Abstract
Microglia maintain homeostasis in the central nervous system (CNS) through phagocytic clearance of protein aggregates and cellular debris. This function deteriorates during aging and neurodegenerative disease, concomitant with cognitive decline. However, the mechanisms of impaired microglial homeostatic function and the cognitive effects of restoring this function remain unknown. We combined CRISPR-Cas9 knockout screens with RNA-seq to discover age-related genetic modifiers of microglial phagocytosis. These screens identified CD22, a canonical B-cell receptor, as a negative regulator of phagocytosis that is upregulated on aged microglia. CD22 mediates the anti-phagocytic effect of α2–6-linked sialic acid, and inhibition of CD22 promotes the clearance of myelin debris, amyloid-β oligomers, and α-synuclein fibrils in vivo. Strikingly, long-term CNS-delivery of a CD22 function-blocking antibody reprograms microglia towards a homeostatic transcriptional state and improves cognitive function in aged mice. These findings elucidate a mechanism of age-related microglial impairment and a strategy to restore homeostasis in the aging brain.
Microglia are among the longest-lived mammalian cell types, residing in the brains of mice for years1 and humans for decades2. During this period, microglia peruse the brain parenchyma with ramified processes to sense perturbations3 and respond with specialized compensatory functions. One of the core microglial functions necessary for homeostasis in the CNS is the recognition, engulfment, and degradation of extracellular material via phagocytosis4. While microglial phagocytosis is crucial throughout an organism’s lifespan, the targets of engulfment change depending on age. For example, complement-mediated synaptic pruning is essential for developmental circuit refinement5,6, yet inappropriate activation of this pathway in neurodegenerative diseases may contribute to deleterious synapse elimination7,8. With normal aging and age-related disease, microglia encounter unique perturbations in the CNS including abundant myelin debris9 and protein aggregates. A complex system of extrinsic and intrinsic inhibitory molecules normally regulates microglial phagocytosis to enable efficient elimination of these targets without damaging surrounding tissue10. However, microglia in the aged brain are hypo-motile11, burdened with lysosomal cargo9,12,13, and chronically express pro-inflammatory signaling molecules14,15 indicative of impaired homeostatic function. We sought to understand the molecular mechanisms governing this impairment, and to determine the consequences of restoring homeostatic microglial phagocytosis on age-related brain dysfunction.
Combined CRISPR-Cas9 screens and RNA-seq analysis identify an age-associated genetic modifier of phagocytosis
Microglia undergo significant transcriptional changes with age16,17. However, the functional consequences of these molecular changes are not well understood. We aimed to identify genes that 1) modulate microglial phagocytosis, 2) are amenable to therapeutic intervention, and 3) are differentially expressed with age. To do so, we used CRISPR-Cas9 knockout screens18,19 to discover genetic modifiers of phagocytosis in the microglia-derived cell line, BV2. Given cell number limitations20,21, primary microglia screens are not currently feasible. Although BV2 cells do not accurately recapitulate all features of microglia in vivo22, the two cell types express similar phagocytic machinery (Extended Data Fig. 1a). Therefore, genetic modifiers of phagocytosis in BV2 cells are suitable candidates for subsequent validation in vivo. To identify genes that might be druggable, we used libraries of single-guide RNAs (sgRNAs) targeting 954 genes coding for transmembrane proteins and 2,015 genes coding for known drug targets, kinases, and phosphatases, with 10 sgRNAs per gene along with ~3,000 negative control sgRNAs.
Cas9-expressing BV2 cells were infected with these libraries and selected for stable sgRNA expression to obtain a pool of single-knockout cells (Fig. 1a). We incubated the cells with pH-sensitive fluorescent latex particles and sorted phagocytic and non-phagocytic populations by fluorescence-activated cell sorting (FACS) (Extended Data Fig. 1b, c). Next, we sequenced genomic DNA from each population, analyzed the sgRNA distributions between populations (Supplemental Table 1), and estimated the effect size and P-value of each gene knockout using casTLE23. These screens identified 286 significant hits, including multiple known negative and positive regulators of phagocytosis (Supplementary Table 2; Extended Data Fig. 1d, e, f, g, h).
Figure 1. Combined CRISPR-Cas9 screens and RNA-seq analysis identify an age-associated genetic modifier of phagocytosis.
a, Screening strategy for age-related genetic modifiers of phagocytosis. Scale bar = 50 microns. Screens were performed in technical duplicate.
b, RNA-seq differential expression analysis between aged (20 m.o., n=6) and young (3 m.o., n=6) primary microglia overlaid with significant hits from CRISPR-Cas9 screen of BV2 cells. Red dots represent hits that promote phagocytosis, blue dots represent hits that inhibit phagocytosis, and gray dots are knockouts that showed no effect. Dashed line represents adjusted P-value of 0.001 (Benjamini-Hochberg method).
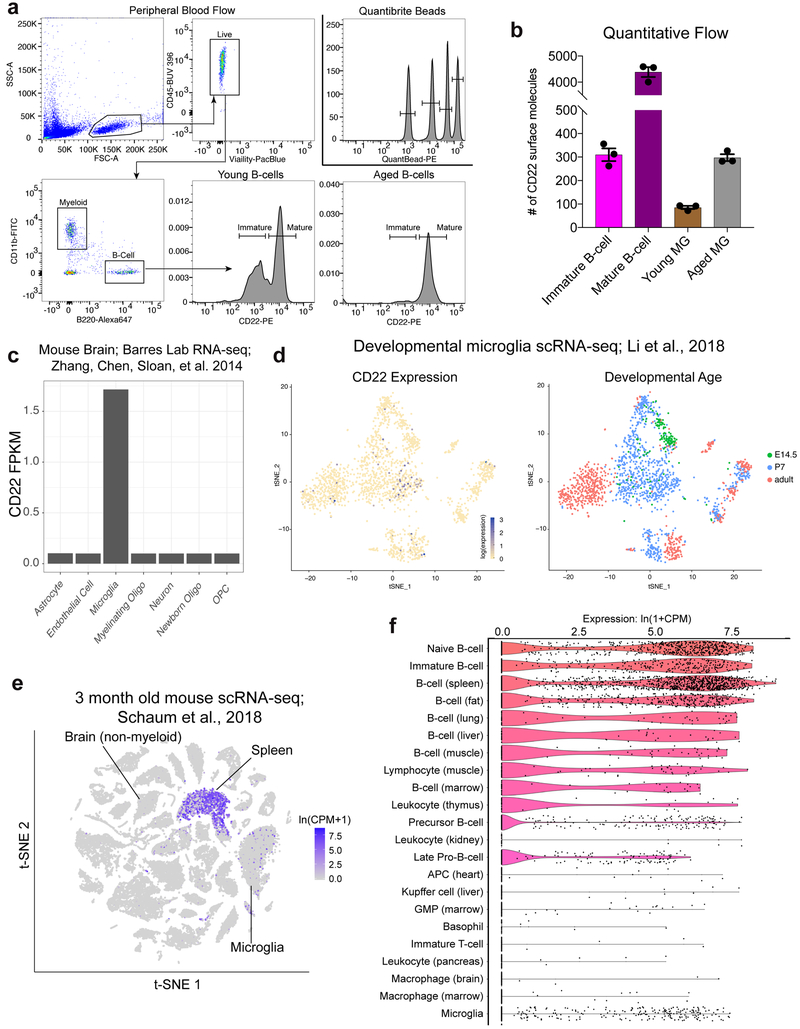
c, qPCR analysis of CD22 expression in acutely isolated primary microglia from young (2-3 m.o.) and aged (20-22 m.o.) mice, normalized to a housekeeping gene (β-actin); data represent fold change relative to young microglia (n=4, ****P<0.00005, two-sided t-test, mean +/− s.e.m.).
d, Flow cytometry quantification (mean fluorescence intensity (MFI) – fluorescence minus one (FMO) background intensity) of CD22 expression in acutely isolated primary microglia from young (2-3 m.o.) and aged (20-22 m.o.) mice (n=5, ***P<0.0005, two-sided t-test, mean +/− s.e.m.).
e, Normalized phagocytosis (fluorescent area / confluence) monitored over 24 hours, imaged every hour. Control BV2 cells were infected with a “safe-targeting” sgRNA and KO BV2 cells were infected with a sgRNA targeting CD22 (n=6, ****P<0.00005, two-sided t-test; mean +/− s.e.m.).
f, Representative images of young and aged cerebella probed for Tmem119 (green), CD22 (red), and nuclei (DAPI, blue). Green arrows indicate Tmem119+ microglia, and red-green arrows indicate Tmem119+CD22+ microglia. Scale bar = 30 microns.
g, Percentage of Tmem119+ microglia that co-express CD22 in multiple brain regions of young (3 m.o., brown) and aged (22 m.o., gray) mice (n=3, *P<0.05, ***P<0.0005, ****P<0.00005, 2-way ANOVA with Sidak correction, mean +/− s.e.m.).
Data in c-g were replicated in at least 2 independent experiments.
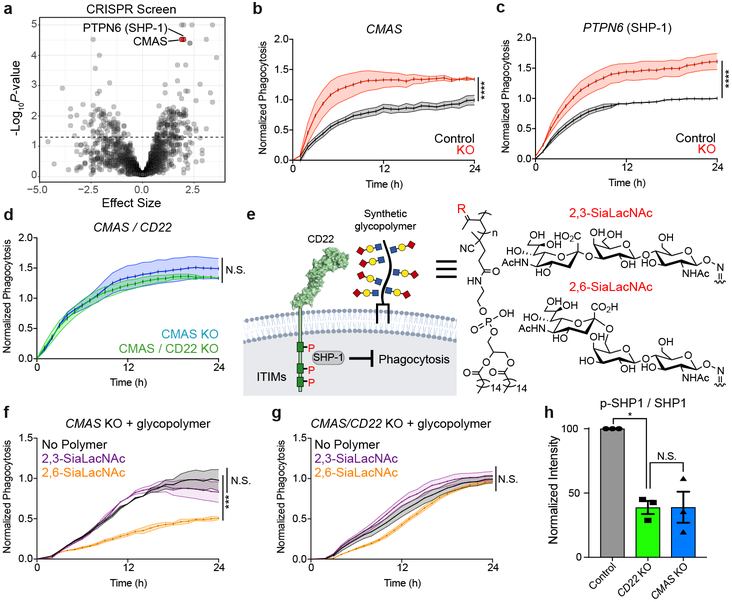
To filter this list for genes that might contribute to age-related phagocytic decline, we performed RNA-seq on hippocampal microglia from young and aged mice and compared hits from our BV2 screen to genes differentially expressed with age in primary microglia. Within the subset of genes targeted by our CRISPR-Cas9 screen, we searched for age-upregulated microglial genes whose corresponding BV2 knockout promotes phagocytosis, or age-downregulated microglial genes whose corresponding BV2 knockout inhibits phagocytosis (Fig. 1a). These genes might have a functional role in age-related microglial impairment, either through increased inhibition or decreased activation of phagocytosis. Strikingly, CD22, a sialic acid-binding immunoglobulin-like lectin (Siglec) typically expressed on B-cells24, was the only gene that fit these criteria (Fig. 1b). Specifically, CD22 is significantly upregulated in aged microglia (Fig. 1b, c), corroborating previously published datasets16,17, and knocking it out in BV2 cells promotes phagocytosis (Fig. 1d; Extended Data Fig. 1i, j, k; Supplementary Video 1, 2). CD22 negatively regulates B-cell receptor (BCR) signaling25, and has been reported to modulate leukocyte phagocytosis in fish26. However, its role in the regulation of mammalian phagocytosis is completely unknown.
