Abstract
Prostaglandin E2 (PGE2) mediates the stimulatory effect of norepinephrine (NE) on the secretion of luteinizing hormone-releasing hormone (LHRH), the neuropeptide controlling reproductive function. In rodents, this facilitatory effect requires previous exposure to estradiol, suggesting that the steroid affects downstream components in the cascade that leads to PGE2-induced LHRH release. Because astroglia are the predominant cell type contacting LHRH-secreting nerve terminals, we investigated the involvement of hypothalamic astrocytes in the estradiol facilitation of PGE2-induced LHRH release. A subpopulation of LHRH neurons was found to express the mRNA encoding the PGE2 receptor subtype EP1-R, which is coupled to calcium mobilization. The LHRH-producing cell line GT1–1 also contains EP1-R mRNA and, to a lesser extent, the three alternatively spliced forms of EP3-R mRNA (α, β, and γ) that encode receptors linked to inhibition and stimulation of cAMP formation. Hypothalamic astrocytes treated with estradiol produced a conditioned medium that when applied to GT1–1 cells resulted in a selective upregulation of EP1-R and EP3γ-R mRNAs. The conditioned medium also enhanced the LHRH response to EP1-R and EP3-R agonists and the cAMP response to EP3-R activation. Thus, one mechanism by which estradiol facilitates the effect of neurotransmitters acting via PGE2 to stimulate LHRH release is by enhancing the glial production of substances that upregulate PGE2 receptors on LHRH neurons. The existence of such a mechanism underscores the emerging importance of glial–neuronal communication in the control of brain neurosecretory activity.
Keywords: gonadal steroids, astrocytes, hypothalamus, neuropeptide secretion, prostaglandin receptors, glial–neuronal interactions
The secretory activity of luteinizing hormone-releasing hormone (LHRH) neurons is controlled by both neuronal and glial inputs (Ramirez et al., 1984; Kalra, 1986;Ojeda, 1994; Terasawa, 1995). Although neuronal inputs are conveyed via trans-synaptic mechanisms, glial influences are exerted via substances able to effect cell–cell communication in a trans-synaptic-independent manner (Ojeda et al., 1990; Gallo et al., 1995; Melcangi et al., 1995;Voigt et al., 1996; Ma et al., 1997). Among the neurotransmitters affecting LHRH release, norepinephrine (NE) is, perhaps, one of the best characterized, because its effects on LHRH secretion have been thoroughly documented using both in vivo and in vitro approaches (for review, see Barraclough and Wise, 1982;Ramirez et al., 1984; Kalra, 1986). The stimulatory effect of NE on LHRH release requires the intermediacy of prostaglandin E2(PGE2) (Ojeda et al., 1979, 1982), and the intracellular mechanisms underlying the actions of PGE2include mobilization of calcium from intracellular stores (Ojeda and Negro-Vilar, 1985) and cAMP formation (Ojeda et al., 1985).
Surprisingly, central administration of NE to ovariectomized rats or stimulation of a mesencephalic noradrenergic pathway in these animals suppresses LH release, in contrast to the increase seen in estrogen-primed rats (Gallo and Drouva, 1979; Leung et al., 1981,1982). It is possible that in the absence of estrogen, NE no longer sets in motion stimulatory mechanisms but instead activates pathways inhibitory to LHRH secretion. Notwithstanding any action of estradiol on the neuronal circuitry synaptically connected to LHRH neurons, it seems clear that a significant part of the facilitatory effect of estradiol is exerted at the median eminence (ME), where the LHRH neuronal axons converge to release the neuropeptide into the portal vasculature. Median eminence fragments from ovariectomized rats respond less not only to NE but also to PGE2 (Ojeda et al., 1986), indicating that estradiol is required for the manifestation of events initiated by the prostaglandin.
In earlier reports, we and others (Ojeda et al., 1982; Gearing and Terasawa, 1991) postulated that NE stimulates PGE2 by binding to α1-adrenergic receptors located on LHRH neurons and that, consequently, PGE2 was an intracellular messenger acting within the LHRH neuron itself. There are, however, several findings indicating the need to revise this concept. First, noradrenergic neurons do not seem to contact LHRH neuronal perikarya directly (Leranth et al., 1988). Second, noradrenergic axons projecting to the ME terminate in the internal rather than the external layer where most of the LHRH nerve terminals are located (McNeill and Sladek, 1978; Jennes et al., 1982). Third, NE induces LHRH release from the LHRH-producing neuronal cell lines GT1 by activating β1-adrenoreceptors (Martinez de la Escalera et al., 1992) instead of the α1 receptors shown to mediate its effectin vivo. Fourth, the actions of PGE2 are initiated by binding to the extracellular domain of membrane-anchored receptors (Coleman et al., 1990; Narumiya, 1994). This localization suggests that after α-adrenoreceptor-mediated NE stimulation, the prostaglandin is not synthesized within LHRH neurons themselves but rather by an intermediate cell type. After release from these cells, PGE2 would bind to specific receptors located on the LHRH neuron cell membrane.
Astroglia represent the cell type most intimately associated with LHRH nerve terminals in the ME (Kozlowski and Coates, 1985; Ugrumov et al., 1989; Silverman et al., 1991). The findings that both glial cells of the median eminence and isolated hypothalamic astrocytes are targets of estradiol action (Witkin et al., 1991; Langub and Watson, 1992; King and Letourneau, 1994; Ma et al., 1994a) and that astrocytes release PGE2 in response to growth factor stimulation (Ma et al., 1997) prompted us to consider the possibility that the sensitizing effect of estradiol on PGE2-induced LHRH release is, at least in part, mediated by glial cells.
The present experiments provide evidence of this concept. Partial reports of the results in this paper have been published previously (Rage et al., 1994, 1995).
MATERIALS AND METHODS
Animals
Pregnant Sprague Dawley rats were purchased from B & K Universal (Fremont, CA) and housed in a room with a controlled photoperiod (14:10 hr light/dark cycle; lights on from 5:00 A.M. to 7:00 P.M.) and temperature (23–25°C). They were fed pelleted food and tap waterad libitum. Two to three days after birth, the pups were used to prepare astrocyte cultures. Immature (27–28-d-old) female mice of the CD strain were used for detection of prostaglandin receptor mRNA on hypothalamic LHRH neurons in situ. They were purchased from Charles River Laboratories (Wilmington, MA) and housed under the same conditions outlined above but in special quarters for mice.
Reagents
The selective EP1-R agonist 17-phenyl trinor PGE2and the EP3-R and EP1-R agonist sulprostone were purchased from Cayman Chemical (Ann Arbor, MI). The EP2-R agonist butaprost was the generous gift of Dr. P. J. Gardiner (Bayer PLC-Stoke Court Research, Berkshire, United Kingdom). The EP1-R antagonist AH-6809 was generously provided by Dr. Simon G. Lister (Glaxo Wellcome Medicines Research Centre, Hertfordshire, United Kingdom). Isobutyl methylxanthine was from Sigma (St. Louis, MO).
Cell culture
Hypothalamic astrocytes were purified by the method of McCarthy and de Vellis (1980), as reported by Ma et al. (1994a). In brief, hypothalamic cells were first cultured to confluency in T-75 flasks (8–10 d). At this time, contaminating cells (neurons and oligodendrocytes) were removed by shaking the cultures at 37°C for 6 hr at 250 rpm, followed by replacement of the medium and a second shaking period of 18 hr. The purified astrocytes were then seeded on six-well plates at 800,000 cells per well and grown in a mixture of DMEM and F-12 medium (1/1, v/v) containing 10% bovine calf serum plus penicillin (100 U/ml) and streptomycin (100 μg/ml). After reaching 80–90% confluency, the serum-containing medium was replaced by an astrocyte-defined medium (ADM) consisting of a glutamate-free DMEM without phenol red and supplemented with l-glutamine (2 mm), HEPES (15 mm), insulin (5 μg/ml), and putrescine (100 μm). The experiments were initiated 48 hr later by treating the cultures with 17β-estradiol (17β-E2), its inactive stereoisomer 17α-estradiol (17α-E2), or ethanol (ETOH) (see below).
The immortalized LHRH-producing cells GT1–1 (kindly provided by Dr. R. Weiner, University of California, San Francisco, CA) were grown in DMEM containing 10% fetal calf serum and the antibiotics indicated above under an atmosphere of 5% CO2/95% air at 37°C. For experiments involving measurement of LHRH or cAMP release, the cells were seeded at 100,000 cells/well in 24-well plates and used at 50–60% confluency. For experiments involving RNA measurements, the cells were seeded at 500,000 cells/well in six-well plates and used at 70–80% confluency. At this time, the medium was replaced by the above described ADM. The experiments were performed 24 hr later in either ADM or astrocyte-conditioned media.
Treatments
Astrocytes
After 48 hr in ADM, the astrocytes were exposed for 12, 24, and 48 hr to ADM containing 17β-E2 (1 nm), 17α-E2 (1 nm), or the diluent ethanol (5 μl/ml). The conditioned media (CM-17β-E2, CM-17α-E2, and CM-ETOH, respectively) were collected at the end of the treatments and used to determine potential effects on EP receptor gene expression in GT1–1 cells (see below). Initial experiments demonstrated that the most effective treatment was the CM from a 24 hr exposure of the astrocytes to 17β-E2. This interval was then selected to prepare CM for all subsequent experiments.
GT1–1 cells
To determine the effect of the different CMs on EP-R mRNA content of GT1–1 cells, we exposed the cells to the CMs for 1, 2, 4, 6, and 8 hr before RNA extraction. Direct estradiol effects were controlled for by treatment of GT1–1 cells with the hormone at 1 nm.
