Abstract
ZC3H12B is the most enigmatic member of the ZC3H12 protein family. The founding member of this family, Regnase-1/MCPIP1/ZC3H12A, is a well-known modulator of inflammation and is involved in the degradation of inflammatory mRNAs. In this study, for the first time, we characterized the properties of the ZC3H12B protein. We show that the biological role of ZC3H12B depends on an intact NYN/PIN RNase domain. Using RNA immunoprecipitation, experiments utilizing actinomycin D and ELISA, we show that ZC3H12B binds interleukin-6 (IL-6) mRNA in vivo, regulates its turnover, and results in reduced production of IL-6 protein upon stimulation with IL-1β. We verified that regulation of IL-6 mRNA stability occurs via interaction of ZC3H12B with the stem–loop structure present in the IL-6 3′UTR. The IL-6 transcript is not the only target of ZC3H12B. ZC3H12B also interacts with other known substrates of Regnase-1 and ZC3H12D, such as the 3′UTRs of IER3 and Regnase-1, and binds IER3 mRNA in vivo. Using immunofluorescence, we examined the localization of ZC3H12B within the cell. ZC3H12B forms small, granule-like structures in the cytoplasm that are characteristic of proteins involved in mRNA turnover. The overexpression of ZC3H12B inhibits proliferation by stalling the cell cycle in the G2 phase. This effect of ZC3H12B is also NYN/PIN dependent. The analysis of the ZC3H12B mRNA level reveals its highest expression in the human brain and the neuroblastoma cell line SH-SY5Y, although the factors regulating its expression remain elusive. Down-regulation of ZC3H12B in SH-SY5Y cells by specific shRNAs results in up-regulation of ZC3H12B-target mRNAs.
Keywords: IL-6, Regnase-1/MCPIP1, ZC3H12B, proliferation, transcript turnover
INTRODUCTION
The regulation of gene expression is usually associated with the activation of transcription. However, recently, many well-documented reports have emphasized the importance of the regulation of transcript turnover in this phenomenon (Hao and Baltimore 2009; Carpenter et al. 2014). The fate of mRNA is mainly determined by the cis-regulatory elements present in its 3′-untranslated region (3′UTR). These elements serve as a platform for trans-acting RNA-binding proteins (RBP). The most intensely studied sequences that participate in mRNA half-life regulation are adenine and uridine-rich elements (ARE), which provide binding sites for ARE-RBPs, such as tristetraprolin (TTP), AUF-1, KSRP, HuR, TIA-1, and TIAR (for reviews, see Anderson 2008; García-Mauriño et al. 2017). mRNA stability is also regulated by another set of RBPs that recognize stem–loop structures present in 3′UTRs, such as Roquin and Regnase-1 (Reg-1, also known as ZC3H12A or MCPIP1). Although Roquin and Regnase-1 have overlapping substrate specificities, the mechanisms of their action are distinct. Roquin localizes to P bodies and stress granules and mainly participates in the degradation of translationally inactive mRNAs by recruiting CCR4-NOT deadenylase complexes. In contrast, Regnase-1 localizes to the endoplasmic reticulum and cytoplasm and directly degrades translationally active transcripts. Interestingly, the presence of helicase UPF1 involved in the nonsense-mediated decay is essential for Regnase-1-mediated mRNA turnover (Lykke-Andersen and Jensen 2015; Mino et al. 2015). The enzymatic activity of Regnase-1 solely depends on the presence of an intact RNase NYN/PIN-like domain (Xu et al. 2012; Wilamowski et al. 2018). This domain along with the carboxy-terminal proline-rich domain is also involved in the Regnase-1-dependent degradation of pre-microRNAs (Suzuki et al. 2011).
Regnase-1 belongs to a protein family that includes three other members named ZC3H12B-D (MCPIP2-4) (Liang et al. 2008). All members of this family possess an NYN/PIN-like domain and a CCCH-zinc finger domain. The homologies of the NYN domain of Regnase-1 among ZC3H12B-D are 83.2%, 84.4%, and 75.3%, whereas the homologies of their CCCH domains are 76%, 76%, and 72% (Liang et al. 2008; Suzuki et al. 2011). Regnase-1 is involved not only in RNA degradation but also in the modulation of cell signaling. By removing ubiquitin moieties from TRAF2, TRAF3, and TRAF6, Regnase-1 negatively regulates the JNK and NF-κB signaling pathways (Liang et al. 2010). Regnase-1 also participates in the antiviral cellular response by degrading the genomic nucleic acids of both positive-sense and negative-sense RNA viruses and DNA viruses (Lin et al. 2013, 2014). Since Regnase-1 mRNA is rapidly induced by proinflammatory factors and Regnase-1 contributes to the degradation of proinflammatory cytokine transcripts and influences inflammatory signaling and the antiviral response, Regnase-1 has emerged as a key regulator of inflammation (Uehata and Takeuchi 2017).
Our knowledge of the biological roles played by other members of the ZC3H12 family is very limited, although it appears that these members are also involved in inflammatory processes. ZC3H12D (MCPIP4) participates in the degradation of proinflammatory mRNAs, such as the mRNAs of IL-2, IL-6, IL-10, TNF, IER3, and Regnase-1 (Zhang et al. 2015; Wawro et al. 2017). Similar to Regnase-1, the overexpression of ZC3H12D attenuates global protein ubiquitination and leads to the inhibition of the TLR2- and TLR4-induced activation of JNK, ERK, and NF-κB signaling and TLR4-induced inflammatory gene expression in macrophages (Huang et al. 2012). Recently, ZC3H12D and Regnase-1 have been reported to form a complex but act independently in the regulation of IL-6 mRNA degradation (Huang et al. 2015). ZC3H12D was originally identified as a tumor suppressor gene. The overexpression of MCPIP4 suppresses tumor cell growth both in vitro and in vivo (Minagawa et al. 2009). The inhibition of proliferation has also been observed in cells with the overexpression of Regnase-1 (Marona et al. 2017; Lichawska-Cieslar et al. 2018). Another member of this family, that is, ZC3H12C (MCPIP3), suppresses NFκB activation by decreasing TNF-induced IKKα/β and IκBα phosphorylation and p65 nuclear translocation. The overexpression of ZC3H12C attenuates the TNF-induced expression of chemokines and adhesive molecules in endothelial cells (Liu et al. 2013). In this report, we focused on the fourth member of the ZC3H12 family, that is, ZC3H12B (MCPIP2), since the biological role of this protein is entirely unknown.
RESULTS
NYN/PIN domain of ZC3H12B contains all residues important for RNA binding and cleavage
To determine whether the ZC3H12B protein contains a functional nuclease domain, we aligned the sequence of the NYN/PIN domain of ZC3H12B with that of the NYN/PIN domain of Regnase-1/ZC3H12A/MCPIP1, which is the most thoroughly characterized protein of the ZC3H12 family (Fig. 1). The crystallographic studies of Regnase-1 revealed that its NYN/PIN domain contains four aspartic acid residues (D141, D225, D226, and D244) that are responsible for magnesium ion binding and crucial for its ribonucleolytic activity (Xu et al. 2012). All four residues are conserved in the NYN/PIN domain of ZC3H12B, indicating that the nuclease domain of this protein is potentially functional. Additionally, the characteristics of the positively charged residues that supposedly play a role in the binding of Regnase-1 to the negatively charged mRNA backbone are also conserved. Moreover, similar to Regnase-1, ZC3H12B contains a CCCH-type zinc finger adjacent to the NYN/PIN domain, which is important for binding target RNAs. The conservation of residues crucial for RNA binding and cleavage indicates that ZC3H12B may function as a nuclease that is involved in the regulation of the mRNA half-life.
FIGURE 1.
NYN/PIN domain of ZC3H12B contains all features required for nuclease activity. Alignment of the NYN/PIN domain and CCCH-zinc finger of Regnase-1/ZC3H12A/MCPIP1 and ZC3H12B. The triangles indicate the aspartic acid residues engaged in magnesium ion binding. The hollow triangle indicates one of the four residues that was mutated to alanine for the NYN/PIN domain inactivation and used in this study (D141 and D196 for Regnase-1 and ZC3H12B, respectively). The dashed line indicates the CCCH-zinc finger domain with three cysteine and one histidine residues indicated with squares. The asterisks denote the positively charged amino acid residues engaged in the binding of the negatively charged backbone of the substrate RNA. The alignment was performed using ClustalW2 (Larkin et al. 2007) using the default settings and visualized using Adobe Illustrator CS4 (Adobe).
ZC3H12B overexpression results in a decrease in luciferase activity dependent on the presence of the IL-6 3′UTR
Given the high conservation of the NYN/PIN domain and CCCH-type zinc finger, we tested whether the ZC3H12B protein is indeed functional. We used a luciferase reporter system in which the 3′UTR of IL-6, which is a well-known target of two other proteins belonging to the ZC3H12 family (Regnase-1 and MCPIP4), is attached to the luciferase coding sequence (CDS). In HeLa cells we coexpressed luciferase mRNA with the attached IL-6 3′UTR (pmirGLO_IL-6-3′UTR) and the expression vector of ZC3H12B or Regnase-1, the latter used as a reference (Fig. 2A). The obtained results show that ZC3H12B overexpression leads to a decrease in luciferase activity, although the effect was weaker than the effect observed using the same amount of Regnase-1 expression plasmid. This milder effect of ZC3H12B is probably due to the fact that the level of ZC3H12B protein is much lower than Regnase-1 when the same amounts of expression plasmids are used (Fig. 2A). One of the reasons of this phenomenon may be the difference in the size of the proteins (94 kDa for ZC3H12B compared to 66 kDa for Regnase-1). The regulation was dose-dependent and required the presence of the IL-6 3′UTR as no decrease in luciferase activity was observed when this region was absent from the luciferase mRNA (pmirGLO) even when increased amounts of the ZC3H12B expression vector were used. The specificity of the regulation was further confirmed using a construct expressing luc mRNA with IL-6 3′UTR lacking a conserved element that contains a stem–loop structure. This structure was found to be crucial for the cleavage of IL-6 mRNA by Regnase-1 and MCPIP4. As indicated in Figure 2B, no regulation of the IL-6 3′UTR was observed following the removal of the investigated structure. The 3′UTRs of VEGFA and IL-17, which we previously demonstrated as not regulated by either Regnase-1 or ZC3H12D, were not influenced by ZC3H12B, highlighting the clear 3′UTR-driven substrate specificity of the investigated protein (Fig. 2C).
