Abstract
We have identified and characterized a novel murine gene,Ectoderm-Neural Cortex-1 (ENC-1), that is an early and highly specific marker of neural induction in vertebrates.ENC-1, which encodes a kelch family related protein, is expressed during early gastrulation in the prospective neuroectodermal region of the epiblast and later in development throughout the nervous system (NS). ENC-1 expression is highly dynamic and, after neurulation, preferentially defines prospective cortical areas. The only apparent expression of ENC-1 outside the NS is restricted to the rostral-most somitomere of the presomitic mesoderm, at the times corresponding to the epithelialization that precedes somite formation. Cellular expression of epitope-tagged ENC-1 shows extensive co-localization of ENC-1 with the actin cytoskeleton, and immunoprecipitation studies demonstrate a physical association between ENC-1 and actin. ENC-1 functions as an actin-binding protein that may be important in the organization of the actin cytoskeleton during neural fate specification and development of the NS.
Keywords: ENC-1, kelch repeats, epiblast, nervous system, actin, neuron
The complexity of the nervous system (NS) presents major challenges for understanding how its neural components are formed and organized. Only a few molecular markers are available to monitor the earliest events of NS development. Among the earliest markers identified in vertebrates are homeobox genes Otx-2(Simeone et al., 1992, 1993), XlHbox 6 (Wright et al., 1990), Nkx-2.1 and Nkx-2.2 (Shimamura et al., 1995), and XlPOU 2 (Witta et al., 1995); the basic helix-loop-helix gene XASH-3 (Zimmerman et al., 1993; Turner and Weintraub, 1994); the cell adhesion molecule N-CAM (Kintner and Melton, 1987); class II β-tubulin (Richter et al., 1988; Oschwald et al., 1991); and the carbohydrate epitope L5 (Roberts et al., 1991). As development proceeds, an increasing number of genes are expressed in restricted domains of the neural tube. These expression domains define the following two types of boundaries: (1) transverse boundaries perpendicular to the longitudinal axis that segregate transverse or neuromeric domains and (2) longitudinal boundaries parallel to the longitudinal axis segregating longitudinal domains extending through multiple transverse domains. In embryonic hindbrain and forebrain, the expression pattern of a large number of candidate regulatory genes, including homeobox genes, suggests a neuromeric organization (Bulfone et al., 1993; Puelles and Rubenstein, 1993; Rubenstein et al., 1994,Shimamura et al., 1995). Similarly, genes marking areas of regional specification in the cortex have been identified (Bulfone et al., 1995), although little is known of the mechanism regulating such regionalization.
Most genes defining early events or domains of CNS development are transcription factors, the expression of which is temporally and spatially restricted. This is compatible with data indicating that NS development involves a combination of signaling by soluble cytokines, cell–cell interactions, and interactions with the extracellular matrix that regulate gene expression in a lineage-specific manner (for review, see Simpson, 1995; Calof, 1995). Other observations suggest that changes in cell shape leading to reorganization of the cytoskeleton alters gene expression, possibly by interacting with nuclear matrix or by activating and facilitating the transport of regulatory factors to the nucleus (Ben-Ze’ev, 1991; Li et al., 1994; Rosette and Karin, 1995; Weitzer et al., 1995).
We have identified and characterized a novel gene, the expression of which is highly specific for neural tissue. ENC-1, named for its expression pattern in Ectoderm and Neural Cortex, is expressed early during embryogenesis in the prospective neuroectodermal region of the epiblast and continues to be expressed during development throughout the NS. Analysis ofENC-1 indicates that it is a homolog of kelch, aDrosophila gene essential for oogenesis (Xue and Cooley, 1993). Members of the kelch family have been identified in several species and are important for cytoskeletal organization and function (Varkey et al., 1995; Way et al., 1995b). ENC-1 is the only member of this family expressed in the NS and encodes a cytoplasmic protein that interacts with the actin cytoskeleton. Based on ENC-1’s pattern of expression, primary structure, subcellular localization, and in vivo interaction with actin, we propose that ENC-1 functions as an actin-binding protein important for organization of the cytoskeleton during neural fate specification and development of the NS.
MATERIALS AND METHODS
Cloning and sequence analysis of ENC-1 cDNA.Total RNA was isolated from embryonic day 17 (E17), postnatal day 1, and adult mouse brains by the guanidinium–CsCl method (Ausubel et al., 1994). Total RNA (5 μg) was used for the synthesis of cDNA (SuperScript preamplification system, Life Technologies, Gaithersburg, MD) that served as a template for a PCR amplification using primers specific for Id molecules. A cDNA fragment of 732 bp was amplified with the oligonucleotide 5′-AAGGAGCTGGTGCCCACC-3′, then cloned into the PCRII (Invitrogen, San Diego, CA) plasmid (clone p2x) and its DNA sequence determined. To obtain a full-length ENC-1 cDNA clone corresponding to the mRNA recognized by p2x DNA, we screened a previously described mouse brain cDNA library (Porteus et al., 1992) using standard colony hybridization procedures (Sambrook et al., 1989). Several positive clones were identified and analyzed. Both strands of the longest cDNA (clone p10.2x) were sequenced as double-stranded plasmids with synthetic primers by the dideoxy-nucleotide chain termination method using the Sequenase enzyme (United States Biochemicals, Cleveland, Ohio) and [α-35S]dATP (Amersham, Arlington Heights, IL). A XbaI fragment of 598 bp corresponding to the 5′ end of ENC-1 cDNA was subcloned in the M13mp19 RF vector (Boehringer Mannheim, Indianapolis, IN) to facilitate sequencing, because this region is rich in guanine and cytosine nucleotides. Sequence analyses were performed using the Wisconsin Genetics Computer Group Sequence Analysis Software Package.
Northern blot analysis. Northern blot analysis was performed as described previously in Ausubel et al. (1994). A blot containing 1 μg of poly A+ RNA isolated from a variety of tissues of adult mice (Clontech, Palo Alto, CA) was hybridized to a32P-radiolabeled DNA probe prepared by random primed (Amersham) DNA synthesis using clone p2x (732 bp fragment of a coding region of ENC-1 cDNA). Blots were subsequently hybridized with a β-actin cDNA probe used as an RNA loading and transfer control.
Construction of tagged ENC-1. Epitope-tagged ENC-1 was prepared by introduction of a DNA sequence encoding an 11 amino acid peptide (MASMTGGQQMG) corresponding to the major capsid protein of T7 (Tsai et al., 1992) at the initiation codon ofENC-1. A 54 bp oligonucleotide encoding the 11 amino acids of the T7 epitope and amino acids 2–8 of ENC-1 was synthesized as the upstream primer and 5′-CCTGCCTTCCTAATGTAGAGC-3′ as the downstream primer located at the 3′ end of the ENC-1 coding region. Clone p10.2x served as a template to amplify the coding region ofENC-1 fused to the DNA encoding the T7 epitope at the 5′ end by PCR. The expected 1800 bp PCR fragment was cloned into the PCR3 eukaryotic expression plasmid (T/A cloning kit, Invitrogen). DNA sequence of this insert was determined and found to be unchanged.
In vitro transcription and translation of ENC-1.To prepare ENC-1 and T7tagENC-1 proteins,SalI-linearized DNA from plasmid p10.2x andEcoRI-linearized DNA from plasmid pT7tagENC-1 were used as templates in in vitro transcription reactions (Stratagene, La Jolla, CA) and in vitro translation using [35S]methionine (Amersham) and a rabbit reticulocyte translation kit, as recommended by the supplier (Promega, Madison, WI). The protein products were boiled in SDS loading buffer, separated on SDS-PAGE, and visualized by autoradiography.
Cell culture, transfection, and immunofluorescence. National Institutes of Health (NIH) 3T3 cells and Daoy cells were obtained from the American Type Culture Collection (Rockville, MD) (catalog #ATCC CRL 1658 and #ATCC HTB 186, respectively). SNB40 cells were kindly provided by Dr. R. Youle, NIH, Bethesda, MD. NIH 3T3 cells were cultured in high-glucose DMEM containing 100 μg/ml penicillin G, 100 μg/ml streptomycin, and 5% FBS (all from Life Technologies) and maintained in a humidified 5% CO2/95% air incubator at 37°C. A transient transfection protocol modified from that described by Felgner and Ringold (1989) was used to examine ENC-1 expression. Briefly, cells were grown to 70–80% confluence in a two-well tissue culture chamber slide (Nunc, InterMed, Naperville, IL) and washed once with calcium- and magnesium-free PBS (CM-PBS) and once with OptiMEM medium (Life Technologies). A lipofectin-DNA mixture containing 2 μg of pT7tagENC-1 DNA or pCR3 DNA and 3 μg of lipofectin (Life Technologies) diluted in 1 ml of OptiMEM was then added to each well. After incubation at 37°C for 6–7 hr, the medium was replaced with 1 ml of culture medium containing 10% FBS. The cells were examined between 12 and 48 hr after transfection. Medulloblastoma cell lines Daoy and SNB40 were cultured in Eagle’s Minimal Essential Medium with Earle’s BSS supplemented with nonessential amino acids, 1 mm sodium pyruvate, 100 μg/ml penicillin G, 100 μg/ml streptomycin, and 10% FBS (all from Life Technologies). The cultures were maintained in a humidified 5% CO2/95% air incubator at 37°C. Stable transfections were performed in cells grown to 70–80% confluence in 35 mm tissue culture plates by adding a lipofectin-DNA mixture containing 8 μg of pT7tagENC-1 DNA or pCR3 DNA and 10 μg of lipofectin diluted in 2 ml of OptiMEM. After incubation at 37°C for 6–12 hr, the transfection medium was replaced with 2 ml of culture medium, and cells were incubated for an additional 48 hr. Cells were then divided between two 100 mm tissue culture plates and selected for 3 weeks in culture medium containing 500 μg/ml G418 (Geneticin, Life Technologies). Four pools of transfectant clones were obtained and subsequently maintained in presence of G418.