Other Siglecs have been shown to regulate microglial phagocytosis27,including suppression of amyloid-beta (Aβ) clearance by CD33 in Alzheimer’s disease (AD)28. Based on our RNA-seq analysis and flow cytometry profiling of primary microglia, CD22 is the only mouse Siglec that is significantly upregulated with age in the hippocampus (Supplementary Table 3; Extended Data Fig. 2). Using a quantitative flow cytometry-based assay (Extended Data Fig. 3a), we determined that aged microglia display approximately three times more CD22 surface molecules than young microglia, comparable to the number of molecules on immature B-cells (Extended Data Fig. 3b). To assess the regional distribution of microglial CD22 expression, we performed multiplexed fluorescence RNA in situ hybridization (RNAscope) on five brain regions from young and aged mice. We probed for CD22 as well as Tmem119, a microglia specific marker29. Whereas CD22+Tmem119+ microglia were almost completely absent in the young brain, the aged brain contained a large proportion of these cells in every region that we assessed (Fig. 1f, g), particularly the thalamus and cerebellum. We did not observe CD22+ puncta outside of Tmem119+ microglia, corroborating previously published RNA-seq datasets30 that show CD22 is expressed exclusively by microglia in the mouse CNS (Extended Data Fig. 3c, e, f).
CD22 mediates the anti-phagocytic effect of α2–6-linked sialic acid
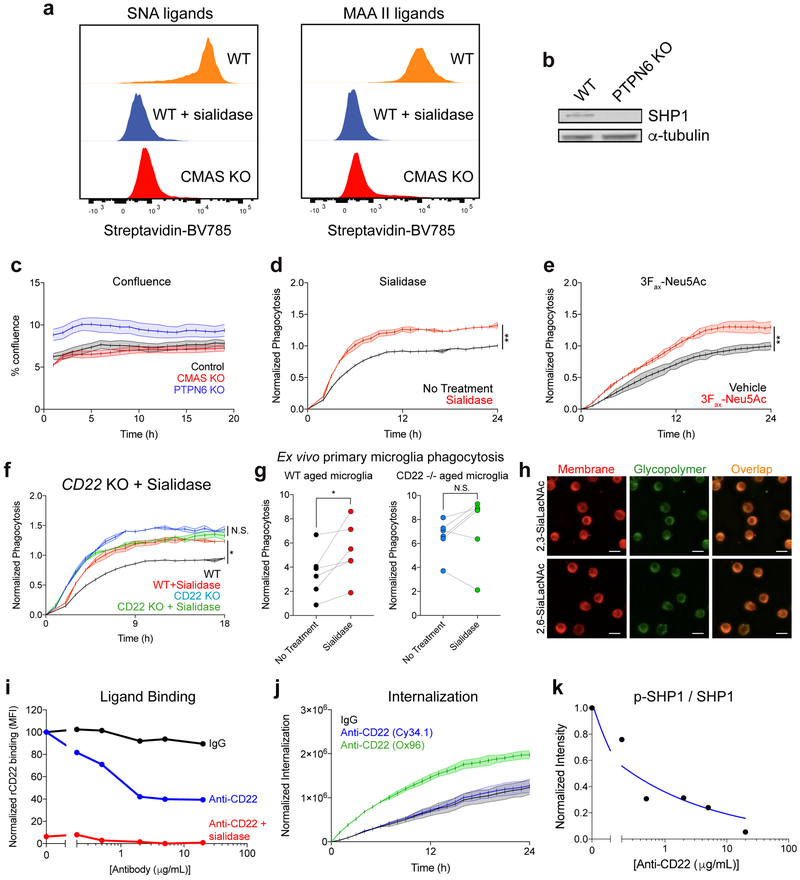
CD22 is canonically expressed on B-cells, where it negatively regulates BCR signaling by binding sialic acid and recruiting SHP-1 or SHIP-1 via immunoreceptor tyrosine-based inhibitory motifs (ITIMs)31. To search for possible signaling partners of CD22 on microglia, we re-analyzed our initial CRISPR-Cas9 screen for hits related to CD22 function. Surprisingly, CMAS, a key enzyme in sialic acid synthesis, and PTPN6, which codes for SHP-1, were among the most significant hits (Fig. 2a). Time-lapse microscopy confirmed that knocking out CMAS or PTPN6, or removal of sialic acid via treatment with sialidase or 3Fax-Neu5Ac, a sialic acid biosynthesis inhibitor, robustly promotes phagocytosis (Fig. 2b, c; Extended Data Fig. 4a, b, c, d, e), phenocopying CD22 ablation. However, genetic or pharmacological inhibition of both CD22 and sialic acid simultaneously did not produce an additive phagocytic effect (Fig. 2d; Extended Data Fig. 4f, g), suggesting that sialic acid is involved in CD22-mediated inhibition of phagocytosis.
Figure 2. CD22 mediates the anti-phagocytic effect of α2-6-linked sialic acid.
a, Results from CRISPR-Cas9 screen targeting 2,015 drug targets, kinases, and phosphatases in BV2 cells (screen performed in technical duplicate; dashed line, P=0.05, two-sided t-test).
b, c, d, Phagocytosis of pH-sensitive fluorescent beads by control (black) vs CMAS KO (red) BV2 cells (b), control (black) vs PTPN6 KO (red) cells (c), and CMAS KO (blue) vs CMAS/CD22 double KO (green) cells (d) (n=3, ****P<0.00005, N.S. not significant, two-sided t-test; mean +/− s.e.m.).
e, Cell-surface glycan engineering using lipid tail-functionalized glycopolymers bearing sialic acid α2,3- or α2–6-linked to N-acetyllactosamine. ITIMs = immunoreceptor tyrosine-based inhibitory motifs, SHP-1 = Src homology region 2 domain-containing phosphatase-1.
f, g, Phagocytosis of pH-sensitive fluorescent beads by CMAS KO (f) or CMAS/CD22 double KO (g) BV2 cells coated with no polymer (black), α2,3-linked sialic acid (purple) or α2,6-linked sialic acid (orange) (n=3, ***P<0.0005, N.S. not significant, one-way ANOVA with Tukey’s correction; mean +/− s.e.m.).
h, Western blot quantification of ratio of active phosphorylated SHP1 (p-SHP1) to total SHP1 in control (gray), CD22 KO (green), and CMAS KO (blue) BV2 cells, normalized to a loading control protein (α-tubulin) (n=3, *P<0.05, N.S. not significant, one-way ANOVA with Tukey’s correction). For raw source image, see Supplementary Figure 1.
Data in b-h were replicated in at least 2 independent experiments.
To probe CD22-sialic acid receptor-ligand interactions, we utilized synthetic glycopolymers functionalized with lipid tails to introduce defined glycans onto the cell-surface32 (Fig. 2e). We decorated CMAS knockout cells, devoid of endogenous sialic acid, with synthetic glycopolymers bearing sialic acid linked to N-acetyllactosamine in either an α2–3- or α2–6-configuration and confirmed membrane incorporation (Extended Data Fig. 4h). Addition of α2–6-linked sialic acid, the preferred ligand of CD2233, but not α2–3-linked sialic acid, inhibited phagocytosis in CMAS KO cells (Fig. 2f). Strikingly, the anti-phagocytic effect of α2–6-linked sialic acid was abrogated in a CMAS/CD22 double knockout background (Fig. 2g). These findings demonstrate that α2–6-linked sialic acid inhibits phagocytosis in a CD22-dependent manner.
To assess inhibitory signaling downstream of CD22, we measured basal SHP-1 activity in BV2 cells and found that CD22 KO cells express a reduced ratio of active phosphorylated SHP-1 to total SHP-1 protein, comparable to CMAS KO cells (Fig. 2h). Furthermore, treatment with a CD22 ligand-blocking antibody (Extended Data Fig. 4i, j) decreased SHP-1 activity in a dose-dependent manner (Extended Data Fig. 4k). Taken together, these data support a model in which CD22 binds an α2–6-sialylated ligand and promotes inhibitory SHP-1 signaling to restrain the phagocytic capacity of microglia.
CD22 inhibition promotes microglial phagocytosis in vivo
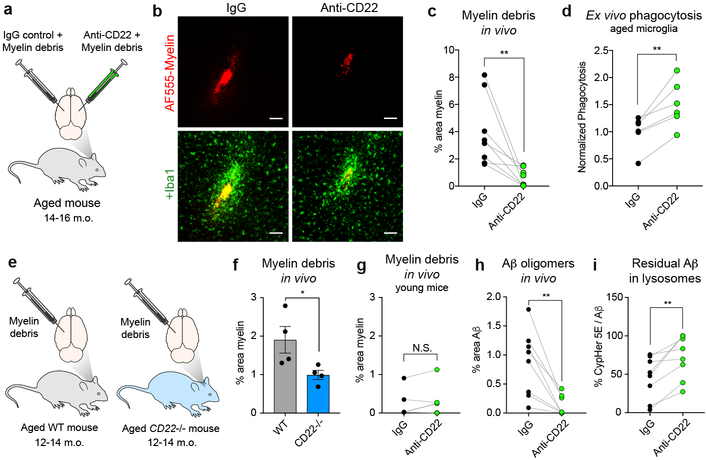
To assess the functional significance of age-related microglial CD22 upregulation in vivo, we stereotactically co-injected myelin debris, a phagocytic substrate that accumulates in the aging brain, with a CD22 blocking antibody or an IgG isotype control antibody into opposite hemispheres of the same aged mouse (Fig. 3a). We labeled the myelin debris with a constitutively fluorescent dye to assess clearance and with a pH-sensitive dye to confirm engulfment and lysosomal trafficking within Iba1+ cells (Extended Data Fig. 5a, b).
Figure 3. CD22 inhibition restores microglial phagocytosis in vivo.
a, Myelin debris labeled with a fluorescent dye (AF555) was stereotactically co-injected with anti-CD22 or IgG into the cortex on opposite hemispheres of the same aged (14-16 m.o.) mouse.
b, Representative images of AF555-labed myelin (red, top row) overlaid with the myeloid marker Iba1 (green, bottom row) at the injection sites of IgG (left) or anti-CD22 (right) treated hemispheres of the same aged brain. Scale bar = 100 microns.
c, Clearance of myelin debris in IgG (black) or anti-CD22 (green) treated hemispheres of aged mice assessed 48 hours post-injection (n=8, **P<0.005, paired two-sided t-test).
d, Flow cytometry quantification of ex vivo phagocytosis of pH-sensitive beads by aged microglia pretreated with IgG or anti-CD22 (n=6, **P<0.005, paired two-sided t-test).
e, Labeled myelin debris was stereotactically injected into the cortices of aged (12–14 m.o.) WT or CD22−/− mice.
f, Clearance of myelin debris in cortices of aged WT (black) vs CD22−/− (green) mice assessed 48 hours post-injection (n=4, *P<0.05, two-sided t-test, mean +/− s.e.m.).
g, Clearance of myelin debris in IgG (black) or anti-CD22 (green) treated hemispheres of young mice assessed 48 hours post-injection (n=4, N.S. not significant, paired two-sided t-test).
h, Clearance of Aβ oligomers in IgG (black) or anti-CD22 (green) treated hemispheres of aged mice assessed 48 hours post-injection (n=8, **P<0.005, paired two-sided t-test).
i, Percent area of residual Aβ oligomers that were CypHer5E+, indicating localization to acidified lysosomes (n=8, **P<0.005, paired two-sided t-test).
