) the distribution of NGF protein in transgenic mouse lines with NGF gene alterations (Carlson et al., 1995; Ma et al., 1995), and (4) changes in the in situdistribution of NGF protein after various experimental manipulations (Conner and Varon, 1992, 1995; Conner et al., 1994; Holtzman and Lowenstein, 1995).
In the present investigation, we have used the sensitive immunohistochemical protocol developed for NGF, along with a well characterized preparation of affinity-purified polyclonal antibodies to BDNF (Yan et al., 1997), to generate a detailed mapping of BDNF protein in adult rat brain and spinal cord. A parallel analysis of BDNF mRNA distribution was also carried out. These combined data revealed several regions containing high densities of BDNF-immunoreactive processes but no detectable mRNA, thereby suggesting that BDNF may have been distributed to these regions by way of anterograde axonal transport. This hypothesis was tested in two different systems using lesion paradigms to destroy specific cell populations supplying afferent innervation to regions containing abundant BDNF protein and no detectable mRNA.
MATERIALS AND METHODS
BDNF antibodies and cDNA probes. Affinity-purified rabbit polyclonal antibodies to BDNF used in this investigation were generated and characterized as described previously (Conner et al., 1996; Yan et al., 1997). This antibody preparation was shown to be specific for BDNF by several criteria. (1) In a Western blot, the antibody was capable of recognizing as little as 0.1 ng of BDNF per lane but did not cross-react with NGF, NT-3, or NT-4/5 at concentrations of even 100-fold greater; (2) in a chick dorsal root ganglion bioassay, the antibody specifically interfered with the survival-promoting activity of BDNF but was not capable of inhibiting the actions of either NGF or NT-3; and (3) the specific pattern of BDNF-ir observed in CNS tissues with this antibody was not detected in mice in which the BDNF gene was deleted but was present in mice with a deletion of the NGF gene (J. Conner, unpublished observations). Antisense BDNF mRNA was generated from the rat recombinant plasmid pR1112–8 with T3 TNA polymerase after digestion with PvuII. This cRNA probe contains 384 bases complementary to the mRNA encoding mature rat BDNF protein (Isackson et al., 1991). Radiolabeled sense mRNA was generated from the plasmid DNA as described previously (Gall and Isackson, 1989).
BDNF immunohistochemistry. Adult male and female Sprague Dawley rats (Simonsen, Gilroy, CA, and Harlan, San Diego, CA) were used (n = 5 for normal distribution; n = 14 for analysis of lesion effects). All animals were perfused under deep anesthesia with ∼50 ml PBS followed with ∼250 ml 2% paraformaldehyde + 0.2% parabenzoquinone in 0.07 mphosphate buffer, pH 7.2. Brains were removed, post-fixed for 2 hr in the same fixative, and cryoprotected overnight in 30% sucrose in 0.1m phosphate buffer (all solutions at 4°C). Coronal sections (40 μm) were cut on a sliding microtome and stored in Millonig’s buffer until they were processed for BDNF-ir as has been described (Conner et al., 1996). In brief, sections (taken every 240 μm) were washed in 0.1 m Tris-buffered saline (TBS), pH 7.4, incubated in TBS containing 0.25% Triton X-100, and incubated in TBS + 2% BSA + 5% normal goat serum. Staining was performed by incubating sections with primary antibodies (50 ng/ml anti-BDNF) for 48 hr at 4°C, with secondary antibodies (1.5 μg/ml biotinylated goat anti-rabbit; Vector Labs, Burlingame, CA) for 3 hr at room temperature, and with an avidin–biotin–peroxidase reagent (1:250 dilution, ABC Elite; Vector Labs) for 90 min at room temperature. Sections were then reacted with a solution containing 0.04% diaminobenzidine tetrahydrochloride, 0.06% nickel chloride, and 0.06% H2O2 in Tris-HCl buffer.
In situ hybridization for BDNF mRNA. Sprague Dawley rats (Simonsen) (n = 5) were anesthetized by intraperitoneal injection with sodium pentobarbital and killed by perfusion with 50 ml 0.9% saline followed by 350 ml 4% paraformaldehyde in 0.1 m phosphate buffer (PPB). Brains were removed from the cranium and post-fixed in PPB for 24 hr at 4°, cryoprotected in 20% sucrose in PPB for 24–48 hr, and then sectioned on a freezing microtome at a thickness of 25 μm in the coronal plane. Tissue sections were processed free-floating as described previously (Lauterborn et al., 1991). Briefly, coronal sections were permeabilized with proteinase K, treated with acetic anhydride, and then hybridized with cRNA probe (1 × 104 cpm/μl) at 60°C for 24–36 hr. Hybridization was followed by two rinses in 4× SSC at 60°C, treatment with ribonuclease A (1.2 kU/ml) (Sigma, St. Louis, MO) for 30 min at 45°C, and washes through descending concentrations of SSC to a final stringency of 0.1× SSC at 60°C. Dithiothreitol was added to all washes at a final concentration of 5 mm. Tissue was then mounted onto gelatin-coated slides and air-dried. For emulsion autoradiography, slides were dehydrated in ethanol and defatted in chloroform. Hybridization was visualized by both film (Amersham β-max) and emulsion (Kodak NTB2) autoradiography, with exposure times of 3–4 d at room temperature and 4–6 weeks at 4°C, respectively. After autoradiographic development, emulsion-coated slides were counterstained with cresyl violet, coverslipped with Permount, and analyzed by bright- and dark-field microscopy.
Lesions. Unilateral (n = 4) or bilateral (n = 4) aspirative lesions of the septofimbrial and triangular septal nuclei were accomplished by removing a 3 × 3 mm area of skull on either side of Bregma and immediately lateral to the sagittal sinus. After the dura was opened, successive aspiration of part of the parietal and cingulate cortices, the supracallosal stria, and the corpus callosum exposed the fimbria and supracommissural septal nuclei. Aspiration of the fimbria and supracommissural septal nuclei was verified visually, and the resulting gap was filled with Gelfoam soaked in sterile PBS. Unilateral (n = 4) or bilateral (n = 2) lesions of the pontine parabrachial nuclei were accomplished by passing a 1 mA current for 20 sec through a tungsten electrode placed 11.5 mm caudal to bregma, 1.9 mm lateral from the midline, and 7.6 mm down from the skull surface (with the electrode carrier angled 20° anterior). For both lesion paradigms, animals survived for 2 weeks after surgery and were then perfused under deep anesthesia. The precise location of all lesions and the extent of the damage was assessed histologically.
RESULTS
Immunostaining for BDNF revealed specific labeling throughout the full extent of the CNS that was confined to select populations of cells or fibers/terminals in well defined regions (for an overview, see Fig.1). The anatomical nomenclature used throughout this description was taken from Paxinos and Watson (1988). Results presented in the tables are a summary of data obtained by the microscopic observation (made independently by two different investigators) of histological slides from many animals, and they may differ slightly from what appears in any single photograph. BDNF cellular staining always appeared as a diffuse reaction product distributed throughout the perikaryal cytoplasm and occasionally extended into the most proximal processes, but it did not occupy the cell nucleus (Fig. 2A). The assessment of fiber/terminal staining was made on the basis of high-magnification examination of the tissue and was characterized as (1) coarse, with distinctly defined individual fibers (e.g., spinal cord and spinal trigeminal nucleus) (Fig. 2B), (2) diffuse and finely punctate with individual fibers not distinguishable (e.g., hypothalamus) (Fig. 2C), or (3) heavy punctate, in what appeared to be noded processes that formed pericellular baskets around unstained neuronal cell bodies (e.g., bed nucleus of the stria terminalis, lateral septum, central nucleus of amygdala) (Fig.2D). Throughout the CNS, the cellular localization of BDNF immunostaining and mRNA was restricted to cells possessing a neuronal morphology. In all cases, the specificity of the immunostaining was confirmed by omitting the primary antibody or through the use of preadsorption controls (Fig. 3).
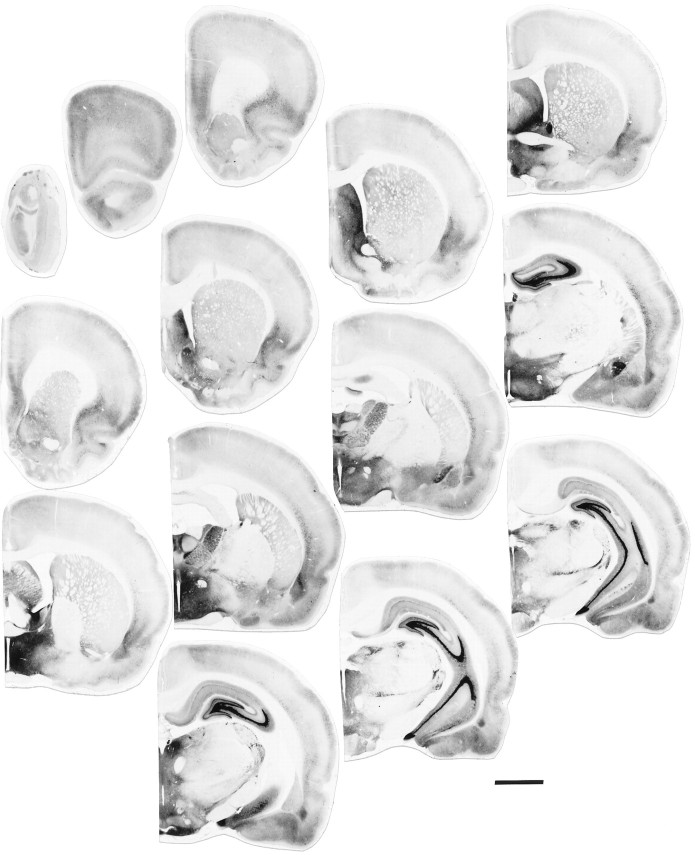
Fig. 1.
Distribution of BDNF-ir in the adult rat brain. Both plates show at low magnification a rostrocaudal series of sections (left to right) that were obtained from two female rats immunostained using affinity-purified antibodies specific for BDNF (bright-field illumination). BDNF immunostaining in male rats (not shown) was indistinguishable from that seen in females. Scale bar, 3 mm.
