Abstract
We have identified a protein, Rat O/E-1-associated zinc finger protein (Roaz), that plays a role in regulating the temporal and spatial pattern of olfactory neuronal-specific gene expression. This protein functions by interacting with the olfactory factor O/E-1 and modulating its transcriptional activity. Roaz, isolated via a yeast two-hybrid screen, encoded a protein containing 29 C2H2 zinc fingers of the TFIIIA type. The Roaz mRNA was found in brain, eye, olfactory epithelium, spleen, and heart.In situ hybridization data indicated that Roaz was expressed in the basal layer, consisting of neural precursor cells and immature sensory neurons of the olfactory epithelium, but not in the mature receptor cells. We showed that the Roaz protein bound specifically to O/E-1 by using the yeast two-hybrid system. The two proteins formed a stable complex in coimmunoprecipitation and in vitro binding assays. Introduction of Roaz and O/E-1 into cells containing an olfactory promoter-driven luciferase reporter demonstrated that Roaz abolished O/E-1-mediated transcriptional activation. We propose that the function of Roaz is to modulate negatively the transactivational activity of O/E-1 and to act as a switch protein in the coordination of olfactory sensory neuron differentiation.
Keywords: coregulator, sensory neuron, zinc finger, transcription factors, neurodevelopment, olfactory epithelium
The olfactory epithelium uses a specialized G-protein-mediated signal transduction pathway to detect a wide spectrum of odors in a highly specific and sensitive way (Reed, 1990;Bakalyar and Reed, 1991; Breer and Boekhoff, 1992). Several components involved in this sensory signaling pathway have been cloned and characterized (Danciger et al., 1989; Jones and Reed, 1989; Bakalyar and Reed, 1990; Dhallan et al., 1990; Buck and Axel, 1991; Levy et al., 1991). Each of these components is expressed preferentially or exclusively in the receptor neurons of the olfactory epithelium.
The olfactory receptor neuronal population possesses a remarkable ability to undergo continuous replenishment throughout adult life (Graziadei and Graziadei, 1979a,b). During neuronal differentiation stem cells undergo asymmetric division and give rise to progenitor cells (Stemple and Mahanthappa, 1997). The progenitor cells divide symmetrically and give rise to postmitotic immature neurons, which migrate apically as they undergo terminal differentiation. The mature olfactory receptor neuron (ORN) derives from the globose basal cells that may represent a neural stem cell population in the pseudostratified epithelium (Calof and Chikaraishi, 1989; Caggiano et al., 1994; Hunter et al., 1994).
The establishment of tissue-specific phenotypes in several systems is controlled by specific transcriptional activators (Weintraub et al., 1991; Kaushal et al., 1994). An effort to find factors responsible for ORN cell fate determination and the coordinated regulation of olfactory-specific genes led to the identification of the Olf-1 gene (hereafter called O/E-1 for Olf-1/EBF) (Hagman et al., 1993; Wang and Reed, 1993). The O/E-1 protein is the first member of a novel class of transcription factors containing a complex structural organization composed of an extended region of 350 residues responsible for protein dimerization and DNA binding (Hagman et al., 1993, 1995; Wang and Reed, 1993). It has the ability to homodimerize efficiently and bind to the pseudosymmetric cis-acting regulatory sequence YTCCCYRGGGAR found in proximity to the promoter regions of several olfactory-specific genes, including olfactory marker protein (OMP), type III adenylyl cyclase (ACIII), Golf, olfactory cyclic nucleotide-gated channel (OcNC), and two genes, 50.06 and 50.11, of unknown functions (Kudrycki et al., 1993; Wang et al., 1993). O/E-1 immunoreactivity can be detected in the nasal turbinate as early as embryonic day 11 and throughout adult life in the neural precursors and the receptor neurons.
An independent effort to identify DNA binding proteins involved in the regulation of the pre-B lymphocyte-specific gene mb-1resulted in the identification of early B-cell factor (EBF), apparently encoded by the same gene as O/E-1 (Hagman et al., 1993). In O/E-1-deficient mice the B-cell lineage failed to develop normally, but the olfactory epithelium appeared to be unaffected (Lin and Grosschedl, 1995). We recently have identified a family of O/E-1-like proteins (the O/E proteins), displaying redundant biochemical and functional activities, which are expressed in olfactory epithelium, but not in tissues in which B-cell progenitors reside. These observations could account for the lack of an obvious phenotype in the olfactory epithelium for the O/E-1 knock-out mice (Wang et al., 1997).
Expression of the O/E family members significantly precedes activation of known target genes, both temporally during embryonic development and spatially in the adult olfactory epithelium. These observations suggest that, in addition to maintaining the cellular phenotype of terminally differentiated ORNs, O/E proteins may have an additional role in the early stage of neuronal differentiation (Kudrycki et al., 1993). Similar dual functions also have been described in several other systems such as twist, distal-less-2, andfushi tarazu (Price et al., 1991; Turner et al., 1994;Baylies and Bate, 1996; Spicer et al., 1996).
In this paper we have used O/E-1 as the canonical member of the O/E transcription factor family to identify factors that might regulate its activity. Using a yeast two-hybrid system, we identified a protein, Rat O/E-1-associated zinc finger protein (Roaz), which was expressed predominantly in the basal layer, consisting of neural precursor cells and immature ORNs. We further demonstrated that coexpression of O/E-1 and Roaz had a dramatic effect on expression of a reporter in cell-based transactivation assays. The properties of this zinc finger protein suggest that it may act to alter the expression of two groups of target genes during cell lineage determination and differentiation.
MATERIALS AND METHODS
Yeast two-hybrid screen and plasmid construction.All experiments were performed with the yeast strain y190 (MATa gal4 gal80 his3 trp1–901 ade2–101 ura3–52 leu2–3,−112 + URA3:: GAL→lacZ, LYS2:: GAL→HIS3 cyhr; Steven J. Elledge, University of Texas Southwestern Medical School) (Harper et al., 1993). Yeast was grown in yeast extract, peptone, and dextrose (YPD), Synthetic Complete (SC), or drop-out medium.
A 2.3 kb SalI/NotI fragment containing the full-length O/E-1 was obtained by restriction digestion of Y11 plasmid (Wang and Reed, 1993) and subcloned into the bait vector pPC97 containing GAL4 DNA binding domain and Leu selection marker (Chevray and Nathans, 1992). The resulting construct, pPC97-O/E-1, was transformed into y190 by selecting for Leu+ colonies. An individual isolate was transformed with the rat olfactory cDNA library (3.6 × 106 independent transformants) fused to GAL4 transactivator domain in the prey vector, pPC86 (Wang and Reed, 1993). The 1.9 × 106 double transformants were plated on SC-Leu,-Trp,-His minimal plates containing 50 mm of 3-aminotriazole (Sigma, St. Louis, MO) to reduce leaky expression of HIS+ phenotype. HIS+ colonies from the primary screen were restreaked on secondary plates and screened for both HIS+ phenotype and β-galactosidase activity.
The expression vectors pCIS-XPRoazD86 and pCIS-XPIC2 were made by subcloning the SalI/NotI inserts from the pPC86 vectors containing RoazD86 and IC2 inserts into pEBVHisB (Invitrogen, San Diego, CA). An EcoRI/XbaI fragment, containing the insert sequence fused in-frame with a 5′ eukaryotic ATG, His6, and XPress epitope tag, then was subcloned into pCIS vector. The pCIS-GST-RoazD86 expression vector was made by subcloning the SalI/NotI fragment, isolated from RoazD86, into a pCIS-GST vector. pCIS-GST-Roaz was constructed by subcloning a 4.5 kb EcoRV/NotI fragment isolated from pBS-Roaz into a pCIS-GST vector. The pBS-Roaz composite cDNA clone was reconstituted from three different cDNA clones by using uniquePmlI and BspEI sites to ligate between JBOZ2.2 and JBOZ1.3, and JBOZ1.3 and RoazD86, respectively. The pCIS-OED5(NcoI) vector was made by subcloning theSalI/NotI insert derived from pPC86-OED5 into pCIS-GST and removing the last 69 amino acids, including the antisera JH1132 epitope, by NcoI/NotI digestion, Klenow fill-in, and blunt end ligation.