All data were replicated in at least 2 independent experiments.
After 48 hours, anti-CD22 treatment robustly promoted clearance of myelin debris compared to IgG (Fig. 3b, c). Because microglia appear to be the only cell type in the mouse CNS that express CD22 (Extended Data Fig. 3c), this effect is likely microglia-specific. Importantly, CD22 blockade did not influence the recruitment of microglia to the injection site relative to IgG (Extended Data Fig. 5c) or a stab wound control (Extended Data Fig. 5d, e), and IgG itself did not influence myelin clearance (Extended Data Fig. 5f). Nearly all Iba1+ cells at the injection site were Tmem119+ regardless of treatment (Extended Data Fig. 5g, h), suggesting that the pro-phagocytic effect of CD22 blockade is mainly mediated by resident microglia and not infiltrating peripheral macrophages29. We confirmed the pro-phagocytic effect of anti-CD22 treatment ex vivo using freshly isolated microglia from aged mice and pH-sensitive fluorescent latex particles (Fig. 3d). Next, we injected labeled myelin debris into the brains of aged CD22−/− mice or age-matched wild-type mice (Fig. 3e). We observed comparable clearance of myelin debris with genetic ablation of CD22 as with antibody-based blockade of CD22 (Fig. 3f). Furthermore, CD22 blockade (Fig. 3g) or genetic ablation (Extended Data Fig. 5i) had no significant effect in the young mouse CNS, where myelin debris is efficiently cleared at baseline and CD22 is minimally expressed.
Next, we evaluated the effect of anti-CD22 treatment on microglial phagocytosis of oligomeric Aβ, a particularly neurotoxic protein aggregate that accumulates in the AD brain34. Compared to IgG, anti-CD22 treatment efficiently cleared injected Aβ oligomers in vivo (Fig. 3h; Extended Data Fig. 5j, k, l). Interestingly, a larger percentage of residual Aβ in anti-CD22 treated hemispheres was contained in acidified lysosomes (Fig. 3i), suggesting that CD22 blockade promotes degradation of engulfed debris. In an analogous in vivo phagocytosis assay, we found that anti-CD22 treatment promotes the clearance of extracellular α-synuclein fibrils (Extended Data Fig. 5m, n, o), a pathological hallmark of Parkinson’s disease. Taken together, these data suggest that CD22 is a broad negative regulator of microglial phagocytosis in the aged CNS.
Long-term CD22 blockade restores microglial homeostasis and improves cognitive function in aged mice
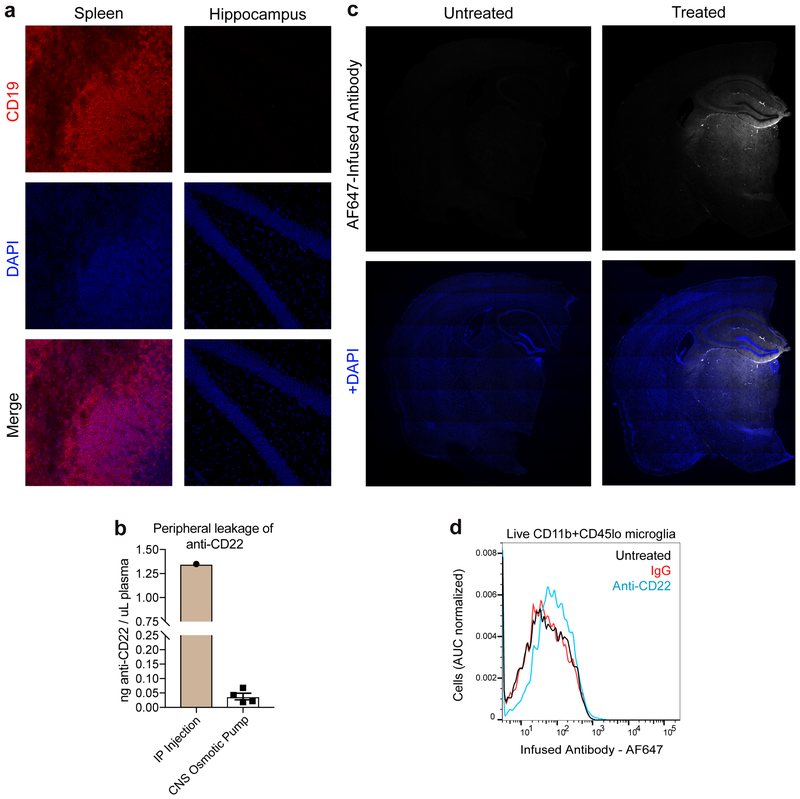
Aging and disease overwhelm the homeostatic function of microglia, leading to a distinctive transcriptional state35 characterized by the downregulation of resting microglial genes and the upregulation of activated microglial genes. To assess the transcriptional effects of CD22 blockade, we implanted aged mice with osmotic pumps to continuously infuse a CD22 blocking antibody or an IgG control antibody directly into the cerebrospinal fluid for one month (Fig. 4a). As opposed to systemic antibody administration or CD22−/− mice, this intervention enabled CNS-specific targeting of CD22 without B-cell infiltration into the brain (Extended Data Fig. 6a) or significant leakage into the periphery (Extended Data Fig. 6b). We labeled the respective antibodies with a fluorescent dye in order to assess drug distribution within the brain parenchyma (Extended Data Fig. 6c) and target engagement with CD22 on microglia (Extended Data Fig. 6d).
Figure 4. Long-term CD22 blockade restores microglial homeostasis and improves cognitive function in aged mice.
a, Schematic of long-term CNS-targeted CD22 blockade.
b, c, Correlation of gene fold changes between microglia from anti-CD22 (n=7) and IgG (n=7) treated mice and between microglia from aged (22 m.o., n=3) and young (3 m.o., n=3) mice36 (b) (R = −0.47, P=1.7e-12) or between microglia from 5xFAD (8.5 m.o., n=5) and age-matched WT (8.5 m.o., n=5) mice37 (c) (R = −0.27, P=8.5e-6, blue line, linear regression, Spearman correlation). Genes differentially expressed (P<0.05, two-sided t-test) in either dataset (union) are shown.
d, Hierarchical clustering of gene fold changes between microglia from anti-CD22 (n=7) and IgG (n=7) treated mice, between microglia from aged (22 m.o., n=3) and young (3 m.o., n=3) mice36, and between microglia from 5xFAD (8.5 m.o., n=5) and age-matched WT (8.5 m.o., n=5) mice37. Genes differentially expressed (P-value<0.05, two-tailed t-test) in all three datasets (intersection) are shown.
e, f, g, h, Percentage of time spent in the novel arm of the forced alternation Y-maze (e, g) or displaying freezing behavior in a contextual fear conditioning test (f, h) for aged (18 m.o.) WT and CD22−/− mice (e, f, n=6) or aged (18 m.o.) WT mice treated with IgG or anti-CD22 (g, h, independent cohorts of mice in g (n=7 IgG, 9 anti-CD22) and h (n=7 for both groups)) (**P<0.005, *P<0.05, two-sided t-test; mean +/− s.e.m.).
i, Representative images of dentate gyri stained for Prox1 (white) and c-Fos (red) from IgG and anti-CD22 treated mice. White arrows indicate cFos+Prox1+ active granule neurons. Scale bar = 100 microns.
j, Total number of Prox1+c-Fos+ neurons in the dentate gyrus quantified over 5 tissue sections (n=7, *P<0.05, two-sided t-test; mean +/− s.e.m.).
Data in a, g, h, i, j were replicated in at least 2 independent experiments.
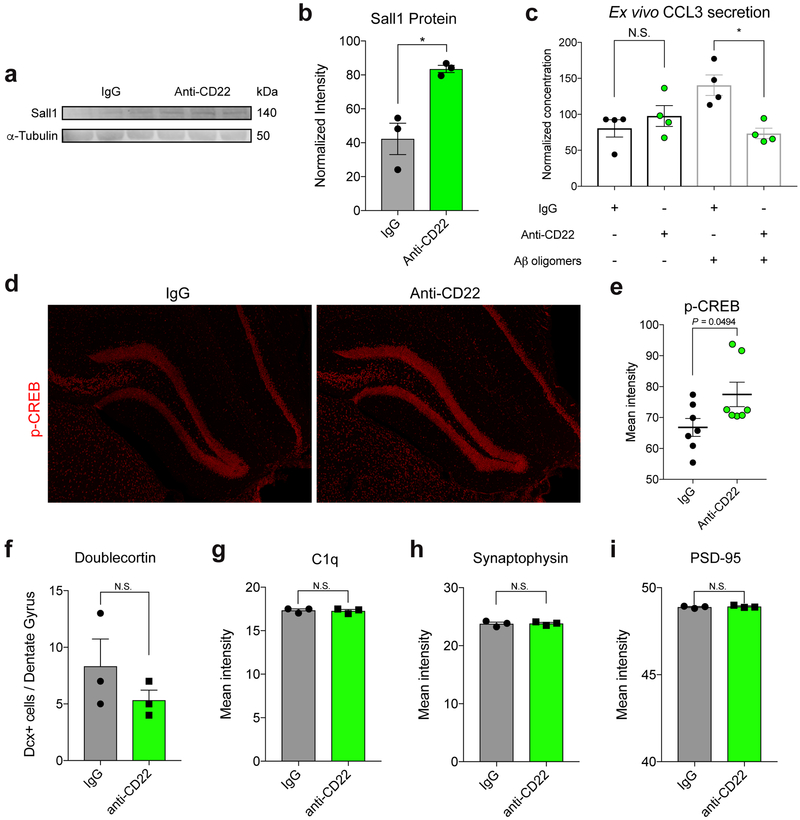
Following one month of continuous infusion, we performed RNA-seq on purified microglia from the hemi-brains of these mice contralateral to the cannulation site to minimize injury-induced confounding factors. We found very few transcriptional changes between microglia from untreated and IgG-infused mice (Extended Data Fig. 7a). In contrast, we found 315 differentially expressed genes between IgG and anti-CD22 treated microglia meeting a false discovery rate (FDR) cutoff of 10% (Supplementary Table 4; Extended Data Fig. 7b). We compared these genes to those that are differentially expressed between microglia from young and aged mice36, and between microglia from wild-type and end-stage 5xFAD AD-model mice37 (Fig. 4b, c). Remarkably, anti-CD22 treatment promoted the upregulation of homeostatic microglial genes (e.g. Tgfbr1, P2ry13, Sall1, Il10ra, Mef2a; Fig. 4d, Extended Data Fig. 7c, Extended Data Fig. 8a, b) and the downregulation of activated and disease-associated microglial genes (e.g. H2-K1, Tspo, Lgals3, Tnfsf13b, Ccl3; Fig. 4d, Extended Data Fig. 7d-h), partially reversing the transcriptional hallmarks of aging and disease. We confirmed downregulation of CCL3, a chemokine implicated in hippocampal impairment38, by ex vivo secretome profiling (Extended Data Fig. 8c). Of note, CD22 blockade abrogated CCL3 secretion in the presence of oligomeric Aβ, but had no effect on basal levels.
