Abstract
Glial cell line-derived neurotrophic factor (GDNF), the most potent trophic factor yet described for both dopaminergic neurons of the substantia nigra and spinal motorneurons, has recently been shown to signal through a multireceptor complex composed of a novel glycosylphosphatidylinositol-anchored GDNF receptor-α (GDNFR-α) and the receptor tyrosine kinase product of the c-ret proto-oncogene (RET). Despite its importance, the individual expression patterns and the relationships between domains of expression of the different components of this trophic system are not understood. We show here by in situ hybridization that GDNF mRNA is expressed in the normal adult rat brain in several targets of substantia nigra neurons, including striatum, nucleus accumbens, thalamic nuclei, olfactory tubercle, hippocampus, cerebellum, and cingulate cortex as well as in the internal granular cell layer of the olfactory bulb. Within the basal ganglia we observe a pronounced segregation of regions expressing GDNF from those expressing GDNF receptors, suggesting that within these structures GDNF is functioning in its anticipated role as a target-derived trophic factor. In addition, the expression of GDNF and both GDNF receptors within the cerebellum, hippocampus, and olfactory bulb may indicate a paracrine mode of action. Importantly, we also see expression of RET mRNA in cellular populations within the cerebellum and the glomerular layer of the olfactory bulb, as well as in the subthalamic nucleus, which lack GDNFR-α expression, indicating that RET functions either independently of GDNFR-α or with GDNFR-α presented in trans. Conversely, GDNFR-α is widely expressed in many regions in which RET expression is absent, suggesting that GDNFR-α may associate with additional signaling receptors. Finally, RET and GDNFR-α show distinct patterns of regulated expression in the brain after kainic acid stimulation and in the sciatic nerve after nerve transection. Taken together these findings indicate that GDNF, RET, and GDNFR-α utilize multiple mechanisms to comprise physiologically relevant trophic circuits for different neuronal populations.
Keywords: in situ hybridization, GDNF, ret, GDNFR-α, mRNA regulation, cerebellum, olfactory, kainate-induced activation, sciatic nerve transection, motorneuron regeneration
Although the initial purification and cloning of glial cell line-derived neurotrophic factor (GDNF) generated a great deal of interest centered around its pronounced effects on cultured dopaminergic neurons of the substantia nigra (Lin et al., 1993), subsequent studies have shown that GDNF has similarly potent effects on a wide range of neuronal populations. First characterized were the survival-promoting activities of GDNF on primary cultures of spinal motorneurons (Henderson et al., 1994) and lesioned motorneuronsin vivo (Li et al., 1995; Oppenheim et al., 1995; Yan et al., 1995). Additionally, GDNF was seen to have pronounced effects on cultures of dissociated neurons from various chick peripheral ganglia—sympathetic, sensory, and enteroceptive (Bujbello et al., 1995; Ebendal et al., 1995; Trupp et al., 1995). Further in vitro studies identified Purkinje cells as a population that responds to GDNF in primary cultures (Mount et al., 1995), and severalin vivo studies have indicated that noradrenergic neurons of the locus coeruleus (Arenas et al., 1995) and cholinergic neurons of the basal forebrain (Williams et al., 1996), as well as thalamic and hippocampal neurons (Martin et al., 1995), all benefit from treatment with GDNF prior or subsequent to experimental lesioning. While GDNF has widespread effects in the adult CNS, its critical role as a morphogen in the developing excretory and enteric nervous systems is highlighted by the phenotype of GDNF null-mutated mice, which die shortly after birth (Moore et al., 1996; Pichel et al., 1996; Sanchez et al., 1996).
The diverse actions of GDNF may be better understood in light of the finding that GDNF signaling is mediated by the receptor tyrosine kinase encoded by the c-ret proto-oncogene (RET) (Durbec et al., 1996; Trupp et al., 1996). Indeed, although very little is known about the expression of RET mRNA in the adult mammalian nervous system, several of the populations responsive to GDNF express RET. These include spinal motorneurons (Pachnis et al., 1993; Tsuzuki et al., 1995), dopaminergic neurons of the adult substantia nigra (Trupp et al., 1996), and various subpopulations of peripheral ganglia—sympathetic, nodose, enteric, and some sensory (Pachnis et al., 1993; Tsuzuki et al., 1995). Strikingly, the RET protein is known to be expressed in the ureteric buds of the metanephric kidney, where it is absolutely required for ureteric branching induced by GDNF from the adjacent mesoderm (Schuchardt et al., 1994; Durbec et al., 1996).
Although it is clear that reciprocal RET/GDNF interactions are necessary for normal development, a newly cloned glycosylphosphatidylinositol (GPI)-anchored GDNF binding protein, GDNF receptor-α (GDNFR-α), is believed to enhance the affinity of GDNF for its signaling receptor (Jing et al., 1996; Treanor et al., 1996). These studies also revealed that GDNFR-α can assist in GDNF binding to RET even when administered in a soluble form. Preliminary studies indicate some overlap between GDNFRα and RET expression patterns such that both transcripts appear in embryonic rat motor neurons, ureteric buds of developing nephrons, and adult substantia nigra neurons (Treanor et al., 1996).
We have sought to further characterize possible GDNF/RET/GDNFR-α interactions in vivo by analyzing expression of these mRNA species using in situ hybridization and RNase protection assays. We first show that some of these interactions may be physiologically relevant in the normal adult rat brain by describing constitutive expression of GDNF in several targets of substantia nigra neurons. We also describe RET mRNA expression patterns that suggest novel neuronal populations that may respond to GDNF. Finally, we see distinct patterns of regulation for the GDNF receptors after experimental manipulations indicating novels modes of action for GDNF that may be amenable to therapeutic manipulations.
MATERIALS AND METHODS
In situ hybridization. Fourteen micrometer sections of spinal cord or brain were thawed onto 3-aminopropyl ethoxysilane-coated slides for hybridization with radiolabeled probes as follows. After fixation in 4% paraformaldehyde for 15 min, slides were rinsed once in PBS and twice in distilled water. Tissue was deproteinated in 0.2 m HCl for 10 min, acetylated with 0.25% acetic anhydride in 0.1 m ethanolamine for 20 min, and dehydrated with increasing concentrations of ethanol. Slides were incubated 16 hr in a humidified chamber at 58° with 8 × 105 cpm of probe in 300 μl of hybridization cocktail (50% formamide, 20 mm Tris-HCl, pH 7.6, 1 mmEDTA, pH 8.0, 0.3 m NaCl, 0.1 m dithiothreitol, 0.5 mg/ml yeast tRNA, 0.1 mg/ml polyA RNA, 1× Denhardt’s solution, and 10% dextran sulfate). Slides were first washed at room temperature in Formamide/SSC (1:1) followed by 30 min at 65° in 1× SSC. Single-stranded RNA was digested by RNase treatment (10 mg/ml) for 30 min at 37° in 0.5m NaCl, 20 mm Tris-HCl, pH 7.5, 2 mm EDTA. Tissue was washed twice with 1× SSC in 65° for 30 min before it was dehydrated in ethanol and air-dried. Slides were either exposed to β-max x-ray film (Amersham, Buckinghamshire, UK) for 10–20 d or dipped in NTB-2 photoemulsion diluted 1:1 in water (Eastman Kodak, Rochester, NY) exposed at 4° for 3–5 weeks, developed with D19 (Eastman Kodak), fixed with Al-4 (Agfa Gevaert, Kista, Sweden), and counterstained with cresyl violet.
