Abstract
Alcohol dependence (alcoholism) is accompanied by evidence of tolerance, withdrawal (physiological dependence), or compulsive behavior related to alcohol use. Studies of strain and individual differences using animal models for acute physiological dependence liability are useful means to identify potential genetic determinants of liability in humans. Behavioral and quantitative trait analyses were conducted using animal models for high risk versus resistance to acute physiological dependence. Using a two-step genetic mapping strategy, loci on mouse chromosomes 1, 4, and 11 were mapped that contain genes that influence alcohol withdrawal severity. In the aggregate, these three risk markers accounted for 68% of the genetic variability in alcohol withdrawal. Candidate genes in proximity to the chromosome 11 locus include genes encoding the α1, α6, and γ2 subunits of type-A receptors for the inhibitory neurotransmitter, GABA. In addition, suggestive linkage is indicated for two loci on mouse chromosome 2, one near Gad1encoding glutamic acid decarboxylase, and the other near theEl2 locus which influences the seizure phenotype in the neurological mutant strain El. The present analyses detect and map some of the loci that increase risk to develop physiological dependence and may facilitate identification of genes related to the development of alcoholism. Syntenic conservation between human and mouse chromosomes suggests that human homologs of genes that increase risk for physiological dependence may localize to 1q21–q32, 2q24–q37/11p13, 9p21–p23/1p32–p22.1, and 5q32–q35.
Keywords: ethanol, QTL, quantitative trait locus, recombinant inbred strain, selective breeding, GABAA receptor, glutamic acid decarboxylase, physiological dependence, withdrawal, seizure, handling-induced convulsions
Alcoholism is clearly a multifactorial disorder. A genetic contribution to alcoholism is supported by half-sibling and adoption studies that demonstrate an increased risk for severe alcohol-related problems in children of alcoholics who were adopted out, even if they had been raised without knowledge of their biological parents’ problems (Schuckit et al., 1972; Goodwin et al., 1974;Bohman, 1978; Cadoret et al., 1980). Four large twin studies published in the 1990s also substantiate the conclusion that alcoholism is >50% heritable and that heritability for alcoholism in females and males is similar (Goldman, 1993). Evidence also exists for genetic heterogeneity and the influence of multiple genes on alcoholism vulnerability (Gilligan et al., 1987; Aston and Hill, 1990; Goldman, 1993), but the specific genes related to alcoholism are still unknown.
Recent official diagnostic manuals indicate that alcohol dependence is associated with evidence of tolerance, withdrawal, or a maladaptive pattern of alcohol use (DSM-IV, 1994). Diagnostic criteria for withdrawal include two or more of the following, developing within several hours to a few days after cessation of alcohol use: autonomic hyperactivity, tremor, insomnia, nausea, hallucinations, psychomotor agitation, anxiety, seizures. A majority of individuals who have alcohol dependence experience clinically relevant levels of withdrawal, although only ∼5% experience severe late developing manifestations of the disorder (DSM-IV, 1994). Previous studies show that genetic differences in acute withdrawal liability may be related to high or low risk for onset of alcoholism. Family history positive males report greater withdrawal effects, measured 3–8 hr after administration of 1 gm/kg ethanol, as compared with family history negative males (McCaul et al., 1991). Sons of alcoholics also report greater hangover symptoms, which are thought to represent an acute withdrawal syndrome, as compared with sons of nonalcoholics (Newlin and Pretorius, 1990).
Because family members who carry genes that increase risk do not always develop alcoholism, linkage studies may not be an optimal step until markers or candidate genes are identified (Schuckit, 1994). The present studies were performed to identify markers and candidate genes for genetic variation in degree of acute physiological dependence using animal models that differ in withdrawal liability, because physiological dependence is defined as the manifestation of withdrawal (e.g., seizures) after alcohol administration is suspended. Among the many signs of physiological dependence, withdrawal seizures are a particularly useful index of withdrawal, because they are displayed in all species tested, including humans (Friedman, 1980). Behavioral and quantitative trait analyses were conducted using populations derived from the C57BL/6J (B6) and DBA/2J (D2) inbred strains of mice [i.e., their BXD recombinant inbred (RI) strains, a B6D2 F2 intercross, and mice selectively bred for high vs low alcohol withdrawal liability]. The D2 strain is a well characterized animal model with severe withdrawal seizures, whereas the B6 strain has mild withdrawal reactions (Crabbe et al., 1983; Belknap et al., 1993). By studying animal models, complications imposed by environmental variation, which has a strong effect on human studies, are minimized. Moreover, by studying inbred strains and their crosses, the problem of genetic heterogeneity is eliminated. By using a sufficient number of animals, one can achieve the statistical power required to detect the influence of the individual genes at quantitative trait loci/locus (QTL) affecting a trait with polygenic inheritance (Lander and Botstein, 1989). QTL mapping methods using rodent models have previously allowed dramatic progress toward the detection and chromosome mapping of minor and major gene loci involved in complex traits such as hypertension, diabetes, and epilepsy (Jacob et al., 1991; Rise et al., 1991; Todd et al., 1991). In the present analyses, QTL mapping using a rodent model for physiological dependence successfully mapped loci associated with increased risk for withdrawal on mouse chromosomes 1, 4, and 11 and detected suggestive linkage on chromosome 2.
MATERIALS AND METHODS
Animals. The BXD RI strains were originally purchased from The Jackson Laboratory (Bar Harbor, ME) and were bred and maintained in our colonies at the Portland VA Medical Center Veterinary Medical Unit. The B6D2 F2 population was generated from crosses of B6D2 F1 hybrids purchased from The Jackson Laboratory. A distinct B6D2 F2 intercross (not included in our B6D2 F2 analysis) served as founders used for short-term selection of lines of mice bred for differences in acute alcohol withdrawal [High (HAW) and Low Alcohol Withdrawal (LAW) ] (Metten and Crabbe, 1996). The 10 males and 10 females with the highest scores for withdrawal convulsion severity were selected to serve as breeders of the HAW line, whereas the 10 males and 10 females with the lowest scores were bred to form the LAW line. In subsequent generations, the highest of the HAW line and the lowest of the LAW line were chosen to perpetuate the HAW and LAW lines, respectively (10 breeding pairs per line, using individual selection but excluding sibling matings).
Quantitation of withdrawal using the handling-induced convulsion. Physiological dependence is operationally defined as the manifestation of physical disturbances (withdrawal syndrome) after alcohol administration is suspended. McQuarrie and Fingl (1958) first demonstrated a state of withdrawal CNS hyperexcitability after acute alcohol administration. To index acute withdrawal, we used the handling-induced convulsion, a sensitive index of alcohol withdrawal severity (Goldstein and Pal, 1971). Genetic variation in withdrawal was examined using populations derived from the D2 and B6 strains by monitoring changes in convulsions induced by handling after withdrawal from alcohol. Adult mice (8–15 weeks) were scored for baseline handling-induced convulsions immediately before administration of a high dose of ethanol (4 gm/kg, 20% v/v in saline, i.p.) and between 2 and 12 hr after alcohol administration. Details of the methods and scoring system have been published previously (Crabbe et al., 1991). Briefly, each mouse is picked up gently by the tail and, if necessary, spun gently through 180°, and handling-induced convulsions scored as follows: 7, severe, tonic-clonic convulsion, with quick onset and long duration: spontaneous, or elicited by mild environmental stimulus (e.g., lifting cage top); 6, severe, tonic-clonic convulsion when lifted by the tail: quick onset and long duration, often continuing for several seconds after the mouse is released; 5, tonic-clonic convulsions when lifted by the tail: onset often delayed by up to 1–2 sec; 4, tonic convulsion when lifted by the tail; 3, tonic-clonic convulsion after gentle 180° spin; 2, no convulsion when lifted by the tail, but tonic convulsion elicited by gentle 180° spin; 1, facial grimace only after a gentle 180° spin; 0, no convulsion. Individual mice and/or different inbred strains can differ in baseline (pre-injection) handling-induced convulsion scores. Therefore, to characterize withdrawal severity, scores were first computed as the area under the curve (AUC), between 4 and 12 hr after alcohol administration. We regressed AUC on baseline scores and used the regression residual as our index of withdrawal severity (these two variables were genetically correlated, r = 0.66 in the 21 RI strains). This method yielded withdrawal scores that were normally distributed and guarantees that the derived scores have a correlation of exactly 0 with baseline scores. Therefore, this is our best estimate of the contribution of alcohol withdrawal to AUC scores independent of baseline score differences.