Other studies were performed to determine the ability of EP1 and EP3 receptor agonists to stimulate LHRH release after exposure of GT1–1 cells to astrocyte CMs. In trial experiments, the cells were exposed to the CMs for 1, 2, or 3 hr before addition of the receptor agonist, and the GT1–1 culture medium was removed for LHRH measurement after both 30 and 60 min exposures to the agonists. Based on the results from these preliminary experiments, a protocol was adopted in which the cells were first treated with the different CMs for 3 hr, the medium was then removed, and the cells were exposed for 30 min to different doses of the EP1-R agonist 17-phenyl trinor PGE2 (Coleman et al., 1990; Watabe et al., 1993) or the EP3-R and EP1-R agonist sulprostone (Coleman et al., 1990). The specificity of EP1-R activation by 17-phenyl trinor PGE2 was verified by exposing the cells to the agonist in the presence of the selective EP1-R antagonist AH-6809 (Coleman et al., 1990; Watabe et al., 1993). The antagonist was added to the cultures 30 min before the agonist. The ability of sulprostone to activate EP3 receptors positively linked to the cAMP-generating system (Irie et al., 1993) was assessed by measuring cAMP release to the culture medium of GT1–1 cells treated with the agonist. The medium of cultures treated with sulprostone was supplemented with isobutyl methylxanthine (IBMX; 0.5 mm) to inhibit phosphodiesterase activity. At the end of these treatments, LHRH and cAMP released to the culture medium were measured by RIA (see below).
Finally, it was important to determine whether GT1–1 cells have functional EP2 receptors. The presence of these receptors in GT1–1 cells was assessed by treating the cells with different concentrations of the EP2-R agonist butaprost (Nishigaki et al., 1995) diluted in ADM.
RNA extraction
Total RNA was extracted by the method of Peppel and Baglioni (1990) as modified by Salvatori et al. (1992) for RNA extraction from cultured cells.
Probes
EP1-R mRNA was detected with a cRNA complementary to nucleotides (nt) 1065–1312 in mouse EP1-R mRNA (Watabe et al., 1993). The cDNA template for transcription was obtained by subcloning an appropriatePstI–EcoRI fragment from a 1.3 kb cDNA (Watabe et al., 1993) into the corresponding sites of the riboprobe vector pGEM-3Z. After linearizing the vector with EcoRI, transcription with SP6 RNA polymerase yields a 268 nt cRNA, of which 21 nt correspond to vector sequences. Sense EP1-R mRNA, used to construct standard curves for RNase protection assays, was synthesized from the same template linearized with HindIII and using T7 RNA polymerase.
EP3-R mRNAs were detected with a cRNA derived from the 3′-end of a mouse EP3β-R cDNA (Sugimoto et al., 1992). The DNA template used for transcription was prepared by subcloning a blunt-endedBstEII–HindIII fragment (nt 774–1282 in EP3β-R mRNA) into the SmaI site of pGEM-3Z. After linearization of the vector with HindIII, T7 polymerase-directed transcription yields a probe of 577 nt, of which 69 nt correspond to vector sequences.
To detect EP4-R mRNA, we excised a SmaI–SmaI DNA fragment complementary to nt 703–1362 in EP4-R mRNA from a 2.3 kb mouse EP4-R cDNA (Honda et al., 1993) and subcloned this fragment into the SmaI site of pGEM-3Z. Transcription of theEcoRI-linearized template yields a 728 nt transcript, of which 69 nt correspond to vector sequences.
Cyclophilin mRNA, which is constitutively expressed in many cells (Danielson et al., 1988), was used in RNase protection assays to correct for procedural variability and, thus, to normalize the EP1-R mRNA values obtained. The labeled cyclophilin cRNA was transcribed from a cDNA template generated by PCR amplification of a 158 bp fragment corresponding to nt 265–422 in the rat cyclophilin mRNA sequence (Danielson et al., 1988). Because of the similarity of the rat to the mouse sequence, mouse cyclophilin mRNA protects 110 nt of this probe (Voigt et al., 1996). The cDNA was cloned into the SmaI site of pGEM-3Z, linearized with XbaI, and transcribed using T7 RNA polymerase.
Estrogen receptor (ER) mRNA was detected by RNase protection assay in GT1–1 cells using a 608 nt cRNA probe complementary to nt 1470–2078 in the 3′-end of mouse ER mRNA (White et al., 1987). The cRNA was transcribed, using SP6 RNA polymerase, from a 608 bp cDNA template generated by linearizing a 2.1 kb mouse ER cDNA (kindly provided by Dr. R. White, Imperial Cancer Research Fund, London, United Kingdom) withBglII.
When cRNAs were used in RNase protection assays, the probes were transcribed using 32P-UTP as the radiolabeled nucleotide. When used for in situ hybridization experiments, the radiolabeled nucleotide was 35S-UTP.
Measurement of EP3-R mRNA variants
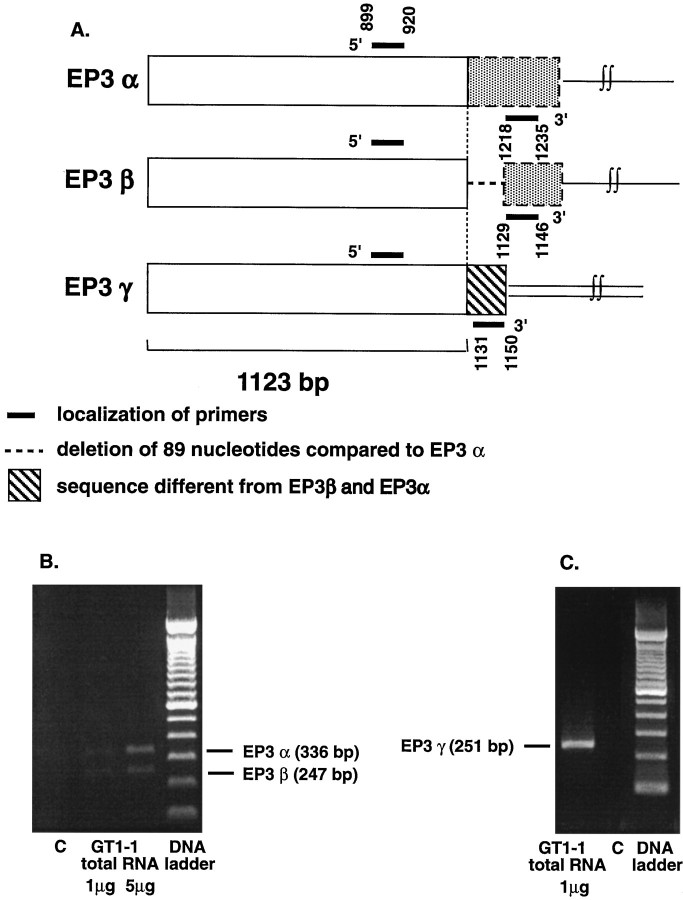
The EP3-R gene can be alternatively spliced to produce at least three mRNA variants, which differ in portions of their 3′-ends (Irie et al., 1993; Sugimoto et al., 1993). Because the EP3β-R cRNA used in RNase protection assays does not differentiate between the EP3α-R and EP3γ-R forms, it was necessary to detect them by a different method. We chose the method of reverse transcription (RT)–PCR because of its resolution capabilities, sensitivity, and our previous experience with the technique (Ma et al., 1994b; Dissen et al., 1995).
Primers
All oligodeoxynucleotides used were synthesized in an Applied Biosystems 391 DNA synthesizer (Foster City, CA). To amplify EP3α-R and EP3β-R DNA fragments, we used a 21 mer sense primer (5′-ATG-GGG-ATC-ATG-TGT-GTG-CTG-3′) corresponding to nt 899–920 of the EP3-R common sequence (see Fig. 3) and an antisense 18 mer primer (5′-GTC-CAC-TTC-AGG-TTG-TTC-3′) corresponding to nt 1218– 1235 in EP3α-R mRNA and 1129–1146 in EP3β-R mRNA (Sugimoto et al., 1993) (see Fig. 3). An EP3γ-R DNA fragment was amplified using the same 5′-sense oligodeoxynucleotide and a 3′-primer corresponding to nt 1131–1150 in EP3γ-R mRNA (Irie et al., 1993) (see Fig. 3). We also synthesized an internal primer corresponding to nt 1064–1081 in the EP3-R common sequence (Irie et al., 1993; Sugimoto et al., 1993). This oligodeoxynucleotide was used as a probe in Southern blots to identify the amplified EP3-R cDNA forms. Finally, a set of cyclophilin-specific primers (sense, 5′-GGG-AAG-TCC-ATC-TAC-GGA-3′; antisense, 5′-CAC-TGC-TTG-CCA-TCC-AAC-3′), corresponding to nt 265–282 and 405–422, respectively, in rat cyclophilin mRNA, was synthesized for the simultaneous amplification of cyclophilin mRNA and EP3-R mRNA forms in the quantitative RT–PCR assays (see below).
Fig. 3.
PCR cloning of cDNAs encoding the alternatively spliced products of the EP3-R gene from GT1–1 cells. A, mRNA location of the deoxyoligonucleotide primers used for amplification of the α, β, and γ forms of EP3-R mRNA.B, Ethidium bromide staining of the PCR products derived from the amplification of GT1–1 RNA using a pair of primers common for the mRNA sequences encoding the EP3α-R and EP3β-R mRNA forms.C, Ethidium bromide staining of the single PCR product derived from the amplification of GT1–1 RNA using a 5′-primer complementary to the mRNA sequence common to all forms of EP3-R mRNA and a 3′-primer complementary to the unique 3′-sequence in EP3γ-R mRNA. C, PCR control, no RNA input.