RNA isolation, cRNA probes, and RNase protection assays.Total RNA was isolated from frozen samples homogenized in guanidine isothiocyanate and β-mercaptoethanol by the addition of 1:10 vol of 2m sodium acetate followed by phenol/chloroform extraction and ethanol precipitation. cRNA probes, generated from linearized plasmids using T3 or T7 RNA polymerase (Promega Biotec, Madison, WI) and 35S-αUTP or 32P-αCTP (Amersham), respectively, were used for in situ hybridization as above, and RNase protection assays (RPA) were performed according to manufacturer’s instructions (Ambion, Austin, TX). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) riboprobe for RPA has been described previously (Trupp et al., 1995). GDNF cRNA probes were transcribed from a new fragment (nucleotides 279–700 of the originally published rat GDNF cDNA) subcloned for use as an in situhybridization probe containing 422 nucleotides homologous to both published GDNF transcripts. The rat RET riboprobe has been described previously (Trupp et al., 1996). GDNFR-α probes of 250 (RPA) and 400 nucleotides (in situ hybridization) were generated by RT-PCR using primers designed from the previously described cDNA sequence (Treanor et al., 1996). Control sense probes for in situhybridization were generated from all plasmids using the reciprocal RNA polymerase.
Kainic acid stimulation. Adult male Wistar rats (150–170 gm) received bilateral intracerebroventricular injections of kainic acid (0.35 μg/0.5 μl; Sigma, St. Louis, MO), because in our experience this offers the advantage of a standardized latency of initiation and intensity of seizure activity with abrupt termination of status epilepticus, which contributes notably to a reduction of interanimal variability. Animals were mounted in a stereotactic frame and injected using the following stereotaxic coordinates: anteroposterior 0.2, lateral 1.5, ventral 4.2 from the bregma. At the indicated time points animals were decapitated after ether anesthesia, and the brains were removed, frozen in −40° isopentane, stored at −70°, and cryosectioned for in situhybridization.
Sciatic nerve transection. Adult male Sprague Dawley rats (200–250 gm) were anesthetized with sodium pentobarbital (50 mg/kg, i.p.) before transection of the right sciatic nerve, distal to the obturator tendon. To minimize religation of nerve segments, both the proximal and distal ends of the transected nerve were reflected. At the indicated times after axotomy, the animals were sacrificed, and the gastrocnemius muscle, lumbosacral portion of the spinal cord, and four segments (distal/distal; distal/proximal; proximal/distal; proximal/proximal) of the sciatic nerve were dissected out. All tissues were frozen immediately on dry ice and stored at −70° before preparation of RNA.
Sciatic nerve cultures. Sciatic nerves dissected out from decapitated early postnatal rat pups (P1–5) were incubated in PBS containing 0.2% collagenase II (Sigma) for 15 min at 37°, followed by an additional 15 min incubation after the addition of trypsin to 0.25% final concentration. Nerves were washed, triturated, and plated in DMEM supplemented with 10% fetal calf serum (Life Technologies, Gaithersburg, MD), glutamine, penicillin, and streptomycin. Cultures were washed with the above media twice for 2 hr before affinity-labeling with iodinated recombinant GDNF, as described previously (Trupp et al., 1996).
RESULTS
Physiological trophic circuits in the basal ganglia
Despite the several critical brain regions that respond to GDNF treatment in the adult rodent brain, endogenous GDNF expression has been verified only at very low levels using a highly sensitive RNase protection assay (Arenas et al., 1995), and one in situhybridization study reported the absence of GDNF mRNA expression in adult striatum (Strömberg et al., 1993), raising the possibility that the rescue of lesioned adult neurons by exogenous GDNF may not represent the endogenous activities of this factor. We therefore have sought to resolve more clearly the expression of GDNF by performingin situ hybridization using radiolabeled cRNA probes. Several regions of the adult brain are labeled with GDNF cRNA probes, with the general pattern being moderate labeling of only scattered cells. Most striking is the number of regions that are innervated by the various projections of substantia nigra neurons, which have been shown to express both GDNF receptors. Importantly, we see strong labeling over scattered, medium-sized neurons throughout the caudate nucleus, putamen, and internal and external segments of the globus pallidus (Fig.1A,B), whereas neither GDNFR-α nor RET mRNA were detected anywhere in the striatum (Fig. 1, C and D, respectively). As with all regions studied, no labeling was detected with radiolabeled sense probes complementary to those used to assay mRNA expression (not shown). In addition, slightly stronger labeling for GDNF is seen in several thalamic nuclei—ventromedial, anteroventral, and ventropostero-lateral and -medial nuclei (Fig. 1E; and data not shown), none of which show expression of either receptor (Fig. 1G,H; and data not shown). Conversely, the only RET labeling of thalamic nuclei appears at high levels in the reticular thalamic nucleus and zona incerta (Fig. 1H) and the lateral habenula (Fig. 6), all of which express GDNFR-α (Fig.1G). With the noted exception of GDNF-expressing populations, GDNFR-α mRNA is seen at moderate to high levels throughout the thalamus, with the most pronounced labeling appearing in cells of the zona incerta and reticular thalamic nucleus. Generally, within the basal ganglia there appears a clear-cut separation of populations that express GDNF from those expressing mRNA encoding the GDNF receptors (Fig. 1). These observations suggest that GDNF is functioning as a target-derived factor to maintain neuronal circuits within the basal ganglia. We observe GDNF expression in several targets of efferent projections of substantia nigra, namely olfactory tubercle, nucleus accumbens, and the second layer of the cingulate cortex (Fig.1A), indicating that external sources of GDNF may also have effects on neurons in the basal ganglia.
Fig. 1.

Segregated patterns of expression within the basal ganglia. In situ hybridization of 14 μm coronal sections through striatal (A–D) and thalamic (E–H) levels of the basal ganglia demonstrates a clear separation of domains of expression of mRNA encoding GDNF and its receptors. A, Autoradiogram indicating GDNF expression in scattered cells throughout the striatum (str), nucleus accumbens (na), and cingulate cortex (cc), and stronger labeling throughout the olfactory tubercle (ot). B, Bright-field photomicrograph showing that medium-sized neurons throughout the striatum express low levels of GDNF, whereas some scattered neurons express considerably higher levels. C, Although no GDNFR-α is expressed in the striatum, very high levels are seen throughout the lateral septum (ls) and in some peripheral regions of the medial septum. Strong labeling is also seen in the claustrum (cl), in the dorsal endopiriform nucleus (ep), and in scattered cells in all layers of the cortex. D, Again, although no RET is expressed in the striatum, low levels are seen throughout the medial septum (ms). E, GDNF mRNA is detected at high levels in ventromedial and ventrolateral thalamic nuclei (th) and at low levels throughout the hippocampus.F, Bright-field photomicrograph of the ventrolateral thalamic nucleus indicates that a large proportion of medium-sized neurons express GDNF mRNA. G, Although thalamic nuclei that produce GDNF express neither of its receptors, high levels of GDNFR-α mRNA are seen in the reticular thalamic nucleus (rt), zona incerta (zi), and medial habenula (mh). In addition, GDNFR-α labeling can be seen throughout the amygdala (am) and hypothalamus (ht). H, Similarly, RET mRNA is detected in the zona incerta and reticular thalamic nucleus. Scale bar (shown inF): B, F, 93 μm.
Fig. 6.

Segregated domains of expression within the olfactory bulb. A, An autoradiogram of a sagittal section through the olfactory bulb indicates that RET mRNA is expressed at high levels throughout the glomerular layer (gl) and at lower levels in some regions of the internal granular layer (igl). B, C, Conversely, GDNF mRNA is detected in the internal granular cell layer, whereas GDNFR-α is most prominent within the external plexiform layer (epl). D, Dark-field photomicrograph showing detail of red boxed region in A indicates that RET mRNA is expressed in cells surrounding glomeruli. E, Detail ofgreen boxed region in C indicates that GDNFR-α is expressed at high levels in cells throughout the external plexiform layer and at low levels surrounding some glomeruli on the periphery of the external plexiform layer. F, G, Bright-field photomicrograph of one glomerulus (F) and scattered cells in the external plexiform layer (G). H, Superimposition of colorized autoradiograms A–C. RET labeling was coloredred, GDNF blue, and GDNFR-αgreen. Scale bar (shown in G): D, E, 150 μm; F, G, 30 μm.