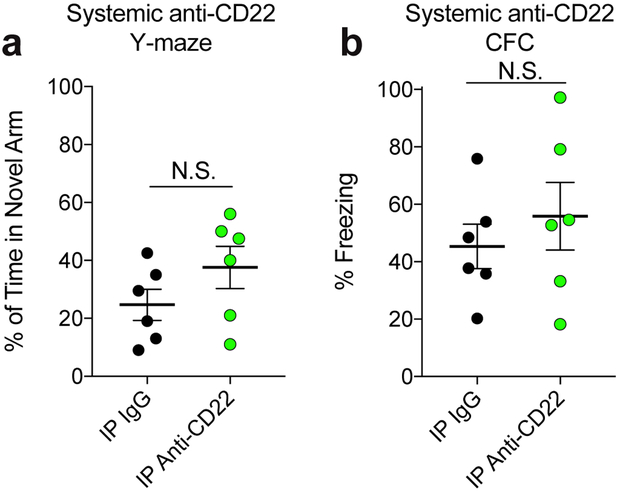
To determine the effects of CD22 inhibition on age-related cognitive dysfunction, we assessed hippocampal-dependent learning and memory performance in aged WT and CD22−/− mice. Surprisingly, CD22−/− mice exhibited improved spatial memory in the forced-alternation Y-maze test (Fig. 4e) and improved associative memory in the contextual fear conditioning test (Fig. 4f). To more specifically assess the contribution of microglial CD22 to this behavioral phenotype, we performed the same tests in aged mice infused with IgG or anti-CD22 directly into the CNS via osmotic pump. Remarkably, blocking CD22 specifically in the CNS phenocopied the learning and memory improvements seen in CD22−/− mice (Fig. 4g, h), suggesting that microglial CD22 negatively and reversibly regulates cognitive function in the aging brain. Of note, we did not observe a significant cognitive improvement in aged mice treated systemically with anti-CD22 via intraperitoneal injection, suggesting that peripheral B-cells are unlikely to contribute to this effect (Extended Data Fig. 9a, b).
These hippocampal-dependent behavioral phenotypes were accompanied by an increase in the number of hippocampal Prox1+ dentate granule neurons expressing c-Fos, an immediately early gene induced by neuronal activation39 (Fig. 4i, j). In addition, anti-CD22 treatment promoted a modest increase in phosphorylated-CREB (Extended Data Fig. 8d, e), a plasticity-related marker40, but did not alter hippocampal neurogenesis (Extended Data Fig. 8f). We observed no difference in C1q deposition (Extended Data Fig. 8g) or synaptic density, as assessed by the intensity of pre- and post-synaptic markers, synaptophysin and PSD-95 (Extended Data Fig. 8h, i).
Discussion
Here, we show that CD22 is a negative regulator of phagocytosis that is upregulated on aged microglia. CD22 mediates the anti-phagocytic effect of α2–6-linked sialic acid, and inhibition of CD22, either via antibody blockade or genetic ablation, promotes the clearance of myelin debris, Aβ oligomers, and α-synuclein fibrils in vivo. Long-term CD22 inhibition partially reverses the transcriptional signature of age- and disease-related microglia, increases markers of neuronal activation, and, ultimately, improves cognitive function in aged mice (Extended Data Fig. 10).
These findings raise important questions. First, what causes CD22 upregulation in the aging brain? Recent studies have highlighted the importance of environmental cues in shaping microglial identity41,42,43. Based on our recent scRNA-seq atlas of microglia during development44, CD22 is enriched in a subpopulation of postnatal day 7 microglia (Extended Data Fig. 3D) that engulf apoptotic oligodendrocytes and resemble disease-associated microglia of aging and neurodegenerating brains. Therefore, it is possible that CD22 is upregulated as a negative feedback mechanism to restrain excessive phagocytosis in response to overwhelming cellular debris in the developing brain. During aging, this protective developmental program might be inappropriately re-activated in response to increased myelin fragmentation and protein aggregation.
Correspondingly, how does CD22 inhibition improve cognitive function in aged mice? Our RNA-seq analysis revealed that anti-CD22 reduces the microglial expression of various neuromodulatory pro-inflammatory molecules. Age-related impairments in perivascular glymphatic45 and meningeal lymphatic46 systems have been shown to affect cognitive function, and CD22-regulated phagocytosis may cooperate with these clearance pathways to control debris accumulation and local cytokine concentration in the aging brain.
Similar to their dysfunction during normal aging, microglia in the AD brain display signs of impaired phagocytosis, and allelic variants in microglia-specific AD risk genes modify phagocytic function28,47,48,49,50. Several human RNA-seq datasets show that CD22 is upregulated in the AD brain51, enriched in subpopulations of microglia correlated with clinicopathological decline52, and, anomalously, expressed on human oligodendrocytes53. Furthermore, a variant in the INPP5D locus, which codes for the CD22 signaling partner SHIP-1, is associated with AD risk54. Determining whether CD22 function in the CNS is conserved between mice and humans will be an important step prior to translational studies.
In addition to AD, recent studies have found that CD22 is drastically upregulated in mouse models of amyotrophic lateral sclerosis55, a motor neuron disease, and Niemann Pick Type C56, a lysosomal storage disease. Future investigations of CD22 on microglia may illuminate therapeutic strategies to treat or prevent these neurodegenerative diseases via restoration of homeostasis in the CNS.
Methods
Animals
Aged C57BL/6 mice (18–24 months old) were obtained from the National Institute on Aging rodent colony. Young male C57BL/6 mice (2–4 months old) were obtained from Jackson Laboratories or Charles River Laboratories. CD22−/− mice were originally generated by L. Nitschke (University of Erlangen) and generously provided by James Paulson (Scripps Institute) and Romain Ballet and Eugene Butcher (Stanford University). All experiments used male mice, except aged WT vs CD22−/− behavioral experiments contained balanced groups of male and female mice. All animal care and procedures complied with the Animal Welfare Act and were in accordance with institutional guidelines and approved by the V.A. Palo Alto Committee on Animal Research and the institutional administrative panel of laboratory animal care at Stanford University.
Cell culture
BV2 cells were originally obtained from E. Blasi (Università di Modena e Reggio Emilia) and expanded in DMEM supplemented with 10% FBS, penicillin/streptomycin, and GlutaMAX (Thermo Fisher Scientific). HEK293T cells were cultured in DMEM supplemented with 10% FBS, penicillin/streptomycin, and GlutaMAX. All cells were maintained in a humidified incubator containing 5% CO2 at 37 °C.
CRISPR-Cas9 Screen
The 10-sgRNA-per-gene CRISPR/Cas9 deletion library was synthesized, cloned, and infected into Cas9-expressing BV2 cells as previously described18. Briefly, ~12 million (for Membrane Proteins sub-library) or ~24 million (for Drug Targets, Kinases, Phosphatases sub-library) BV2 cells stably expressing EF1alpa-Cas9-BLAST were infected with the 10 guide/gene sgRNA sub-libraries (Addgene cat nos. 1000000122 and 1000000124) at an MOI < 1. Infected cells underwent puromycin selection (1.5 μg/mL) for 5 days after which point puromycin was removed and cells were resuspended in normal growth media without puromycin. After selection, sgRNA infection was confirmed by flow cytometry, which indicated >90% of cells expressed the mCherry reporter. Sufficient sgRNA library representation was confirmed by deep sequencing after selection. Cells were cultured and maintained at 1,000x coverage for one week. Phagocytic screening prey were prepared by labeling 3 μm amino-coated polystyrene particles (Polysciences) with CypHer5E pH-dependent fluorescent dye (GE Life Sciences). For the membrane proteins screen, 12.5 million cells were sorted per replicate (>1000x coverage) with 55.2% in the non-phagocytic gate and 11.2% in the phagocytic gate. For the drug target, kinase, phosphatase screen, 24 million cells were sorted per replicate (>1000x coverage) with 53.1% in the non-phagocytic and 19.4% in the phagocytic gate. At the end of each screen, genomic DNA was extracted for all populations separately using a QIAGEN Blood Midi Kit. Deep sequencing of sgRNA sequences on an Illumina Nextseq was used to monitor library composition. Guide composition was analyzed and compared to the plasmid library and between conditions using casTLE (https://bitbucket.org/dmorgens/castle). Briefly, casTLE compares each set of gene-targeting guides to the negative controls, comprising non-targeting and non-genic (“safe-targeting”) sgRNAs, which have been shown to more aptly control for on-target toxicity due to endonuclease-induced DNA damage. The enrichment of individual guides was calculated as the log ratio between phagocytic and non-phagocytic populations, and gene-level effects were calculated from ten guides targeting each gene. P values were then calculated by permutating the targeting guides as previously described23.
Primary Microglia Isolation
Primary mouse microglia were isolated as previously described20. Briefly, mice were transcardially perfused with ice-cold HBSS containing glucose and HEPES, and brains were removed into the same medium containing DNAse I. On ice, brains were minced with a razor blade and a single cell suspension was obtained by gentle Dounce homogenization. The suspension was filtered through a 100 micron strainer. Myelin was removed using magnetic myelin removal beads (Miltenyi Biotec). The remaining myelin-depleted cell suspension was stained with antibodies to distinguish microglia by flow cytometry. Previous studies have demonstrated the intense autofluorescent signal of microglia in the aging brain, likely due to the accumulation of the age-related pigment, lipofuscin14. In order to measure the expression of proteins on aged microglia by flow cytometry, we developed a gating scheme (Extended Data Figure 3F) to exclude autofluorescent cells (~10% of total microglia) from our analysis. Specifically, we excluded cells with high side-scatter signal which coincided with the autofluorescent population. Whenever possible, we used fluorophores with emission spectra in far red channels (e.g. APC-Cy7). Although this technique excludes an interesting age-specific population from our analyses, it enables confident cell-surface profiling of the majority of aged microglia by flow cytometry. For RNA-seq, all microglia (including autofluorescent cells) were collected.
RNA-sequencing and analysis
20,000 microglia were isolated from either hemibrains or micro-dissected hippocampi as described above, and sorted into RLT Plus buffer (Qiagen) containing beta-mercaptoethanol. RNA was extracted using a RNeasy Micro Plus kit (Qiagen) according the manufacturer’s protocol. RNA integrity was assessed on a Bioanalyzer (Agilent), and high quality samples were used for library preparation. cDNA synthesis and amplification was performed using the SmartSeq v4 Ultra-low input kit (Takara), and libraries were tagmented, adaptor ligated, and indexed using the Nextera XT kit (Illumina). After normalization and pooling, libraries were sequenced on a Hiseq 4000 (Illumina) using paired-end 100bp reads. Libraries were sequenced to a depth of >30 million reads per sample. Raw sequencing files were demultiplexed with bcl2fastq, reads were aligned using STAR, the count matrix was generated using SummarizedExperiment, and differential expression analysis was performed using DESeq2 with standard settings.