FIGURE 2.
ZC3H12B is involved in the 3′UTR-dependent destabilization of transcripts. (A) HeLa cells were transfected with vectors encoding luciferase with attached IL-6 3′UTR (pmirGLO_IL-6-3′UTR) or without any additional 3′UTRs (pmirGLO) and varying amounts (10 and 20 ng/well) of Regnase-1 and ZC3H12B expression vectors (left panels), increasing amounts of ZC3H12B expression vectors (middle panels, twofold increments from 10 to 160 ng/well) or an empty control vector (pcDNA3). The expression level of Regnase-1 and ZC3H12B was examined by western blotting. Representative image of three independent experiments with similar results is shown (right panel). (B) HeLa cells were transfected with vectors encoding luciferase with the attached 3′UTR of wild-type IL-6 (pmirGLO_IL-6-3′UTR) or IL-6 with a deletion of a conserved element containing a stem–loop structure (pmirGLO_IL-6-3′UTRΔCE) and varying amounts (40 and 80 ng/well) of the ZC3H12B expression vector or an empty control vector (pcDNA3). (C) HeLa cells were transfected with vectors encoding luciferase with the attached 3′UTRs of VEGFA or IL-17 (pmirGLO_VEGFA-3′UTR and pmirGLO_IL-17-3′UTR, respectively) and increasing amounts of the ZC3H12B expression vector (40 and 80 ng/well) or an empty control vector (pcDNA3). The graphs show the mean results of four (A) or three (B,C) independent experiments ±SD. The data were analyzed using one-way ANOVA (A,C) or two-way-ANOVA (B) and Bonferroni's post-hoc test ([*] P < 0.05; [**] P < 0.01; [***] P < 0.001).
IL-6 3′UTR-dependent decrease in luciferase activity results from NYN/PIN-dependent transcript destabilization
The ZC3H12B-dependent changes in luciferase activity could be caused by recombinant transcript destabilization or translation inhibition. To test these hypotheses, we performed real-time PCR to quantify the amount of luciferase (luc) mRNA in parallel to luciferase activity. As shown in Figure 3A, the overexpression of ZC3H12B results in a decrease in the relative amount of luc-IL-6 3′UTR mRNA that approximately corresponds to the changes observed in the luciferase activity assay. Moreover, the half-life of the luc-IL-6 3′UTR mRNA is visibly shortened when ZC3H12B is expressed (Fig. 3B) pointing that the observed effect of ZC3H12B is the result of transcript destabilization and not other post-transcriptional mechanisms. Because the changes in luciferase activity are caused by a decrease in the amount of transcript, we examined whether the decrease depends on the presence of an active NYN/PIN nuclease domain. Using site-directed mutagenesis, we introduced a point mutation into the NYN/PIN domain of ZC3H12B that changed one of the crucial aspartic acid residues required for magnesium ion binding to alanine (D196A). As shown in Figure 3C, the introduction of the mutation completely abolished the observed decrease in luciferase activity. Simultaneously, we assessed the level of expression of the ZC3H12B D196A mutein and its wild-type counterpart. Both proteins are expressed at approximately the same level (Fig. 3D); thus, the absence of luciferase regulation by ZC3H12B D196A is due to the inactivation of the nuclease domain rather than the improper synthesis of the mutein. This finding clearly indicates that a functional NYN/PIN domain is crucial for the observed ribonucleolytic activity of ZC3H12B.
FIGURE 3.

Decrease in luciferase activity following ZC3H12B overexpression results from changes in luc transcript levels. (A) HeLa cells were transfected with vector encoding luc mRNA with the 3′UTR of IL-6 (pmirGLO_IL-6-3′UTR) and increasing amounts (40 and 80 ng/well) of the ZC3H12B expression vector or an empty control vector (pcDNA3), and the luciferase protein activity (left panel) and luc mRNA transcript level (right panel) were assayed. (B) HeLa cells were transfected with vector encoding wild-type ZC3H12B or an empty control plasmid (empty vector). After transfection the cells were treated with actinomycin D (10 µg/mL) for the indicated time periods, and the luc_IL-6-3′UTR mRNA levels were measured by qRT-PCR. (C) HeLa cells were transfected with the same luciferase vector as described in A and increasing amounts (40 and 80 ng/well) of vectors encoding wild-type ZC3H12B protein or the D196A mutein (ZC3H12B WT and ZC3H12B D196A, respectively). (D) Western blot analysis of HeLa cells transfected with vectors encoding the wild-type ZC3H12B protein or the D196A mutein showing the expression of both proteins (a representative result of three independent experiments is shown). The graphs show the mean results of three independent experiments ±SD. The data were analyzed using one-way ANOVA and Bonferroni's post-hoc test ([*] P < 0.05; [**] P < 0.01; [***] P < 0.001).
Known substrates of Regnase-1 are also substrates of ZC3H12B
IL-6 is not the only substrate of Regnase-1 or ZC3H12D. We and other groups have previously shown that both proteins can decrease luciferase activity when the 3′UTRs of IER3 or Regnase-1 (Reg-1) are attached to its mRNA. We examined whether these two transcripts are also regulated by ZC3H12B. The addition of the 3′UTR fragment of Regnase-1 and IER3 to luc mRNA results in a ZC3H12B-dependent decrease in luciferase activity (Fig. 4).
FIGURE 4.

ZC3H12B, Regnase-1, and ZC3H12D destabilize the same mRNA substrates. HeLa cells were transfected with vectors encoding luciferase mRNA with 3′UTRs of the known targets of the two other ZC3H12-family proteins (Regnase-1 and ZC3H12D), that is, Regnase-1 and IER3 (pmirGLO_Reg-1-3′UTR and pmirGLO_IER3-3′UTR) and increasing amounts (40 and 80 ng/well) of the ZC3H12B expression vector or an empty control vector (pcDNA3). The graphs show the mean results of three independent experiments ±SD. The data were analyzed using one-way ANOVA and Bonferroni's post-hoc test ([*] P < 0.05; [**] P < 0.01; [***] P < 0.001).
ZC3H12B binds and destabilizes endogenous IL-6 transcripts
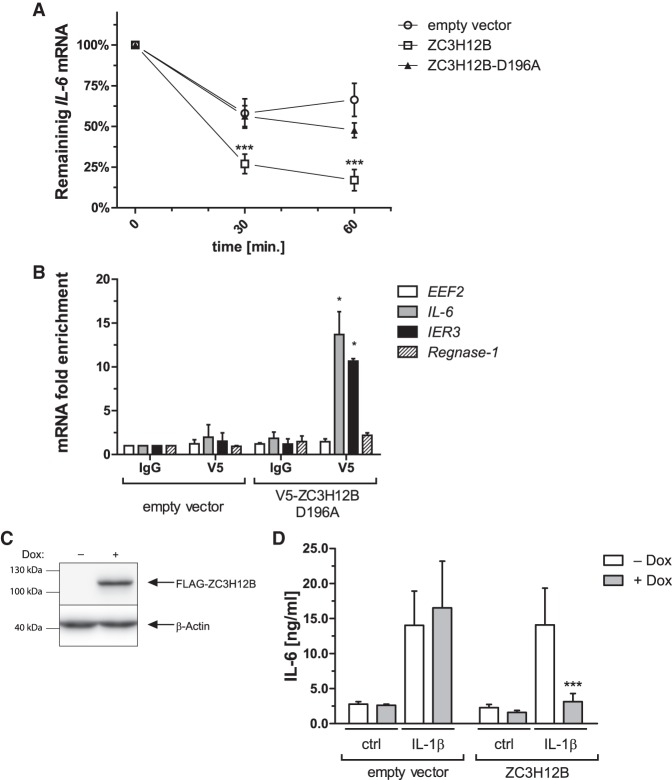
Because ZC3H12B can destabilize luc transcripts containing the 3′UTRs of IL-6, IER3 or Regnase-1 mRNAs, we investigated whether ZC3H12B could bind and destabilize endogenous substrate mRNAs. Thus, we transfected HeLa cells with ZC3H12B wild-type or D196A mutant expression plasmids or an empty control vector, enriched the cultures of the transfected cells and performed endogenous IL-6 mRNA stability assays using actinomycin D. As shown in Figure 5A, the overexpression of the wild-type, but not the D196A mutant, is correlated with a faster degradation of IL-6 mRNA. Compared to the zero time point, a 1 h incubation with actinomycin D results in a 83% decrease in IL-6 mRNA, while the decrease in the cells expressing the D196A mutein or empty control plasmid was 52% and 34%, respectively (data not shown). To determine whether ZC3H12B binds the IL-6 transcript in vivo, we performed a ribonucleoprotein immunoprecipitation (RIP) experiment. HeLa cells were transfected with a V5-tagged ZC3H12B D196A expression plasmid or an empty control plasmid, and after 24 h, RIP was performed (Fig. 5B). Compared to the IgG control samples, enrichment of IL-6 and IER3 transcripts was observed in the samples immunoprecipitated with the anti-V5 antibody from the ZC3H12B D196A expressing cells, while no such enrichment was observed with the nonregulated EEF2 mRNA. Surprisingly, no substantial enrichment of the Regnase-1 transcript was observed, indicating that this mRNA may not be the preferred substrate of ZC3H12B in vivo in HeLa cells.
FIGURE 5.
ZC3H12B directly binds and destabilizes endogenous IL-6 mRNA. (A) HeLa cells were transfected with vectors encoding either wild-type ZC3H12B or the D196A mutein (ZC3H12B WT and ZC3H12B D196A, respectively) or an empty control plasmid (empty vector); after enriching the transfected cells, the cells were treated with actinomycin D (10 µg/mL) for the indicated time periods, and the IL-6 mRNA levels were measured by qRT-PCR. (B) HeLa cells were transfected with vectors encoding the ZC3H12B D196A mutein (which contains an amino-terminal V5 tag) or an empty control vector and, after UV-crosslinking, the riboimmunoprecipitation procedure (RIP) was performed. The obtained RNA was used to analyze the level of the indicated mRNAs (EEF2, IL-6, IER3, and Regnase-1). (C,D) HeLa cells with Sleeping Beauty (SB) transposon–based doxycycline-inducible expression of amino-terminally Flag-tagged wild-type ZC3H12B or luciferase (control) were treated with doxycycline (1 µg/mL) or left untreated for 24 h, and the expression of induced protein was examined by western blotting. Representative image of three independent experiments with similar results is shown (C). After induction of transgene expression, the cells were stimulated with IL-1β (10 ng/mL) or left untreated. The following day the media were collected and IL-6 protein levels were measured using ELISA (D). The graphs show the mean results of four (D), three (A,C), or two (B) independent experiments ±SD. The data were analyzed using two-way ANOVA and Bonferroni's post-hoc test ([*] P < 0.05; [***] P < 0.001).