Step 1: QTL analysis using BXD RI strains. Of the 26 BXD strains, 21 were available in our colonies in sufficient numbers for testing; a total of 288 adult male BXD mice were tested and scored for alcohol withdrawal. For each of 1522 markers in the MAP MANAGER BXD marker data base, an arbitrary value of 0 was assigned to each BXD strain bearing two copies of the B6 allele, and a value of 1 was assigned to each strain bearing two copies of the D2 allele. Correlation coefficients (r) were determined between each marker and the BXD strain means for residual alcohol withdrawal severity. Two-tailed p values for associations detected using the BXD strains are given. Rather than using overly stringent α levels at this stage, which could lead to inflated type II errors and potential failure to identify important QTL, we chose in step 2 of our analysis to test putative BXD-implicated QTL, using F2 and selection line experiments. This two-step approach avoids the type I errors expected using the RI data alone. In a preliminary BXD analysis, fewer RI strains were tested, fewer genetic markers were available for mapping, and peak measures of withdrawal without regression residuals were calculated (Belknap et al., 1993).
Step 2: Verification testing of putative QTL using a B6D2 F2 intercross. B6D2 F2 mice (n = 451 mice) of approximately equal numbers of either sex were tested and scored for alcohol withdrawal as described above. Genomic DNA was isolated from individual mouse spleens using a salting-out method adapted from Miller et al. (1988). Mice were genotyped using microsatellite markers from the MIT series using a method adapted from Dietrich et al. (1992) andSerikawa et al. (1992). Alternatively, for the Tyrp1(tyrosinase-related protein) locus, also known as the b(brown) locus, mice were genotyped based on coat color. The D2 allele produces brown coat color and is recessive to the B6 allele, which produces black coat color (Silvers, 1979). For each marker tested, individual B6D2 F2 mice were assigned a genotypic score of 0, 1, or 2 based on gene dosage (the number of D2 alleles at that locus). Correlation coefficients (Pearson’s r) were determined and are equivalent to regressing withdrawal scores on gene dosage for each marker. For two linked loci on chromosome 2 with opposing effects,r values were determined for F2 mice recombinant betweenD2Mit9 and D2Mit17 (n = 127 recombinant mice) to increase the power of our analysis to detect either locus with greater confidence. One-tailed p values are given for the F2 and HAW/LAW experiments, because the direction of effect (whether the B6 or D2 allele was associated with greater withdrawal intensity) was predicted from the RI results.
Verification testing using lines selectively bred for differential risk for alcohol withdrawal. Mice from the HAW and LAW lines of approximately equal numbers of either sex were tested and scored for alcohol withdrawal and genotyped as described above. The D2 allelic frequency (q) in the LAW and HAW lines, and the difference between the lines (qLAW−qHAW), was determined for the Tyrp1 locus using generations one to four of selective breeding (S1–S4). MIT markers were examined using generations S2 and S4. The p values given are for the S2 (n = 113 mice) and are based on z (normal variate) calculated asqLAW−qHAW divided by the SE of qLAW-qHAW expected from genetic drift (random mating without selection) and qestimation error (Lebowitz et al., 1987; Falconer, 1989; Belknap et al., 1997). This SE was calculated as the square root of the following: 2poqoFt+ [(ptLAW)(qtLAW)]/ntLAW+ [(ptHAW)(qtHAW)]/ntHAW, representing the drift and q estimation error variances, respectively. F is the inbreeding coefficient at generationt of selection, and po andqo are the founding F2 allelic frequencies (for B6 and D2 alleles, respectively), and n is the number of genotyped mice per line (Belknap et al., 1997). This provides a valuable test that is statistically independent from analyses using RI or F2 mice.
p Values and estimated LOD scores. Individualp values for the RI, F2, and S2 experiments were determined using linear least squares statistical analyses rather than maximum likelihood methods. The latter are applicable only for F2 data, and there is little loss of statistical power when markers are within about 10 centiMorgans (cM) of the QTL (Darvasi et al, 1993). For each QTL, a combined P value (and estimated logarithm of the likelihood for linkage, or LOD) was determined using Fisher’s method (Fisher, 1958; Sokal and Rohlf, 1981) for combining p values fromt experiments testing the same hypothesis: −2∑lnp[df = 2t]. LOD scores were estimated from P values based on the asymptotic distribution of LOD as χ2[df = 1] using the expression: LOD = 1/2(log10e)χ2for an additive (i.e., df = 1) model (Lander and Botstein, 1989;Lander and Kruglyak, 1995). Guidelines recommended by Lander and Kruglyak (1995) were used for interpreting and reporting the linkage results.
QTL contributions to phenotypic and genetic variance in risk for physiological dependence. For all three experiments, the proportion of the phenotypic variance contributed by each QTL was calculated as estimated for an F2 population to allow direct comparison of RI, F2, and selection line results (Belknap et al., 1996), and the average for all three experiments was taken as the overall estimate of the proportion of phenotypic variance contributed by each QTL. For the F2 experiment, phenotypic variance was estimated asr2 (the square of the correlation coefficient). For the RI data, one-half of the variance attributable to each QTL in the RI strains (r2/2) was used to estimate the phenotypic variance expected in a comparable F2 population (Belknap et al., 1996), because 50% of an F2 population are heterozygotes, which are absent in RI strains. Parallel estimates of phenotypic variance from the HAW/LAW data were based on divergence in allelic frequencies in the two oppositely selected lines (Belknap et al., 1997). The heritability for alcohol withdrawal was estimated from the RI data (additive effects) (Belknap et al., 1996), and from the HAW/LAW data by dividing the selection response by the selection differential for the divergence in the two lines in the S2 generation (Falconer, 1989). These two estimates were 0.19 and 0.275, respectively, for an average overall estimate of heritability of h2 = 0.23. This compares well with the heritability estimateh2 = 0.26 from an earlier experiment selecting for chronic alcohol withdrawal severity (Crabbe et al., 1985). The percentage of genetic variance contributed by each QTL was estimated by the percentage of phenotypic variance divided by 0.23.
Maximum likelihood analysis. Estimated 1.0 LOD confidence intervals (∼90% confidence intervals) for the positions of QTL on chromosomes 1, 2 (proximal), 4, and 11 were estimated based on interval analysis of the B6D2 F2 data using the MAPMAKER/QTL analysis program (Lincoln et al., 1992). Maximum likelihood analysis using MAPMAKER/QTL was also used to examine additive and dominant models of inheritance using the B6D2 F2 data.
Glutamic acid decarboxylase (GAD) activity. Adult (70–85 d) male B6 and D2 mice (The Jackson Laboratory) were killed by cervical dislocation. The whole brain was dissected and retained and homogenized using a glass Teflon homogenizer in 15.5–17.5 ml ice-cold 0.32m sucrose containing 10 mm glutathione to inhibit oxidation of GAD and to stabilize enzyme activity. Brain homogenates from B6 and D2 mice were prepared in parallel on the same day and exposed to the same experimental conditions. GAD activity was assayed using a modification of the method of Deaizpurua et al. (1992)based on the method of Albers and Brady (1959). Each reaction included 0.4 μCi l-[14C]-glutamic acid, 1 mm cold glutamic acid, 1 mm2-aminoethylisothiouroniium bromide, and potassium phosphate buffer, pH 7.3, and was initiated by the addition of D2 or B6 brain tissue (0.08–0.1 mg of protein) and was performed under N2 for 60 min in a shaking 37°C incubator. After this incubation, 150 μl of 5N H2SO4 was injected into each reaction tube to inactivate GAD and release [14C]CO2liberated from the aqueous reaction mixture. The tubes were then shaken for 60 min at 37°C to facilitate release of the liberated [14C]CO2 and allow absorption to hyamine base soaked filter disks supported above the aqueous phase. Liberation of [14C]CO2 is linear using these conditions (e.g., <0.2 mg protein per reaction, incubation <90 min) (data not shown). The amount of radioactivity absorbed to the filters was determined by liquid scintillation spectrometry. The amount of radioactivity absorbed to the filters in the absence of membranes (no-tissue blank) was subtracted from all values.
RESULTS
Detection of putative QTL for alcohol withdrawal using the BXD strains
QTL analysis using the BXD RI strains was used for a genome-wide screen for associations between susceptibility to alcohol withdrawal and 1522 genetic markers in our BXD data base. Many individuals of each genotype (i.e., each RI strain) could be tested, permitting a high degree of accuracy in determining the relationship between genotype and risk for alcohol withdrawal. The progenitor B6 and D2 strains and the BXD RI strains showed a range of postethanol withdrawal severities and baseline convulsion scores (Fig. 1). QTL analyses of the distribution of BXD RI strain means for residual alcohol withdrawal severity indicated that markers associated with increased risk for withdrawal clustered within seven discrete chromosomal regions on chromosomes 1, 2, 4, 6, 8, and 11 (p <0.05, indicating potential linkage)
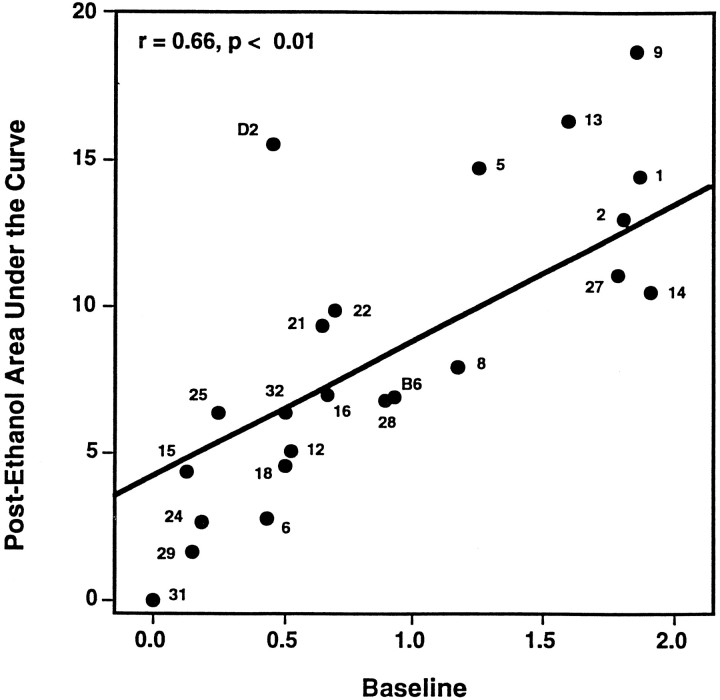
Fig. 1.