RT-PCR for DNA cloning
RT was performed for 2 hr at 37°C in a 20 μl volume containing 1 or 5 μg of total RNA from GT1–1 cells, 1× RT buffer (50 mm Tris-HCl, pH 8.3; 75 mm KCl; and 3 mm MgCl2), 0.01 mdithiothreitol, 0.5 mm each dNTP, 20 U RNasin, 25 pmol of an oligo-dT primer, and 200 U of Moloney murine leukemia virus reverse transcriptase (Life Technologies, Gaithersburg, MD). Two μl of this reaction were then used for PCR amplification. The PCR reaction was performed in a 75 μl volume consisting of two parts. Part A contained the RT mixture: 7.5 μl of 10× Taq buffer (Promega, Madison, WI), 3 μl of 25 mm MgCl2, and 1 μl of 10 mm dNTPs in a 60 μl volume. Part B contained 50 pmol of each EP3-R specific sense and antisense primer and 0.5 U ofTaq polymerase. Part A was dispensed into 0.6 ml tubes, overlaid with oil, and held at 94°C for 7 min in a thermal cycler (MJ Research, Watertown, MA) to inactivate the reverse transcriptase. Part B was added through the oil, while the tubes were at 72°C to diminish nonspecific annealing of the primers (“hot” start). The subsequent PCR reaction consisted of 35 cycles of denaturing at 94°C for 15 sec, annealing at 55°C for 1 min, and extension at 72°C for 2 min, followed by a final extension of 7 min at 72°C.
Southern blot and sequencing
The PCR products were separated by gel electrophoresis on a 2% agarose gel, transferred to Nytran membranes (Stratagene, La Jolla, CA), cross-linked, and hybridized with the internal primer described above with the 5′-end labeled with γ-32P-dATP, as reported previously (Ma et al., 1994b). After identification and isolation of the appropriate bands, they were cloned into the pGEM-T vector (Promega) and sequenced by the dideoxynucleotide termination method of Sanger et al. (1977) using Sequenase T7 DNA polymerase and a kit (Sequenase Version 2.0) purchased from United States Biochemicals (Cleveland, OH).
Quantitative RT–PCR
Preparation of polyadenylated mRNA standards.Synthetic mRNA standards used to quantitate the changes in EP3-R mRNA (α, β, and γ forms) were prepared exactly as described (Ma et al., 1994b; Dissen et al., 1995). In brief, the corresponding cDNAs were first subcloned into the SmaI site of the pSP64 A+ vector and then transcribed using SP6 RNA polymerase to generate transcripts having a polyadenylated tail. RNA yields were estimated by absorbance at 260 nm and by comparison with known amounts of RNA in ethidium bromide-stained gels.
Assay. The conditions for both reverse transcription and PCR amplification were the same as those described for the cloning of EP3-R cDNA fragments (see above). In this case, however, each reaction tube contained two sets of primers, one to amplify the EP3-R mRNA form of interest (20 pmol of each primer) and another (10 pmol of each primer) to amplify a fragment from cyclophilin mRNA to be used as an internal standard for normalization of the EP3-R mRNA values obtained. Different amounts of standard mRNA were reverse transcribed and amplified at the same time as the unknowns.
Quantitative analysis. The assay used is designed to minimize the two main sources of variability in quantitative PCR, i.e., variability caused by differences in RT and primer efficiency and variability caused by “tube” effects and sample-to-sample processing errors (Wang et al., 1989). The former is minimized by referring the experimental values to mRNA standards identical to the target sequences and processed in the same assay; the latter is reduced by coamplifying a fragment of cyclophilin, a constitutively expressed gene (Danielson et al., 1988).
Aliquots of each PCR reaction were electrophoresed in a 2% agarose gel containing ethidium bromide, photographed, and analyzed by computer densitometry exactly as described previously (Ma et al., 1994b). The mRNA values obtained were normalized according to the cyclophilin content of each sample and are expressed as fg/sample.
RNase protection assay
The presence of EP1- and EP3-R and of estrogen receptor mRNA in GT1–1 cells was detected by RNase protection assay using a procedure described previously (Ma et al., 1996). To quantitate the changes in EP1-R mRNA observed after estradiol treatment, we first scanned the hybridization signals using an Agfa flatbed scanner and then analyzed them with an edited version of the computer program National Institutes of Health-Image (Correa-Rotter et al., 1992). The optical density of each protected band was compared with that of a standard curve generated by different amounts of in vitro synthesized EP1-R mRNA. The values obtained were then normalized using as a normalizing unit the cyclophilin mRNA values detected in each sample (Ma et al., 1994a, 1996).
Combined immunohistochemistry and in situ hybridization
Because the PGE2 receptor subtypes EP1 and EP3 were identified and studied in GT1–1 cells, which are of murine origin, mouse brains were used to determine whether the mRNAs encoding these receptors are also expressed in normal LHRH neurons. The brains of four immature female mice were fixed by transcardiac perfusion of 4% paraformaldehyde in borate buffer, pH 9.5, as reported (Berg-von der Emde et al., 1995). After an overnight post-fixation in the same fixative plus 10% sucrose, blocks of tissue containing the septum, diagonal band of Broca, the preoptic area, and the rostral part of the anterior hypothalamic area were frozen on dry ice and stored at −85°C until sectioned. Thirty micrometer sections were obtained with a frozen sliding microtome and stored in cryoprotectant (Simmons et al., 1989) at −20°C.
On the day of the immunohistochemical procedure, the sections were rinsed extensively in 0.02 m potassium phosphate buffer, pH 7.4 (KPBS); incubated in KPBS containing 2% BSA, 5 mm DTT, and 0.3% Triton X-100 for 2–4 hr at 4°C; and then incubated overnight at 4°C with the polyclonal LHRH antibody HU60 diluted at 1:3200 in KPBS. The specificity of this antibody has been described previously (Urbanski, 1990). On the next day, the sections were rinsed in KPBS and incubated for 45 min with a goat anti-rabbit gamma globulin (Vector Laboratories, Burlingame, CA) diluted 1:250 and then for 1 hr with ABC (Vector Laboratories), before developing the reaction to a brown color with diaminobenzidine (Nilaver and Kozlowski, 1989). After completion of the reaction, the sections were mounted on Superfrost Plus glass slides (Fisher Scientific, Houston, TX) and dried overnight under vacuum before being subjected to the hybridization histochemistry procedure.
Hybridization histochemistry was performed as described (Berg-von der Emde et al., 1995) according to the procedure reported by Simmons et al. (1989). LHRH neurons were considered to contain EP1-R or EP3-R mRNA when the density of the silver grains overlying the cell was at least twice that of an area devoid of other cells.
Radioimmunoassays
LHRH was measured as described using the same antibody used for immunohistochemistry but at a 1:25,000 dilution. cAMP was measured as reported earlier (Ojeda et al., 1988) using a rabbit anti-3′,5′-cAMP-BSA antibody (ICN Biomedicals, Costa Mesa, CA) at a 1:200 dilution. The samples and the standards were acetylated before the assay to enhance the sensitivity of detection (Brooker et al., 1979). Under these conditions, the sensitivity of the assay is 2 fmol/tube, and the standard curve is linear between 4 and 200 fmol/tube.
Statistics
Results were analyzed using a one-way ANOVA followed by the Student–Neumann–Keuls multiple comparison test for unequal replications.
RESULTS
LHRH neurons express the gene encoding EP1-R
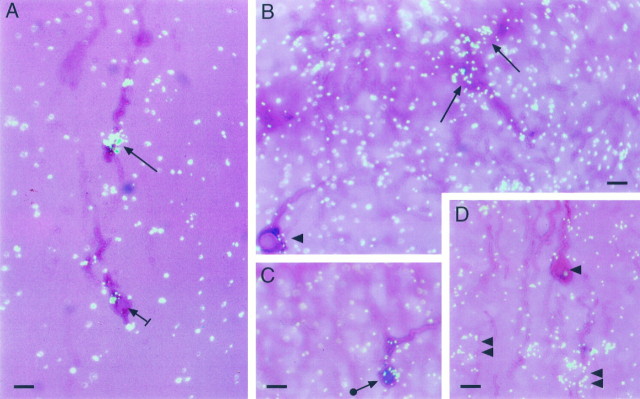
Combined immunohistochemistry–in situ hybridization revealed that the basal forebrain of prepubertal (28-d-old) female mice contains immunoreactive LHRH neurons expressing the mRNA encoding EP1-R (Fig. 1). A minority of LHRH neurons exhibited strong (Fig. 1A) to moderate (Fig.1B) EP1-R mRNA hybridization; in others, the content of EP1-R mRNA was more discrete (Fig. 1C, arrowwith circle); and still, in many others there was either no hybridization (Fig. 1B,D,arrowheads) or a questionable signal (Fig.1A, short arrow). EP1-R mRNA was also expressed in LHRH-negative cells present in the immediate vicinity of EP1-R mRNA-negative LHRH neurons (Fig. 1D,double arrowheads). Whereas the neuronal nature of these EP1-R-containing, LHRH-negative cells was evident, there were other regions, particularly near the OVLT, that exhibited a more diffuse pattern of hybridization (Fig. 1B). Although this pattern could be interpreted as hybridization background, its unevenness suggests the presence of low levels of EP1-R mRNA in astrocytes. A similar hybridization profile has been reported for epidermal growth factor receptor mRNA, which is also expressed in astrocytes (Ma et al., 1994c). Ten percent or less of the LHRH neurons identified by immunohistochemistry in the four brains examined contained detectable EP1-R mRNA levels. In contrast to EP1-R, we did not find any LHRH neurons showing a distinct EP3-R mRNA hybridization signal; the neurons exhibited either no hybridization (Fig.1D) or a weak signal difficult to resolve from background hybridization without quantitative analysis. Because combined immunohistochemistry–in situ hybridization inevitably results in loss of mRNA content and/or decreased accessibility of the 35S-labeled cRNA to cellular mRNA, no attempts were made to perform such an analysis. The presence of a “borderline” hybridization signal and the detection of EP3-R mRNA in GT1–1 LHRH-producing cells (see below) do, however, suggest the presence of EP3-R in LHRH neurons in situ.