RET mRNA expression indicates novel GDNF-responsive populations
In addition to the various neuronal populations of the basal ganglia, some of which have been shown to respond to GDNF in vivo, we see mRNA encoding receptors for GDNF in several other brain regions. We have shown previously that lesioned noradrenergic neurons of the locus coeruleus can be rescued by GDNF (Arenas et al., 1995) and show here that both RET and GDNFR-α mRNAs are expressed by these neurons (Fig. 2A,B,D). We also detected both RET and GDNFR-α mRNA in sensory neurons of the trigeminal mesencephalic nucleus (Fig. 2A,C). Also as expected, brain stem motor nuclei, including facial (Fig.3A,B), hypoglossal (Fig. 3C,D), trigeminal (Fig. 3E,F), and oculomotor (Fig.4A–D), are seen to express high levels of both RET and GDNFR-α. Within the motor nucleus of the trigeminal, both large and small neurons were labeled with RET and GDNFR-α (Fig. 3F; data not shown). Cholinergic neurons of the basal forebrain have also been described as responsive to the survival-promoting effects of GDNF (Williams et al., 1996); however, although GDNFR-α is seen to be expressed at high levels in neurons of the lateral septum (Figs. 1C, 2E), labeling was observed only on the periphery of the medial septum (Fig.1C). Conversely, RET mRNA was detected in scattered neurons in the medial septum (Figs. 1D, 2F) and not at all in the lateral septum (Fig. 1D). Thus, there appears to be some segregation of RET and GDNFR-α expression within the basal forebrain.
Fig. 2.

RET and GDNFR-α coexpression correlates within vivo responsiveness to GDNF. Previous studies of GDNFin vivo activities have identified noradrenergic neurons of the locus coeruleus and cholinergic neurons of the basal forebrain as responsive to GDNF (see introductory remarks). A, Dark-field photomicrograph showing intense labeling of sensory neurons of the trigeminal mesencephalic (mt) nucleus and lower, diffuse labeling of the locus coeruleus (lc), which is shown encircled in dashed white lines. B, Bright-field photomicrograph of the medial-dorsal edge of the locus coeruleus showing diffuse, specific labeling over the densely packed neurons of the lc. C, Intense labeling for RET mRNA is observed over large proprioceptive neurons of the trigeminal mesencephalic nucleus. D, GDNFR-α mRNA is also observed throughout the locus coeruleus. E, Although intense labeling for GDNFR-α is observed over large neurons of the lateral septum, less strong labeling is also seen over smaller neurons. F, In the medial septum, RET mRNA is detected in scattered neurons. Scale bar (shown in F):A, 250 μm; B, C, E, F, 21 μm;D, 27 μm.
Fig. 3.

RET and GDNFR-α are highly expressed in motor nuclei of cranial nerves. Motor neurons of the rodent brainstem, including facial (VII) and hypoglossal (XII), have been shown previously to respond to GDNF survival-promoting effects.A, Dark-field photomicrograph depicting high levels of RET mRNA in the facial motor nucleus. B, Bright-field image showing that the RET labeling is over the large motorneurons.C, D, In addition, motor neurons of the hypoglossal nucleus express mRNA encoding GDNFR-α. E, Dark-field image of the motor nucleus of the trigeminal (V) indicates that RET is expressed widely and at varying levels within the nucleus.F, Higher magnification bright-field image of this section reveals that RET is expressed in both small and large neurons within this nucleus. GDNFR-α mRNA is also detected in both small and large neurons within the motor nucleus of the trigeminal (not shown). Scale bar (shown in F): A, C, E, 230 μm; B, D, 20 μm; F, 25 μm.
Fig. 4.

Coexpression of GDNFR-α and RET suggests novel GDNF-responsive populations. A, Dark-field photomicrograph showing high GDNFR-α mRNA levels in caudal linear (CLi) and dorsal Raphe (DR) nuclei and lower levels in motor nuclei of the oculomotor nerve (III).B, Conversely, hybridization of an adjacent section with an RET cRNA probe indicates that RET is more highly expressed in oculomotor (OM) neurons and at lower levels in both caudal linear and dorsal Raphe. C, D, Higher-magnification bright-field images of the dorsal Raphe indicate that expression of GDNFR-α mRNA is more widespread than that of RET mRNA. Lower levels of transcripts encoding both receptors were also detected in the medial Raphe nucleus (not shown). E, F, Dark-field photomicrographs depicting GDNFR-α mRNA expression in the dorsal cochlear nucleus (E) and the lateral vestibular nucleus (F). G, H, Bright-field micrographs of adjacent sections hybridized with RET antisense cRNA probes indicate that RET is expressed in neurons of both the dorsal cochlear (G) and lateral vestibular (H) nuclei. Although expression of both GDNFR-α and RET mRNA is confined to the dorsal region of the cochlear nucleus, both transcripts were detected in medial and superior vestibular nuclei in addition to the lateral region (not shown). Scale bar (shown inH): A, B, 107 μm; C, D, 21 μm; E, F, 160 μm; G, 11 μm; H, 16 μm.
In addition to these previously characterized GDNF-responsive populations, we find novel neuronal populations, which, based on the expression of both GDNF receptor components, are likely to benefit from the neurotrophic activities of GDNF. Of considerable interest is the finding that serotonergic neurons of the dorsal Raphe nucleus express both GDNF receptors (Fig. 4A–D). As this population has been shown to degenerate in some cases of Parkinson’s disease, this suggests that GDNF may be an even more efficient therapeutic agent for this disorder. In support of this notion, preliminary results indicate that GDNF can promote the survival of a portion of lesioned serotonergic neurons of the dorsal Raphe (P. Åkerud and E. Arenas, personal communication). Additional populations that express both RET and GDNFR-α mRNAs include dorsal cochlear (Fig.4E,G), lateral vestibular (4F,H), medial and lateral habenular (see Fig.6J,L), and several hypothalamic nuclei, including arcuate and dorsomedial compacta (see Fig.6H,M). We also see many populations of neurons that express GDNFR-α in the absence of RET, including all cortical layers, lateral geniculate nucleus, and the superior colliculus (Table1; and data not shown).
Table 1.
Summary of expression of GDNF, GDNFR-α, and RET mRNA
| Brain structure | GDNF | GDNFR-α | RET |
|---|---|---|---|
| Olfactory system | |||
| Olfactory tubercule | ++ | + | − |
| Internal granular layer of olfactory bulb | + | + | + |
| External plexiform layer of olfactory bulb | − | + | − |
| Glomerular layer | − | ± | ++ |
| Olfactory nerve layer | − | + | + |
| Cingulate cortex | + | + | − |
| Neocortex | ± | ++ | − |
| Hippocampus | + | ++ | (+) |
| Amygdala | − | ++ | ± |
| Caudate-putamen | + | − | − |
| Accumbens | + | − | − |
| Lateral septal nucleus | − | +++ | − |
| Septum medialis | − | ± | ++ |
| Hypothalamic nuclei | − | ++ | + |
| Thalamus | |||
| Reticular thalamic nucleus | − | +++ | ++ |
| Zona incerta | − | +++ | + |
| Subthalamic nucleus | − | − | ++ |
| Ventroposterolateral/medial nucleus | + | − | − |
| Anteromedial and dorsal thal nucl | + | + | − |
| Anteroventral thalamic nucleus | ++ | − | − |
| Ventromedial thalamic nucleus | + | − | − |
| Lateral habenular nucleus | − | +++ | ++ |
| Medial habenular nucleus | − | +++ | ++ |
| Mesencephalon | |||
| Substantia nigra compacta and scattered cells of SN reticulata | − | +++ | +++ |
| Ventral tegmental area | − | +++ | +++ |
| Interpeduncular nucleus | − | +++ | +++ |
| Supramammilary nucleus | − | ++ | +++ |
| Red nucleus | − | ++ | +++ |
| Dorsal raphe nucleus | − | ++ | ++ |
| Cerebellum | |||
| Granular layer (not granule cells) | + | − | + |
| Purkinje layer (not Purkinje cells) | (+) | ++ | ++ |
| Molecular layer | − | ++ | ++ |
| Deep cerebellar nuclei | (+) | + | ++ |
| Pons | |||
| Pontine reticular nucleus | − | ++ | + |
| Pontine nucleus | − | ± | ++ |
| Motor trigeminal nucleus | − | +++ | +++ |
| Inferior olive nuclei | − | + | ++ |
| Locus coeruleus | − | + | + |
| Dorsal cochlear nucleus | − | +++ | + |
| Facial nucleus | − | +++ | +++ |
| Vestibular nucleus | − | + | + |
| Hypoglossal nucleus | − | +++ | + |
Semiquantitative analysis depicted as high levels, +++; moderate, ++; low, +; marginally detectable, (+); scattered populations at generally very low levels, ±.
