Abstract
Multiple sites of extraretinal photoreception are present in vertebrates, but the molecular basis of extraretinal phototransduction is poorly understood. This study reports the cloning of the first opsin specifically expressed in the directly photosensitive pineal and parapineal of cold-blooded vertebrates. This opsin, identified in channel catfish and termed parapinopsin, defines a new gene family of vertebrate photopigments and is expressed in a majority of parapinealocytes and a subset of pineal photoreceptor cells. Parapinopsin shows a caudal–rostral gradient of expression within the pineal organ. This study also reports the cloning of partial cDNAs encoding the channel catfish orthologues of rhodopsin and the red cone pigment—the full complement of retinal opsins in the species.In situ hybridization studies using probes derived from these retinal opsins, together with parapinopsin, reveal no expression of retinal opsins in pineal and parapineal organ and no expression of any opsin tested in the “deep brain,” iris, or dermal melanophores. These data imply that phototransduction in these sites of extraretinal photoreception must be mediated by novel opsins.
Keywords: pineal, parapineal, parapinopsin, opsin, deep brain, iris, skin, taste bud, extraretinal, in situhybridization
Molecular aspects of phototransduction in the retina have been characterized exhaustively (cf. Palczewski, 1994; Koutalos and Yau, 1996). Photoreception can take place in vertebrates in extraretinal sites, as has been documented extensively for the parapineal and pineal organs, deep regions in the brain, the iris, and melanophores (Oksche, 1965; Wurtman et al., 1968;Eakin, 1973; Hartwig and Oksche, 1982; Weber, 1983; Barr, 1989). These extraretinal photoreceptors seem to play important roles in circadian rhythmicity, camouflage and color change, detection of ambient light conditions and seasonal changes in photoperiod, and, especially in juveniles, negative phototaxis. Very little is known about the molecular features of extraretinal phototransduction, although on the basis of physiological studies (cf. Dodt and Heerd, 1962; Dodt, 1963;Dodt and Scherer, 1968; Roberts, 1978; Marchiafava and Kusmic, 1993;Solessio and Engbretson, 1993) it is likely that novel opsins are involved. Immunohistochemical studies with antibodies generated to bovine rhodopsin and chicken cone pigments and to selected mammalian phototransduction enzymes have reported immunoreactivity in these regions (Vigh et al., 1980; Vigh-Teichmann et al., 1982, 1983; Korf, et al., 1985; van Veen, 1986; Kuo et al., 1988; Tamotsu et al., 1990,1994; Vigh-Teichmann and Vigh, 1990; Masuda et al., 1994; Grace et al., 1996), although whether these correspond to the retinal antigens themselves is unclear. Pinopsin, a novel opsin cloned from the chicken pineal gland, is sensitive to blue light and occurs in certain other birds (Okano et al., 1994; Max et al., 1995; Kawamura and Yokoyama (1996).
In the present study we describe the identification and molecular cloning of parapinopsin, a novel opsin predominantly localized to the parapineal organ of the channel catfish. We also describe retinal and extraretinal localizations of parapinopsin and the full complement of retinal opsins.
MATERIALS AND METHODS
Cloning of catfish opsins. Catfish parapinopsin and red cone pigment first were identified via degenerate PCR from random primed cDNA obtained from poly(A+) RNA isolated from the barbel (taste organ) of adult channel catfish. The primers ofLibert et al. (1989) were used to identify parapinopsin and were used with identical cycling conditions as described, with the exception that only one round of 35 cycles was performed. The red cone pigment was cloned with the following primers: 5′, AAGAAGCTICGIACICCICTIAA, and 3′, RMAIAMRTADAT- IAYIGGRTTRRA.
These were used with the following cycling protocol: 94°C for 1 min, 55°C for 2 min, and 72°C for 2 min for 35 cycles. PCR products were subcloned into the pCR vector (Invitrogen) and sequenced manually. A partial-length cDNA encoding the catfish rhodopsin gene was obtained from random primed retina cDNA by using the primers and cycling conditions of Fitzgibbon et al. (1995). Full-length parapinopsin cDNA was obtained via a combination of genomic sequencing, 5′ and 3′ rapid amplification of cDNA ends (RACE), and reverse transcription-PCR (RT-PCR; Frohman et al., 1990). A genomic library was constructed from Sau3A partially digested genomic DNA obtained from catfish liver and subcloned into EMBL3 phage, which gave a total of 700,000 pfu on first plating. This was screened with a probe derived from the initial parapinopsin PCR product, resulting in the isolation of four positively hybridizing plaques, at least one of which contained all translated exons of parapinopsin. Double-strand sequencing of genomic DNA yielded the complete sequences of exons 1–3 and introns 1 and 2. The sequence of the 5′ end of the gene was confirmed by 5′ RACE (Frohman et al., 1990), whereas the 3′ end of the gene containing exon 4 was obtained via 3′ RACE, both conducted on random primed cDNA obtained from whole catfish fry (5–10 mm in length), and confirmed via genomic sequencing. A cDNA corresponding to the full-length coding sequence of parapinopsin was obtained by using the following primers designed to the initial and final coding residues in parapinopsin: 5′, CGGAATTCATCCCAATAGACCTCGTCAGTGT, and 3′, CGGGATCCTTATAATGGTGACAC TGAGGTGTTCTT.
PCR was conducted with Pfu polymerase for 35 cycles on cDNA obtained from whole juvenile fish (<1 cm in length), using the following cycling protocol: 94°C for 1 min, 60°C for 1 min, and 72°C for 2 min. The sequence of the product was confirmed by complete double-stranded sequencing of two independent clones.
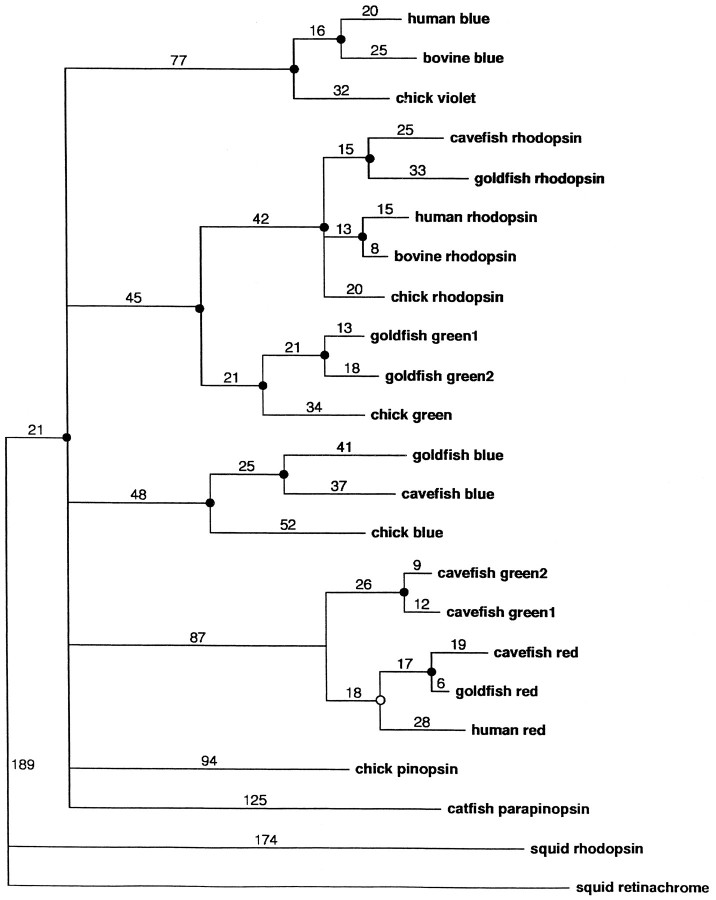
Phylogenetic analysis. Phylogenetic analysis was performed with the Phylogeny Analysis Using Parsimony (PAUP) program (pre-release version 4d51, generously provided by Dr. David Swafford, Smithsonian Institute, Washington, DC). We constructed an inferred phylogenetic tree between parapinopsin and an assortment of other vertebrate opsins, along with squid rhodopsin and squid retinachrome. To construct the tree, we aligned all opsin sequences and eliminated character positions containing any gaps. Parsimony analysis was used to construct a tree that required the minimal number of evolutionary changes to account for the differences among the opsins at each amino acid position. The tree is unrooted, because no ancestral opsin is known to define an outgroup. In an unrooted tree such as this, there is no root node and branch lengths specify relationships among the opsins without defining a primordial evolutionary path. To gain a statistical measure of confidence in the tree, we performed bootstrap analysis. One hundred trees were generated from the initial data set, and the percentage of trees containing a particular clade was measured. (A clade is a group of opsins containing a common ancestor that is not shared by any opsin outside the group.) Bootstrap values >70% were associated with statistical significance at the p < 0.05 level (Hillis and Bull, 1993).