ZC3H12B overexpression results in reduced production of IL-6 protein in response to IL-1β
As ZC3H12B binds to endogenous IL-6 transcript and causes faster degradation of this mRNA we decided to check whether ZC3H12B overexpression results in reduced production of IL-6 protein in response to proinflammatory stimuli as well. To do this we took advantage of the SB transposon system allowing for rapid generation of stable cell lines (Mátés et al. 2009; Kowarz et al. 2015). We inserted the ZC3H12B CDS into the pSBtet-GP vector (pSBtet-GP-ZC3H12B), cotransfected it with the SB transposase expression vector [pCMV(CAT)T7-SB100] into HeLa cells and after a week of puromycin selection obtained a polyclonal population of cells with doxycycline inducible expression of ZC3H12B (Fig. 5C). As shown in Figure 5D stimulation of HeLa cells with the proinflammatory cytokine IL-1β results in increased secretion of IL-6 cytokine to the culture medium. However, when expression of ZC3H12B was induced prior to IL-1β stimulation the secretion of IL-6 cytokine was significantly diminished (Fig. 5D). This observation in conjunction with the results showed above clearly indicates that ZC3H12B may account for reduced production of proinflammatory target molecules.
ZC3H12B is a cytoplasmic protein whose overexpression results in the inhibition of proliferation
To gain some insight into the biological function of ZC3H12B, we examined its localization within the cell. We transfected HeLa cells with a V5-tagged ZC3H12B expression plasmid and, 24 h after transfection, immunofluorescent staining was performed. As shown in Figure 6A, ZC3H12B is localized in the cytoplasm, but not in the nucleus, and forms small, granule-like structures characteristic of proteins involved in mRNA turnover control (Buchan 2014). Because we were unable to prepare HeLa cells demonstrating stable, constitutive overexpression of ZC3H12B, we hypothesized that ZC3H12B may negatively influence cell conditions and/or proliferation. To test this hypothesis, we examined whether ZC3H12B overexpression is correlated with a decrease in cell count and whether this effect depends on the presence of an active NYN/PIN domain. In these experiments, we used 293T cells since these cells can maintain episomal plasmids bearing the SV40 ori. The 293T cells were transfected with expression plasmids encoding wild-type ZC3H12B or the D196A mutein or an empty control plasmid or left untreated. After transfection, the cells were counted every 24 h up to 72 h post-transfection. As shown in Figure 6B, the ZC3H12B overexpression negatively influenced the cell number. This effect is most visible 72 h posttransfection and depends on the presence of an intact NYN/PIN domain as no effect was observed with the D196A mutein.
FIGURE 6.

ZC3H12B is a cytoplasm-localized negative regulator of cell count. (A) (Upper and middle panels) HeLa cells were transfected with vectors encoding the ZC3H12B protein (containing an amino-terminal V5 tag). After 24 h the cells were fixed and stained using antibodies against the V5 tag (green). The DNA was counterstained using DAPI (blue). Scale bar, 10 µm. (Lower panel) Zoom-in of the cell presented in the middle panel showing frontal (xz) and transverse (yz) sections revealing the exclusion of the ZC3H12B protein from the nucleus. Scale bars, 5 µm. (B) 293T cells were transfected with vectors encoding ZC3H12B proteins (wild-type or the D196A mutein; ZC3H12B WT and ZC3H12B D196A, respectively) or empty control plasmids (empty vector) or left untreated. Every 24 h after transfection (up to 72 h posttransfection), the cells were counted. The graph shows the mean results of three independent experiments ±SD. The data were analyzed using two-way ANOVA with Bonferroni's post-hoc test ([*] P < 0.05; [**] P < 0.01; [***] P < 0.001).
ZC3H12B overexpression causes decreased cell proliferation and increased cell cytotoxicity
As transient overexpression often results in the massive production of the expressed protein, we verified these results under more physiological conditions, that is, when the level of the overexpressed ZC3H12B protein is lower. Thus, we established 293FlipInTrex cell lines with doxycycline-inducible expression of either the wild-type ZC3H12B or the D196A mutein (293FlipInTrex-ZC3H12B or 293FlipInTrex-ZC3H12BD196A, respectively; Fig. 7A). The use of 293FlipInTrex cells allows for the incorporation of a transgene expression cassette into a single site in the genome, allowing for a more uniform expression of the transgene at much lower and more physiological levels than transient transfection. As shown in Figure 7B, the doxycycline induction resulted in a significant decrease in the number of 293FlipInTrex-ZC3H12B cells compared to that of noninduced cells. This effect is almost fully reversed in the 293FlipInTrex-ZC3H12BD196A cells, indicating that the observed biological effect is the result of ZC3H12B ribonucleolytic activity. The observed decrease in cell number following ZC3H12B overexpression could be due to cell death and/or inhibition of proliferation. To examine this hypothesis, we stained the 293FlipInTrex-ZC3H12B cell lines (both WT and D196A mutant) with carboxyfluorescein succinimidyl ester (CFSE) and cultured the cells for 72 h with or without doxycycline induction. CFSE is a fluorescent dye that diffuses into live cells, is cleaved by intracellular esterases and then covalently binds intracellular molecules. As the cell components are spread during division to daughter cells, the fluorescent signal decreases approximately twofold per division, which allows for the tracking of cell division. Figure 7C clearly shows that following the induction of ZC3H12B expression, the cells proliferate more slowly as demonstrated by a shift toward higher fluorescence intensities at the peak of the fluorescence histogram of the doxycycline-induced cells compared to that of the noninduced cells (an approximately twofold change in the median between the noninduced and induced cells with the wild-type ZC3H12B). As no such shift is observed in the 293FlipInTrex-ZC3H12BD196A cells, the decrease in cell proliferation strongly depends on ZC3H12B nuclease activity. Although we clearly saw that ZC3H12B inhibits cell proliferation we could not exclude that expression of this protein may also cause a cytotoxic effect. To verify that in addition to the proliferation test with CFSE described above we performed a cytotoxicity test. We cultured the 293FlipInTrex-ZC3H12B cell lines (both WT and D196A mutant) in presence or absence of doxycycline and examined the levels of LDH released to the culture medium each 24 h after induction up to 72 h post-induction (Fig. 7D). We observed an increase of LDH activity in culture media only in doxycycline-induced cells expressing the wild-type ZC3H12B. The cytotoxic effect revealed in the LDH assay is visible not earlier than 48 h post-induction and is most significant 72 h after induction of ZC3H12B expression. Because of the potent negative effect of ZC3H12B expression on proliferation and increase in cytotoxicity we decided to verify whether the decrease in the luciferase activity in the luciferase assay is not caused by cell stress. We transfected the 293FlipInTrex-ZC3H12B cell lines (both WT and D196A mutant) with pmirGLO_IL-6-3′UTR vector and treated them with doxycycline for 24 h (or left untreated) and checked the luciferase activity in doxycycline-induced cells in comparison to noninduced cells (Fig. 7E). We chose the 24 h induction as no negative effect on proliferation and no cytotoxicity was observed in these cells at this timepoint (Figs. 6B, 7D). As shown in Figure 7E, induction of 293FlipInTrex-ZC3H12B wild-type cells with doxycycline causes a decrease in luciferase activity and this effect is not visible in the case of cells expressing the D196A mutein.
FIGURE 7.
Negative influence of ZC3H12B on cell proliferation depends on the presence of a functional NYN/PIN domain. (A) Flp-In T-REx-293 cells with doxycycline-inducible expression of amino-terminally V5-tagged wild-type ZC3H12B or D196A mutein (ZC3H12B WT and ZC3H12B D196A, respectively) were treated with doxycycline (1 µg/mL) or left untreated for 24 h, and the expression of induced protein was examined by western blotting. Representative image of three independent experiments with similar results is shown. (B) ZC3H12B WT and ZC3H12B D196A Flp-In T-REx-293 cells were treated with doxycycline (1 µg/mL) or left untreated and counted 72 h after the addition of doxycycline. The data are presented as the ratios of the induced to the noninduced cells. The graph shows the mean results of three independent experiments ±SD. The data were analyzed using two-way ANOVA and Bonferroni's post-hoc test ([***] P < 0.001). (C) ZC3H12B WT and ZC3H12B D196A Flp-In T-REx-293 cells were labeled with CFSE and divided into two dishes. On the following day, one dish was induced with doxycycline (1 µg/mL) and the other dish was left untreated. After 72 h postinduction, the fluorescence of the cells was analyzed by flow cytometry. The graphs show histogram overlays normalized to the mode of CFSE fluorescence in the noninduced (gray filling) and induced (black line) cells. Representative results of three independent experiments are shown. (D) ZC3H12B WT and ZC3H12B D196A Flp-In T-REx-293 cells were treated with doxycycline (1 µg/mL) or left untreated, and cell cytotoxicity (using LDH cytotoxicity assay) was measured every 24 h, up to 72 h postinduction with doxycycline. The data are presented as the ratios of the induced to the noninduced ZC3H12B WT cells. The graph shows the mean results of three independent experiments ±SD. The data were analyzed using two-way ANOVA and Bonferroni's post-hoc test ([***] P < 0.001). (E) 293FlipInTrex-ZC3H12B cell lines (both WT and D196A mutant) were transfected with pmirGLO_IL-6-3′UTR vector and treated with doxycycline for 24 h (or left untreated). The graph shows the mean results of three independent experiments ±SD. The data were analyzed using one-way ANOVA and Bonferroni's post-hoc test ([**]) P < 0.01).