Fig. 1.
Detection of EP1-R mRNA on LHRH neurons of the mouse brain by combined immunohistochemistry–in situhybridization. The LHRH decapeptide was first detected by immunohistochemistry using a polyclonal antibody (see Materials and Methods). After completion of this procedure, the sections were hybridized with an 35S-UTP-labeled EP1-R cRNA; the reaction was developed after a 4 week exposure to NTB-2 emulsion.A, LHRH neurons dorsal to the anterior recess of the third ventricle showing a low (short arrow) to abundant (arrow) content of EP1-R mRNA. B, LHRH neurons in the vicinity of the organum vasculosum of the lamina terminalis (OVLT) showing either moderate EP1-R mRNA levels (arrows) or an undetectable hybridization signal (arrowhead). C, An LHRH neuron in the preoptic area showing moderate levels of EP1-R mRNA (arrow with circle). D, An LHRH neuron devoid of detectable EP1-R mRNA (arrowhead) located in the vicinity of unidentified cells containing EP1-R mRNA (double arrowheads). Scale bar, 10 μm.
The LHRH-producing cell line GT1–1 expresses the mRNAs encoding both the EP1-R and EP3-R genes
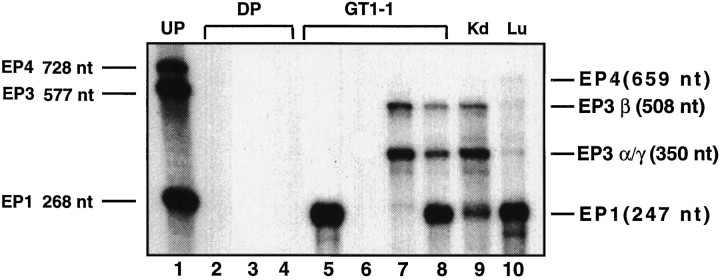
RNase protection assay demonstrated that GT1–1 cells contain EP1-R and EP3β-R mRNAs (Fig. 2). The protected fragments were identical in size to those protected by total RNA derived from kidney, a tissue known to express the EP1 and EP3β receptors (Sugimoto et al., 1993; Watabe et al., 1993). GT1–1 cells also contain mRNA species protecting a portion of the probe corresponding to the expected size of EP3α-R and EP3γ-R mRNAs. They do not, however, express EP4-R mRNA, which (as shown in Fig. 2) is present at low levels in lung.
Fig. 2.
Detection of prostaglandin E2 receptor mRNAs in GT1–1 cells by RNase protection assay. Lane 1, Undigested probes (UP); lanes 2, 3, 4, digested EP1-R, EP3β-R, and EP4-R cRNA probes, respectively (DP); lane 5, GT1–1 RNA hybridized to the EP1-R cRNA probe alone; lane 6, GT1–1 mRNA hybridized to the EP4-R cRNA probe alone; lane 7, GT1–1 RNA hybridized to the EP3β-R cRNA probe alone; lane 8, GT1–1 RNA hybridized to both the EP1-R and EP3β-R probes;lane 9, kidney (Kd) RNA hybridized to both probes; and lane 10, lung (Lu) RNA hybridized to all three probes.
The EP3β-R cRNA used for the RNase protection assay does not differentiate between the EP3α-R and EP3γ-R alternatively spliced forms of the EP3-R gene, because each of these species protects a portion of the probe corresponding to the common EP3-R mRNA sequence (of ∼350 nt), whereas EP3α-R protects an additional fragment of 160 nt (not shown in Fig. 2). To determine whether the α and/or the γ forms are expressed in GT1–1 cells, we subjected total mRNA from these cells to RT–PCR amplification using specific oligodeoxynucleotide primers (Fig. 3A; see Material and Methods for details). The common primers for EP3α-R and EP3β-R mRNA amplified two fragments of the expected size (Fig. 3B). Sequencing of each DNA fragment verified that the 336 bp cDNA contains the EP3α-R sequence, whereas the 247 bp fragment corresponds to the EP3β-R sequence. The EP3γ-R primers amplified a 251 bp fragment (Fig. 3C), which corresponded to the EP3γ-R mRNA sequence. Thus, GT1–1 cells express all three alternatively spliced mRNA forms known for the EP3-R gene.
Astrocytes treated with estradiol secrete substances that selectively increase EP1-R and EP3γ-R mRNA levels in GT1–1 cells
To study the effect of estradiol on EP receptor expression, we treated cultures of GT1–1 cells with CM from astrocytes exposed to 17β-estradiol, the inactive stereoisomer 17α-estradiol, or the diluent ethanol. The changes in EP1-R mRNA were measured by RNase protection assay; the changes in EP3-R alternatively spliced mRNA forms were detected by quantitative RT–PCR.
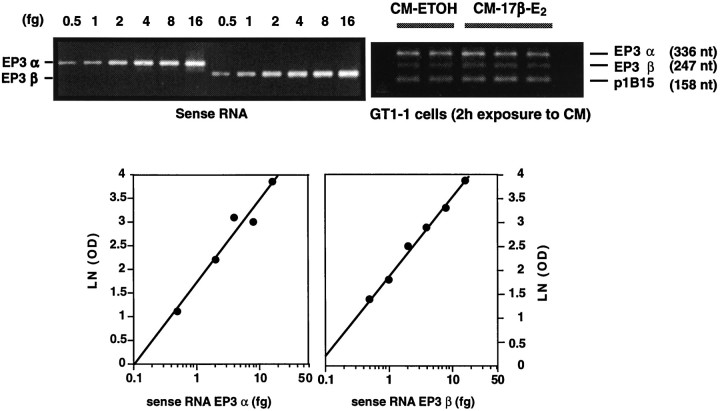
Figure 4 illustrates the characteristics of this quantitative assay. PCR amplification of increasing amounts ofin vitro transcribed EP3α-R and EP3β-R mRNAs resulted in a concentration-dependent increase in the amount of PCR product detected by ethidium bromide staining (top left). Regression analysis of these PCR signals demonstrates the linearity of the amplification reactions for both EP3α-R and EP3β-R mRNAs (bottom). As shown in the top right, the method allows one to measure simultaneously three mRNAs (EP3α-R, EP3β-R, and that encoding the housekeeping protein cyclophilin, p1B15) in a single assay. This experiment (top right) illustrates the inability of CM-17β-E2 to affect the abundance of either EP3α-R or EP3β-R mRNA in GT1–1 cells (see below).
Fig. 4.
Quantitation of EP3-R mRNA alternatively spliced forms by quantitative RT–PCR. Top left, Ethidium bromide staining of PCR products obtained from the amplification of increasing amounts of in vitro transcribed EP3α-R and EP3β-R mRNAs, corresponding to the same cellular sequence targeted for amplification. Bottom, Standard curves generated by regression analysis of the PCR signals (top left) and used to estimate the content of EP3α and EP3β-R mRNAs in GT1–1 cells. Top right, Example of an RT–PCR assay demonstrating the ability of the assay to detect simultaneously three mRNAs in GT1–1 cells [in this case EP3α-R, EP3β-R, and cyclophilin (p1B15)]. Cyclophilin mRNA was used to normalize the EP3-R mRNA values detected in the assay (see Materials and Methods); in all cases the amount of total RNA reverse transcribed for PCR amplification was 100 ng. LN (OD), Natural logarithm of optical density.
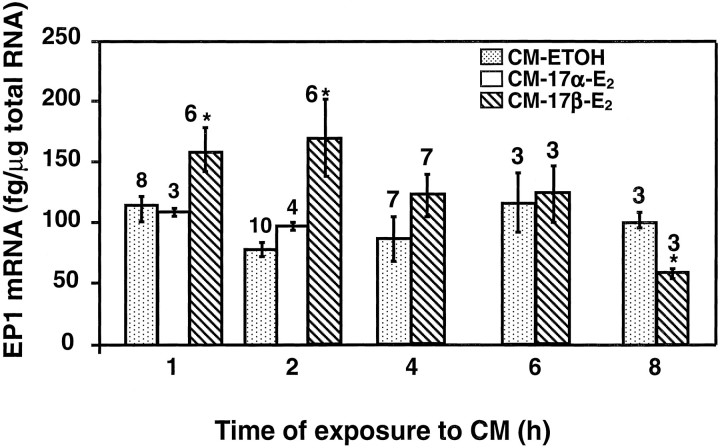
EP1-R mRNA levels, measured by RNase protection assay, increased within 1 hr of exposure to CM-17β-E2, remained significantly elevated for the next hour, and decreased thereafter to control values (Fig. 5). By 8 hr, the levels seemed to be lower than that in ethanol-treated cells. Of the three alternatively spliced forms of the EP3-R gene expressed in GT1–1 cells, only EP3γ-R mRNA levels were affected by the CM-17β-E2 treatment (Fig.6). As seen with EP1-R mRNA, EP3γ-R mRNA levels increased rapidly after CM-17β-E2 exposure (within 2 hr) and returned to control values thereafter.
Fig. 5.
Increase in EP1-R mRNA levels in GT1–1 cells by exposure to culture medium derived from astrocytes treated for 24 hr with 17β-estradiol (1 nm;CM-17β-E2).CM-17α-E2, Medium from astrocytes treated with the inactive stereoisomer 17α-estradiol;CM-ETOH, medium from astrocytes treated with ethanol. Numbers above the bars are the number of independent observations per group; error bars indicate SEM; and *p < 0.05, 17β-estradiol versus control group(s).
Fig. 6.
Selective increase in EP3γ-R mRNA levels in GT1–1 cells by exposure to culture medium derived from astrocytes treated for 24 hr with 17β-estradiol (1 nm;CM-17β-E2).CM-17α-E2, Medium from astrocytes treated with 17α-estradiol; CM-ETOH, medium from astrocytes treated with ethanol. Numbers above the barsare the number of independent observations per group; error bars indicate SEM; and *p < 0.01, 17β-estradiol versus control group(s).