Reverse Transcription-PCR. RT-PCR was performed by using 50 ng of random primed cDNA from each of the designated tissues. The primers used were 5′, GTAGTATGCAGACCATTAGGTGCA, and 3′, TTAT- AATGGTGACACTGAGGTGTTCTT.
Primers were chosen so that the expected PCR product would span introns 2 and 3, thereby eliminating the possibility of genomic DNA contamination. The cycling protocol used was 94°C for 1 min, 60°C for 1 min, and 72°C for 1.5 min for the designated number of cycles.
In situ hybridization. The probes used forin situ hybridization were digoxigenin cRNA probes corresponding to the full-length coding sequence of parapinopsin and the partial-length cDNAs encoding catfish rhodopsin and red cone pigment. Parapinopsin was cloned into pBluescript KS for the purpose, whereas rhodopsin and the red cone pigment were subcloned into the pCR vector. DNA for probe synthesis was prepared by Perfect Prep (5′→3′, Boulder, CO), digested with the appropriate enzymes, checked for completeness of digestion on minigel, and then protease K-digested, phenol/chloroform-extracted four times, chloroform-extracted two times, and collected via ethanol precipitation. Probes were synthesized with T3, T7, or Sp6 polymerase, checked on denaturing gels, and purified via LiCl precipitation. The in situ protocol used was a modification of that of Schaeren-Wiemers and Gerfin-Moser (1993). The following modifications were made: albino channel catfish 70–90 mm in length were decapitated, and the brains, eyes, and portions of the skin were removed. These were post-fixed overnight at 4°C in 4% paraformaldehyde in PBS, cryoprotected overnight at 4°C in 30% sucrose in PBS, and then immersed overnight at 4°C in O.C.T. mounting medium. Then tissue samples were embedded and frozen at −80°C until use. Ten micrometer sections were cut onto Superfrost Plus slides (Fisher Scientific, Pittsburgh, PA). In each case in which pineal and brain were examined, a series of sections was cut from the anterior portion of the nucleus of the facial nerve to the anterior tip of the telencephalon. In cases in which parapinopsin, rhodopsin, and red cone pigment were examined together, adjacent sections were cut onto a series of three slides in the manner 1 = 1a, 2 = 2a, 3 = 3a, 4 = 1b, etc. As sets of three were filled, new sets were started. Sections were allowed to dry 1–3 hr before processing. All sections used through the hybridization step were DEPC-treated and RNase-free. Sections were post-fixed for 5 min in 4% paraformaldehyde/PBS, washed three times for 3 min in PBS, permeabilized for 30 min in 0.1% Triton X-100 in PBS, washed three times for 3 min in PBS, acetylated for 10 min in 0.1 mtriethanolamine, pH 8.0, and 0.25% acetic anhydride, washed three times for 3 min in PBS, and prehybridized for 2–6 hr in hybridization buffer containing 50% formamide, 5× SSC, 5× Denhardt’s solution, 500 μg/ml sonicated, denatured herring sperm DNA, and 250 μg/mlE. coli MRE 600 tRNA. This buffer was removed, and 0.1 ml of fresh buffer containing 30–40 ng of probe was added to the slide, covered with a siliconized coverslip, and hybridized overnight at 72°C. Coverslips were removed by soaking in 5× SSC at 72°C, and slides were washed two times for 1 hr in 0.2× SSC at 72°C. Sections were washed for 5 min in 0.2× SSC at room temperature, for 5 min in TBS, and then blocked for 1 hr at room temperature in 5% heat-inactivated normal goat serum in TBS. Slides were incubated overnight in this solution containing a 1:5000 dilution of sheep anti-digoxigenin Fab fragments conjugated to alkaline phosphatase. On the next day the slides were washed three times for 5 min in TBS and one time for 5 min in alkaline phosphatase buffer containing 0.1m Tris, pH 9.5, 0.1 m NaCl, and 50 mm MgCl2. Slides were placed in light-tight boxes, and 1–2 ml of color development solution, containing 3.375 μg/ml nitroblue tetrazolium, 3.5 μg/ml 5-bromo-4-chloro-3-indoyl-phosphate (BCIP), and 0.24 mg/ml levamisole, was placed on the slide. The color reaction was allowed to run from 20 min (for retina sections) to 1–2 d (for all others) at room temperature. The reaction was stopped in Tris/EDTA buffer and ddH20, and the slides were sealed in Aquapolymount (Polysciences, Warrington, PA).
RESULTS
Molecular cloning of parapinopsin reveals a new gene family
In an effort to identify G-protein-coupled taste receptors, we conducted a PCR analysis of first-strand cDNA, using a variety of primers for G-protein-coupled receptors. Among a large number of candidate molecules we identified a partial cDNA, the sequence of which indicated that it encodes a red cone pigment. We also identified a novel cDNA that displays only 40% amino acid identity to known retinal opsins, thus placing it outside any known family of vertebrate retinal opsins. RNA–slot blot analysis indicates that the red cone pigment expression is localized exclusively to the retina, whereas the novel opsin is not detected readily in any tissue. We obtained a full-length cDNA for the novel clone, subsequently named parapinopsin, by a combination of genomic library screening and 5′ and 3′ RACE (Fig.1). The full-length coding sequence for parapinopsin is 1036 base pairs interrupted by three introns located at positions 342, 676, and 913 bp. Although the localization of the intron–exon junctions within the coating sequence is identical to that of retinal opsins, no intron is present at the end of transmembrane domain 4, differing from the structure of all known vertebrate retinal rod and cone opsins, with the exception of the rhodopsin genes of some teleosts that have no introns (Fitzgibbon et al., 1995). The intron found in the N-terminal sequence of many long-wave opsins is also absent in parapinopsin. Moreover, parapinopsin, like many invertebrate opsins and chick pinopsin, shows a deletion of two amino acids in the second extracellular loop when it is aligned to vertebrate retinal opsins, with the location of this gap being identical to both invertebrate opsins and chick pinopsin. The parapinopsin amino acid sequence is 40–45% identical to chick pinopsin, long-wave retinal opsins, and the recently identified VA opsin (Boni and Foster, 1997), excluding the N- and C-terminal domains, which show little primary sequence homology to any known opsin. The N-terminal domain of parapinopsin contains a consensus N-linked glycosylation site, whereas the C-terminal domain possesses a conserved cystine residue implicated in palmitoylation as well as multiple serine and threonine residues, which represent potential sites of opsin kinase activity.