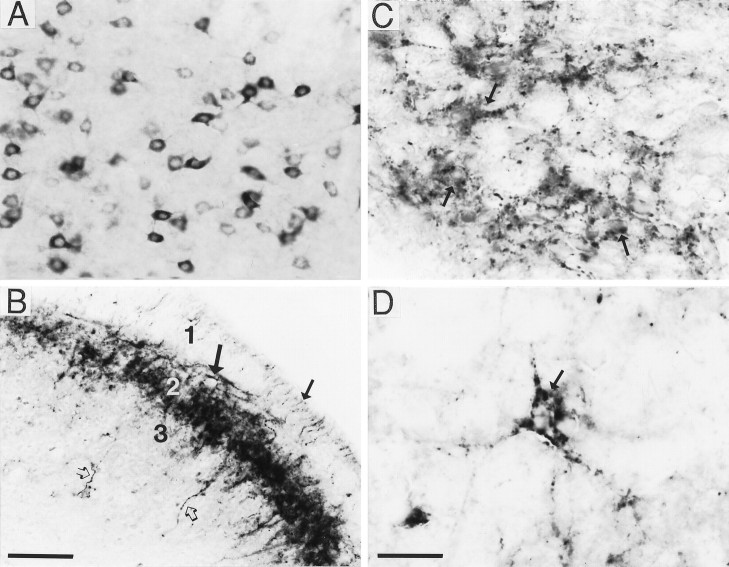
Fig. 2.
Characteristic patterns of BDNF immunostaining in the rat CNS. Panels show examples of BDNF immunostaining in basolateral amygdaloid nucleus (A), spinal cord (B), hypothalamus (C), and lateral septum (D) that are representative of the quality of immunolabeling seen throughout the CNS (bright-field illumination). As shown inA, perikaryal labeling was characterized by a diffuse reaction product distributed throughout the cell cytoplasm but excluding the cell nucleus. B–D, Various types of fiber/terminal staining. As seen in spinal cord lamina 1 (B, small arrow) and laminae 3 (B, open arrows), numerous individual fibers were immunolabeled; immunoreactive fibers were also concentrated in lamina 2 (B, large arrow). Other fields contained punctate immunostaining that appeared either diffuse or clustered around unlabeled cell bodies (C, arrows), or well defined, with discrete pericellular baskets around unlabeled cells (D, arrow). Scale bars (shown inB): A, B, 100 μm; (shown inD) C, D, 50 μm.
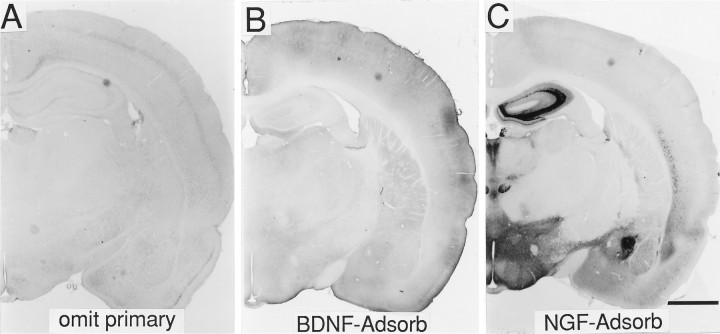
Fig. 3.
Specificity of BDNF immunostaining.A–C, Tissue sections processed for immunohistochemistry with either the BDNF antibody omitted (A), the BDNF antibody solution preadsorbed against purified recombinant human BDNF (B), or the BDNF antibody solution preadsorbed against mouse β-NGF (C) (bright-field illumination). As seen in A and B, when the BDNF antibody was omitted or preadsorbed with BDNF protein, no specific staining was present. In C, preadsorbing against mouse β-NGF revealed a pattern of immunostaining that was identical to sections reacted with the BDNF antibody alone (see Fig. 1). Scale bar, 2 mm.
In the following paragraphs the distributions of both BDNF-ir and BDNF cRNA hybridization throughout the CNS will be described. Although tissue from separate sets of rats was processed for the two techniques, these data will be presented together for the sake of comparison within a field.
The distribution of BDNF protein and mRNA in the olfactory bulb, cortex, and hippocampal formation is summarized in Table1. In the main olfactory bulb, a few periglomerular cells were lightly labeled for both BDNF mRNA and protein. In the mitral cell layer, a few cells were lightly labeled for BDNF-ir but not for mRNA, and in the granule cell layer, a faint haze of autoradiographic grains, indicative of low mRNA content, was distributed over the entire region, but no BDNF-ir was detected. In contrast, granule cells in the accessory olfactory bulb were lightly immunostained but not labeled by the cRNA. In the anterior olfactory nucleus (Fig. 4A–C), all subregions contained numerous cells that were well labeled for BDNF mRNA and moderately labeled for BDNF-ir. Moderate BDNF immunostaining and hybridization were detected in nearly all cells within the rostral portion of the tenia tecta but were nearly absent from more caudal sections. In contrast, many cells containing BDNF-ir and BDNF mRNA were distributed within the intermediate lateral septal nucleus adjacent to the tenia tecta at this level (Fig. 4D–F). Throughout the rostrocaudal extent of the claustrum and endopiriform nucleus, many cells were heavily labeled for BDNF mRNA and moderately heavily labeled for BDNF-ir (Fig. 4G,H).
Table 1.
BDNF in the olfactory bulb, cortex, and hippocampal formation
| BDNF-ir fibers | BDNF-ir cells | BDNF mRNA | |
|---|---|---|---|
| Mitral cell layer | − | 2/l | 0 |
| Internal granule layer | − | 0 | 4/l |
| Glomerular layer (periglomerular) | − | 1/l | 2/l |
| External plexiform/tufted cells | − | 0 | 0 |
| Granule cell layer of AOB | ± | 2/l–m | 0 |
| Anterior olfactory nucleus | + | 3/m | 3–4/h |
| Orbital cortex | − | 3/l | 2–3/m |
| Insular cortex (granular + agranular) | − | 3/l–m | 3/m–h |
| Frontal cortex | − | 3/m | 3/m |
| Cingulate cortex | − | 3/l–m | 3–4/h |
| Piriform cortex | − | 3/m | 3–4/h |
| Parietal cortex | − | 3/l | 2–3/m |
| Perirhinal cortex | − | 3/l–m | 3/m–h |
| Retrosplenial cortex | − | 3/l | 3/m |
| Occipital cortex | − | 2–3/l | 2–3/m |
| Temporal cortex | − | 3/l | 2–3/m |
| Entorhinal cortex | − | 3/m–h | 3/m–h |
| Claustrum | ± | 3–4/h | 3–4/h |
| Endopiriform nucleus (dorsal/ventral) | − | 2–3/m | 3/h |
| Tenia tecta (rostral part) | − | 3/m | 4/h |
| Tenia tecta (caudal part) | − | 0 | 4/l |
| Indusium griseum | − | 2/l–m | 1/m |
| CA1-stratum oriens | ++ | 1/l | 0 |
| CA1-stratum pyramidale | − | 2/l–m | 3–4/m |
| CA1-stratum radiatum | ++ | 0 | 0 |
| CA1-stratum lacunosum moleculare | ++ | 0 | 0 |
| CA2-stratum oriens | ++ | 1/m | 0 |
| CA2-stratum pyramidale | − | 3/l–m | 4/h |
| CA2-stratum lucidum | +++++ | 0 | 0 |
| CA3-stratum oriens | ++ | 1/m | 0 |
| CA3-stratum pyramidale | − | 4/m–h | 4/h |
| CA3-stratum lucidum | +++++ | 0 | 0 |
| Dentate gyrus-polymorph layer/hilus | +++++ | 1-160 | 1–2/h |
| Dentate gyrus-granule cell layer | − | 4/l–m | 4/m |
| Dentate gyrus-inner molecular layer | ++++ | 0–1/m | 0 |
| Dentate gyrus-middle molecular layer | + | 0 | 0 |
| Dentate gyrus-outer molecular layer | − | 0 | 0 |
| Subiculum | ± | 2/m | 3–4/m |
| Presubiculum | − | 0 | 2/l |
| Parasubiculum | − | 0 | 2/l |
Fibers: −, none; +, very light staining; ++, light staining; +++, moderate staining; ++++, heavy staining; +++++, extremely heavy staining. Cells: 0, no cells observed; 1, occasional cells; 2, few scattered cells; 3, moderate number of cells; 4, densely packed cells;
F1-160: , could not be determined. l, Lightly stained; m, moderately stained; h, heavily stained.
Fig. 4.
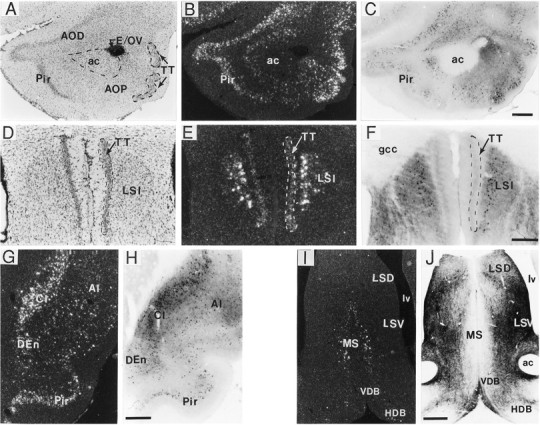
BDNF-ir and mRNA in forebrain regions.A–J, Sections through the anterior olfactory nucleus (A–C), caudal portion of the tenia tecta (D–F), claustrum and piriform cortex (G, H), and septal region (I, J) processed for immunohistochemistry (C, F, H, J; bright field) and in situ hybridization (B, E, G, I; dark field). Photomicrographs inA and D are of Nissl-stained tissue. Patterns of BDNF-ir and cRNA labeling were very similar in the anterior olfactory nucleus and rostral tenia tecta (TT) (A–C) and, more caudally, in the claustrum (Cl), endopiriform nucleus (DEn), and piriform cortex (Pir) (G, H). In the caudal portion of the tenia tecta, very light hybridization but no immunostaining were observed (D–F); notably heavy hybridization and immunostaining were present in the adjacent intermediate lateral septum (LSI) (D–F). In septal regions, cRNA-labeled cells were seen only in the medial septum (MS), whereas intense BDNF-ir was localized mostly to fibers/terminals in the lateral septum (I, J). Scale bars (shown inC): A–C, 500 μm; (shown inF): D–F, 200 μm; (shown inH): G, H, 500 μm; (shown inJ): I, J, 500 μm.ac, Anterior commissure; AI, agranular insular cortex; AOD, anterior olfactory nucleus, dorsal part; AOP, anterior olfactory nucleus, posterior part;E/OV, ependymal layer/olfactory ventricle;gcc, genu of the corpus callosum; HDB, horizontal limb of the diagonal band; LSD, lateral septum, dorsal part; LSV, lateral septum, ventral part;lv, lateral ventricle; VDB, vertical limb of the diagonal band.
In cortex, BDNF-ir and cRNA-labeled cells were distributed throughout all rostrocaudal levels with both hybridization and immunostaining densities varying markedly across cortical regions (Fig.5, Table 1). In nearly all cortical regions, immuno- and hybridization-labeling were observed in layers II, III, V, and VI, with only a few lightly labeled cells in layer IV. The most robust neocortical labeling was often seen deep in layer VI adjacent to the corpus callosum, especially in parietal cortex (Fig. 5A–C). By comparison, fewer labeled cells were present in layer VI of temporal cortex (Fig. 5D–F). Within a given field, the laminar patterns of hybridization and immunolabeling were generally the same, as shown for entorhinal cortex in Figure 5G–I. Layer I of all cortical regions was completely lacking BDNF immunostaining or cRNA hybridization.