Phenotypic relationship between baseline and postethanol withdrawal convulsions in BXD RI strains. Each data point identifies the strain mean ± SEM for baseline (x-axis) and postethanol withdrawal convulsions (area under the curve,y-axis) for the BXD RI strains (e.g., 9 represents strain BXD-9) or the B6 or D2 progenitor strains. The least squares linear regression of postethanol withdrawal severity on baseline convulsion scores using 21 BXD RI strains is shown (r = 0.66, p < 0.01) and did not include the two progenitor strains. Residual alcohol withdrawal severities for the BXD RI strains and their progenitor strains are measured as the vertical distance between theregression line and the strain means for postethanol withdrawal area under the curve.
(Table 1). Two putative QTL were detected on chromosome 2, one identified by several markers located 28–38 cM from the centromere (referred to as 2proximal), and the other associated with markers located ∼75–85 cM distal to the centromere (2distal). Because of statistical limitations imposed by the limited number of BXD strains, QTL identified using only the RI strains were considered putative and were subjected to more testing in additional populations derived from the D2 and B6 strains: an F2 intercross, and in HAW and LAW lines of mice selectively bred from B6D2 F2 founders for differences in alcohol withdrawal. This two-step approach explicitly considers the consequences of type I error (identification of a false QTL) and type II error (failure to detect an actual QTL) (Belknap et al., 1996). The results of these analyses identify significant linkage with QTL on chromosomes 1, 4, and 11 that contribute to differences in alcohol withdrawal severity, indicate suggestive linkage with two QTL on chromosome 2, and disconfirm putative loci detected on chromosomes 6 and 8. No gender differences were detected using the HAW/LAW lines or in the F2 analysis [F(1,376) = 2.48, p = 0.12, not significant].
Table 1.
Marker loci identified by QTL analysis using BXD RI mice
| Marker | Location (Chr:cM) | Correlation coefficent | pvalue |
|---|---|---|---|
| D1Ncvs55 | 1:73 | +0.46 | 0.05 |
| D1Ncvs12 | 1:75 | +0.76 | 0.0003 |
| D1Byu7 | 1.82 | +0.52 | 0.02 |
| D2Ncvs50 | 2:∼35 | +0.58 | 0.006 |
| D2Ncvs51 | 2:∼35 | +0.61 | 0.005 |
| Pmv7 | 2:35 | +0.46 | 0.03 |
| Scn2a | 2:36 | +0.48 | 0.025 |
| B2m | 2:69 | −0.50 | 0.02 |
| D2Mcl | 2:83 | −0.65 | 0.005 |
| Iapls2-4 | 2:86 | −0.52 | 0.02 |
| D4Ncvs78 | 4:14 | +0.45 | 0.04 |
| D4Bir5 | 4:39 | +0.45 | 0.04 |
| Iapls1-10 | 4:39 | +0.45 | 0.04 |
| D4Ncvs82 | 4:41 | +0.51 | 0.02 |
| D6Mit9 | 6:37 | +0.59 | 0.005 |
| Tgfa | 6:38 | +0.59 | 0.005 |
| D6Nds2 | 6:40 | +0.59 | 0.02 |
| Rho | 6:41 | +0.52 | 0.04 |
| D8Ncvs61 | 8:32 | +0.58 | 0.009 |
| D8Ncvs60 | 8:32 | +0.49 | 0.04 |
| D11Byu2 | 11:16 | −0.52 | 0.015 |
| D11Ncvs69 | 11:16 | −0.71 | 0.0004 |
| D11Mit53 | 11:16 | −0.52 | 0.015 |
| Ebf | 11:∼19 | −0.44 | 0.04 |
Genetic markers, chromosome number, distance from the centromere in centiMorgans (cM) (e.g., 1:73), and associations with acute alcohol withdrawal liability are given. Locations indicate the chromosome and the distance from the centromere in centiMorgans. Values in the table indicate point biserial correlation coefficients (r) and the corresponding p values (two-tailed) using the BXD RI strains. Boldfaced associations indicate the most highly correlated marker in each BXD-implicated region; nearby clusters of markers associated at p ≤ 0.05 are also shown. All boldfaced associations are significant at p ≤ 0.01, except forD4Ncvs82 on chromosome 4, which is associated at p= 0.02. Positive correlation coefficent (r) values indicate that the D2 allele is associated with greater withdrawal liability.
Identification of a QTL on chromosome 4 linked to Tyrp1
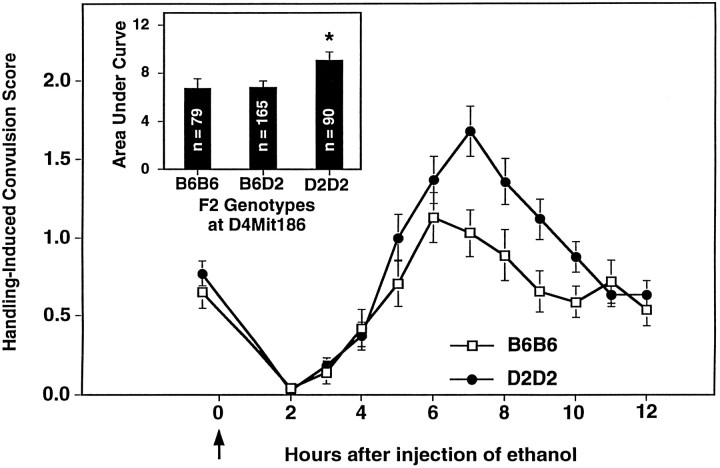
The present studies identify a QTL on chromosome 4 that influences risk for alcohol withdrawal. QTL analysis using the RI strain means for withdrawal severity identified several markers located ∼40 cM from the centromere associated with increased susceptibility to alcohol withdrawal (Table 1). Testing this BXD-implicated locus using a B6D2 F2 intercross and the HAW and LAW selectively bred lines verified that a QTL influencing risk for alcohol withdrawal is in proximity toTyrp1 and D4Mit186. These data are summarized in Table 2. In the F2 intercross, alcohol withdrawal severity was associated with genotypic status at Tyrp1 (38 cM; p = 0.004, Table 2) and several MIT markers, including D4Mit186. Figure 2 shows that F2 mice homozygous for the D2 allele at D4Mit186 show greater withdrawal liability than B6B6 homozygotes or heterozygotes (p = 0.02). Maximum likelihood analysis of the F2 data indicated maximum LOD scores of 2.147 and 1.056 using unconstrained and B6 dominant models, respectively, and excluded a B6 dominant model of inheritance for this QTL. Selective breeding rapidly produced highly divergent high and low lines for alcohol withdrawal severity, and by generation S2, the HAW and LAW lines differed threefold in withdrawal intensity (Fig.3A). Differences between the HAW and LAW lines in allelic frequencies at Tyrp1significantly exceeded those expected from genetic drift and estimation error, providing additional supporting evidence for the presence of a QTL linked to Tyrp1 (Fig. 3B). Within just two generations of selective breeding (i.e., S2), allelic frequency of the D2 progenitor allele (q) at Tyrp1 was highly correlated with alcohol withdrawal severity (p = 1 × 10−5). Allelic frequencies for MIT markers located 31–56 cM from the centromere (D4Mit55, D4Mit178, D4Mit186, D4Mit37, D4Mit12) also differed between the HAW and LAW lines (p < 0.001, data not shown). Taken together, our data indicate that a locus in proximity toTyrp1 is associated with higher risk for physiological dependence (P = 3 × 10−7, LOD = 5.6). This QTL accounts for ∼6% of the total phenotypic variance in alcohol withdrawal between the D2 and B6 strains, representing 26% of the genetic variance. In each of the populations tested, risk for alcohol withdrawal was most highly correlated with markers located 38–42 cM from the centromere (Fig. 4). This region of mouse chromosome 4 is syntenic with human 9p21–p23 and 1p32–p22.1 (Silver et al., 1996).
Table 2.