For immunofluorescence, the transfected cells were fixed in 4% paraformaldehyde in CM-PBS for 10 min at room temperature. Cells were permeabilized, and nonspecific binding was blocked by incubating the cells for 15 min at room temperature in CM-PBS-Tween (CM-PBS/0.1% Tween 20) containing 0.05% Nonidet P-40, 10% goat serum, and 3% BSA. The same solution without Nonidet P-40 was used for the following incubations with antibodies. Initially, an incubation with mouse T7tag monoclonal antibody (1:300 dilution) (Novagen, Madison, WI) was performed at room temperature for 1 hr. After washing in PBS, cells were incubated for 1 hr at room temperature with a 1:50 dilution of FITC-conjugated goat anti-mouse IgG (Boehringer Mannheim). To visualize the F-actin cytoskeleton, cells were washed and incubated further for 40 min with TRITC-labeled phalloidin (Sigma, St. Louis, MO) at a final concentration of 0.2 μg/ml in PBS. Cells were finally washed and mounted, and preparations were examined and photographed using a Zeiss Axioplan epifluorescence microscope (Zeiss, Oberkochen, Germany).
Immunoblot analysis and immunoprecipitation. Transfected cell lines were lysed in EBC buffer (50 mm Tris-HCl, pH 8.0, 120 mm NaCl, 0.5% Nonidet P-40, 1 mmEDTA) containing 10 μg/ml aprotinin and leupeptin and 1 mm PMSF (Sigma). The protein concentration of the lysates was quantitated by the Bradford protein assay (Bio-Rad, Hercules, CA), and 100 μg of total protein was resolved by SDS-PAGE using standard methods. The proteins were then transferred to nitrocellulose membranes by electroblotting and processed for immunodetection. Nitrocellulose membranes were incubated for 3 hr at room temperature in TBS-Tween (50 mm Tris, pH 7.6, 1.5% NaCl, 0.1% Tween 20) containing 6% nonfat milk (dilution buffer) and overnight at 4°C with the T7tag monoclonal antibody diluted 1:10,000 in dilution buffer. Then the membranes were rinsed briefly, washed three times for 5 min each in TBS-Tween, and incubated for 1 hr at room temperature with the secondary antibody (peroxidase-labeled goat anti-mouse IgG, Life Technologies) diluted 1:15,000 in dilution buffer. Finally, the membranes were washed in TBS-Tween for 1 hr at room temperature and the labeled bands visualized with an ECL detection system (Amersham).
To immunoprecipitate actin and actin-associated proteins from stably transfected Daoy cells, 85% confluent cultures in 150 mm dishes were lysed at 4°C in 1.5 ml of EBC buffer containing 10 μg/ml aprotinin and leupeptin and 1 mm PMSF. Lysates were clarified by centrifugation, and the supernatants were gently agitated at 4°C for 3 hr with an affinity-purified rabbit anti-actin antibody (Sigma) or with normal rabbit Igs (Dako, Carpinteria, CA). This was followed by the addition of 60 μl of protein A-Sepharose beads (Life Technologies) and incubation further for 1 hr. Immunoprecipitates were then collected by centrifugation, washed five times with EBC buffer at 4°C, and denatured in SDS loading buffer for electrophoresis. Samples were separated on 10% SDS-PAGE and transferred to nitrocellulose membranes. The membranes were treated as above with T7tag monoclonal antibody.
In situ hybridization. ENC-1 expression was detected in tissue sections from whole mouse embryos and fetuses, as well as in postnatal brains using radioactive in situ RNA hybridization (Bulfone et al., 1993). A radiolabeled antisense ENC-1riboprobe was transcribed from the 732 bp cDNA clone p2x. In situ hybridization to whole mouse embryos (E8.0–E10.5) and whole embryonic brains (E11.5 and E12.5) was performed using nonradioactive probes according to the methods of Conlon and Rossant (1992) andShimamura et al. (1994), with modifications described in Bulfone et al. (1995). For these experiments, the antisense ENC-1 riboprobe was synthesized using a digoxigenin RNA labeling kit (Boehringer Mannheim) and DNA from the clone p10.2x. The hybridized probe was detected by anti-digoxigenin antibody using an alkaline phosphatase reaction. As a control, radioactive and nonradioactive riboprobes ofTbr-1 and Pax6 genes were used in adjacent sections and in whole mouse embryos at the same stage of development in parallel, simultaneous experiments. These probes detected the previously reported Tbr-1 and Pax6 patterns of expression (data not shown).
RESULTS
Molecular cloning of ENC-1
To better understand mammalian NS development, we sought to identify novel genes, the expression for which was limited to the NS. For this purpose, we used PCR amplification of cDNA prepared from mRNA of mouse brain at different stages of development. These reactions were primed with oligonucleotides designed to reflect conserved regions of transcription factors likely to be important in NS development. Without additional characterization, we then screened the reaction products for differential expression in the developing NS. Among the reaction products of an RT-PCR amplification designed to identify Id family members (Benezra et al., 1990) in mRNA isolated from the brain of a 1-d-old mouse, we isolated a cDNA fragment of 732 bp. This fragment was used as a probe in Northern blot analysis of poly(A+) RNA from adult mouse tissues. This Northern blot analysis revealed abundant transcripts in the brain, but expression was not apparent in any other mouse tissue examined (Fig. 1). Overexposure of the autoradiogram revealed a very weak signal in testis tissue (data not shown). Abundant transcripts were also detected in total RNA isolated from brains of E17 and postnatal day 1 mice (data not shown). The transcript we detected is ∼4.5 kb. The 732 bp fragment was cloned in the PCRII vector (clone p2x) and then used to screen a mouse brain cDNA library (Porteus et al., 1992). We analyzed two of several cDNA clones obtained, p7.2x and p10.2x, which had a size of ∼1.9 and 2.4 kb, respectively.
Fig. 1.
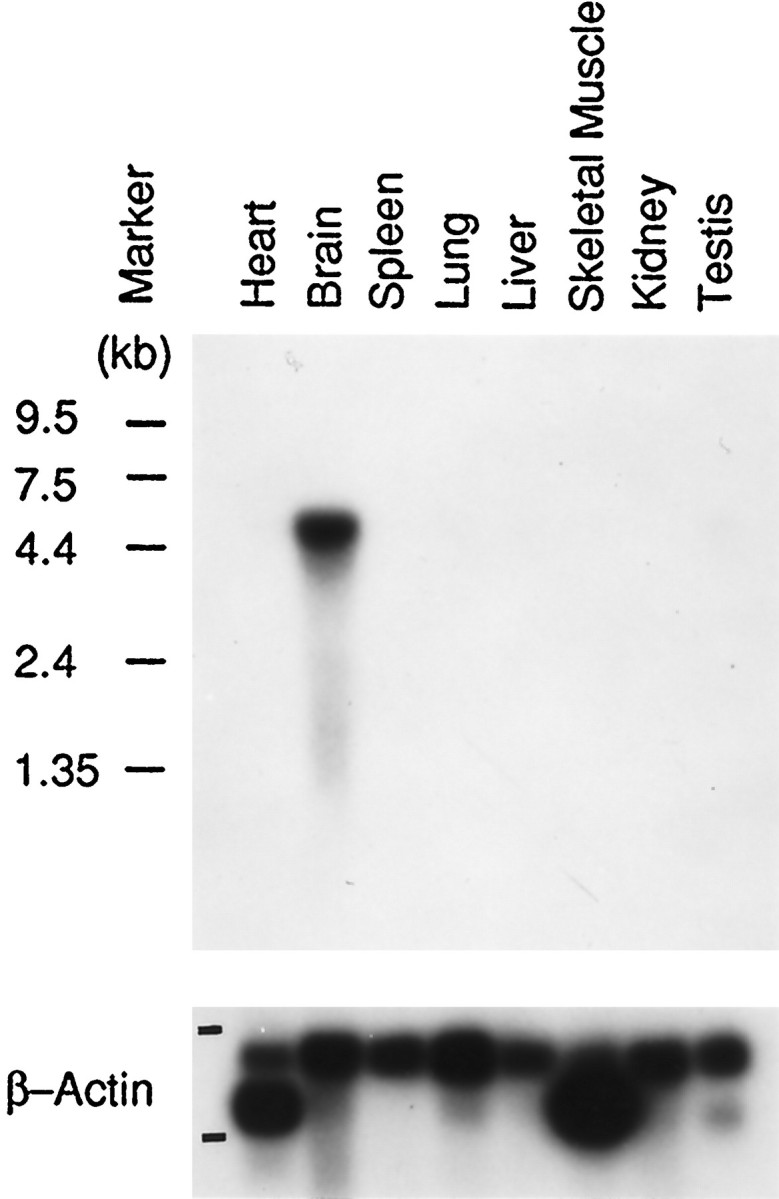
Northern blot analysis of ENC-1mRNA in mouse tissues. ENC-1 transcript distribution in adult mouse tissues shows high levels of tissue-specificENC-1 expression in the brain. Positions of molecular size markers are indicated on the left. Bottom panel, β-Actin cDNA hybridization as an RNA loading and transfer control.
Sequence analysis of ENC-1
Figure 2 presents the nucleotide sequence of theENC-1 cDNA clone p10.2x and the predicted amino acid sequence of the polypeptide it encodes. This cDNA clone of 2390 bp is a partial representation of the 4.5 kb mRNA (see above), but it contains a long open reading frame (ORF) of 1767 bp. A stop codon is found 9 nt upstream of the ATG that marks the 5′ end of the longest ORF of this cDNA clone; therefore, we assume that this is the initial methionine of the translational product. The predicted protein has 589 amino acids and a calculated molecular weight of 66,043 Da. We performed in vitro transcription–translation assays using both p7.2x and p10.2x DNA as a template and detected a protein of ∼67 kDa in agreement with the predicted molecular weight (see Fig.4A) (data not shown).