17β-E2 does not act directly on GT1–1 cells to affect EP-R gene expression
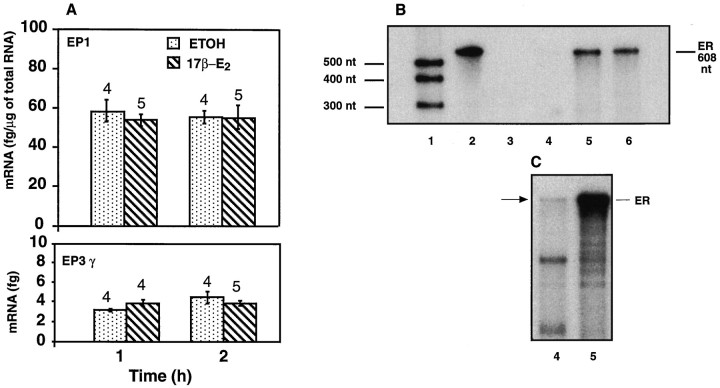
Other investigators have shown the presence of functional estrogen receptors on GT1 cells (Poletti et al., 1994), raising the possibility that the increases in EP-R mRNAs caused by CM-17β-E2 are because of a direct effect of the steroid on GT1–1 cells. Experiments addressing this issue demonstrated, however, that direct exposure of GT1–1 cells to 17β-E2, for 1–2 hr, affected neither EP1-R nor EP3γ-R mRNA abundance, as determined by RNase protection assay and quantitative RT–PCR, respectively (Fig.7A). Thus, the effect of CM-17β-E2 seems to be mainly because of substances produced by astrocytes in response to 17β-E2 and not a direct effect of the steroid on the GT1–1 cells. Nevertheless, RNase protection assay revealed that GT1–1 cells do contain very low levels of estrogen receptor mRNA. As shown in Figure 7B, a protected band was not detected in GT1–1 cells after 19 hr of film exposure, a duration that resulted in a strong signal when using RNA from the suprachiasmatic region of the brain. A weak band was detected only after an interval (96 hr) that resulted in overexposure of the suprachiasmatic signal (Fig. 7C).
Fig. 7.
A, Inability of 17β-estradiol to directly affect EP1-R and EP3γ-R mRNA levels in GT1–1 cells.B, Absence of detectable estrogen receptor (ER) mRNA in GT1–1 cells as assessed by RNase protection assay after a 19 hr film exposure. Lane 1, Radiolabeled RNA standards; lane 2, undigested ER cRNA probe; lane 3, digested probe; lane 4, GT1–1 RNA (10 μg); lanes 5, 6, RNA (10 μg) from the suprachiasmatic region of the mouse brain. C, Detection of low ER mRNA levels (arrow) in GT1–1 cells after a longer (96 hr) film exposure. Notice the presence of more abundant, unidentified protected bands of lower molecular size.
The increase in EP1-R mRNA caused by CM-17β-E2 in GT1–1 cells is accompanied by an increased LHRH response to EP1-R activation
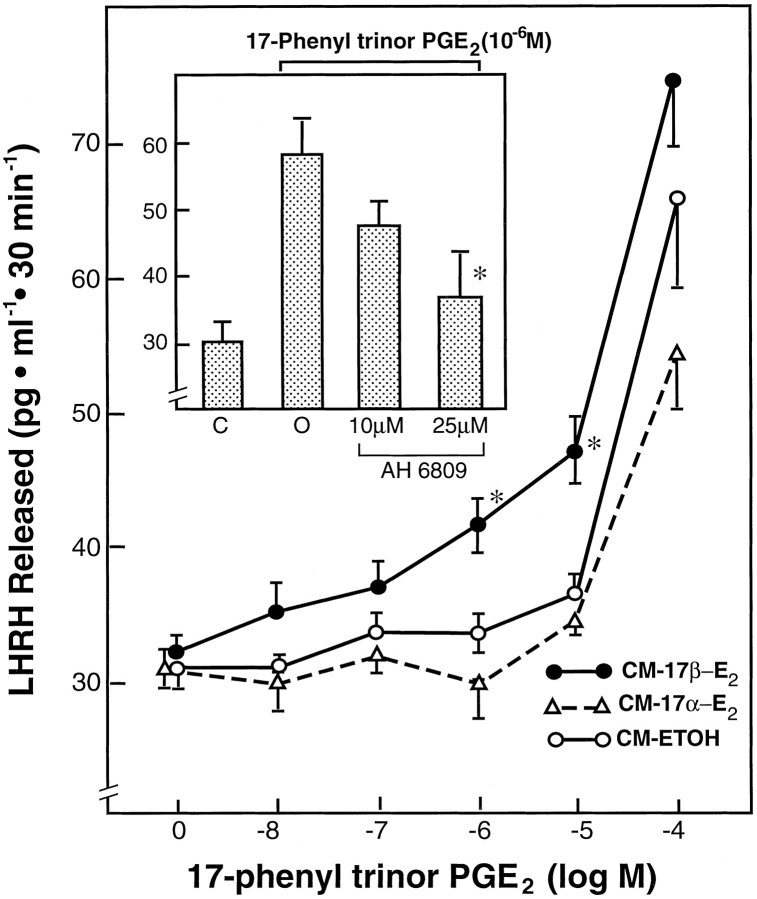
A 3 hr pretreatment of GT1–1 cells with CM-ETOH or CM-17α-E2 followed by a 30 min exposure of the cells to different doses of the EP1-R selective agonist 17-phenyl trinor PGE2 resulted in a shallow LHRH response that was only significant at the 10−5 and 10−4m doses (Fig.8). Pretreatment with CM-17β-E2, however, significantly enhanced the response of the cells to the EP1-R agonist, which was now effective in stimulating LHRH release at 10−6m. The increase in LHRH levels caused by this dose after CM-17β-E2 treatment was blocked by the EP1-R antagonist AH-6809 (Fig. 8, inset), demonstrating that the enhanced effect of 17-phenyl trinor PGE2 on LHRH release after exposure of the cells to CM-17β-E2 is indeed mediated by EP1-R activation.
Fig. 8.
Facilitatory effect of astrocyte culture medium conditioned by a 24 hr exposure to 17β-estradiol (1 nm;CM-17β-E2) on the LHRH release induced by 17-phenyl trinor PGE2, a selective EP1-R agonist, from GT1–1 cells. CM-17α-E2, Astrocyte medium conditioned by 17α-estradiol; CM-ETOH, Astrocyte medium with diluent. The cells were treated with the different CMs for 3 hr before a 30 min exposure to the agonist.Inset, Blockade of the facilitatory effect of CM-17β-E2 on 17-phenyl trinor PGE2-induced LHRH release by the EP1-R antagonist AH-6809. Data are mean ± SEM; each group consists of 6–12 independent observations; *p < 0.05, CM-17β-E2 group versus either the untreated or the other two CM-treated control groups;inset *p < 0.02, AH-6809 group versus group treated with only 17-phenyl trinor PGE2.
The increase in EP3γ-R mRNA caused by CM-17β-E2 in GT1–1 cells is accompanied by an increased cAMP and LHRH response to EP3-R activation
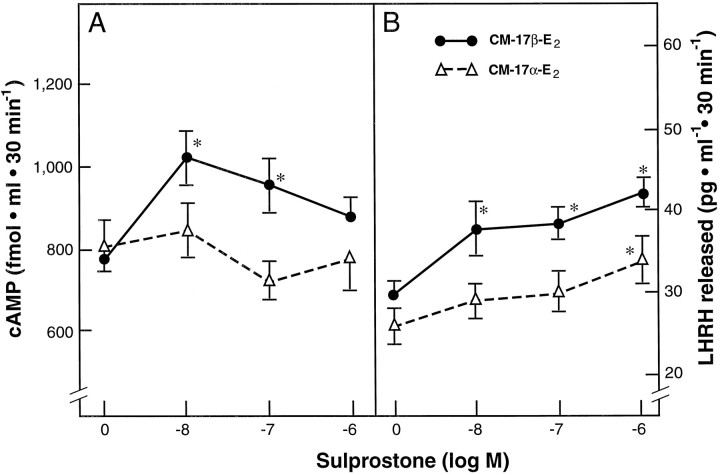
As observed in previous experiments, CM-ETOH and CM-17α-E2 were similarly ineffective in altering EP-R gene expression or LHRH release; thus only the latter treatment was used as a control for these experiments. Treatment of GT1–1 cells pre-exposed to CM-17α-E2 with different doses of the mixed EP3 and EP1 agonist sulprostone failed to increase cAMP levels with respect to the values observed in untreated controls (Fig.9A). In contrast, the response of GT1–1 cells pre-exposed to CM-17β-E2 to sulprostone was more complex, with cAMP levels increasing after the two lower doses of the agonist (10−8 and 10−7m) and failing to change significantly after the highest dose (10−6m). LHRH levels, on the other hand, were increased by sulprostone in cultures pre-exposed to either CM-17β-E2 or CM-17α-E2, but the responsiveness of two groups was markedly different. Whereas CM-17α-E2 pretreated cultures responded only to the highest dose of sulprostone (10−6m) with a significant increase in LHRH release, the response of cultures pretreated with CM-17β-E2 to the agonist was significant at all doses tested (10−8 to 10−6m; Fig. 9B).
Fig. 9.
Facilitatory effect of astrocyte culture medium conditioned by a 24 hr exposure to 17β-estradiol (CM-17β-E2) on cAMP formation (A) and LHRH release (B) induced by the treatment of GT1–1 cells with the EP3-R and EP1-R agonist sulprostone. CM-17α-E2, Medium from astrocytes treated with 17α-estradiol. The GT1–1 cells were pretreated with the CMs for 3 hr before a 30 min exposure to sulprostone; *p < 0.05,CM-17β-E2-pretreated groups versus control groups pretreated with CM-17α-E2and the control group not exposed to sulprostone. In the case ofCM-17α-E2-pretreated cultures versus controls not challenged with sulprostone, *p < 0.05.
