Abstract
Drosophila Belle (human ortholog DDX3) is a conserved DEAD-box RNA helicase implicated in regulating gene expression. However, the molecular mechanisms by which Belle/DDX3 regulates gene expression are poorly understood. Here we performed systematic mutational analysis to determine the contributions of conserved motifs within Belle to its in vivo function. We found that Belle RNA-binding and RNA-unwinding activities and intrinsically disordered regions (IDRs) are required for Belle in vivo function. Expression of Belle ATPase mutants that cannot bind, hydrolyze, or release ATP resulted in dominant toxic phenotypes. Mechanistically, we discovered that Belle up-regulates reporter protein level when tethered to reporter mRNA, without corresponding changes at the mRNA level, indicating that Belle promotes translation of mRNA that it binds. Belle ATPase activity and amino-terminal IDR were required for this translational promotion activity. We also found that ectopic ovary expression of dominant Belle ATPase mutants decreases levels of cyclin proteins, including Cyclin B, without corresponding changes in their mRNA levels. Finally, we found that Belle binds endogenous cyclin B mRNA. We propose that Belle promotes translation of specific target mRNAs, including cyclin B mRNA, in an ATPase activity-dependent manner.
Keywords: DEAD-box, ATPase, helicase, RNA-binding, posttranscriptional gene regulation, Drosophila
INTRODUCTION
DEAD-box (DDX) proteins are the largest family of evolutionarily conserved RNA helicases that play critical roles in RNA biology. The DDX helicase core contains highly conserved residues required for ATP-binding, hydrolysis, and release—all of which are required for ATPase activity—including the DEAD (Asp-Glu-Ala-Asp) motif (Linder et al. 1989). The helicase core also includes RNA-binding and RNA-unwinding motifs (Putnam and Jankowsky 2013). DDX helicases utilize ATP hydrolysis to undergo conformational changes and remodel structures of RNAs and/or RNA–protein complexes (Liu et al. 2008; Del Campo and Lambowitz 2009; Nielsen et al. 2009). DDX helicase cores are often flanked by amino- and carboxy-terminal regions, which may have distinct functional roles for unique DDX helicases (Kim and Myong 2016; Samatanga et al. 2017).
DEAD-box helicases regulate gene expression—including regulating mRNA transcription, splicing, export, translation, and decay (Rocak and Linder 2004; Linder and Jankowsky 2011). Many DDX helicases are multifunctional and are thus involved in multiple steps in gene expression regulation. One example of a multifunctional DDX helicase is Drosophila Vasa (human ortholog DDX4), which is involved in posttranscriptional gene regulation through its roles in translational regulation and small silencing RNA biogenesis (Carrera et al. 2000; Liu et al. 2009; Xiol et al. 2014).
Drosophila Belle (human ortholog DDX3) is a multifunctional DDX helicase and the closest Drosophila Vasa paralog. Belle is required for viability and development (Johnstone et al. 2005). Belle is also required for both male and female fertility and is most highly expressed in gonadal tissues (Johnstone et al. 2005; Kotov et al. 2016). Like Vasa, Belle/DDX3 has been proposed to regulate multiple aspects of gene expression. Interestingly, Belle/DDX3 was proposed to function as both a translational repressor (Shih et al. 2008; Yarunin et al. 2011; Ihry et al. 2012; Götze et al. 2017) and activator (Lai et al. 2010, 2016; Guenther et al. 2018). Previous studies also suggested that Belle/DDX3 may function in small silencing RNA pathways (Zhou et al. 2008; Pek and Kai 2011; Kasim et al. 2013). DDX3 is implicated in cancer, where it has been suggested to have both oncogenic and tumor suppressor functions (Botlagunta et al. 2008; Chen et al. 2015, 2018; Heerma van Voss et al. 2015; Oh et al. 2016).
Deletion of Ded1p, the yeast ortholog of Belle/DDX3, causes lethality (Mamiya and Worman 1999). Viability can be rescued by expression of either Drosophila Belle or human DDX3, underscoring the conserved functionality across diverse species (Mamiya and Worman 1999; Johnstone et al. 2005).
Despite the essential biological role and conserved functionality of Belle/DDX3, the molecular functions and mechanisms of Belle/DDX3 are not well understood. Importantly, many previous Belle/DDX3 studies were conducted in tissue cell culture and thus in vivo functions and mechanisms remain poorly understood.
Here we sought to determine the contributions of conserved Belle motifs to Belle functions in vivo by designing and characterizing Belle mutants in flies. Belle ATPase activity, RNA-binding, RNA-unwinding, and intrinsically disordered regions (IDRs) were required for fly viability. Interestingly, we found that Belle ATPase mutants, which cannot bind, hydrolyze, or release ATP, cause dominant toxic phenotypes in both somatic and germline cells, decreasing cyclin protein (i.e., Cyclin B) levels and fertility. When tethered to reporter mRNA, Belle increased reporter protein level using its ATPase activity without a change in the reporter mRNA level. Belle directly bound endogenous cyclin B mRNA, a key cell cycle factor required for mitosis and meiosis. Thus, we defined the conserved motifs and regions required for Belle in vivo functions and showed that Belle binds a subset of target mRNAs to promote their translation using its ATPase activity.
RESULTS
Overview of experimental design
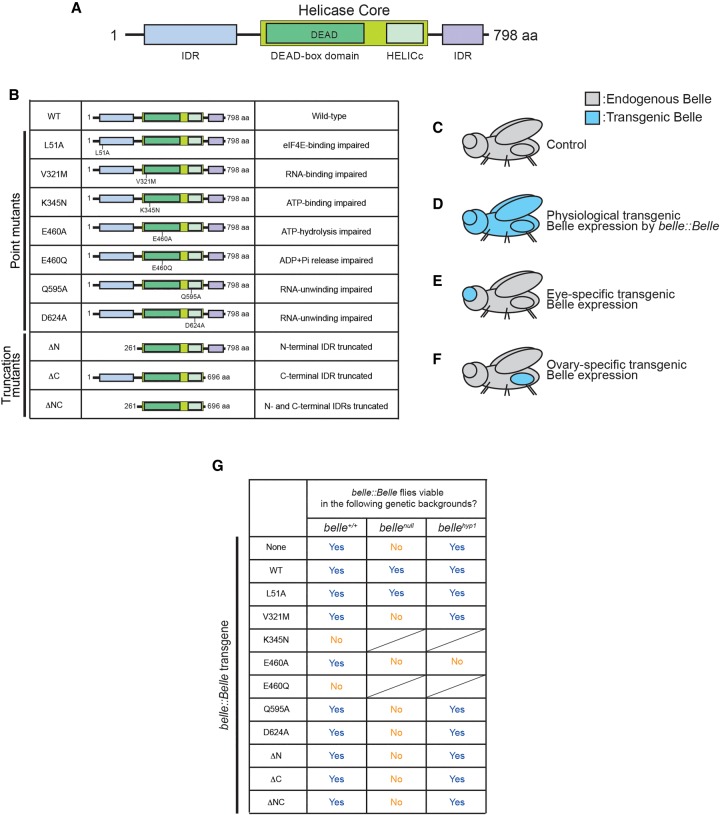
To understand the biological and molecular functions of Belle, we designed and studied a series of Belle mutants modeled on previous studies on Belle orthologues in yeast (Ded1p) (Iost et al. 1999), Caenorhabditis elegans (LAF-1) (Kim and Myong 2016) and human (DDX3) (Shih et al. 2008; Valentin-Vega et al. 2016) and Drosophila Vasa (Sengoku et al. 2006; Xiol et al. 2014). We designed seven point mutations which disrupt residues required for binding or catalytic activity: (i) BelleL51A, which cannot bind eIF4E, (ii) BelleV321M, which cannot bind RNA, (iii) BelleK345N, which cannot bind ATP, (iv) BelleE460A, which can bind but cannot hydrolyze ATP, (v) BelleE460Q, which can bind and hydrolyze ATP into ADP and Pi but cannot release ADP and Pi, (vi) BelleQ595A, and (vii) BelleD624A, both of which can bind RNA but cannot unwind bound RNA (“uncoupler”). We also designed three amino- and carboxy-terminal truncation mutants which lack intrinsically disordered regions (IDRs): (viii) BelleΔN, which lacks the amino-terminal IDR, (ix) BelleΔC, which lacks the carboxy-terminal IDR, and (x) BelleΔNC, which lacks both amino- and carboxy-terminal IDRs (Fig. 1B).
FIGURE 1.
Belle constructs and transgenic expression strategy and results showing that transgenic Belle ATPase mutants under belle promoter causes dominant toxicity. (A) Domain structure of Drosophila Belle. Belle has a DDX core with a DEAD motif and a HELICc domain, flanked by amino- and carboxy-terminal IDRs. (B) Description of Belle mutants used in this study. (C–F) Transgenic expression strategy used in this study. (C) Control flies have endogenous Belle. (D) Expression of transgenic Belle under endogenous Belle promoter (belle-Belle). (E) Eye-specific transgenic Belle expression achieved by using UAST-Belle and eye-specific Gal4 drivers (ey-Gal4 > Belle and longGMR-Gal4 > Belle). (F) Ovary-specific transgenic Belle expression achieved by using UASP-Belle and ovary-specific Gal4 drivers (MTD-Gal4 > Belle, tj-Gal4 > Belle, and MAT67-Gal4 > Belle). (G) Viability of belle::Belle transgenic flies in the genetic backgrounds of belle+/+, bellenull, and bellehyp1.