Flow cytometry
Antibodies to CD11b (clone M1/70, BioLegend) and CD45 (clone 30-F11, Biolegend) were used for microglia identification (CD11b+CD45lo). For primary microglia immunophenotyping, the following lectins and antibodies were used: biotinylated Sambucus nigra agglutinin (SNA, Vector Labs), biotinylated Maackia amurensis agglutinins (MAA-1 and MAA-2, Vector), biotinylated Erythrina cristagalli lectin (ECL, EY Labs), biotinylated wheat germ agglutinin (WGA, Vector), recombinant mouse Siglec-E (R&D), recombinant mouse Siglec-F (R&D), antibodies to mouse Siglec-1 (clone REA197, Miltenyi Biotec), CD22 (clone OX-97, Biolegend), Siglec-E (clone M1304A01, Biolegend), Siglec-F (clone ES22–10D8, Miltenyi Biotec), Siglec-G (clone SH1, BD Biosciences), Siglec-H (clone 551, Biolegend), CD33 (clone 9A11, eBioscience). For immunostaining, cells were passed through a 100 micron strainer, blocked for 10 minutes on ice with mouse Fc-blocking reagent (BD), and stained for 30 minutes on ice in PBS supplemented with 0.5% bovine serum albumin. When biotinylated antibodies or lectins were used, cells were stained with APC-Cy7-conjugated streptavidin (Biolegend) for 15 minutes on ice following primary stain. When recombinant Siglecs were used, they were precomplexed with APC-Cy7-conjugated anti-human IgG Fc (Biolegend) on ice at 10 micrograms/mL each. The complex was used at 1:2 for a final staining concentration of 5 micrograms/mL. Live cells were identified using Sytox Blue viability dye. Flow cytometry analysis was performed on a BD LSRFortessa and sorting was performed on a BD FACSAria III. Data was analyzed using FlowJo software (TreeStar).
Quantitative flow cytometry
Primary microglia from young and aged mice were isolated as described above. For the same mice, peripheral blood was collected with EDTA anticoagulant by terminal intracardial bleeding. Whole blood was mixed 1:1 with PBS and layered over Ficoll-Paque Plus (GE), spun at 400 x g for 30 minutes with no break, and the peripheral blood mononuclear fraction was collected. Microglia and peripheral blood cells were stained with immunophenotyping antibodies as well as PE-conjugated anti-CD22 (Ox97, Biolegend). Quantibrite-PE (BD) beads were resuspended in FACS buffer and analyzed on a BD LSRFortessa. Using the same laser powers and detector voltages, peripheral blood and microglia samples were analyzed for CD22 expression. A standard curve was constructed to correlate PE intensity with number of PE molecules per Quantibrite bead. Finally, CD22 molecule numbers on microglia and peripheral blood cells were calculated by interpolation.
qPCR
Microglia were isolated by FACS and RNA was isolated as described above. cDNA synthesis and amplification were performed with a SMART-seq V4 Ultra-Low Input RNA Kit (Clontech). Samples were diluted and mixed with SYBR green master mix before loading as technical triplicates for qPCR on a LightCycler 480 (Roche). ΔΔCT values normalized to β-actin were used to assess relative gene expression between samples. The following validated primer pairs for murine CD22 were used: 5’-CCA CTC CTC AGG CCA GAA ACT-3’ (forward) and 5’-TGC CGA TGG TCT CTG GAC TG-3’ (reverse).
In vitro phagocytosis assay
For in vitro phagocytosis assays, BV2 cells were split into a 24-well plate 24 hours prior to feeding at a density of 50,000 cells/well in serum free medium. Following specific treatments, wells were washed twice with PBS and fed pHrodo red-labeled 3 micron amino-coated polystyrene particles (Polysciences) at a ratio of 10 particles per cell. 4 phase and red fluorescent images per well were acquired every hour for 24 hours using the Incucyte S3 live cell analysis system (Essen Bioscience). For each time point, we calculated normalized phagocytosis using the following formula: red object area / phase confluence. To combine data from independent experiments, we calculated phagocytosis relative to control by setting phagocytosis at zero hours in the control condition to 0, and this value at 24 hours to 1. Technical triplicates for each experiment were averaged, and the average of three independent experiments was reported.
RNAScope in situ hybridization
RNA in situ hybridization was performed on fresh frozen brain tissue using the Multiplex Fluorescence v2 kit (Advanced Cell Diagnostics) according to the manufacturer’s protocol. Probes for CD22 and Tmem119 were commercially available from the manufacturer.
Lentivirus production and infection
HEK293T cells were transfected with packaging plasmids and sgRNA-containing plasmids. Supernatant was harvested at 48 hours and 72 hours and concentrated with Lenti-X solution (Clontech). BV2 cells stably expressing Cas9 endonuclease under blasticidin (1 μg/mL) selection were spin infected with lentivirus containing sgRNA plasmids under puromycin selection. Puromycin selection (1.5 μg/mL) was started 48 hours after infection and maintained for 7–14 days. Antibiotic selection was subsequently removed for expansion and freezing. CMAS and CD22 KO BV2 cells were sub-cloned by single-cell sorting to obtain a monoclonal knockout population. Other single-knockout cell lines were assayed as a polyclonal population.
Sialidase Expression
Sialidase from Vibrio cholerae was prepared as previously described57. In brief, Escherichia coli C600 transfected with the pCVD364 vector were grown in 2xYT media supplemented with ampicillin (100 mg/mL) for 12 hours at 37 °C. Cells were collected by centrifugation at 4,500 x g for 10 minutes. The pellet was resuspended in osmotic shock buffer (20% sucrose, 1 mM EDTA, 30 mM Tris-HCl, pH 8), incubated for 10 min at RT, then spun at 13,000 x g for 10 minutes. The pellet was resuspended in ice cold ddH2O, incubated at 4 °C for 10 min, and then spun at 13,000 x g. The supernatant was concentrated using a tangential flow filtration system (Pall Corporation). The concentrate was loaded onto a HiTrap Q HP anion exchange column (GE Healthcare Life Sciences) and eluted using a gradient of NaCl in 20 mM Tris, pH 7.6. Fractions containing sialidase were assessed by SDS-PAGE and pooled. Endotoxins were removed using a high capacity endotoxin removal kit (Thermo Fisher Scientific).
Sialic Acid Inhibition
For sialidase treatment, cells were incubated with 400nM Vibrio cholerae sialidase for 1 hour at 37 °C in serum-free DMEM prior to phagocytosis. 3Fax-Neu5Ac (R&D) was used as previously described. Briefly, the inhibitor was used at a concentration of 32 μM serially diluted in DMSO and used at a final concentration of 0.1% DMSO in DMEM. 3Fax-Neu5Ac was added with serum treatments for 48 hours prior to phagocytosis assays, and replaced in the feeding medium.
Glycopolymer Decoration
Aminooxy glycan-conjugated polymers were based on a poly(methyl vinyl ketone) backbone coupled to a dipalmitoylphosphatidylethanolamine lipid anchor. Glycopolymers were conjugated at 70–85% of ketone sites to either Neu5Ac-α2–3-LacNAc or Neu5Ac-α2–6-LacNAc through oxime chemistry. Cells were decorated with aminooxy glycan-conjugated polymers as previously described32. Briefly, cells were harvested and resuspended in PBS at 107 cells/mL. Labeling was carried out by incubating the cell suspension with 1 μM polymer for 30 minutes at room temperature with constant gentle agitation. Cells were washed with PBS and split into a 24-well plate at 50,000 cells/well for subsequent phagocytosis assays.
Western blot
Protein lysates were prepared by incubating cell pellets on ice in Pierce IP Lysis Buffer or RIPA Lysis Buffer with cOmplete protease inhibitor cocktail (Thermo Fisher Scientific) and spun at 13,000 x g for 10 minutes. The supernatant was collected and protein concentration was measured with the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). An aliquot containing 5–20ug of protein from each sample was mixed with 4X loading buffer (Thermo Fisher Scientific) and boiled for 5 minutes at 95 °C before being subjected to SDS-PAGE and transferred to a nitrocellulose membrane (Bio-Rad). The membrane was first blocked with Odyssey Blocking Buffer (LI-COR) and stained overnight at 4°C with primary antibodies at the designated concentrations: mouse anti-α-tubulin (1:10,000, T9026, Sigma), rabbit anti-pSHP-1 (1:5,000, D11G5, Cell Signaling), rabbit anti-Sall1 (1:1000, ab31526, Abcam). The membrane was washed, stained with IRDye conjugated secondary antibodies (1:15,000, LI-COR) and imaged on the Odyssey CLx (LI-COR). The membrane was stripped by incubation with ReBlot Plus Strong Antibody Stripping Solution (Millipore) for 15 minutes before blocking, staining with anti-SHP-1 (1:10,000, C14H6, Cell Signaling) then an IRDye conjugated secondary antibody, and reimaged as described. Images were analyzed for band intensities with the ImageStudio software (LI-COR).
Antibody-mediated inhibition assay
WT BV2 cells were incubated in serum-free DMEM containing varying concentrations of a monoclonal anti-CD22 antibody (Cy34, BioXCell) from 20ug/mL to 0ug/mL for 8 hours at 37 °C. The p-SHP-1:total SHP-1 ratio, indicative of signaling activity downstream of CD22, was determined by phospho-western blot as described above.
Ligand-binding assay
Recombinant mouse CD22-human Fc fusion protein (R&D) was pre-complexed to an AF647-conjugated anti-human IgG secondary antibody on ice for 30 minutes at equal concentrations (10 μg/mL). The complex was then incubated with various concentrations of a mouse monoclonal anti-CD22 antibody (Cy34, BioXCell) or a mouse IgG1 isotype control antibody (MOPC21, BioXCell) on ice for 30 minutes. Next, the antibody-treated complexes were incubated with WT or sialidase treated BV2 cells for 30 minutes on ice. The cells were washed and rCD22 binding was assessed by flow cytometry on a BD Accuri C6.
Antibody internalization assay
WT BV2 cells were split into a 24-well plate 24 hours prior to antibody treatment at a density of 50,000 cells/well. Cells were treated with rat anti-mouse CD16/CD32 Fc Blocker (2.4G2, BD) then incubated with the following antibodies at 1ug/mL: mouse IgG1 isotype control antibody (MOPC21, BioXCell), mouse anti-CD22 (Cy34, BioXCell) and rat anti-CD22 (OX-96, Bio-Rad). All antibodies were pre-labeled with Incucyte FabFluor-pH Red (Essen Bioscience) according to the manufacturer’s protocol. 9 phase and red fluorescent images per well were acquired every hour for 24 hours using the Incucyte S3 live cell analysis system (Essen Bioscience). For each time point, we calculated antibody internalization using the following formula: red object area / phase confluence. Technical triplicates for each experiment were averaged and reported.
Immunohistochemistry
Mice were euthanized with 2.5% (v/v) Avertin and transcardially perfused with ice-cold HBSS containing glucose and HEPES, or with 4% PFA. Hemibrains were post-fixed in 4% PFA at 4 °C overnight before preservation in 30% sucrose in PBS. Hemibrains were sectioned into 40 micron coronal slices on a microtome and stored in cryoprotective solution at −20 °C. Free-floating sections were permeabilized, blocked, and stained overnight at 4°C with the following primary antibodies at the designated concentrations: goat anti-Iba1 (1:500, ab5076, Abcam), goat anti-Prox1 (1:500, AF2727, R&D), rabbit anti-c-Fos (1:400, 9F6, Cell Signaling), rabbit anti-Aβ (1:200, D54D2, Cell Signaling), rabbit anti-alpha-synuclein (1:200, MJFR1, Abcam), rabbit anti-p-CREB (1:500, 06–519, Millipore), rabbit anti-synaptophysin (1:500, D8F6H, Cell Signaling), rabbit anti-PSD95 (1:500, D27E11, Cell Signaling), rabbit anti-C1q (1:1200, ab182451, Abcam), goat anti-doublecortin (1:500, SC8066, Santa Cruz Biotech), rabbit anti-CD19 (1:500, D4V4B, Cell Signaling), rabbit anti-Tmem119 (1:100, 28–3, Abcam). Sections were washed, stained with Alexa Fluor-conjugated secondary antibodies (1:250), mounted and set under a coverslip before imaging on a confocal laser-scanning microscope (Zeiss LSM880). Thioflavin S staining was performed after secondary antibody staining. Briefly, sections were stained in a 0.1% (w/v) solution for 5 minutes, followed by 3 washes in 50% EtOH, and a 30 minute rehydration in diH2O prior to mounting.