Fig. 1.
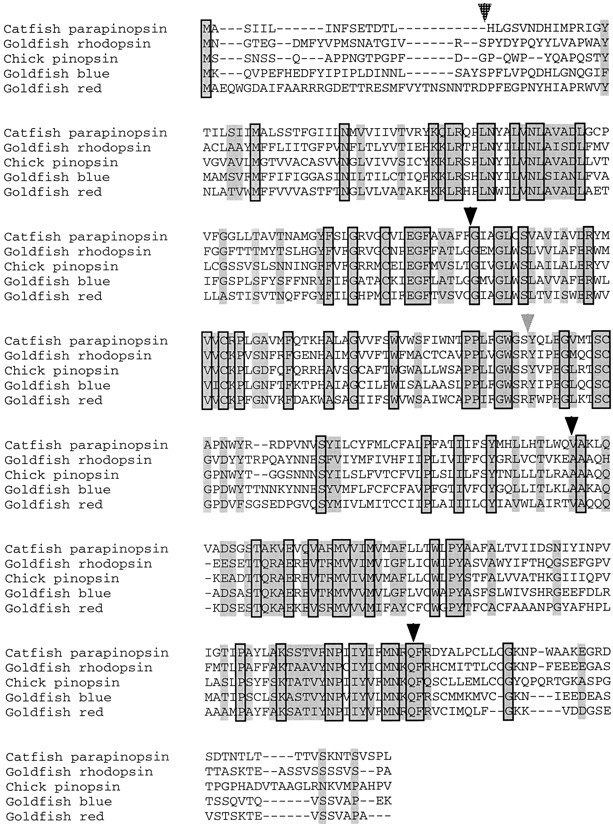
Primary sequence of catfish parapinopsin aligned to chick pinopsin and selected goldfish retinal opsins. Residues conserved in all sequences are boxed. Residues similar in all sequences are shaded. Black arrowheads indicate introns present in all genes. Thehatched arrow indicates the position of the N-terminal intron found in the red/green-sensitive opsins. The gray arrowhead indicates the position of the intron found at the end of transmembrane domain 4, which is absent in parapinopsin but found in all other vertebrate opsins studied to date.
Parapinopsin likely is involved in phototransduction, because it contains all of the residues within the transmembrane domain that are essential for contacting retinal as well as most of those implicated in spectral tuning. Thus, the lysine at residue 296 (numbers as per bovine rhodopsin) is conserved, as is the glutamate counter-ion at residue 113, along with many regions in the transmembrane domains implicated in spectral tuning (Chang et al., 1995) and most of the residues in the cytoplasmic domains that are implicated in activating transducin (Franke et al., 1992; Fahmy and Sakmar, 1993). Attempts to reconstitute parapinopsin biochemically in transfected mammalian cells for spectral analysis and efforts to to demonstrate light-activated inositol 1,4,5-trisphosphate generation in cells cotransfected with parapinopsin and Gα16 (Offermann and Simon, 1995) so far have proven unsuccessful.
Alignment with a wide assortment of vertebrate opsins reveals that parapinopsin defines a new gene family of photopigments (Fig.2). It clearly is not an orthologue of avian pinopsin. Parapinopsin seems to be the most highly divergent of any known vertebrate opsin, with the exception of the Vertebrate Ancient (VA) opsin recently cloned from salmon eyes (Boni and Foster, 1997), and likely diverged from the retinal opsins very early in vertebrate evolution. Degenerate PCR reveals highly conserved parapinopsin orthologues in several other species of bony fish (data not shown).
Fig. 2.
A space-filling dendrogram plot of catfish parapinopsin aligned to other vertebrate and invertebrate opsins. The PAUP sequence alignment program is used. Statistically significant groupings (p < 0.05) are indicated by afilled circle at the node, whereas those that are not statistically significant (p > 0.05) are indicated by an open circle.
Localization of parapinopsin in the parapineal organ and a caudal–rostral gradient in parapinopsin-expressing pineal photoreceptors
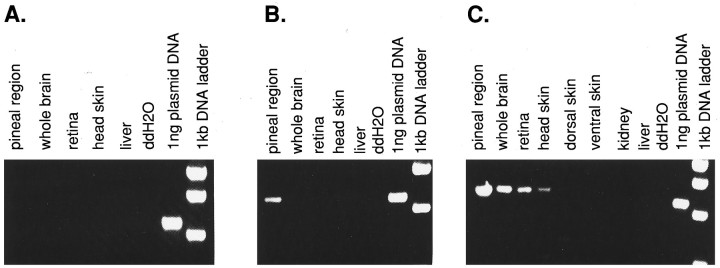
Using Northern blot and RNase protection analysis, we have been unable to detect parapinopsin expression because of the small amount of tissue in the pineal and related tissues. Instead, we used a semiquantitative RT-PCR analysis, which reveals discrete localizations of parapinopsin expression (Fig. 3). At 30 cycles no parapinopsin expression is evident in any tissues that were examined. At 35 cycles we observed parapinopsin expression in the pineal region, which includes the pineal and parapineal organs as well as some associated brain and meningeal tissue. The dimensions of the pineal in the adult fish are ∼40 × 200 μm, whereas those of the parapineal are ∼40 × 40 μm, making clean dissection very difficult, even with the use of a dissecting microscope. At 40 cycles we also observed expression in whole brain tissue, which includes the pineal and parapineal organs that might account for the brain signal. We detected expression in the retina and head skin, but not in ventral and dorsal skin, kidney, or liver. Primers to evolutionarily conserved regions of β actin indicated equal levels of amplification from each tissue tested (data not shown).
Fig. 3.
RT-PCR analysis of parapinopsin expression. Primers and conditions are described in Materials and Methods.A, PCR conducted for 30 cycles. B, PCR conducted for 35 cycles. C, PCR conducted for 40 cycles.