Activation of EP2-R increases LHRH release from GT1–1 cells
To determine whether GT1–1 cells have functional EP2 receptors, we treated the cells with butaprost, a selective EP2-R agonist. Consistent with the reported low potency of butaprost (10–100 × less potent than PGE2) (Coleman et al., 1994), butaprost stimulated LHRH release only at the highest dose tested (10−4 M) (20.5 ± 1 vs 46.7 ± 2.2 pg LHRH/ml, control vs butaprost-treated cells; n = 10 in each group; p < 0.01).
DISCUSSION
The present study demonstrates that GT1–1-immortalized LHRH neurons express a complement of PGE2 receptors known to be encoded by two different genes and to be linked to both calcium mobilization and cAMP formation. In addition, the results show that at least the PGE2 receptor subtype linked to calcium mobilization is also present in normal LHRH neurons. In all, the study provides evidence of two related concepts: (1) that glial cells are involved in the central mechanism by which estradiol facilitates the response of LHRH neurons to neurotransmitters acting via PGE2 and (2) that a significant portion of the estradiol-directed glial influence on LHRH neurons may be exerted by modifying the complement of PGE2 receptors expressed in these neurons.
The actions of PGE2 are initiated by its binding to a family of guanine nucleotide-binding protein (G-protein)-coupled receptors (Narumiya, 1994). Four classes of PGE2 receptors, EP1, EP2, EP3, and EP4, have been described based on their pharmacological properties (Coleman et al., 1990), and cDNAs encoding each class have been isolated and characterized (Sugimoto et al., 1992,1993; Honda et al., 1993; Irie et al., 1993; Watabe et al., 1993; Regan et al., 1994; Nishigaki et al., 1995). The EP3-R is the most complex of the known PGE2 receptors, because its gene can be alternatively spliced to generate at least three different isoforms (EP3α, EP3β, and EP3γ) that differ in their C-terminal tails (Irie et al., 1993; Sugimoto et al., 1993) and in their efficiency for activation and coupling to the G-protein/adenylate cyclase signal transduction pathway. Although the EP3α-R and EP3β-R isoforms are coupled exclusively to inhibition of adenylate cyclase via the inhibitory G-protein Gi (Sugimoto et al., 1993), the EP3γ isoform is coupled to both inhibition and stimulation of adenylate cyclase, probably via activation of Gi and the cholera toxin-sensitive protein Gs (Irie et al., 1993).
The presence of EP1-R mRNA in LHRH neurons of immature mice in situ and in GT1–1 cells, plus the detection of less abundant amounts of all three alternatively spliced forms of EP3-R mRNA in these cells, is consistent with earlier findings demonstrating the involvement of calcium (Ojeda and Negro-Vilar, 1985) and cAMP (Ojeda et al., 1985) in the intracellular mechanism underlying the stimulatory effect of PGE2 on LHRH release. Although the sensitivity of the combined immunohistochemistry and in situ hybridization procedure used was insufficient to detect EP3-R mRNA in LHRH neuronsin vivo, the presence of all three alternatively spliced forms of the EP3-R gene in GT1–1 cells in vitro and the appearance of borderline hybridization signals in some LHRH neuronsin situ suggest that the EP3-R gene is indeed expressed in LHRH neurons of the intact hypothalamus. The ability of cholera toxin and pertussis toxin to stimulate LHRH release from median eminence fragments in vitro (Ojeda et al., 1985) further supports this view, because it demonstrates that modification of the signal transduction pathway used by the EP3 receptor to affect cellular function (i.e., activation of stimulatory G-proteins or inhibition of inhibitory G-proteins coupled to adenylate cyclase) results in LHRH release from LHRH nerve terminals derived from normal animals.
The finding that only a small subpopulation of LHRH neurons contain detectable EP1-R and the low prevalence of EP3-R throughout the LHRH neuronal network suggest that at any given time only selected LHRH neurons are responsive to PGE2 and, hence, to those neurotransmitters that, such as NE, use the prostaglandin to affect LHRH secretion. It also seems that the response of LHRH neurons to PGE2 would depend on the relative prevalence of each receptor subtype in a particular physiological or pathological situation. Circumstances may be envisioned in which a predominance of PGE2 receptors coupled to inhibition of adenylate cyclase (EP3α-R and EP3β-R, for example) will lead to inhibition of LHRH release after NE stimulation (as it occurs in ovariectomized rats). In contrast, a predominance of EP1-R and EP3γ-R may lead to stimulatory responses (as seen in estradiol-treated animals). To date, very little, if anything, is known about the factors that may regulate the expression of PGE2 receptors in any system. Our results indicate that hypothalamic astrocytes exposed to estradiol produce substances able to increase the expression of the EP1-R gene and selectively facilitate the accumulation of EP3γ-R mRNA, one of the alternatively spliced forms of EP3 mRNA. Because these two mRNAs encode PGE2 receptors associated with activation of intracellular signaling pathways mediating the stimulatory effect of PGE2on LHRH release, it would seem that this is one of the cell–cell signaling mechanisms used by the steroid to facilitate the LHRH response to neurotransmitters acting via PGE2.
The enhanced LHRH response to EP1-R and EP3-R selective agonists after exposure of GT1–1 cells to CM-17β-E2 suggests that the transient increases in EP receptor mRNA levels induced by this conditioned medium are associated with an increase in functional receptors. Our experiments, however, do not provide direct evidence of this view. In addition, they identify neither the substance(s) responsible for the increase in receptor expression nor the mechanism by which such a substance(s) may act.
The PGE2 receptor agonists and antagonists currently available are not absolutely specific for the different receptor subtypes. Thus, 17-phenyl trinor PGE2, widely used as an EP1-R agonist, can also activate EP2 and EP3 receptors (Coleman et al., 1990). Sulprostone, on the other hand, is a mixed EP3 and EP1 agonist (Coleman et al., 1990), although it has been shown to be 10 times more potent on EP3 than on EP1 receptors (Coleman et al., 1994). The increase in basal cAMP release elicited by sulprostone in CM-17β-E2-treated GT1–1 cells indicates that, regardless of any stimulatory effect on EP1-R, this dose of sulprostone activates EP3γ-R in these cells, i.e., the EP3-R positively coupled to cAMP formation. On the other hand, the inability of the higher dose of sulprostone to increase cAMP release in cells pre-exposed to CM-17β-E2, coupled with its effectiveness to stimulate LHRH release, suggests that at this dose sulprostone not only activates EP3γ-R but also EP3-Rs coupled to inhibition of adenylate cyclase and EP1-R.
In addition to EP1-R and EP3-Rs, GT1–1 cells may also contain EP2-R, as evidenced by the ability of butaprost to stimulate LHRH release. Butaprost has been shown to be selective for EP2-R but to have a much lower potency than PGE2 (Coleman et al., 1994). Detection of EP2-R mRNA will be required to conclusively demonstrate the presence or absence of this receptor in GT1–1 cells.
Our results clearly show that the effect of estradiol on PGE2 receptor gene expression is not directly exerted on the LHRH neurons. In fact, RNase protection assays show that under our culture conditions, GT1–1 cells contain very low levels of ER mRNA, an observation consistent with the finding of very low levels of specific estradiol binding sites on GT1–1 cells (Poletti et al., 1994). Perhaps the use of a much higher dose of estradiol may have allowed us to detect an effect on EP3-R mRNA levels, because exposing the cells to a micromolar dose of the steroid resulted in a measurable effect on the content of another recognition molecule, the androgen receptor (Poletti et al., 1994).
Taken altogether, the present observations identify some of the cellular mechanisms that may underlie the facilitatory effect of estradiol on PGE2-induced LHRH release and provide initial insights into the basic processes underlying the central component of estradiol-positive feedback. By demonstrating the ability of estradiol to activate a glia-to-neuron signaling pathway that leads to upregulation of PGE2 receptors on LHRH neurons, the results also suggest that activation of this pathway may be important for the integrated response of the LHRH neuronal network during the preovulatory, estradiol-dependent surge of LHRH secretion. Because the ability of estradiol to facilitate LHRH secretory responses also occurs in systems that affect LHRH release via PGE2-independent pathways (Kalra and Crowley, 1992; Woller and Terasawa, 1992), a different mechanism must underlie the effect of estradiol in these systems.
Considering that hypothalamic astrocytes are a significant source of PGE2 (Ma et al., 1997) and that LHRH neurons contain PGE2 receptors, it may be postulated that the neuronal–glial mechanisms underlying the facilitatory effect of estradiol on neurotransmitter-induced LHRH release involve a two-step process, the release of PGE2 from astrocytes (as a consequence of a neurotransmitter-mediated activation) and the estradiol-dependent production of glial substances able to act on LHRH neurons to selectively enhance the expression of PGE2receptors linked to stimulation of LHRH release.
Footnotes
This work was supported by National Institutes of Health Grants HD-25123, P-30 Population Center Grant HD-18185, and RR-00163 for the operation of the Oregon Regional Primate Research Center. F.R. is a postdoctoral research fellow supported by Institut National de la Santé et de la Recherche Médicale France, and HD-25123. B.J.L. is a visiting professor supported by the Korea Science and Engineering Foundation through the Hormone Research Center (96-K1-0405-01-02-2). We are indebted to Dr. Yukihiko Sugimoto (Faculty of Pharmaceutical Sciences, Kyoto University, Japan) for providing us with the EP receptor cDNAs used in this study.
Correspondence should be addressed to Sergio Ojeda, Division of Neuroscience, Oregon Regional Primate Research Center, 505 Northwest 185th Avenue, Beaverton, OR 97006.