Mechanism of ZC3H12B overexpression-induced inhibition of proliferation appears to differ from that of Regnase-1
We have recently shown that Regnase-1 overexpression inhibits the proliferation of Caki-1 cells by down-regulating mRNA coding for DDB1 E3 ligase and increasing both the transcript and protein level of p21Cip1 (Lichawska-Cieslar et al. 2018). To determine whether the same mechanism is responsible for the effects observed following ZC3H12B overexpression in 293 cells, we measured the DDB1 and p21 transcripts levels in 293FlipInTrex cells with the inducible expression of either wild-type ZC3H12B or the D196A mutant. The wild-type- or D196A-ZC3H12B expressing 293FlipInTrex cells were treated with doxycycline for 24 h or left untreated, and the mRNA level was analyzed by qRT-PCR (Fig. 8A). The doxycycline induction of the wild-type ZC3H12B cells results in an ∼50% decrease in the DDB1 transcript level. However, the p21 transcript level is also decreased by almost the same extent, which contradicts the effect observed in Caki-1 cells following Regnase-1 overexpression. The decrease in the levels of both transcripts is due to the effect of ZC3H12B nuclease activity as no difference in the DDB1 or p21 mRNA levels is observed following the ZC3H12B D196A mutein overexpression. The changes in the p21 protein levels approximately correspond to the changes observed in the transcript levels (Fig. 8B). As the mechanism of the inhibition of proliferation by ZC3H12B in 293 cells appears to differ from the mechanism of Regnase-1 we saw in Caki-1 cells, we analyzed the cell cycle in 293FlipInTrex cells with inducible ZC3H12B wild-type or D196A mutant expression. Our results indicate that the overexpression of the wild-type, but not the D196A mutant, stalls the cells in the G2 phase of the cell cycle (Fig. 8C). However, the transcripts involved in the ZC3H12B-induced G2 block remain to be determined.
FIGURE 8.
Mechanism underlying ZC3H12B-induced inhibition of proliferation differs from that of Regnase-1. (A,B) ZC3H12B WT and ZC3H12B D196A Flp-In T-REx-293 cells were treated with doxycycline (1 µg/mL) or left untreated for 24 h, and the levels of the indicated mRNAs (A) and proteins (B) were examined using qRT-PCR and western blotting, respectively. The graphs show the mean results of seven independent experiments ±SD. The data were analyzed using one-way ANOVA with Bonferroni's post-hoc test ([**] P < 0.01; [***] P < 0.001). For the western blot analysis, representative results of three independent experiments are shown. (C) ZC3H12B WT and ZC3H12B D196A Flp-In T-REx-293 cells were treated with doxycycline (1 µg/mL) or left untreated for 72 h, and a cell cycle analysis was performed. The graphs show histogram overlays of propidium iodide (DNA content) fluorescence in noninduced (gray filling) and induced (black line) cells. Representative results of three independent experiments are shown. The table below shows the percentages of cells in each cell cycle phase. The numbers in bold indicate statistically significant differences (P < 0.05) in the percentage of cells in the specified cell cycle phase between the noninduced and induced cells.
Highest expression of ZC3H12B is found in the human brain and the neuroblastoma cell line SH-SY5Y
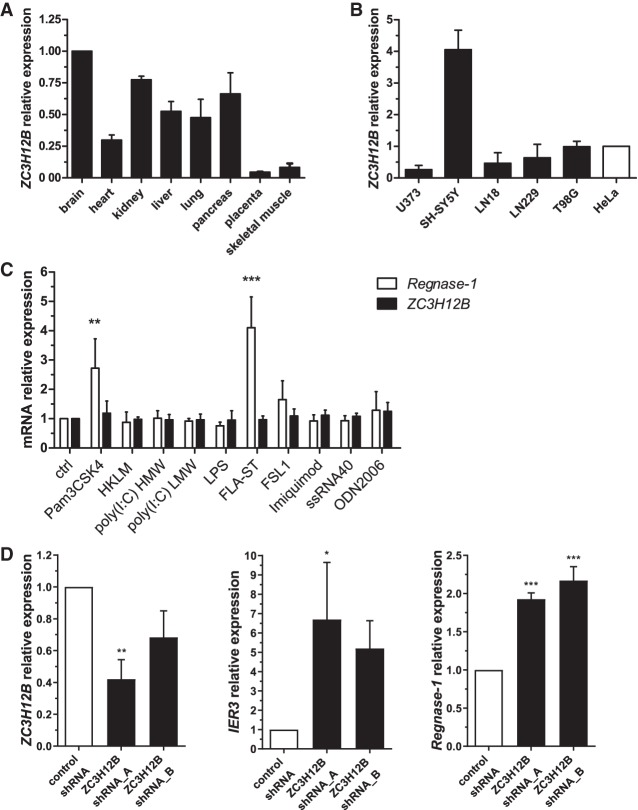
The results shown in this report, together with previous data, indicate that ZC3H12B shares substrates with Regnase-1 and ZC3H12D. Taking into consideration the differences between the phenotypes of knockout mice lacking genes encoding proteins from the ZC3H12 family this overlap may be surprising. While Regnase-1 deficient mice die soon after birth, the Zc3h12d knockout mice develop without visible disorders. No data is available on the viability and development of mice lacking Zc3h12b and Zc3h12c genes, however the big difference in phenotypes of Regnase-1 and Zc3h12d indicates that the function of one protein cannot be substituted by the other. The expression data presented by other groups for mouse tissues indicate that the reason for these discrepancies may be differential expression across tissues of each member of the ZC3H12 family with ZC3H12B being expressed at high levels in the mouse brain. Using the commercially available normalized human tissue cDNA panel (Human MTC panel I, Clontech) we decided to examine whether ZC3H12B mRNA is also abundant in human brain. Figure 9A shows that from the tissues present within the analyzed panel, brain is the one with the highest expression of ZC3H12B. Knowing that, we decided to further analyze the mRNA levels of ZC3H12B in human cell lines of neural origin (HeLa cells served as a reference). From the analyzed cell lines only the SH-SY5Y cells express ZC3H12B at higher levels than the reference HeLa cells (Fig. 9B).
FIGURE 9.
ZC3H12B is expressed at the mRNA level in human brain and the neuroblastoma cell line SH-SY5Y. ZC3H12B mRNA levels were analyzed by qRT-PCR in selected human tissues using the commercially available human tissue cDNA (A) or in human cell lines of neural origin with HeLa cells as reference (B). (C) SH-SY5Y cells were stimulated with TLR ligands: Pam3CSK4 (100 ng/mL), HKLM (107 cells/mL), poly (I:C) HMW (10 µg/mL), poly(I:C) LMW (10 µg/mL), LPS (100 ng/mL), FLA-ST (100 ng/mL), FSL1 (10 ng/mL), Imiquimod (250 ng/mL), ssRNA40 (250 ng/mL), ODN2006 (250 ng/mL) for 24 h, and the levels of the investigated transcripts were measured by qRT-PCR. (D) SH-SY5Y cells were transduced with lentiviruses expressing shRNAs targeting two different sites in ZC3H12B mRNA (shRNA_A and shRNA_B) and control lentiviruses expressing scrambled shRNA. Levels of the investigated transcripts (ZC3H12B, IER3 or Regnase-1) were measured by qRT-PCR. Graphs show mean results from two (A,B) or three (C,D) independent experiments ±SD. For C and D, the data were analyzed using two-way ANOVA with Bonferroni's post-hoc test ([*] P < 0.05; [**] P < 0.01; [***] P < 0.001).
In order to gain more insight into the regulation of ZC3H12B expression we treated SH-SY5Y with proinflammatory agents as we supposed that ZC3H12B being a negative regulator of inflammatory molecules may, similarly to Regnase-1, be induced by inflammation. We treated SH-SY5Y cells with TLR ligands for 24 h and examined the level of ZC3H12B and Regnase-1 mRNAs by qRT-PCR (Fig. 9C). Although we saw a clear induction of Regnase-1 transcript in response to two of the used TLR ligands (Pam3CSK and FLA-ST) we did not observe any increase of ZC3H12B mRNA. Moreover we checked the expression of ZC3H12B in SH-SY5Y cells upon stimulation with TNF and IL-1β and also did not see any increase in ZC3H12B transcript (data not shown). This indicates that ZC3H12B may be regulated by a different set of agents than Regnase-1 but further studies are needed to unveil which stimulants are important for ZC3H12B expression.
Finally, knowing that the highest expression of ZC3H12B is found in SH-SY5Y cells we decided to silence its expression and examine the levels of its mRNA targets. To do this we prepared lentiviruses expressing shRNAs targeting two different sites in ZC3H12B mRNA and control lentiviruses expressing scrambled shRNA, transduced SH-SY5Y cells with the lentiviruses (MOI = 2) and after blasticidin selection checked the levels of ZC3H12B mRNA and its target mRNAs (IER3 and Regnase-1 as SH-SY5Y cells do not express IL-6) using qRT-PCR. As shown on Figure 9D both shRNAs down-regulated ZC3H12B expression on the mRNA level (shRNA_A to 42% and shRNA_B to 68%). The down-regulation of ZC3H12B was accompanied by an increase in the level of IER3 and Regnase-1 mRNAs.
DISCUSSION
The ZC3H12 family of proteins consists of four members. Despite the great interest in the characterization of its founding member, Regnase-1/ZC3H12A/MCPIP1, research investigating the other three members has proceeded at a much more modest level, and ZC3H12B is the most enigmatic member of the entire family. In this paper, for the first time, we show that ZC3H12B is a functional member of the ZC3H12 nuclease family that can bind and destabilize target mRNAs. This finding is particularly important, as two previous reports raise some doubt regarding the functionality of this protein. Suzuki et al. (2011) showed that ZC3H12B lacks the miRNA silencing activity present in Regnase-1, which was attributed to the lack of the proline-rich domain important for this activity. In a second report, Huang et al. (2015) used a luciferase-based system similar to that used in this report. Huang and coworkers investigated the role of MCPIP4 in IL-6 transcript turnover. Using a single assay, the authors also examined the possible influence of ZC3H12B on IL-6 3′UTRs containing luc mRNA; however, for an unknown reason, no effect was observed. The different cell line used by Huang and coworkers likely did not cause the ZC3H12B inactivity because we observed the ZC3H12B effect described in this study in other cell lines (U251-MG and HEK 293, data not shown). Nevertheless, our data clearly indicate that IL-6 mRNA is a direct target of ZC3H12B.