Fig. 5.
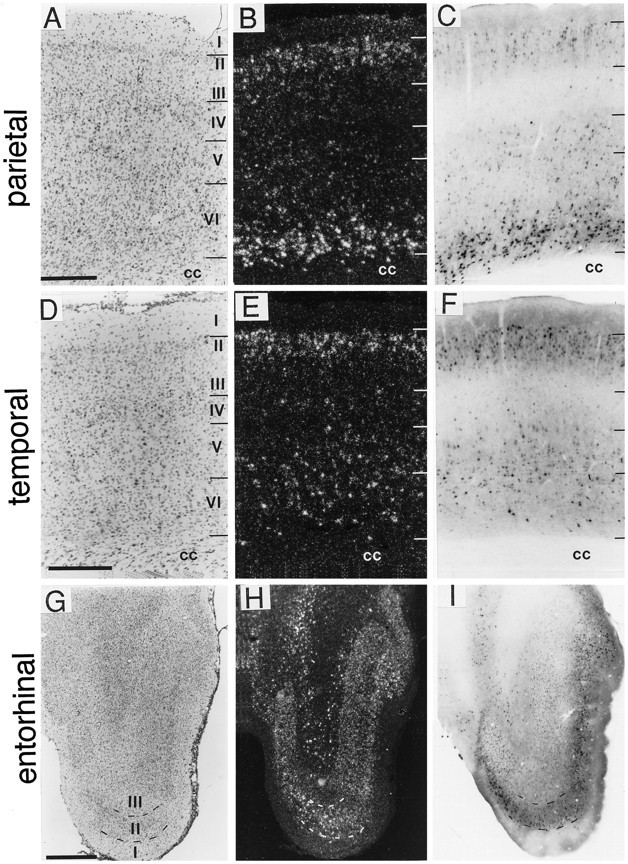
Comparison of the distributions of BDNF-ir and mRNA in cortical fields. Sections through parietal (A–C), temporal (D–F), and entorhinal (G–I) cortices were processed forin situ hybridization to localize BDNF mRNA (B, E, H; dark field) or immunohistochemistry to localize BDNF-ir (C, F, I; bright field). Sections through similar planes were Nissl-stained (A, D, G; bright field). In parietal cortex (A–C), the BDNF cRNA heavily labeled neurons in layer VI, moderately in layersII/III, and lightly in layer V; BDNF-ir was very heavy in deep layer VI, lighter in layersII, III, V, and superficial layer VI, and almost undetectable in layer IV. In temporal cortex (D–F), BDNF mRNA was moderately labeled in layers II/III and V/VI, with lighter labeling in layer IV. BDNF-ir in temporal cortex was heaviest in layers II/III, moderate in layersV/VI, and light in layer IV. In entorhinal cortex (G–I), the patterns of BDNF cRNA hybridization and immunoreactivity were similar. Scale bars (shown in A): A–C, 500 μm; (shown inD): D–F, 500 μm; (shown inG): G–I, 1 mm. cc, Corpus callosum.
In the hippocampus, BDNF immunostaining was detected in both cell bodies and fibers/terminals (Fig. 6). In the pyramidal cell layer, the intensity of cRNA hybridization and immunostaining and the number of cells labeled varied across subfields (Fig.6B,C). In CA1, a few scattered cells were cRNA-labeled with a moderate density of autoradiographic grains, and a few cells had detectable immunoreactivity. In CA2, most cells had moderate to heavy cRNA labeling and light to moderate levels of immunostaining. In CA3, both cRNA labeling and BDNF-ir were relatively dense in nearly all cells (although it was often difficult to distinguish immunolabeled CA3 pyramidal cells because of the intense BDNF immunostaining within the mossy fiber zone). Only an occasional immunolabeled cell was detected in stratum oriens of CA1–CA3, and no mRNA-positive cells were observed in this layer. Immunoreactive fibers were distributed throughout regions CA1–CA3. As illustrated for region CA1 in Figure 6D, the density of fiber staining varied across the hippocampal laminae, with greater immunostaining in strata oriens and radiatum and less BDNF-ir in strata lacunosum moleculare and pyramidale. In the dentate gyrus, nearly all cells in stratum granulosum were mRNA-positive and BDNF-immunoreactive (Fig.6B,C,E). In the dentate molecular layer, a distinctive laminar pattern of BDNF-ir was seen (Fig.6E). Immunostaining was moderate in the inner molecular layer, light in the middle molecular layer, and not detectable in the outer molecular layer. In the hilus, some cells were heavily labeled by the cRNA, and a few immunoreactive perikarya were visible; however, the dense immunohistochemical labeling of fibers in the deeper hilus prevented identification of immunoreactive cells in this field (Fig. 6C). The field of fiber labeling in the deep hilus was the most dense and well stained in the entire CNS and corresponded to the location of the mossy fiber axons of the granule cells.
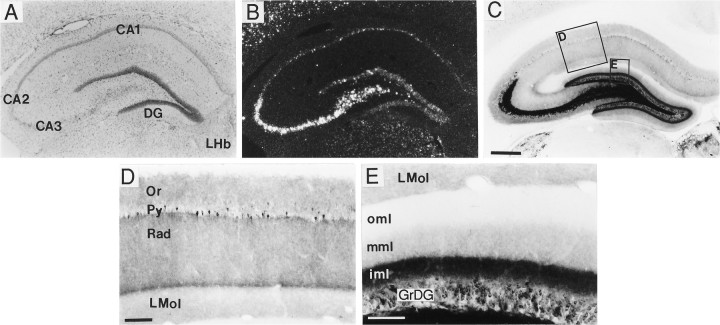
Fig. 6.
Distribution of BDNF-ir and mRNA in hippocampus. Low-magnification (A–C) and high-magnification (D, E) photomicrographs of sections through hippocampus that were either Nissl-stained (A, bright field) or processed for in situ hybridization (B, dark field) or immunohistochemistry (C–E, bright field). In hippocampus, the BDNF cRNA densely labeled pyramidal cells in CA3, CA2, and the hilus, and less densely the CA1 pyramidal cells and the dentate gyrus granule cells (GrDG) (B). BDNF immunostaining densely labeled processes within the mossy fiber system (C). At higher magnification, one could see that in region CA1 BDNF immunostaining was diffuse in strata oriens (Or) and radiatum (Rad) and weaker in stratum lacunosum moleculare (LMol) (D). Scattered neurons in the CA1 pyramidal cell layer (Py) were immunolabeled (D). In the dentate gyrus (DG) molecular layer, a trilaminar pattern of BDNF-ir was seen, with moderately dense labeling in the inner molecular layer (iml), light labeling in the middle molecular layer (mml), and no detectable staining in the outer molecular layer (oml). Scale bars (shown in C):A–C, 500 μm; (shown in D): D, E, 100 μm and 75 μm, respectively. LHb, Lateral habenular nucleus.
The distributions of BDNF-ir and mRNA within the basal forebrain and basal ganglia are summarized in Table 2. In the basal forebrain, many densely immunostained and BDNF mRNA-positive cells were distributed in the rostral part of the intermediate lateral septum (Fig. 4D–F). In other basal forebrain regions, such as medial septal nucleus (Fig. 4I), vertical limb of the diagonal band (Fig. 4I), magnocellular preoptic nucleus, medial preoptic area, and septohypothalamic nucleus, there were a few BDNF mRNA-positive cells. In these regions, only an occasional BDNF-immunoreactive cell was seen (Fig. 4J). In addition, many lightly immunostained cells were observed in the triangular septal nucleus and the septofimbrial nucleus (not shown). Immunoreactive fibers also were detected in many basal forebrain regions. In the dorsal and ventral portions of the lateral septum, fibers were well labeled and often appeared to form pericellular baskets around unstained perikarya (Figs.2D, 4J). In contrast, the medial septum was relatively devoid of fiber staining (Fig.4J). Moderate BDNF-immunoreactive fiber labeling also was observed in the septohypothalamic nucleus and the subfornical organ. In the basal ganglia, no BDNF mRNA and little BDNF-ir was detected in either cells or fibers. Single, well labeled BDNF-immunoreactive cells with a neuronal morphology were only occasionally observed in the dorsal most aspect of the striatum and in the adjacent corpus callosum.
Table 2.
BDNF in the basal forebrain, amygdala, and basal ganglia
| BDNF-ir fibers | BDNF-ir cells | BDNF mRNA | |
|---|---|---|---|
| Lateral septal nucleus, intermediate | ++ | 3 /m–h | 3/m–h |
| Lateral septal nucleus, dorsal/ventral | +++ | 0 | 0 |
| Medial septal nucleus | + | 1/m | 2–3/l–m |
| Diagonal band nucleus, horizontal limb | +/++ | 1/m | 0 |
| Diagonal band nucleus, vertical limb | +/++ | 1/m | 2/l–m |
| Nucleus basalis of Meynert | ++ | 0 | 0 |
| Ventral pallidum | − | 0 | 0 |
| Islands of Calleja | − | 0 | 0 |
| Lateral preoptic nucleus | ++ | 1/l | 0 |
| Magnocellular preoptic nucleus | − | 1/l | 2/l |
| Medial preoptic area | ++ | 1/l | 2–3/m |
| Medial preoptic nucleus | +++ | 2/h | 2–3/m |
| Substantia inominata | + | 1/l–m | 0 |
| Bed nucleus of the stria terminalis, medial/lateral/ventral/intermediate | ++/+++ | 0 | 0 |
| Bed nucleus of the stria terminalis, juxtacapsular/lateral dorsal | ++++ | 0 | 0 |
| Septohypothalamic nucleus | +++ | 1/m | 2/m |
| Olfactory tubercle | + | 0 | 0 |
| Optic chiasm | − | 0 | 0 |
| Suprachiasmatic nucleus | + | 0 | 0 |
| Triangular septal nucleus | ± | 3/l | 0 |
| Septofimbrial nucleus | ++ | 3/l | 3–4/m |
| Subfornical organ | ++/+++ | 0 | 0 |
| Corpus callosum | − | 1/m | 0 |
| Caudate putamen | ± | 1/m | 0 |
| Globus pallidus | − | 0 | 0 |
| Accumbens nucleus | + | 0 | 0 |
| Nucleus of the lateral olfactory tract, part 1 | − | 0 | 0 |
| Nucleus of the lateral olfactory tract, part 2 | − | 2–3/l | 3/l |
| Nucleus of the lateral olfactory tract, part 3 | − | 2–3/m | 2/m–h |
| Central amygdaloid nucleus, lateral | ++++ | 0 | 0 |
| Central amygdaloid nuclues, medial | ++ | 0 | 0 |
| Basolateral amygdaloid nucleus, anterior | − | 3/h | 3–4/h |
| Basolateral amygdaloid nucleus, ventral | − | 2/m | 2/m–h |
| Basolateral amygdaloid nucleus, posterior | − | 2/m–h | 3/h |
| Medial amygdaloid nucleus, ventral | + | 1/l–m | 3/m–h |
| Cortical amygdaloid nucleus | + | 2/l–m | 3/m |
| Basomedial amygdaloid nucleus | + | 1/l–m | 3/l–m |
| Lateral amygdaloid nucleus | − | 1/l–m | 2/m–h |
| Anterior amygdaloid area | + | 1/m | 1/l |
| Amygdalohippocampal area | + | 2/m | 2–3/m–h |
| Amygdalostriatal transition area | ++ | 0 | 0 |
Fibers: −, none; +, very light staining; ++, light staining; +++, moderate staining; ++++, heavy staining; +++++, extremely heavy staining. Cells: 0, no cells observed; 1, occasional cells; 2, few scattered cells; 3, moderate number of cells; 4, densely packed cells;
F2-160: , could not be determined. l, Lightly stained; m, moderately stained; h, heavily stained.