Fig. 2.

Nucleotide and deduced amino acid sequence of the mouse ENC-1 cDNA (GenBank accession number U65079). The nucleotide sequence of the 2390 bp cDNA (clone p10.2x) is shown in thetop lines, and its predicted amino acid sequence is shown below the cDNA sequence in single-letter code. Numbers on the right refer to the last nucleotide or last amino acid in each corresponding line. The ORF extends from nucleotide 1 to 1767 and encodes a protein of 589 amino acids. The terminator codon is indicated by an asterisk in the protein sequence.
Fig. 4.
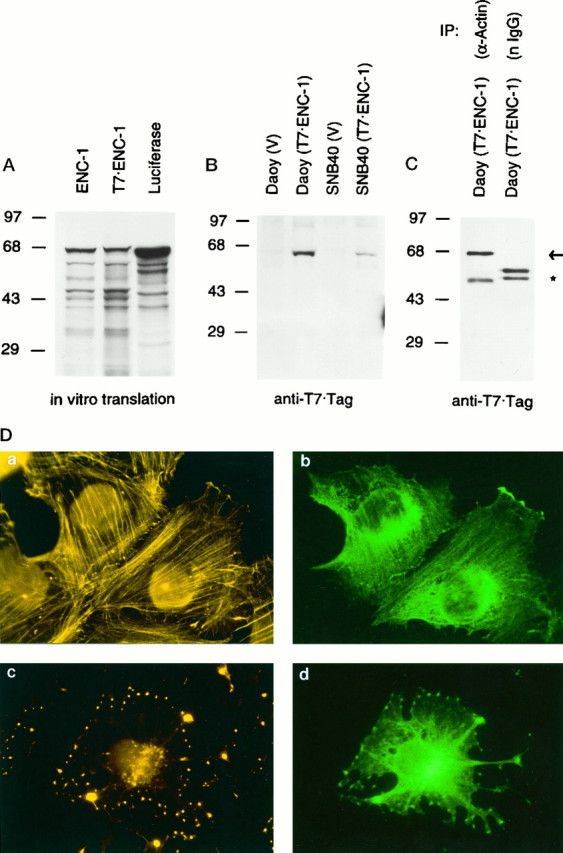
In vivo association between ENC-1 and actin, and cellular localization of ENC-1. A,In vitro synthesis of 35S-radiolabeled full-length ENC-1 from ENC-1 and T7-tagged ENC-1 expression vectors. Autoradiogram of SDS-PAGE resolved ENC-1 and T7-tagged ENC-1 proteins after synthesis by in vitro transcription–translation assays. Luciferase was used as a control. Positions of the molecular mass markers in kilodaltons are shown at left.B, Stable expression of T7-tagged ENC-1 in Daoy and SNB40 cell lines. Immunoblot analysis with T7tag monoclonal antibody was performed from stably transfected Daoy and SNB40 cell lines with T7-tagged ENC-1 expression vector and corresponding vector control. Positions of the molecular mass markers in kilodaltons are shown atleft. C, In vivoassociation between ENC-1 and actin. Lysates from Daoy cells stably transfected with T7-tagged ENC-1 were immunoprecipitated with a polyclonal antibody against actin or with normal rabbit Igs. Immunoprecipitates were then analyzed by immunoblotting with T7 tag monoclonal antibody. The T7-tagged ENC-1 (67 kDa) was present in the anti-actin immunoprecitate, but not in the control (arrow). Cross-reactivity with rabbit heavy-chain IgG is indicated by an asterisk. Normal rabbit Igs were used in an excess concentration in relation to the affinity-purified anti-actin antibody, which accounts for a stronger signal of the heavy-chain IgG in the control immunoprecipitate. Positions of the molecular mass markers in kilodaltons are shown at left.D, Co-localization of ENC-1 with the actin cytoskeleton. T7-tagged ENC-1 was stably expressed in Daoy cells and localized by immunofluorescence with a mouse T7tag monoclonal antibody followed by FITC-conjugated goat anti-mouse Ig (b,d). In the same cells, actin filaments were visualized with TRITC-labeled phalloidin (a,c). Cells fixed after a 30 min treatment with cytochalasin D to disrupt actin filaments were also examined (c, d). Photomicrographs were taken from the same field using filters for rhodamine (left) and fluorescein (right). Magnification 630×.
Searches for homology in protein databases revealed that ENC-1 shares a significant degree of homology (28% overall identity and 50% overall similarity) with the 76.5 kDa kelch protein, a component of ring canals in Drosophila egg chambers (Fig.3A,B) (ORF1) (Xue and Cooley, 1993). Sequence analysis of ENC-1 also indicated the presence of a 50 amino acid stretch consecutively repeated six times in the C-terminal half of the protein (Fig. 3B, shaded boxes). This tandemly repeated motif was first identified in the kelch protein and subsequently defined a family of proteins containing highly similar repeats (Chang-Yeh et al., 1991; Xue and Cooley, 1993;Bork and Doolittle, 1994). Other members of this family that share significant sequence identity with ENC-1 are calicin, a major basic protein of the mammalian sperm head cytoskeleton (von Bülow et al., 1995); SPE-26, a Caenorhabditis elegans protein expressed throughout the testis in both spermatogonial cells and spermatides (Varkey et al., 1995); α-scruin, an actin-bundling protein found in the acrosomal process of Limulus polyhemussperm (Way et al., 1995b); β-scruin, a homolog of α-scruin that is localized to the acrosomal vesicle of Limulus sperm (Way et al., 1995a); and MIPP, a protein encoded by a intracisternal A-particle-promoted placenta-expressed gene (Chang-Yeh et al., 1991). Additional protein sequences that contain this repeated motif include several ORFs of the poxvirus family such as A55R, C2L, F15, and F3L of vaccinia virus (Goebel et al., 1990); C4L and C13L of swinepox virus (Massung et al., 1993); and P65 of entromelia virus (Senkevich et al., 1993).
Fig. 3.
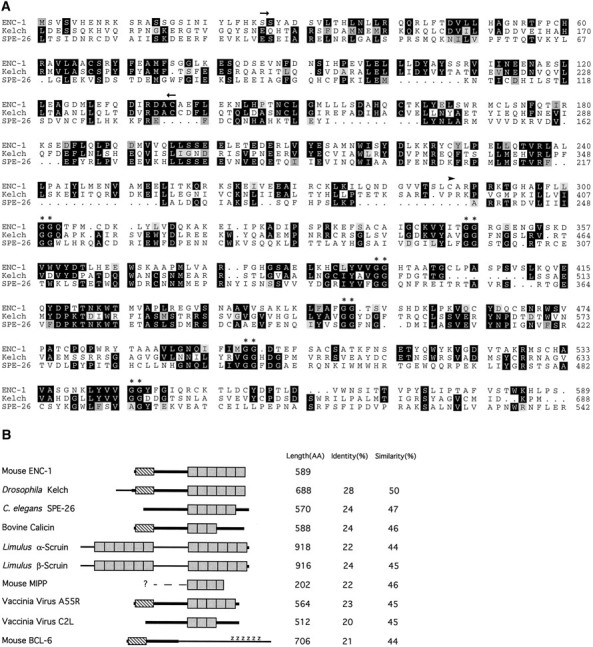
Amino acid comparison of ENC-1 with kelch-related proteins. A, Alignment of ENC-1 and two homologous proteins, kelch and SPE-26, generated by the Wisconsin Genetics Computer Group Sequence Analysis Software Package modules PILE UP and PRETTYBOX. The arrows above the aligned sequences denote a region of homology (BTB/POZ domain) present in ENC-1, kelch, and a number of zinc finger proteins. The arrowhead indicates the start position of the first repeat, based on structural analysis of the repeat motif (kelch motif) by Bork and Doolittle (1994). Thedouble asterisks indicate the double-glycine motif found in all kelch repeats. B, Schematic representation of proteins that share homology to ENC-1. The homology between the proteins is represented by thicker parts oflines representing each sequence. The shaded boxes represent the repeated segments of ∼50 amino acids (kelch motif). The number and location of repeats were based strictly on the presence of two adjacent glycine residues. The hatched box represents the BTB/POZ domain. Z represents the zinc finger regions in the BCL-6 protein. Protein length in amino acids and percentages of amino acid identity and similarity are shown on the right.
An additional domain of significant homology near the N terminus of ENC-1, kelch, calicin, and VA55 comprises ∼120 amino acids (Fig.3B, hatched box). This domain is homologous to a domain found in several zinc finger proteins such as the mammalian BCL-6 protein (Ye et al., 1993) (Fig. 3B) and proteins ofDrosophila such as those encoded by tramtrack(TTK) (Harrison and Travers, 1990) and Broad Complex (BR-C) (DiBello et al., 1991). This domain has previously been named the BTB box (for Broad Complex,tramtrack, and bric à brac) (Godt et al., 1993) or POZ domain (for poxviruses and zinc finger) (Bardwell and Treisman, 1994).
ENC-1 associates in vivo with actin and co-localizes with the actin cytoskeleton
To study the subcellular localization of ENC-1, we transfected Daoy and SNB40 cells, which normally express the ENC-1 gene, and NIH 3T3 cells, which do not express ENC-1 (data not shown), with an expression vector encoding a T7 epitope-taggedENC-1 cDNA (pT7tag ENC-1). Before transfection, we confirmed by in vitro transcription–translation assays that a protein of the predicted full length was synthesized from this vector. As shown in Figure 4A, the band corresponding to the T7-tagged ENC-1 fusion protein migrated in SDS-PAGE slightly slower than ENC-1, presumably as a result of the 11 amino acid insert at its N-terminal region. Using a T7tag monoclonal antibody, the T7-tagged ENC-1 fusion protein was also detected by immunoblot analysis in cell extracts from stably transfected Daoy and SNB40 cells, but not from NIH 3T3 cells (Fig. 4B) (data not shown).