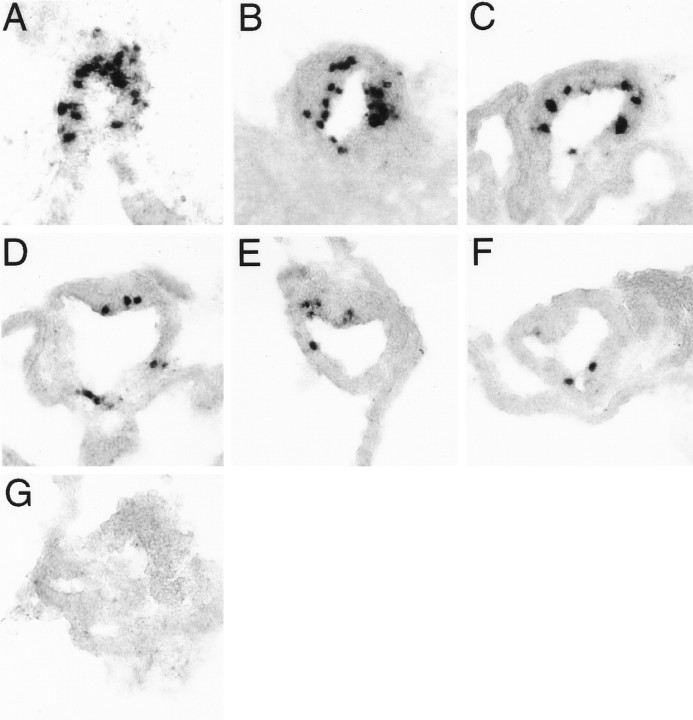
In situ hybridization reveals highly selective localizations of parapinopsin in the pineal complex (Fig.4). A high level of expression of parapinopsin is evident in the majority of cells in the parapineal organ. Nonexpressing cells in the parapineal may represent afferent parapineal neurons, a cell type that is abundant in the parapineal (Ekstrom et al., 1983, 1987). In the pineal organ a smaller subset of cells express parapinopsin. These localizations have suggested the designation parapinopsin (parapineal and pineal opsin). During development the pineal organ begins as a paired structure with one structure moving toward the midline and becoming the pineal organ, whereas the other remains on a single side and is designated the parapineal. In frogs and lizards the structure designated parapineal in fish moves rostrally and dorsally to become the parietal eye. The channel catfish parapineal organ is found in an asymmetric location adjacent to the pineal, much like that of other species of bony fish (Hafeez and Merhige, 1977; Vigh et al., 1980; Ekstrom et al., 1983; Vigh-Teichman et al., 1983; Yanez et al., 1996). This explains the selective localization of parapinopsin in the parapineal organ on only the right side of the coronal section of Figure 4. In nine catfish examined, we have observed the parapineal organ on the right side in eight instances and on the left side in a single instance. Although different teleost species vary in parapineal sideness (cf. Ekstrom et al., 1983; Vigh-Teichmann et al., 1983; Yanez et al., 1996), ours is the first reported example of within-species individual variation.
Fig. 4.
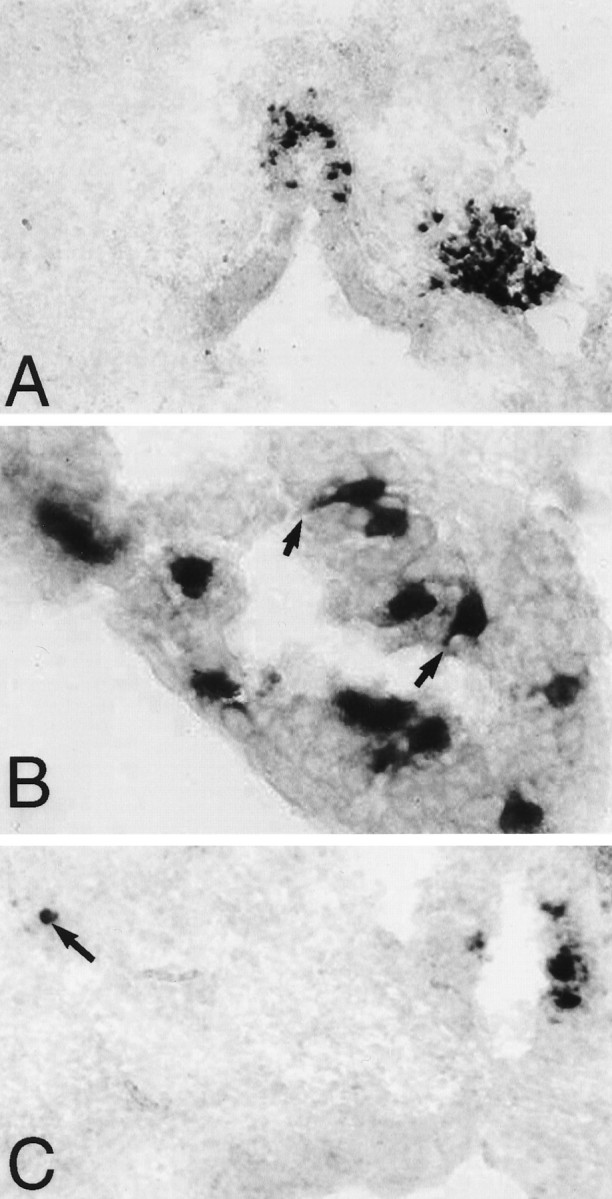
Parapinopsin expression, as determined byin situ hybridization. A, Expression in pineal stalk and right-sided parapineal organ (magnification, 200×).B, Expression in pineal photoreceptor cells.Black arrows indicate selected cells with prominent inner segments that show parapinopsin expression (magnification, 1000×). C, Expression in pineal organ and a displaced cell of the lateral habenula, which is indicated by a black arrow (magnification, 200×). Sections are counterstained with Fast Green.
We wondered whether parapinopsin is selectively localized to photoreceptive cells. Photoreceptive cells in the retina and pineal gland possess distinctive inner and outer segments. Although parapineal cells are thought to be photoreceptive, they typically do not possess readily identifiable inner and outer segments (Ekstrom et al., 1983; Vigh-Teichman et al., 1983), whereas such segments do exist in the pineal organ (Oksche, 1965; Eakin, 1973). At high magnification, parapinopsin in the pineal is clearly localized to cells with prominent inner segments that contact the pineal lumen (Fig. 4). Whereas at least one-half of all cells in the parapineal express parapinopsin, in the pineal organ parapinopsin occurs in a much smaller fraction of photoreceptive cells.
Besides localizations in the parapineal and pineal, we observed parapinopsin expression in scattered cells of the lateral habenula (Fig. 4). In catfish the parapineal organ is contiguous with the habenula, suggesting that parapinopsin expression in the habenula reflects displaced parapineal cells. In salmon a small number of opsin and transducin-positive cells have been detected by immunohistochemistry in the lateral habenula (Ekstrom, 1987).
In rodents, cone pigments display gradients of expression across the retina. Thus red cones are most concentrated in the ventral and blue in the dorsal halves of the retina, a feature that is thought to enhance color discrimination in different portions of the visual field (Wang et al., 1992). In contrast, in many teleosts the different cone pigments are expressed in a mosaic of repeating units with no gradient throughout the retina (Cepko, 1996). We wondered whether catfish pineal photoreceptors would be organized in regular arrays or in gradients and so determined the expression pattern of parapinopsin within the pineal. Serial sections through the pineal reveal a caudal–rostral gradient in the number of cells expressing parapinopsin (Fig.5). Thus, parapinopsin-positive cells, encircling the lumen of the pineal organ, occur in highest density in caudal areas (Fig. 5A) with a gradual diminution in more rostral sections until the most rostral tip, which contains no parapinopsin cells (Fig. 5G). The number of parapinopsin-expressing cells in the pineal ranges from 8% of all cells in the pineal stalk to <1% of cells in the more rostral portions of the pineal. By contrast, in the parapineal a large fraction of cells express parapinopsin, and we cannot detect any gradients of expression.
Fig. 5.