Summary of evidence for QTLs involved in acute alcohol withdrawal
| Chr | pvalues | Combined | ||||
|---|---|---|---|---|---|---|
| RI | F2 | S2 | P | LOD | % VG | |
| 4 | 0.02 | 0.004 | 0.00001 | 3 × 10−7 | 5.6* | 26 |
| 1 | 0.0003 | 0.004 | 0.001 | 3 × 10−7 | 5.6* | 26 |
| 11 | 0.0004 | 0.018 | 0.008 | 1 × 10−5 | 4.1* | 12 |
| 2proximal | 0.005 | 0.025 | 0.12 | 0.001 | 2.3 | — |
| 2distal | 0.0046 | 0.025 | 0.22 | 0.002 | 2.1 | — |
| 6 | 0.005 | 0.5 | 0.07 | 0.01 | 1.4 | — |
| 8 | 0.009 | 0.3 | 0.5 | 0.02 | 1.2 | — |
The p values from the BXD RI (21 strains), B6D2 F2 (n = 451 mice), and HAW/LAW (n = 113 mice, generation S2) experiments are shown. All chromosome regions reachingp < 0.01 in the BXD QTL results were searched in the F2 and S2 experiments for supporting evidence. The most significantly associated marker in each chromosomal region is shown. Entries arep values for each of the three independent experiments, plus a combined P value for all three experiments (and estimated LOD score). For all three experiments, the D2 allele was consistently associated with greater withdrawal severity for all chromosomal regions shown, except for loci on chromosome 11 and the distal region of chromosome 2, where the B6 allele was consistently associated with more severe withdrawal.
QTL detected on chromosomes 1, 4, and 11 exceed theLander and Kruglyak (1995) criteria for significant linkage. In the aggregate, these three QTL account for ∼64% of the genetic variance in acute alcohol withdrawal severity, representing about 15% of the total variance in these genetic populations. The percentage of the genetic variance (% VG) contributed by each QTL showing significant linkage is also shown. Suggestive linkage was also detected for two QTL on chromosome 2.
Fig. 2.
Linkage analysis using B6D2 F2 intercross mice provides evidence for a QTL influencing alcohol withdrawal on chromosome 4. Alcohol withdrawal was indexed using the handling-induced convulsion. The mice were scored for baseline handling-induced convulsions immediately before administration of 4 gm/kg ethanol (thearrow marks ethanol injection at time 0), and hourly between 2 and 12 hr after alcohol administration. Data represent the mean raw scores ± SEM for baseline and postethanol handling-induced convulsions. Alcohol administration initially lowers convulsion scores (0–4 hr). Later, convulsion scores increase above baseline, indicating a state of withdrawal hyperexcitability, which peaks ∼6–7 hr after alcohol administration. From 451 F2 intercross mice tested for physiological dependence, we genotyped 167 mice with the highest withdrawal scores and 167 mice with the lowest withdrawal scores. B6D2 F2 mice homozygous for the D2 allele atD4Mit186, a marker located 42.6–45.5 cM from the centromere, showed more severe withdrawal than B6B6 homozygous F2 mice.Inset, Gene dosage at D4Mit186 has a significant influence on alcohol withdrawal severity calculated as area under the curve [F(2,331) = 3.3,p = 0.02). *D2D2 homozygotes have more severe withdrawal than B6B6 homozygotes or B6D2 heterozygotes (Tukey HSD test,p < 0.05).
Fig. 3.
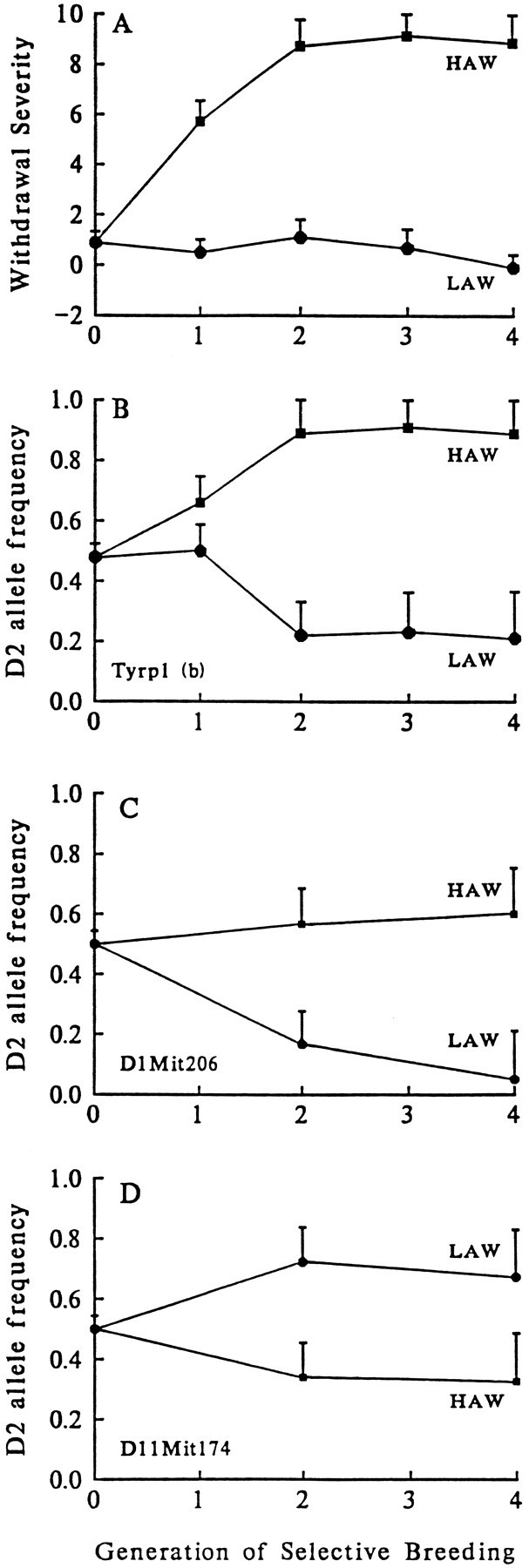
Allelic frequencies at loci on chromosomes 1, 4, and 11 cosegregate with phenotypic selection for acute alcohol withdrawal severity. A, The selection response [mean ± SEM is shown for the first four generations of selective breeding (S1–S4)] for differences in acute withdrawal liability (HAW and LAW). On they-coordinate, alcohol withdrawal severity is shown as the computed area under the curve(AUC), calculated on the basis of the time course for handling-induced convulsions measured between 4 and 12 hr after alcohol administration. B, In generations S1–S4, gene frequency at Tyrp1 for the D2 allele diverged in the two oppositely selected lines approximately in parallel with the trait under selection. By generation S2, the HAW and LAW lines differed in their gene frequencies (q) for the D2 allele for Tyrp1(qHAW = 0.89,qLAW = 0.22, z = 4.18,p = 1 × 10−5). Similarly, in generations S2 and S4, allelic frequencies at C,D1Mit206 (qHAW = 0.57,qLAW = 0.17, z = 2.47,p = 0.006 using generation S2) andD, D11Mit174(qHAW = 0.39,qLAW = 0.76, z = 2.34,p = 0.008 in generation S2) diverged approximately in parallel with alcohol withdrawal severity. Because allelic frequencies of the D2 and B6 alleles at the markers shown diverged in the HAW and LAW lines approximately in parallel with divergence of the trait under selection, these data indicate that QTL underlying differences in alcohol withdrawal between the HAW and LAW lines are linked to the markers Tyrp1 (chromosome 4, 38 cM),D1Mit206 (chromosome 1, 96 cM), andD11Mit174 (chromosome 11, 20 cM).
Fig. 4.
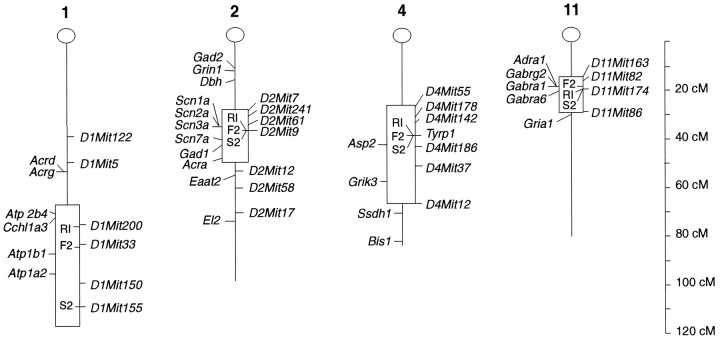
Estimated confidence intervals and candidate genes for alcohol withdrawal QTL identified on chromosomes 1, 2, 4, and 11. The markers tested using the B6D2 F2 population are shown, and their map positions (Silver et al., 1996) indicated in centiMorgans from the centromere (at 0 cM). Estimated 1.0 LOD confidence intervals for the positions of QTL on mouse chromosomes 1, 2 (proximal), and 11 are shown (boxed regions), based on interval analysis of our B6D2 F2 data using MAPMAKER/QTL. For chromosome 4, the boxed regionindicates the range of markers examined in our F2 data, but the 1.0 LOD confidence interval for this QTL actually extends beyond the range of markers examined. For chromosome 2, the 1.0 LOD confidence interval is shown only for the proximal QTL, because MAPMAKER/QTL interval analysis cannot resolve the influence of two linked QTL with opposite effects on a phenotype. The position of the best correlated marker for each separate experiment (i.e., RI, F2, or S2) is also indicated within eachboxed region. Candidate genes located within or near the 1.0 LOD confidence intervals are also shown (from Silver et al., 1996).El2, an epilepsy quantitative trait locus, is located ∼75 cM (between 53 and 80 cM) distal to the centromere of chromosome 2 (Frankel et al., 1995).