Complete trophic circuits within the cerebellum
In correlation with the recent finding that endogenous RET protein in the cerebellum is constitutively phosphorylated on tyrosine residues (Colucci-D’Amato et al., 1996), we see GDNF expression in scattered cells of the granular layer of the cerebellum (Fig.5A,D). In addition, strong labeling for both GDNFR-α and RET mRNAs is detected in a narrow band at the external edge of the granular cell layer (Fig. 5B,C) over cells directly adjacent to Purkinje cells (Fig. 5E,F). The strong labeling for RET in this region dissipates gradually both toward the granular layer and into the molecular layer, creating gradients of RET expression (Fig. 5C). Importantly, this demonstrates a population of cells within the granular layer, which do not appear to be granule cells (Fig. 5G), that express RET but not GDNFR-α. Interestingly, although GDNF has been shown to have effects on Purkinje cells in mixed cultures of embryonic cerebellum (Mount et al., 1995), we do not detect mRNA for either of the previously characterized GDNF receptors in Purkinje cells of adult animals (Fig.5D,F); however, we do observe GDNFR-α and RET mRNAs as well as weak labeling for GDNF mRNA in deep cerebellar nuclei (Fig. 5A–C,H,I). The pattern of RET mRNA expression detected in the cerebellar cortex suggests that this receptor is activated by GDNF produced within the structure. GDNF secreted by granule cells may be acting on afferent mossy fibers as well. It is interesting to note in this context the expression of GDNF receptors in the pontine, red, cochlear, and vestibular nuclei (Table 1), all of which extend projections to the cerebellar granular layer.
Fig. 5.
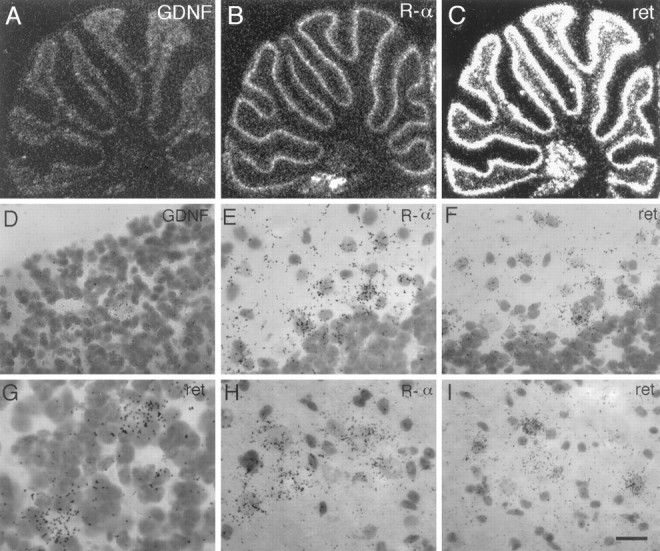
Complete trophic circuits within the cerebellum.A–C, Autoradiograms of hybridized sagittal sections through the cerebellum show that GDNF, RET, and GDNFR-α mRNAs are all expressed in the cerebellar cortex as well as the deep cerebellar nuclei. D, Bright-field photomicrograph of the granular layer reveals that scattered cells within this layer that do not seem to be granule cells express GDNF mRNA. E, GDNFR-α mRNA is seen to be expressed most highly in the Purkinje layer in cells directly adjacent to Purkinje cells (possibly basket cells) and also at lower levels reaching into the molecular layer.F, RET mRNA is also observed in cells similarly placed next to Purkinje cells, as well as within the molecular layer.G, High-magnification bright-field photomicrograph of the granular layer of the cerebellum reveals that RET mRNA is highly expressed in cells that stain very lightly with cresyl violet.H, I, Neurons of deep cerebellar nuclei are seen to express both GDNF receptors. Scale bar (shown inI): D, 29 μm; E, F, H, I, 20 μm; G, 14 μm.
Segregated domains of expression of GDNF and its receptors within the olfactory bulb
Analysis of the expression patterns of the GDNF receptors within the olfactory bulb indicates that RET mRNA is present in cells that express no or very low levels of GDNFRα; labeling for RET mRNA is seen at high levels within the olfactory nerve layer and the glomerular layer of the olfactory bulb (Fig.6A,D,F). This suggests that RET is using another co-receptor or collaborating with GDNFR-α expressed on the surface of an adjacent cell or as a soluble protein. Indeed, either of these latter mechanisms seems possible, because GDNFR-α expression is pronounced in the external plexiform layer of the bulb (Fig. 6C), juxtaposed to the nerve and glomerular layers. In this region, GDNFR-α is detected at high levels in scattered cells (Fig. 6E,G). In addition, we see GDNF mRNA expression in the internal granular layer of the olfactory bulb (Fig.6B), which suggests that GDNF could activate RET expressed within the olfactory bulb but could also act on RET-containing projections from afferent nuclei, such as the dorsal Raphe or locus coeruleus. Superimposition of the expression patterns of the three molecules reveals unexpectedly segregated domains of expression for GDNF and its receptors within the olfactory bulb (Fig.6H).
Kainic acid-induced regulation of GDNF and its receptors in the adult brain
RET-expressing populations within the septum could be physiologically activated by GDNF expressed in the normal adult hippocampus at low levels (Figs. 1E, 7A). We have studied the effects of kainic acid-induced activation on the expression of GDNF and its receptors in adult hippocampus. Similar to previous studies (Humpel et al., 1994), we see an upregulation of GDNF in dentate gyrus after 2 hr that peaks at 4 hr (Fig.7B,C). Twelve hours after kainic acid treatment, increased GDNF mRNA levels are observed in all regions of the hippocampus (Fig. 7D), whereas after 24 hr, labeling remains elevated in only the CA3 and hilar regions (Fig.7E). In view of the present results, the previously described kainate-induced induction of GDNF mRNA in granule cells of the dentate gyrus should now be reconsidered as an upregulation of endogenous GDNF mRNA levels. This increase of ligand is in close correlation with that which we observe for the receptors. As with GDNF, the upregulation of GDNFR-α mRNA in dentate gyrus is pronounced and rapid, already increasing after 2 hr (Fig. 7G), reaching peak levels at 4 hr (Fig. 7H), and returning to basal levels after 48 hr (not shown). Although this time course of upregulation is similar in the CA3 region, GDNFR-α expression is greatly increased in CA1 only after 24 hr (Fig. 7J) and remains high after 48 hr (data not shown). Finally, an increase in CA4 is seen to begin at 12 hr and peak at 24 hr (Fig.7I,J). Although moderate levels of GDNFR-α are seen in all regions of the normal adult hippocampus, RET mRNA is expressed only at very low levels (Fig. 7K). Also, distinct patterns of expression for these receptors are observed after kainic acid treatment: GDNFR-α mRNA is observed in all cells, whereas RET mRNA is seen to be strongly expressed in only a subset of neurons scattered throughout the pyramidal layer (Fig. 7N). In addition, RET mRNA is seen to be upregulated after 12 and 24 hr in scattered extrapyramidal cells (Fig. 7N,O). Furthermore, upregulation of transcripts encoding both RET and GDNFR-α is observed to begin in the piriform cortex after 12 hr (Fig.7I,N), peak at 24 hr (Fig. 7J,O), and subside after 48 hr (not shown).