Terminally truncated constructs were made by using Erase-a-base (Promega, Madison, WI). N-terminal deletion of pPC97-O/E-1 was made by protecting the SalI site (using Klenow fill-in with α-phosphorothioate deoxyribonucleotides) and deleting from theBspEI site. C-terminal deletion of pPC97-O/E-1 was made by protecting the SacI site and deleting from theNotI site. All deletion constructs were verified by DNA sequence analysis.
cDNA library screen and 5′ rapid amplification of cDNA ends (RACE)-PCR. Random primed 32P-labeled DNA fragments were used to screen an oligo-dT-primed λZAP rat olfactory cDNA library (1 × 106 independent clones) and a random-primed λZAP rat forebrain cDNA library (3 × 106 independent clones; Jim Boulter, Salk Institute, San Diego, CA). The 5′ RACE-PCR procedure was performed with a Clontech Marathon cDNA amplification kit (Palo Alto, CA). For the first and second RACE reactions, a sequence derived from JBOZ1.3 was used to make a gene-specific primer (RT198, 5′-AGTCAGGTGTAG-3′) for the reverse transcriptase/cDNA synthesis reaction. PCR reactions were performed with an internal antisense primer (RT200, 5′-GGGGGAATTCCAGGTGTAGCACTGCTCATGGAAG-3′) and a nested primer (RT197, 5′-CTGTCCAGGTGGCAGTAC-3′) for the first round PCR and an internal primer (RT205, 5′-TCCTGTCTTCCA-GCGCACGGCTGG-3′) for the second round PCR. Six clones were sequenced. Four clones from the second round RACE-PCR, RE2.7, RE2.9, RE2.11, and RE2.16, represented the same PCR products extending 822 bp 5′ of JBOZ1.3. Two clones from the first round RACE-PCR, RE1.2 and RE1.3, were 363 and 75 bp shorter than RE2.11, respectively. For the third round of RACE-PCR, a gene-specific RT primer derived from RE2.11 (RT241, 5′-AGGTCTGCCAGAGACTCGAAGTCC-3′) and an internal antisense PCR primer (RT205, 5′-TCCTGTCTTCCAGCGCACGGC-TGG-3′) were used. Thermostable rTth polymerase was used for first-strand cDNA synthesis at 70°C. For each round of RACE-PCR, products were blunt-ended and subcloned into pBluescript KS. The sizes of the inserts were estimated by PCR, and 5–10 clones with the longest lengths of inserts were sequenced.
Reverse transcription-PCR. Total RNA (2 μg) prepared from tissues of Sprague Dawley rats by using RNAzol B was reverse-transcribed into first-strand cDNA with 500 ng of gene-specific primer (RT163, 5′-TGGTTCTGCAACTCTGTC-3′) for Roaz or random hexamers for O/E-1 and RNA polymerase II control and M-MLV reverse transcriptase (Life Technologies, Gaithersburg, MD) in 20 μl reactions. For Roaz expression the PCR reaction was performed with 2 μl of the first-strand cDNA solution and 1 μm each of sense (RT250, 5′-ACTCCCTCACTGGTTTCCGC-TGTG-3′) and antisense primer (RT251, 5′-CGAAGGTCATCTGGCA-CTTGATGC-3′) for 35 cycles of denaturation (94°C, 30 sec), annealing (65°C, 1 min), and extension (72°C, 1 min). The expression pattern of O/E-1 and RNA polymerase II was examined under similar conditions with the following pairs of primers: RT216, 5′-CCTGGCCCTCTACGACAGAC-3′; SW9, 5′-CCTATGATGATCACAGTCGCG-3′ with 52°C annealing temperature for O/E-1; and RT56, 5′-GCCATGCAGAAGTCTGGCCGTCCCCTCAAG-3′; RT57, 5′-CTTATAGCCAGTCTGCAGATGAAGGT-CAC-3′ with 65°C annealing temperature for RNA polymerase.
Tissue preparation and in situ hybridization.Mice, C57B6/J (Jackson Laboratories, Bar Harbor, ME), were anesthetized with pentobarbital and perfused intracardially with ice-cold PBS, followed by Bouin’s solution (Sigma). Tissues were harvested and post-fixed in the same fixative for 2 hr before immersion in 30% sucrose/1× PBS at 4°C overnight. Then tissues were embedded in OCT (Tissue-Tek, Torrance, CA), and sections (18 μm) were collected onto Superfrost Plus (Fisher, Pittsburgh, PA) glass slides.In situ hybridization was performed as described (Schaeren and Gerfin, 1993), using digoxigenin-labeled RNA probes synthesized from plasmid JBOZ1.3 for Roaz, pBS-O/E-1 (containing full O/E-1 coding sequence) for O/E-1, and pBS-OMP (containing full OMP coding region) for OMP.
Affinity chromatography. RoazD86 was expressed as glutathione S-transferase (GST) fusion protein by transient transfection into human embryonic kidney cell line (HEK293) cells. Four 100 mm plates of 293 cells were collected, washed two times with 1× PBS, and extracted with 4 ml of extraction buffer (1× PBS, 1% Triton X-100, 1 mm PMSF, 1 μg/ml leupeptin, 2 μg/ml aprotinin, and 1 μg/ml pepstatin A) for 30 min at 4°C. After preclearing the extract by centrifugation at 15,600 × g for 20 min, we added 200 μl of 50% slurry of glutathione-bound agarose beads to the supernatant and rocked it at 4°C for 1 hr. The beads were washed one time with 1× PBS, one time with 1× PBS plus 500 mm NaCl, and three times with 1× PBS. The GST protein was isolated in a similar way. Whole-cell extracts for binding studies were isolated as described above from HEK293 cells transfected with pCIS-O/E-1, O/E-2, or O/E-3 plasmid (Wang et al., 1997). The extracts (400 μg) were mixed with 6–8 μg of GST-RoazD86 fusion protein bound to 30 μl of glutathione-agarose beads and rocked for 2 hr at 4°C. The beads were washed one time with 1× PBS, two times with 1× PBS plus 500 mm NaCl, and three times with 1× PBS. Sample buffer (30 μl) was added, and a fraction (4 μl) was resolved on a 10% SDS-PAGE and analyzed by Western blotting with anti-O/E-1 antibody that recognized all three of the O/E proteins [JH1132, antipeptide antibody (Davis and Reed, 1996)].
The study of interaction between full-length Roaz and O/E-1 was performed essentially as described above, with the following modification: 700 ng of GST-Roaz fusion protein was mixed with 400 μg of pCIS-O/E-1-transfected whole-cell extract in the binding assay. Retained proteins were resolved on a 10% SDS-PAGE and analyzed by Western blotting with anti-O/E-1 antibody.
Coimmunoprecipitation. RoazD86 was tagged with an engineered eukaryotic ATG, His6 epitope tag, and XPress sequence (XPRoazD86) and subcloned into the pCIS expression vector. pCIS-XPRoazD86 and pCIS-XPIC2, containing an irrelevant sequence, were transfected into HEK293 cells. A soluble fraction of whole-cell protein (200 μg) was mixed with a similar amount of protein extracted from cells transfected with pCIS-O/E-1 or pCIS. Protein A-Sepharose (80 μl of a 50% slurry) was mixed with 15 μl of anti-O/E-1 crude serum (JH865, anti-O/E-1 fusion protein antibody) or preimmune serum. The volume was adjusted to 1 ml with extraction buffer and rocked at 4°C for 2 hr. After incubation the protein A-Sepharose beads were washed one time with 1× PBS, two times with 1× PBS plus 500 mmNaCl, and three times with 1× PBS. Sample buffer (50 μl) was added, and a 20 μl aliquot was fractionated on a 10% SDS-PAGE and subjected to Western blot analysis with anti-XPress antibody (1:5000, Invitrogen, San Diego, CA).
Cell culture and transfection. The HEK293 was grown in DMEM (Life Technologies) supplemented with 10% fetal calf serum, penicillin (100 U/ml), streptomycin (100 μg/ml), andl-glutamine (2 mm). Calcium phosphate-mediated transfections were performed in a 100 mm Petri dish with 5 μg of construct DNA, 5 μg of pBluescript DNA, and 1 μg of pRSV-T antigen expression vector. After 5 hr of incubation at 37°C, the cells were treated with 10% DMSO in 1× HBS (140 mm NaCl, 25 mm HEPES, and 1.4 mmNa2HPO4) and incubated for 48 hr before the preparation of protein extracts (Levin and Reed, 1995).