Verification of a second QTL on chromosome 1
We also identified a locus on chromosome 1 that accounts for ∼6% of the total phenotypic variance in alcohol withdrawal, representing 26% of the genetic variance between the D2 and B6 strains. QTL analysis using the BXD RI strains implicated markers within the distal region of chromosome 1 (Table 1). To test the involvement of a QTL within this region, we tested B6D2 F2 mice using six microsatellite markers located 37–109 cM from the centromere (Fig.4); gene dosage at D1Mit206 (96 cM) showed the strongest association with alcohol withdrawal severity (p= 0.004, Table 2). This marker is located more distally than markers associated with alcohol withdrawal in the BXD analysis, but some variation in QTL map location among the RI, F2, and HAW/LAW experiments is expected, because the 95% confidence limits in all three experiments span at least 20 cM (Silver, 1985; Darvasi and Soller, 1995; Visscher et al., 1996). Allelic frequencies atD1Mit206 were correlated with withdrawal severity in the HAW and LAW lines, providing additional evidence for the presence of a QTL linked to D1Mit206 (Fig. 3C). Allelic frequencies also differed between the HAW and LAW lines for D1Mit155(qHAW = 0.66, qLAW = 0.16, z = 3.11; p = 0.001, Table 2), as well as several other markers (D1Mit200, D1Mit33, D1Mit150) located 73–98 cM distal to the centromere (p < 0.01, data not shown). Taken together, these three experiments show that D2 alleles in the distal region of chromosome 1 are associated with increased withdrawal severity (P = 3 × 10−7, LOD = 5.6).
A third QTL is in close proximity to GABAA receptor genes on chromosome 11
The present RI analysis indicates that markers located ∼16–19 cM from the centromere of chromosome 11 are correlated with risk for alcohol withdrawal (Table 1). In the B6D2 F2 intercross, risk was strongly correlated with gene dosage at D11Mit174 (20 cM,p = 0.018, Table 2). Maximum likelihood analysis using the B6D2 F2 data excluded B6 dominant and additive models of inheritance for this locus (maximum LOD = 2.146, 1.117, and 1.142 using unconstrained, additive, and B6 dominant models, respectively). The HAW and LAW lines also differed in their allelic frequencies forD11Mit174 (Fig. 3D), and for D11Mit163and D11Mit82 located about 16 cM from the centromere (p < 0.01, data not shown). These data indicate that a QTL influencing alcohol withdrawal liability is located in proximity to D11Mit174 (P = 1 × 10−5, LOD = 4.1). In each of the populations tested, the most highly correlated marker is located 16–20 cM from the centromere (Fig. 4), within a region corresponding to human chromosome 5q32-q35. This QTL accounts for ∼3% of the phenotypic variance or 12% of the genetic variance in risk for physiological dependence between the D2 and B6 strains. The BXD strain distribution pattern forD11Mit174 is identical to that for restriction fragment length polymorphisms for Gabra1 and Gabra6, indicating no recombinations among these three loci (Garrett et al., 1997). GABAA receptors have previously been implicated in physiological dependence on alcohol (Buck et al., 1991a,b). The present studies implicate specific genes encoding the α1, α6, and γ2 subunits of GABAAreceptors (19–23 cM) (Fig. 4).
Detection of suggestive QTL for alcohol withdrawal on chromosome 2
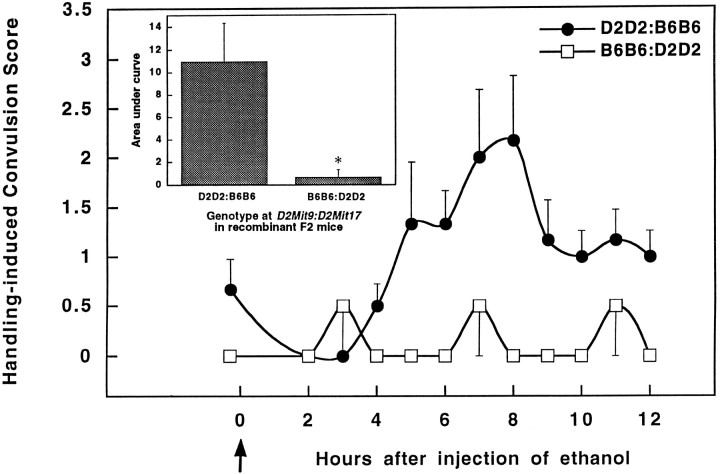
Our data also identify two suggestive QTL on chromosome 2. The 2proximal locus was detected in the BXD QTL analysis by several markers located 25–38 cM from the centromere, whereas the 2distal locus was associated with markers located ∼68–85 cM distal to the centromere. These two QTL have opposing effects on alcohol withdrawal, with D2 alleles at the 2proximal locus associated with higher withdrawal scores (r = 0.61,p = 0.005), and D2 alleles at the 2distallocus instead associated with lower risk for withdrawal (r = − 0.65, p = 0.0046). Of the 21 BXD RI strains tested, 16 were recombinant between D2Mit9(near the 2proximal locus) and D2Mit17 (near 2distal). Because possession of positive alleles at one QTL, but negative alleles at the other QTL, would tend to cancel each other in terms of their phenotypic effects, the BXD RI series was particularly well suited to detect the influence of linked QTL at which D2 alleles have opposing effects. Moreover, the RI strains are homozygous at each marker, which further increases the power to detect both loci. The utility of recombinants for elucidating linked QTL is clearly prescribed in recent discussions of QTL mapping (Tanksley, 1993). The negative relationship between the two QTL also predicts that F2 mice that are nonrecombinants would be uninformative for QTL mapping, because their predicted withdrawal scores would be near the median value for all F2 mice. To increase the power of our analysis to detect either QTL with greater confidence, their association with withdrawal was determined using the 127 F2 mice that showed recombination between D2Mit9 and D2Mit17. These data indicate that withdrawal is more severe in recombinant F2 mice with genotypes associated with increased risk for withdrawal (D2 allelic dosage at D2Mit9/D2Mit17 = 2:0, 2:1, or 1:0) as compared with mice with protective alleles (D2Mit9/D2Mit17gene dosage = 0:2, 0:1, or 1:2) (Table 2). However, 118 of the recombinant F2 mice were heterozygous at one or both putative QTL, which greatly diminished the statistical power to detect the simultaneous influence of both loci. In recombinant F2 mice that are homozygous at D2Mit9 and D2Mit17, withdrawal severity is strikingly different between mice with the D2D2:B6B6 genotype as compared with B6B6:D2D2 mice (Fig. 5). No correlation with any chromosome 2 marker even approached significance in nonrecombinant F2 mice. The most plausible explanation for this marked difference between recombinant and nonrecombinant outcomes is the presence of two linked QTL with offsetting influences on withdrawal severity. A trend toward divergence between the HAW and LAW lines inD2Mit9 and D2Mit17 allelic frequencies was suggested in generation S2 (p = 0.12 andp = 0.22, respectively). In the aggregate, our data suggest that D2 alleles at the 2proximal QTL are associated with more severe withdrawal (p = 0.001, LOD = 2.3) and that D2 alleles at the 2distal locus are instead associated with decreased withdrawal severity (p= 0.002, LOD = 2.1). These LOD scores indicate suggestive linkage, but are not sufficient to confirm linkage (Lander and Kruglyak, 1995). More definitive confirmation of these suggestive QTL will likely require verification using additional recombinant F2 mice that are strictly homozygous at D2Mit9 and D2Mit17.
Fig. 5.
Alcohol withdrawal severity in F2 mice recombinant between D2Mit9 and D2Mit17. Data represent the mean ± SEM for F2 mice recombinant betweenD2Mit9 (37 cM) and D2Mit17 (69 cM), markers associated with two opposing QTL detected within the proximal and distal regions of chromosome 2. Recombinant F2 mice with risk alleles at both QTL (i.e., D2D2 at D2Mit9 and B6B6 atD2Mit17, n = 6 mice) showed higher handling-induced convulsion scores during withdrawal than recombinant F2 mice that possess protective alleles at both QTL (i.e., B6B6 atD2Mit9 and D2D2 at D2Mit17,n = 3 mice). Inset, Genotype atD2Mit9 and D2Mit17 has a significant influence on alcohol withdrawal severity calculated as area under the curve (*p < 0.05, two-tailedt test).