As seen in Table 3, BDNF-ir and mRNA were found in many areas of the thalamus and mesencephalon, with distinctions between regions containing labeled cells and immunoreactive fibers. For example, the parafascicular thalamic nucleus (Fig. 7G,H) contained numerous mRNA-positive and immunoreactive perikarya but no immunostained fibers. In contrast, in the anteroventral and anteromedial nuclei there was moderate fiber immunostaining, but there was almost no cellular hybridization or immunoreactivity (Fig. 7A,B). Within the dorsal and ventral lateral geniculate nuclei many immunoreactive fibers were present, but immunoreactive or cRNA-labeled cell bodies were not (Fig.8I,J). In some fields the distributions of hybridization and immunolabeling were similar. For example, in the medial geniculate, the dorsal and ventral portions were devoid of immunolabeling and hybridization, whereas the medial portion of this nucleus and the suprageniculate nucleus contained moderate perikaryal hybridization and immunolabeling (Fig.7M,N). In the superior colliculus, cRNA hybridization and immunoreactivity also overlapped in layers deep to the superficial gray layer (Fig. 7I,J).
Table 3.
BDNF in the thalamus/mesencephalon
| BDNF-ir fibers | BDNF-ir cells | BDNF mRNA | |
|---|---|---|---|
| Paratenial thalamic nucleus | + | 0 | 0 |
| Paraventricular thalamic nucleus | ++ | 1–2/l–m | 3/m |
| Central medial thalamic nucleus | ++ | 3/l–m | 3/l–m |
| Central lateral thalamic nucleus | ± | 1–2/l | 2/l |
| Intermediodorsal thalamic nucleus | ++ | 3/m | 3/l–m |
| Interanterodorsal/interanteromedial thalamic nucleus | ++ | 0 | Light haze |
| Anterodorsal thalamic nucleus | ± | 3–4/l | 3–4/l |
| Anteroventral thalamic nucleus | ++/+++ | 0 | 0 |
| Anteromedial thalamic nucleus | ++ | - | Light haze |
| Mediodorsal thalamic nucleus | + | 1/l–m | 0 |
| Laterodorsal thalamic nucleus, dorsomedial | + | 0 | 2/l |
| Laterodorsal thalamic nucleus, ventrolateral | + | 0 | 0 |
| Ventrolateral/ventromedial thalamic nucleus | − | 0 | 0 |
| Ventral posterior/posterior | − | 0 | 0 |
| Reticular thalamic nucleus | − | 0 | 0 |
| Reuniens thalamic nucleus | ++ | 0 | 2/m |
| Rhomboid thalamic nucleus | + | 2/l–m | 2/m |
| Zona incerta | +/++ | 0 | 0 |
| Parafascicular thalamic nucleus | − | 4/l | 3/m |
| Posterior intralaminar thalamic nucleus | ++ | 2/m | 2/m |
| Lateral posterior thalamic nucleus, mediocaudal | ++ | 0 | 0 |
| Lateral geniculate nucleus (dorsal/ventral) | ++/+++ | 0 | 0 |
| Medial geniculate nucleus (dorsal/ventral) | − | 0 | 0 |
| Medial geniculate nucleus, medial | + | 3/m | 3/m |
| Suprageniculate thalamic nucleus | + | 3/m | 3/m |
| Subthalamic nucleus | − | 3–4/l | 3/l |
| Peripeduncular nucleus | ++ | 2/m | 2–3/m |
| Medial pretectal area | ++ | 2/m–h | 2–3/m–h |
| Parabigeminal nucleus | − | 3/m | 3/l–m |
| Ventral tegmental area | +/++ | 2/l | 3/m–h |
| Substantia nigra, compacta | +/++ | 2/l | 2–3/m–h |
| Substantia nigra, reticular | − | 0 | 0 |
| Substantia nigra, lateral | ++ | 2/m–h | 2/m–h |
| Nucleus of the posterior commissure | ++ | 1/m | 2/m |
| Retroethmoid nucleus | ++ | 0 | 0 |
| Medial accessory occulomoter nucleus | + | 1/m | 1/m |
| Superior colliculus, superficial gray layer | + | 0 | 0 |
| Superior colliculus, optic nerve layer | − | 0 | 3–4/l |
| Superior colliculus, intermediate and deep gray layer | + | 3/l | 0 |
| Precommissural nucleus | ++ | 0 | 2–3/l–m |
Fibers: −, none; +, very light staining; ++, light staining; +++, moderate staining; ++++, heavy staining; +++++, extremely heavy staining. Cells: 0, no cells observed; 1, occasional cells; 2, few scattered cells; 3, moderate number of cells; 4, densely packed cells;
F3-160: , could not be determined. l, Lightly stained; m, moderately stained; h, heavily stained.
Fig. 7.
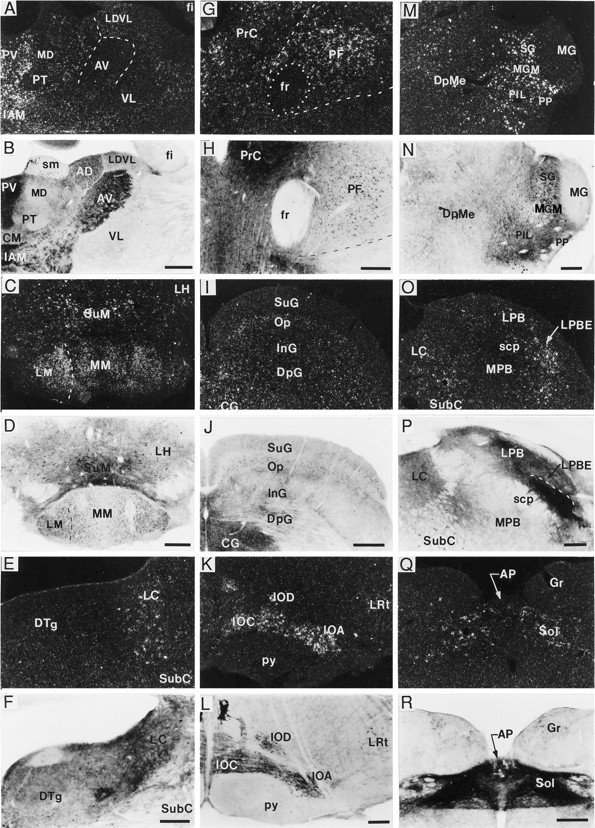
Localization of BDNF-ir and mRNA in thalamic and brainstem regions. Dark-field and bright-field photomicrographs showing BDNF cRNA hybridization (A, C, E, G, I, K, M, O, Q) and BDNF immunolabeling (B, D, F, H, J, L, N, P, R), respectively, in thalamus (A, B), mammillary region (C, D), locus coeruleus (E, F), parafascicular nucleus (PF) (G, H), superior colliculus (I, J), inferior olivary complex (K, L), medial geniculate nucleus (M, N), parabrachial region (O, P), and the nucleus of the solitary tract (Q, R). In many thalamic (A, B, G, H, M, N) and hypothalamic (C, D) areas, the distributions of BDNF mRNA and perikaryal BDNF-ir overlapped; although in some regions [e.g., anteroventral thalamic nucleus (AV)] there was heavy fiber/terminal BDNF-ir in the absence of hybridization (A, B). In other regions, such as the locus coeruleus (LC) (E, F) and nucleus of the solitary tract (Sol) (Q, R), cRNA-labeled cells could be detected, but it was difficult to identify immunostained cells because of the fiber/terminal immunolabeling. Weak labeling with both the cRNA and antibody also overlapped in most layers of superior colliculus, excluding the superficial gray layer, which did not contain cRNA labeling (I, J). In contrast, heavy hybridization and BDNF-ir were detected throughout the inferior olivary complex (K, L). Moderate to heavy BDNF cRNA labeling was found in the external portion of the lateral parabrachial nucleus (LPBE), whereas heavy fiber/terminal BDNF-ir was present in most subregions of the lateral parabrachial subfield (LPB) (O, P); neither cRNA nor antibody labeled the medial portion of the parabrachial nucleus (MPB) (O, P). Scale bars (shown inB, D, J, N): A–D, I, J, M, N, 500 μm, respectively; (shown in F): E, F, 200 μm; (shown in H, P, R): G, H, O, P–R, 250 μm, respectively. AP, Area postrema;CG, central (periaqueductal) gray; DpG, superior colliculus, deep gray layer; DpMe, deep mesencephalic nucleus; DTg, dorsal tegmental nucleus;fr, fasciculus retroflexus; Gr, gracile nucleus; IAM, interanteromedial thalamic nucleus;InG, superior colliculus, intermediate gray layer;IOA, inferior olive, subnucleus A; IOC, inferior olive, subnucleus C; IOD, inferior olive, dorsal nucleus; LDVL, laterodorsal thalamic nucleus, ventrolateral; LH, lateral hypothalamic area;LM, lateral mammillary nucleus; LRt, lateral reticular nucleus; MG, medial geniculate nucleus; MGM, medial geniculate nucleus, medial portion;MM, medial mammillary nucleus; Op, superior colliculus, optic nerve layer; PIL, posterior intralaminar nucleus; PrC, precommissural nucleus; PP, peripeduncular thalamic nucleus;PT, paratenial thalamic nucleus; PV, paraventricular thalamic nucleus; py, pyramidal tract;scp, superior cerebellar peduncle; SG, suprageniculate nucleus; SubC, subcoeruleus nucleus;SuG, superior colliculus, superficial gray layer;SuM, supramammillary nucleus; VL, ventrolateral thalamic nucleus.