To study the effects of these mutations on Belle functions in vivo, we expressed each of these Belle mutants in different fly tissues using several genetic strategies (Fig. 1C–F). To express transgenic Belle proteins at the same spatiotemporal pattern and comparable expression level as endogenous Belle, Belle transgenes were expressed under a belle promoter (Fig. 1D). To express transgenic Belle proteins specifically in eyes, UAST-Belle transgenes were expressed using eye-specific Gal4 drivers (Fig. 1E). To express transgenic Belle proteins specifically in ovaries, UASP-Belle transgenes were expressed using ovary-specific Gal4 drivers (Fig. 1F).
ATP-hydrolysis, RNA-binding, RNA-unwinding, and IDRs of Belle are required for fly viability
We cloned wild-type and mutant Belle transgenes with a carboxy-terminal 3xFlag tag under a belle promoter (hereafter belle::Belle). We attempted to make belle::Belle transgenic flies of wild-type and mutant Belle in endogenous belle+/+ background (Fig. 1G). We successfully generated belle::Belle strains for wild-type Belle and several Belle mutants. We could not obtain belle::BelleK345N and belle::BelleE460Q strains, demonstrating that these two transgenes cause dominant lethality.
Belle is essential for fly viability; the belle null (belleL4740/belle47110 = bellenull) fly is not viable (Johnstone et al. 2005). To determine which Belle motifs/regions are required for viability, we tested which belle::Belle transgenes can rescue fly viability in the bellenull background. As expected, belle::BelleWT rescued viability in the bellenull background (Fig. 1G). Among our mutant belle::Belle transgenes, only belle::BelleL51A, but not belle::BelleV321M, belle::BelleE460A, belle::BelleQ595A, belle::BelleD624A, belle::BelleΔN, belle::BelleΔC, or belle::BelleΔNC, rescued viability in the bellenull background. Thus, while Belle eIF4E-binding activity is dispensable for fly viability, Belle ATP-hydrolysis, RNA-binding, and RNA-unwinding activities and its amino-terminal and carboxy-terminal IDRs are required for fly viability.
Of the Belle strains that we established in the belle+/+ background, belle::BelleE460A was the most difficult to maintain as a stock because the flies were weaker. In fact, belle::BelleE460A, but not the other viable belle::Belle transgenes, caused fly lethality in belle hypomorph background (belleneo30/belle47110 = bellehyp1 [Fig. 1G; Johnstone et al. 2005; Poulton et al. 2011]). These observations show that the belle::BelleE460A transgene causes dominant toxicity. Thus, all three ATPase mutant belle::Belle transgenes (belle::BelleK345N, belle::BelleE460A, and belle::BelleE460Q) exhibited dominant lethality or toxicity, revealing that Belle ATPase mutants cause dominant toxic effects.
We confirmed that belle::Belle transgenic proteins were expressed at a level similar to endogenous Belle as expected (Supplemental Fig. S1). We also confirmed that bellehyp1 expressed less endogenous Belle than control as expected.
Belle RNA-binding, RNA-unwinding, and IDRs are important for somatic development
The Drosophila eye is commonly used for functional analysis of genes involved in somatic development. The normal Drosophila eye develops as a smooth lattice of cells (Ready et al. 1976). Loss of functions of genes required for somatic eye development result in aberrant eye phenotypes, such as rough texture, misdistribution of bristles, or decreased eye size (Iyer et al. 2016).
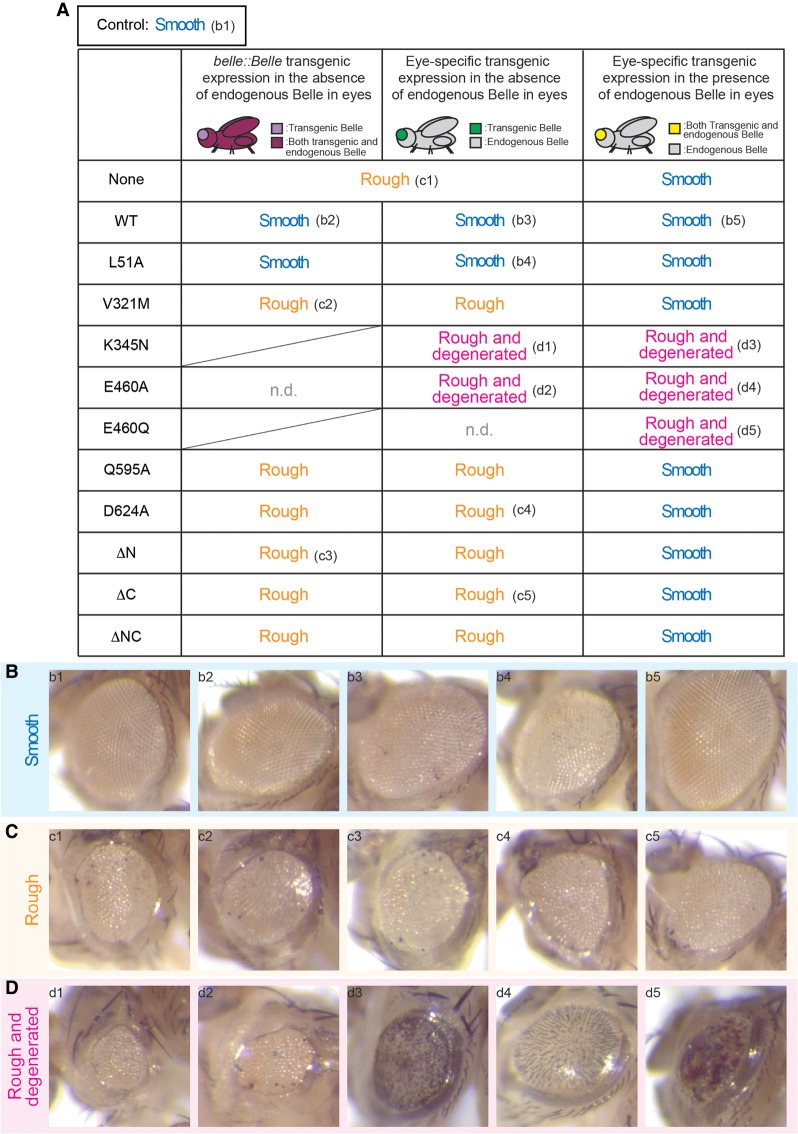
First, we examined the effects of eye-specific belle knockout on eye development using the Gal4-UAS-FLP-FRT system combined with GMR-hid (Fig. 2; Supplemental Fig. S2). We found that eyes lacking endogenous belle have rough textures (Fig. 2A,C-c1; Supplemental Fig. S2B) in contrast to the smooth textures of control flies (Fig. 2A,B-b1; Supplemental Fig. S2A; Jenny and Basler 2016). The eye phenotype in the absence of endogenous Belle in eyes was rescued by the belle::BelleWT transgene (Fig. 2A,B-b2; Supplemental Fig. S2D), which expresses transgenic Belle in a physiological expression pattern. belle::BelleL51A, but not the other mutant belle::Belle, rescued eye phenotype in the absence of endogenous Belle in eyes (Fig. 2; Supplemental Fig. S2D). The ATPase hydrolysis mutant belle::BelleE460A exhibited lethality in this genetic background, showing again its dominant toxicity (Fig. 2A). These results revealed that while Belle eIF4E-binding activity is dispensable, Belle RNA-binding and RNA-unwinding activities and Belle amino-terminal and carboxy-terminal IDRs are important for normal somatic eye development.
FIGURE 2.
RNA-binding, RNA-unwinding, and IDRs of Belle are important for normal eye development and ectopic expression of Belle ATPase mutants has dominant toxic effects in eyes. (A) Summary of eye phenotypes of control and Belle transgenic flies. belle::Belle transgenic expression in the absence of endogenous Belle in eyes was achieved using flies wIR; ey-Gal4, UAS-FLP/belle-Belle; FRT82B, belle47110/FRT82B, GMR-hid (left column). Eye-specific transgenic Belle expression in the absence of endogenous Belle in eyes was achieved using flies wIR; ey-Gal4, UAS-FLP/UAST-3xFlag-Belle; FRT82B, belle47110/FRT82B, GMR-hid (middle column). Eye-specific transgenic Belle expression in the presence of endogenous Belle in eyes was achieved using flies wIR; UAST-3xFlag-Belle/CyO; longGMR-Gal4/TM3, Sb (right column). Control flies, which express endogenous Belle but no transgenic Belle, have normal “Smooth” eyes (representative image shown in panel B-b1). Flies that lack endogenous belle specifically in eyes and do not express any transgenic Belle (wIR; ey-Gal4, UAS-FLP/+; FRT82B, belle47110/FRT82B, GMR-hid) have “Rough” eyes (representative image shown in panel C-c1). (B–D) Representative images of (B) smooth, (C) rough, and (D) rough and degenerated eyes. Each panel shows the eye image of flies with a corresponding annotation in the table shown in (A).
In the eye assay above, we also tested whether belle is required for small interfering RNA (siRNA)-mediated silencing using the white inverted repeat (wIR) RNA-silencing reporter (Lee and Carthew 2003). wIR produces an inverted repeat hairpin RNAs corresponding to white exon 3. In a wIR background, flies with functional siRNA-mediated silencing activity have white eyes since Dicer-2 processes the wIR hairpin into siRNAs and Argonaute2 binds those siRNAs and cleaves white mRNAs (Fig. 2B-b1; Supplemental Fig. S2A). In contrast, flies lacking siRNA-mediated silencing activity, such as the dicer-2null mutant, have red eyes due to failure to silence white expression in a wIR background (Supplemental Fig. S2C). We found that in a wIR background, eyes that lack endogenous belle are white, showing that belle is not required for siRNA-mediated silencing (Fig. 2C-c1; Supplemental Fig. S2B).