The data presented in this report along with our and other researchers’ previous reports indicate that at least three proteins belonging to the ZC3H12 family (Regnase-1, ZC3H12B, and ZC3H12D) regulate a common set of substrates. The luciferase-based assays prove that all three proteins regulate the transcripts of IL-6, Regnase-1, and IER3. Regnase-1 and ZC3H12D also participate in the regulation of TNF mRNA, which, according to our results, remains ambiguous in the case of ZC3H12B (data not shown). Such an overlap in substrate specificity is common among proteins engaged in RNA decay. For example, all three members of the ZFP36 family (ZFP36/TTP, ZFP36L1, and ZFP36L2) bind and destabilize TNF and GM-CSF transcripts (Baou et al. 2009). However, further characterization of how the reporter system–based results correspond to the specificity of each ZC3H12 family protein in vivo is required. The results obtained from the RIP experiments indicate that ZC3H12B is associated with endogenous IL-6 and IER3 transcripts but is not associated or weakly associated with Regnase-1 transcripts in HeLa cells. However, all three mRNAs showed a similar response to ZC3H12B in the luciferase assays. Moreover, knockdown of ZC3H12B in SH-SY5Y neuroblastoma cells correlates with increased levels of IER3 and Regnase-1 mRNAs. The obtained results suggest that the binding affinity of ZC3H12B to target mRNAs may be affected by some other, cell-specific factors.
Previous reports indicate that two proteins of the ZC3H12 family, that is, Regnase-1 and ZC3H12D, are negative regulators of proliferation. We showed that Regnase-1 inhibits growth and viability in the clear cell carcinoma cell line Caki-1, which correlates with the up-regulation of the transcript and protein levels of p21. The elevated levels of p21 in the Regnase-1-overexpressing cells were correlated with the diminished expression level of the DDB1 transcript encoding the component of the E3 ligase that negatively regulates p21 (Lichawska-Cieslar et al. 2018). We have also shown that Regnase-1 overexpression decreases the viability of HeLa, but not HepG2, cells (Skalniak et al. 2013). Simultaneously, the low level of Regnase-1 is correlated with increased proliferation, tumor outgrowth, and vascularity in patient-derived tumors and xenografts established in NOD-SCID and nude mice (Marona et al. 2017). Moreover, Regnase-1 is implicated in the destabilization of antiapoptotic transcripts, including those of Bcl2L1, Bcl2A1, RelB, Birc3, and Bcl3, and acts as a tumor suppressor in breast cancer (Lu et al. 2016). ZC3H12D is also a negative regulator of proliferation and was first discovered as a tumor suppressor that inhibits tumor growth both in vivo and in vitro (Wang et al. 2007). ZC3H12D was found to inhibit G1 to S phase progression via suppression of the phosphorylation of retinoblastoma protein (Rb) (Minagawa et al. 2009). Here, for the first time, we show that similar to Regnase-1 and ZC3H12D, ZC3H12B acts as a negative regulator of proliferation, and this regulation is NYN/PIN-dependent. However, the mechanism of the effect of ZC3H12B on cell proliferation seems to differ from that of Regnase-1 and ZC3H12D. Although we observed a decrease in the DDB1 transcript level following ZC3H12B overexpression, this decrease was accompanied by a similar decrease in the p21 mRNA level, which contradicts the effects observed after Regnase-1 overexpression. The effects of ZC3H12B also differ from the effects of ZC3H12D. The cell cycle analysis presented in this report indicates that ZC3H12B causes cells to stall in the G2 phase, while previous reports regarding the mode of action of ZC3H12D indicate that ZC3H12D blocks G1 to S phase progression. Our data indicate that the mechanism by which ZC3H12B inhibits proliferation may be unique among the ZC3H12 family, but the determination of the ZC3H12B mRNA substrates that are associated with this effect requires further investigation.
Our data reveal the role of ZC3H12B in the regulation of the course of inflammation. We have confirmed its involvement in the control of the level of transcripts, which are known targets of Regnase-1 and ZC3H12D. The possible explanation of the overlapping substrate specificity of the members of the ZC3H12 protein family could be a different pattern of their tissue expression. ZC3H12B is expressed mainly in the brain whereas the highest level of Regnase-1 is observed in the bone marrow, and ZC3H12D in the appendix, lymph node, and spleen. ZC3H12C seems to be present in most tissues at quite a similar level. Moreover, the regulation of expression of the members of the ZC3H12 protein family seems to differ: Regnase-1 and ZC3H12C are induced by proinflammatory factors, as well as ZC3H12D although with a different time-course. The only as yet proinflammatory factor noninducible gene from this family is ZC3H12B. Interesting is also the phenomenon of the regulation of Regnase-1 level by other members of the ZC3H12 family. All proteins from this family (including Regnase-1 itself) are able to degrade Regnase-1 transcript (Iwasaki et al. 2011; Wawro et al. 2016, 2017; M Wawro, A Solecka, W Sowinska, et al., unpubl.). In the present study, we have shown that ZC3H12B interacts with Regnase-1 3′UTR although we could not confirm its direct interaction with Regnase-1 mRNA using RIP. However, we have found that in human primary astrocytes down-regulation of ZC3H12B results in the increased level of Regnase-1 mRNA (A Kasza, W Sowinska, DD Biswass, et al., unpubl.). Despite intensive studies concentrated mainly on Regnase-1, a comprehensive picture of the role of proteins from ZC3H12 family requires further studies, which would include the crosstalk of the proteins from this family and the mechanisms of the regulation of their expression.
MATERIALS AND METHODS
Cell culture
The HeLa cell line was cultured in Dulbecco's modified Eagle's medium (DMEM) with 1.0 g/L d-glucose (Lonza) supplemented with 2 mM l-glutamine (Sigma-Aldrich) and 10% fetal bovine serum (Biowest). 293T, SH-SY5Y, T98G, LN18, LN229, and U251-MG cell lines were maintained in DMEM with 4.5 g/L d-glucose (Lonza) supplemented with 10% fetal bovine serum. The Flp-In T-REx-293 cell line (Invitrogen) was cultured in DMEM with 4.5 g/L d-glucose supplemented with 10% tetracycline-free fetal bovine serum (Biowest), 100 µg/mL zeocin (Invitrogen) and 15 µg/mL blasticidin S (Invivogen). In stimulation experiments for qRT-PCR the cells were grown in reduced serum medium (0.5% FBS) 24 h prior to stimulation. Cell lines were maintained at 37°C in a humidified atmosphere with 5% CO2. All cell cultures were examined on a regular basis for mycoplasma contamination using PCR (van Kuppeveld et al. 1993).
Plasmid construction
Human ZC3H12B (NM_001010888.3) CDS was amplified with the following primers: eZC3H12B-nTC-for (CACCACGGCCACAGCTGAGGTAG) and eZC3H12B-nTC-rev (TCAACGTGCAGCCCTAAGCTTAG) using the PfuUltra II Fusion HS polymerase (Agilent) with human cDNA mix as a template. The appropriate PCR product was cloned into the entry vector pENTR/D-TOPO using the pENTR/D-TOPO Cloning Kit (Invitorgen). The insert from the selected vector was subsequently subcloned into the destination vector pcDNA6.2/nTC-Tag-DEST (Invitrogen) using the Gateway LR Clonase II enzyme mix obtaining the destination vector for expression of N-terminally tagged (TC and V5 tags) ZC3H12B (pcDNA6.2-V5-ZC3H12B). pcDNA6.2/nTC-Tag-DEST–based human Regnase-1 (NM_025079.2) expression vector was prepared similarly using the following primers: MCPIP1-nTC-Tag-V5_FOR (CACCAGTGGCCCCTGTGGAGAGAAG) and MCPIP1-nTC-Tag-V5_REV (TTACTCACTGGGGTGCTGGGAC) and the previously described pcDNA3.1/myc-His A-ZC3H12A construct as template (Kochan et al. 2015). The ZC3H12B D196A mutein expression vector (pcDNA6.2-V5-ZC3H12B-D196A) was prepared by whole plasmid site directed-mutagenesis using the PfuUltra II Fusion HS polymerase, the pcDNA6.2-V5-ZC3H12B vector as template and the following primers: eMCPIP2_D196As (GAGGCCAGTTGTCATTGCTGGAAGTAATGTGGCAA) and eMCPIP2_ D196Aas (TTGCCACATTACTTCCAGCAATGACAACTGGCCTC). The pcDNA5/FRT/TO-ZC3H12B and pcDNA5/FRT/TO-ZC3H12B-D196A vectors used for the generation of Flp-In T-REx-293 cell lines with inducible expression of ZC312B wild-type or D196A mutein were prepared by traditional cloning techniques. Firstly, PCR products were obtained on pcDNA6.2-V5-ZC3H12B and pcDNA6.2-V5-ZC3H12B-D196A templates using the PfuUltra II Fusion HS polymerase and the following primers: TetKpnNTCV5M2for (ATGGTACCCACCATGGCTGGTGGCTGTTGTC) and TetXhoNTCV5M2rev (ATTACTCGAGTCAACGTGCAGCCCTAAGCTTAGC). The PCR products were digested with KpnI and XhoI (New England Biolabs) and inserted into the pcDNA5/FRT/TO vector (Invitrogen) linearized with the same enzymes using T4 DNA ligase (New England Biolabs). Construct for doxycycline inducible ZC3H12B expression in the SB transposon system (pSBtet-GP-ZC3H12B) was prepared through substitution of the luciferase CDS with ZC3H12B CDS by ligation of SfiI (EurX) digested PCR product into SfiI linearized pSBtet-GP vector. The PCR product was obtained on the template of pcDNA5/FRT/TO-SF-ZC3H12B using the following primers: SFMCPIP2SfiFor (AGGCCTCTGAGGCCACCCACCATGGCAAGCTGGAGC) and SFMCPIP2SfiRev (AGGCCTGACAGGCCTCAACGTGCAGCCCTAAGC). pcDNA5/FRT/TO-SF-ZC3H12B was prepared by substitution of the V5-TC tag present in the original pcDNA5/FRT/TO-ZC3H12B vector by StrepTagII-Flag tag. Briefly, the V5-TC tag was excised with KpnI and NotI and the STreptagII-Flag tag was inserted by ligation of two phosphorylated and reannealed oligos (StrepFlagM2s: CCACCATGGCAAGCTGGAGCCACCCGCAGTTCGAAAAGGGTGCAGACTACAAAGACGATGACGACAAGCTTGC; StrepFlagM2s: GGCCGCAAGCTTGTCGTCATCGTCTTTGTAGTCTGCACCCTTTTCGAACTGCGGGTGGCTCCAGCTTGCCAT GGTGGGTAC spSBtet-GP was a gift from Eric Kowarz (Addgene plasmid #60495) (Kowarz et al. 2015). The luciferase constructs (pmirGLO_IL-6-3′UTR, pmirGLO_IL-6-3′UTRΔCE, pmirGLO_ZC3H12A-3′UTR, pmirGLO_VEGFA-3′UTR, and pmirGLO_IER3-3′UTR) were described previously (Kochan et al. 2016; Wawro et al. 2016, 2017). The sequences of all used constructs were verified by Sanger sequencing (Genomed) and the expression of the recombinant proteins was confirmed by western blotting analysis.