The subcellular localization of the T7-tagged ENC-1 fusion protein in transiently or stably transfected Daoy, SNB40, and NIH 3T3 cells was investigated by immunofluorescence. We used the T7tag monoclonal antibody followed by FITC-conjugated anti-mouse Ig. We were unable to detect ENC-1 expression in NIH 3T3 cells except when transiently transfected cells were examined 12–18 hr after transfection. We noticed that fewer cells were labeled at 24 hr, and fewer yet at 48 hr. ENC-1-positive cells at 24–48 hr showed obvious morphological evidence of cytotoxicity including areas of detachment from the tissue culture surface and contraction of the cytoplasm (data not shown). We observed qualitatively similar expression in the medulloblastoma cell lines SNB40 and Daoy. In the pools we studied, the level of expression was highest in cell lines derived from Daoy. The staining in Daoy-positive cells was distributed throughout the cytoplasm and was particularly intense in the perinuclear area (Fig. 4D,b). Because kelch-related proteins are known to be associated with actin filaments, we performed dual labeling with phalloidin, a fungal toxin specific for filamentous actin, and found extensive co-localization of ENC-1 with the actin cytoskeleton (Fig.4D, a,b). Co-localization of ENC-1 with actin was maintained after treatment of the cells with cytochalasin D, an actin depolymerizing agent (Fig.4D, c,d). These results strongly suggested the possibility that ENC-1 might be associated with the actin filament network.
To address this question more critically, we sought to co-immunoprecipitate these proteins. Lysates from Daoy cells stably transfected with T7-tagged ENC-1 were immunoprecipitated with an affinity-purified anti-actin antibody or with normal rabbit Igs as a control and then analyzed by immunoblotting with T7tag monoclonal antibody. As shown in Figure 4C, the 67 kDa T7-tagged ENC-1 was specifically detected by the anti-T7tag monoclonal antibody in the actin immunoprecipitates, but it was absent in the immunoprecipitation from normal rabbit Igs. Although quantitation of the amount of ENC-1 associated with actin under physiological conditions awaits the availability of an antibody that recognizes ENC-1, these results demonstrate a physical association between ENC-1 and actin in these transfected cells.
Expression of ENC-1
To study the expression of ENC-1 during mouse development, we used in situ RNA hybridization to histological sections and whole-mount preparations. ENC-1expression was highly dynamic over the course of development but restricted almost exclusively to the NS. Expression was detected at the preneurulation stage of mouse embryos (E6.5) in the prospective neuroectodermal region of the epiblast. In the neural plate (E8.0–E9.0), there was diffuse expression. The highest levels of expression were observed in lateral areas, particularly in the neural ridge. Early neural tube expression (E9.0–E10.5) was primarily localized to the alar plate. At later stages, ventral domains ofENC-1 expression appeared in the prosencephalon, rhombencephalon (RH), and spinal cord (SC). To describeENC-1 expression, we used the terminology of the prosomeric model (Bulfone et al., 1993; Puelles and Rubenstein, 1993; Rubenstein et al., 1994).
Early expression of ENC-1 (E6.5–E10.5)
ENC-1 expression was detected at E6.5 in the anterior and distal region of the egg cylinder. This is the ectodermal region of the epiblast that later differentiates predominantly into neuroectodermal cells (Lawson et al., 1991; Quinlan et al., 1995) (Fig.5a,b). No expression was detected in any extraembryonic tissue. At E8.0,ENC-1 expression was detected in ectodermal derivatives. The neural plate was labeled rostrally in the lateral areas of the forebrain anlage and neural ridge (Fig. 5c). The cranial neural crest, which is derived from the neural ridge (Couly and Le Douarin, 1987), was also labeled (Fig.5d,h). We observed weak or no expression in the anterior neural ridge (Fig. 5c); cranial neural crest does not form from this region (Osumi-Yamashita et al., 1994). The prospective areas of the midbrain (MB) and rostral hindbrain, including the ventral neuroepithelial zones (Fig. 5c), expressedENC-1. Caudally, the expression was localized to the ectoderm lateral to the primitive streak.
Table 1.
List of anatomical abbreviations
| AP | Alar plate | MES | Intra-embryonic mesoderm |
| BP | Basal plate | MGE | Medial ganglionic eminence |
| CB | Cerebellar plate | ML | Mantle layer |
| CN | Caudal neuropore | MX | Maxillary process |
| CP | Cortical plate | NE | Neuroepithelial zone |
| Cx | Cortex | OC | Optic chiasma |
| CxA | Cortical telencephalic anlage | OP | Optic eminence |
| D | Diencephalon | OS | Optic stalk |
| DRG | Dorsal root ganglion | OV | Optic vesicle |
| DT | Dorsal thalamus | P1–P6 | Prosomeres 1–6 |
| E | Embryonic ectoderm | POA | Preoptic area |
| EC | Ectoplacental cone | PP | Primordial plexiform layer |
| EMT | Eminentia thalami | PRA | Prorhombomere A |
| EN | Primitive endoderm | PS | Primitive streak region |
| ET | Epithalamus | PT | Pretectal area |
| FB | Forebrain | R1–R8 | Rhombomeres 1–8 |
| HB | Hindbrain | RH | Rhombencephalon |
| HP | Hippocampus | S | Somite |
| Hy | Hypothalamus | SC | Spinal cord |
| I | Isthmus | ST | Striatum |
| IC | Inferior colliculus | SuC | Superior colliculus |
| IF | Isthmic fosa | T | Telencephalon |
| IN | Infundibulum | TF | Trophoblast |
| LGE | Lateral ganglionic eminence | TG | Trigeminal ganglion |
| M | Mesoderm | V | Ventricle |
| MA | Mammillary area | VT | Ventral thalamus |
| MB | Midbrain | YSC | Yolk sac cavity |
| MD | Mandibular process | ZL | Zona limitans |
Fig. 5.
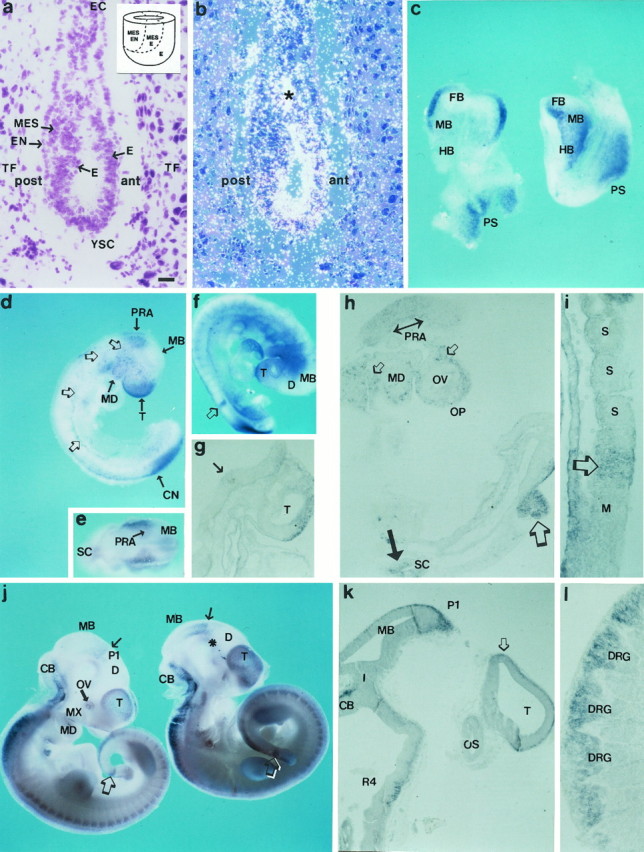
ENC-1 expression at early stages of mouse development: E6.5–E10.5. a, b, Bright- and dark-field photomicrographs of the same sagittal section of an E6.5 mouse embryo after in situ hybridization forENC-1 mRNA. The anterior (ant) and posterior (post) regions of the embryo are indicated. a, Cresyl violet stain showing the localization of the epiblast. The insert is a schematic diagram showing the fate map of the mouse epiblast during the early gastrula stage of development (based on the work of Lawson et al., 1991and Quinlan et al., 1995) and identifies the approximate locations within the epiblast of precursor cells for each of the germ layers: ectoderm (E), mesoderm (MES), and endoderm (EN). b, In situ RNA hybridization with a radiolabeled probe shows the expression of ENC-1 in the prospective neuroectodermal region of the epiblast. The asterisk indicates an artifact produced during the process of autoradiography.c, Dorsal (left) and lateral (right) views of E8.0 mouse embryos processed by nonradioactive whole-mount in situ RNA hybridization.ENC-1 expression is localized in neuroectodermal derivatives. d, Lateral view of an E9.0 mouse embryo processed by whole-mount in situ hybridization. Thearrows show the expression of the gene in the telencephalic vesicle (T), midbrain (MB), prorhombomere A (PRA), mandibular process (MD), and caudal neuropore (CN). The open arrows show the neural crest cells that are labeled in their migration pathway to the branchial arches and body segments. e, Dorsal view of the same embryo as in d showing the segmental expression in the rostral RH, in coincidence with PRA. f, Whole-mount preparation of an E9.5 mouse embryo; g,h, sections of the same embryo. The open arrow in f and the large open arrow in h show the restricted mesodermalENC-1 expression to the rostral-most somitomere of the presomitic mesoderm. The arrow in g and the bidirectional arrow in h show expression in PRA neuroepithelium. The dorsal telencephalic neuroepithelium is also labeled in g. Inh, the small open arrows show expression in migrating neural crest cells, and the large arrowshows the expression in the alar plate of the SC. i, Sagittal section showing the caudal paraxial mesoderm of an E9.5 mouse embryo that was processed by whole-mount in situhybridization. The open arrow shows thatENC-1 is segmentally expressed in the paraxial mesoderm just before the somitic segmentation is morphologically recognized, but not in already-formed somites (S). j, Lateral view of two E10.5 mouse embryos processed by whole-mountin situ hybridization. k,l, Sagittal sections of the embryo shown inj, right. The thin arrowin j shows the boundary between midbrain (MB) and diencephalon (D). Theasterisk indicates that some labeling is apparent in the prosencephalic basal plate. The open arrows indicate the somitomeres that express ENC-1, which at this stage appears in the tail bud. The open arrow ink shows the prospective neuroepithelium of the HP.l, Photomicrograph showing the expression ofENC-1 in cells of the dorsal root ganglia. For abbreviations, see Table 1. Scale bars: a,b, 30 μm; c, 150 μm;d, e, 280 μm; f, 300 μm; j, 320 μm. g–iare enlarged fivefold more than f;k and l are enlarged threefold more thanj.