Dr. Rage’s present address: Lab de Neurobiologie Endocrinologique, Université de Montpellier-2, Unité de Recherche Associeé, 34095 Montpellier, France.
Dr. Lee’s present address: Department of Biology, College of Natural Sciences, University of Ulsan, Ulsan, Korea.
REFERENCES
- 1.Barraclough CA, Wise PM. The role of catecholamines in the regulation of pituitary-luteinizing hormone and follicle-stimulating hormone secretion. Endocr Rev. 1982;3:91–119. doi: 10.1210/edrv-3-1-91. [DOI] [PubMed] [Google Scholar]
- 2.Berg-von der Emde K, Dees WL, Hiney JK, Hill DF, Dissen GA, Costa ME, Moholt-Siebert M, Ojeda SR. Neurotrophins and the neuroendocrine brain: different neurotrophins sustain anatomically and functionally segregated subsets of hypothalamic dopaminergic neurons. J Neurosci. 1995;15:4223–4237. doi: 10.1523/JNEUROSCI.15-06-04223.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooker G, Harper JF, Terasaki WL, Moylan RD. Radioimmunoassay of cyclic AMP and cyclic GMP. In: Brooker G, Greengard P, editors. Advances in cyclic nucleotide research, Vol 10. Raven; New York: 1979. pp. 1–32. [PubMed] [Google Scholar]
- 4.Coleman RA, Kennedy I, Humphrey PPA, Bunce K, Lumley P. Prostanoids and their receptors. In: Hansch C, Sammes PG, Taylor JB, Emmett JC, editors. Comprehensive medicinal chemistry, Vol 3. Pergamon; New York: 1990. pp. 643–714. [Google Scholar]
- 5.Coleman RA, Smith WL, Narumiya S. International union of pharmacology classification of prostanoid receptors: properties, distribution and structure of the receptors and their subtypes. Pharmacol Rev. 1994;46:205–229. [PubMed] [Google Scholar]
- 6.Correa-Rotter R, Mariash CN, Rosenberg ME. Loading and transfer control for Northern hybridization. Biotechniques. 1992;12:154–158. [PubMed] [Google Scholar]
- 7.Danielson PE, Forss-Petter S, Brow MA, Calavetta L, Douglass J, Milner RJ, Sutcliffe JG. p1B15: a cDNA clone of the rat mRNA encoding cyclophilin. DNA. 1988;7:261–267. doi: 10.1089/dna.1988.7.261. [DOI] [PubMed] [Google Scholar]
- 8.Dissen GA, Newman Hirshfield A, Malamed S, Ojeda SR. Expression of neurotrophins and their receptors in the mammalian ovary is developmentally regulated: changes at the time of folliculogenesis. Endocrinology. 1995;136:4681–4692. doi: 10.1210/endo.136.10.7664689. [DOI] [PubMed] [Google Scholar]
- 9.Gallo F, Morale MC, Avola R, Marchetti B. Cross-talk between luteinizing hormone-releasing hormone (LHRH) neurons and astroglial cells: developing glia release factors that accelerate neuronal differentiation and stimulate LHRH release from GT1–1 neuronal cell line and LHRH neurons induce astroglia proliferation. Endocrine. 1995;3:863–874. doi: 10.1007/BF02738891. [DOI] [PubMed] [Google Scholar]
- 10.Gallo RV, Drouva SV. Effect of intraventricular infusion of catecholamines on luteinizing hormone release in ovariectomized and ovariectomized, steroid-primed rats. Neuroendocrinology. 1979;29:149–162. doi: 10.1159/000122917. [DOI] [PubMed] [Google Scholar]
- 11.Gearing M, Terasawa E. The alpha-1-adrenergic neuronal system is involved in the pulsatile release of luteinizing hormone-releasing hormone in the ovariectomized female rhesus monkey. Neuroendocrinology. 1991;53:373–381. doi: 10.1159/000125744. [DOI] [PubMed] [Google Scholar]
- 12.Honda A, Sugimoto Y, Namba T, Watabe A, Irie A, Negishi M, Narumiya S, Ichikawa A. Cloning and expression of a cDNA for mouse prostaglandin E receptor EP2 subtype. J Biol Chem. 1993;268:7759–7762. [PubMed] [Google Scholar]
- 13.Irie A, Sugimoto Y, Namba T, Harazono A, Honda A, Watabe A, Negishi M, Narumiya S, Ichikawa A. Third isoform of the prostaglandin-E-receptor EP3 subtype with different C-terminal tail coupling to both stimulation and inhibition of adenylate cyclase. Eur J Biochem. 1993;217:313–318. doi: 10.1111/j.1432-1033.1993.tb18248.x. [DOI] [PubMed] [Google Scholar]
- 14.Jennes L, Beckman WC, Stumpf WE, Grzanna R. Anatomical relationships of serotoninergic and noradrenergic projections with the GnRH system in septum and hypothalamus. Exp Brain Res. 1982;46:331–338. doi: 10.1007/BF00238628. [DOI] [PubMed] [Google Scholar]
- 15.Kalra SP. Neural circuitry involved in the control of LHRH secretion: a model for preovulatory LH release. In: Ganong WF, Martini L, editors. Frontiers in neuroendocrinology, Vol 9. Raven; New York: 1986. pp. 31–75. [Google Scholar]
- 16.Kalra SP, Crowley WR. Neuropeptide Y: a novel neuroendocrine peptide in the control of pituitary hormone secretion, and its relation to luteinizing hormone. In: Ganong WF, Martini L, editors. Frontiers in neuroendocrinology, Vol 13. Raven; New York: 1992. pp. 1–46. [PubMed] [Google Scholar]
- 17.King JC, Letourneau RL. Luteinizing hormone-releasing hormone terminals in the median eminence of rats undergo dramatic changes after gonadectomy, as revealed by electron microscopic image analysis. Endocrinology. 1994;134:1340–1351. doi: 10.1210/endo.134.3.8119174. [DOI] [PubMed] [Google Scholar]
- 18.Kozlowski GP, Coates PW. Ependymoneuronal specializations between LHRH fibers and cells of the cerebroventricular system. Cell Tissue Res. 1985;242:301–311. doi: 10.1007/BF00214542. [DOI] [PubMed] [Google Scholar]
- 19.Langub MC, Jr, Watson RE., Jr Estrogen receptor-immunoreactive glia, endothelia, and ependyma in guinea pig preoptic area and median eminence: electron microscopy. Endocrinology. 1992;130:364–372. doi: 10.1210/endo.130.1.1727710. [DOI] [PubMed] [Google Scholar]
- 20.Leranth C, MacLusky NJ, Shanabrough M, Naftolin F. Catecholaminergic innervation of luteinizing hormone-releasing hormone and glutamic acid decarboxylase immunopositive neurons in the rat medial preoptic area. Neuroendocrinology. 1988;48:591–602. doi: 10.1159/000125068. [DOI] [PubMed] [Google Scholar]
- 21.Leung PCK, Arendash GW, Whitmoyer DI, Gorski RA, Sawyer CH. Electrical stimulation of mesencephalic noradrenergic pathway: effects on luteinizing hormone levels in blood of ovariectomized and ovariectomized, steroid-primed rats. Endocrinology. 1981;109:720–728. doi: 10.1210/endo-109-3-720. [DOI] [PubMed] [Google Scholar]
- 22.Leung PCK, Arendash GW, Whitmoyer DI, Gorski RA, Sawyer CH. Differential effects of central adrenoceptor agonists on luteinizing hormone release. Neuroendocrinology. 1982;34:207–214. doi: 10.1159/000123301. [DOI] [PubMed] [Google Scholar]
- 23.Ma YJ, Berg-von der Emde K, Moholt-Siebert M, Hill DF, Ojeda SR. Region-specific regulation of transforming growth factor α (TGFα) gene expression in astrocytes of the neuroendocrine brain. J Neurosci. 1994a;14:5644–5651. doi: 10.1523/JNEUROSCI.14-09-05644.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma YJ, Costa ME, Ojeda SR. Developmental expression of the genes encoding transforming growth factor alpha (TGFα) and its receptor in the hypothalamus of female rhesus macaques. Neuroendocrinology. 1994b;60:346–359. doi: 10.1159/000126769. [DOI] [PubMed] [Google Scholar]
- 25.Ma YJ, Hill DF, Junier M, Costa ME, Felder SE, Ojeda SR. Expression of epidermal growth factor receptor changes in the hypothalamus during the onset of female puberty. Mol Cell Neurosci. 1994c;5:246–262. doi: 10.1006/mcne.1994.1029. [DOI] [PubMed] [Google Scholar]
- 26.Ma YJ, Dissen GA, Rage F, Ojeda SR. RNase protection assay. Methods. 1996;10:273–278. doi: 10.1006/meth.1996.0102. [DOI] [PubMed] [Google Scholar]
- 27.Ma YJ, Berg-von der Emde K, Rage F, Wetsel WC, Ojeda SR. Hypothalamic astrocytes respond to transforming growth factor alpha with secretion of neuroactive substances that stimulate the release of luteinizing hormone-releasing hormone. Endocrinology. 1997;138:19–25. doi: 10.1210/endo.138.1.4863. [DOI] [PubMed] [Google Scholar]
- 28.Martinez de la Escalera G, Choi ALH, Weiner RI. β1-Adrenergic regulation of the GT1 gonadotropin-releasing hormone (GnRH) neuronal cell lines: stimulation of GnRH release via receptors positively coupled to adenylate cyclase. Endocrinology. 1992;131:1397–1402. doi: 10.1210/endo.131.3.1354602. [DOI] [PubMed] [Google Scholar]