Fig. 8.
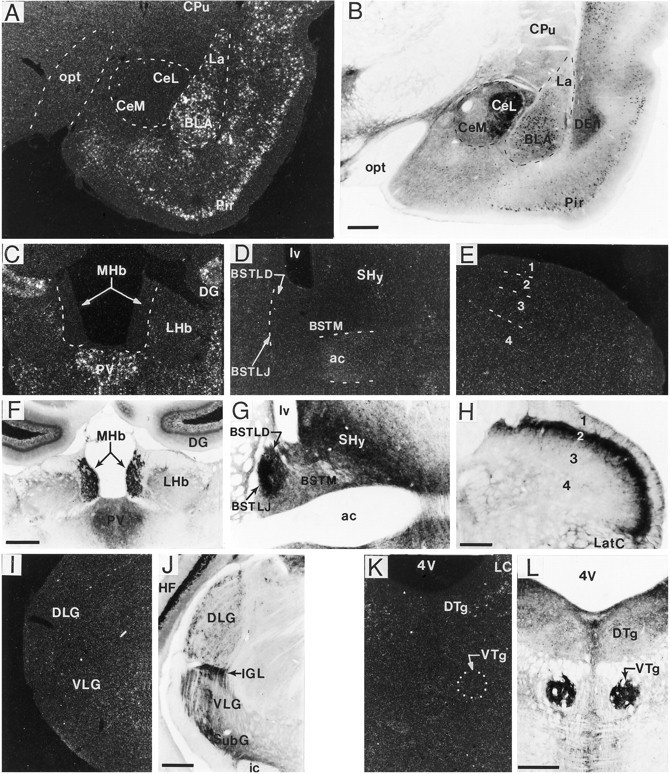
Dense fiber/terminal BDNF-ir was present in many regions lacking BDNF mRNA expression. A–L, Dark-field and bright-field photomicrographs of sections processed for in situ hybridization (A, C–E, I, K) and immunohistochemistry (B, F–H, J, L), respectively, through the amygdala (A, B), habenula (C, F), bed nucleus of stria terminalis (D, G), spinal cord (E, H), lateral geniculate nucleus (I, J), and dorsal/ventral tegmental nucleus (K, L). As seen in Aand B, in the basolateral (BLA) and lateral amygdala (La), many cells were heavily labeled for both mRNA and BDNF-ir, whereas in the central amygdala many immunolabeled fibers/terminals were present but cRNA hybridization was not. Similarly, in the medial habenular nucleus (MHb) (C, F) subregions of the bed nucleus of stria terminalis (D, G), the dorsal (DLG) and ventral regions (VLG) of the lateral geniculate nucleus (I, J), the intergeniculate leaf (IGL) (I, J), and ventral tegmental nucleus (VTg) (K, L), heavy fiber/terminal BDNF-ir was found without detectable mRNA. In spinal cord (cervical level shown), heavily immunostained fibers were present primarily in lamina 2 (H), which lacked BDNF mRNA (E). Scale bars (shown in B): A, B, 500 μm; (shown in F): C, D, F, G, 500 μm; (shown in H): E, H, 150 μm; (shown in J, L):I–L, 500 μm. ac, Anterior commissure; BSTLD, bed nucleus of stria terminalis, lateral dorsal; BSTLJ, bed nucleus of the stria terminalis, juxtacapsular; BSTM, bed nucleus of stria terminalis, medial; CeL, central amygdaloid nucleus, lateral division; CeM, central amygdaloid nucleus, medial division; CPu, caudate putamen (striatum); DEn, dorsal endopiriform nucleus;DG, dentate gyrus; DTg, dorsal tegmental nucleus; HF, hippocampal formation; ic, internal capsule; LatC, lateral cervical nucleus;LC, locus coeruleus; LHb, lateral habenular nucleus; lv, lateral ventricle;opt, optic tract; Pir, piriform cortex;PV, paraventricular thalamic nucleus;SHy, septohypothalamic nucleus; SubG, subgeniculate nucleus; 4V, 4th ventricle.
As illustrated in Figure 8A,B and summarized in Table2, striking patterns of BDNF mRNA and BDNF-ir were seen in the amygdaloid complex. The basolateral, medial, and basomedial amygdaloid nuclei contained many heavily cRNA-labeled and BDNF-immunoreactive cell bodies but very few immunoreactive fibers. In contrast, the central nucleus of the amygdala (especially the lateral subdivision) contained absolutely no BDNF mRNA or BDNF-immunoreactive cells but did contain high densities of immunolabeled fibers, which often formed pericellular baskets around unlabeled perikarya. Within the bed nucleus of the stria terminalis (Fig. 8D,G), heavy pericellular fiber labeling was observed in the lateral dorsal and juxtacapsular regions, whereas other aspects of this nucleus had moderate densities of immunoreactive fibers. BDNF mRNA was not detected in any portion of the bed nucleus of the stria terminalis.
In the hypothalamus, varying degrees of cellular labeling for BDNF-ir and mRNA were observed and a low to moderate level of immunoreactive fiber labeling was present in many areas (Table4, Fig. 1). A few lightly cRNA-labeled cells were scattered in the lateral and lateroanterior hypothalamic nuclei and extended into the tuber cinereum area. More heavily cRNA-labeled and immunoreactive cells were distributed in the ventromedial hypothalamic nucleus and in the posterior hypothalamic area. Within the paraventricular hypothalamic nucleus, numerous cells of the medial and ventral parvocellular region were well labeled with the cRNA, whereas cells in the lateral magnocellular region were not labeled. Within the mammillary and supramammillary region, many cells were positive for both mRNA and immunoreactivity (Fig. 7C,D). Interestingly, BDNF-immunoreactive fibers but no immunoreactive or cRNA-labeled cell bodies were observed in the median eminence and the infundibular stem. A similar pattern was also seen in the medial habenular nucleus, which contained a very high density of immunoreactive fibers but lacked detectable cellular labeling (immunoreactivity or hybridization) (Fig.8C,F).
Table 4.
BDNF in the hypothalamus and epithalamus
| BDNF-ir fibers | BDNF-ir cells | BDNF mRNA | |
|---|---|---|---|
| Medial habenular neucleus | +++++ | 0 | 0 |
| Lateral habenular nucleus | + | 1–2/l | Light haze |
| Anterior hypothalamic area, anterior | ++ | 1/l | 0 |
| Lateroanterior hypothalamic nucleus | ++ | 1/l | 2–3/l |
| Lateral hypothalamic area | ++ | 1/m | 2/l |
| Anterior hypothalamic area, central | ++ | 0 | 0 |
| Paraventricular hypothalamic nucleus (parvocellular) | ++/+++ | 1/m | 3/m |
| Paraventricular hypothalamic nucleus, lateral (magnocellular) | + | 0 | 0 |
| Medial tuberal nucleus | ++ | 1/l–m | 2–3/m |
| Tuber cinereum area | ++ | 1/l–m | 2–3/l–m |
| Ventromedial hypothalamic nucleus | ++ | 2/m–h | 3/h |
| Arcuate hypothalamic nucleus | +++ | 0 | 0 |
| Median eminance | +/++ | 0 | 0 |
| Dorsomedial hypothalamic nucleus, diffuse | ++ | 1/m | 3/l–m |
| Dorsal hypothalamic area | ++ | 1/m | 1/l |
| Perifornical nucleus | ++ | 0 | 1/m |
| Premammillary nucleus | ++ | 0 | 2–3/m–h |
| Supramammillary nucleus | ++ | 2/m | 3/h |
| Medial mammillary nucleus | − | 4/l–m | 3–4/m–h |
| Lateral mammillary nucleus | − | 4/l | 3–4/m |
| Interpeduncular nucleus | + | 0 | 0 |
| Posterior hypothalamic area | ++ | 2/l–m | 3/m |
| Infundibular stem | +/++ | 0 | 0 |
Fibers: −, none; +, very light staining; ++, light staining; +++, moderate staining; ++++, heavy staining; +++++, extremely heavy staining. Cells: 0, no cells observed; 1, occasional cells; 2, few scattered cells; 3, moderate number of cells; 4, densely packed cells;
F4-160: , could not be determined. l, Lightly stained; m, moderately stained; h, heavily stained.
In the brainstem, many BDNF cRNA-labeled cells were observed, and with a few exceptions the distribution of these cells strongly correlated with the distribution of immunoreactive cell bodies (Table5). Some notable exceptions were the lateral parabrachial nucleus (Fig. 7O,P), locus coeruleus (Fig.7E,F), and nucleus of the solitary tract (Fig.7Q,R), which contained densely cRNA-labeled cells but no detectable immunoreactive perikarya. Fiber immunostaining was present in all three regions, although in the lateral parabrachial nucleus (especially within the external region) and nucleus of the solitary tract, the high density and unique characteristics of the fiber staining may have obscured immunoreactive cell bodies. Conversely, in some brainstem nuclei, such as spinal vestibular nucleus, central nucleus of the inferior colliculus, and gigantocellular reticular nucleus, cell bodies were lightly immunostained but BDNF mRNA was not detected. Very heavy fiber immunolabeling was noted in many brainstem regions, such as the lateral parabrachial nucleus, nucleus of the solitary tract (Fig. 7R), area postrema, inferior olive (Fig. 7L), and ventral tegmental nucleus (Fig.8L). As seen in Table 5, no cRNA hybridization or immunostaining was detected in the cerebellum.
Table 5.