A caudal–rostral gradient exists in the number of cells expressing parapinopsin in the catfish pineal.A–G, Shown are consecutive 10 μm sections, starting from the most caudal region of the pineal stalk and taken 20 μm apart, in a typical pineal organ (magnification, 400×). Sections are counterstained with Fast Green.
Parapinopsin, rhodopsin, and red cone pigment are differentially expressed in extraretinal and retinal tissues
We wondered whether known visual pigments might be expressed in the pineal and, conversely, whether parapinopsin might be expressed in the retina. The channel catfish possesses only one cone, sensitive to red, and one rod (Naka, 1969; Liebman, 1973), thus making it easy to obtain the full complement of retinal opsins. To develop appropriate probes, we cloned a partial cDNA encoding catfish rhodopsin. We used the catfish red cone pigment that we had cloned in our initial screens. Catfish rhodopsin and red cone pigments display ∼80% amino acid sequence identity with comparable pigments in the goldfish over the regions cloned (Fig. 6).
Fig. 6.

Alignment of catfish rhodopsin and red cone pigment partial-length cDNAs to their goldfish orthologues. Residues identical in both genes are boxed; similar residues areshaded. The catfish rhodopsin cDNA runs from the middle of the N-terminal domain to the middle of the C-terminal domain of the gene and shows 79% identity and 84% similarity to its goldfish orthologue. The catfish red cone pigment cDNA runs from the middle of the second cytoplasmic domain to the middle of transmembrane domain 7 and shows 78% identity and 82% similarity to its goldfish orthologue.
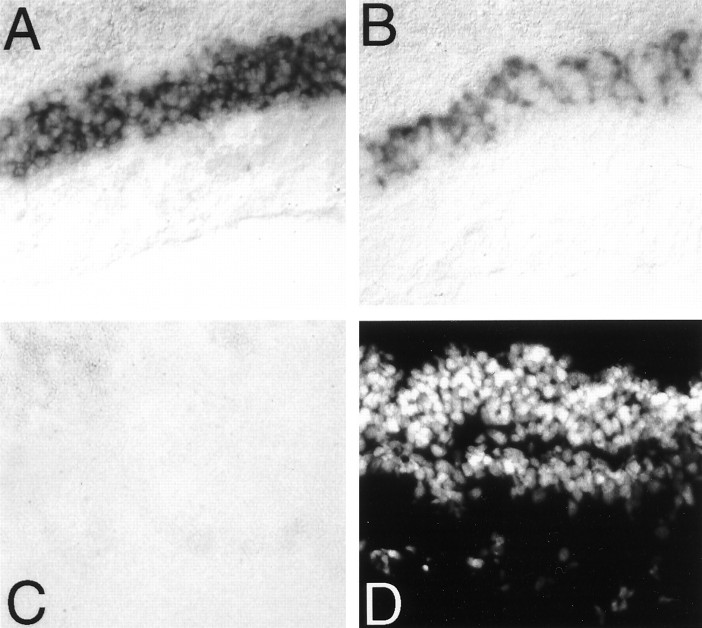
Rhodopsin and the red cone pigment showed strong expression in retinal photoreceptor cell layers (Fig.7). By contrast, parapinopsin was not expressed in the retina at detectable levels. These initial experiments used 20 min color reactions. After 3 d of color reaction, we did observe very faint expression of parapinopsin in the retina (data not shown). This expression may account for the band of parapinopsin observed in RT-PCR experiments (see Fig. 3). In contrast to the pronounced expression of rhodopsin and red cone pigment in the retina, we did not detect significant expression of these pigments in the pineal gland under conditions in which robust staining of parapinopsin was evident (Fig. 8). Color reaction for 3 d still failed to reveal any expression of rhodopsin or red cone pigment in the pineal.
Fig. 7.
Parapinopsin is not expressed in the retina.A, Rhodopsin expression in catfish retina.B, Red cone pigment expression in catfish retina.C, Parapinopsin expression in catfish retina.D, DAPI stain of the section used in C. Color reaction in these sections was allowed to run for 20 min. A faint level of parapinopsin expression was observed in the photoreceptor cell layer of the retina after 3 d of exposure (data not shown). All photographs were taken at 200× magnification.
Fig. 8.
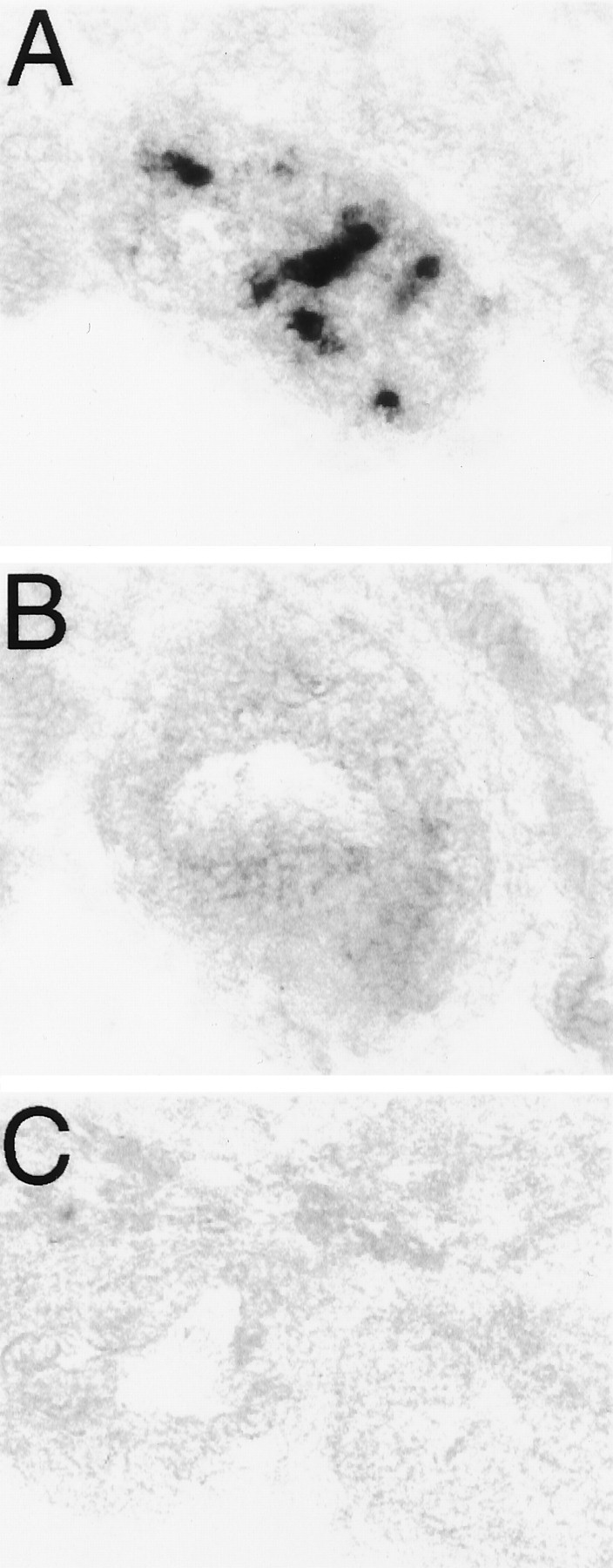
Retinal opsins are not expressed in the pineal complex. A, Parapinopsin expression in a catfish pineal organ. B, Rhodopsin expression in caudally adjacent section of catfish pineal organ. C, Red cone pigment in a rostrally adjacent section of catfish pineal organ. Three different pineal complexes were analyzed in this manner, and no rhodopsin or red cone pigment expression was observed in either the pineal or parapineal organs.