Fig. 7.
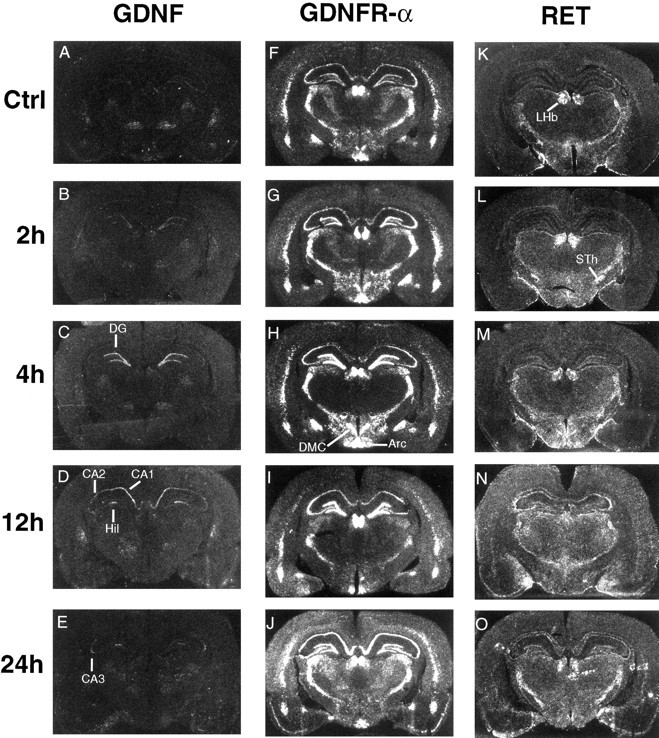
Kainate-induced activation upregulates GDNF, RET, and GDNFR-α mRNAs. GDNF mRNA upregulation is detected in the dentate gyrus (DG), CA1, CA2, CA3, and hilar (Hil) regions of the hippocampus. In addition to an increase in all hippocampal regions, GDNFR-α mRNA levels also increase in the piriform cortex. RET mRNA increases are most pronounced in DG after 4 hr and in CA3 and piriform cortex after 12 hr. Also, extrapyramidal cells of the hippocampus increase RET mRNA expression after 12 and 24 hr of induced activation. In contrast to GDNFR-α and GDNF patterns, RET mRNA appears to be expressed by only a subset of neurons scattered within and outside of the pyramidal layers. Arcuate hypothalamic nucleus (Arc), dorsal-medial hypothalamic nucleus, compacta (DMC), lateral habenular nucleus (LHb), subthalamic nucleus (STh).
A role for endogenous GDNF and GDNFR-α in the repair of lesioned motorneurons
We have shown previously that GDNF mRNA is upregulated dramatically in distal segments of the adult rat sciatic nerve after mechanical transection (Trupp et al., 1995). We have now analyzed the expression of GDNFR-α in similarly treated nerve segments by RNase protection assay. We observe a very dramatic upregulation of GDNFR-α mRNA appearing at 3 d, with a maximum at 7 d (Fig.8A). Importantly, this regulation is most pronounced in the more distal segments of the sciatic nerve (Fig. 8B), resulting in a distally increasing gradient of GDNFR-α mRNA expression. Although no RET mRNA was detected anywhere in the sciatic nerve (data not shown), expression is detected in the sacrolumbar spinal motorneurons that contribute axons to this nerve (Fig. 9A). We observe an upregulation of RET mRNA expression in spinal motorneurons after transection of the sciatic nerve similar to that shown previously in chick spinal motorneurons after axotomy (Nakamura et al., 1996). Likewise, levels of GDNFR-α mRNA were seen to increase in primary motorneurons at the L5 level after axotomy (Fig. 9A). In addition, we observe an increase of GDNF mRNA expression in axotomized muscle 3 d after nerve transection (Fig. 9B), as well as a distally increasing gradient of GDNF mRNA expression in cells of the lesioned sciatic nerve (data not shown) similar to that seen for GDNFR-α mRNA. Taken together, these expression patterns suggest a novel mechanism of trophic support for degenerating motorneurons.
Fig. 8.
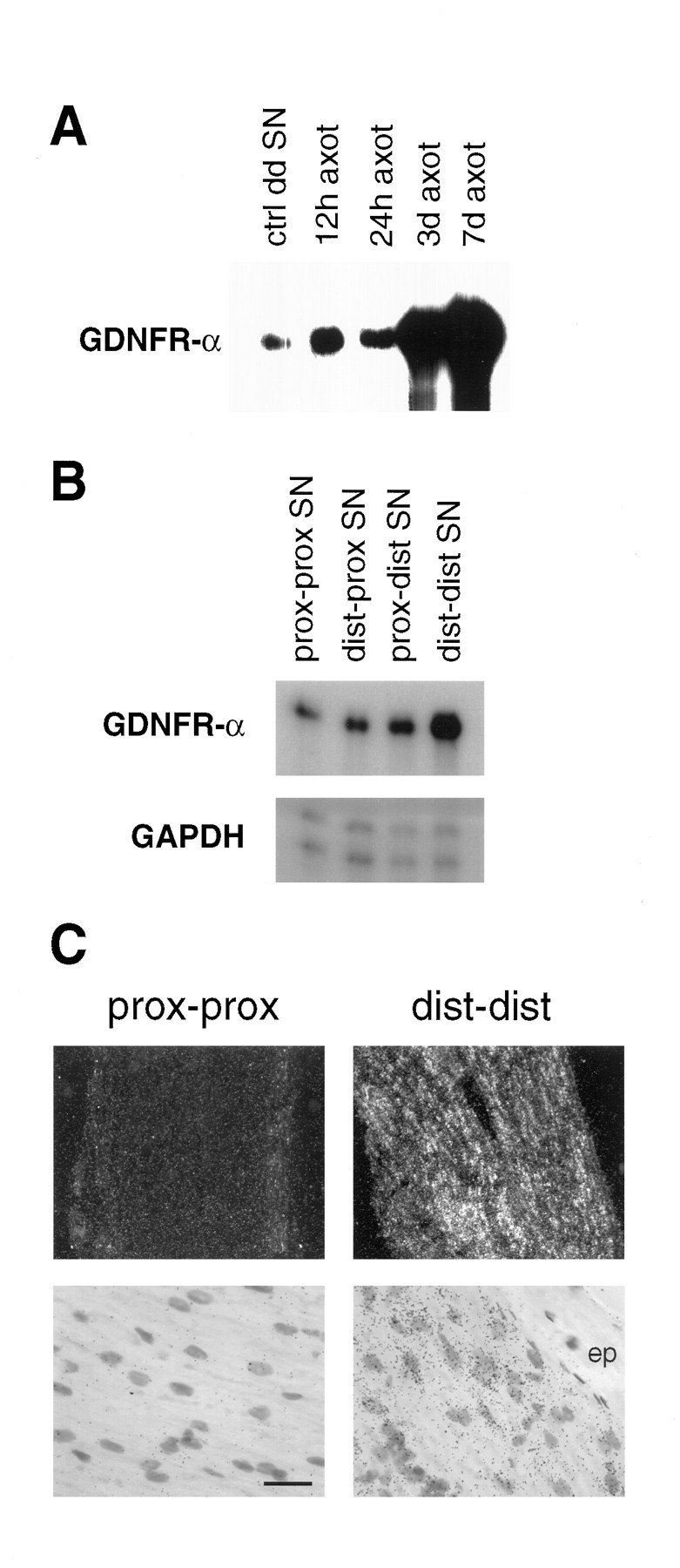
Upregulation of GDNFR-α mRNA after sciatic nerve transection. A, Autoradiogram of an RPA revealing a pronounced increase in GDNFR-α mRNA in the distal segment of transected sciatic nerve 3 and 7 d after surgery.B, A distally increasing gradient of GDNFR-α mRNA is demonstrated in an autoradiogram of an RPA of sciatic nerve samples collected 3 d after nerve transection. C, Photomicrographs of GDNFR-α in situ hybridization of nerve segments from animals sacrificed 3 d after nerve transection. The GDNFR-α mRNA-expressing cells are clearly present in the Schwann cell layer, whereas the epineurium (ep) is devoid of labeling. For analysis, the transected nerve was dissected into four parts: moving laterally from the cord, the proximal segment of the proximal stump (prox-prox), distal segment of the proximal stump (dist-prox), proximal segment of the distal stump (prox-dist), and distal segment of the distal stump (dist-dist). Scale bar (shown inC): dark field, 134 μm; bright field, 18 μm.