BDNF in the brainstem, cerebellum, and spinal cord
| BDNF-ir fibers | BDNF-ir cells | BDNF mRNA | |
|---|---|---|---|
| Central gray | +++ | l/m | 3/m |
| Dorsal raphe nucleus | ++ | 1/l | 0 |
| Deep mesencephalic nucleus | + | 2/m | 2/l |
| Pedunculopontine tegmental nucleus | − | 3/m | 3/m |
| Pontine nuclei | − | 4/l–m | 4/l–m |
| Spinal vestibular nucleus | − | 2/m | 0 |
| Ventral tegmental nucleus | ++++ | 0 | 0 |
| Cuniform nucleus | ++ | 1/m | 3/m |
| Peritrigeminal zone | + | 2/m | 0 |
| Laterodorsal tegmental nucleus | + | 0 | 0 |
| Laterodorsal tegmental nucleus, ventral | + | 0 | 0 |
| Dorsal tegmental nucleus, central/pericentral | + | 0 | 0 |
| Periolivary nucleus, medioventral/lateroventral | +++ | 0 | 0 |
| Inferior colliculus, nucleus brachium | + | 2/l | 1–2/l |
| Inferior colliculus, central nucleus | + | 2–3/l | 0 |
| Inferior colliculus, dorsal cortex | + | 1–2/l | 0 |
| Inferior colliculus, external cortex | − | 3/l–m | 2/l |
| Locus coeruleus | ++ | 0 | 3/l–m |
| Subcoeruleus nucleus | + | 2–3/m | 2–3/l–m |
| Lateral parabrachial nucleus, internal/dorsal/central | +++ | 0 | 3/m |
| Lateral parabrachial nucleus, external | ++++ | 0 | 3/h |
| Medial parabrachial nucleus | ± | 0 | 0 |
| Principal sensory trigeminal nucleus, ventrolateral | − | 2/m | 3/m |
| Nucleus of the solitary tract | ++++ | 1/m | 3/m–h |
| Cuneate/external cuneate nucleus | − | 2–3/l | 2–3/l |
| Gracile nucleus | + | 0 | 0 |
| Spinal trigeminal nucleus, interpolar and caudal | +++(cap) | 2/l | 1/m |
| Lateral reticular nucleus | ++ | 0 | 2–3/l–m |
| Inferior olive | +++/++++ | 3/m | 3/m–h |
| Dorsal paragigantocellular nucleus | − | 3/m | 2–3/m |
| Gigantocellular reticular nucleus | + | 2/l | 0 |
| Ambiguus nucleus | − | 0 | 0 |
| Area postrema | +++ | 3/m | 0 |
| Cerebellum | − | 0 | 0 |
| Lamina 1 spinal cord | + | 0 | 0 |
| Lamina 2 | +++ | 0 | 0 |
| Lamina 3 | + | 2/l | 0 |
| Lamina 4/5 | − | 0 | 0 |
| Lamina 6 | − | 1/l | 0 |
| Lamina 7 | − | 1/l | 1/l |
| Lamina 8/9 | − | 0 | 0 |
| Lateral cervical nucleus | + | 0 | 0 |
Fibers: −, none; +, very light staining; ++, light staining; +++, moderate staining; ++++, heavy staining; +++++, extremely heavy staining. Cells: 0, no cells observed; 1, occasional cells; 2, few scattered cells; 3, moderate number of cells; 4, densely packed cells;
F5-160: , could not be determined. l, Lightly stained; m, moderately stained; h, heavily stained.
In the spinal cord, fiber labeling was observed in lamina 1–3 and was especially heavy in lamina 2 (Table 5, Fig.8E,H). In addition, occasional BDNF-immunoreactive cells were scattered in lamina 3, 6, and 7, and occasional hybridized cells were present in lamina 7.
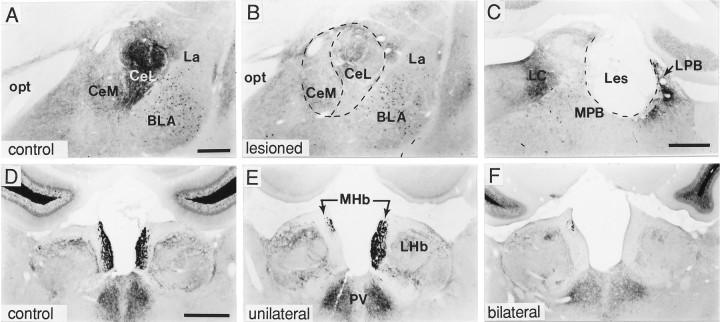
Although axonal BDNF-ir could arise via anterograde or retrograde transport, the presence of immunostaining in what appeared to be terminal arbors (i.e., noded processes) in fields lacking BDNF mRNA (Fig. 8) suggested that BDNF within these regions was likely derived from anterograde axonal transport by afferent systems. To test this hypothesis, we selected two regions where this mismatch was particularly striking and where adequate knowledge of afferent systems had been documented: namely, the central nucleus of the amygdala/bed nucleus of the stria terminalis and the medial habenular nucleus. Afferents to the central amygdala and juxtacapsular and lateral dorsal portion of the bed nucleus are known to arise from the pontine parabrachial nucleus (Bernard et al., 1993; Alden et al., 1994). Specific ablation of the parabrachial nucleus (either unilaterally or bilaterally) (Fig. 9C) resulted in nearly a complete loss of BDNF-immunoreactive fibers in the central amygdala (Fig. 9B) and juxtacapsular and lateral dorsal portions of the bed nucleus (not shown) on the side ipsilateral to the lesion. Similarly, selective destruction of the supracommissural septal neurons that provide afferent innervation to the medial habenular nucleus (Sutherland, 1982) resulted in the loss of nearly all BDNF-ir within the ipsilateral medial habenular nucleus (Fig.9D–F). In these animals, special care was taken to avoid damage to more ventral fields, including the stria medularis where these and other afferents to the medial habenulae course (Sutherland, 1982).
Fig. 9.
Anterograde axonal transport of BDNF by afferent systems. Bright-field photomicrographs showing sections processed for BDNF immunohistochemistry through amygdala (A, B), parabrachial region (C), and habenula (D–F) in control rats (A, D) or rats that received either an electrolytic lesion of the lateral parabrachial nucleus (C) or an aspirative lesion of the septofimbrial and triangular nuclei (E, F; lesion site is not shown). As seen in A–C, BDNF immunolabeling in the central nucleus (Ce) of the amygdala (A) is almost completely eliminated (B) after ablation of cell bodies in the lateral parabrachial nucleus (LPB) (C); in C, the lesion destroyed much of the LPB but spared the medial parabrachial nucleus (MPB). D–F show that BDNF-ir in the medial habenular nucleus (MHb) (D) is lost ipsilateral to an unilateral septofimbrial/triangular septal nuclei lesion (E) or bilaterally after bilateral lesions of these fields (F). Scale bars (shown inA): A, B, 500 μm; C, 500 μm; (shown in D): D–F, 750 μm.BLA, Basolateral amygdala; CeL, central amygdaloid nucleus, lateral division; CeM, central amygdaloid nucleus, medial division; La, lateral amygdala; LC, locus coeruleus; LHb, lateral habenular nucleus; opt, optic tract;PV, paraventricular thalamic nucleus.
DISCUSSION
In recent years, several investigators have documented sites of neurotrophin synthesis using in situ hybridization techniques. Studies aimed at localizing neurotrophin proteins within the CNS by immunohistochemical methods are less prevalent. Although it may be perceived initially that the localization of a protein will yield information redundant to the identification of its mRNA, this is not necessarily the case. As was observed in investigations of NGF immunoreactivity in the rat CNS (Conner and Varon, 1992; Conner et al., 1992), cells synthesizing NGF did not accumulate enough antigen under normal circumstances to permit detection by immunohistochemical methods (although pretreatment with colchicine, to force the accumulation of normally exported proteins within the cell body, did permit NGF-producing cells to be detected). Moreover, some cells that donot produce NGF, such as the basal forebrain cholinergic neurons (Lauterborn et al., 1995), do contain NGF immunoreactivity under normal circumstances (Conner et al., 1992). This immunoreactivity is presumed to originate from the retrograde transport and accumulation of NGF from distant sites of synthesis; this is consistent with the observation that accumulation is reduced after colchicine treatment. Finally, NGF protein has been identified in locations other than within cell bodies (i.e., the hippocampal mossy fiber region), indicating that the site where a protein is stored need not be within the cell body where synthesis occurs. Accumulation of a protein at extrasomal sites may result from either its anterograde transport down axonal or dendritic arbors of the producing cell, or by the selective binding or uptake and accumulation of protein released into the extracellular space.
In the current study, BDNF protein was localized in two basic compartments: within neuronal cell bodies and within fibers or axon terminals. Immunostaining for BDNF within cell bodies was characterized by a diffuse cytoplasmic label and was associated almost exclusively with cell populations containing BDNF mRNA, suggesting that this pool of BDNF-ir represents protein retained by cells producing BDNF. We did not observe any neuronal populations with robust, punctate BDNF immunostaining similar to what was seen in the case of NGF immunostaining of basal forebrain cholinergic neurons (Conner et al., 1992). This suggests that in the CNS, neuronal perikarya do not accumulate endogenous BDNF by way of retrograde transport as do the basal forebrain neurons with NGF, or if they do accumulate BDNF, they do not store it in sufficient quantities to permit detection of these punctate bodies. Alternatively, the ultrastructural localization of retrogradely accumulated BDNF (diffuse) may be different from NGF (punctate).
The distribution of BDNF-producing cells observed in the present study is consistent, for the most part, with the cumulative findings of others (Ernfors et al., 1990; Phillips et al., 1990; Ceccatelli et al., 1991; Friedman et al., 1991; Guthrie and Gall, 1991; Gall et al., 1992;Castrén et al., 1995). A few distinctions between the current and previous studies include (1) our inability to confirm BDNF expression (hybridization or immunolabeling) by rat cerebellar granule cells (Castrén et al., 1995); (2) the present lack of BDNF cRNA labeling within magnocellular neurons of the hypothalamic paraventricular nucleus (Castrén et al., 1995), although hybridization and immunostaining did label neurons in the parvocellular region of the hypothalamic paraventricular nucleus; and (3) our inability to detect BDNF-ir or mRNA in neurons of the facial motor nucleus or in motor neurons of the spinal cord (Ceccatelli et al., 1991). In regard to this last point, however, Ceccatelli et al. (1991)described the distribution of BDNF mRNA after colchicine treatment, whereas in the present study BDNF expression was examined in untreated rats. Finally, we detected BDNF mRNA within many neuronal populations not previously reported to express this neurotrophin, including cells located between the lateral geniculate and the substantia nigra (e.g., the suprageniculate nucleus, peripeduncular nucleus, medial division of the medial geniculate nucleus), cells in the septohypothalamic nucleus, septofimbrial nucleus, medial pretectal nucleus, nucleus of the lateral olfactory tract, and subcoeruleus nucleus, and cells located in various subgroups of the lateral parabrachial nucleus (especially within the external part).
The immunohistochemical data presented here reveal widespread but discrete localization of BDNF protein throughout the rostrocaudal extent of rat brain and in spinal cord and are in partial agreement with previous immunohistochemical studies of select forebrain fields, including hippocampus (Wetmore et al., 1991, 1993, 1994; Humpel et al., 1993; Dugich-Djordjevic et al., 1995) and several thalamic and mesencephalic/brainstem nuclei (Dugich-Djordjevic et al., 1995). The present results, however, show features not reported previously, such as the identification of numerous neuronal populations with moderate to heavy perikaryal BDNF-ir (and high BDNF mRNA content) including, but not limited to, cells in anterior olfactory nucleus, supramammillary nucleus, various hypothalamic nuclei, endopiriform nucleus, central gray, subgeniculate/peripeduncular region, pedunculopontine tegmental nucleus, nucleus of the solitary tract, and inferior olive. More importantly, one of the most striking differences between the present investigation and others is the intense fiber labeling seen throughout many brain regions in the present material that has not been described previously. The apparent lack of immunostaining of the hippocampal mossy fibers in previous studies is one of the most notable differences. This discrepancy in fiber staining may have resulted from either differences in the characteristics of the antibody preparations or fixation protocols used in the various studies or differences in the scope of previous investigations, whereby some structures simply may not have been commented on.