Preparation of fluorescent phagocytic material
Purified CNS myelin was isolated as previously described58. Briefly, mice were euthanized and their brains homogenized. Crude brain homogenate was subjected to a series of sucrose gradients under ultracentrifugation followed by osmotic shocks in hypotonic buffers to remove myelin sheaths from severed axons. Aβ oligomers were prepared as previously described59. Briefly, Aβ1–42 (Anaspec) was treated with hexafluoroisopropanol (HFIP) to achieve a monomeric solution. Oligomerization was induced by diluting a 5mM monomeric Aβ DMSO stock to 200μM in ice-cold PBS and incubated overnight at 4 degrees. Pre-formed α-synuclein fibrils were prepared as previously described60. Phagocytic material was conjugated to fluorescent dyes as previously described21. For double-labeled myelin, 50 mg/mL of myelin prep was incubated with equimolar concentrations of AlexaFluor555-NHS ester and CypHer5E-NHS ester in PBS with 0.1M sodium bicarbonate for 45 minutes at room temperature. Myelin was washed 4 times with PBS and centrifugation to remove free dye. For Aβ and α-synuclein, 100μL of the 200μM protein aggregates were incubated with 4μL CypHer5E-NHS ester in PBS with 0.1M sodium bicarbonate for 30 minutes at room temperature. Free dye was removed using BioSpin Micro P-6 desalting columns (BioRad).
In vivo phagocytosis assay
CypHer5E and AlexaFluor555 double-labeled myelin (25mg/mL), CypHer5E-labeled Aβ oligomers (1mg/mL), or CypHer5E-labeled alpha-synuclein fibrils (1mg/mL) were mixed with a mouse IgG1 isotype control antibody (MOPC21, BioXCell, 1mg/mL) or anti-CD22 (Cy34, BioXCell, 1mg/mL) and injected into opposite hemispheres of aged mice using a stereotaxic apparatus (Kopf instruments). Mice were anesthetized using isofluorane, their skulls were exposed and a hole was drilled at the injection site using aseptic technique. One microliter of the antibody/phagocytic target material mixture was injected at +/− 2mm lateral, 0mm anterior-posterior, and −1.5mm deep relative to the intersection of the coronal and sagittal suture (bregma) at a rate of 200nL/minute. The needle was left in place for an additional 3 minutes to allow for diffusion, then slowly withdrawn. Mice received post-surgical buprenorphine and baytril for pain and infection prevention, respectively. After 48 hours, mice were anesthetized and transcardially perfused with 4% PFA. The entire injection site was sectioned, stained, and imaged as described above, and 6–8 consecutive sections, spaced 40 μm apart, were quantified to assess total target clearance over the entire area of the injection site. Sections at the margins of the injection site without noticeable Iba1+ microgliosis (which takes >48 hours to resolve and is not influenced by treatment) were excluded.
Ex vivo microglial phagocytosis assay
Whole brain myelin-depleted single-cell suspensions were prepared as described above. Next, a rough estimation of microglia counts from each brain was determined by strictly gating a volume-designated aliquot of the suspensions by forward scatter/side scatter on a flow cytometer (BD Accuri C6). Each suspension was adjusted to contain equal microglia per unit volume. Then, single cell brain suspensions were treated accordingly for 1 hour in serum free DMEM-F12 at 37 °C with periodic agitation. Treatments were washed out and cells were resuspended in FACS buffer containing pHrodo- or CypHer5E-conjugated particles at a ratio of 50 particles per microglia. The cell-particle mixture was incubated for 1–2 hours at 37 °C with periodic agitation. Phagocytosis was stopped by transferring the suspensions to ice, where the cells were stained with antibodies to distinguish microglia. Microglial phagocytosis was assessed by flow cytometry by pre-gating live CD11b+CD45lo cells and assessing pH-dependent particle fluorescence within this population. The eating gate was determined using an unfed control, and confirmed by sorting an aliquot of the sample into non-eating and eating populations by FACS followed by microscopic assessment.
Osmotic pump intracerebroventricular delivery of anti-CD22
A monoclonal anti-CD22 antibody (Cy34, BioXCell, 1mg/mL) or mouse IgG1 isotype control antibody (MOPC21, BioXCell, 1mg/mL) were conjugated to AlexaFluor647 (ThermoFisher) using NHS chemistry at a dye:protein molar ratio of 4. Free dye was removed and buffer was exchanged with artificial CSF (Tocris) using 40kD MWCO Zeba spin desalting columns (ThermoFisher). The labeled antibodies were loaded into 200 microliter osmotic pumps (Alzet/Durect) with a 28-day infusion rate of 0.25mL per hour. An infusion rate was chosen to maintain a steady state concentration of 10 μg/mL, a dose shown to promote phagocytosis with this particular antibody ex vivo. Osmotic pumps were connected to a cannula (Brain infusion kit III, Alzet) inserted at +1mm lateral, −0.3mm anterior-posterior, and −3mm deep relative to bregma in order to target the right lateral ventricle. The pump was placed subcutaneously and mice received post-surgical buprenorphine and Baytril.
Quantification of antibody leakage into the periphery following intracerebroventricular administration.
1 mg of CD22 antibody was incubated with 30x molar ratio of Trans-Cyclooctene-NHS ester (Click Chemistry Tools) overnight at 4°C before being quenched with 1M Tris pH 8.0 and desalted (Zeba, 40K MWCO, Thermo). Mice were administered unlabeled or Trans-Cyclooctene (TCO)-labeled CD22 antibody via osmotic pump intracerebroventricularly as detailed above, or with equimolar amounts intravenously (retro-orbital) or intraperitoneally. Seven days later, blood was collected with EDTA anticoagulant by terminal intracardial bleeding. EDTA-plasma was isolated by centrifugation at 1,000g for 15 min at 4 °C before aliquoting, flash freezing, and storage at −80 °C. Protein concentrations of plasma samples and unlabeled and TCO-labeled CD22 antibodies were measured with a BCA Protein Assay Kit (Pierce). 75 μg of plasma was aliquoted, denatured in 1% SDS, and incubated for 90 minutes in the dark at room temperature with 6 μM SiR-tetrazine (Spirochrome) for subsequent click chemistry detection by in-gel fluorescence. 10x decreasing amounts of unlabeled and TCO-labeled CD22 antibody—from 1 μg to 0.1 ng—were processed similarly as a fluorescence standard. A 4x stock solution of NuPAGE LDS (Thermo) was added to each sample before heating at 95°C for 5 minutes. Proteins were briefly spun and separated by electrophoresis in 12% Bis-Tris polyacrylamide gels (Invitrogen). Gels were washed twice in distilled water for 15 minutes before SiR imaging in the 700-nm channel of an Odyssey CLx (LI-COR). To ensure accurate protein loading, gels were incubated with GelCode Blue Stain Reagent (Thermo) overnight before destaining in distilled water for at least 3 hours and imaged in the 800-nm channel of an Odyssey CLx (LI-COR). CD22 fluorescence signals were quantified in accompanying Image Studio™ software (LI-COR).
Behavioral assays
The forced alternation Y-maze was performed as previously described with minor alterations61. Briefly, the test consisted of a 5 minute training trial followed by a 5 minute retrieval trail, with a 1 hour inter-trial interval. For the training trial, one arm of the Y-maze was blocked off and mice were allowed to explore the two open arms. One hour later, the mouse was again placed in the Y-maze with all three arms open and a black and white pattern placed at the end of the novel arm. Between mice and trials, the maze was wiped with ethanol to remove odor cues. For analysis, video was analyzed by a blinded observer and both number of arm entries and time spent in each arm was quantified. Mice with less than two arm entries in the first minute of the retrieval trial were excluded from the analysis.
The fear-conditioning paradigm was performed as previously described62. Briefly, mice were trained to associate cage context or an audiovisual cue with an aversive stimulus (foot-shock). The test was administered over two days. On day 1, mice were placed in a cage and exposed to two periods of 30s of paired cue light and 1,000-Hz tone followed by a 2-s foot shock (0.6 mA), with a 180-s interval. On day 2, mice were subjected to two trials. In the first trial assessing contextual memory, mice were re-exposed to the same cage context, and freezing behavior was measured during minute 1–3 using a FreezeScan tracking system (Cleversys). In the second trial measuring cued memory, mice were placed in a novel context and exposed to the same cue light and tone from day 1 after 2 minutes of exploration. Freezing behavior was measured for 1–3 minutes following the cue. No significant differences in cued fear conditioning were observed between groups, consistent with previous rejuvenation studies from our lab62.
Assessment of the cognitive effects of systemically administered anti-CD22
Aged (18 m.o.) mice were intraperitoneally injected with 300μg of anti-CD22 (Cy34, BioXCell) or mouse IgG1 isotype control (MOPC21, BioXCell) twice weekly for one month, followed by cognitive assessment using the forced alternation Y-maze and contextual fear conditioning paradigms described above.
Gene Ontology Analysis
For gene ontology analysis of CRISPR-Cas9 screen hits, Panther was used to assess statistical overrepresentation of hits in the Reactome pathway database given a reference list containing all genes targeted by the sgRNA library. For analysis of RNA-seq data, Enrichr was used to assess enrichment of significantly up- or down-regulated genes in KEGG, BioCarta, WikiPathways, and Reactome databases. Pre-ranked gene set enrichment analysis was performed using GSEA software v3 (Broad Institute) with default settings and signed log-normalized p-values as a ranking metric.
Ex vivo microglia secretome profiling
Microglia from aged (20–24 m.o.) mice were isolated as described above, with minor modifications. Namely, following myelin depletion, cells were incubated with CD11b-positive enrichment beads (Miltenyi), isolated by magnetic selection using LS-columns, and plated at 30,000 cells / well of a 96-well plate. Microglia were maintained in serum-free defined medium as previously described21, and treated with either IgG (MOPC21, BioXCell, 20 μg/mL) or anti-CD22 (Cy34, BioXCell, 20 μg/mL) in the presence or absence of Aβ oligomers (5 μM) for 8 hours. Following incubation, supernatant was collected, spun down to remove cells, and flash frozen on dry ice. A bead-based immunoassay for mouse cytokines and chemokines (Eve Technologies) was used for protein detection.
Data Availability.
RNA-seq datasets have been deposited online in the Gene Expression Omnibus (GEO) under accession numbers GSE127542 and GSE127543.