At E9.0 and E9.5, ENC-1 RNA expression was detected in the neuroepithelium of the caudal pole of the telencephalic vesicle (T) (Fig. 5d,f,g), the rostral RH [prorhombomere A (PRA)] (Fig.5d–h), and the alar plate of the SC (Fig.5h). Mesenchymal cells (probably neural crest cells) in the head and body were also labeled (Fig.5d,h). Caudally, ENC-1 was expressed in the margins of the caudal neuropore (Fig.5d).
In E10.5 mouse embryos, ENC-1 expression was observed in the T, where the cortical anlage showed more intense labeling than the basal ganglia anlage (Fig. 5j). The caudal diencephalon showed scattered cells expressing ENC-1 in ventral areas of the alar plate of prosomere 1 (P1). The mesencephalon (MB) showed a rostro-caudal gradient of expression that started at the mesencephalic–diencephalic boundary (Fig. 5j,arrows). ENC-1 was also expressed in the cerebellar plate and in dorsal areas of the RH and SC. Cells expressingENC-1 in the dorsal domains were primarily localized in the mantle layer (Fig. 5k). In addition, at this stage, weak expression of ENC-1 appeared in the basal plate of P1 and P2 (Fig. 5j, asterisk). ENC-1 expression was also localized to the neural crest-derived cells of the dorsal root ganglia (Fig. 5l).
Late embryonic expression of ENC-1 (E11.5–E16.5)
At E11.5, additional domains of expression, as well as those present at earlier stages, were observed. ENC-1 expression was strong in the cortical anlage of the T and in the medial ganglionic eminence (MGE) (Fig. 6a). A transverse zone of ENC-1 expression between P1 and P2, in the region of the retroflexus tract (Fig. 6a), was recognizable. In the basal plate of the prosencephalon, the expression extended from P1 to P4, where the mammillary region (MA) was more intensely labeled (Fig. 6a). More posteriorly,ENC-1 was expressed in the cerebellar plate and in the alar plate of RH and SC.
Fig. 6.
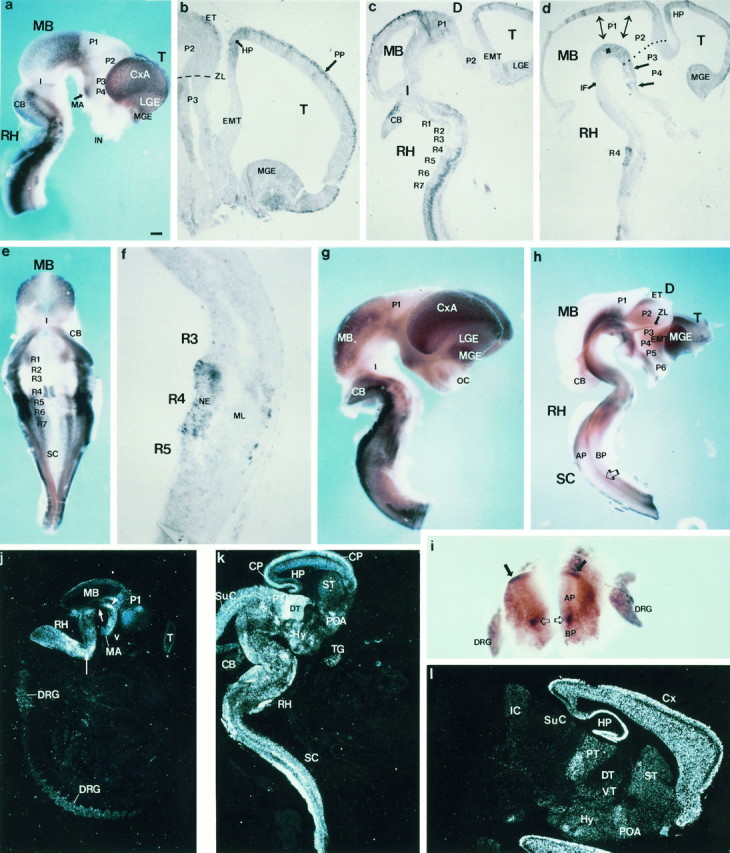
Expression of ENC-1 in mouse CNS at late embryonic stages of development and adult brain.a–f, Isolated neural tube from an E11.5 mouse embryo processed by whole-mount in situhybridization. a, Lateral view; e, dorsal view, after opening of the rhombencephalic roof;b–d, f, sagittal sections of the same embryo. The arrow ina indicates the mammillary region (MA). Inb, the large arrow shows that the telencephalic expression of ENC-1 is localized in the mantle layer of the cortex [primordial plexiform layer (PP)], and the short arrow shows that the unique ventricular expression is localized in the anlage of the HP. The dashed line shows the location of the zona limitans (ZL). The asterisk in dshows the ventricular expression in the diencephalic basal plate from the rostral MB to the MA region. The bidirectional arrows show the position of interneuromeric boundaries. Thelarge arrows show the basal plate of P3 and P4. Thedotted line labels the position of the zona limitans.g, Lateral view and h, ventricular view after midline section of whole-mount processed neural tubes from E12.5 mouse embryos. The small arrow in h shows that the zona limitans express ENC-1, and thelarge open arrow shows a band of expression localized between basal and alar plate limits extending throughout the SC.i, Detail of a thick section of SC from the same embryo as in g. The arrows show the expression in the dorsal area of alar plate, and the open arrows in basal plate ventral to the sulcus limitans.j–l, ENC-1 expression detected by in situ RNA hybridization. j, Sagittal sections of an E14.5 mouse embryo. The plane of section only cut a small tangential area of the telencephalic vesicle (T). The expression is visible in prosencephalic and mesencephalic basal plates, from the MA to the isthmic negative gap (short arrow). The interstitial nucleus of Cajal appears intensely labeled (arrowhead). In the RH, the migrating cells from the rhombic lip to ventral areas are positive (long arrow). k, Sagittal section of an E16.5 mouse embryo. In the telencephalon, the cortical plate (CP) appears intensely positive, and some scattered expressing cells are present in the POA and the ST. The mamillary nuclear complex at the basal plate express ENC-1. DT and PT show intense positive cells. The SuC cortex express ENC-1. The isthmic segment are negative to ENC-1 expression, whereas scattered cells fill the rhombencephalic basal plate. Intense labeling is observed in the rhombic lip derivatives, the pontine, and the inferior olivary nuclei. l, Sagittal section of adult mouse brain. ENC-1 is strongly expressed in the HP and neocortex. Cells from the striatal and POA also expressedENC-1. Caudally, transverse domains of expression are observed in ventral thalamic area (VT), caudal hypothalamus (Hy), and pretectal area (PT). The dorsal thalamus (DT) and the collicular plate do not expressENC-1 at this stage. For abbreviations, see Table 1. Scale bars: a, g, h, 300 μm; e, 220 μm; i, 100 μm;j, 600 μm; k, 480 μm;l, 360 μm; b is enlarged threefold more than a; c and d are enlarged 1.2-fold more than a; f is enlarged fivefold more than a.
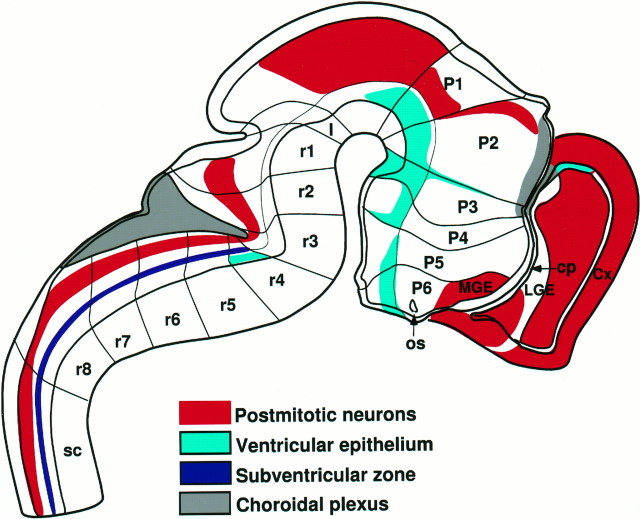
In some regions, ENC-1 was expressed in the proliferative layer (the ventricular zone), whereas in other domains, its expression was restricted to mantle layers that contain postmitotic cells (Fig.7, schema). The telencephalic expression ofENC-1 was primarily restricted to the mantle layer of the cortex (primordial plexiform layer) and MGE (Fig. 6b). Mantle expression was also detected in P1, MB, CB, and the dorsal rhombomeric regions (Fig. 6c,e). In RH, two new domains of ENC-1 expression were detected, a longitudinal domain limited by the basal and alar plates extending throughout RH and SC with the exception of rhombomeres 2 and 3 (r2 and r3) (Fig.6c,e). The second domain was localized segmentally in the paramedian area of the floor plate on both sides of r4 (Fig. 6d,e). Ventricular zone expression was found in the anlage of the hippocampus (HP) (Fig. 6b), the diencephalic basal plate (from the rostral MB to MA) (Fig.6d, asterisk), and in the r4 paramedian area (Fig. 6d–f).