Belle ATPase mutants cause dominant toxicity in somatic cells
Since we were unable to establish viable belle::BelleK345N and belle::BelleE460Q strains (Fig. 1G) and belle::BelleE460A showed lethality in the genetic background of the eye phenotype assay (Fig. 2A), we could not test the effects of these Belle ATPase mutants on eye development using the belle::Belle strategy. Therefore, we created UAST-3xFlag-Belle transgenic flies and expressed Belle transgenes using an eye-specific ey-Gal4 driver (ey-Gal4 > Belle), which we thought may avoid the lethality issue seen with belle::Belle. Consistent with the results using belle::Belle transgenes, eye-specific expression of BelleWT and BelleL51A, but not BelleV321M, BelleQ595A, BelleD624A, BelleΔN, BelleΔC, or BelleΔNC, rescued normal smooth eye phenotype in the absence of endogenous Belle in eyes (Fig. 2; Supplemental Fig. S3). Interestingly, eyes expressing transgenic BelleK345N and BelleE460A in the absence of endogenous Belle exhibited a more severe phenotype than eyes without endogenous or transgenic Belle; eyes in these mutants were not only rough but also degenerated (Fig. 2A,D-d1,d2; Supplemental Fig. S3). Eye-specific BelleE460Q expression in the genetic background lacking endogenous Belle resulted in fly lethality (Fig. 2A). These results suggest that these Belle ATPase mutants have toxic effects in eyes. All viable flies examined here had white eyes (Supplemental Fig. S3), showing again that belle is not required for small RNA-mediated silencing.
To test whether Belle ATPase mutants exhibit toxic effects in a dominant manner, we expressed Belle transgenes in eyes using the UAST-Gal4 system with an eye-specific longGMR-Gal4 driver (longGMR-Gal4 > Belle) in the presence of endogenous wild-type Belle. Interestingly, eye-specific expression of BelleK345N, BelleE460A, and BelleE460Q in the presence of endogenous wild-type Belle caused rough and degenerated eyes (Fig. 2A,D-d3–d5) while eyes expressing BelleWT or the other mutant Belle were smooth and normal (Fig. 2; Supplemental Fig. S4). These results showed that Belle ATPase-binding, hydrolysis, and release mutants have dominant toxic effects in eyes. All these flies had white eyes (Supplemental Fig. S4), showing that ATPase mutant BelleK345N, BelleE460A, and BelleE460Q do not show a dominant negative effect on siRNA-mediated silencing.
Belle RNA-binding, RNA-unwinding, and IDRs are required for male and female fertility
Both male and female bellehyp1 flies have reduced fertility. Using belle::Belle, we tested which Belle features are required to rescue bellehyp1 fertility.
bellehyp1 is completely male-sterile and male fertility was rescued by belle::BelleWT, showing the essential role of belle in male fertility (Supplemental Fig. S5A). Only belle::BelleL51A, but not belle::BelleV321M, belle::BelleQ595A, belle::BelleD624A, belle::BelleΔN, belle::BelleΔC, or belle::BelleΔNC, rescued bellehyp1 male fertility. These results revealed that while Belle eIF4E-binding activity is dispensable, Belle RNA-binding and RNA-unwinding activities and Belle amino-terminal and carboxy-terminal IDRs are crucial for male fertility.
Next, we examined female fertility. bellehyp1 female produced comparable numbers of eggs compared with controls (Supplemental Fig. S5B). However, a significantly lower ratio of the eggs laid by bellehyp1 could hatch (Supplemental Fig. S5C). The bellehyp1 egg hatching rate was rescued by belle::BelleWT. These results show that belle is important for female fertility (Johnstone et al. 2005). belle::BelleL51A, but not belle::BelleV321M, belle::BelleQ595A, belle::BelleD624A, belle::BelleΔN, belle::BelleΔC, or belle::BelleΔNC, fully rescued bellehyp1 egg hatching rate. belle::BelleQ595A, belle::BelleD624A only partially rescued bellehyp1 egg hatching rate. These results revealed that while Belle eIF4E-binding activity is dispensable, Belle RNA-binding activitiy and Belle amino-terminal and carboxy-terminal IDRs are required for female fertility. Belle RNA-unwinding activity contributes to female fertility.
Belle ATPase mutants have dominant toxic effects on female fertility
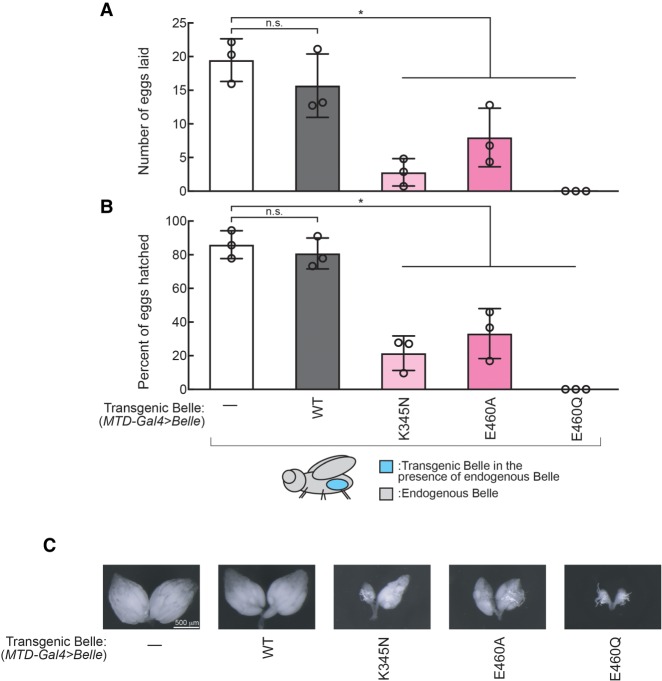
We sought to determine if Belle ATPase mutants are also dominant toxic in the female germline since Belle is most highly expressed in ovaries (Johnstone et al. 2005). We created UASP-3xFlag-Belle transgenic flies and used the germline-specific MTD-GAL4 driver to express BelleWT, BelleK345N, BelleE460A, and BelleE460Q in ovary germlines in the endogenous belle+/+ genetic background (MTD-Gal4 > BelleWT, MTD-Gal4 > BelleK345N, MTD-Gal4 > BelleE460A, MTD-Gal4 > BelleE460Q). Expressing transgenic Belle ATPase mutants in female germlines resulted in significantly decreased fecundity (number of eggs laid) and fertility (number of eggs hatched) compared with controls, while expressing transgenic wild-type Belle affected neither fecundity nor fertility (Fig. 3A,B). We also found that germline expression of transgenic Belle ATPase mutants resulted in decreases in ovary mass (Fig. 3C). Transgenic Belle wild-type and ATPase mutants all exhibited cytoplasmic localization in oocytes (Supplemental Fig. S6), revealing that the ATPase mutations do not affect Belle localization.
FIGURE 3.
Ectopic expression of Belle ATPase mutants has dominant toxic effects on female fertility. Female fertility assays using flies expressing Belle transgene in germlines (MTD-Gal4 > Belle). (A) The numbers of eggs laid by test females crossed with OregonR wild-type males and (B) the hatching rates from the eggs. Mean ± SD (n = 3 biological replicates). P-value <0.05 (two-sided Student's t-test) are indicated by (*). (C) Representative images of dissected ovaries.
We also tested the effect of expressing transgenic Belle ATPase mutants in ovarian somatic cells (follicle cells) using the tj-GAL4 driver (tj-Gal4 > Belle). Expressing Belle ATPase mutants in ovarian somatic cells resulted in complete sterility with no eggs laid by any of the three ATPase mutants, while expressing transgenic wild-type Belle affected neither fecundity nor fertility (Supplemental Fig. S7).
We conclude that Belle ATPase mutants cause dominant negative effects in both germline and somatic cells in ovaries.
Belle ATPase mutants do not affect small RNA profiles in ovaries
We sought to uncover molecular mechanisms by which germline expression of transgenic Belle ATPase mutants causes the dominant negative effects on female fertility. First, to examine whether expression of dominant Belle ATPase mutants changes small silencing RNA profiles, we performed high-throughput sequencing of ovary small RNAs. miRNA, endo-siRNA (esi-1.1, esi-1.2, and esi-2.1), and piRNA levels were largely unchanged in ovaries expressing wild-type or ATPase mutant of Belle (MTD-Gal4 > Belle) compared with control ovaries (MTD-Gal4 > w1118) (Supplemental Fig. S8). Thus, Belle ATPase mutants do not largely change small RNA profiles in ovaries.