Cell transfections
HeLa cells were transfected with plasmid DNA using PEI MAX 40K (Polysciences) with a 3:1 (w:w) ratio. Concisely, appropriate amounts of DNA and PEI MAX 40K were diluted in Opti-MEM medium (Gibco), vortexed and centrifuged briefly. The Opti-MEM diluted PEI MAX 40K was transferred to the diluted DNA, the solutions were mixed by vortexing, centrifuged briefly and incubated at room temperature for 20 min. After incubation, the solution was applied onto the cells dropwise and the cells were incubated with the transfection solution for 4 h when the solution was removed and replaced with fresh culture medium. 293T cells were transfected using calcium phosphate precipitation. In brief, the appropriate amount of DNA was diluted in sterile water and 2.5 M CaCl2 solution was added to obtain a final concentration of 244 mM. Then an equal volume of 2xHBS (140 mM NaCl,1.5 mM Na2HPO4, 50 mM HEPES, pH 7.05) was added, the solutions were mixed quickly by pipetting and applied onto the cells dropwise within 1 min. The cells were incubated with the DNA/phosphate transfection precipitates for 6 h when the solution was removed and replaced with fresh culture medium. The Flp-In T-REx-293 cell line was transfected using Lipofectamine 2000 reagent (Invitrogen) according to the protocol provided by the manufacturer.
Luciferase assays
Transfection of HeLa cells was performed in 24-well plates. Total amount of DNA used per well was 0.8 µg, including 0.4 µg of pmirGLO dual luciferase expression vector (pmirGLO_IL-6-3′UTR, pmirGLO_IL-6-3′UTRΔCE, pmirGLO_Reg-1-3′UTR, pmirGLO_ VEGFA-3′UTR, pmirGLO_IER3-3′UTR, or empty pmirGLO) and varying amounts of Regnase-1 or ZC3H12B expression vectors (wild-type or aspartic acid to alanine mutants, exact amounts are indicated in figure legends). The amount of DNA per well was equalized using mock plasmid DNA (pcDNA3). Twenty-four hours after transfection the cells were lysed and assayed for firefly and Renilla luciferase activity using the Dual-Luciferase Reporter Assay System (Promega). Renilla luciferase served as internal control.
Luciferase mRNA quantitation and stability assays
Hela cells were seeded in 6-well plates. The following day the cells were transfected with 1.0 µg of pmirGLO_IL-6-3′UTR vector and varying amounts (200 and 400 ng for luc mRNA quantitation or 400 ng for luc mRNA stability) of pcDNA6.2-V5-ZC3H12B vector or empty control plasmid. The amount of DNA per well was equalized using mock plasmid DNA (pcDNA3) to a total of 2.0 µg per well. Twenty-four hours posttransfection the cells were trypsynized, and split into two fractions. For the luc mRNA quantitation assay one fraction (1/5th of the cells) was used for luciferase activity assay and the second fraction (the rest of the cells) was used for mRNA isolation using the Dynabeads mRNA DIRECT Purification Kit (Thermo Fisher) according to the manufacturer's instructions. For luc mRNA stability assay cells after transfection were seeded into 12-well plates and treated with actinomycin D (Sigma-Aldrich) for 30, 60, and 90 min or left untreated and mRNA isolation was performed as for the luc mRNA quantitation assay. After mRNA isolation, the mRNA was reverse-transcribed and the levels of firefly and Renilla luciferase transcripts was measured by qRT-PCR with the following primers: qRT-luc-FOR (TTCGACCGGGACAAAACCAT), qRT-luc-REV (ATCTGGTTGCCGAAGATGGG), qRT-hRluc-for (GTACCTCACCGCTTGGTTCG), qRT-hRluc-rev (TCGTCCCAGGACTCGATCAC).
Establishment of HeLa cell lines with doxycycline inducible expression of ZC3H12B
HeLa cells were seeded in 12-well plates and the following day the cells were transfected using PEI MAX with 900 ng of pSBtet-GP-ZC3H12B, pSBtet-GP-ZC3H12B-D196A, or pSBtet-GP vectors and 100 ng of the pCMV(CAT)T7-SB100 vector. Twenty-four hours later the cells were trypsinized, transferred into 6-well plates and cells with transposon integration were selected with puromycin (1 µg/mL, Invivogen) for 1 wk. pCMV(CAT)T7-SB100 was a gift from Zsuzsanna Izsvak (Addgene plasmid #34879) (Mátés et al. 2009).
IL-6 ELISA
HeLa cells with stable, inducible expression of wild-type ZC3H12B or luciferase (control) were seeded into 12-well plates. The following day the cells were pretreteated with 1 µg/mL doxycycline for induction of transgene expression and then stimulated with IL-1β. After 24 h the media were collected and IL-6 protein levels were measured using the Human IL-6 DuoSet ELISA kit (R&D Systems) according to the manufacturer's protocol.
Establishment of Flp-In T-REx doxycycline inducible cell lines
Flp-In T-REx-293 cells were seeded in 12-well plates and the following day the cells were transfected using Lipofectamine 2000 with 180 ng of pcDNA5/FRT/TO-ZC3H12B or pcDNA5/FRT/TO-ZC3H12B-D196A vectors and 1.42 µg of the pOG44 plasmid (Invitrogen). Twenty-four hours later the cells were trypsinized and split into 100-mm cell culture dishes in selection medium [DMEM with 4.5 g/L d-glucose supplemented with 10% tetracycline-free fetal bovine serum, 15 µg/mL blasticidin S and 100 µg/mL hygromycin B (Invivogen)]. The cells were left until visible formation of colonies. The colonies were transferred into 96-well plates, propagated and examined for expression of V5 tagged ZC3H12B after 24 h of induction with 1 µg/mL of doxycyline (Sigma-Aldrich). Single clones with similar level of induction of the ZC3H12B wild-type and D196A mutation bearing proteins were selected and used in further experiments. Additionally the selected clones were checked for zeocin sensitivity to confirm proper integration.
293T cell count assay
293T cells were seeded in 24-well plates (50,000 cells/well) and the following day the cells were transfected using the calcium phosphate method with 0.91 µg of appropriate plasmid DNA (pcDNA6.2-V5-ZC3H12B, pcDNA6.2-V5-ZC3H12B-D196A, empty control plasmid [pcDNA3]) or left untreated. The cells were trypsynized and counted every 24 h posttransfection up to 72 h.
Cell proliferation assay
Cell proliferation was examined using the CFSE Cell Division Tracker Kit (Biolegend) according to the manufacturer instructions. Briefly, Flp-In T-REx-293-ZC3H12B and Flp-In T-REx-293-ZC3H12B-D196A cells were stained with 5 µM CFSE and the following day the cells were treated with 1 µg/mL doxycycline for induction of recombinant protein expression. After 72 h of induction, the cells (100,000 of FSC/SSC gated cells/sample) were analyzed for CFSE fluorescence intensity by flow cytometry using the BD FACSCalibur cytometer (Becton Dickinson). The BD CellQuest Pro software (Becton Dickinson) was used for data acquisition and FlowJo (v10.4.2, FlowJo) was used for data analysis.
LDH cytotoxicity assay
Cell cytotoxicity was examined using the LDH Cytotoxicity Detection Kit (Clontech) according to the manufacturer's instructions. Briefly, Flp-In T-REx-293-ZC3H12B and Flp-In T-REx-293-ZC3H12B-D196A cells were seeded into 24-well plates and treated in duplicates with doxycycline for 24, 48, and 72 h or left untreated. At each timepoint the media were collected and the cells were lysed with 1% Triton-X-100 for normalization for cell number. After collection the LDH level was measured and normalized to the LDH level after Triton-X-100 treatment. Absorbance measurement was performed using Synergy H1 hybrid reader with Gene5 Software (BIOTEK Instruments).
Cell cycle analysis
Flp-In T-REx-293-ZC3H12B and Flp-In T-REx-293-ZC3H12B-D196A were treated with 1μg/ml doxycycline for induction of recombinant protein expression for 72 h. Then, the cells were trypsynized, washed with PBS two times, resuspended in 2 mL of PBS and fixed by addition of 6 mL of ice-cold 96% ethanol. After fixation the cells were washed two times with PBS and stained with PI/SAP solute ion (0.05 mg/mL propidium iodide, 1 mg/mL RNase A, 0.02% saponin, 0.0001% Triton-X-100 in PBS) by incubation for 15 min at 37°C. Immediately after staining, the cells (200,000 of FSC/SSC gated cells/sample) were analyzed for propidium iodide fluorescence intensity by flow cytometry using the BD FACSCalibur cytometer (Becton Dickinson). BD CellQuest Pro software (Becton Dickinson) was used for data acquisition. The percentage of cells in each cycle was calculated with the FlowJo (v10.4.2, FlowJo) Cell Cycle analysis platform using the Watson (pragmatic) model.
IL-6 mRNA stability assay
HeLa cells were seeded in 100-mm cell culture dishes and the following day the cells were transfected with 9.28 µg of pMACS Kk.II (Miltenyi Biotec) and 13.92 µg of appropriate expression vectors (pcDNA6.2-V5-ZC3H12B-WT/ pcDNA6.2-V5-ZC3H12B-D196A or empty control vector [pcDNA3]). The day after transfection the cells were trypsinized and the transfected cells were enriched using the MACSelect Kk – Transfected Cell Selection Kit (Miltenyi Biotec) and split into three wells of 12-well cell culture plate. The following day cells were treated with 10 µg/mL of actinomycin D (Sigma-Aldrich), lysed at the indicated time points and total RNA isolation was performed.
RNA isolation, DNase digestion, and reverse transcription
Total RNA isolation was performed according to Chomczynski's modified protocol (Chomczynski and Sacchi 2006). RNA quantification and purity were assessed using spectrophotometric measurements using NanoDrop ND-1000 spectrophotometer (Thermo Scientific). RNA integrity was assessed by denaturing, formaldehyde gel electrophoresis. 1.0 µg of RNA was used for DNase digestion with RQ1 RNase-free DNase (Promega) according to the manufacturer's instructions. After DNase treatment, reverse-transcription reaction was performed with M-MLV-Reverse transcriptase (Promega) and 500 ng of oligo(dT) primers (or random hexamers for RNA immunoprecipitation procedure) according to the manufacturer's instructions. The cDNA was used in quantitative real-time PCR (qRT-PCR) for the evaluation of the amounts of the mRNAs of interest.