Because photoreception has been reported in regions of the diencephalon, we stained serial alternating sections of the diencephalon for parapinopsin, red cone pigment, and rhodopsin with color reaction times of up to 3 d. Despite exhaustive analysis of sections throughout the diencephalon, we failed to detect any expression of parapinopsin, red opsin, or red cone pigment. This suggests that the positive signals for parapinopsin in RT-PCR analysis of brain tissue reflect contributions of the pineal and the parapineal organs. Because photoreception has been reported in the iris (Barr, 1989), we also conducted in situ hybridization with color reactions for 3 d and observed no expression of any of the three pigments evaluated.
Melanophores of fish skin are photoreceptive (Wakamatsu et al., 1980). Accordingly, we examined the expression of parapinopsin, rhodopsin, and red pigment in the skin (Fig.9). We did not observe any of the three pigments in the dermis, which contains the presumed photoreceptive melanophores. However, we observed substantial expression of all three pigments in the epidermis. Highest expression occurs in taste buds, whereas the basal layer of the epidermis displays a more intense signal than more superficial layers. Of the three pigments, rhodopsin and parapinopsin stain more intensely than red cone pigment.
Fig. 9.
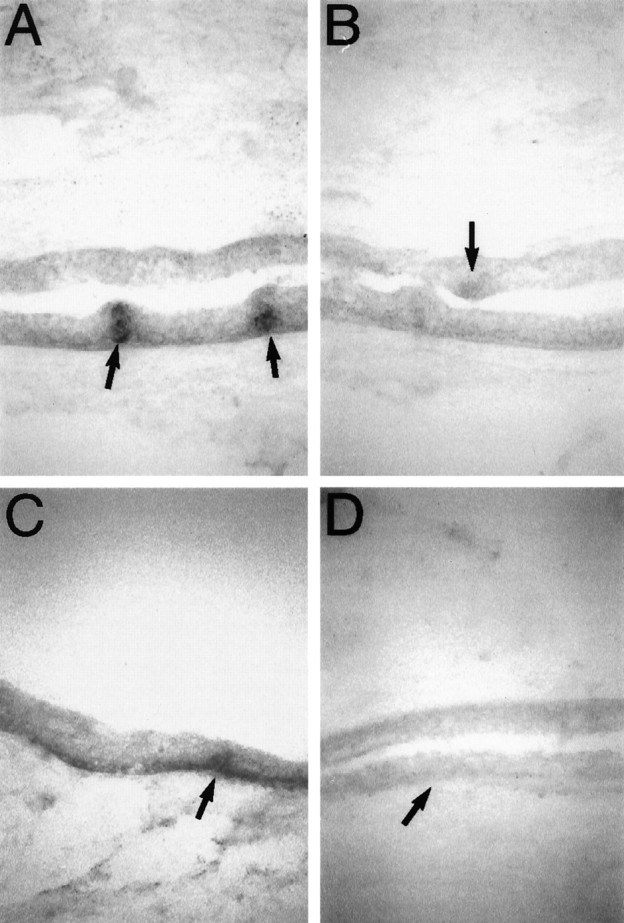
Rhodopsin and parapinopsin are expressed in taste buds and the basal layer of epidermis. A, Expression of rhodopsin in catfish skin. Expression is visible in the basal layer of epidermis and taste buds. Taste buds are indicated by black arrows. B, Expression of red cone pigment in catfish skin. A taste bud is indicated by the black arrow. C, Expression of parapinopsin in catfish skin. Expression is visible in the basal layer of epidermis and taste buds. A taste bud is indicated by the black arrow.D, A sense control probe for parapinopsin hybridized to a section adjacent to C and exposed for an identical period of time. A taste bud is indicated by the black arrow.
DISCUSSION
The main finding of this study is the identification of parapinopsin as the first opsin to be cloned from the pineal gland of cold-blooded vertebrates. It also is the first opsin to be identified in the parapineal organ, strongly supporting the assumption that the parapineal organ is itself a functional site of photoreception. Parapinopsin is the most divergent of all known vertebrate opsins and defines a new gene family within the vertebrate opsins, one that is quite distinct from the only other pineal opsin cloned, avian pinopsin.
Our inability to reconstitute parapinopsin biochemically as a functional photopigment is not surprising, because extraretinal and invertebrate opsins so far have proven resistant to reconstitution. In the initial cloning of the chick pinopsin, only one of the two groups (Okano et al., 1994) was able to reconstitute successfully a spectrum from recombinant protein. Likewise, similar attempts to reconstitute recombinant invertebrate opsins or novel opsins expressed in mammalian retina pigmented epithelium (RPE) have been similarly unsuccessful, although some groups have reported reconstitution either of protein purified from native tissue (Kiselev and Subramaniam, 1994; Hao and Fong, 1996) or of functional opsin produced by expression in the photoreceptors of transgenic Drosophila (Chou et al., 1996).
Parapinopsin is expressed in only a small fraction of pineal organ photoreceptors. This suggests that other hitherto undiscovered opsins exist in the majority of pineal photoreceptor cells. Because we failed to identify parapinopsin, rhodopsin, or red cone photopigment in deep areas of the diencephalon, the iris, or melanophores despite highly sensitive methods of detection, the reported photoreception in these tissues must be subserved by novel opsins.
The caudal–rostral organization of parapinopsin-expressing cells in the pineal organ is notable and surprising, given the regular array of opsin expression in teleost retina. Gradients of visual pigment expression are well known in mammalian retina but are not present in fish retina (Cepko, 1996). In mammalian retina the differential gradients for red and blue pigments are thought to enhance color discrimination in different portions of the visual field. Conceivably, the caudal–rostral organization of parapinopsin plays a similar role in the pineal organ. Perhaps other unrecognized opsins in the pineal gland would display a different type of caudal–rostral variation in expression pattern.
We failed to detect expression of rhodopsin or red cone pigment in the pineal gland. Other workers, using immunohistochemistry with antibodies to bovine rhodopsin and chicken cone pigments, have reported the occurrence of rhodopsin and cone pigment immunoreactivity in the pineal organ of lower vertebrates and other sites of presumed extraretinal photoreception (Vigh et al., 1980; Vigh-Teichmann et al., 1983; Hafeez et al., 1987; Tamotsu et al., 1990, 1994; Vigh-Teichmann and Vigh, 1990; Masuda et al., 1994; Grace et al., 1996). However, many of these studies have used antibodies coming from divergent species at high concentrations and/or long exposure times so that observed immunoreactivity might reflect cross-reactivity with other proteins. By contrast, our in situ hybridization technique with digoxigenin is highly specific as well as sensitive. In light of this evidence, it is likely that the cross-reactivity observed in sites of extraretinal photoreception in lower vertebrates probably represents reactivity with novel, extraretinal opsins.
One of our most striking observations is the substantial expression of the three photopigments in taste buds and the epidermis. The role of these pigments in this tissue is unclear. Conceivably, the epidermis and taste buds possess photoreceptive functions that have not been identified previously. The expression of these pigments in taste buds and epidermis might derive from a similarity in tissue-specific promoter regulation in the retina and epidermis. In support of this notion are the findings of Margolskee and associates (McLaughlin et al., 1994) who have reported the expression of rod transducin in taste bud cells in studies by using RNase protection.