Dominant Belle ATPase mutants reduce ovary cyclin protein levels
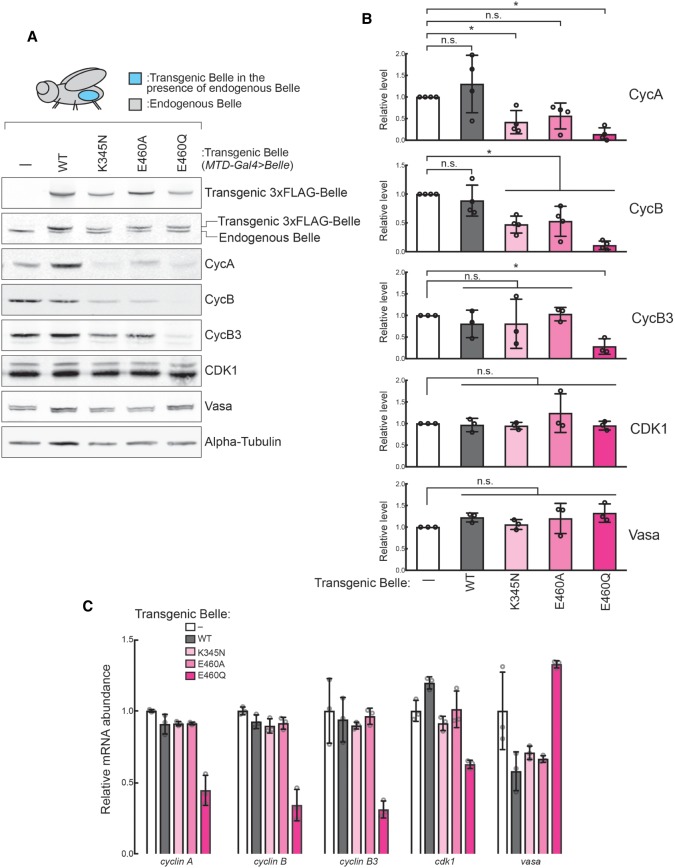
Cyclin proteins Cyclin A, Cyclin B, and Cyclin B3 are crucial regulators of meiosis in oocytes. In ovaries expressing Belle ATPase mutants in the belle+/+ background (MTD-GAL4 > Belle), we found that Cyclin A and Cyclin B protein levels were decreased relative to control ovaries (Fig. 4A,B). Cyclin A protein level was lower in BelleK345M and BelleE460Q mutants. Cyclin B protein level was lower in all three ATPase mutants (BelleK345M, BelleE460A, and BelleE460Q). Cyclin B3 protein level was decreased in the BelleE460Q mutant. In contrast, expressing transgenic wild-type Belle did not affect cyclin protein levels. CDK1 and Vasa protein levels were not changed by transgenic wild-type or ATPase mutant Belle, showing that cyclin protein level decrease is specific. Anti-Belle western blot showed that levels of transgenic Belle proteins were comparable to those of endogenous wild-type Belle. We measured levels of corresponding mRNAs. cyclin A, cyclin B, cyclin B3, cdk1, and vasa mRNA levels were unchanged in BelleK345M and BelleE460A ATPase mutants compared to controls (Fig. 4C). BelleE460Q mutants had decreased levels of cyclin A, cyclin B, cyclin B3, and cdk1 mRNA levels, perhaps because BelleE460Q mutants exhibited the most severe ovary morphology change as compared to control (Fig. 3C). The decrease in Cyclin A and Cyclin B protein levels in BelleK345M and BelleE460A ATPase mutants without a corresponding change in cyclin A and cyclin B mRNA levels suggests that these protein level decreases were due to posttranscriptional dysregulation.
FIGURE 4.
Ectopic expression of Belle ATPase mutants decreases Cyclin B protein level in ovaries. (A) Representative images of western blot using ovary lysates of MTD-Gal4 > w1118 and MTD-Gal4 > Belle flies. (B) Quantification of band intensities of western blot. Mean ± SD (n = 3 or 4 biological replicates). P-value <0.05 (two-sided Student's t-test) are indicated by (*). (C) Relative levels of cyclin A/B/B3, cdk1 and vasa mRNAs in MTD-Gal4 > Belle ovaries compared with controls (MTD-Gal4 > w1118) determined by high-throughput sequencing. Mean ± SD (n = 3 biological replicates).
Belle tethering to a reporter mRNA increases reporter protein level in an ATPase-dependent manner
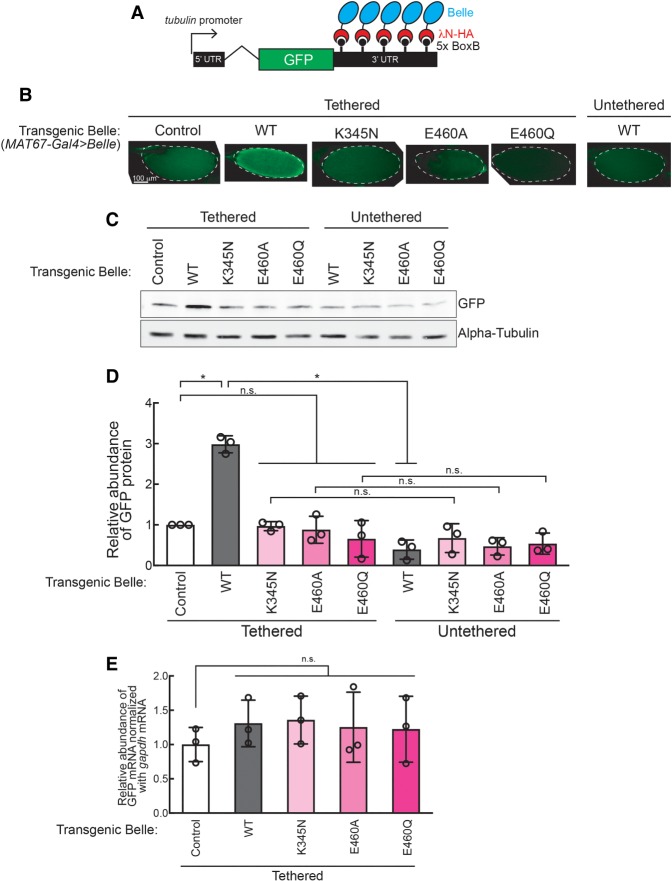
To examine effects of Belle binding to mRNA, we tethered transgenic wild-type and mutant Belle proteins to a reporter GFP mRNA in oocytes using a λN peptide-BoxB hairpin RNA system (Fig. 5A; Sienski et al. 2015). λN-HA-Belle binds BoxB hairpins in the 3′-UTR of the reporter GFP mRNA through the λN peptide. We tested λN-HA tagged wild-type, ATPase mutant, and IDR-truncated Belle. We also tested tethering of λN-HA only as a negative control. As additional negative controls, we tested expression of transgenic Belle proteins without a λN tag. We expressed the transgenic Belle proteins in oocytes using the UASP/MAT67-Gal4 driver system in the GFP reporter background (MAT67-Gal4 > Belle).
FIGURE 5.
Belle posttranscriptionally promotes protein translation in an ATPase activity-dependent manner. (A) GFP-5x BoxB reporter structure, harboring a ubiquitous tubulin promoter, the GFP-coding sequence, and a 3′-UTR containing five BoxB hairpins. λN-HA-fused control peptide and Belle under a UASP promoter were expressed in germline cells using MAT67-Gal4 driver. (B) Confocal images of GFP signal in stage 14 oocytes. (C) Representative images of western blots using ovary lysates. (D) Quantification of western blots band intensities. GFP protein levels normalized with α-tubulin are shown. (E) Relative abundance of GFP-5xBoxB mRNA normalized by gapdh mRNA determined by qRT-PCR.
GFP protein level was significantly increased when wild-type Belle was tethered to the GFP reporter mRNA compared with controls (λN-HA only and BelleWT without a λN tag) as observed by confocal microscopy imaging of oocytes (Fig. 5B) and western blotting of ovary lysates (Fig. 5C,D). In contrast, GFP protein level was not increased when the Belle ATPase mutants were tethered, showing that Belle ATPase activity is required for the reporter GFP protein increase.
We measured GFP reporter mRNA level by qRT-PCR. Tethering of wild-type or ATPase mutant Belle did not change the GFP reporter mRNA level compared to controls (Fig. 5E), revealing that wild-type Belle increased GFP protein level without a corresponding change in GFP mRNA level. We also found that tethering of wild-type Belle did not change the poly(A) tail length of GFP mRNA, showing that increased GFP protein level was not due to a change in poly(A) tail length of GFP mRNA (Supplemental Fig. S9). These results suggest that tethered Belle increases reporter protein levels by promoting translation.
Belle tethering to a reporter mRNA increases reporter protein level in an amino-terminal IDR-dependent manner
Additionally, we tested the tethering of three IDR-truncated Belle mutants. BelleΔC tethering showed up-regulation of GFP protein level, comparable to the level observed in BelleWT tethering, whereas neither BelleΔN nor BelleΔNC tethering increaseed GFP protein levels (Fig. 6A–C). GFP reporter mRNA level was unchanged by tethering of these truncated Belle mutants (Fig. 6D). These results show that the Belle amino-terminal IDR is required for posttranscriptional promotion of reporter protein level.
FIGURE 6.