Suggestive linkage on chromosome 2 near D2Mit9 suggestsGad1 as a plausible candidate gene involved in alcohol withdrawal (Fig. 4). Gad1 encodes the 67 kDa isoform of GAD. GAD is rate-limiting in the synthesis of GABA, the major inhibitory neurotransmitter in the mammalian CNS, and a critical determinant of neural excitability. For supporting evidence for differences in GAD enzyme activity between the B6 and D2 progenitor inbred strains, we examined total brain GAD activity in these strains. Figure6 shows that B6 mouse brain has higher GAD activity as compared with the D2 strain (p < 0.05).
Fig. 6.
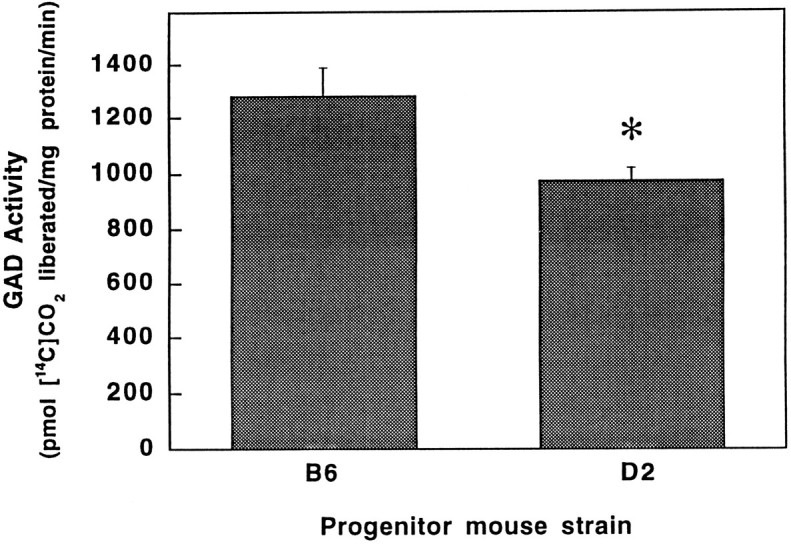
The C57BL/6J strain shows greater GAD activity as compared with DBA/2J mice. Total brain GAD activity was 31% higher in mice from the B6 progenitor strain as compared with the D2 strain (1282 ± 107 and 976 ± 48 pmol [14C]CO2/mg protein per minute were generated, respectively) (*p < 0.05, two-tailedt test). Data represent the mean ± SEM for five independent experiments performed in triplicate.
DISCUSSION
Genetic factors mediate some of the variability between individuals in susceptibility to physiological dependence on alcohol. We analyzed acute alcohol withdrawal severity using the BXD RI strains to detect putative genetic loci contributing to physiological dependence. A genome-wide screen comparing the pattern of strain means for withdrawal severity with polymorphic genetic markers detected seven putative QTL. In the next phase of our analysis, we tested each of these putative loci using a B6D2 F2 intercross and selectively bred HAW and LAW lines. These analyses indicate that QTL detected on chromosomes 1, 4, and 11 demonstrate significant linkage and represent true associations with alcohol withdrawal liability. Two loci, mapped to the proximal and distal regions of chromosome 2, show suggestive linkage and are thought to represent true associations. It is likely that the opposing effects of D2 alleles for these two suggestive QTL reduced the power of our analysis to detect either chromosome 2 locus with sufficient confidence. Analyses using other genetic models, however, also support localization of a QTL to chromosome 2. For example, allelic frequencies at D2Mit9 differ between Withdrawal Seizure Prone (WSP) and Resistant (WSR) mice (two-tailed χ2 = 37.4, p < 0.001), which have been selectively bred from a heterogeneous stock (derived from 8 inbred strains) for severe or mild withdrawal after chronic alcohol inhalation (Crabbe et al., 1985). During the 47 generations since these lines were established, many crossovers have occurred. Consequently, the strong association between withdrawal severity and D2Mit9 allelic frequency would be maintained only if D2Mit9 and a gene affecting withdrawal are closely linked (Darvasi and Soller, 1995). Provisional loci detected on chromosomes 6 and 8 were not associated with withdrawal liability in the F2 and HAW/LAW experiments and were disconfirmed. Consistent with our results, computer simulations suggest that approximately one-half of the loci detected using the BXD RI strains (using p < 0.01) represent true associations (Belknap et al., 1996).
Our results demonstrate the utility of QTL mapping as a powerful hypothesis-generating approach to identify chromosomal regions that influence complex traits such as physiological dependence and thereby aid in the identification of specific candidate genes for study in humans. Behavioral studies have suggested that symptoms of alcohol withdrawal may result from a decrease in GABA-mediated neurotransmission, but the specific genes responsible for adaptive changes in the synaptic actions of GABA are not known. Reduced plasma GABA levels have been reported in chronic alcoholics (Coffman and Petty, 1985), suggesting that decreased GABA synthesis could contribute to physiological dependence. Our data detect a QTL in the proximal region of chromosome 2 and suggest the influence of a candidate gene(s) responsible for GABA synthesis. This QTL maps near Gad1, which encodes the 67 kDa isoform, GAD67. Gad2, a distinct gene encoding GAD65 also maps to the proximal region of chromosome 2. Our data also show that the progenitor D2 and B6 strains differ in brain GAD activity, suggesting that differences in alcohol withdrawal severity among mice derived from these strains could be associated with differences in GAD enzyme activity and/or gene expression. This QTL is also closely linked to a cluster of evolutionarily related genes encoding distinct isoforms of the α-subunit of brain sodium channels (Scn1a, Scn2a, Scn3a) and a glial-specific sodium channel (Scn7a) (Fig. 4). Ethanol has been shown to reduce electrically stimulated uptake into intracellular spaces and neurotoxin-stimulated sodium uptake into rat brain synaptosomes (Hunt, 1985). Interestingly, markers in this region have also been identified by QTL analysis using BXD RI mice tested for withdrawal from nitrous oxide, a gaseous anesthetic (Belknap et al., 1993); alcohol drinking; and withdrawal after chronic inhalation of ethanol vapor (Crabbe et al., 1983). These results suggest that a gene in this region of chromosome 2 might influence withdrawal from a variety of CNS depressants including gaseous anesthetics, pentobarbital, and alcohol.
Our data also detect and verify a QTL on chromosome 11 in proximity to genes encoding the α1, α6, and γ2 subunits of GABAA receptors (Silver et al., 1996) (Fig. 4). The β2 subunit gene may also map to this cluster, because the human homolog, GABRB2, has been mapped to the corresponding gene cluster on human chromosome 5q34–q35 (Russek and Farb, 1994). Previous studies using selectively bred mice suggest that differences in alcohol withdrawal between WSP and WSR lines of mice may be correlated with differences in Gabra1or Gabra6 mRNA content after chronic alcohol treatment (Buck et al., 1991a). Some of the functional properties of GABAAreceptors in these lines are also differentially affected by alcohol treatment (i.e., alcohol-induced sensitization to benzodiazepine receptor inverse agonists), providing additional evidence that differences in GABAA receptor expression or function may contribute to genetic variation in withdrawal (Buck et al., 1991b). In general, these studies suggest that genetic differences in alcohol withdrawal severity may be associated with allelic differences in GABAA receptor genes affecting receptor function and/or expression. The predicted amino acid sequences for the α1and β2 subunits are identical between B6 and D2 mice (Wang et al., 1992; Kamatchi et al., 1995), but allelic differences in α6 or γ2 protein sequences could influence susceptibility to physiological dependence. Alternatively, allelic differences in the genomic regulatory sequences for any of these candidate genes could contribute to differences in GABAA receptor gene expression. In humans, postmortem study of alcoholics has shown an increased number of brain GABA receptors (Tran et al., 1981).
QTL analyses using the BXD RI strains indicate that markers associated with alcohol withdrawal on chromosomes 1 and 11 are also strongly associated with pentobarbital withdrawal liability (K.J. Buck, J.K. Belknap, J.C. Crabbe, unpublished observations). Because alcohol and pentobarbital have extensive pharmacological similarities and are believed to mediate many of their effects through modulation of GABAA receptors, these results may suggest the influence of common genes on chromosomes 1 and 11 that contribute to increased risk for both alcohol and pentobarbital withdrawal. These data further implicate GABAA receptor genes as candidate genes for the QTL on chromosome 11 and suggest that the QTL identified on chromosome 1 may indirectly influence GABAA receptor expression or function, because no GABAA receptor genes have been mapped to chromosome 1. Alternatively, other plausible candidate genes for this QTL include Atp1a2 and Atp1b1 (which encode Na+/K+ ATPase α2 and β1 subunits) and Atp2b4 (encoding a Ca2+ transporting ATPase) (Fig. 4). It might be expected that QTL analyses of alcohol and pentobarbital withdrawal would detect some of the same genes, because mice selectively bred for differences in alcohol withdrawal also differ in withdrawal from pentobarbital and other central depressants, suggesting the influence of common genes on dependence on multiple drugs (Belknap et al., 1987, 1988, 1989; Crabbe et al., 1991).