Fig. 9.

Regulation of GDNF mRNA in muscle and its receptors in spinal motorneurons. A, Dark-field micrographs of transverse sections through L5 spinal motorneurons reveal that GDNFR-α mRNA (top panels) and RET mRNA (bottom panels) are upregulated in spinal motorneurons 3 d after nerve transection. B, Autoradiogram of an RPA showing that sciatic nerve transection induces GDNF mRNA expression in gastrocnemius muscle. Note, this cRNA probe protects two mRNA fragments of slightly different lengths that are present in both previously characterized GDNF transcripts. Scale bar: A, 134 μm in top panels and 107 μm in bottom panels.
GDNFR-α produced by spinal motorneurons is likely to function as a co-receptor with RET in a GDNF binding complex; however, the GDNFR-α expressed in the sciatic nerve in the absence of RET expression must be either acting as a co-receptor for another signaling receptor distinct from RET or alternatively used in trans to cooperate with RET expressed by motorneurons. To address this question, we have investigated GDNF receptor complexes in primary cultures of dissociated sciatic nerve cells by affinity-labeling followed by chemical cross-linking. Although we see pronounced binding of GDNF to a protein of the molecular weight of GDNFR-α, we do not find affinity-labeling of any other proteins (Fig. 10), suggesting that GDNFR-α may be acting in these cells to present GDNF to injured motor neurons.
Fig. 10.
Primary cultures of sciatic nerve affinity-labeled with iodinated GDNF in the absence of any known signaling receptors. Cross-linking of iodinated GDNF to GDNFR-α is demonstrated by affinity-labeling of NIH3T3 fibroblasts and fibroblasts stably transfected with GDNFR-α cDNA (3T3Rα) in a pcDNA3 plasmid (Invitrogen, San Diego, CA), left lanes. Autoradiograms of SDS-PAGE run under reducing conditions showing iodinated GDNF affinity-labeling ofMN1 cells (a motorneuron cell line expressing RET mRNA, which we have previously characterized as responsive to GDNF,center lane) and primary cultures of P5 rat sciatic nerve (1°SN, SN+cold, right lanes) clearly indicate that although GDNF is not binding to RET (155 kDa) in these cells, it does affinity-label a protein of approximately the molecular weight of GDNFR-α (75 kDa, including ligand). Arrowheads indicate bands corresponding to affinity-labeled GDNFR-α or proteins of similar molecular weight.
DISCUSSION
We have sought to answer here the question of whether the activities described previously for exogenous GDNF on various populations of lesioned neurons reflect the presence of endogenous trophic circuits of this factor within the normal adult CNS. The studies reported here indicate that GDNF is expressed in the proper locations to function in the maintenance of various neuronal circuits in the normal adult rat brain. Additionally, the regulation of GDNF and its receptors suggest that these trophic interactions may comprise physiological responses to neurological traumas.
Endogenous target-derived GDNF trophic circuits
The initial characterization of the dopaminotrophic activities of GDNF have generated considerable interest and effort, resulting in the subsequent elucidation of many additional neurotrophic activities for this factor; however, characterization of GDNF expression in the normal adult brain has not been determined satisfactorily to date. We have shown here that GDNF mRNA is expressed in several targets of ventral mesencephalic dopaminergic neurons both within and outside the basal ganglia. Within the basal ganglia, there is a striking segregation of the pattern of expression of GDNF from that of its receptors, which suggests that GDNF is acting as a target-derived trophic factor in the maintenance of several neuronal circuits. As some of these neuronal populations have been shown to respond to GDNF after mechanical or pharmacological lesion, these trophic activities of GDNF may also be used during neuronal degeneration.
Analysis of mRNA expression patterns for the GDNF receptors described here and elsewhere indicates that all neuronal populations shown previously to be responsive to GDNF in vivo express both RET and GDNFR-α. These include lesioned dopaminergic neurons of the substantia nigra and ventral tegmental area (Beck et al., 1995; Tomac et al., 1995), axotomized spinal and facial motorneurons, ischemic thalamic and hippocampal neurons, 6-hydroxydopamine-treated noradrenergic neurons of the locus coeruleus, and transected cholinergic neurons of the basal forebrain. In addition to the several targets of ventral mesencephalic neurons that we have shown to express GDNF mRNA, we also demonstrate the presence of this mRNA in targets of neuronal projections of thalamic nuclei, locus coeruleus, basal forebrain, and hippocampus as well as spinal motor neurons. Together, these data indicate that GDNF may be acting on many responsive populations in a target-derived fashion.
Novel GDNF-responsive populations
The strong correlation between GDNF-responsiveness and GDNF receptor expression, combined with the biochemical studies demonstrating GDNF activation of the RET kinase activity, suggests that novel neuronal populations found to express GDNF receptors are also likely to benefit from the neurotrophic activities of this factor. The Raphe nucleus, the major serotonergic nucleus in the brain, projects to both ventral mesencephalic and striatal targets and as such is anticipated to be the locus responsible for the increase in serotonin levels recorded in striatum and substantia nigra after local administration of GDNF (Beck et al., 1996). Because of the presence of nigral afferent projections in the Raphe, these authors speculated that GDNF was influencing serotonergic neurons indirectly via direct actions on the substantia nigra. Our data suggest that increases of serotonin levels in these paradigms could be attributable to effects of GDNF on serotonergic neurons themselves. This has implications for the possible therapeutic benefits of GDNF administration, because degeneration of Raphe nucleus neurons has been documented in some cases of Parkinson’s disease (Halliday et al., 1990).
RET and GDNFR-α mRNA expression in the pontine and the red nuclei suggests that populations of neurons that send projections to the cerebellar granular layer are likely to respond to GDNF in vivo. In line with the diverse roles of GDNF in establishing and maintaining sensorimotor trophic circuits, we demonstrate that neurons of the dorsal cochlear and lateral vestibular nuclei express mRNA encoding both GDNF receptors. In addition, the mesencephalic nucleus of the trigeminal is seen to express mRNA for both GDNF receptors. The widespread expression of GDNF receptors in the various hypothalamic nuclei could be taken as an indication of a neuroendocrine function for GDNF. Finally, perhaps the most intense labeling for GDNFR-α mRNA in the adult brain is seen in the medial habenular nucleus, where RET mRNA is also detected. The significance of such high levels of GDNFR-α mRNA expression is unclear, considering the high affinity of GDNFR-α for its known ligand.
Neuronal populations expressing RET but not GDNFR-α mRNA
Some recent reports propose the absolute requirement of GDNFR-α for ligand binding to RET (Jing et al., 1996; Treanor et al., 1996). It is therefore of interest to note the several populations of neurons that we find to express RET but not GDNFR-α mRNA. Cells within the granular layer of the cerebellum that do not appear to be granule cells express RET but no GDNFR-α mRNA. Cells that are morphologically very similar are also seen to express GDNF mRNA in the same region. Similarly, within the olfactory bulb, strong labeling of the glomerular layer as well as the olfactory nerve layer is observed for RET but not GDNFR-α. Furthermore, the subthalamic nucleus is also seen to contain neurons that are positive for RET mRNA but negative for GDNFR-α mRNA. Thus, in these cases RET may be using GDNFR-α delivered intrans by other cells, or it may be associating with another molecule related to GDNFR-α for GDNF binding.