Electrophoretic mobility shift assay. A gel shift assay was performed essentially as described (Hagman et al., 1993; Wang et al., 1993), with the following modification. A different amount of protein was mixed with 3–5 × 104 cpm of probe in 20 μl of binding reaction and incubated at 20°C for 30 min. Binding reaction contained 10 mm HEPES, pH 7.9, 70 mm KCl, 1 mm DTT, 4% glycerol, 1 mm EDTA, 2.5 mm MgCl2, 200 μg/ml of poly(dI-dC), and 20 μg/ml of salmon sperm DNA. The mixture was subjected to electrophoresis either on a 6% polyacrylamide gel (acrylamide/bisacrylamide ratio, 59:1) or a 1.5% agarose gel in 0.25× Tris-borate (TBE) buffer at 4°C. Products were detected by autoradiography of the dried gel.
For synthetic oligonucleotides, 150 ng of top-strand primer was labeled with [γ-32P]ATP by using T4 DNA polynucleotide kinase in a 20 μl reaction. After removal of unlabeled free nucleotides through a G-50 column, the primer was annealed with 750 ng of complementary oligonucleotide in annealing buffer (100 mmKCl, 10 mm Tris, pH 8.0, and 1 mm EDTA) at 100°C for 5 min, incubated at 68°C for 1 hr, and slowly cooled to room temperature. DNA fragments were labeled with [α-32P]dCTP by Klenow fill-in.
Luciferase activity assay. One 60 mm plate of HEK293 cells was cotransfected transiently with 1 μg of the indicated pGL2-based reporter plasmid along with 500 ng of pCIS vectors expressing O/E-1, O/E-2, or O/E-3 proteins and/or 500 ng of pCIS-GST-Roaz plasmid. All transfections were adjusted to 5 μg of total DNA with pCIS vector DNA. The luciferase reporter activity was measured from equivalent amounts of protein lysate of each sample with a luciferase assay system (Promega) and Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA). The relative luciferase activity was calculated by comparing the activity measured in each cotransfection experiment with the activity of reporter plasmid plus pCIS expression vector cotransfection control (arbitrarily set at 1.0). All luciferase assays were determined in duplicate.
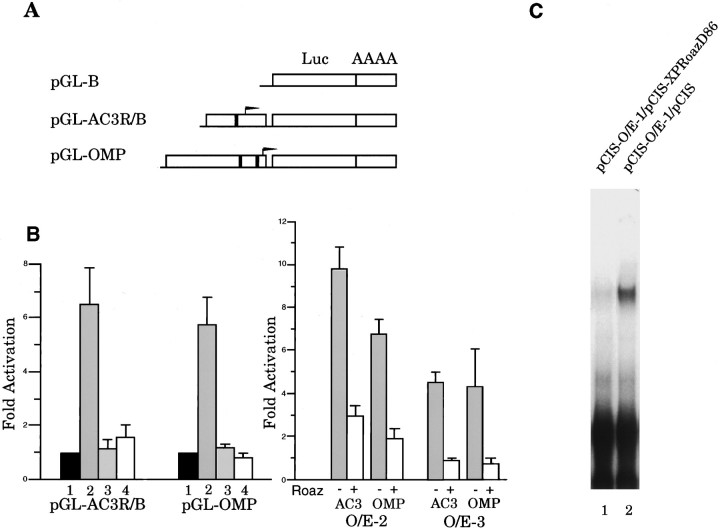
The pGL-AC3R/B vector contained a 1.55 kbEcoRI/BamHI DNA fragment, including 500 bp of 5′-untranslated region (UTR) and an O/E binding site at position −270 cloned into the polylinker region of pGL2-Basic vector (pGL-B) (Promega). The pGL-OMP vector was composed of a 2.7 kb DNA fragment containing 59 bp of 5′-UTR and two O/E binding sites (−180 and −700) cloned into the XhoI site of the pGL-B vector.
RESULTS
Cloning of O/E-1 interacting clones by yeast two-hybrid system
A screen for O/E-1 interacting proteins was performed with the yeast two-hybrid system (Fields and Song, 1989; Chien et al., 1991;Fields and Sternglanz, 1994). The full-length O/E-1 protein fused to GAL4 DNA binding domain [GAL4(DB)] was used as a bait (pPC97-O/E-1) (Chevray and Nathans, 1992) to screen a rat olfactory cDNA library of 3.6 × 106 independent transformants fused to the GAL4 transactivator domain [GAL4(TA)] (Wang and Reed, 1993). The physical association of GAL4(DB) and GAL4(TA) via the interaction of O/E-1 and the prey proteins turned on the expression of His3 and lacZ genes. Selection for HIS+ and β-galactosidase-positive transformants led to the identification of 25 clones from among the 1.9 × 106 transformants. Plasmids isolated from 9 of the 25 colonies displayed O/E-1-dependent expression of the selectable markers. The isolates were grouped according to their strength of interaction with O/E-1 as estimated by both HIS+ phenotype and β-galactosidase activity. Three clones interacted with O/E-1 strongly, two at intermediate levels, and four weakly. The three strongly interacting clones encoded a zinc finger protein. One clone of intermediate strength encoded a partial O/E-1 sequence (OED5) from aa 221 to the stop codon fused in-frame with GAL4 transactivator domain; the other encoded a novel zinc finger protein. We chose to study the three clones that displayed strong and specific interaction with O/E-1.
Identification and characterization of a zinc finger protein, Roaz
Sequence analysis revealed that all of the three strongly interacting clones represented the same cloning event, in which a gene with an open reading frame of 2100 bp was fused in-frame with the GAL4 transactivator domain. The insert, RoazD86, encoded a partial sequence of a C2H2 zinc finger protein of the TFIIIA type.
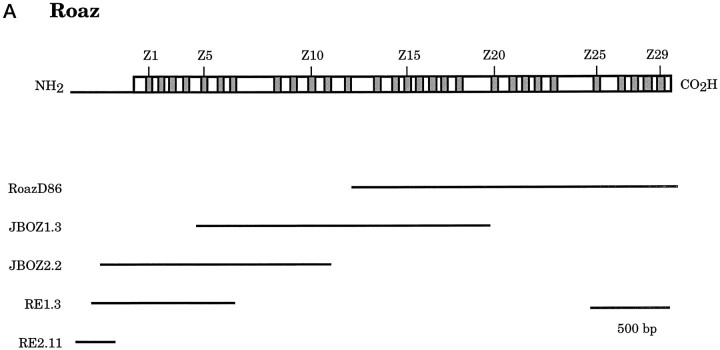
A full-length cDNA of Roaz was obtained by using a combination of cDNA library screen and RACE-PCR (Fig.1A). A rat forebrain random-primed cDNA library was screened by using the complete insert sequence of RoazD86 as a probe, and 12 overlapping clones were identified and sequenced with the longest one, JBOZ1.3, extending 1047 bp further upstream of RoazD86. Then RACE-PCR was performed to characterize further the 5′ end of the mRNA. Two rounds of RACE reactions generated several products, with the longest clone from the first round (RE1.3) extending 747 bp upstream of JBOZ1.3 and the longest clone from the second round (RE2.11) extending 822 bp 5′ upstream of JBOZ1.3. To confirm the sequence obtained from RACE-PCR, we screened the rat forebrain library, using RE1.3 as a probe. Six independent clones were isolated, with the longest one, JBOZ2.2, extending 657 bp upstream of JBOZ1.3. Finally, another round of RACE-PCR was performed with thermostable rTth reverse transcriptase to destabilize possible secondary structures in the first-strand synthesis step. Three additional clones were isolated. One clone was 24 bp shorter than RE2.11. The sequences from the other two clones diverged from RE2.11 from the +74 nucleotide position and extended 21 and 50 bp further upstream, possibly representing spliced variants.
Fig. 1.

Structure and sequence of Roaz cDNA clones.A, Schematic representation of Roaz and various cDNA clones isolated from cDNA library screens and RACE-PCR. Eachshaded box represents one zinc finger structure.RoazD86 is the original clone isolated from the yeast two-hybrid screen. The other clones represent isolates from subsequent cDNA library screens and RACE-PCR, as indicated in the text. The 3′-UTR of Roaz is not shown in this diagram. B, cDNA sequence and predicted open reading frame of Roaz. The amino acid sequences of zinc finger structures areunderlined. Figure continues.