Our data also detect a QTL involved in withdrawal on chromosome 4 and a suggestive QTL within the distal region of chromosome 2. Interestingly, these QTL map to regions that overlap with loci identified for other seizure phenotypes, including several mouse models of epilepsy. For example, QTL analysis using BXD RI mice tested for audiogenic seizures identified Asp2 on chromosome 4 (Neumann and Collins, 1991), and analysis using a D2xEl intercross detected El2 on chromosome 2, a locus involved in the seizure phenotype in the mutant El strain (Rise et al., 1991; Frankel et al., 1995). Reduced risk for withdrawal convulsions could be associated with reduced synaptic or extracellular levels of glutamate, because ethanol treatment enhances glutamate uptake in rat brain (Foley and Rhoads, 1992).Eaat2 encodes an excitatory amino acid transporter that plays a critical role in glutamate uptake in the brain and represents a plausible candidate for the withdrawal QTL localized to the distal region of chromosome 2.
In summary, the phenotype examined in the present studies represents an animal model of acute physiological dependence liability, rather than a model of alcohol dependence. Our results demonstrate that two-step QTL mapping is sufficiently sensitive to detect the influence of some of the individual genes affecting risk for physiological dependence on alcohol, a complex disorder that shows polygenic inheritance and the substantial influence of environmental factors. Because extensive homology exists between human and mouse chromosomes (Copeland et al., 1993), our results suggest that genetic variation for markers within human chromosomes 1q21–q32, 2q24–q37/11p13, 9p21–p23/1p32–p22.1 and 5q32–q35 (syntenic with QTL identified on mouse chromosomes 1, 2, 4, and 11) may be associated with withdrawal liability and that these chromosomal regions may contain some of the genes related to physiological dependence in humans. Other QTL, not detected in BXD mice, may also contribute to genetic variation in withdrawal between B6 and D2 mice and in humans. A major benefit of the two-step approach for detecting QTL is the development of a cumulative data base in the BXD RI strains, which has allowed us to detect QTL that may have a pleiotropic influence on withdrawal from alcohol, pentobarbital, nitrous oxide, and other central depressants with abuse liability.
Footnotes
This research was supported by United States Public Health Service Grants P50AA10760, RO1AA06243, RO1DA05228, a grant from the Alcoholic Beverage Medical Research Foundation, and two grants from the Department of Veterans Affairs. We thank Dr. Steve Mitchell for his help with the QTL analysis; Drs. Adron Harris, Christopher Cunningham, and Michael Forte for their suggestions on the preparation of this manuscript; and Drs. Rosemary Elliot and Kenneth Manly for the MAP MANAGER data base. We also thank Tamara Lischka, Denis Glenn, Diana Wu, Jasper Long, Janet Dorow, Laurie O’Toole, and Emmett Young for technical assistance.
Correspondence should be addressed to Dr. Kari Johnson Buck, Research Service, 151W, Department of Veterans Affairs Medical Center, 3710 SW US Veterans Hospital Road, Portland, OR 97201.
REFERENCES
- 1.Albers RW, Brady RO. The distribution of glutamic decarboxylase in the nervous system of the monkey. J Biol Chem. 1959;234:926–928. [PubMed] [Google Scholar]
- 2.Aston CE, Hill SY. Segregation analysis of alcoholism in families ascertained through a pair of male alcoholics. Am J Human Genet. 1990;46:879–887. [PMC free article] [PubMed] [Google Scholar]
- 3.Belknap JK, Laursen SE, Crabbe JC. Ethanol and nitrous oxide produce withdrawal-induced convulsions by similar mechanisms in mice. Life Sciences. 1987;41:2033–2040. doi: 10.1016/0024-3205(87)90477-2. [DOI] [PubMed] [Google Scholar]
- 4.Belknap JK, Danielson PW, Lame M, Crabbe JC. Ethanol and barbiturate withdrawal convulsions are extensively codetermined in mice. Alcohol. 1988;5:167–171. doi: 10.1016/0741-8329(88)90015-8. [DOI] [PubMed] [Google Scholar]
- 5.Belknap JK, Crabbe JC, Laursen SE. Ethanol and diazepam withdrawal convulsions are extensively codetermined in WSP and WSR mice. Life Sciences. 1989;44:2075–2080. doi: 10.1016/0024-3205(89)90354-8. [DOI] [PubMed] [Google Scholar]
- 6.Belknap JK, Metten P, Helms ML, O’Toole LA, Angeli-Gade S, Crabbe JC, Phillips TJ. Quantitative trait loci (QTL) applications to substances of abuse: physical dependence studies with nitrous oxide and ethanol in BXD mice. Behav Genet. 1993;23:213–222. doi: 10.1007/BF01067426. [DOI] [PubMed] [Google Scholar]
- 7.Belknap JK, Mitchell SR, O’Toole LA, Helms ML, Crabbe JC. Type I and type II error rates for quantitative trait loci (QTL) mapping studies using recombinant inbred mouse strains. Behav Genet. 1996;26:149–160. doi: 10.1007/BF02359892. [DOI] [PubMed] [Google Scholar]
- 8.Belknap JK, Richards SP, O’Toole LA, Helms ML, Phillips TJ (1997) Short-term selective breeding as a tool for QTL mapping: ethanol preference drinking in mice. Behav Genet, in press. [DOI] [PubMed]
- 9.Bohman M. Some genetic aspects of alcoholism and criminality: a population of adoptees. Arch Gen Psychiat. 1978;35:269–276. doi: 10.1001/archpsyc.1978.01770270019001. [DOI] [PubMed] [Google Scholar]
- 10.Buck KJ, Hahner L, Sikela J, Harris RA. Chronic alcohol treatment alters brain levels of γ-aminobutyric acidA receptor subunit mRNAs: relationship to genetic differences in ethanol withdrawal seizure severity. J Neurochem. 1991a;57:1452–1455. doi: 10.1111/j.1471-4159.1991.tb08313.x. [DOI] [PubMed] [Google Scholar]
- 11.Buck KJ, McQuilken SJ, Harris RA. Modulation of γ-aminobutyric acidA receptor-operated chloride channels by benzodiazepine inverse agonists is related to genetic differences in ethanol withdrawal seizure severity. J Neurochem. 1991b;57:2100–2105. doi: 10.1111/j.1471-4159.1991.tb06428.x. [DOI] [PubMed] [Google Scholar]
- 12.Cadoret RJ, Cain CA, Grove WM. Development of alcoholism in adoptees raised apart from alcoholic biologic relatives. Arch Gen Psychiat. 1980;37:561–563. doi: 10.1001/archpsyc.1980.01780180075008. [DOI] [PubMed] [Google Scholar]
- 13.Coffman JA, Petty F. Plasma GABA levels in chronic alcoholics. Am J Psychiat. 1985;142:1204–1205. doi: 10.1176/ajp.142.10.1204. [DOI] [PubMed] [Google Scholar]
- 14.Copeland NG, Jenkins NA, Gilbert DJ, Eppig JT, Maltais LJ, Miller JC, Dietrich WF, Weaver A, Lincoln SE, Steen RG, Stein LD, Nadeau JH, Lander ES. A genetic linkage map of the mouse: current applications and future prospects. Science. 1993;255:57–66. doi: 10.1126/science.8211130. [DOI] [PubMed] [Google Scholar]
- 15.Crabbe JC, Kosobud A, Young ER, Janowsky JS. Polygenic and single-gene determination of response to ethanol in BXD/Ty recombinant inbred mouse strains. Neurobehav Toxicol Terat. 1983;5:181–187. [PubMed] [Google Scholar]
- 16.Crabbe JC, Kosobud A, Young ER, Tam BR, McSwigan JD. Bidirectional selection for susceptibility to ethanol withdrawal seizures in Mus musculus. Behav Genet. 1985;15:521–536. doi: 10.1007/BF01065448. [DOI] [PubMed] [Google Scholar]
- 17.Crabbe JC, Merrill CD, Belknap JK. Acute dependence on depressant drugs is determined by common genes in mice. J Pharmacol Exp Ther. 1991;257:663–667. [PubMed] [Google Scholar]
- 18.Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics. 1995;141:1199–1207. doi: 10.1093/genetics/141.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deaizpurua HJ, Harrison LC, Cram DS. An ELISA for antibodies to recombinant glutamic acid decarboxylase in IDDM. Diabetes. 1992;41:1182–1187. doi: 10.2337/diab.41.9.1182. [DOI] [PubMed] [Google Scholar]
- 20.Dietrich W, Katz H, Lincoln SE, Shin HS, Friedman J, Dracopoli NC, Lander ES. A genetic map of the mouse suitable for typing intraspecific crosses. Genet. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DSM-IV. Diagnostic and statistical manual of mental disorders, 4th Ed (First MB, ed), pp 175–205. American Psychiatric Association; Washington, DC: 1994. [Google Scholar]
- 22.Falconer DS. Introduction to quantitative genetics. Longman; New York: 1989. [Google Scholar]
- 23.Fisher RA. Statistical methods for research workers, 13th Ed, pp 99–101. Hafner; New York: 1958. [Google Scholar]
- 24.Foley TD, Rhoads DE. Effects of ethanol on Na+-dependent amino acid uptake: dependence on rat age and Na+,K+-ATPase activity. Brain Res. 1992;593:39–44. doi: 10.1016/0006-8993(92)91260-l. [DOI] [PubMed] [Google Scholar]
- 25.Frankel WN, Johnson EW, Lutz CM. Congenic strains reveal effects of the epilepsy quantitative trait locus, El2, separate from other El loci. Mamm Genome. 1995;6:839–843. doi: 10.1007/BF00292432. [DOI] [PubMed] [Google Scholar]
- 26.Friedman HJ (1980) Assessment of physical dependence on and withdrawal from ethanol in animals. In: Alcohol tolerance and dependence (Rigter H, Crabbe JC, eds), pp 93–121. North-Holland, Amsterdam: Elsevier.