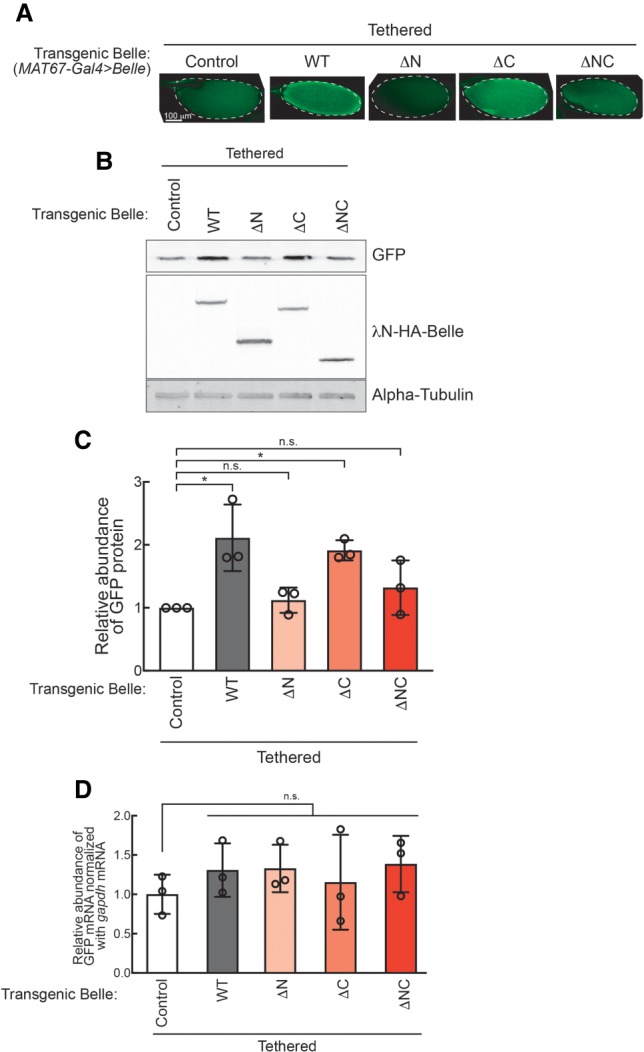
The Belle amino-terminal IDR is required for posttranscriptional promotion of protein translation. (A) Confocal images of GFP signal in stage 14 oocytes. (B) Representative images of western blots using ovary lysates. (C) Quantification of western blots band intensities. GFP protein levels normalized with α-tubulin are shown. (D) Relative abundance of GFP-5xBoxB mRNA normalized by gapdh mRNA determined by qRT-PCR.
Belle binds endogenous cyclin B mRNA
Finally, to identify endogenous Belle target mRNAs, we performed RNA-coimmunoprecipitaion (RIP) followed by qRT-PCR using ovary lysates expressing transgenic wild-type or ATPase mutant Belle in the presence of endogenous wild-type Belle (MTD-Gal4 > Belle). We focused on genes that had decreased protein levels in Belle ATPase mutant ovaries (Fig. 4). Transgenic wild-type Belle did not significantly bind cyclin A, cyclin B, cyclin B3, or cdk1 mRNAs, while it showed a trend to bind cyclin B mRNA (Fig. 7). In contrast, transgenic Belle ATPase mutants bound cyclin B mRNA, but did not significantly bind cyclin A, cyclin B3, or cdk1 mRNAs.
FIGURE 7.
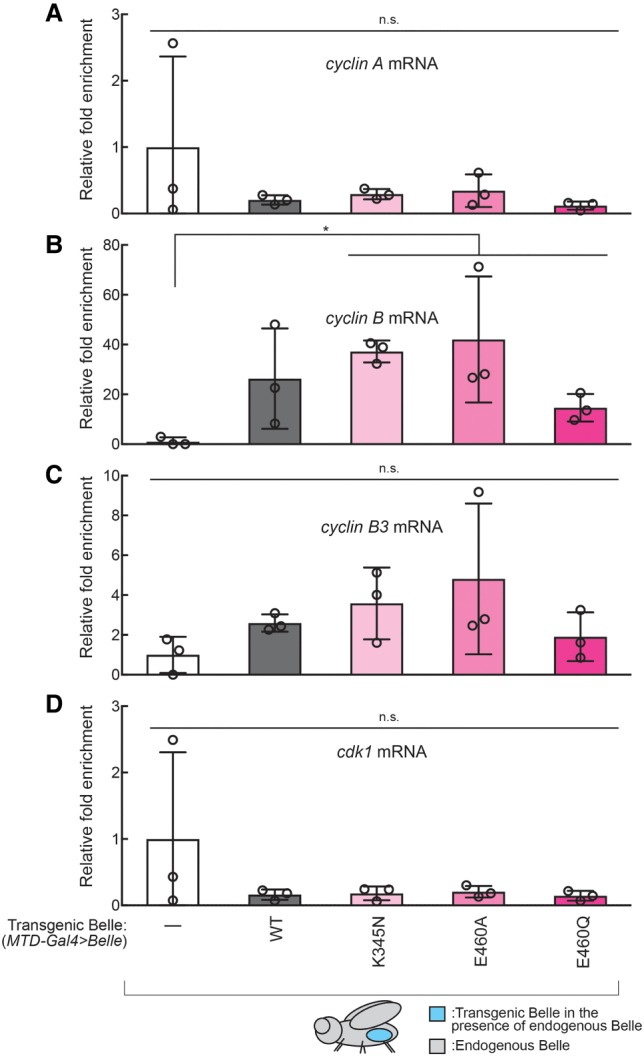
Belle binds endogenous cyclin B mRNA in ovaries. (A–D) Fold enrichment of mRNAs [(A): cyclin A, (B): cyclin B, (C): cyclin B3, (D): cdk1] relative to a control rp49 mRNA that were coimmunoprecipitated with transgenic Belle expressed specifically in germlines in ovaries (MTD-Gal4 > Belle) normalized by w1118 negative control (MTD-Gal4 > w1118), determined by RNA-immunoprecipitation followed by qRT-PCR. Mean ± SD (n = 3 biological replicates). P-value <0.05 (two-sided Student's t-test) is indicated by (*).
DISCUSSION
DEAD-box helicases play important roles in animal development by regulating gene expression, particularly in germline development (Hozumi et al. 2012; Zhang et al. 2018). Previous studies have shown that DDX helicases are involved in developmental gene expression (Hay et al. 1988; Gonzalez-Reyes et al. 1997; Kotov et al. 2014; Bush et al. 2015).
Here we determined the Belle motifs required for fly viability and somatic and germline development. Through Belle mutant transgene expression in varying genetic contexts, we determined that Belle ATPase, RNA-binding, RNA-unwinding, and IDRs are required for fly viability, somatic development, and fertility (Figs. 1, 2; Supplemental Figs. S2–S5). Furthermore, we discovered that Belle mutants impaired in enzymatic ATPase activity (binding, hydrolysis, or release) have dominant negative somatic developmental and fertility outcomes, including degenerated eyes, decreased number of eggs laid and hatched, and smaller ovary size, compared to controls (Figs. 2, 3; Supplemental Figs. S3, S4). To further dissect the molecular mechanism resulting in fertility defects in ovaries expressing dominant Belle ATPase mutants in the presence of endogenous wild-type Belle, we observed decreased levels of cyclin proteins required for meiosis cell cycle regulation in Belle ATPase mutant ovary lysate (Fig. 4). Tethering assay showed that Belle increases a protein level produced from a bound mRNA in its ATPase activity- and amino-terminal IDR-dependent manner (Figs. 5, 6).
Previous studies showed that belle hypomorph testes have decreased mRNA and protein levels of Cyclin A and Cyclin B, implicating Belles’ role in transcriptional and/or posttranscriptional regulation of cyclin A and cyclin B mRNA levels in testes (Kotov et al. 2016). In contrast, we determined that Cyclin A and Cyclin B protein levels were decreased without change in cyclin A and cyclin B mRNA levels in ovaries expressing Belle ATPase mutants in the presence of endogenous wild-type Belle (MTD-Gal4 > Belle) compared with control ovaries, suggesting that Belle functions as a posttranscriptional regulator rather than a transcriptional regulator (Figs. 5, 6). Our Belle–RNA tethering assay results affirmed that Belle functions as a posttranscriptional regulator to increase protein levels without changing corresponding mRNA levels in ovaries in its ATPase activity- and amino-terminal IDR-dependent manner.
We observed direct binding of three Belle ATPase mutants to cyclin B mRNA, further supporting the idea that Belle functions as a posttranscriptional regulator (Fig. 7). In contrast, BelleWT was not observed to bind cyclin B mRNA significantly, while it showed a trend to do so. We propose that this is because Belle ATPase mutants remain bound to target mRNAs while wild-type Belle binds and dissociates from target mRNAs after promoting their translation.
Belle cytoplasmic localization in oocytes was not affected by ATPase mutations (Supplemental Fig. S6). This result excludes the possibility that Belle ATPase mutants have dysregulated subcellular localization resulting in reduced translational promotion activity and further supports our conclusion that Belle ATPase activity is directly required for translational promotion.
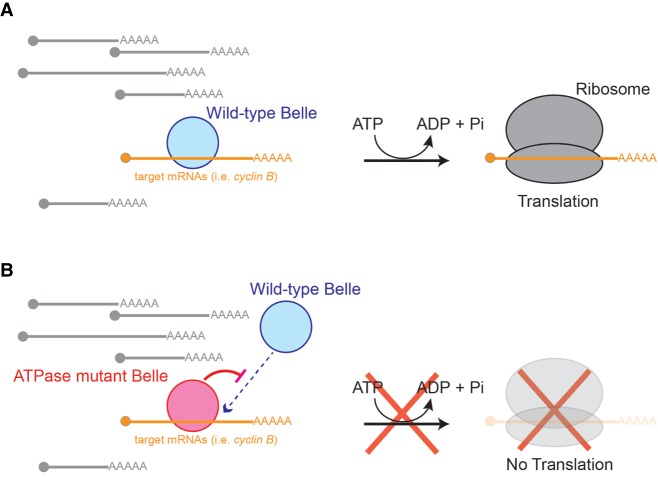
We propose a model for wild-type Belle function and how Belle ATPase mutant expression results in dominant toxicity (Fig. 8). Wild-type Belle binds a subset of target mRNAs including cyclin B mRNAs. Using its ATPase activity and amino-terminal IDR, Belle promotes translation of bound mRNAs, without affecting levels or poly(A) tail length of the bound mRNAs. Belle ATPase mutants occupy target mRNAs and cannot promote their translation due to the lack of ATPase activity. Instead, target mRNA-bound Belle ATPase mutants block access of endogenous wild-type Belle, thus preventing translational promotion of the target mRNAs by endogenous wild-type Belle. This model explains Belle ATPase mutants’ dominant toxic effects in vivo, binding to target mRNAs (cyclin B mRNA), dominant decrease of target protein levels (Cyclin B), and lack of posttranscriptional gene expression promotion activity in the reporter mRNA tethering assay. Consistent with the idea that Belle ATPase mutants remain bound to target mRNAs, Vasa ATPase mutant that cannot release ADP/Pi (corresponding to BelleE460Q mutant) remains bound to piRNA precursor RNAs (Xiol et al. 2014). Furthermore, consistent with our model proposing that Belle ATPase mutants block access of wild-type Belle to cyclin B mRNAs decreasing Cyclin B protein translation, previous studies showed that belle hypomorph ovaries, which have decreased levels of wild-type Belle protein, have decreased Cyclin B protein levels compared with control ovaries (Kotov et al. 2016).