Extended Data
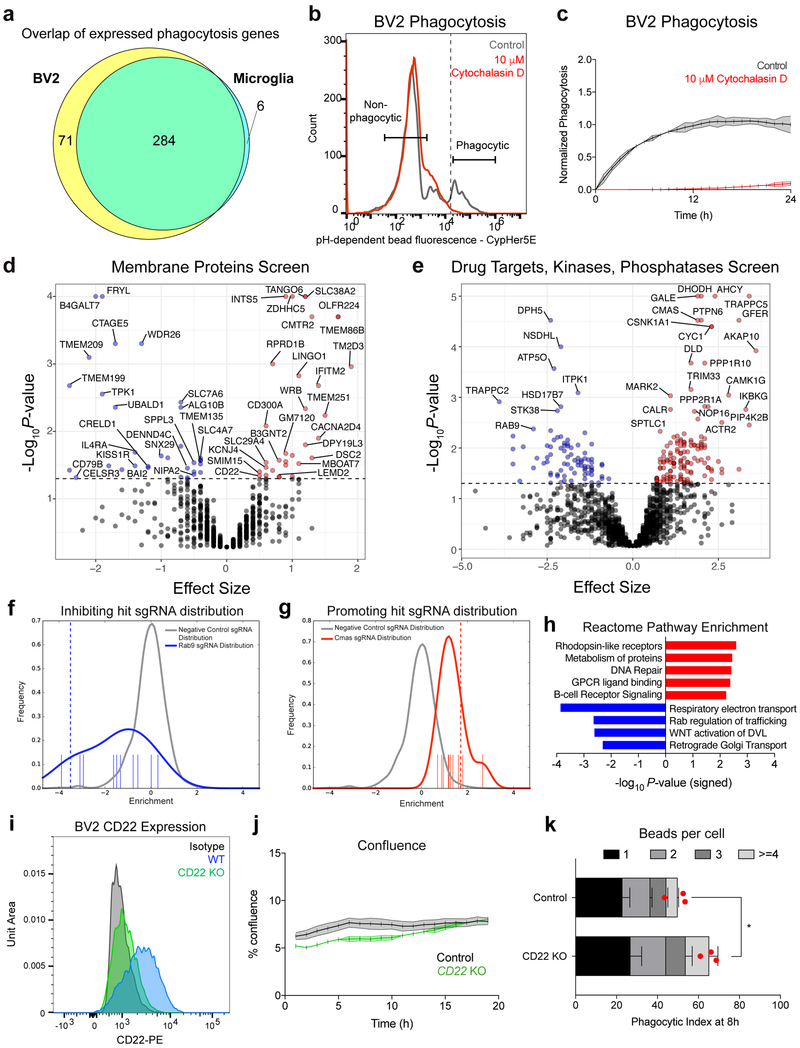
Extended Data Figure 1. CRISPR screen for genetic modifiers of phagocytosis.
a, Intersection and disjunction of 361 genes involved in phagocytosis (Haney et al., 2018) expressed (FPKM>5) by BV2 cells (PRJNA407656) and primary microglia.
b, Gating scheme for FACS separation of phagocytic and non-phagocytic BV2 cells, treated with vehicle (gray) or the actin-polymerization inhibitor, cytochalasin D (red).
c, Time-lapse microscopy readout of phagocytosis by BV2 cells treated with vehicle (gray) or cytochalasin D (red) (n=3, mean +/− s.e.m.).
d, e, Results from CRISPR-Cas9 screen targeting 954 membrane proteins (d) or 2,015 drug targets, kinases, and phosphatases (e) in BV2 cells. Knockouts that promote phagocytosis (red) have a positive effect size and knockouts that inhibit phagocytosis (blue) have a negative effect size (screen performed in technical duplicate; dotted line, P=0.05, two-sided t-test).
f, g, Distributions of negative control sgRNAs (gray) and Rab9-targeting (f) or CMAS-targeting (g) sgRNAs (blue). Positive values indicate enrichment in the phagocytic fraction, and negative values indicate enrichment in the non-phagocytic fraction.
h, Statistical overrepresentation test showing enrichment of Reactome pathway annotations within phagocytosis-promoting (red) and -inhibiting (blue) hits (Fisher’s exact test).
i, CD22 expression in WT (blue), CD22 KO (green), and isotype control stained (black) BV2 cells assessed by flow cytometry.
j, Percent confluence of control (gray) and CD22 KO (green) BV2 cells during time-lapse microscopy phagocytosis assays (n=3, mean +/− s.e.m.).
k, Number of beads ingested per cell were calculated in control and CD22 KO BV2 cells after 8 hours of phagocytosis. While CD22 KO cells display enhanced phagocytosis at a population level (n=3, *P<0.05, two-sided t-test, mean +/− s.e.m.), we observed no significant differences in the number of beads ingested per cell (two-way ANOVA). Red dot represents mean phagocytic index of the entire cell population.
Data in b, c, i, j, k were replicated in at least 2 independent experiments.
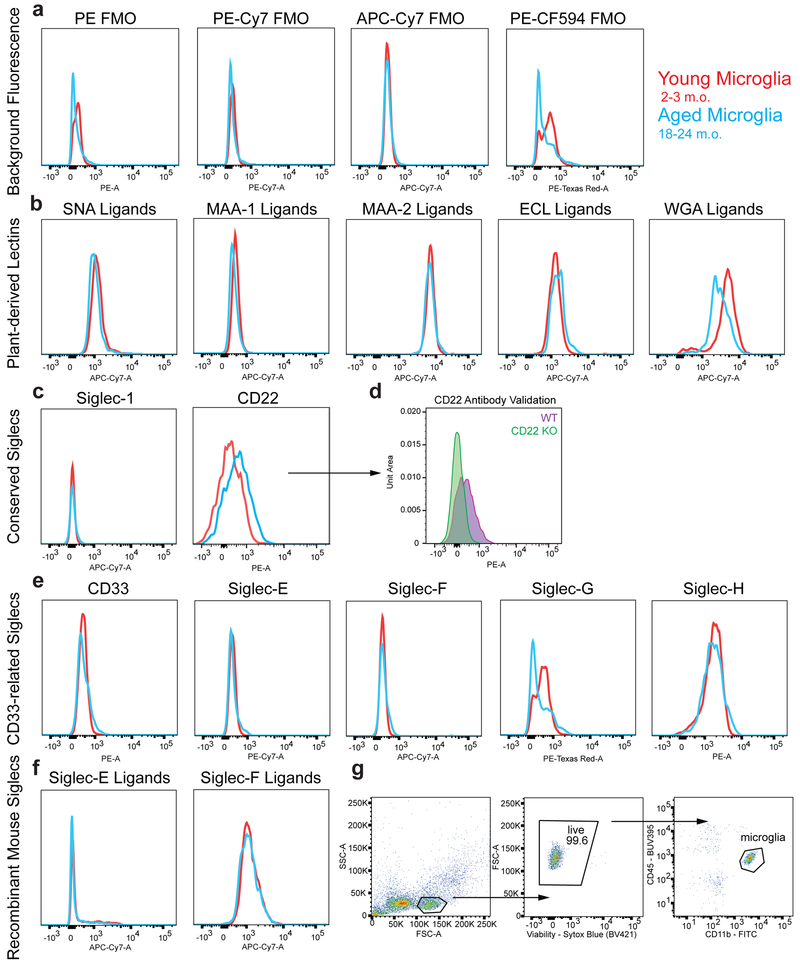
Extended Data Figure 2. Immunophenotyping of sialic acid-related molecules on young and aged microglia.
a, b, c, e, f, Flow cytometry analysis of young (red) and aged (blue) microglia for expression of fluorescence minus one (FMO) background fluorescence (a), plant-derived lectin ligands (b), conserved Siglecs (c), mouse-specific CD33-related Siglecs (e), and recombinant Siglec ligands (f). MFI shown on a biexponential-scale.
d, Microglia from WT or CD22−/− aged mice were stained with the particular anti-CD22 clone (Ox97) used for immunophenotyping. CD22−/− microglia show no staining relative to FMO.
g, Gating strategy to immunophenotype microglia while minimizing autofluorescence.
All data were replicated in at least 2 independent experiments.
Extended Data Figure 3. Protein and transcript expression of CD22 in microglia.
a, Flow cytometry gating scheme for analysis of CD22 expression in peripheral blood-derived myeloid cells (CD45+CD11b+), immature B-cells (CD45+B220+CD22lo), and mature B-cells (CD45+B220+CD22hi). Quantibrite beads are shown in the top right panel.
b, Quantification of flow cytometry analysis showing the number of CD22 molecules on various cell types, interpolated from the Quantibrite bead standard curve (n=3, mean +/− s.e.m.).
c, CD22 expression in various cell types of the young mouse CNS, showing exclusive expression in microglia. Data from Barres lab RNA-seq (http://www.brainrnaseq.org/).
d, t-SNE plot showing single cell RNA-seq (scRNA-seq) analysis of CD22 expression in microglia isolated from E14.5, P7, and adult mouse brains. CD22 is enriched in a subpopulation of P7 microglia. Data from Li et al., 2018 (https://myeloidsc.appspot.com/).
e, t-SNE plot showing scRNA-seq analysis of CD22 expression in cells from 20 different mouse tissues. CD22 is expressed in B-cells and microglia, but absent from non-myeloid brain cells (n=7, young mice). Data from the Tabula Muris Consortium.
f, Violin plots of log-normalized CD22 counts per million reads (CPM) showing high expression in B-cells from multiple organs and in microglia (n=7, young mice). Data from the Tabula Muris Consortium.
Data in a, b were replicated in at least 2 independent experiments.
Extended Data Figure 4. Sialic acid-CD22-SHP1 signaling regulates phagocytosis.
a, Sambucus nigra agglutinin (SNA, recognizes α2,6-linked sialic acid) and Maackia amurensis agglutinin II (MAA II, recognizes α2,3-linked sialic acid) ligand expression in WT BV2 cells (orange), WT BV2 cells pretreated with sialidase (blue), and CMAS KO cells (red) assessed by flow cytometry. Sialidase treatment and CMAS KO reduce sialic acid ligands on the cell surface.
b, Western blot showing SHP1 protein expression in WT and PTPN6 KO BV2 cells. For raw source image, see Supplementary Figure 1.
c, Percent confluence of control (gray), CMAS KO (red), and PTPN6 KO (blue) BV2 cells during time-lapse microscopy phagocytosis assays (n=3, mean +/− s.e.m.).
d, e, Phagocytosis of pH-sensitive fluorescent beads by untreated (black) and sialidase-treated (red) BV2 cells (d) or vehicle-treated and 3Fax-Neu5Ac-treated BV2 cells prior to phagocytosis (n=3, **P<0.005, two-sided t-test; mean +/− s.e.m).
f, Phagocytosis of pH-sensitive fluorescent beads by WT (black), WT + sialidase (red), CD22 KO (blue), or CD22 KO + sialidase (green) BV2 cells (n=3, *P<0.05, N.S. not significant, one-way ANOVA with Tukey’s multiple comparisons correction; mean +/− s.e.m.).
g, Microglia were acutely isolated from the brains of aged (18 m.o.) WT (left) or CD22 −/− (right) mice, treated with or without sialidase, and incubated with pH-sensitive fluorescent latex beads. Microglia specific phagocytosis was measured using flow cytometry (n=6, *P<0.05, N.S. not significant, paired two-sided t-test).
h, Representative images of BV2 cells coated with AlexaFluor 488 conjugated glycopolymers (green) and stained with a plasma membrane-specific dye (CellMask, red) showing overlap (yellow). Scale bar = 25 microns.
i, Recombinant mouse CD22-human Fc fusion protein was pre-complexed with AF647 anti-human Fc secondary antibody, treated with various concentrations of IgG (black) or anti-CD22 (blue, red), and subsequently allowed to bind to ligands on the surface of BV2 cells or BV2 cells pretreated with sialidase (red). Binding was measured by flow cytometry.
j, Internalization of IgG (black), function blocking anti-CD22 (clone Cy34.1, blue), and non-function-blocking anti-CD22 (clone OX96, green) conjugated to a pH-sensitive fluorescent dye by BV2 cells assessed by time-lapse microscopy (n=3, mean +/− s.e.m.).
k, Western blot quantification of ratio of active pSHP1 to total SHP1 protein in BV2 cells pretreated with various concentrations of anti-CD22. Blue line represents the fitted variable slope inhibitor-response curve. For raw source image, see Supplementary Figure 1.