One of the most interesting aspects of the BDNF immunostaining pattern as seen here was the extensive, but anatomically discrete, fiber/terminal labeling. Many regions, such as medial habenulae, central nucleus of the amygdala, ventral tegmental nucleus, and bed nucleus of stria terminalis, contained a high density of BDNF-ir fibers but no detectable BDNF mRNA. Additionally, in regions such as the dentate gyrus molecular layer, laminar differences in the density of BDNF-ir fibers were observed with moderate-to-light staining in the inner third and no detectable staining in the outer third; this laminar pattern corresponds well with the laminar termination of the principal afferents to this region, namely the dentate commissural/associational system, which terminates in the inner molecular layer, and afferents arising from the medial and lateral entorhinal cortex, which terminate in the middle and outer molecular layer, respectively (Gall, 1990). These results suggested that the BDNF protein was distributed to at least some of these regions by anterograde transport and was indeed localized within afferent axonal systems. In the two cases in which this hypothesis was tested, by eliminating neuronal cell bodies supplying afferent innervation to BDNF-ir-rich and mRNA-poor areas, the immunoreactive fibers were eliminated. Although these data do not exclude the possibility that some of the fiber staining within the CNS may reflect retrograde transport, evidence that endogenous BDNF is transported anterogradely in at least two CNS systems is consistent with the recent observations that endogenous BDNF is anterogradely transported by rat peripheral sensory ganglia (Zhou and Rush, 1996) and that exogenous BDNF and NT-3 can be transported anterogradely from retina to tectum in the developing chick (von Bartheld et al., 1996). Together these findings challenge the idea that trophic factor signaling predominantly involves delivery of a factor from a postsynaptic target cell to a presynaptic responsive neuron. Additional studies will be required to determine the extent to which anterograde transport accounts for the fiber staining seen in the present material and, moreover, the fate of the anterogradely transported endogenous BDNF.
Evidence for an axonal polarization of BDNF protein obtained in the current study is consistent with the findings of Goodman et al. (1996), which demonstrate an apical polarization of BDNF in Madin Darby Canine Kidney epithelial cells. Various studies have described the analogy between apical/axonal and basolateral/somatodendritic domains and have suggested that common targeting signals may govern protein trafficking in neuronal and epithelial cells (Dotti and Simons, 1990; for review, see Rodriguez-Boulan and Zurzolo, 1993). The results of the present study, however, are in contrast with the somatodendritic polarization of BDNF and NGF reported recently in cultured hippocampal neurons (Blöchl and Thoenen, 1996; Goodman et al., 1996). This discrepancy may be attributable to differences in the functional properties of mature neurons in vivo as compared with the embryonic neurons used in the in vitro studies. It is possible that molecules required for the appropriate axonal trafficking of neurotrophin proteins were not yet expressed in the embryonic hippocampal neurons. It is also important to point out that the developing hippocampal neurons examined in these previous studies did not normally produce significant amounts of BDNF or NGF protein and were therefore transfected to produce these neurotrophins under the control of an overexpressing promoter.
Another interesting aspect of the fiber/terminal labeling observed here, was that pathways containing the afferent axons presumably delivering BDNF anterogradely were not labeled (such as stria medularis in the case of the septofimbrial/septal triangular–medial habenula projection). This suggests that BDNF protein accumulation was most pronounced in the proximity of the axon terminal. Furthermore, within the region of axonal termination, the immunoreactive product often appeared as discrete puncta surrounding unstained cell bodies, suggesting that BDNF may be preferentially localized within presynaptic terminal boutons. Although additional studies using electron microscopic techniques will be needed to identify the precise subcellular location of this BDNF-ir, the functional significance underlying the accumulation of this neurotrophin within axon terminals remains to be elucidated. It is possible that the storage of neurotrophins within axonal terminals may provide a means for the rapid activity-dependent release of these factors (Blöchl and Thoenen, 1995, 1996; Humpel et al., 1995; Goodman et al., 1996), whereby they have the opportunity to modulate synaptic events. This prospect is particularly intriguing in light of recent studies indicating that BDNF may have acute (Kang and Schuman, 1995) and enduring effects (Lohof et al., 1993; Korte et al., 1995) on the potentiation of synaptic transmission in brain.
Footnotes
This work was supported by National Institute of Neurological Communicative Disorders and Stroke Grants NS16349 (S.V.) and NS26748 (C.M.G.), and National Institute of Mental Health Grant RSDA-MH00974 (C.M.G.).
Correspondence should be addressed to Dr. James M. Conner, Department of Biology, 0506, University of California, San Diego, La Jolla, CA 92093-0506.
REFERENCES
- 1.Alden M, Besson J-M, Bernard J-F. Organization of the efferent projections from the pontine parabrachial area to the bed nucleus of the stria terminalis and neighboring regions: a PHA-L study in the rat. J Comp Neurol. 1994;341:289–314. doi: 10.1002/cne.903410302. [DOI] [PubMed] [Google Scholar]
- 2.Berkemeier LR, Winslow JW, Kaplan DR, Nikolics K, Goeddel DV, Rosenthal A. Neurotrophin-5: a novel neurotrophic factor that activates trk and trkB. Neuron. 1991;7:857–866. doi: 10.1016/0896-6273(91)90287-a. [DOI] [PubMed] [Google Scholar]
- 3.Bernard J-F, Alden M, Besson J-M. The organization of the efferent projections from the pontine parabrachial area to the amygdaloid complex: a phaseolus vulgaris leucoagglutinin (PHA-L) study in the rat. J Comp Neurol. 1993;329:201–229. doi: 10.1002/cne.903290205. [DOI] [PubMed] [Google Scholar]
- 4.Blöchl A, Thoenen H. Characterization of nerve growth factor (NGF) release from hippocampal neurons: evidence for a constitutive and an unconventional sodium-dependent regulated pathway. Eur J Neurosci. 1995;7:1220–1228. doi: 10.1111/j.1460-9568.1995.tb01112.x. [DOI] [PubMed] [Google Scholar]
- 5.Blöchl A, Thoenen H. Localization of cellular storage compartments and sites of constitutive and activity-dependent release of nerve growth factor (NGF) in primary cultures of hippocampal neurons. Mol Cell Neurosci. 1996;7:173–190. doi: 10.1006/mcne.1996.0014. [DOI] [PubMed] [Google Scholar]
- 6.Carlson SL, Albers KM, Beiting DJ, Parish M, Conner JM, Davis BM. NGF modulates sympathetic innervation of lymphoid tissues. J Neurosci. 1995;15:5892–5899. doi: 10.1523/JNEUROSCI.15-09-05892.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castrén E, Thoenen H, Lindholm D. Brain-derived neurotrophic factor messenger RNA is expressed in the septum, hypothalamus and in adrenergic brain stem nuclei of adult rat brain and is increased by osmotic stimulation in the paraventricular nucleus. Neuroscience. 1995;64:71–80. doi: 10.1016/0306-4522(94)00386-j. [DOI] [PubMed] [Google Scholar]
- 8.Ceccatelli S, Ernfors P, Villar MJ, Persson H, Hökfelt T. Expanded distribution of mRNA for nerve growth factor, brain-derived neurotrophic factor, and neurotrophin 3 in the rat brain after colchicine treatment. Proc Natl Acad Sci USA. 1991;88:10352–10356. doi: 10.1073/pnas.88.22.10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conner JM, Varon S. Distribution of nerve growth factor-like immunoreactive neurons in the adult rat brain following colchicine treatment. J Comp Neurol. 1992;326:347–362. doi: 10.1002/cne.903260304. [DOI] [PubMed] [Google Scholar]
- 10.Conner JM, Varon S. Effects of exogenous nerve growth factor upon sympathetic sprouting into the hippocampal formation. Exp Neurol. 1995;136:123–135. doi: 10.1006/exnr.1995.1089. [DOI] [PubMed] [Google Scholar]
- 11.Conner JM, Muir D, Varon S, Hagg T, Manthorpe M. The localization of nerve growth factor-like immunoreactivity in the adult rat basal forebrain and hippocampal formation. J Comp Neurol. 1992;319:454–462. doi: 10.1002/cne.903190310. [DOI] [PubMed] [Google Scholar]
- 12.Conner JM, Fass-Holmes B, Varon S. Changes in nerve growth factor immunoreactivity following entorhinal cortex lesion: a possible molecular mechanism regulating cholinergic sprouting. J Comp Neurol. 1994;345:409–418. doi: 10.1002/cne.903450307. [DOI] [PubMed] [Google Scholar]
- 13.Conner JM, Yan Q, Varon S. Distribution of brain-derived neurotrophic factor in the rat pituitary gland. NeuroReport. 1996;7:1937–1940. doi: 10.1097/00001756-199608120-00014. [DOI] [PubMed] [Google Scholar]
- 14.Dotti CG, Simons K. Polarized sorting of viral glycoproteins in the axon and dendrites of hippocampal neurons in culture. Cell. 1990;62:63–72. doi: 10.1016/0092-8674(90)90240-f. [DOI] [PubMed] [Google Scholar]
- 15.Dugich-Djordjevic MM, Peterson C, Isono F, Ohsawa F, Widmer HR, Denton TL, Bennett GL, Hefti F. Immunohistochemical visualization of brain-derived neurotrophic factor in the rat brain. Eur J Neurosci. 1995;7:1831–1839. doi: 10.1111/j.1460-9568.1995.tb00703.x. [DOI] [PubMed] [Google Scholar]
- 16.Ernfors P, Wetmore C, Olson L, Persson H. Identification of cells in rat brain and peripheral tissues expressing mRNA for members of the nerve growth factor family. Neuron. 1990;5:511–526. doi: 10.1016/0896-6273(90)90090-3. [DOI] [PubMed] [Google Scholar]