Fig. 7.
Schematic diagram illustrating theENC-1 expression domains in a medial view of the mouse brain at E12.5. The medial wall of the telencephalon is opened to show the internal ganglionic eminences. The transverse (neuromeric) subdivisions are indicated by solid lines that are perpendicular to the principal longitudinal subdivision that divides the alar and basal zones and defines the longitudinal axis of the brain. Other longitudinal zones are indicated by black lines that are parallel to the longitudinal axis. Four longitudinal zones are shown in the SC; from dorsal to ventral, they are the roof plate, alar plate, basal plate, and floor plate. These four zones extend rostrally. For the definitions of the letter codes, see Table 1. The rhombomeres (r1–r8) and theoretical prosomeres (P1–P6) are labeled.
Expression of ENC-1 in the E12.5 neural tube continued to be strong in the forebrain. Expression in the MGE and cortical anlage was separated by a zone of weaker expression in lateral ganglionic eminence (LGE) (Figs. 6g,h, 7,schema). Two interprosomeric boundary zones expressedENC-1, the P2/P3 and P1/P2 boundaries (Figs. 6h,7). The longitudinal domain of ENC-1 expression that extended along RH and SC was also present at this stage (Fig.6h, open arrow). In SC, ENC-1 was expressed in the dorsal area of alar plate and in the subventricular zone of the sulcus limitans (Fig. 6h,i).
At E14.5 and E16.5, ENC-1 continued to be expressed in the maturing tissues of the CNS, in patterns very similar to those observed in earlier stages (Fig. 6j,k). In the T, the cortical plate, including the hippocampal anlage (HP), was clearly labeled (Fig. 6k). Much less expression was found in LGE-derived striatum (ST), whereas the preoptic area (POA) expressedENC-1. Hypothalamic (Hy) expression extended through infundibular and MA domains. The alar diencephalon showed expression ofENC-1 in superficial areas of the dorsal thalamus (DT) and pretectum (PT) (Fig. 6k). MB expression included the superior colliculus (SuC) and some tegmental areas. There was widespread expression of ENC-1 in the hindbrain, including rhombic lip derivatives (pontine and inferior olivary nuclei). The isthmic region had little or no expression (Fig. 6j,short arrow).
ENC-1 expression in adult brain
ENC-1 continued to be strongly expressed in HP and neocortex, except in a thin layer that corresponds to the deep area of layer I (Fig. 6l). Some cells in the ST and POA were also positive for ENC-1. Expression was present in alternating transverse domains of the ventral thalamus (P3), PT (P1), and inferior colliculus. The Hy expressed ENC-1 in the MA and infundibular regions (Fig. 6l). In the adult, caudal regions of the brain had undetectable levels of ENC-1expression (Fig. 6l).
ENC-1 is expressed at E9.5 and E10.5 in the rostral-most somitomere of the presomitic mesoderm
ENC-1 is limited to only a somite-sized domain at the anterior end of the presomitic mesoderm. This region corresponds to the rostral-most somitomere in the paraxial mesoderm (Tam and Trainor, 1994) (Fig. 5h, large open arrow). Figure5i shows a sagittal section from an E9.5 mouse embryo that contains the caudal paraxial mesoderm region, where ENC-1was segmentally expressed just before somitic segmentation could be morphologically recognized (Fig. 5i, open arrow). Because the embryonic axis developed and somites and somitomeres are formed in a strict rostro-caudal sequence (Tam and Trainor, 1994), the new rostral-most somitomere that express ENC-1 at E10.5 was localized in the tail bud (Fig. 5j, open arrows).
DISCUSSION
ENC-1 is a member of the kelch family of proteins and interacts with the actin cytoskeleton
We found significant amino acid homology between mouse ENC-1 and proteins of the kelch family. These proteins are characterized by the presence of a motif of ∼50 amino acids, which is repeated two to seven times and invariably contains two adjacent glycine residues. This motif is called the kelch repeat (Bork and Doolittle, 1994; Cooley and Theurkauf, 1994). Among members of this family, kelch, SPE-26, calicin, and α-scruin are synthesized as cytoskeletal components during germ cell differentiation, and they occur in membrane-associated dense structures (Xu and Cooley, 1993; Varkey et al., 1995; von Bülow et al., 1995; Way et al., 1995b). Kelch co-localizes with actin filaments that form ring canals that regulate nutrient transport from the nurse cells and oocyte. Mutations in the kelch gene affect this cytoplasm transport, producing a female sterile phenotype (Xue and Cooley, 1993; Knowles and Cooley, 1994). Mutations inSPE-26 cause sterility in C. elegans males and hermaphrodites by disrupting the intracellular segregation of components necessary to form spermatids (Varkey et al., 1995). Five of six loss-of-function SPE-26 mutations were in the tandem repeats, and one of the most severe mutations was a substitution in a highly conserved glycine. In the case of α-scruin, there are six kelch repeats at the N terminus and six kelch repeats at the C terminus that are responsible for α-scruin-actin cross-linking activity that stabilizes Limulus sperm acrosomal microfilaments (Owen and De Rosier, 1993; Schmid et al., 1993, 1994). Because it has been suggested that kelch repeats identify a family of actin-binding proteins (Cooley and Theurkauf, 1994; Knowles and Cooley, 1994), and ENC-1 contains six of these repeats in the C-terminal half of the protein, we sought to examine the ability of ENC-1 to interact with the actin cytoskeleton. ENC-1 associates with the actin cytoskeleton (Fig.4D, a,b) in a transfected medulloblastoma cell line, which is a tumor arising in primitive cells of the developing NS that have evidence of neuronal differentiation (Jacobsen et al., 1985). Additional evidence supporting the close association of ENC-1 and the actin cytoskeleton is our finding that ENC-1 and actin co-localize even after depolymerization of the actin filaments by treatment with cytochalasin D (Fig. 4D,c,d). Moreover, immunoprecipitation studies using anti-actin antibodies demonstrated that ENC-1 exists in a complex with actin (Fig. 4C).
ENC-1 also shares significant homology to kelch repeats found in a large number of ORFs within the genome of poxviruses. The functional role of these repeats is unknown, but the poxviruses do associate with actin as part of their intracellular movement (Hiller et al., 1979,1981; Krempien et al., 1981). Recently, it was shown that the intracellular, enveloped form of vaccinia virus induces the formation of actin tails as a mechanism to facilitate direct spread between cells by exploiting the actin cytoskeleton, which directs virions to the cell surface (Cudmore et al., 1995). Kelch-related proteins might mediate such viral interactions.
Interestingly, another domain of ∼120 amino acids is present at the N terminus of ENC-1 and several members of the kelch family. This domain, found primarily in zinc finger proteins, is called BTB/POZ and defines a newly characterized protein–protein interaction interface (Godt et al., 1993; Bardwell and Treisman, 1994; Zollman et al., 1994). This domain mediates both dimer and heterodimer formation in vitro (for review, see Albagli et al., 1995) (Chen et al., 1995). The presence of the BTB/POZ domain could allow ENC-1 homodimerization resulting in a complex with two actin-binding domains that could cross-link and stabilize actin filaments. Alternatively, ENC-1 could interact with other proteins by forming heterodimers through the BTB/POZ domain.
ENC-1 is expressed throughout neural development and in the adult brain
ENC-1 is the only member of the kelch gene family that is primarily expressed in the NS. Northern blot analysis showed high expression levels of ENC-1 in brain, but not in any other mouse tissue examined (Fig. 1). To gain insight about the functional role of ENC-1, we examined the expression ofENC-1 during mouse development. We have detectedENC-1 expression in the epiblast of E6.5 mouse embryos. At this stage, ENC-1 expression appears to be localized to the anterior and distal region of the egg cylinder (Fig.5a,b). Fate-mapping studies demonstrate that neuroectoderm is derived from these epiblast regions (Lawson et al., 1991; Quinlan et al., 1995), which is consistent with the observation that later in development, ENC-1 expression is primarily restricted to the neural plate. The expression of ENC-1 in early gastrula stage embryos makes it one of the earliest markers of neural induction. Others genes recognized to be expressed in early precursors of the developing vertebrate NS, including Otx-2,XlHbox 6, XIPOU 2, and N-CAM, are distinguishable from ENC-1 in that their expression can also be detected in a variety of different lineages (Kintner and Melton, 1987; Wright et al., 1990; Simeone et al., 1992, 1993; Pannese et al., 1995; Witta et al., 1995). For example, N-CAM, which is the most commonly used general marker of neural induction, is first detected at E8.0, and it is also expressed in other body regions such as somites, unsegmented mesoderm, and developing heart (Probstmeier et al., 1994). In contrast, other genes such as type II β-tubulin and midsize neurofilament, which are specifically detected within neural structures, are not expressed before NS differentiation is morphologically recognizable.ENC-1 expression before development of NS tissues as well as its restricted expression within these tissues strongly suggests a role in the regulation of NS development.