- 27.Garrett KM, Haque D, Berry D, Niekrasz I, Gan J, Rotter A, Seale TW (1997) The GABAA receptor α6subunit gene (GABRA6) is tightly linked to the α1-γ2 subunit cluster on mouse chromosome 11. Mol Brain Res, in press. [DOI] [PubMed]
- 28.Gilligan SB, Reich T, Cloninger CR. Etiologic heterogeneity in alcoholism. Genet Epidem. 1987;4:395–414. doi: 10.1002/gepi.1370040602. [DOI] [PubMed] [Google Scholar]
- 29.Goldman DM. Alcoholism: Genetic transmission and genetic loci, In: Recent developments in alcoholism (Galanter M, ed), 11:231–248. Plenum; New York: 1993. [PubMed] [Google Scholar]
- 30.Goldstein DB, Pal N. Alcohol dependence produced in mice by inhalation of ethanol: grading the withdrawal reaction. Science. 1971;172:288–290. doi: 10.1126/science.172.3980.288. [DOI] [PubMed] [Google Scholar]
- 31.Goodwin DW, Schulsinger F, Moller N, Hermansen L, Winokur G, Guze SB. Drinking problems in adopted and nonadopted sons of alcoholics. Arch Gen Psychiat. 1974;31:164–169. doi: 10.1001/archpsyc.1974.01760140022003. [DOI] [PubMed] [Google Scholar]
- 32.Hunt WA. Alcohol and biological membranes. Guilford; New York: 1985. [Google Scholar]
- 33.Jacob HJ, Lindpaintner K, Lincoln SE, Kusumi K, Bunker RK, Mao YP, Ganten D, Dzau VJ, Lander ES. Genetic mapping of a gene causing hypertension in the stroke-prone spontaneously hypertensive rat. Cell. 1991;67:213–224. doi: 10.1016/0092-8674(91)90584-l. [DOI] [PubMed] [Google Scholar]
- 34.Kamatchi GL, Kofuji P, Wang JB, Fernando JC, Liu Z, Mathura JR, Burt DR. GABAA receptor β1, β2, and β3 subunits: comparisons in DBA/2J and C57BL/6J mice. Biochim Biophys Acta. 1995;1261:134–142. doi: 10.1016/0167-4781(95)00009-6. [DOI] [PubMed] [Google Scholar]
- 35.Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lander ES, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nature Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 37.Lebowitz RJ, Solle M, Beckmann JS. Trait-based analyses for the detection of linkage between marker loci and quantitative trait loci in crosses between inbred lines. Theor Appl Genet. 1987;73:556–562. doi: 10.1007/BF00289194. [DOI] [PubMed] [Google Scholar]
- 38.Lincoln S, Daly M, Lander ES (1992) Mapping genes controlling quantitative traits with MAPMAKER/QTL 1.1. (Technical report) Cambridge, MA: Whitehead Institute.
- 39.McCaul ME, Turkkan JS, Svikis DS, Bigelow GE (1991) Alcohol and secobarbital effects as a function of familial alcoholism: extended intoxication and increased withdrawal effects. Alcoholism: Clin Exp Res 15:94–101. [DOI] [PubMed]
- 40.McQuarrie DG, Fingl E. Effects of single doses and chronic administration of ethanol on experimental seizures in mice. J Pharmacol Exp Ther. 1958;124:264–271. [PubMed] [Google Scholar]
- 41.Metten P, Crabbe JC. Dependence and withdrawal, In: Pharmacological effects of ethanol on the nervous system (Deitrich RA, Erwin VG, eds), pp 269–290. CRC; Boca Raton, FL: 1996. [Google Scholar]
- 42.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucl Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neumann PE, Collins RL. Genetic dissection of susceptibility to audiogenic seizures in inbred mice. Proc Natl Acad Sci USA. 1991;88:5408–5412. doi: 10.1073/pnas.88.12.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newlin DB, Pretorius MB (1990) Sons of alcoholics report greater hangover symptoms than sons of nonalcoholics: a pilot study. Alcoholism: Clin Exp Res 14:713–716. [DOI] [PubMed]
- 45.Rise ML, Frankel WN, Coffin JM, Seyfried TN. Genes for epilepsy mapped in the mouse. Science. 1991;253:669–673. doi: 10.1126/science.1871601. [DOI] [PubMed] [Google Scholar]
- 46.Russek SJ, Farb DH. Mapping of the beta 2 subunit gene (GABRB2) to microdissected chromosome 5q34–q35 defines a gene cluster for the most abundant GABAA receptor isoform. Genomics. 1994;23:528–533. doi: 10.1006/geno.1994.1539. [DOI] [PubMed] [Google Scholar]
- 47.Schuckit MA, Goodwin DA, Winokur GA. A study of alcoholism in half siblings. Amer J Psychiat. 1972;128:1132–1136. doi: 10.1176/ajp.128.9.1132. [DOI] [PubMed] [Google Scholar]
- 48.Schuckit MA. A clinical model of genetic influences in alcohol dependence. J Stud Alcohol. 1994;55:5–17. doi: 10.15288/jsa.1994.55.5. [DOI] [PubMed] [Google Scholar]
- 49.Serikawa T, Kuramoto T, Hilbert P, Mori M, Yamada J, Dubay CJ, Lindpainter K, Ganten D, Guenet JL, Lathrop GM, Beckmann JS. Rat gene mapping using PCR-analyzed microsatellites. Genet. 1992;131:701–721. doi: 10.1093/genetics/131.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silver J. Confidence limits for estimates of gene linkage based on analysis of recombinant inbred strains. J Hered. 1985;76:436–444. doi: 10.1093/oxfordjournals.jhered.a110140. [DOI] [PubMed] [Google Scholar]
- 51.Silver LM, Nadeau JH, Goodfellow PN. Encyclopedia of the Mouse Genome. Mammal Genome. 1996;6:S1–S352. [Google Scholar]
- 52.Silvers WK. The coat colors of mice. Springer; New York: 1979. [Google Scholar]
- 53.Simpson SP. Detection of linkage between quantitative trait loci and restriction fragment length polymorphisms using inbred lines. Theoret Appl Genet. 1989;77:815–819. doi: 10.1007/BF00268332. [DOI] [PubMed] [Google Scholar]
- 54.Sokal RR, Rohlf FJ. Biometry. Freeman; San Francisco: 1981. [Google Scholar]
- 55.Tanksley SD. Mapping polygenes. Ann Rev Genet. 1993;27:205–233. doi: 10.1146/annurev.ge.27.120193.001225. [DOI] [PubMed] [Google Scholar]
- 56.Todd JA, Aitman TJ, Cornall RJ, Ghosh S, Hall JR, Hearne CM, Knight AM, Love JM, McAleer MA, Prins JB, Rodrigues N, Lathrop M, Pressey A, DeLarato NH, Peterson LB, Wicker LS. Genetic analysis of autoimmune type 1 diabetes mellitus in mice. Nature. 1991;351:542–547. doi: 10.1038/351542a0. [DOI] [PubMed] [Google Scholar]
- 57.Tran VT, Snyder SH, Major FL, Hawley RJ. GABA receptors are increased in the brains of alcoholics. Ann Neurol. 1981;9:289–292. doi: 10.1002/ana.410090312. [DOI] [PubMed] [Google Scholar]
- 58.Visscher PM, Thompson R, Haley CS. Confidence intervals in QTL mapping by bootstrapping. Genet. 1996;143:1013–1020. doi: 10.1093/genetics/143.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang JB, Kofuji P, Fernando JC, Moss SJ, Huganir RL, Burt DR. The α1, α2 and α3 subunits of the GABAA receptors: comparison in seizure-prone and -resistant mice during development. J Mol Neurosci. 1992;3:177–184. [PubMed] [Google Scholar]