Note added in proof: The sequences of catfish parapinopsin, red cone pigment, and rhodopsin have been deposited in GenBank and are available via accession numbers AF028014, AF028015, andAF028016, respectively.
Footnotes
This work was supported by United States Public Health Service Grant DA-00266 and Research Scientist Award DA00074 to S.H.S. S.B. was a Howard Hughes Medical Institute Predoctoral fellow. We thank R. Vassar and R. Axel for advice concerning nonradioactive in situhybridization, H. Sun for assistance with biochemical reconstitution, and J. Pevsner for assistance with phylogenetic analysis.
Correspondence should be addressed to Dr. Solomon H. Snyder, Johns Hopkins University School of Medicine, Department of Neuroscience, 725 North Wolfe Street, Baltimore, MD 21205.
REFERENCES
- 1.Barr L. Photomechanical coupling in the vertebrate sphincter pupillae. CRC Crit Rev Neurosci. 1989;4:325–366. [PubMed] [Google Scholar]
- 2.Boni SG, Foster RG. A novel and ancient vertebrate opsin. FEBS Lett. 1997;406:279–283. doi: 10.1016/s0014-5793(97)00287-1. [DOI] [PubMed] [Google Scholar]
- 3.Cepko CL. The patterning and onset of opsin expression in vertebrate retina. Curr Opin Neurol. 1996;6:542–546. doi: 10.1016/s0959-4388(96)80062-6. [DOI] [PubMed] [Google Scholar]
- 4.Chang BSW, Crandall KA, Carulli JP, Hartl DL. Opsin phylogeny and evolution: a model for blue shifts in wavelength regulation. Mol Phylogenet Evol. 1995;4:31–43. doi: 10.1006/mpev.1995.1004. [DOI] [PubMed] [Google Scholar]
- 5.Chou W-H, Hall KJ, Wilson DB, Wideman CL, Townson SM, Chadwell LV, Britt SG. Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron. 1996;17:1101–1115. doi: 10.1016/s0896-6273(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 6.Dodt E. Photosensitivity of the pineal organ in the teleost, Salmo irideus (Gibbons). Experientia. 1963;19:642–643. doi: 10.1007/BF02151295. [DOI] [PubMed] [Google Scholar]
- 7.Dodt E, Heerd E. Mode of action of pineal nerve fibers in frogs. J Neurophysiol. 1962;25:405–429. doi: 10.1152/jn.1962.25.3.405. [DOI] [PubMed] [Google Scholar]
- 8.Dodt E, Scherer E. Photic responses from the parietal eye of the lizard Lacerta sicula campestris (De Betta). Vision Res. 1968;8:61–72. [Google Scholar]
- 9.Eakin RM. The third eye. University of California; Berkeley, CA: 1973. [Google Scholar]
- 10.Ekstrom P, Borg B, van Veen T. Ontogenetic development of the pineal organ, parapineal organ, and retina of the three-spined stickleback, Gasterosteus aculeatus: development of photoreceptors. Cell Tissue Res. 1983;233:593–609. doi: 10.1007/BF00212227. [DOI] [PubMed] [Google Scholar]
- 11.Ekstrom P, Foster RG, Korf H-W, Schalken JJ. Antibodies against retinal photoreceptor-specific protein reveal axonal projections from the photosensory pineal organs of teleosts. J Comp Neurol. 1987;265:25–33. doi: 10.1002/cne.902650103. [DOI] [PubMed] [Google Scholar]
- 12.Fahmy K, Sakmar TP. Regulation of the rhodopsin–transducin interaction by a highly conserved carboxylic acid group. Biochemistry. 1993;32:7229–7236. doi: 10.1021/bi00079a020. [DOI] [PubMed] [Google Scholar]
- 13.Fitzgibbon J, Hope A, Slobodyanyuk SJ, Bellingham J, Bowmaker JK, Hunt DM. The rhodopsin-encoding gene of bony fish lacks introns. Gene. 1995;164:273–277. doi: 10.1016/0378-1119(95)00458-i. [DOI] [PubMed] [Google Scholar]
- 14.Franke RR, Sakmar TP, Graham RM, Khorana HG. Structure and function in rhodopsin. Studies of the interaction between the rhodopsin cytoplasmic domain and transducin. J Biol Chem. 1992;267:14767–14774. [PubMed] [Google Scholar]
- 15.Frohman MA. RACE-PCR. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. Academic; San Diego: 1990. pp. 28–38. [Google Scholar]
- 16.Grace MS, Alones V, Menaker M, Foster RG. Light perception in the vertebrate brain: an ultrastructural analysis of opsin- and vasoactive intestinal polypeptide-immunoreactive neurons in iguanid lizards. J Comp Neurol. 1996;367:575–594. doi: 10.1002/(SICI)1096-9861(19960415)367:4<575::AID-CNE8>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 17.Hafeez MA, Merhige ME. Light and electron microscopic study on the pineal complex of the coelacanth, Latimeria chalumnae Smith. Cell Tissue Res. 1977;178:249–265. doi: 10.1007/BF00219052. [DOI] [PubMed] [Google Scholar]
- 18.Hafeez MA, Korf H-W, Oksche A. Immunocytochemical and electron microscopic investigations of the pineal organ in adult agamid lizards, Uromastix hardwicki. Cell Tissue Res. 1987;250:571–578. doi: 10.1007/BF00218948. [DOI] [PubMed] [Google Scholar]
- 19.Hao W, Fong HKW. Blue and ultraviolet light-absorbing opsin from the retinal pigment epithelium. Biochemistry. 1996;35:6251–6256. doi: 10.1021/bi952420k. [DOI] [PubMed] [Google Scholar]
- 20.Hartwig H-G, Oksche A. Neurobiological aspects of extraretinal photoreceptive systems: structure and function. Experientia. 1982;38:991–996. doi: 10.1007/BF01955341. [DOI] [PubMed] [Google Scholar]
- 21.Hillis DM, Bull JJ. An empirical test of bootstrapping as a method of assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- 22.Kawamura S, Yokoyama S. Molecular characterization of the pigeon P-opsin gene. Gene. 1996;182:213–214. doi: 10.1016/s0378-1119(96)00476-3. [DOI] [PubMed] [Google Scholar]
- 23.Kiselev A, Subramaniam S. Activity and regeneration of rhodopsin in the insect visual cycle. Science. 1994;266:1369–1373. doi: 10.1126/science.7973725. [DOI] [PubMed] [Google Scholar]
- 24.Korf H-W, Foster RG, Ekstrom P, Schalken JJ. Opsin-like immunoreactivity in the retinae and pineal organs of four mammalian species. Cell Tissue Res. 1985;242:645–648. doi: 10.1007/BF00225432. [DOI] [PubMed] [Google Scholar]
- 25.Koutalos Y, Yau K-W. Regulation of sensitivity in vertebrate rod photoreceptors by calcium. Trends Neurosci. 1996;19:73–81. doi: 10.1016/0166-2236(96)89624-x. [DOI] [PubMed] [Google Scholar]
- 26.Kuo CH, Tamotsu S, Morita Y, Shinozawa T, Akiyama M, Miki N. Presence of retina-specific proteins in the lamprey pineal complex. Brain Res. 1988;442:147–151. doi: 10.1016/0006-8993(88)91442-4. [DOI] [PubMed] [Google Scholar]