The overlapping clones resulted in a composite insert of 4.7 kb containing a predicted open reading frame of 1186 amino acids initiating at the 5′-most methionine and followed by 693 bp of 3′-UTR with a polyadenylation signal and poly(A) tail (Fig.1B). The deduced polypeptide sequence consisted of a protein encoding 29 C2H2 zinc finger motifs. A search of available databases with the Roaz sequence revealed extensive homology with an unpublished partial cDNA fragment isolated from rat aorta (gi/207695).
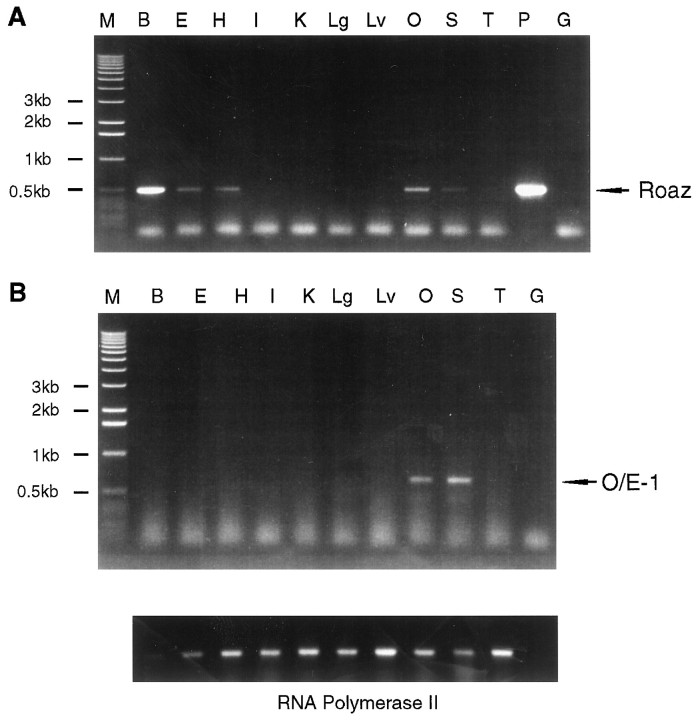
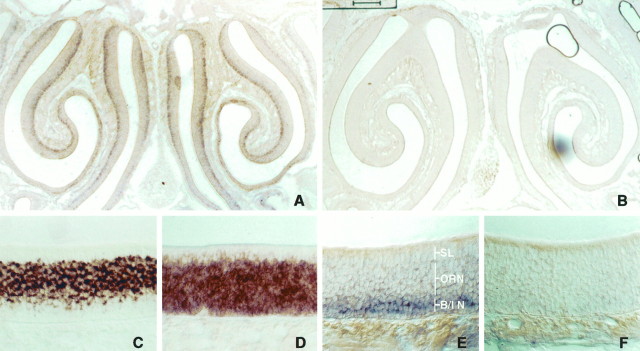
Northern blot analysis of 30 μg of poly A+ RNA isolated from rat forebrain identified a major 4.7–5.1 kb transcript and a minor 5.8–6 kb transcript (our unpublished data). Both of these transcripts were of low abundance. The expression pattern of Roaz in different tissues was examined by RT-PCR, and the Roaz transcripts were found to be present in the olfactory epithelium, spleen, brain, eye, and heart (Fig. 2A). No message was detected in the intestine, kidney, lung, liver, or testis. The O/E-1 mRNA was expressed preferentially in the olfactory epithelium and spleen, the site of B-cell development (Fig. 2B). To determine the cell type(s) expressing Roaz mRNA in the olfactory epithelium, we performed in situ hybridization with digoxigenin-labeled riboprobe (Fig. 3). Roaz was found mainly in the basal cell layer and the lower one-third of the pseudostratified epithelium containing the immature ORNs.
Fig. 2.
Expression pattern of Roaz and O/E-1.A, RT-PCR of Roaz expression in tissues with the use of a gene-specific oligonucleotide. M, 1 kb marker;B, forebrain; E, eye; H, heart; I, intestine; K, kidney;Lg, lung; Lv, liver; O, olfactory epithelium; S, spleen; T, testis; P, 5 ng of Roaz-containing plasmid DNA as a positive control; G, 100 ng of genomic DNA.B, RT-PCR of O/E-1 and RNA Polymerase II.
Fig. 3.
In Situ hybridization. Digoxigeninin situ hybridization of Roaz, O/E-1, and OMP in adult olfactory epithelium. The pseudostratified epithelium is composed of the sustentacular cell layer (SL), the basal/immature cells (B/IN) and mature olfactory receptor neurons (ORN). Sections were hybridized with Roaz antisense probe (A, E), Roaz sense probe (B, F), OMP antisense probe (C), and O/E-1 antisense probe (D). Images in A and B are low-magnification coronal images of the mouse epithelium. Scale bar, 200 μm. Images inC–F are from the same sections at high magnification.
In vitro binding assay and coimmunoprecipitation
The direct interaction between Roaz and O/E-1 was demonstrated by an in vitro binding assay and coimmunoprecipitation. Full-length O/E-1 protein expressed in HEK293 cells was retained specifically on the GST-RoazD86 affinity column, but not on a GST-bound affinity column (Fig. 4A, left,lanes 1, 2). The previously documented ability of O/E-1 to homodimerize was confirmed in this assay (lane 4), although the amount of retained O/E-1 protein was less than that observed for the Roaz and O/E-1 heteromeric interaction. This observation was consistent with the result from the original yeast screen in which Roaz displayed stronger interactions than the O/E-1 protein with the O/E-1 bait. The ability of full-length Roaz protein, expressed as a GST fusion and immobilized on a column, to retain the soluble O/E-1 protein was assessed (Fig. 4A, right, lane 1). The results of this experiment confirm that the Roaz/O/E-1 interaction is maintained with the native protein. Further supporting the specificity of this interaction is the ability of the other O/E proteins to interact with RoazD86 (Fig.4B).
Fig. 4.
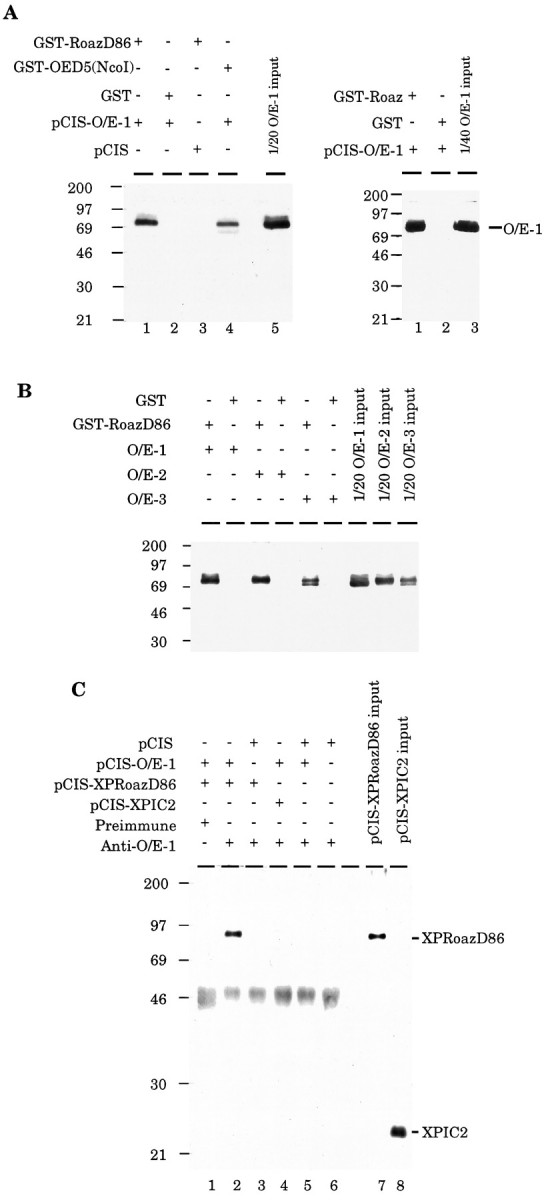
Biochemical characterization of Roaz/O/E-1 interaction. A, Left, Demonstration of Roaz interaction with O/E-1 by affinity chromatography. Purified proteins (GST or GST fusions) were mixed with whole-cell protein isolated from HEK293 cells transfected with pCIS-O/E-1 or pCIS vector. Bound protein was extracted with 30 μl of sample buffer, and 20 μl was fractionated on a 10% SDS-PAGE and detected by Western blotting with the use of anti-O/E-1 antibody (JH1132); one-twentieth of the input was loaded for comparison. Right, Interaction of O/E-1 with full-length Roaz protein. Proteins were purified as described above except that pCIS-GST-Roaz was transfected. Protein yields for the full-length construct were consistently lower than those observed for the pCISRoazD86 vector. B, Demonstration of Roaz interaction with O/E family proteins by affinity chromatography. Purified proteins (GST or GST fusions, 9 μg) were mixed with whole-cell proteins (400 μg) isolated from HEK293 cells transfected with pCIS-O/E-1, pCIS-O/E-2, or pCIS-O/E-3. Bound proteins were extracted with 60 μl of sample buffer. One-fifth (12 μl) was fractionated on a 10% SDS-PAGE and detected by Western blotting with the use of the anti-O/E-1 antibody (JH1132); one-twentieth of the input was loaded for comparison. C, Coimmunoprecipitation of Roaz and O/E-1 with anti-O/E-1 antibody (JH865). Whole-cell extract from HEK293 cells transfected with different constructs was mixed with anti-O/E-1 antibody (JH865) or preimmune serum and protein A-Sepharose. After being washed, resins were extracted with 50 μl of sample buffer; 20 μl was loaded on a 10% SDS-PAGE and detected after Western blotting with the use of anti-XPress antibody. One-fortieth of the input was loaded as a reference.