GDNF receptors exhibit distinct regulation of expression patterns
It should be noted that although several GDNF-responsive populations have been shown to express both previously characterized receptors, some nuclei exhibit only very low levels of receptor expression. It has been noted in previous studies that GDNF has more pronounced biological effects on lesioned neurons than on intact neurons. Indeed, it was shown that although normal basal forebrain cholinergic neurons do not alter choline acetyltransferase (ChAT) mRNA levels after stimulation with GDNF, cells that had been transected mechanically did upregulate ChAT mRNA in response to GDNF (Williams et al., 1996). Furthermore, we have shown previously that lesioned noradrenergic neurons of the locus coeruleus increase tyrosine hydroxylase mRNA levels and exhibit more pronounced sprouting after administration of GDNF than unlesioned neurons do (Arenas et al., 1995). It has been shown recently in the chicken that RET mRNA expression is upregulated in both facial and spinal motorneurons after nerve transection (Colucci-D’Amato et al., 1996; Nakamura et al., 1996). We have shown here that GDNFR-α mRNA is also upregulated in spinal motorneurons after sciatic nerve transection and that hippocampal RET mRNA expression increases in response to excitotoxins. Because very limited regulation of GDNF itself has been detected under various stimulations and insults, these results suggest that trophic circuits of GDNF may be regulated dynamically at the level of the receptors.
GDNFR-α mRNA is often expressed in the absence of RET mRNA
The widespread and high levels of GDNFR-α mRNA expression, in particular in areas devoid of RET labeling, are striking. Notable among these populations are the lateral geniculate nucleus, superior colliculus, and extensive regions of the cerebral cortex. GDNFR-α may be associating with additional signaling receptors for GDNF in these cells. Alternatively, GDNFR-α might function in these populations to capture and concentrate diffusible GDNF from the extracellular space to present the factor in a target-derived fashion to afferent GDNF-responsive cells. We have found one possible example of such a mechanism in the lesioned sciatic nerve.
A possible role for GDNF and GDNFR-α in an intrinsic mechanism of motorneuron regeneration
We have shown that GDNFR-α mRNA levels are upregulated in nerve cells in the distal segments of the lesioned sciatic nerve, which expresses no known GDNF signaling receptors. The localization and overall morphology of these cells resemble those of Schwann cells, which have been shown previously to express GDNFR-α mRNA by RT-PCR analysis. We propose that lesion-induced GDNFR-α may be functioning in conjunction with GDNF to activate RET on regenerating motor axons. In this model (Fig. 11), we envision GDNFR-α capturing and presenting soluble GDNF to function as a membrane-anchored trophic signal for regenerating motorneurons. This model is supported further by the observation of distally increasing gradients of both GDNF and GDNFR-α mRNAs in the lesioned sciatic nerve as well as increased levels of GDNF mRNA in skeletal muscle. Thus, regenerating motor axons might grow through tracts of increasing concentration of GDNF associated with GDNFR-α in a preformed complex ready to activate the RET tyrosine kinase. Because GPI-anchored proteins are known to be shed by the cells that produce them, a good amount of the GDNF released by cells in the peripheral nerve may in fact already be in a complex with GDNFR-α as a co-ligand. A similar mechanism could be envisioned in regions of the brain in which GDNFR-α is expressed at high levels.
Fig. 11.
GDNF/RET/GDNFR-α interactions constitute a novel mechanism of trophic support for degenerating motorneurons. Our current data support the well characterized upregulation of RET mRNA in spinal and facial motorneurons after axotomy (see Discussion). We have shown here that GDNFR-α is also upregulated in motorneurons after transection. In addition, we see that GDNF mRNA is upregulated in the denervated muscle as well as the axotomized distal nerve segments. Of particular interest is the finding that GDNFR-α is expressed in a distally increasing gradient within the sciatic nerve, suggesting that this accessory receptor acts to generate an insoluble gradient of GDNF for presentation to regenerating motorneurons.
Conclusions
Our expression studies indicate that GDNF is an endogenous target-derived trophic factor in the adult brain. Importantly, most populations that have been shown to respond to exogenous GDNF are seen to express mRNA for both GDNF receptor components. RET mRNA expression in the absence of GDNFR-α, however, indicates that RET functions alone, uses another co-receptor, or uses GDNFR-α in trans. Indeed, expression patterns of GDNFR-α mRNA indicate that it may also be available in trans or as a target-derived co-ligand in a complex with GDNF. The regulation after sciatic nerve lesion of all the components of this trophic circuit suggests an endogenous mechanism of spinal motorneuron regeneration.
Footnotes
These studies were funded in part by The Swedish Cancer Society and The Swedish Medical Research Council. N.B. was supported by Consiglio Nazionale delle Ricerche. We sincerely thank Carina Raynoschek for generating the GDNFR-α expressing cell line. We gratefully acknowledge the stimulating discussions and critical readings of Ernest Arenas, Michael Fainzilber, and Mikael Rydén.
Correspondence should be addressed to Miles Trupp or Carlos Ibáñez, Division of Molecular Neurobiology, Department of Neuroscience, Karolinska Institute, Doktorsringen 12, 171 77 Stockholm, Sweden. Miles or Carlos@cajal.mbb.ki.se
Dr. Belluardo’s present address: Department of Physiological Sciences, University of Catania, 95125 Catania, Italy.
Dr. Funakoshi’s present address: Division of Biochemistry, Biomedical Research Center, Osaka University, Medical School, Osaka 565, Japan.