FIGURE 8.
Model of Belle mechanism. (A). Wild-type Belle binds a specific subset of mRNAs. Using its ATPase activity, Belle promotes translation of bound mRNAs. (B) Belle ATPase mutants remain bound to a specific subset of mRNAs and therefore blocking access of endogenous wild-type Belle. Without its ATPase activity, Belle cannot promote translation of bound mRNAs.
Interestingly, the amino-terminal IDR of C. elegans LAF-1 (Belle ortholog), was shown to be necessary and sufficient for phase separation of LAF-1 in liquid droplets (Elbaum-Garfinkle et al. 2015). Our results show that the Belle amino-terminal IDR is required for translational promotion (Fig. 8). Therefore, phase separation and translational promotion may be linked by amino-terminal IDR of LAF-1/Belle/DDX3. We suggest that Belle posttranscriptionally regulates gene expression by promoting translation of direct targets. While expressing Belle ATPase mutants decreased Cyclin A protein levels (Fig. 4), Belle did not bind cyclin A mRNAs (Fig. 7). One potential explanation for these results could include that our RNA-binding assay was not sensitive enough to detect weaker or transient interaction. Another explanation is that a decrease in Cyclin A protein is caused in an indirect mechanism rather than Belle directly binds cyclin A mRNA and promote its translation. For example, a decrease in Cyclin B protein level, which is more likely to be a direct consequence of Belle ATPase mutant expression considering their binding to cyclin B mRNA (Fig. 7), may be causing the observed decrease in Cyclin A protein level.
While we found that Belle promotes translation of target mRNAs without affecting their poly(A) tail length in ovaries, previous studies in early embryos showed that Belle forms a large protein–RNA complex that includes Smaug, Cup, Me31B, Trailer hitch, eIF4E, and PABPC, and is involved in poly(A) tail shortening and thus translational repression of a target mRNA (nanos mRNA) (Götze et al. 2017). It remains unknown whether Belle ATPase activities and amino-terminal IDR are required for this translational repression activity as we showed in this study that they are required for translational promotion of target mRNAs in ovaries. The consequences of Belle binding on target mRNAs (translational promotion vs. repression) may depend on protein partners that Belle binds, which may differ among different tissues.
While we focused on Belle's regulation of cyclin protein expression levels, we suspect that Belle may in fact bind several other endogenous target mRNAs to promote their translation similarly to other DDX helicases. For example, the Belle paralog Vasa promotes translation of several embryonic mRNAs, such as mei-P26 and gurken (Styhler et al. 1998; Tomancak et al. 1998; Liu et al. 2009). Determining proteome-wide changes in gene expression in belle mutants is required to identify the full repertoire of genes whose protein expression is regulated by Belle. RNA-immunoprecipitation followed by high-throughput sequencing can further dissect which mRNAs Belle directly binds, versus indirect regulation. Furthermore, identifying Belle protein binding partners and how the interaction is regulated by Belle ATPase activity and IDRs will be important to better understand the molecular mechanism by which Belle regulates gene expression posttranscriptionally. Our studies suggest that Belle ATPase mutants allow capturing intermediate or transient Belle–RNA interactions and thus will be useful for the future studies of Belle.
MATERIALS AND METHODS
Fly strains
The belle transgenic expression constructs under endogenous belle promoter (P[belle-Belle-3xFlag], belle::Belle) were produced by inserting the coding sequences of wild-type and mutant Belle with a carboxy-terminal 3xFlag tag flanked by ∼1.8 kb upstream region and ∼0.8 kb downstream region of belle gene in a pattB plasmid vector 40. UAST- and UASP-based belle transgenic constructs (P[UAST-3xFlag-Belle], P[UASP-3xFlag-Belle], P[UASP-3xHA-Belle], P[UASP-LambdaN-HA-Belle], and P[UASP-3xHA-HRV3Csite-3xFlag-Belle]) were generated by inserting the coding sequences of wild-type and mutant Belle with an amino-terminal 3x-Flag, 3xHA, λN-HA, or 3xHA-HRV3Csite-3xFlag tag into a pUASTattB or pUASPattB plasmid vector (Bischof et al. 2007; Takeo et al. 2012; Kandasamy and Fukunaga 2016; Kandasamy et al. 2017; Zhu et al. 2018a,b). The pUASTattB-transgenes were integrated at the position 51C1 on the second chromosome of the BDRC fly strain 24482 using the PhiC31 system40. The mini-white gene (w+mC) derived from the integrated plasmids and the RFP gene originally present in the fly strain to mark the landing site were removed using Cre-Lox. The pUASPattB-transgenes were integrated at the attP40 site on the position 25C6 on the second chromosome of fly using the PhiC31 system.
Eye phenotype assay
Eye-specific belle null phenotype was analyzed using fly strain wIR; ey-Gal4, UAS-FLP/+; FRT82B, belle47110/FRT82B, GMR-hid. Phenotypes of eye-specific belle null in the presence of genomic rescue belle transgenes were analyzed using fly strains wIR; ey-Gal4, UAS-FLP/gBelle; FRT82B, belle47110/FRT82B, GMR-hid. Phenotypes of eye-specific belle null in the presence of eye-specific expression of transgenic Belle were analyzed using fly strains wIR; ey-Gal4, UAS-FLP/UAST-3xFlag-Belle; FRT82B, belle47110/FRT82B, GMR-hid. Phenotypes of eye-specific expression of transgenic Belle in the presence of endogenous wild-type belle were analyzed using fly strains wIR; UAST-3xFlag-Belle/CyO; longGMR-Gal4/TM3,Sb. Eye images were taken using Leica M125 Stereomicroscope.
Fertility assay
For the male fertility assay, one test male was mated with five virgin wild-type (OregonR) females in a vial (Fukunaga et al. 2012; Zhu et al. 2018b). After 3 d, the five OregonR females were transferred to a new vial (vial 1). Every 2 d, the five OregonR females were transferred to a new vial until a total of four vials were obtained. The five OregonR females were removed from the last vial (vial 4) after 2 d. The total number of progenies in these four vials was counted. For the female fertility assay, five test virgin females were mated with three wild-type (OregonR) males in a cage with a 6-cm grape juice agar plate with wet yeast paste (Genesee) (Fukunaga et al. 2012; Zhu et al. 2018b). The grape juice agar plate was exchanged with a fresh one every day. The number of eggs laid on the third grape juice agar plate (set up on Day 3 and recovered on Day 4) was counted. Then, this grape juice agar plate was kept for one more day at 25°C and the number of hatched eggs was counted.
Western blot
Lysates of hand-dissected ovaries were prepared by homogenizing in RIPA buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% [v/v] IGEPAL CA-630, 0.1% [w/v] sodium dodecyl sulfate [SDS], 0.5% [w/v] sodium deoxycholate, 1 mM ethylenediaminetetraacetic acid [EDTA], 5 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) (Kandasamy and Fukunaga 2016; Kandasamy et al. 2017; Zhu et al. 2018b). The homogenates were clarified by centrifugation at 21000g at 4°C for 10 min, and the protein concentration was determined using the BCA protein assay kit (Pierce). Twenty micrograms of total protein was loaded per lane for western blot. The sources and dilutions of the primary antibodies were as below. Rabbit anti-Tubulin (1/1000, Sigma, T3526), mouse anti-α-tubulin (1/1500, Sigma, T9026), mouse anti-GFP (1/1000, Pierce, MA515256), mouse anti-HA (1/1000, Sigma, H3663), rat anti-Vasa (1/2000, Developmental Studies Hybridoma Bank, AB_760351), mouse anti-Cyclin A (1/1,000, Developmental Studies Hybridoma Bank, AB_528188), goat anti-Cyclin A (1/1,000, Santa Cruz Biotechnology, sc-15869, dN-15), mouse anti-Cyclin B (1/1000, Santa Cruz Biotechnology, sc-166152, B-6), rabbit anti-Cyclin B3 (1/2000 [Jacobs et al. 1998]), mouse anti-CDK1 (1/5000, Sigma, P7962), rabbit anti-Belle (1/2000 [Pek and Kai 2011]). IRDye 800CW goat anti-mouse IgG, IRDye 800CW goat anti-rat IgG, IRDye 800CW goat anti-rabbit IgG, IRDye 680RD goat anti-mouse IgG, and IRDye 680RD goat anti-rabbit IgG (Licor) were used as secondary antibodies. The membranes were scanned on an Odyssey imaging system (Licor).
Immunostaining
Stereomicroscope images of dissected ovaries and oocytes were taken using a Leica M125 stereomicroscope (Zhu et al. 2018b).