In parallel, the interaction of Roaz and O/E-1 was examined in solution by anti-O/E-1 antiserum coimmunoprecipitation of an XPress epitope-tagged Roaz protein (XPRoazD86) in the presence of O/E-1 protein (Fig. 4C). XPRoazD86 could be coimmunoprecipitated in the presence of O/E-1 protein and anti-O/E-1 antiserum (lane 2), but not by preimmune serum (lane 1) or when O/E-1 was replaced by pCIS-transfected whole-cell extract (lane 3). XPIC2, an irrelevant polypeptide, could not be coimmunoprecipitated with O/E-1 by anti-O/E-1 antibody (lane 4), which confirmed the specific interaction between O/E-1 and Roaz.
The region of O/E-1 that interacts with Roaz includes the helix-loop-helix domain
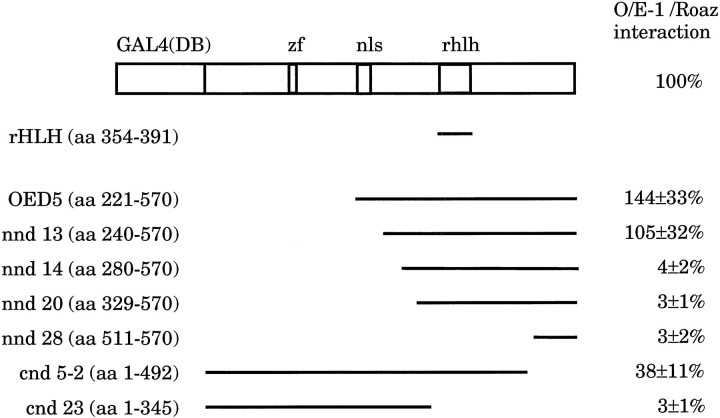
Regions of O/E-1 involved in specific intermolecular interactions with Roaz were identified in the yeast two-hybrid interacting assay by using a series of N-terminal and C-terminal deletion constructs of the O/E-1/Gal4(DB) fusion. The strength of interaction was determined by a β-galactosidase assay in yeast liquid cultures (Ausubel et al., 1995) and expressed as a percentage of the activity induced by intact RoazD86 and O/E-1 (Fig. 5). The interacting domain of O/E-1 was localized to a 253 amino acid region (aa 240–492), which included the helix-loop-helix domain (aa 354–391). The level of protein expression was assessed for each construct by using an anti-Gal4(DB) antibody and was found to vary less than twofold.
Fig. 5.
Localization of interaction domain of O/E-1 with Roaz. Yeast strain Y190 expressing GAL4(TA)-RoazD86 was transformed with constructs encoding C-terminal deletions (cnd), N-terminal deletions (nnd) of O/E-1, orOED5 (a partial sequence of O/E-1 cDNA isolated from original yeast two-hybrid screen) as GAL4(DB) fusions. Double transformants were grown in Leu/Trp drop-out medium before assaying for β-galactosidase activity. The strength of interaction for an individual mutant is expressed as a percentage relative to that of intact proteins, which was set at 100% and corresponded to 43 ± 7 U. All measurements were determined from at least four independent colonies. Numbers in parentheses indicate the corresponding starting and ending amino acids of the O/E-1 sequence. Previously characterized protein motifs in O/E-1 are indicated. Zf, Zinc finger; nls, nuclear localization signal; rhlh, repeat helix-loop-helix motif.
Roaz repressed O/E-1-mediated transactivation
To investigate the role of Roaz in O/E-1 transactivation of olfactory-specific gene expression, we performed transient cotransfection and luciferase assays in HEK293 cells. The OMP 2.7 kb promoter fragment and a 1.6 kb fragment from the ACIII promoter were cloned into the pGL2-Basic luciferase reporter vectors (pGL-OMP and pGL-AC3R/B) (Fig. 6A), and activity was measured when cells were cotransfected with pCIS (lane 1), pCIS-O/E-1 (lane 2), pCIS-GST-Roaz (lane 3), or both (lane 4) (Fig. 6B, left). We found that O/E-1 alone had moderate transactivational effect on OMP and ACIII promoter sequences (fold activation above reporter construct and expression vector cotransfection: 6.5 ± 1.3 for ACIII, 5.8 ± 0.9 for OMP; mean ± SEM,n = 6). Remarkably, GST-Roaz completely abolished this effect when cotransfected with pCIS-O/E-1 (1.6 ± 0.5 for ACIII and 0.8 ± 0.1 for OMP) (Fig. 6B left,lane 4). There was no difference in luciferase activity detected when pCIS-GST-Roaz or pCIS was introduced into cells transfected with the pGL-OMP or pGL-AC3R/B reporter constructs. The ability of Roaz to inhibit O/E-mediated transcriptional activation extended to the other members of the O/E family (Fig.6B, right). The presence of Roaz led to a greater than threefold inhibition of activation on the AC3 and OMP promoters.
Fig. 6.
Transcriptional activation of O/E binding site-containing reporter constructs. A, Schematic representation of reporter constructs containing the type III adenylyl cyclase promoter (pGL-AC3R/B) and the OMP promoter (pGL-OMP) is shown. The filled boxes represent O/E binding sites, and thearrows represent the transcriptional start sites.Luc, Luciferase; AAAA, polyadenylation sequence. B, Luciferase activity assay of the O/E family proteins on ACIII and OMP promoters. Left panel shows inhibitory effect of Roaz on O/E-1-mediated transactivation.Lane 1, pCIS transfection; lane 2, pCIS-O/E-1 transfection; lane 3, pCIS-GST-Roaz transfection; lane 4, pCIS-O/E-1 and pCIS-GST-Roaz cotransfection. Right panel shows a similar effect of Roaz on O/E-2 and O/E-3-mediated activation. The data are derived from six independent transfection experiments. Relative luciferase activity was calculated by dividing the relative light units (RLU) of each transfection with that of the pCIS expression vector control. The absolute luciferase activity for each reporter construct cotransfected with pCIS vector was pGL-AC3R/B, 126,542 RLU; pGL-OMP, 156,119 RLU.C, Roaz interaction with O/E-1 protein/DNA complex. The pCIS-XPRoazD86 or pCIS-transfected whole-cell extracts (120 ng) were mixed with 312.5 ng of pCIS-O/E-1-transfected whole-cell lysate in an EMSA assay, using synthetic O/E binding site as a probe, and fractionated on a 6% SDS-PAGE. Quantitative assessment of binding revealed a 76% reduction of complex formation in the presence of XPRoazD86 (lane 1).
The transcriptional repression observed could have been attributable to the loss of O/E-1 DNA binding ability at its cognate site on formation of a heteromeric complex with Roaz or, alternatively, a direct repression of O/E-1 transactivational activity by Roaz. To distinguish among these possibilities, we performed an electrophoretic mobility shift assay (EMSA) with an O/E binding site-containing oligonucleotide derived from the OcNC promoter region as a probe. Whole-cell extracts were prepared from HEK293 cells transfected with pCIS-O/E-1, pCIS-XPRoazD86, and pCIS. In the presence of XPRoazD86 protein, the intensity of the O/E-1-containing complex was decreased, and no additional supershifted complex reflecting a Roaz/O/E-1 heteromeric complex bound to DNA was detected (Fig. 6C). Additionally, there was no evidence that XPRoazD86 could bind to the O/E binding site DNA (our unpublished data).