Quantitative real-time PCR (qRT-PCR)
qRT-PCR was performed using RT-HS-PCR-Mix-SYBR-A (A&A Biotechnology). Levels of the mRNAs of interest in each sample were analyzed in duplicates and the expression level was normalized to EEF2. Expression levels were analyzed by the ΔΔCt method. The following primers were used: qRT-EEF2-for (GACATCACCAAGGGTGTGCAG), qRT-EEF2-rev (TTCAGCACACTGGCATAGAGGC), qRT-IL-6-for (GACAGCCACTCACCTCTTCA), qRT-IL-6-rev (AGTGCCTCTTTGCTGCTTTC), qRT-DDB1-for (TGCCTACAAGCCCATGGAAG), qRT-DDB1-rev (TGGCAGCGCTATCCTTTTGA), qRT-p21-for (CATGACAGATTTCTACCACTCCA), qRT-p21-rev (GGCCAGGGTATGTACATGAGG), qRT-MCPIP2-2-for (CCTCGTGGGCCCAGCTCCAG), qRT-MCPIP2-2-rev (TCCCATGGCTCATTGCCACATTACT). Statistical analysis of qRT-PCR obtained results was performed on the logarithm-transformed relative quantification (RQ) values (Rieu and Powers 2009).
Western blot analysis
HeLa cells were transfected with V5-ZC3H12B wild-type or aspartic acid to alanine mutant expression vectors or empty control vector (pcDNA3) in 12-well plates. Twenty-four hours after transfection the cells were lysed in RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS with cOmplete ULTRA protease inhibitors [Roche] and 1 mM PMSF [Sigma-Aldrich]) and 20–30 µg of protein extracts were analyzed by western blot. For the doxycycline inducible cell lines, the cells were seeded in 12-well plates and the following day the cells were treated with 1 µg/mL doxycycline for recombinant protein expression. After induction the cells were lysed and analyzed as described above for HeLa cells. The following antibodies were used: anti-V5 tag (R96025, Thermo Scientific), anti-p21 Waf1/Cip1 (12D1, Cell Signaling Technology), anti-β-actin, anti-β-tubulin, anti-mouse-HRP and anti-rabbit HRP (Cell Signaling). Luminescence was detected using Clarity Western ECL Substrate (Bio-Rad) and recorded using the Fusion-Fx documentation system (Vilber Lourmat).
Immunofluorescence
HeLa cells were plated on glass coverslips in 12-well culture plates. The following day the cells were transfected with plasmid encoding V5-ZC3H12B (pcDNA6.2-V5-ZC3H12B). Twenty-four hours post-transfection the cells were fixed for 15 min at −20°C in ice-cold 100% methanol. After fixation the cells were washed three times with PBS, blocked and permeabilized for 60 min at room temperature in blocking buffer (5% fetal bovine serum, 0.3% Triton X-100 in PBS). Directly after blocking, cells were incubated overnight in a humidified chamber , in the dark, at 4°C with anti-V5 tag antibodies (R96025, Thermo Scientific) diluted in blocking buffer. The following day, after washing with PBS the cells were incubated for 90 min, in the dark, at room temperature with Alexa Fluor 488-conjugated donkey anti-mouse antibody (715-546-150, Jackson ImmunoResearch) diluted in blocking buffer. After incubation the cells were washed three times with PBS and the samples were mounted onto slides in VECTASHIELD Mounting Medium with DAPI (Vector Laboratories). After sealing with nail polish, the samples were imaged using Leica DMI6000B (AF7000 version) inverted widefield fluorescence microscope (Leica Microsystems). All images were acquired using a high numerical aperture 100× oil immersion objective (HCX PL APO 100.0 × 1.47 oil; Leica Microsystems) and a 14-bit Hamamatsu 9100-02 EM-CCD High Speed Set cooled CCD camera (Hamamatsu Photonics) with Leica LAS X image acquisition software. The following filter sets (Leica Microsystems) were used: A4 for detection of DAPI and GFP-T ET for detection of Alexa Fluor 488 Dye. After deconvolution from about 60 z-sections with 0.15–0.20 µm spacing, images were deconvolved using Huygens Professional Software (Scientific Volume Imaging). Final image adjustments were performed using ImageJ 1.48v (National Institutes of Health), Adobe Photoshop CS4 Extended Version 11.0.2, and Adobe Illustrator CS4 Version 14.0.0 (Adobe Systems).
RNA immunoprecipitation (RIP)
RIP was performed as described previously (Kochan et al. 2016; Wawro et al. 2017). HeLa cells were seeded on 150-mm cell culture dishes and the following day the cells were transfected with plasmid encoding V5-ZC3H12B-D196A mutein (pcDNA6.2-V5-ZC3H12B-D196A) or empty vector (pcDNA3.1/MycHisA). Twenty-four hours later the cells were washed twice with PBS, covered with 1 mL of PBS per dish and the RNA–protein complexes were crosslinked by UV-irradiation of the cells (150 mJ/cm2) at 254 nm (Hoefer UVC 500 Ultraviolet Crosslinker, Hoefer). Cells were scraped in 1 mL of PBS, transferred to an eppendorf tube and centrifuged at 300g for 5 min at 4°C. Then, PBS was removed and the cell pellets were lysed in 1 mL of RIP lysis buffer (10 mM HEPES pH 7.5, 10 mM KCl, 1 mM DTT, 0.5% NP-40) supplemented with 1 mM PMSF (Sigma-Aldrich), 1× cOmplete protease inhibitors (Roche) and 1 U/µL of murine RNase inhibitor (New England Biolabs), incubated for 5 min on ice and then cell nuclei was sedimented by centrifugation for 5 min at 300g at 4°C. After centrifugation the supernatant containing the cytoplasmic fraction was transferred to a fresh eppendrof tube and used for downstream processing. Four hundred microliters of the samples were used for immunoprecipitation and 100 µL were frozen as inputs. Before addition of antibodies each sample was diluted 1:1 with RIP dilution buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 2 mM EDTA and 2% Triton X-100 supplemented with inhibitors as described above). mRNA–protein complexes were immunoprecipitated overnight at 4°C with gentle rotation using 4 µg of anti-V5 antibody (R96025, Thermo Scientific) or 4 µg of normal mouse IgG (Thermo Scientific) used as a control of specificity. The following day, the antibody captured protein–RNA complexes were recovered by incubation for 60 min at 4°C with Dynabeads Protein G (Novex Thermo Scientific) magnetic beads. After incubation the beads were washed with 1 mL of each of the following buffers in the presented order: RIP low salt wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.25% sodium deoxycholate), RIP high salt wash buffer 1 (20 mM HEPES pH 7.5, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.25% sodium deoxycholate), RIP high salt wash buffer 2 (20 mM HEPES pH 7.5, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.01% SDS) and TE buffer (10 mM Tris ph 8.0, 1 mM EDTA). Each wash was performed at room temperature for 5 min and the buffers were removed from the beads collected on a magnet (Dynal MPC-S, Thermo Scientific). Finally, the beads were resuspended in Proteinase K digestion buffer (10 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% SDS) and treated with 200 µg/mL Proteinase K (New England Biolabs) for 1 h at 50°C. Input samples were diluted 1:4 with Proteinase K digestion buffer and similarly treated with Proteinase K. Following digestion, RNA was isolated by phenol–chloroform extraction and precipitated with isopropanol and Pellet Paint Co-Precipitant (Novagen, Merck Millipore) overnight at −20°C. The RNA pellets were dissolved in nuclease-free water (Sigma-Aldrich), reverse transcribed—for input samples 1/7th of the obtained RNA was used—and analyzed by qRT-PCR. The following primers were used (primers for EEF2 and IL-6 were already described in the qRT-PCR section): RIP-ZC3H12Afor (GGTAACGGCGTCGGTCCGTG), RIP-ZC3H12Arev (CGGTGCCCAACTAGCCAGCC), RIP-IER3for (AATGAGATCCGTGAGATCCTTCC), RIP-IER3rev (TTTGTGTTCACAGAACATACTAGGC). The relative amounts of each transcript was calculated using the ΔΔCt normalized to input samples.
Lentivirus production and titration
The lentiviral vectors for silencing of ZC3H12B were constructed based on the LeGO-G/BSD vector. Oligos for the expression of shRNAs targeting ZC3H12B (two different sites were used; shRNA_Asense: AACTGCCTAGATCGTCCAAGTTTCCTCGAGGAAACTTGGACGATCTAGGCATTTTTC; shRNA_Aantisense: TCGAGAAAAATGCCTAGATCGTCCAAGTTTCCTCGAGGAAACTTGGACGATCTAGGCAGTT; shRNA_Bsense: AACTGTCCACCCTTTGGGTAATAACTCGAGTTATTACCCAAAGGGTGGACATTTTTC; shRNA_Bantisense: TCGAGAAAAATGTCCACCCTTTGGGTAATAACTCGAGTTATTACCCAAAGGGTGGACAGTT;) or scrambled shRNA (shRNA_SCsense: AACCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTTC; shRNA_ SCantisense: TCGAGAAAAACAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGGTT) were phosphorylated with T4 PNK, annealed and inserted into HpaI and XhoI digested LeGO-G/BSD vector. Vectors containing the desired inserts were verified by Sanger sequencing. 1.64 pmol of the obtained vectors were transfected together with 1.3 pmol of psPAX and 0.72 pmol of pMD2.G plasmids using PEI MAX into 293T cells seeded 24 h earlier into 100-mm dishes. Twenty-four hours posttransfection the medium was exchanged and the cells were left for 48 h allowing for lentivirus production. After 48 h the lentivirus-containing media were collected and the lentivirus was concentrated on a sucrose gradient as described by Jiang et al. (2015). After concentration the virus pellets were resuspended in OptiMEM medium, aliquoted, snap-frozen in liquid nitrogen and stored at −80°C. The titer of the obtained virus was estimated by transduction of the SH-SY5Y cell line with serial dilutions of the obtained virus preparations followed by the analysis of the number of GFP-expressing cells after transduction using flow cytometry. LeGO-G/BSD was a gift from Boris Fehse (Addgene plasmid #27354). psPAX2 and pM2D.G were a gift from Didier Trono (Addgene plasmid #12260 and #12259, respectively).