Ovaries from 2- to 5-d-old yeast-fed and mated females were hand-dissected in Robb's buffer (55 mM NaOAc, 8 mM KOAc, 100 mM sucrose, 10 mM glucose, 1.2 mM MgCl2, 1 mM CaCl2, 100 mM HEPES-KOH [pH 7.4]) at room temperature. Stage 14 oocytes and earlier stage oocytes were separated by forceps and were separately placed in fixative containing 4% formaldehyde in PBS (137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8.1 mM Na2HPO4, pH 7.4). Fixation was done at room temperature on a rocker for 30 min. For GFP signal detection in stage 14 oocytes, fixed oocytes were rinsed three times with PBS. For HA-Belle signal detection in earlier stage oocytes, fixed oocytes were rinsed three times with PBS containing 0.05% (v/v) TritonX-100. The rinsed oocytes were incubated in blocking buffer (2% [v/v] donkey serum, 3% [w/v] BSA, 0.02% [w/v] NaN3, 0.05% [v/v] TritonX-100 in PBS) for 1 h at room temperature. Then oocytes were incubated in blocking buffer containing mouse anti-HA (1/100, Sigma, H3663) and rat anti-Vasa (1/100, Developmental Studies Hybridoma Bank, AB_760351) at 4°C overnight. Oocytes were rinsed three times with PBS containing 0.05% (v/v) TritonX-100. Oocytes were incubated in blocking buffer containing AlexxaFluor-conjugated anti-mouse (1/100, ThermoFisher, A21202) and anti-rat (1/100, ThermoFisher, A21209) secondary IgG antibodies at at 4°C overnight. Oocytes were rinsed three times with PBS containing 0.05% (v/v) TritonX-100. All oocytes were mounted in mounting reagent containing DAPI (ProLong Diamond Antifade Mountant with DAPI, ThermoFisher). Confocal images were collected on a Zeiss LSM700 confocal microscope at the Johns Hopkins University School of Medicine Microscope Facility.
RNA-coimmunoprecipitation
RNA-coimmunoprecipitation was performed as previously described (Zhu et al. 2018b). Hand-dissected ovaries of w1118 and MTD-Gal4 > 3xHA-HRV3Csite-3xFlag-Belle flies were homogenized in lysis buffer containing RNase inhibitor (50 U/µL, Thermo Fisher Scientific). After centrifugation, cleared supernatant was incubated with 10% volume of Pierce anti-HA magnetic beads (Thermo Fisher Scientific) at 4°C for 2 h. After washing five times with the lysis buffer, 3xFlag-Belle protein–RNA complex was released by HRV3C protease cleavage in cleavage solution (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5% [v/v] glycerol, 2 mM EDTA, 200 nM HRV3C protease) at 4°C overnight. RNA was purified from the supernatant using TRIzol LS (Sigma).
qRT-PCR
RNA from ovaries was prepared using miRVana (Thermo Fisher Scientific). RNAs extracted from ovaries and RNAs coimmunoprecipitated with proteins were treated with Turbo DNase (Thermo Fisher Scientific), and then were reverse-transcribed into cDNA using a random hexamer primer and AMV Reverse Transcriptase (NEB). qPCR was performed using SsoAdvanced Universal SYBR Green Supermix on CFX96 (Biorad). The primers used were as follows. cyclin A, 5′-AAGAGTCAAGGAGCTTCCGC-3′ and 5′-TGTTTCTTCTCGCTCTCCCG-3′. cyclin B, 5′-CCACTGTAGAACCCACTAAAGTTAC-3′ and 5′-GGTCAGCGACTTCTTCGACA-3′. cyclin B3, 5′-ACCCTGGCTCGATACATCCT-3′ and 5′-TACGCAGTGCCATGAACAGT-3′. cdk1, 5′-CGTGGTGTATAAGGGTCGCA-3′ and 5′-ACGAAATTTCTCTGATCGCGG-3′. GFP, 5′-GAGCTGAAGGGCATCGACTT-3′ and 5′-TTCTGCTTGTCGGCCATGAT-3′. gapdh, 5′-TGATGAAATTAAGGCCAAGGTTCAGGA-3′ and 5′-TCGTTGTCGTACCAAGAGATCAGCTTC-3′. rp49, 5′-CTGCCCACCGGATTCAAG-3′ and 5′-CGATCTCGCCGCAGTAAAC-3′.
ePAT
ePAT assay was performed as previously described (Janicke et al. 2012; Zhu et al. 2018b) using the forward primer GTCTCGATTCTACGCGTACCGG with 6-fluorescein amidite and the reverse primer GAGCTCCGCGGCCGCG. The fragment analysis data was analyzed using Peak Scanner 2 (Thermo Fisher Scientific).
Small RNA and mRNA sequencing
small RNA and poly(A)+ mRNA libraries were prepared and sequenced on Hiseq4000 (Illumina) as previously described (Fukunaga et al. 2012; Liao et al. 2018; Vakrou et al. 2018; Zhu et al. 2018b).
DATA DEPOSITION
The SRA accession number for the small RNA and mRNA libraries reported in this paper is PRJNA494685.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Wu-Min Deng (Florida State University), Dr. Toshie Kai (Osaka University), Dr. Paul Lasko (McGill University), Dr. Christian Lehner (University of Zurich), Dr. Julius Brennecke (Institute of Molecular Biotechnology, Vienna), and Dr. Richard Carthew (Northwestern University) for their kind gifts of fly strains and antibodies. We thank Bloomington Drosophila Stock Center, Drosophila Genomics and Genetic Resources, Kyoto Stock Center, and Vienna Drosophila Resource Center for fly strain stocks. We thank the Johns Hopkins University School of Medicine Microscope Facility for use of the Zeiss LSM700, supported by National Institutes of Health grant S10OD016374 awarded to Dr. Scot C. Kuo. This work was supported by grants from the American Heart Association (15SDG23220028) and the National Institutes of Health (National Institute of General Medical Sciences, R01GM116841) to R.F.
Author contributions: Conceptualization, S.E.L., S.K.K, and R.F.; methodology, S.E.L., S.K.K, and R.F.; investigation, S.E.L., S.K.K., L.Z., and R.F.; writing—original draft, S.E.L. and R.F; writing—review and editing, S.E.L., S.K.K., L.Z., and R.F.; funding acquisition, R.F.; supervision, R.F.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.070268.118.
REFERENCES
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. 2007. An optimized transgenesis system for Drosophila using germ-line-specific ϕC31 integrases. Proc Natl Acad Sci 104: 3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok A, Winnard P, Mukadam S, Van Diest P, Chen JH, Farabaugh P, et al. 2008. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene 27: 3912–3922. 10.1038/onc.2008.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush MS, Crowe N, Zheng T, Doonan JH. 2015. The RNA helicase, eIF4A-1, is required for ovule development and cell size homeostasis in Arabidopsis. Plant J 84: 989–1004. 10.1111/tpj.13062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera P, Johnstone O, Nakamura A, Casanova J, Jäckle H, Lasko P. 2000. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol Cell 5: 181–187. 10.1016/S1097-2765(00)80414-1 [DOI] [PubMed] [Google Scholar]
- Chen HH, Yu HI, Cho WC, Tarn WY. 2015. DDX3 modulates cell adhesion and motility and cancer cell metastasis via Rac1-mediated signaling pathway. Oncogene 34: 2790–2800. 10.1038/onc.2014.190 [DOI] [PubMed] [Google Scholar]
- Chen HH, Yu HI, Yang MH, Tarn WY. 2018. DDX3 activates CBC-eIF3–mediated translation of uORF-containing oncogenic mRNAs to promote metastasis in HNSCC. Cancer Res 78: 4512–4523. 10.1158/0008-5472.CAN-18-0282 [DOI] [PubMed] [Google Scholar]
- Del Campo M, Lambowitz AM. 2009. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell 35: 598–609. 10.1016/j.molcel.2009.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP. 2015. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci 112: 7189–7194. 10.1073/pnas.1504822112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R, Han BW, Hung JH, Xu J, Weng Z, Zamore PD. 2012. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell 151: 533–546. 10.1016/j.cell.2012.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, Elliott H, St Johnston D. 1997. Oocyte determination and the origin of polarity in Drosophila: the role of the spindle genes. Development 124: 4927–4937. [DOI] [PubMed] [Google Scholar]
- Götze M, Dufourt J, Ihling C, Rammelt C, Pierson S, Sambrani N, Temme C, Sinz A, Simonelig M, Wahle E. 2017. Translational repression of the Drosophila nanos mRNA involves the RNA helicase Belle and RNA coating by Me31B and Trailer hitch. RNA 23: 1552–1568. 10.1261/rna.062208.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther UP, Weinberg DE, Zubradt MM, Tedeschi FA, Stawicki BN, Zagore LL, Brar GA, Licatalosi DD, Bartel DP, Weissman JS, et al. 2018. The helicase Ded1p controls use of near-cognate translation initiation codons in 5′ UTRs. Nature 559: 130–134. 10.1038/s41586-018-0258-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. 1988. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 55: 577–587. 10.1016/0092-8674(88)90216-4 [DOI] [PubMed] [Google Scholar]
- Heerma van Voss MR, Vesuna F, Trumpi K, Brilliant J, Berlinicke C, de Leng W, Kranenburg O, Offerhaus GJ, Burger H, van der Wall E, et al. 2015. Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget 6: 28312–28326. 10.18632/oncotarget.4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozumi S, Hirabayashi R, Yoshizawa A, Ogata M, Ishitani T, Tsutsumi M, Kuroiwa A, Itoh M, Kikuchi Y. 2012. DEAD-box protein Ddx46 is required for the development of the digestive organs and brain in zebrafish. PLoS One 7: e33675 10.1371/journal.pone.0033675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihry RJ, Sapiro AL, Bashirullah A. 2012. Translational control by the DEAD Box RNA helicase belle regulates ecdysone-triggered transcriptional cascades. PLoS Genet 8: e1003085 10.1371/journal.pgen.1003085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I, Dreyfus M, Linder P. 1999. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem 274: 17677–17683. 10.1074/jbc.274.25.17677 [DOI] [PubMed] [Google Scholar]