Roaz and O/E-1 heteromeric complex can transactivate luciferase activity driven by a minimal SV40 early promoter
In the course of these experiments we used a control plasmid containing a minimal SV40 early promoter and the luciferase sequence (pGL-P) (Fig. 7A, top). Surprisingly, we found that cotransfection of the O/E-1 and GST-Roaz expression vectors with the pGL-P reporter construct increased luciferase activity 67-fold over basal levels. Neither O/E-1 nor Roaz individually elicited a similar effect. The same transactivational effect on the SV40 early promoter also was observed for other O/E family members cotransfected with Roaz and was dependent on the presence of both factors (Fig.7A). The observed transcriptional activation suggested that the heteromeric complex recognized DNA sequences in the SV40 minimal promoter region. Therefore, a 200 bp XhoI/HindIII DNA fragment comprising this promoter was isolated from the pGL-P vector, labeled with [α-32P]dCTP, and used in an EMSA with purified GST fusion protein of Roaz and O/E-1 (Fig. 7B) The GST-Roaz protein displayed specific binding to this fragment, which could be mapped further to the distal 110 bp region by using additional restriction fragments (Fig. 7B, arrow). The GST-O/E-1 protein (lane 2) failed to produce a complex of the mobility expected for such a complex. The fast mobility complex (Fig. 7B, bracket) probably derived from other proteins that were present in the GST-O/E-1 preparation, which was less pure than the GST protein as a result of the lower yield of the former in the expression system. No supershifted bands were observed when purified GST-Roaz and GST-O/E-1 were used in an EMSA with the promoter fragment probe P1, possibly because the trimolecular complexes of DNA/Roaz/O/E-1 are not sufficiently stable to be detected in this assay (our unpublished data).
Fig. 7.
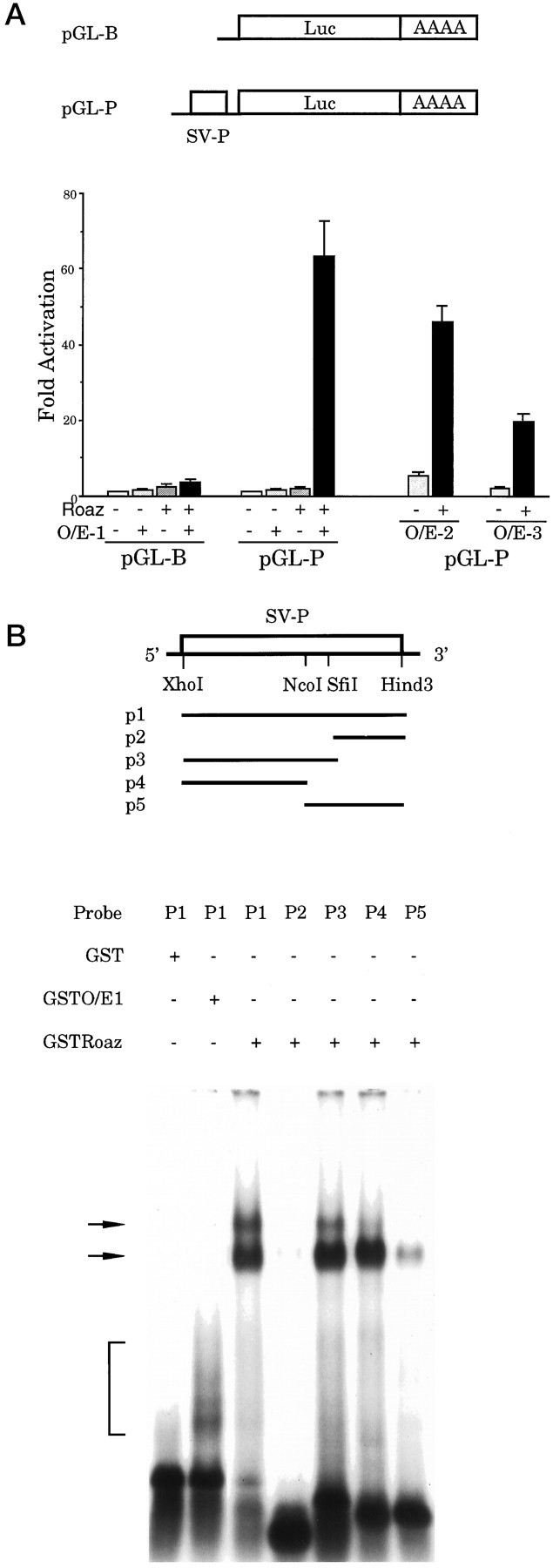
O/E-1 and Roaz interactions with the SV40 early promoter. A, Top, The pGL-P plasmid contains a 200 bp SV40 early promoter lacking the enhancer sequence.SV-P, SV40 early promoter. Bottom, Transcriptional activation of the pGL-P reporter construct was assessed after cotransfection with pCIS-GST-Roaz and/or the pCIS-O/E expression vectors. The absolute luciferase activity for each reporter construct cotransfected with pCIS vector waspGL-B, 22,975 RLU; pGL-P, 304,316 RLU. B, DNA binding ability of Roaz and O/E-1 on SV40 early promoter. GST-purified O/E-1 or Roaz (200 ng) was mixed with the indicated probes (p1–p5) in an EMSA, as shown in this figure. Shifted complexes were fractionated on a 1.5% agarose gel in 0.25× TBE running buffer.
DISCUSSION
The C2H2 zinc finger-containing protein, Roaz, interacts with O/E-1
Dimerization of proteins into homomeric and heteromeric multimers plays a pivotal role in the control of gene expression and cell function. Previous studies have demonstrated that characteristic structural motifs, including leucine zipper (Landschulz et al., 1988;Turner and Tjian, 1989), helix-loop-helix (Murre et al., 1989;Anthony-Cahill et al., 1992), ankyrin (Blank et al., 1992), PAS domain (Huang et al., 1993), Cys4 zinc finger, and Lim/double zinc finger motifs (Feuerstein et al., 1994), often mediate these protein–protein interactions. In a search for proteins capable of interacting with O/E-1 using a yeast two-hybrid screen, we have identified a rat cDNA, Roaz, encoding a 29 C2H2zinc finger protein of the TFIIIA type. Here we demonstrate that Roaz and O/E-1 display specific intermolecular interaction in vitro and in an eukaryotic cell-based in vivo system. Moreover, the Roaz protein displays comparable interactions with other O/E family proteins as assessed in the in vitro binding and transcriptional activation assays.
A region containing the HLH motif of O/E-1 is necessary to mediate Roaz/O/E-1 interaction, and a zinc finger motif previously described in the O/E-1 sequence lies outside of the essential region (Hagman et al., 1995). A distinct domain of the Roaz protein containing the last 85 amino acids and three zinc fingers is essential for heterodimerization with O/E-1, further confirming the specificity of the Roaz/O/E-1 interactions (our unpublished data). All of the interactions described here occur in the absence of DNA and therefore are likely to represent direct association of the two proteins.
Roaz serves as a coregulator of O/E-1
The O/E-1 protein has a moderate transactivational effect on promoter fragments isolated from several olfactory-specific genes containing one or more O/E binding sites and strong activational activity on five concatemeric O/E binding sites and a minimal promoter (25-fold) (Wang and Reed, 1993). When Roaz is cotransfected with O/E-1, it completely abolishes the activational effect of O/E-1 on these native promoters. The other O/E family members display similar levels of transcriptional activation on these promoters, and elimination of transcriptional activation is observed on cotransfection with Roaz. Roaz may function as a corepressor of O/E-1 by binding to the transactivational domain of O/E-1, forming a ternary or higher order complex, as is the case for twist (Spicer et al., 1996) and p270 (Horlein et al., 1995). An alternative possibility is that formation of a heteromeric complex of Roaz and O/E-1 prevents the assembly of active O/E-1 homodimers, which recognizes an O/E binding site. The mapping of the O/E-1 homomeric and heteromeric interaction domains to an identical region that is distinct from the C-terminal transactivational domain, combined with the disruption of the O/E-1 protein–DNA complex by Roaz in EMSA, supports the latter hypothesis. These observations also argue against a role for Roaz as an O/E-1 coactivator.