Lentiviral transduction
SH-SY5Y cells were seeded into 24-well plates and 24 h later transduced with lentiviruses (MOI = 2) allowing for expression of shRNAs targeting ZC3H12B mRNA or scrambled shRNA in the presence of 5 µg/mL polybrene (Merck-Millipore). Forty-eight hours posttransduction the cells were trypsinized and seeded into 12-well plates in selection medium containing 15 µg/mL blasticidin S. After a week of selection, the cells were analyzed for expression of selected genes by qRT-PCR.
Statistics
Statistical analysis was performed using Graph Pad Prism software (version 5.01, GraphPad Software Inc.). If not specified differently the asterisks denote statistical significance of the indicated data point versus the control sample. The exact tests performed are indicated in figure descriptions.
ACKNOWLEDGMENTS
This work was funded by the National Science Centre, Poland (NCN, OPUS8 Funding scheme, project number UMO-2014/15/B/NZ2/03379 to A.K.) and the Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University in Kraków (Grant for Young Researchers number BMN 23/2018 to M.W.). The Faculty of Biochemistry, Biophysics, and Biotechnology of Jagiellonian University is a partner of the Leading National Research Center (KNOW) supported by the Ministry of Science and Higher Education.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.071381.119.
REFERENCES
- Anderson P. 2008. Post-transcriptional control of cytokine production. Nat Immunol 9: 353–359. 10.1038/ni1584 [DOI] [PubMed] [Google Scholar]
- Baou M, Jewell A, Murphy JJ. 2009. TIS11 family proteins and their roles in posttranscriptional gene regulation. J Biomed Biotechnol 2009: 634520 10.1155/2009/634520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR. 2014. mRNP granules. RNA Biol 11: 1019–1030. 10.4161/15476286.2014.972208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA. 2014. Post-transcriptional regulation of gene expression in innate immunity. Nat Rev Immunol 14: 361–376. 10.1038/nri3682 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. 2006. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc 1: 581–585. 10.1038/nprot.2006.83 [DOI] [PubMed] [Google Scholar]
- García-Mauriño SM, Rivero-Rodríguez F, Velázquez-Cruz A, Hernández-Vellisca M, Díaz-Quintana A, De la Rosa MA, Díaz-Moreno I. 2017. RNA binding protein regulation and cross-talk in the control of AU-rich mRNA fate. Front Mol Biosci 4: 71 10.3389/fmolb.2017.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Baltimore D. 2009. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol 10: 281–288. 10.1038/ni.1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Qi D, Liang J, Miao R, Minagawa K, Quinn T, Matsui T, Fan D, Liu J, Fu M. 2012. The putative tumor suppressor Zc3h12d modulates toll-like receptor signaling in macrophages. Cell Signal 24: 569–576. 10.1016/j.cellsig.2011.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Liu S, Fu JJ, Wang TT, Yao X, Kumar A, Liu G, Fu M. 2015. Monocyte chemotactic protein-induced protein 1 and 4 form a complex but act independently in regulation of interleukin-6 mRNA degradation. J Biol Chem 290: 20782–20792. 10.1074/jbc.M114.635870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Takeuchi O, Teraguchi S, Matsushita K, Uehata T, Kuniyoshi K, Satoh T, Saitoh T, Matsushita M, Standley DM, et al. 2011. The IκB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR–IL-1R by controlling degradation of regnase-1. Nat Immunol 12: 1167–1175. 10.1038/ni.2137 [DOI] [PubMed] [Google Scholar]
- Jiang W, Hua R, Wei M, Li C, Qiu Z, Yang X, Zhang C. 2015. An optimized method for high-titer lentivirus preparations without ultracentrifugation. Sci Rep 5: 13875 10.1038/srep13875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochan J, Wawro M, Kasza A. 2015. Simultaneous detection of mRNA and protein in single cells using immunofluorescence-combined single-molecule RNA FISH. Biotechniques 59: 209–212. 10.2144/000114340 [DOI] [PubMed] [Google Scholar]
- Kochan J, Wawro M, Kasza A. 2016. IF-combined smRNA FISH reveals interaction of MCPIP1 protein with IER3 mRNA. Biol Open 5: 889–898. 10.1242/bio.018010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowarz E, Löscher D, Marschalek R. 2015. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol J 10: 647–653. 10.1002/biot.201400821 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Liang J, Wang J, Azfer A, Song W, Tromp G, Kolattukudy PE, Fu M. 2008. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J Biol Chem 283: 6337–6346. 10.1074/jbc.M707861200 [DOI] [PubMed] [Google Scholar]
- Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, Kolattukudy PE, Fu M. 2010. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-κB signaling. J Exp Med 207: 2959–2973. 10.1084/jem.20092641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichawska-Cieslar A, Pietrzycka R, Ligeza J, Kulecka M, Paziewska A, Kalita A, Dolicka DD, Wilamowski M, Miekus K, Ostrowski J, et al. 2018. RNA sequencing reveals widespread transcriptome changes in a renal carcinoma cell line. Oncotarget 9: 8597–8613. 10.18632/oncotarget.24269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Chien HL, Lin SY, Chang BL, Yu HP, Tang WC, Lin YL. 2013. MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation. Nucleic Acids Res 41: 3314–3326. 10.1093/nar/gkt019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R-J, Chu J-S, Chien H-L, Tseng C-H, Ko P-C, Mei Y-Y, Tang W-C, Kao Y-T, Cheng H-Y, Liang Y-C, et al. 2014. MCPIP1 suppresses hepatitis C virus replication and negatively regulates virus-induced proinflammatory cytokine responses. J Immunol 193: 4159–4168. 10.4049/jimmunol.1400337 [DOI] [PubMed] [Google Scholar]
- Liu L, Zhou Z, Huang S, Guo Y, Fan Y, Zhang J, Zhang J, Fu M, Chen YE. 2013. Zc3h12c inhibits vascular inflammation by repressing NF-κB activation and pro-inflammatory gene expression in endothelial cells. Biochem J 451: 55–60. 10.1042/BJ20130019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Ning H, Gu L, Peng H, Wang Q, Hou R, Fu M, Hoft DF, Liu J. 2016. MCPIP1 selectively destabilizes transcripts associated with an antiapoptotic gene expression program in breast cancer cells that can elicit complete tumor regression. Cancer Res 76: 1429–1440. 10.1158/0008-5472.CAN-15-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Jensen TH. 2015. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol 16: 665–677. 10.1038/nrm4063 [DOI] [PubMed] [Google Scholar]
- Marona P, Górka J, Mazurek Z, Wilk W, Rys J, Majka M, Jura J, Miekus K. 2017. MCPIP1 downregulation in clear cell renal cell carcinoma promotes vascularization and metastatic progression. Cancer Res 77: 4905–4920. 10.1158/1538-7445.AM2017-4905 [DOI] [PubMed] [Google Scholar]
- Mátés L, Chuah MKL, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, et al. 2009. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet 41: 753–761. 10.1038/ng.343 [DOI] [PubMed] [Google Scholar]
- Minagawa K, Katayama Y, Nishikawa S, Yamamoto K, Sada A, Okamura A, Shimoyama M, Matsui T. 2009. Inhibition of G1 to S phase progression by a novel zinc finger protein P58TFL at P-bodies. Mol Cancer Res 7: 880–889. 10.1158/1541-7786.MCR-08-0511 [DOI] [PubMed] [Google Scholar]
- Mino T, Murakawa Y, Fukao A, Vandenbon A, Wessels H-H, Ori D, Uehata T, Tartey S, Akira S, Suzuki Y, et al. 2015. Regnase-1 and Roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell 161: 1058–1073. 10.1016/j.cell.2015.04.029 [DOI] [PubMed] [Google Scholar]
- Rieu I, Powers SJ. 2009. Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell Online 21: 1031–1033. 10.1105/tpc.109.066001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalniak L, Koj A, Jura J. 2013. Proteasome inhibitor MG-132 induces MCPIP1 expression. FEBS J 280: 2665–2674. 10.1111/febs.12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, Sugimoto K, Miyazono K. 2011. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell 44: 424–436. 10.1016/j.molcel.2011.09.012 [DOI] [PubMed] [Google Scholar]
- Uehata T, Takeuchi O. 2017. Regnase-1 is an endoribonuclease essential for the maintenance of immune homeostasis. J Interf Cytokine Res 37: 220–229. 10.1089/jir.2017.0001 [DOI] [PubMed] [Google Scholar]
- van Kuppeveld FJ, van der Logt JT, Angulo AF, van Zoest MJ, Quint WG, Niesters HG, Galama JM, Melchers WJ. 1993. Genus- and species-specific identification of mycoplasmas by 16S rRNA amplification. Appl Environ Microbiol 59: 655 10.1007/bf01716704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Vikis HG, Wang Y, Jia D, Wang D, Bierut LJ, Bailey-Wilson JE, Amos CI, Pinney SM, Petersen GM, et al. 2007. Identification of a novel tumor suppressor gene p34 on human chromosome 6q25.1. Cancer Res 67: 93–99. 10.1158/0008-5472.CAN-06-2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawro M, Kochan J, Kasza A. 2016. The perplexities of the ZC3H12A self-mRNA regulation. Acta Biochim Pol 63: 411–415. 10.18388/abp.2016_1325 [DOI] [PubMed] [Google Scholar]
- Wawro M, Kochan J, Krzanik S, Jura J, Kasza A. 2017. Intact NYN/PIN-like domain is crucial for the degradation of inflammation-related transcripts by ZC3H12D. J Cell Biochem 118: 487–498. 10.1002/jcb.25665 [DOI] [PubMed] [Google Scholar]
- Wilamowski M, Gorecki A, Dziedzicka-Wasylewska M, Jura J. 2018. Substrate specificity of human MCPIP1 endoribonuclease. Sci Rep 8: 7381 10.1038/s41598-018-25765-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Peng W, Sun Y, Wang X, Xu Y, Li X, Gao G, Rao Z. 2012. Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase. Nucleic Acids Res 40: 6957–6965. 10.1093/nar/gks359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang W-C, Chen J-K, Zhou L, Wang M, Wang Z-D, Yang B, Xia Y-M, Lei S, Fu E-Q, et al. 2015. ZC3H12D attenuated inflammation responses by reducing mRNA stability of proinflammatory genes. Mol Immunol 67: 206–212. 10.1016/j.molimm.2015.05.018 [DOI] [PubMed] [Google Scholar]