- 29.McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNeill TH, Sladek JR. Fluorescence–immunocytochemistry: simultaneous localization of catecholamines and gonadotropin-releasing hormone. Science. 1978;200:72–74. doi: 10.1126/science.345442. [DOI] [PubMed] [Google Scholar]
- 31.Melcangi RC, Galbiati M, Messi E, Piva F, Martini L, Motta M. Type 1 astrocytes influence luteinizing hormone-releasing hormone release from the hypothalamic cell line GT1–1: is transforming growth factor-β the principle involved?. Endocrinology. 1995;136:679–686. doi: 10.1210/endo.136.2.7835301. [DOI] [PubMed] [Google Scholar]
- 32.Narumiya S. Prostanoid receptors: structure, function, and distribution. Ann NY Acad Sci. 1994;744:126–138. doi: 10.1111/j.1749-6632.1994.tb52729.x. [DOI] [PubMed] [Google Scholar]
- 33.Nilaver G, Kozlowski GP. Comparison of the PAP and ABC immunocytochemical techniques. In: Bullock GR, Petrusz P, editors. Techniques in immunocytochemistry, Vol 4. Academic; New York: 1989. pp. 199–215. [Google Scholar]
- 34.Nishigaki N, Negishi M, Honda A, Sugimoto Y, Namba T, Narumiya S, Ichikawa A. Identification of prostaglandin E receptor “EP2” cloned from mastocytoma cells as EP4 subtype. FEBS Lett. 1995;364:339–341. doi: 10.1016/0014-5793(95)00421-5. [DOI] [PubMed] [Google Scholar]
- 35.Ojeda SR. The neurobiology of mammalian puberty: has the contribution of glial cells been underestimated?. J NIH Res. 1994;6:51–56. [Google Scholar]
- 36.Ojeda SR, Negro-Vilar A. Prostaglandin E2-induced LHRH release involves mobilization of intracellular Ca+2. Endocrinology. 1985;116:1763–1770. doi: 10.1210/endo-116-5-1763. [DOI] [PubMed] [Google Scholar]
- 37.Ojeda SR, Negro-Vilar A, McCann SM. Release of prostaglandin E (PGEs) by hypothalamic tissue: evidence for their involvement in catecholamine-induced LHRH release. Endocrinology. 1979;104:617–624. doi: 10.1210/endo-104-3-617. [DOI] [PubMed] [Google Scholar]
- 38.Ojeda SR, Negro-Vilar A, McCann SM. Evidence for involvement of α-adrenergic receptors in norepinephrine-induced PGE2 and LHRH release from the median eminence. Endocrinology. 1982;110:409–412. doi: 10.1210/endo-110-2-409. [DOI] [PubMed] [Google Scholar]
- 39.Ojeda SR, Urbanski HF, Katz KH, Costa ME. Stimulation of cyclic adenosine 3′,5′-monophosphate production enhances hypothalamic luteinizing hormone-releasing hormone release without increasing prostaglandin E2 synthesis: studies in prepubertal female rats. Endocrinology. 1985;117:1175–1178. doi: 10.1210/endo-117-3-1175. [DOI] [PubMed] [Google Scholar]
- 40.Ojeda SR, Urbanski HF, Katz KH, Costa ME. Activation of estradiol positive feedback at puberty: estradiol sensitizes the LHRH releasing system at two different biochemical steps. Neuroendocrinology. 1986;43:259–265. doi: 10.1159/000124535. [DOI] [PubMed] [Google Scholar]
- 41.Ojeda SR, Urbanski HF, Katz KH, Costa ME. Prostaglandin E2 releases luteinizing hormone-releasing hormone from the female juvenile hypothalamus through a Ca2+-dependent, calmodulin-independent mechanism. Brain Res. 1988;441:339–351. doi: 10.1016/0006-8993(88)91412-6. [DOI] [PubMed] [Google Scholar]
- 42.Ojeda SR, Urbanski HF, Costa ME, Hill DF, Moholt-Siebert M. Involvement of transforming growth factor α in the release of luteinizing hormone-releasing hormone from the developing female hypothalamus. Proc Natl Acad Sci USA. 1990;87:9698–9702. doi: 10.1073/pnas.87.24.9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peppel K, Baglioni C. A simple and fast method to extract RNA from tissue culture cells. Biotechniques. 1990;9:711–713. [PubMed] [Google Scholar]
- 44.Poletti A, Melcangi RC, Negri-Cesi P, Maggi R, Martini L. Steroid binding and metabolism in the luteinizing hormone-releasing hormone-producing neuronal cell line GT1–1. Endocrinology. 1994;135:2623–2628. doi: 10.1210/endo.135.6.7988451. [DOI] [PubMed] [Google Scholar]
- 45.Rage F, Ma YJ, Ojeda SR. Different prostaglandin E2 (PGE2) receptor subtypes are expressed in immortalized luteinizing hormone releasing hormone (LHRH) neurons. Soc Neurosci Abstr. 1994;20:637. [Google Scholar]
- 46.Rage F, Costa ME, Ojeda SR. Estradiol upregulates prostaglandin E2 (PGE2) receptor gene expression in LHRH neurons via glial intermediacy. Soc Neurosci Abstr. 1995;21:265. [Google Scholar]
- 47.Ramirez VD, Feder HH, Sawyer CH. The role of brain catecholamines in the regulation of LH secretion: a critical inquiry. In: Martini L, Ganong WF, editors. Frontiers in neuroendocrinology, Vol 8. Raven; New York: 1984. pp. 27–84. [Google Scholar]
- 48.Regan JW, Bailey TJ, Pepperl DJ, Pierce KL, Bogardus AM, Donello JE, Fairbairn CE, Kedzie KM, Woodward DF, Gil DW. Cloning of a novel human prostaglandin receptor with characteristics of the pharmacologically defined EP2 subtype. Mol Pharmacol. 1994;46:213–220. [PubMed] [Google Scholar]
- 49.Salvatori R, Bockman RS, Guidon PT., Jr A simple modification of the Peppel/Baglioni method for RNA isolation from cell culture. Biotechniques. 1992;13:510–512. [PubMed] [Google Scholar]
- 50.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silverman RC, Gibson MJ, Silverman A. Relationship of glia to GnRH axonal outgrowth from third ventricular grafts in hpg hosts. Exp Neurol. 1991;114:259–274. doi: 10.1016/0014-4886(91)90152-3. [DOI] [PubMed] [Google Scholar]
- 52.Simmons DM, Arriza JL, Swanson LW. A complete protocol for in situ hybridization of messenger RNAs in brain and other tissues with radiolabeled single-stranded RNA probes. J Histotechnol. 1989;12:169–181. [Google Scholar]
- 53.Sugimoto Y, Namba T, Honda A, Hayashi Y, Negishi M, Ichikawa A, Narumiya S. Cloning and expression of a cDNA for mouse prostaglandin E receptor EP3 subtype. J Biol Chem. 1992;267:6463–6466. [PubMed] [Google Scholar]
- 54.Sugimoto Y, Negishi M, Hayashi Y, Namba T, Honda A, Watabe A, Hirata M, Narumiya S, Ichikawa A. Two isoforms of the EP3 receptor with different carboxyl-terminal domains: identical ligand binding properties and different coupling properties with Gi proteins. J Biol Chem. 1993;268:2712–2718. [PubMed] [Google Scholar]
- 55.Terasawa E. Mechanisms controlling the onset of puberty in primates: the role of GABAergic neurons. In: Plant TM, Lee PA, editors. The neurobiology of puberty. Journal of Endocrinology; Bristol, U.K.: 1995. pp. 139–151. [Google Scholar]
- 56.Ugrumov M, Hisano S, Daikoku S. Topographic relations between tyrosine hydroxylase- and luteinizing hormone-releasing hormone-immunoreactive fibers in the median eminence of adult rats. Neurosci Lett. 1989;102:159–164. doi: 10.1016/0304-3940(89)90072-4. [DOI] [PubMed] [Google Scholar]
- 57.Urbanski HF. A role for N-methyl-d-aspartate receptors in the control of seasonal breeding. Endocrinology. 1990;127:2223–2228. doi: 10.1210/endo-127-5-2223. [DOI] [PubMed] [Google Scholar]
- 58.Voigt P, Ma YJ, Gonzalez D, Fahrenbach WH, Wetsel WC, Berg-von der Emde K, Hill DF, Taylor KG, Costa ME, Seidah NG, Ojeda SR. Neural and glial-mediated effects of growth factors acting via tyrosine kinase receptors on LHRH neurons. Endocrinology. 1996;137:2593–2605. doi: 10.1210/endo.137.6.8641214. [DOI] [PubMed] [Google Scholar]
- 59.Wang AM, Doyle MV, Mark DF. Quantitation of mRNA by the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watabe A, Sugimoto Y, Honda A, Irie A, Namba T, Negishi M, Ito S, Narumiya S, Ichikawa A. Cloning and expression of cDNA for a mouse EP1 subtype of prostaglandin E receptor. J Biol Chem. 1993;268:20175–20178. [PubMed] [Google Scholar]
- 61.White R, Lees JA, Needham M, Ham J, Parker M. Structural organization and expression of the mouse estrogen receptor. Mol Endocrinol. 1987;1:735–744. doi: 10.1210/mend-1-10-735. [DOI] [PubMed] [Google Scholar]
- 62.Witkin JW, Ferin M, Popilskis SJ, Silverman A. Effects of gonadal steroids on the ultrastructure of GnRH neurons in the rhesus monkey: synaptic input and glial apposition. Endocrinology. 1991;129:1083–1092. doi: 10.1210/endo-129-2-1083. [DOI] [PubMed] [Google Scholar]
- 63.Woller MJ, Terasawa E. Estradiol enhances the action of neuropeptide Y on in vivo luteinizing hormone-releasing hormone release in ovariectomized rhesus monkey. Neuroendocrinology. 1992;56:921–925. doi: 10.1159/000126325. [DOI] [PubMed] [Google Scholar]