ENC-1 is expressed in complex temporal and spatial patterns in the developing and adult CNS and neural crest. It appears to have distinct modules of expression that are delimited by longitudinal and transverse boundaries (Fig. 7) consistent with the prosomeric model (Bulfone et al., 1993; Puelles and Rubenstein, 1993; Rubenstein et al., 1994; Shimamura et al., 1995). The fact that ENC-1 is expressed in dividing and postmitotic neural cells (Fig. 7) implies that its function is linked not only to properties of undifferentiated neural cells, such as a high proliferative potential, active migration, or process outgrowth, but also to properties of differentiated neural cells. During the early stages of development, alar plate expression ofENC-1 is localized primarily in domains that later develop a cortical cytoarchitecture: the laterodorsal areas of the T, the anlage of the cerebral cortex; the rostral MB, the anlage of the SuC; and the dorsal areas of the SC, the anlage of the dorsal horn of the SC. The expression of ENC-1 in these cortical postmitotic neurons suggests that this gene plays a role in the histogenesis of cortical CNS tissues. From E12.5 to E16.5, ENC-1 is expressed in cells of the rhombencephalic mantle layer that appear grouped in columnar clusters at different levels along the migration pathways defined by radial glia. The alternation of theseENC-1-positive clusters with columns of cells that do not express ENC-1 suggests the possibility of a clonal relationship among the cells that express ENC-1 and migrate along the radial glia. Interaction of ENC-1 with actin could be important for changes in the cytoskeleton associated with this cellular migration and differentiation (Sadler et al., 1982; Ostrovsky et al., 1983; Hatten, 1993; Lin et al., 1994).
ENC-1 expression in the rostral-most somitomere of the presomitic mesoderm
We detected expression of ENC-1 in the rostral-most somitomere of the presomitic mesoderm. It is the only apparent expression of ENC-1 outside of the NS (Fig. 5i). Somitomeres are spherical clusters of mesenchymal cells in the presomitic mesoderm that presage the segmentation of somites in the paraxial mesoderm (Tam and Trainer, 1994). Interestingly,Noch-1 and Dll1, which are mammalian homologs of the Drosophila neurogenic genes Notch andDelta, are expressed both in the NS and in the paraxial mesoderm (Reaume et al., 1992; Bettenhausen et al., 1995), and recentlyNoch-1 expression has been shown to be required for coordinated segmentation of somites (Conlon et al., 1995). The somitomere matures with a concomitant increase in cell number and cell-packing density, reverting to an epithelial structure at the time of segmentation (Tam and Trainer, 1994). This epithelialization process depends on cell–cell and cell–matrix interactions (Christ and Ordahl, 1995). Our finding that ENC-1 is expressed in the rostral-most somitomere of the presomitic mesoderm, before its conversion into somites, but not in somitomeres in the cranial mesoderm, which never segment into somites, suggests thatENC-1 is expressed at the time of epithelialization that precedes the formation of somites. Actin distribution is diffuse and random before this morphogenetic event, but during epithelialization, actin becomes prominent in the apical regions of the epithelial cells (Ostrovsky et al., 1983). It is possible that ENC-1 plays a role in the actin rearrangement that accompanies this cellular reorganization that takes place in the epithelialization process during somite formation.
Footnotes
This work was supported by National Institutes of Health Grant 1 U01 CA64898, the Nissen Family, and the Preuss Foundation. P.J.A.B. is a Research Fellow of the Spanish Ministerio de Educacion y Ciencia. We thank Patrick P. L. Tam and Roger A. Pedersen for helpful discussions, Ingeborg Holt for technical assistance, and Lucy Avila and Norma Shipp for help with the manuscript.
Correspondence should be addressed to Dr. Mark A. Israel, Preuss Laboratory for Molecular Neuro-Oncology, Brain Tumor Research Center, Department of Neurological Surgery, HSE 722, University of California San Francisco, 513 Parnassus Avenue, San Francisco, California 94143-0520.
REFERENCES
- 1.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 2.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. Wiley; New York: 1994. [Google Scholar]
- 3.Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Ze’ev A. Animal cell shape changes and gene expression. BioEssays. 1991;13:207–212. doi: 10.1002/bies.950130502. [DOI] [PubMed] [Google Scholar]
- 5.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 6.Bettenhausen B, Hrabe de Angelis M, Simon D, Guenet J-L. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila delta. Development. 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- 7.Bork P, Doolittle RF. Drosophila kelch motif is derived from a common enzyme fold. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 8.Bulfone A, Puelles L, Porteus MH, Frohman MA, Martin GR, Rubenstein JLR. Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci. 1993;13:3155–3172. doi: 10.1523/JNEUROSCI.13-07-03155.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulfone A, Smiga SM, Shimamura K, Peterson A, Puelles L, Rubenstein JLR. T-brain-1: a homolog of Brachyury whose expression defines molecularly distinct domains within the cerebral cortex. Neuron. 1995;15:63–78. doi: 10.1016/0896-6273(95)90065-9. [DOI] [PubMed] [Google Scholar]
- 10.Calof AL. Intrinsic and extrinsic factors regulating vertebrate neurogenesis. Curr Opin Neurobiol. 1995;5:19–27. doi: 10.1016/0959-4388(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 11.Chang-Yeh A, Mold DE, Huang CC. Identification of a novel murine IAP-promoted placenta-expressed gene. Nucleic Acids Res. 1991;19:3667–3672. doi: 10.1093/nar/19.13.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W, Zollman S, Couderc J-L, Laski FA. The BTB domain of bric à brac mediates dimerization in vitro. Mol Cell Biol. 1995;15:3424–3429. doi: 10.1128/mcb.15.6.3424. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Christ B, Ordahl CP. Early stages of chick somite development. Anat Embryol. 1995;191:381–396. doi: 10.1007/BF00304424. [DOI] [PubMed] [Google Scholar]
- 14.Conlon RA, Rossant J. Exogenous retinoic acid rapidly induces anterior ectopic expression of murine Hox-2 gene in vivo. Development. 1992;116:357–358. doi: 10.1242/dev.116.2.357. [DOI] [PubMed] [Google Scholar]
- 15.Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 16.Cooley L, Theurkauf WE. Cytoskeletal functions during Drosophila oogenesis. Science. 1994;266:590–596. doi: 10.1126/science.7939713. [DOI] [PubMed] [Google Scholar]
- 17.Couly GF, Le Douarin NM. Mapping of the early neural primordium in quail-chick chimeras. The prosencephalic neural folds: implications for the genesis of cephalic human congenital abnormalities. Dev Biol. 1987;120:198–214. doi: 10.1016/0012-1606(87)90118-7. [DOI] [PubMed] [Google Scholar]
- 18.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 19.DiBello PR, Withers DA, Bayer CA, Fristrom JW, Guild GM. The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics. 1991;129:385–397. doi: 10.1093/genetics/129.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felgner PL, Ringold GM. Cationic liposome-mediated transfection. Nature. 1989;337:387–388. doi: 10.1038/337387a0. [DOI] [PubMed] [Google Scholar]
- 21.Godt D, Couderc J-L, Cramton SE, Laski FA. Pattern formation in the limbs of Drosophila: bric à brac is expressed in both a gradient and a wave-like pattern and is required for specification and proper segmentation of the tarsus. Development. 1993;119:799–812. doi: 10.1242/dev.119.3.799. [DOI] [PubMed] [Google Scholar]
- 22.Goebel SJ, Johnson GP, Perkus ME, Davis SW, Winslow JP, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 23.Harrison SD, Travers AA. The tramtrack gene encodes a Drosophila finger protein that interacts with the ftz transcriptional regulatory region and shows a novel embryonic expression pattern. EMBO J. 1990;9:207–216. doi: 10.1002/j.1460-2075.1990.tb08097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatten ME. The role of migration in central nervous system neuronal development. Curr Opin Neurobiol. 1993;3:38–44. doi: 10.1016/0959-4388(93)90033-u. [DOI] [PubMed] [Google Scholar]
- 25.Hiller G, Weber K, Schneider L, Parajsz C, Jungwirth C. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology. 1979;98:142–153. doi: 10.1016/0042-6822(79)90533-6. [DOI] [PubMed] [Google Scholar]
- 26.Hiller G, Jungwirth C, Weber K. Fluorescence microscopical analysis of the life cycle of vaccinia virus in chick embryo fibroblast. Virus-cytoskeleton interactions. Exp Cell Res. 1981;132:81–87. doi: 10.1016/0014-4827(81)90085-9. [DOI] [PubMed] [Google Scholar]
- 27.Jacobsen PF, Jenkyn DJ, Papadimitriou JM. Establishment of a human meduloblastoma cell line and its heterotransplantation into nude mice. J Neuropathol Exp Neurol. 1985;44:472–485. doi: 10.1097/00005072-198509000-00003. [DOI] [PubMed] [Google Scholar]
- 28.Kintner CR, Melton DA. Expression of Xenopus NCAM RNA is an early response of ectoderm to induction. Development. 1987;99:311–325. doi: 10.1242/dev.99.3.311. [DOI] [PubMed] [Google Scholar]
- 29.Knowles BA, Cooley L. The specialized cytoskeleton of the Drosophila egg chamber. Trends Genet. 1994;10:235–241. doi: 10.1016/0168-9525(94)90170-8. [DOI] [PubMed] [Google Scholar]