- 27.Libert F, Parmentier M, Lefort A, Dinsart C, van Sande J, Maenhaut C, Simons M-J, Dumont JE, Vassart G. Selective amplification and cloning of four new members of the G-protein-coupled receptor family. Science. 1989;244:569–572. doi: 10.1126/science.2541503. [DOI] [PubMed] [Google Scholar]
- 28.Liebman PA. Biochemistry and physiology of visual pigments (Langer H, ed), pp 299–306. Springer; Berlin: 1973. [Google Scholar]
- 29.Marchiafava PL, Kusmic C. The electrical responses of the trout pineal photoreceptors to brief and prolonged illumination. Prog Brain Res. 1993;95:3–13. doi: 10.1016/s0079-6123(08)60352-0. [DOI] [PubMed] [Google Scholar]
- 30.Masuda H, Oishi T, Ohtani M, Michinomae M, Fukuda Y, Shichida Y, Yoshizawa T. Visual pigments in the pineal complex of the Japanese quail, Japanese grass lizard, and bullfrog—immunocytochemistry and HPLC analysis. Tissue Cell. 1994;26:101–113. doi: 10.1016/0040-8166(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 31.Max M, McKinnon PJ, Seidenman KJ, Barrett RK, Applebury ML, Takahashi JS, Margolskee RF. Pineal opsin: a nonvisual opsin expressed in chick pineal. Science. 1995;267:1502–1506. doi: 10.1126/science.7878470. [DOI] [PubMed] [Google Scholar]
- 32.McLaughlin SK, McKinnon PJ, Spickofsky N, Danho W, Margolskee RF. Molecular cloning of G-proteins and phosphodiesterases from taste cells. Physiol Behav. 1994;56:1157–1164. doi: 10.1016/0031-9384(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 33.Naka KI. Computer assisted analysis of S-potentials. Biophys J. 1969;9:855–899. doi: 10.1016/S0006-3495(69)86422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Offermann S, Simon M. Gα15 and Gα16 couple a wide variety of receptors to phospholipase C. J Biol Chem. 1995;270:15175–15180. doi: 10.1074/jbc.270.25.15175. [DOI] [PubMed] [Google Scholar]
- 35.Okano T, Yoshizawa T, Fukuda Y. Pinopsin is a chicken pineal photoreceptive molecule. Nature. 1994;372:94–97. doi: 10.1038/372094a0. [DOI] [PubMed] [Google Scholar]
- 36.Oksche A. Survey of the development and comparative morphology of the pineal organ. Prog Brain Res. 1965;10:3–28. doi: 10.1016/s0079-6123(08)63445-7. [DOI] [PubMed] [Google Scholar]
- 37.Palczewski K. Is vertebrate phototransduction solved? New insights into the molecular mechanism of phototransduction. Invest Ophthalmol Vis Sci. 1994;35:3577–3581. [PubMed] [Google Scholar]
- 38.Roberts A. Pineal eye and behavior in Xenopus tadpoles. Nature. 1978;273:774–775. doi: 10.1038/273774a0. [DOI] [PubMed] [Google Scholar]
- 39.Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and culture cells: in situ hybridization using digoxygenin-labeled probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 40.Solessio E, Engbretson GA. Antagonistic chromatic mechanisms in photoreceptors of the parietal eye of lizards. Nature. 1993;364:442–445. doi: 10.1038/364442a0. [DOI] [PubMed] [Google Scholar]
- 41.Tamotsu S, Korf H-W, Morita Y, Oksche A. Immunocytochemical localization of serotonin and photoreceptor-specific proteins (rod-opsin, S-antigen) in the pineal complex of the river lamprey, Lampetra japonica, with special reference to photoneuroendocrine cells. Cell Tissue Res. 1990;262:205–216. doi: 10.1007/BF00309875. [DOI] [PubMed] [Google Scholar]
- 42.Tamotsu S, Oishi T, Nakao K, Fukuda Y, Shichida Y, Yoshizawa T, Morita Y. Localization of iodopsin and rod-opsin immunoreactivity in the retina and pineal complex of the river lamprey, Lampetra japonica. Cell Tissue Res. 1994;278:1–10. [Google Scholar]
- 43.van Veen T, Ostholm T, Gierschik P, Spiegel A, Somers R, Korf H-W, Klein DC. α-Transducin immunoreactivity in retinae and sensory pineal organs of adult vertebrates. Proc Natl Acad Sci USA. 1986;83:912–916. doi: 10.1073/pnas.83.4.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vigh B, Vigh-Teichmann I, Rohlich P, Oksche A. Comparison of the pineal complex, retina, and cerebrospinal fluid-contacting neurons by immunocytochemical antirhodopsin reaction. Z Mikrosk Anat Forsch. 1980;94:623–640. [PubMed] [Google Scholar]
- 45.Vigh-Teichmann I, Vigh B. Opsin immunocytochemical characterization of different types of photoreceptors in the frog pineal organ. J Pineal Res. 1990;8:323–333. doi: 10.1111/j.1600-079x.1990.tb00892.x. [DOI] [PubMed] [Google Scholar]
- 46.Vigh-Teichmann I, Korf H-W, Oksche A, Vigh B. Opsin-immunoreactive outer segments and acetylcholinesterase-positive neurons in the pineal complex of Phoxinus phoxinus. Cell Tissue Res. 1982;227:351–369. doi: 10.1007/BF00210891. [DOI] [PubMed] [Google Scholar]
- 47.Vigh-Teichmann I, Korf H-W, Nurnberger F, Oksche A, Vigh B, Olsson R. Opsin-immunoreactive outer segments in the pineal and parapineal organs of the lamprey (Lampetra fluviatilis), the eel (Anguilla anguilla), and the rainbow trout (Salmo gairdneri). Cell Tissue Res. 1983;230:289–307. doi: 10.1007/BF00213806. [DOI] [PubMed] [Google Scholar]
- 48.Wakamatsu Y, Kawamura S, Yoshizawa T. Light-induced pigment aggregation in cultured fish melanophores: spectral sensitivity and inhibitory effects of theophylline and cyclic adenosine-3′,5′-monophosphate. J Cell Sci. 1980;41:65–74. doi: 10.1242/jcs.41.1.65. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Macke JP, Merbs SL, Zack DJ, Klaunberg B, Bennett J, Gearhart J, Nathans J. A locus control region adjacent to the human red and green visual pigment genes. Neuron. 1992;9:429–440. doi: 10.1016/0896-6273(92)90181-c. [DOI] [PubMed] [Google Scholar]
- 50.Weber W. Photosensitivity of chromatophores. Am Zool. 1983;23:495–506. [Google Scholar]
- 51.Wurtman RJ, Axelrod J, Kelly DE. The pineal. Academic; New York: 1968. [Google Scholar]
- 52.Yanez J, Meissl H, Anadon R. Central projections of the parapineal organ of the adult rainbow trout (Oncorhynchus mykiss) Cell Tissue Res. 1996;285:69–74. doi: 10.1007/s004410050621. [DOI] [PubMed] [Google Scholar]