- 17.Freidman WJ, Olson L, Persson K. Cells that express brain-derived neurotrophic factor mRNA in the developing postnatal rat brain. Eur J Neurosci. 1991;3:688–697. doi: 10.1111/j.1460-9568.1991.tb00854.x. [DOI] [PubMed] [Google Scholar]
- 18.Gall C. Comparative anatomy of hippocampus with special reference to differences in the distributions of neuroactive peptides. In: Jones EG, Peters A, editors. Cerebral cortex, Vol 8. Plenum; New York: 1990. pp. 167–213. [Google Scholar]
- 19.Gall CM, Isackson PJ. Limbic seizures increase neuronal production of messenger RNA for nerve growth factor. Science. 1989;245:758–761. doi: 10.1126/science.2549634. [DOI] [PubMed] [Google Scholar]
- 20.Gall CM, Gold SJ, Isackson PJ, Seroogy KB. Brain-derived neurotrophic factor and neurotrophin-3 mRNAs are expressed in ventral midbrain regions containing dopaminergic neurons. Mol Cell Neurosci. 1992;3:56–63. doi: 10.1016/1044-7431(92)90009-q. [DOI] [PubMed] [Google Scholar]
- 21.Goodman LJ, Valverde J, Lim F, Geschwind MD, Federoff HJ, Geller AI, Hefti F. Regulated release and polarized localization of brain-derived neurotrophic factor in hippocampal neurons. Mol Cell Neurosci. 1996;7:222–238. doi: 10.1006/mcne.1996.0017. [DOI] [PubMed] [Google Scholar]
- 22.Gotz R, Koster R, Winkler C, Raulf F, Lottspeich F, Schartl M, Thoenen H. Neurotrophin-6 is a new member of the nerve growth factor family. Nature. 1994;372:266–269. doi: 10.1038/372266a0. [DOI] [PubMed] [Google Scholar]
- 23.Guthrie KM, Gall CM. Differential expression of mRNAs for the NGF family of neurotrophic factors in the adult rat central olfactory system. J Comp Neurol. 1991;313:95–102. doi: 10.1002/cne.903130107. [DOI] [PubMed] [Google Scholar]
- 24.Holtzman DM, Lowenstein DH. Selective inhibition of axonal outgrowth by antibodies to NGF in a model of temporal lobe epilepsy. J Neurosci. 1995;15:7062–7070. doi: 10.1523/JNEUROSCI.15-11-07062.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humpel C, Wetmore C, Olson L. Regulation of brain-derived neurotrophic factor messenger RNA and protein at the cellular level in pentylenetetrazol-induced epileptic seizures. Neuroscience. 1993;53:909–918. doi: 10.1016/0306-4522(93)90476-v. [DOI] [PubMed] [Google Scholar]
- 26.Humpel C, Lindqvist E, Soderstrom S, Kylberg A, Ebendal T, Olson L. Monitoring release of neurotrophic activity in the brains of awake rats. Science. 1995;269:552–554. doi: 10.1126/science.7624780. [DOI] [PubMed] [Google Scholar]
- 27.Ibañez CF. Structure-function relationships in the neurotrophin family. J Neurobiol. 1994;25:1349–1361. doi: 10.1002/neu.480251104. [DOI] [PubMed] [Google Scholar]
- 28.Isackson PJ, Huntsman MM, Murray KD, Gall CM. BDNF mRNA expression is increased in adult rat forebrain after limbic seizures: temporal patterns of induction distinct from NGF. Neuron. 1991;6:937–948. doi: 10.1016/0896-6273(91)90234-q. [DOI] [PubMed] [Google Scholar]
- 29.Kang H, Schuman EM. Long-lasting neurotrophin-induced enhancement of synaptic transmission in the adult hippocampus. Science. 1995;267:1658–1662. doi: 10.1126/science.7886457. [DOI] [PubMed] [Google Scholar]
- 30.Korsching S, Auburger G, Heumann R, Scott J, Thoenen H. Levels of nerve growth factor and its mRNA in the central nervous system of the rat correlate with cholinergic innervation. EMBO J. 1985;4:1389–1393. doi: 10.1002/j.1460-2075.1985.tb03791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korte M, Carroll P, Wolf E, Brem G, Thoenen H, Bonhoeffer T. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc Natl Acad Sci USA. 1995;92:8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauterborn JC, Gall CM. Postnatal ontogeny of NT-3 and NGF expression in rat forebrain. Mol Cell Neurosci. 1994;5:46–62. doi: 10.1006/mcne.1994.1005. [DOI] [PubMed] [Google Scholar]
- 33.Lauterborn JC, Isackson PJ, Gall CM. Nerve growth factor messenger RNA-containing cells are distributed within regions of cholinergic neurons in the rat basal forebrain. J Comp Neurol. 1991;306:439–446. doi: 10.1002/cne.903060308. [DOI] [PubMed] [Google Scholar]
- 34.Lauterborn JC, Bizon JL, Tran TM, Gall CM. NGF mRNA is expressed by GABAergic but not cholinergic neurons in the rat basal forebrain. J Comp Neurol. 1995;360:454–462. doi: 10.1002/cne.903600307. [DOI] [PubMed] [Google Scholar]
- 35.Lohof AM, Ip NY, Poo MM. Potentiation of developing neuromuscular synapses by the neurotrophins NT-3 and BDNF. Nature. 1993;363:350–353. doi: 10.1038/363350a0. [DOI] [PubMed] [Google Scholar]
- 36.Ma W, Ribeiro-da-Silva A, Noel G, Julien J-P, Cuello AC. Ectopic substance P and calcitonin gene-related peptide immunoreactive fibers in the spinal cord of transgenic mice over-expressing nerve growth factor. Eur J Neurosci. 1995;7:2021–2035. doi: 10.1111/j.1460-9568.1995.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 37.Maisonpierre PC, Belluscio L, Friedman B, Alderson RF, Weigand SJ, Furth ME, Lindsay RM, Yancopoulos GD. NT-3, BDNF, and NGF in the developing rat nervous system: parallel as well as reciprocal patterns of expression. Neuron. 1990;5:501–509. doi: 10.1016/0896-6273(90)90089-x. [DOI] [PubMed] [Google Scholar]
- 38.Mufson EJ, Conner JM, Varon S, Kordower JH. Nerve growth factor immunoreactive profiles in the primate basal forebrain and hippocampal formation. J Comp Neurol. 1994;341:507–519. doi: 10.1002/cne.903410407. [DOI] [PubMed] [Google Scholar]
- 39.Mufson EJ, Conner JM, Kordower JH. Nerve growth factor in Alzheimer’s disease: defective retrograde transport to nucleus basalis. NeuroReport. 1995;6:1063–1066. doi: 10.1097/00001756-199505090-00028. [DOI] [PubMed] [Google Scholar]
- 40.Narhi LO, Rosenfeld R, Talvenheimo J, Prestrelski SJ, Arakawa T, Lary JW, Kolvenbach CG, Hecht R, Boone T, Miller JA, Yphantis DA. Comparison of the biophysical characteristics of human brain-derived neurotrophic factor, neurotrophin-3 and nerve growth factor. J Biol Chem. 1993;268:13309–13317. [PubMed] [Google Scholar]
- 41.Paxinos G, Watson C. The rat brain in stereotaxic coordinates. Academic; Orlando, FL: 1986. [DOI] [PubMed] [Google Scholar]
- 42.Phillips HS, Hains JM, Laramee GR, Rosenthal A, Winslow JW. Widespread expression of BDNF but not NT3 by target areas of basal forebrain cholinergic neurons. Science. 1990;250:290–294. doi: 10.1126/science.1688328. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez-Boulan E, Zurzolo C. Polarity signals in epithelial cells. J Cell Sci [Suppl] 1993;17:9–12. doi: 10.1242/jcs.1993.supplement_17.2. [DOI] [PubMed] [Google Scholar]
- 44.Seroogy KB, Gall CM. Neurotrophin expression by dopaminergic midbrain neurons. Exp Neurol. 1993;124:119–128. doi: 10.1006/exnr.1993.1182. [DOI] [PubMed] [Google Scholar]
- 45.Shelton DL, Reichardt LF. Studies on the expression of the β nerve growth factor (NGF) gene in the central nervous system: level and regional distribution of NGF mRNA suggest that NGF functions as a trophic factor for several distinct populations of neurons. Proc Natl Acad Sci USA. 1986;83:2714–2718. doi: 10.1073/pnas.83.8.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spillantini MG, Aloe L, Alleva E, de Simone R, Goedert M, Levi-Montalcini R. Nerve growth factor mRNA and protein increase in hypothalamus in a mouse model of aggression. Proc Natl Acad Sci USA. 1989;86:8555–8559. doi: 10.1073/pnas.86.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutherland RJ. The dorsal diencephalic conduction system: a review of the anatomy, functions of the habenular complex. Neurosci Biobehav Rev. 1982;6:1–13. doi: 10.1016/0149-7634(82)90003-3. [DOI] [PubMed] [Google Scholar]
- 48.Timmusk T, Belluardo N, Metsis M, Persson H. Widespread and developmentally regulated expression of neurotrophin-4 mRNA in rat brain and peripheral tissues. Eur J Neurosci. 1993;5:605–613. doi: 10.1111/j.1460-9568.1993.tb00526.x. [DOI] [PubMed] [Google Scholar]
- 49.Wetmore C, Cao YH, Pettersson RF, Olson L. Brain-derived neurotrophic factor-subcellular compartmentalization and interneuronal transfer as visualized with anti-peptide antibodies. Proc Natl Acad Sci USA. 1991;88:9843–9847. doi: 10.1073/pnas.88.21.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wetmore CJ, Cao Y, Pettersson RF, Olson L. Brain-derived neurotrophic factor (BDNF) peptide antibodies: characterization using a Vaccinia virus expression system. J Histochem Cytochem. 1993;41:521–533. doi: 10.1177/41.4.8450192. [DOI] [PubMed] [Google Scholar]
- 51.Wetmore CJ, Olson L, Bean AJ. Regulation of brain-derived neurotrophic factor (BDNF) expression and release from hippocampal neurons is mediated by non-NMDA type receptors. J Neurosci. 1994;14:1688–1700. doi: 10.1523/JNEUROSCI.14-03-01688.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whittemore SR, Ebendal T, Lärkfors L, Olson L, Seiger A, Strömberg I, Persson H. Developmental and regional expression of β nerve growth factor messenger RNA and protein in the rat central nervous system. Proc Natl Acad Sci USA. 1986;83:817–821. doi: 10.1073/pnas.83.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.vonBartheld CS, Byers MR, Williams R, Bothwell M. Anterograde transport of neurotrophins and axodendritic transfer in the developing visual system. Nature. 1996;379:830–833. doi: 10.1038/379830a0. [DOI] [PubMed] [Google Scholar]
- 54.Yan Q, Rosenfeld RD, Matheson CR, Hawkins N, Lopez OT, Bennet L, Welcher AA (1997) Expression of brain-derived neurotrophic factor (BDNF) protein in the adult rat central nervous system. Neuroscience, in press. [DOI] [PubMed]
- 55.Zhou X-F, Rush RA. Endogenous brain-derived neurotrophic factor is anterogradely transported in primary sensory neurons. Neuroscience. 1996;74:945–951. doi: 10.1016/0306-4522(96)00237-0. [DOI] [PubMed] [Google Scholar]