- 30.Krempien U, Schneider L, Hiller G, Weber K, Katz E, Jungwirth C. Conditions for pox virus-specific microvilli formation studied during synchronized virus assembly. Virology. 1981;113:556–564. doi: 10.1016/0042-6822(81)90183-5. [DOI] [PubMed] [Google Scholar]
- 31.Lawson KA, Meneses JJ, Pedersen RA. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Choudhary SK, Milner DJ, Munir MI, Kuisk IR, Capetanaki Y. Inhibition of desmin expression blocks myoblast fusion and interferes with the myogenic regulators MyoD and myogenin. J Cell Biol. 1994;124:827–841. doi: 10.1083/jcb.124.5.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin C-H, Thompson CA, Forscher P. Cytoskeletal reorganization underlying growth cone motility. Curr Opin Neurobiol. 1994;4:640–647. doi: 10.1016/0959-4388(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 34.Massung RF, Jayarama V, Moyer RW. DNA sequence analysis of conserved and unique regions of swinepox virus: identification of genetic elements supporting phenotypic observations including a novel G-protein-coupled receptor homologue. Virology. 1993;197:511–528. doi: 10.1006/viro.1993.1625. [DOI] [PubMed] [Google Scholar]
- 35.Oschwald R, Richter K, Grunz H. Localization of a nervous system-specific class II beta-tubulin gene in Xenopus laevis embryos by whole-mount in situ hybridization. Int J Dev Biol. 1991;35:399–405. [PubMed] [Google Scholar]
- 36.Ostrovsky D, Sanger JW, Lash JW. Light microscope observations on actin distribution during morphogenetic movements in the chick embryo. J Embryol Exp Morphol. 1983;78:23–32. [PubMed] [Google Scholar]
- 37.Osumi-Yamashita N, Ninomiya Y, Doi H, Eto K. The contribution of both forebrain and midbrain crest cells to the mesenchyme in the frontonasal mass of the mouse embryos. Dev Biol. 1994;164:409–419. doi: 10.1006/dbio.1994.1211. [DOI] [PubMed] [Google Scholar]
- 38.Owen C, DeRosier D. A 13 Å map of the actin-scruin filament from the Limulus acrosomal process. J Cell Biol. 1993;123:337–344. doi: 10.1083/jcb.123.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pannese M, Polo C, Andreazzoli M, Vignali R, Kablar B, Barsacchi G, Boncinelli E. The Xenopus homologue Otx2 is a maternal homeobox gene that demarcates and specifies anterior body regions. Development. 1995;121:707–720. doi: 10.1242/dev.121.3.707. [DOI] [PubMed] [Google Scholar]
- 40.Porteus MH, Brice AEJ, Bulfone A, Usdin TB, Ciaranello RD, Rubenstein JLR. Isolation and characterization of a library of cDNA clones that are preferentially expressed in the embryonic telencephalon. Mol Brain Res. 1992;12:7–22. doi: 10.1016/0169-328x(92)90063-h. [DOI] [PubMed] [Google Scholar]
- 41.Probstmeier R, Bilz A, Schneider-Schaulies J. Expression of the neural cell adhesion molecule and polysialic acid during early mouse embryogenesis. J Neurosci Res. 1994;37:324–335. doi: 10.1002/jnr.490370305. [DOI] [PubMed] [Google Scholar]
- 42.Puelles L, Rubenstein JLR. Expression patterns of homeobox and other putative regulatory genes in the embryonic mouse forebrain suggest a neuromeric organization. Trends Neurosci. 1993;16:472–479. doi: 10.1016/0166-2236(93)90080-6. [DOI] [PubMed] [Google Scholar]
- 43.Quinlan GA, Williams EA, Tan S-S, Tam PPL. Neuroectodermal fate of epiblast cells in the distal region of the mouse egg cylinder: implication for body plan organization during early embryogenesis. Development. 1995;121:87–98. doi: 10.1242/dev.121.1.87. [DOI] [PubMed] [Google Scholar]
- 44.Reaume AG, Conlon RA, Zirngibl R, Yamaguchi TP, Rossant J. Expression analysis of a Notch homologue in the mouse embryo. Dev Biol. 1992;154:377–387. doi: 10.1016/0012-1606(92)90076-s. [DOI] [PubMed] [Google Scholar]
- 45.Richter K, Grunz H, Dawid IB. Gene expression in the embryonic nervous system of Xenopus laevis. Proc Natl Acad Sci USA. 1988;85:8086–8090. doi: 10.1073/pnas.85.21.8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts C, Platt N, Streit A, Schachner M, Stern CD. The L5 epitope: an early marker for neural induction in the chick embryo and its involvement in inductive interactions. Development. 1991;112:959–970. doi: 10.1242/dev.112.4.959. [DOI] [PubMed] [Google Scholar]
- 47.Rosette C, Karin M. Cytoskeletal control of gene expression: depolymerization of microtubules activates NF-kB. J Cell Biol. 1995;128:1111–1119. doi: 10.1083/jcb.128.6.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubenstein JLR, Martinez S, Shimamura K, Puelles L. The embryonic vertebrate forebrain: the prosomeric model. Science. 1994;266:578–580. doi: 10.1126/science.7939711. [DOI] [PubMed] [Google Scholar]
- 49.Sadler TW, Greenberg D, Coughlin P. Actin distribution patterns in the mouse neural tube during neurulation. Science. 1982;215:172–174. doi: 10.1126/science.7031898. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, Ed 2. Cold Spring Harbor Laboratory; New York: 1989. [Google Scholar]
- 51.Schmid MF, Jakana J, Matsudaira P, Chiu W. Imaging frozen, hydrated acrosomal bundle from Limulus sperm at 7 Å resolution with a 400 kV electron cryomicroscope. J Mol Biol. 1993;230:384–386. doi: 10.1006/jmbi.1993.1155. [DOI] [PubMed] [Google Scholar]
- 52.Schmid MF, Agris JM, Jakana J, Matsudaira P, Chiu W. Three-dimensional structure of a single filament in the Limulus acrosomal bundle: scruin binds to homologous helix-loop-beta motifs in actin. J Cell Biol. 1994;124:341–350. doi: 10.1083/jcb.124.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Senkevich TG, Muravnik GL, Pozdnyakov SG, Chizhikov VE, Ryazankina OI, Shchelkunov SN, Koonin EV, Chernos VI. Nucleotide sequence of XhoI fragment of ectromelia virus DNA reveals significant differences from vaccinia virus. Virus Res. 1993;30:73–88. doi: 10.1016/0168-1702(93)90017-h. [DOI] [PubMed] [Google Scholar]
- 54.Shimamura K, Hirano S, McMahon AP, Takeichi M. Wnt-1-dependent regulation of local E-cadherin and N-catenin expression in the embryonic mouse brain. Development. 1994;120:2225–2234. doi: 10.1242/dev.120.8.2225. [DOI] [PubMed] [Google Scholar]
- 55.Shimamura K, Hartigan DJ, Martinez S, Puelles L, Rubenstein JLR. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121:3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- 56.Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nested expression domains of four homeobox genes in developing rostral brain. Nature. 1992;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- 57.Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D’Apice MR, Nigro V, Boncinelli E. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. EMBO J. 1993;12:2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simpson P. Positive and negative regulators of neural fate. Neuron. 1995;15:739–742. doi: 10.1016/0896-6273(95)90163-9. [DOI] [PubMed] [Google Scholar]
- 59.Tam PPL, Trainor PA. Specification and segmentation of the paraxial mesoderm. Anat Embryol. 1994;189:275–305. doi: 10.1007/BF00190586. [DOI] [PubMed] [Google Scholar]
- 60.Tsai DE, Kenan DJ, Keene JD. In vitro selection of an RNA epitope immunologically cross-reactive with a peptide. Proc Natl Acad Sci USA. 1992;89:8864–8868. doi: 10.1073/pnas.89.19.8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 62.Varkey JP, Muhlrad PJ, Minniti AN, Do B, Ward S. The Caenorhabditis elegans SPE-26 gene is necessary to form spermatids and encodes a protein similar to the actin-associated proteins kelch and scruin. Genes Dev. 1995;9:1074–1086. doi: 10.1101/gad.9.9.1074. [DOI] [PubMed] [Google Scholar]
- 63.von Bülow M, Heid H, Hess H, Franke WW. Molecular nature of calicin, a major basic protein of the mammalian sperm head cytoskeleton. Exp Cell Res. 1995;219:407–413. doi: 10.1006/excr.1995.1246. [DOI] [PubMed] [Google Scholar]
- 64.Way M, Sanders M, Chafel M, Tu Y-H, Knight A, Matsudaira P. β-scruin, a homolog of the actin crosslinking protein scruin, is localized to the acrosomal vesicle of Limulus sperm. J Cell Sci. 1995a;108:3155–3162. doi: 10.1242/jcs.108.10.3155. [DOI] [PubMed] [Google Scholar]
- 65.Way M, Sanders M, Garcia C, Sakai J, Matsudaira P. Sequence and domain organization of scruin, an actin-cross-linking protein in the acrosomal process of Limulus sperm. J Cell Biol. 1995b;128:51–60. doi: 10.1083/jcb.128.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weitzer G, Milner DJ, Kim JU, Bradley A, Capetanaki Y. Cytoskeletal control of myogenesis: a desmin null mutation blocks the myogenic pathway during embryonic stem cell differentiation. Dev Biol. 1995;172:422–439. doi: 10.1006/dbio.1995.8070. [DOI] [PubMed] [Google Scholar]
- 67.Witta SE, Agarwal VR, Sato SM. XIPOU 2, a noggin-inducible gene, has direct neuralizing activity. Development. 1995;121:721–730. doi: 10.1242/dev.121.3.721. [DOI] [PubMed] [Google Scholar]
- 68.Wright CVE, Morita EA, Wilkin DJ, De Robertis EM. The Xenopus XlHbox 6 homeo protein, a marker of posterior neural induction, is expressed in proliferating neurons. Development. 1990;109:225–234. doi: 10.1242/dev.109.1.225. [DOI] [PubMed] [Google Scholar]
- 69.Xue F, Cooley L. Kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- 70.Ye BH, Lista F, Coco FL, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 71.Zimmerman K, Shih J, Bars J, Collazo A, Anderson DJ. XASH-3, a novel Xenopus achaete-scute homolog, provides an early marker of planar neural induction and position along the mediolateral axis of the neural plate. Development. 1993;119:221–232. doi: 10.1242/dev.119.1.221. [DOI] [PubMed] [Google Scholar]
- 72.Zollman S, Godt D, Prive GG, Couderc J-L, Laski FA. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci USA. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]