The transactivational effect of Roaz and O/E-1 on the SV40 early promoter-driven reporter plasmid strongly suggests that Roaz/O/E-1 heteromultimer possesses DNA binding and transactivational activity at distinct DNA binding sites. It seems likely that this DNA binding ability is contributed by Roaz in a heteromeric complex with O/E-1, whereas the transactivational activity comes from O/E-1.
The physiological roles of Roaz
Several lines of evidence suggest that coordinated expression of genes essential for the terminally differentiated olfactory neuronal phenotype is achieved via O/E proteins acting at O/E binding sites. The importance of these sites has been demonstrated in transgenic mice, in which 300 bp containing the O/E site in OMP promoter is sufficient to drive olfactory neuron-specific reporter expression, and mutation of that site disrupts the olfactory-specific pattern (Walters et al., 1996). The existence of a family of related O/E proteins expressed in the olfactory epithelium that display a high degree of functional redundancy (Wang et al., 1997) is consistent with an important role for these factors in olfactory neuronal gene expression and explains the lack of an observable phenotype when expression of one member is abolished by gene disruption (Lin and Grosschedl, 1995). Although we have described in this paper the interactions between Roaz and the canonical member of the O/E protein family O/E-1, the existence of similar interactions with the other members of the O/E family expressed in the olfactory epithelium is consistent with the importance of Roaz in the pathway of O/E-regulated gene expression.
The onset of O/E-1 expression at E11.5 in the developing mouse olfactory epithelium significantly precedes OMP expression (E16) as well as the appearance of other terminally differentiated markers (Margalit and Lancet, 1993). In adult olfactory tissue O/E-1 message and protein are found in the immature cells (NCAM+, OMP−) (Calof and Chikaraishi, 1989) as well as in the mature receptor neurons (OMP+) (Wang and Reed, 1993). In addition, O/E binding activity is higher in the young animal and increases after bulbectomy (Kudrycki et al., 1993). These observations suggest that O/E-1 expression is regulated with a time course different from OMP during embryonic development and in the continual proliferation of ORNs in the adult tissue. These observations suggest that additional factors interacting with O/E proteins might account for these differences either by negatively regulating O/E activity in precursor cells or by functioning as a coactivator in the mature neurons.
The properties of Roaz, including its interaction with O/E proteins and its ability to inhibit the O/E-mediated transactivation, suggest that it functions as an inhibitor. It is particularly striking that the expression of Roaz in the olfactory epithelium (see Fig. 3) is found in a population of cells that contain O/E-1 message but fail to express the targets of this transcription factor. Thus, Roaz resolves the paradox between the expression of O/E-1 in the epithelium and the delayed expression of target genes. Negative regulators such as mammalian hairy and enhancer of split homolog-1 (HES-1) andtwist are involved in switching cells from a proliferative to differentiated state in the developing cortex and myoblast lineage, respectively (Baylies and Bate, 1996; Nakao and Campos-Ortega, 1996;Tomita et al., 1996). They share the common features of being expressed in the dividing precursor cells and functioning to repress the transactivational activity of proteins in the bHLH family. Negative regulators are important not only for preventing premature differentiation but also for coordinating the timing of differentiation. The induction of O/E protein expression before the onset of its known target genes has additional advantages. It allows build-up of protein in the cell so that once the inhibitor protein is removed, transcriptional activation can proceed readily. The induction of transactivation by O/E-1 and Roaz heterocomplex on the SV40 early promoter further suggests that it may act as a positive regulator at distinct sites during neurogenesis. A similar effect also has been described for twist in Drosophila (Baylies and Bate, 1996; Spicer et al., 1996).
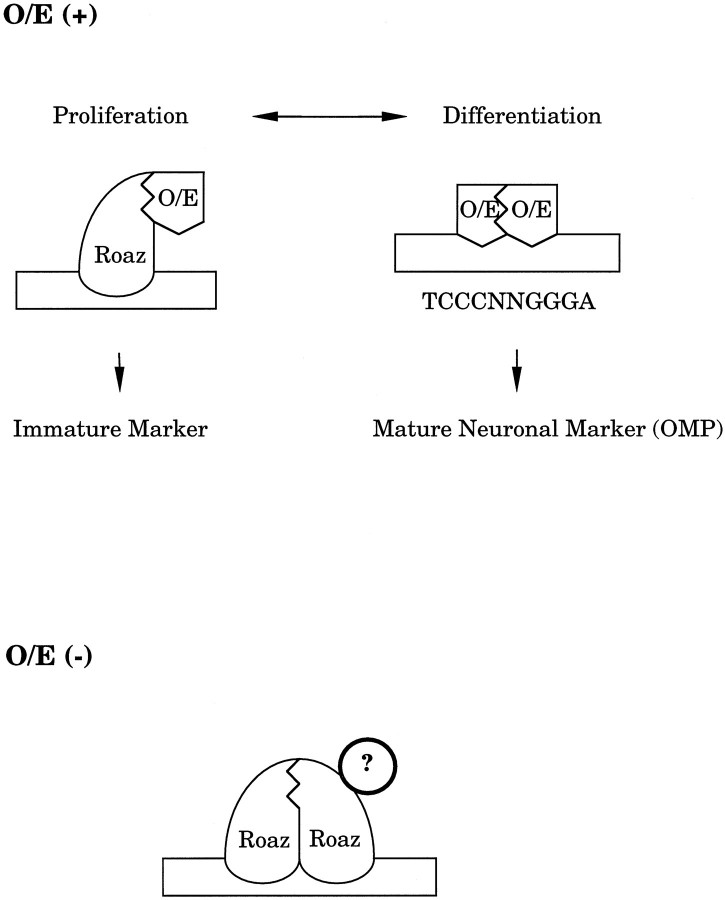
The ability of Roaz to mediate specific interactions with DNA and with heterologous proteins suggests a model in which Roaz serves several distinct roles in transcriptional regulation (Fig. 8). The transcriptional activator complex composed of O/E homodimers functions at mature neuronal marker gene promoters. In immature neurons, Roaz complexes with the O/E factors and inhibits transcriptional activation at the O/E DNA recognition site by sequestering the O/E protein and preventing formation of an active DNA-binding O/E homodimer. The heteromeric complex of O/E and Roaz possesses transcriptional activation activity at distinct sites where DNA binding is mediated by the Roaz protein. The expression of Roaz in adult tissues that do not express O/E protein suggests that, in addition to switching cells from a proliferative to differentiated state, Roaz may have distinct activities arising from the action of Roaz binding as a homodimer at specific DNA sequences (our unpublished data), where it may function as a repressor or as a transcriptional activator by association with coactivating factors present in particular cell types.
Fig. 8.
A model for the transregulatory function of Roaz.
In conclusion, the C2H2 zinc finger protein Roaz can dimerize with a helix-loop-helix protein O/E-1 both in vitro and in cell-based expression systems. The Roaz/O/E-1 protein complex functions as a negative regulator on native O/E-binding site-containing promoters while retaining the ability to bind to other DNA sequences and to function as a transcriptional activator. These activities suggest that Roaz may play a pivotal role as a switch protein in the regulation of O/E-1 transcriptional activity and the control of olfactory neuronal differentiation.
Footnotes
We thank Mengqing Xiang for his helpful suggestions in the course of this work; Jim Boulter and Mike M. Wang for their generous gifts of cDNA libraries; Steven J. Elledge for kindly providing y190 yeast strain and Song S. Wang for O/E-2 and O/E-3 plasmids; and Janine S. Davis, Jeremy Nathans, Stephen Desiderio, and other members of the Reed laboratory for their invaluable comments on this manuscript.
Correspondence should be addressed to Dr. Randall R. Reed, Room 800 Preclinical Teaching Building, The Johns Hopkins University School of Medicine, 725 North Wolfe Street, Baltimore, MD 21205.