- Iyer J, Wang Q, Le T, Pizzo L, Grönke S, Ambegaokar SS, Imai Y, Srivastava A, Troisí BL, Mardon G, et al. 2016. Quantitative assessment of eye phenotypes for functional genetic studies using Drosophila melanogaster. G3 6: 1427–1437. 10.1534/g3.116.027060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs HW, Knoblich JA, Lehner CF. 1998. Drosophila Cyclin B3 is required for female fertility and is dispensable for mitosis like Cyclin B. Genes Dev 12: 3741–3751. 10.1101/gad.12.23.3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicke A, Vancuylenberg J, Boag PR, Traven A, Beilharz TH. 2012. ePAT: a simple method to tag adenylated RNA to measure poly(A)-tail length and other 3′ RACE applications. RNA 18: 1289–1295. 10.1261/rna.031898.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny FH, Basler K. 2016. Drosophila DDX3/Belle exerts its function outside of the Wnt/Wingless signaling pathway. PLoS One 11: e0166862 10.1371/journal.pone.0166862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone O, Deuring R, Bock R, Linder P, Fuller MT, Lasko P. 2005. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev Biol 277: 92–101. 10.1016/j.ydbio.2004.09.009 [DOI] [PubMed] [Google Scholar]
- Kandasamy SK, Fukunaga R. 2016. Phosphate-binding pocket in Dicer-2 PAZ domain for high-fidelity siRNA production. Proc Natl Acad Sci 113: 14031–14036. 10.1073/pnas.1612393113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy SK, Zhu L, Fukunaga R. 2017. The C-terminal dsRNA-binding domain of Drosophila Dicer-2 is crucial for efficient and high-fidelity production of siRNA and loading of siRNA to Argonaute2. RNA 23: 1139–1153. 10.1261/rna.059915.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasim V, Wu S, Taira K, Miyagishi M. 2013. Determination of the role of DDX3 a factor involved in mammalian RNAi pathway using an shRNA-expression library. PLoS One 8: e59445 10.1371/journal.pone.0059445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Myong S. 2016. RNA remodeling activity of DEAD Box proteins tuned by protein concentration, RNA length, and ATP. Mol Cell 63: 865–876. 10.1016/j.molcel.2016.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotov AA, Akulenko NV, Kibanov MV, Olenina LV. 2014. Dead-box RNA helicases in animal gametogenesis. Mol Biol 48: 22–35. 10.1134/S0026893314010063 [DOI] [PubMed] [Google Scholar]
- Kotov AA, Olenkina OM, Kibanov MV, Olenina LV. 2016. RNA helicase Belle (DDX3) is essential for male germline stem cell maintenance and division in Drosophila. Biochim Biophys Acta 1863: 1093–1105. 10.1016/j.bbamcr.2016.02.006 [DOI] [PubMed] [Google Scholar]
- Lai MC, Chang WC, Shieh SY, Tarn WY. 2010. DDX3 regulates cell growth through translational control of cyclin E1. Mol Cell Biol 30: 5444–5453. 10.1128/MCB.00560-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Sun HS, Wang SW, Tarn WY. 2016. DDX3 functions in antiviral innate immunity through translational control of PACT. FEBS J 283: 88–101. 10.1111/febs.13553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Carthew RW. 2003. Making a better RNAi vector for Drosophila: use of intron spacers. Methods 30: 322–329. 10.1016/S1046-2023(03)00051-3 [DOI] [PubMed] [Google Scholar]
- Liao SE, Ai Y, Fukunaga R. 2018. An RNA-binding protein Blanks plays important roles in defining small RNA and mRNA profiles in Drosophila testes. Heliyon 4: e00706 10.1016/j.heliyon.2018.e00706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. 2011. From unwinding to clamping—the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12: 505–516. 10.1038/nrm3154 [DOI] [PubMed] [Google Scholar]
- Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, Schnier J, Slonimski PP. 1989. Birth of the D-E-A-D box. Nature 337: 121–122. 10.1038/337121a0 [DOI] [PubMed] [Google Scholar]
- Liu F, Putnam A, Jankowsky E. 2008. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci 105: 20209–20214. 10.1073/pnas.0811115106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Han H, Lasko P. 2009. Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3′ UTR. Genes Dev 23: 2742–2752. 10.1101/gad.1820709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamiya N, Worman HJ. 1999. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem 274: 15751–15756. 10.1074/jbc.274.22.15751 [DOI] [PubMed] [Google Scholar]
- Nielsen KH, Chamieh H, Andersen CB, Fredslund F, Hamborg K, Le Hir H, Andersen GR. 2009. Mechanism of ATP turnover inhibition in the EJC. RNA 15: 67–75. 10.1261/rna.1283109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Flynn RA, Floor SN, Purzner J, Martin L, Do BT, Schubert S, Vaka D, Morrissy S, Li Y, et al. 2016. Medulloblastoma-associated DDX3 variant selectively alters the translational response to stress. Oncotarget 7: 28169–28182. 10.18632/oncotarget.8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek JW, Kai T. 2011. DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc Natl Acad Sci 108: 12007–12012. 10.1073/pnas.1106245108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton JS, Huang YC, Smith L, Sun J, Leake N, Schleede J, Stevens LM, Deng WM. 2011. The microRNA pathway regulates the temporal pattern of Notch signaling in Drosophila follicle cells. Development 138: 1737–1745. 10.1242/dev.059352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam AA, Jankowsky E. 2013. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim Biophys Acta 1829: 884–893. 10.1016/j.bbagrm.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. 1976. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol 53: 217–240. 10.1016/0012-1606(76)90225-6 [DOI] [PubMed] [Google Scholar]
- Rocak S, Linder P. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5: 232–241. 10.1038/nrm1335 [DOI] [PubMed] [Google Scholar]
- Samatanga B, Andreou AZ, Klostermeier D. 2017. Allosteric regulation of helicase core activities of the DEAD-box helicase YxiN by RNA binding to its RNA recognition motif. Nucleic Acids Res 45: 1994–2006. 10.1093/nar/gkx136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. 2006. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125: 287–300. 10.1016/j.cell.2006.01.054 [DOI] [PubMed] [Google Scholar]
- Shih JW, Tsai TY, Chao CH, Wu Lee YH. 2008. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 27: 700–714. 10.1038/sj.onc.1210687 [DOI] [PubMed] [Google Scholar]
- Sienski G, Batki J, Senti KA, Dönertas D, Tirian L, Meixner K, Brennecke J. 2015. Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery. Genes Dev 29: 2258–2271. 10.1101/gad.271908.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B, Lasko P. 1998. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125: 1569–1578. [DOI] [PubMed] [Google Scholar]
- Takeo S, Swanson SK, Nandanan K, Nakai Y, Aigaki T, Washburn MP, Florens L, Hawley RS. 2012. Shaggy/glycogen synthase kinase 3β and phosphorylation of Sarah/regulator of calcineurin are essential for completion of Drosophila female meiosis. Proc Natl Acad Sci 109: 6382–6389. 10.1073/pnas.1120367109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Guichet A, Zavorszky P, Ephrussi A. 1998. Oocyte polarity depends on regulation of gurken by Vasa. Development 125: 1723–1732. [DOI] [PubMed] [Google Scholar]
- Vakrou S, Fukunaga R, Foster DB, Sorensen L, Liu Y, Guan Y, Woldemichael K, Pineda-Reyes R, Liu T, Tardiff JC, et al. 2018. Allele-specific differences in transcriptome, miRNome, and mitochondrial function in two hypertrophic cardiomyopathy mouse models. JCI Insight 3: 94493 10.1172/jci.insight.94493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega YA, Wang YD, Parker M, Patmore DM, Kanagaraj A, Moore J, Rusch M, Finkelstein D, Ellison DW, Gilbertson RJ, et al. 2016. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci Rep 6: 25996 10.1038/srep25996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, Coute Y, Conn S, Kadlec J, Sachidanandam R, et al. 2014. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157: 1698–1711. 10.1016/j.cell.2014.05.018 [DOI] [PubMed] [Google Scholar]
- Yarunin A, Harris RE, Ashe MP, Ashe HL. 2011. Patterning of the Drosophila oocyte by a sequential translation repression program involving the d4EHP and Belle translational repressors. RNA Biol 8: 904–912. 10.4161/rna.8.5.16325 [DOI] [PubMed] [Google Scholar]
- Zhang L, Yang Y, Li B, Scott IC, Lou X. 2018. The DEAD-box RNA helicase Ddx39ab is essential for myocyte and lens development in zebrafish. Development 145: dev161018 10.1242/dev.161018 [DOI] [PubMed] [Google Scholar]
- Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. 2008. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell 32: 592–599. 10.1016/j.molcel.2008.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Kandasamy SK, Fukunaga R. 2018a. Dicer partner protein tunes the length of miRNAs using base-mismatch in the pre-miRNA stem. Nucleic Acids Res 46: 3726–3741. 10.1093/nar/gky043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Kandasamy SK, Liao SE, Fukunaga R. 2018b. LOTUS domain protein MARF1 binds CCR4-NOT deadenylase complex to post-transcriptionally regulate gene expression in oocytes. Nat Commun 9: 4031 10.1038/s41467-018-06404-w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.