REFERENCES
- 1.Arenas E, Trupp M, Åkerud P, Ibáñez CF. GDNF prevents degeneration and promotes the phenotype of brain noradrenergic neurons in vivo. Neuron. 1995;15:1465–1473. doi: 10.1016/0896-6273(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 2.Beck K, Valverde J, Alexi T, Poulsen K, Moffat B, Vandlen R, Rosenthal A, Hefti F. Mesencephalic dopaminergic neurons protected by GDNF from axotomy-induced degeneration in the adult brain. Nature. 1995;373:339–341. doi: 10.1038/373339a0. [DOI] [PubMed] [Google Scholar]
- 3.Beck KD, Irwin I, Valverde J, Brennan TJ, Langston JW, Hefti F. GDNF induces a dystonia-like state in neonatal rats and stimulates dopamine and serotonin synthesis. Neuron. 1996;16:665–673. doi: 10.1016/s0896-6273(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 4.Bujbello A, Buchman VL, Horton A, Rosenthal A, Davies AM. GDNF is an age-specific survival factor for sensory and autonomic neurons. Neuron. 1995;15:821–828. doi: 10.1016/0896-6273(95)90173-6. [DOI] [PubMed] [Google Scholar]
- 5.Colucci-D’Amato GL, Dalessio A, Filliatreau G, Florio T, Digiamberardino L, Chiappetta G, Vecchio G, Fusco A, Santoro M, Defranciscis V. Presence of physiologically stimulated ret in adult rat brain: induction of ret expression during nerve regeneration. Cell Growth & Differ. 1996;7:1081–1086. [PubMed] [Google Scholar]
- 6.Durbec P, Marcos-Gutierrez CV, Kilkenny C, Grigoriou M, Suvanto P, Wartiovaara K, Smith D, Ponder B, Costantini F, Saarma M, Sariola H, Pachnis V. Glial cell line-derived neurotrophic factor signalling through the Ret receptor tyrosine kinase. Nature. 1996;381:789–792. doi: 10.1038/381789a0. [DOI] [PubMed] [Google Scholar]
- 7.Ebendal T, Tomac A, Hoffer BJ, Olson L. Glial cell line-derived neurotrophic factor stimulates fiber formation and survival in cultured neurons from peripheral autonomic ganglia. J Neurosci Res. 1995;40:276–284. doi: 10.1002/jnr.490400217. [DOI] [PubMed] [Google Scholar]
- 8.Halliday G, Li Y, Blumbergs P, Joh T, Cotton R, Howe P, Blessing W, Geffen L. Neuropathology of immunohistochemically identified brainstem neurons in Parkinson’s disease. Ann Neurol. 1990;27:373–385. doi: 10.1002/ana.410270405. [DOI] [PubMed] [Google Scholar]
- 9.Henderson CE, Phillips HS, Pollock RA, Davies AM, Lemeulle C, Armanini M, Simpson LC, Moffet B, Vandlen RA, Koliatsos VE, Rosenthal A. GDNF: a potent survival factor for motoneurons present in peripheral nerve and muscle. Science. 1994;266:1062–1064. doi: 10.1126/science.7973664. [DOI] [PubMed] [Google Scholar]
- 10.Humpel C, Hoffer B, Stromberg I, Bektesh S, Collins F, Olson L. Neurons of the hippocampal formation express glial cell line-derived neurotrophic factor messenger RNA in response to kainate-induced excitation. Neuroscience. 1994;59:791–795. doi: 10.1016/0306-4522(94)90284-4. [DOI] [PubMed] [Google Scholar]
- 11.Jing SQ, Wen DZ, Yu YB, Holst PL, Luo Y, Fang M, Tamir R, Antonio L, Hu Z, Cupples R, Louis JC, Hu S, Altrock BW, Fox GM. GDNF-induced activation of the ret protein tyrosine kinase is mediated by GDNFR-alpha, a novel receptor for GDNF. Cell. 1996;85:1113–1124. doi: 10.1016/s0092-8674(00)81311-2. [DOI] [PubMed] [Google Scholar]
- 12.Li LX, Wu WT, Lin LFH, Lei M, Oppenheim RW, Houenou LJ. Rescue of adult mouse motoneurons from injury-induced cell death by glial cell line-derived neurotrophic factor. Proc Natl Acad Sci USA. 1995;92:9771–9775. doi: 10.1073/pnas.92.21.9771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin L-FH, Doherty D, Lile J, Bektesh S, Collins F. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science. 1993;260:1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- 14.Martin D, Miller G, Rosendahl M, Russell DA. Potent inhibitory effects of glial derived neurotrophic factor against kainic acid mediated seizures in the rat. Brain Res. 1995;683:172–178. doi: 10.1016/0006-8993(95)00369-2. [DOI] [PubMed] [Google Scholar]
- 15.Moore MW, Klein RD, Farinas I, Sauer H, Armanini M, Phillips H, Reichardt LF, Ryan AM, Carvermoore K, Rosenthal A. Renal and neuronal abnormalities in mice lacking GDNF. Nature. 1996;382:76–79. doi: 10.1038/382076a0. [DOI] [PubMed] [Google Scholar]
- 16.Mount HTJ, Dean DO, Alberch J, Dreyfus CF, Black IB. Glial cell line-derived neurotrophic factor promotes the survival and morphologic differentiation of Purkinje cells. Proc Natl Acad Sci USA. 1995;92:9092–9096. doi: 10.1073/pnas.92.20.9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura M, Ohta K, Hirokawa K, Fukushima M, Uchino M, Ando M, Tanaka H. Developmental and denervation changes in c-ret proto-oncogene expression in chick motoneurons. Mol Brain Res. 1996;39:1–11. doi: 10.1016/0169-328x(95)00347-u. [DOI] [PubMed] [Google Scholar]
- 18.Oppenheim R, Houenou L, Johnson J, Lin L-F, Li L, Lo A, Newsome A, Prevette D, Wang S. Developing motor neurons rescued from programmed cell death by GDNF. Nature. 1995;373:344–346. doi: 10.1038/373344a0. [DOI] [PubMed] [Google Scholar]
- 19.Pachnis V, Mankoo B, Costantini F. Expression of the c-ret proto-oncogene during mouse embryogenesis. Development. 1993;119:1005–1017. doi: 10.1242/dev.119.4.1005. [DOI] [PubMed] [Google Scholar]
- 20.Pichel JG, Shen LY, Sheng HZ, Granholm AC, Drago J, Grinberg A, Lee EJ, Huang SP, Saarma M, Hoffer BJ, Sariola H, Westphal H. Defects in enteric innervation and kidney development in mice lacking gdnf. Nature. 1996;382:73–76. doi: 10.1038/382073a0. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez MP, Silossantiago I, Frisen J, He B, Lira SA, Barbacid M. Renal agenesis and the absence of enteric neurons in mice lacking gdnf. Nature. 1996;382:70–73. doi: 10.1038/382070a0. [DOI] [PubMed] [Google Scholar]
- 22.Schuchardt A, D’Agati V, Larsson BL, Costantini F, Pachnis V. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature. 1994;367:380–383. doi: 10.1038/367380a0. [DOI] [PubMed] [Google Scholar]
- 23.Strömberg I, Björklund L, Johansson M, Tomac A, Collins F, Olson L, Hoffer B, Humpel C. Glial cell line-derived neurotrophic factor is expressed in the developing but not adult striatum and stimulates developing dopamine neurons in vivo. Exp Neurol. 1993;124:401–412. doi: 10.1006/exnr.1993.1214. [DOI] [PubMed] [Google Scholar]
- 24.Tomac A, Lindqvist E, Lin L-F, Ögren S, Young D, Hoffer B, Olson L. Protection and repair of the nigrostriatal dopaminergic system by GDNF in vivo. Nature. 1995;373:335–339. doi: 10.1038/373335a0. [DOI] [PubMed] [Google Scholar]
- 25.Treanor J, Goodman L, Desauvage F, Stone DM, Poulsen KT, Beck CD, Gray C, Armanini MP, Pollock RA, Hefti F, Phillips HS, Goddard A, Moore MW, Bujbello A, Davies AM, Asai N, Takahashi M, Vandlen R, Henderson CE, Rosenthal A. Characterization of a multicomponent receptor for GDNF. Nature. 1996;382:80–83. doi: 10.1038/382080a0. [DOI] [PubMed] [Google Scholar]
- 26.Trupp M, Rydén M, Jörnvall H, Timmusk T, Funakoshi H, Arenas E, Ibáñez CF. Peripheral expression and biological activities of GDNF, a new neurotrophic factor for avian and mammalian peripheral neurons. J Cell Biol. 1995;130:137–148. doi: 10.1083/jcb.130.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trupp M, Arenas E, Fainzilber M, Nilsson A-S, Sieber BA, Grigoriou M, Kilkenny C, Salazar-Grueso E, Pachnis V, Arumäe U, Sariola H, Saarma M, Ibáñez CF. Functional receptor for glial cell line-derived neurotrophic factor encoded by the c-ret proto-oncogene product. Nature. 1996;381:785–789. doi: 10.1038/381785a0. [DOI] [PubMed] [Google Scholar]
- 28.Tsuzuki T, Takahashi M, Asai N, Iwashita T, Matsuyama M, Asai J. Spatial and temporal expression of the ret proto-oncogene product in embryonic, infant and adult rat tissues. Oncogene. 1995;10:191–198. [PubMed] [Google Scholar]
- 29.Williams LR, Inouye G, Cummins V, Pelleymounter MA. Glial cell line-derived neurotrophic factor sustains axotomized basal forebrain cholinergic neurons in vivo: dose-response comparison to nerve growth factor and brain-derived neurotrophic factor. J Pharmacol Exp Ther. 1996;277:1140–1151. [PubMed] [Google Scholar]
- 30.Yan Q, Matheson C, Lopez O. In vivo neurotrophic effects of GDNF on neonatal and adult facial motor neurons. Nature. 1995;373:341–344. doi: 10.1038/373341a0. [DOI] [PubMed] [Google Scholar]