REFERENCES
- 1.Anthony-Cahill SJ, Benfield PA, Fairman R, Wasserman ZR, Brenner SL, Stafford WD, Altenbach C, Hubbell WL, DeGrado WF. Molecular characterization of helix-loop-helix peptides. Science. 1992;255:979–983. doi: 10.1126/science.1312255. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. Wiley; New York: 1995. [Google Scholar]
- 3.Bakalyar HA, Reed RR. Identification of a specialized adenylyl cyclase that may mediate odorant detection. Science. 1990;250:1403–1406. doi: 10.1126/science.2255909. [DOI] [PubMed] [Google Scholar]
- 4.Bakalyar HA, Reed RR. The second messenger cascade in olfactory receptor neurons. Curr Opin Neurobiol. 1991;1:204–208. doi: 10.1016/0959-4388(91)90079-m. [DOI] [PubMed] [Google Scholar]
- 5.Baylies MK, Bate M. twist: a myogenic switch in Drosophila. Science. 1996;272:1481–1484. doi: 10.1126/science.272.5267.1481. [DOI] [PubMed] [Google Scholar]
- 6.Blank V, Kourilsky P, Israel A. NF-kappa B and related proteins: rel/dorsal homologies meet ankyrin-like repeats [Review]. Trends Biochem Sci. 1992;17:135–140. doi: 10.1016/0968-0004(92)90321-y. [DOI] [PubMed] [Google Scholar]
- 7.Breer H, Boekhoff I. Second messenger signaling in olfaction [Review]. Curr Opin Neurobiol. 1992;2:439–443. doi: 10.1016/0959-4388(92)90177-m. [DOI] [PubMed] [Google Scholar]
- 8.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 9.Caggiano M, Kauer JS, Hunter DD. Globose basal cells are neuronal progenitors in the olfactory epithelium: a lineage analysis using a replication-incompetent retrovirus. Neuron. 1994;13:339–352. doi: 10.1016/0896-6273(94)90351-4. [DOI] [PubMed] [Google Scholar]
- 10.Calof AL, Chikaraishi DM. Analysis of neurogenesis in a mammalian neuroepithelium: proliferation and differentiation of an olfactory neuron precursor in vitro. Neuron. 1989;3:115–127. doi: 10.1016/0896-6273(89)90120-7. [DOI] [PubMed] [Google Scholar]
- 11.Chevray PM, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danciger E, Mettling C, Vidal M, Morris R, Margolis F. Olfactory marker protein gene: its structure and olfactory neuron-specific expression in transgenic mice. Proc Natl Acad Sci USA. 1989;86:8565–8569. doi: 10.1073/pnas.86.21.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis JS, Reed RR. Role of Olf-1 and Pax-6 transcription factor in neurodevelopment. J Neurosci. 1996;16:5082–5094. doi: 10.1523/JNEUROSCI.16-16-05082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhallan RS, Yau KW, Schrader KA, Reed RR. Primary structure and functional expression of a cyclic nucleotide-activated channel from olfactory neurons. Nature. 1990;347:184–187. doi: 10.1038/347184a0. [DOI] [PubMed] [Google Scholar]
- 16.Feuerstein R, Wang X, Song D, Cooke NE, Liebhaber SA. The LIM/double zinc finger motif functions as a protein dimerization domain. Proc Natl Acad Sci USA. 1994;91:10655–10659. doi: 10.1073/pnas.91.22.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fields S, Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 18.Fields S, Sternglanz R. The two-hybrid system: an assay for protein–protein interactions [Review]. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 19.Graziadei GA, Graziadei PP. Neurogenesis and neuron regeneration in the olfactory system of mammals. II. Degeneration and reconstitution of the olfactory sensory neurons after axotomy. J Neurocytol. 1979a;8:197–213. doi: 10.1007/BF01175561. [DOI] [PubMed] [Google Scholar]
- 20.Graziadei PP, Graziadei GA. Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J Neurocytol. 1979b;8:1–18. doi: 10.1007/BF01206454. [DOI] [PubMed] [Google Scholar]
- 21.Hagman J, Belanger C, Travis A, Turck CW, Grosschedl R. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7:760–773. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- 22.Hagman J, Gutch MJ, Lin H, Grosschedl R. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 1995;14:2907–2916. doi: 10.1002/j.1460-2075.1995.tb07290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 24.Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 25.Huang ZJ, Edery I, Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature. 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 26.Hunter DD, Caggiano M, Kauer JS. Lineage analysis of the olfactory epithelium using a replication-incompetent retrovirus. Chem Senses. 1994;19:683–693. doi: 10.1093/chemse/19.6.683. [DOI] [PubMed] [Google Scholar]
- 27.Jones DT, Reed RR. Golf: an olfactory neuron-specific G-protein involved in odorant signal transduction. Science. 1989;244:790–795. doi: 10.1126/science.2499043. [DOI] [PubMed] [Google Scholar]
- 28.Kaushal S, Schneider JW, Nadal GB, Mahdavi V. Activation of the myogenic lineage by MEF2A, a factor that induces and cooperates with MyoD. Science. 1994;266:1236–1240. doi: 10.1126/science.7973707. [DOI] [PubMed] [Google Scholar]
- 29.Kudrycki K, Stein-Izsak C, Behn C, Grillo M, Akeson R, Margolis FL. Olf-1-binding site: characterization of an olfactory neuron-specific promoter motif. Mol Cell Biol. 1993;13:3002–3014. doi: 10.1128/mcb.13.5.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 31.Levin LR, Reed RR. Identification of functional domains of adenylyl cyclase using in vivo chimeras. J Biol Chem. 1995;270:7573–7579. doi: 10.1074/jbc.270.13.7573. [DOI] [PubMed] [Google Scholar]
- 32.Levy NS, Bakalyar HA, Reed RR. Signal transduction in olfactory neurons [Review]. J Steroid Biochem Mol Biol. 1991;39:633–637. doi: 10.1016/0960-0760(91)90262-4. [DOI] [PubMed] [Google Scholar]
- 33.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 34.Margalit T, Lancet D. Expression of olfactory receptor and transduction genes during rat development. Brain Res Dev Brain Res. 1993;73:7–16. doi: 10.1016/0165-3806(93)90040-h. [DOI] [PubMed] [Google Scholar]
- 35.Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 36.Nakao K, Campos-Ortega JA. Persistent expression of genes of the enhancer of split complex suppresses neural development in Drosophila. Cell. 1996;16:275–286. doi: 10.1016/s0896-6273(00)80046-x. [DOI] [PubMed] [Google Scholar]
- 37.Price M, Lemaistre M, Pischetola M, Di LR, Duboule D. A mouse gene related to Distal-less shows a restricted expression in the developing forebrain. Nature. 1991;351:748–751. doi: 10.1038/351748a0. [DOI] [PubMed] [Google Scholar]
- 38.Reed RR. How does the nose know? [Review]. Cell. 1990;60:1–2. doi: 10.1016/0092-8674(90)90706-k. [DOI] [PubMed] [Google Scholar]
- 39.Schaeren WN, Gerfin MA. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labeled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 40.Spicer DB, Rhee J, Cheung WL, Lassar AB. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- 41.Stemple DL, Mahanthappa NK. Neural stem cells are blasting off. Neuron. 1997;18:1–4. doi: 10.1016/s0896-6273(01)80018-0. [DOI] [PubMed] [Google Scholar]
- 42.Tomita K, Ishibashi M, Nakahara K, Ang S, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16:723–734. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- 43.Turner R, Tjian R. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science. 1989;243:1689–1694. doi: 10.1126/science.2494701. [DOI] [PubMed] [Google Scholar]
- 44.Turner EE, Jenne KJ, Rosenfeld MG. Brn-3.2: a Brn-3-related transcription factor with distinctive central nervous system expression and regulation by retinoic acid. Neuron. 1994;12:205–218. doi: 10.1016/0896-6273(94)90164-3. [DOI] [PubMed] [Google Scholar]
- 45.Walters E, Grillo M, Tarozzo G, Stein-Izsak C, Corbin J, Bocchiaro C, Margolis FL. Proximal regions of the olfactory marker protein gene promoter direct olfactory neuron-specific expression in transgenic mice. J Neurosci Res. 1996;43:146–160. doi: 10.1002/(SICI)1097-4547(19960115)43:2<146::AID-JNR3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 46.Wang MM, Reed RR. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- 47.Wang MM, Tsai RY, Schrader KA, Reed RR. Genes encoding components of the olfactory signal transduction cascade contain a DNA binding site that may direct neuronal expression. Mol Cell Biol. 1993;13:5805–5813. doi: 10.1128/mcb.13.9.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang SS, Tsai RYL, Reed RR. The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J Neurosci. 1997;17:4149–4158. doi: 10.1523/JNEUROSCI.17-11-04149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell TK, Turner D, Rupp R, Hollenberg S. The myoD gene family: nodal point during specification of the muscle cell lineage [Review]. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]