All data were replicated in at least 2 independent experiments.
Extended Data Figure 5. Validation of in vivo phagocytosis assay.
a, Representative images of myelin labeled with a pH-sensitive fluorescent dye (CypHer5E, white), a constitutively fluorescent dye (AF555, red) and stained for Iba1 (green). The majority of AF555 overlapping with Iba1 is also positive for CypHer5E, indicating localization to an acidified compartment. Scale bar = 100 microns.
b, 3D reconstruction of microglial cell (Iba1, green) with ingested myelin (CypHer5E and AF555, white and red, yellow arrow) near un-ingested myelin (AF555, red, white arrow). Scale bar = 5 microns.
c, Microgliosis, as assessed by % Iba1+ area at the injection site, was not altered by CD22 blockade (n=8, N.S. not significant, paired two-sided t-test).
d, Representative images of myelin (red) overlaid with the myeloid marker Iba1 (green) at the injection site of IgG (left) or PBS (middle) treated hemispheres of the same aged brain, or an image of a stab wound control (not injected with myelin). Scale bar = 100 microns.
e, Microgliosis, as assessed by % Iba1+ area at the injection site, was not altered by IgG compared to the stab wound control (n=2, N.S. not significant, paired two-sided t-test).
f, Clearance of myelin debris in the IgG (black) or PBS (blue) treated hemispheres assessed 48 hours post-injection (n=4, N.S. not significant, paired two-sided t-test).
g, Representative images of Iba1 (gray), a macrophage marker, and Tmem119 (magenta), a microglia-specific marker, at the injection site in IgG (left) or anti-CD22 (right) treated hemispheres of the same aged brain. Scale bar = 100 microns.
h, Percent of Iba1+ phagocytes expressing Tmem119 at the injection site (n=4, N.S. not significant, paired two-sided t-test).
i, Clearance of myelin debris in young (2.5 m.o.) WT (black) or CD22−/− (green) mice was assessed 48 hours after injection (n=4, N.S. not significant, two-sided t-test; mean +/− s.e.m.).
j, Representative images of total Aβ (white,), Thioflavin S+ fibrillar Aβ (green), and Iba1 (red) in transgenic mice expressing human APP with Swedish and London familial AD mutations (left) or WT mice injected with Aβ oligomers 48 hours prior to analysis (right).
k, Representative images of Aβ (red, left column) and Aβ overlaid with the myeloid marker Iba1 (green, right column) at the injection site (+/− 2mm lateral, 0mm A-P, -1.5mm D-V relative to bregma) of IgG (top row) or anti-CD22 (bottom row) treated hemispheres of the same aged brain. Scale bar = 100 microns.
l, Microgliosis, as assessed by % Iba1+ area at the Aβ oligomer injection site, was not altered by CD22 blockade (n=8, N.S. not significant, paired two-sided t-test).
m, Representative images of α-synuclein and Iba1 at the injection site in IgG and anti-CD22 treated mice. Scale bar = 100 microns.
n, Clearance of α-synuclein fibrils in the IgG (black) or anti-CD22 (green) treated hemispheres assessed 48 hours post-injection (n=7, *P<0.05, paired two-sided t-test).
o, Microgliosis, as assessed by % Iba1+ area at the α-synuclein fibril injection site, was not altered by CD22 blockade (n=7, N.S. not significant, paired two-sided t-test).
All data were replicated in at least 2 independent experiments.
Extended Data Figure 6. Specificity and distribution of long-term, CNS-targeted antibody infusion via osmotic pump.
a, Representative images of CD19+ B-cells (red, top row), DAPI (blue, middle row), and merged (bottom row) in the spleen (left, positive control) and hippocampus (right) of a mouse treated with anti-CD22 via intracerebroventricular osmotic pump.
b, Concentration of trans-cyclooctene-labeled anti-CD22 in the plasma 7 days after administration of 200 μg anti-CD22 via IP injection (n=1) or intracerebroventricular osmotic pump infusion (n=4), assessed by in-gel fluorescence and quantification based on a standard curve (mean +/− s.e.m). For raw source image, see Supplementary Figure 1.
c, Representative images of coronal brain sections of untreated (left column) and IgG treated (right column) mice. IgG was labeled with an AlexaFluor647 NHS-ester (top row, white) to assess antibody distribution throughout the brain (bottom row, DAPI, blue). In addition to the para-ventricular areas, antibodies penetrated the thalamus and hippocampus.
d, Flow cytometry analysis of AF647-labeled antibody on microglia isolated from untreated (black), IgG (red), or anti-CD22 (blue) infused mice. Microglia from anti-CD22 treated mice display elevated AF647 signal, indicative of antibody target engagement.
Data in a, c, d were replicated in at least 2 independent experiments.
Extended Data Figure 7. Anti-CD22 treatment partially reverses age- and disease-related microglia transcriptional signatures.
a, Venn diagram showing the lack of any intersection among 315 genes differentially expressed between IgG (n=7) and anti-CD22 (n=7) treated microglia and 40 genes differentially expressed between untreated (n=2) and IgG (n=7) treated microglia at an FDR cutoff of 10% (Benjamini-Hochberg method).
b, Hierarchical clustering of normalized read counts from IgG and anti-CD22 treated microglia, normalized by row mean. The top-100 differentially expressed genes are shown (n=7).
c, Enrichr gene-ontology analysis of genes upregulated (red) and downregulated (blue) by anti-CD22 treatment (Fisher’s exact test, Benjamini-Hochberg FDR).
d, Gene set enrichment analysis (GSEA) showing normalized enrichment score for microglia genes modulated by anti-CD22 treatment within the gene signature for: aging microglia (this study), disease-associate microglia (DAM, Keren-Shaul, et al. 2017), microglial neurodegenerative phenotype (MGnD, Krasemann, et al. 2017), and microglia from lipopolysaccharide treated mice (LPS, Bennett, et al. 2016) (*FDR<0.05).
e, f, g, h, GSEA showing enrichment distribution for microglia genes modulated by anti-CD22 treatment within the gene signature for aging microglia (e), DAM (f), MGnD (g), and LPS-activated microglia (h).
Extended Data Figure 8. Protein-level assessment of long-term CD22 blockade in the hippocampus.
a, Western blot for Sall1 and α-Tubulin (loading control) in whole hippocampus lysates from IgG and anti-CD22 treated mice. For raw source image, see Supplementary Figure 1.
b, Quantification of (a) showing upregulation of Sall1 protein in anti-CD22 hippocampi (n=3, *P<0.05, two-sided t-test, mean +/− s.e.m.).
c, Protein concentration of CCL3 in the supernatant of acutely isolated aged microglia treated for 8 hours with IgG or anti-CD22 in the absence or presence of Aβ oligomers (n=4, N.S. not significant, *P<0.05, ANOVA with Sidak’s multiple hypothesis correction, mean +/− s.e.m.).
d, Representative images of p-CREB expression (red) in the dentate gyrus of IgG (left) and anti-CD22 (right) treated mice.
e, Quantification of p-CREB mean intensity in the dentate gyrus of IgG (black) and anti-CD22 (green) treated mice (n=7, two-sided t-test, mean +/− s.e.m.).
f, Quantification of total doublecortin-positive cells in three equally-spaced dentate gyrus sections of IgG (black) and anti-CD22 (green) treated mice (n=3, N.S. not significant, two-sided t-test, mean + s.e.m.).
g, h, i, Quantification of C1q (g), synaptophysin (h, pre-synaptic marker), and PSD-95 (i, post-synaptic marker) mean intensity in the hippocampus of IgG (black) and anti-CD22 (green) treated mice (n=3, N.S. not significant, two-sided t-test, mean + s.e.m.).
All data were replicated in at least 2 independent experiments.
Extended Data Figure 9. Cognitive effects of systemically administered anti-CD22.
a, Working memory and exploratory behavior in aged (18 m.o.) mice treated with IgG (black) or anti-CD22 (green) via IP injection weekly for one month as assessed by % of time spent in the novel arm in a forced alternation Y-maze test (n=6, N.S. not significant, two-sided t-test, mean +/− s.e.m.).
b, Contextual memory aged (18 m.o.) mice treated with IgG (black) or anti-CD22 (green) via IP injection weekly for one month as assessed by % of time displaying freezing behavior in a contextual fear conditioning test (n=6, N.S. not significant, two-sided t-test, mean +/− s.e.m.).
Extended Data Figure 10. Graphical abstract.
Microglial phagocytosis declines with age, accompanied by increased CD22 expression. CD22 inhibition restores phagocytosis, promotes a homeostatic transcriptional state, and improves cognitive function in aged mice.
Supplementary Material
Acknowledgments
We thank the members of the Wyss-Coray, Bassik, and Bertozzi labs for feedback and support, M. Macauley, Q. Li, and S. Nagaraja for helpful discussions, M. Bennett and F.C. Bennett for critical reading of the manuscript, H. Zhang and K. Dickey for laboratory management, R. Ballet, E. Butcher, J. Paulson, and L. Nitscke for CD22 −/− mice, B. Lehallier for RNA-seq analysis scripts, L. Zhou for RNAscope advice, H. Yousef for histology protocols, and C. Cain for flow cytometry technical expertise. This work was funded by the Department of Veterans Affairs (T.W.-C.), the National Institute on Aging (R01-AG045034 and DP1-AG053015 to T.W.-C., F30AG060638 to J.V.P.), the National Institute of General Medical Sciences (R01-GM059907 to C.R.B.), the NOMIS Foundation (T.W.-C.), The Glenn Foundation for Aging Research (T.W.-C.), the Stanford University Medical Scientist Training Program (T32GM007365, J.V.P., B.A.H.S., L.B., and M.S.), and the Big Idea Brain Rejuvenation Project from the Wu Tsai Neurosciences Institute (T.W.-C., M.C.B., and C.R.B.). This work used the Stanford Center for Genomics and Personalized Medicine (NIH S10OD020141).
References
- 1.Füger P et al. Microglia turnover with aging and in an Alzheimer’s model via long-term in vivo single-cell imaging. Nat. Neurosci. 20, 1371–1376 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Réu P et al. The lifespan and turnover of microglia in the human brain. Cell Reports 20, 779–784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nimmerjahn A, Kirchhoff F, & Helmchen F Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Arandjelovic S, and Ravichandran KS Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol. 16, 907–917 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schafer DP et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paolicelli RC et al. Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Hong S et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lui H et al. Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell 165, 921–935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safaiyan S et al. Age-related myelin degradation burdens the clearance function of microglia during aging. Nat. Neurosci. 19, 995–998 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deczkowska A, Amit I, & Schwartz M Microglial immune checkpoint mechanisms. Nat. Neurosci. 21, 779–786 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Hefendehl JK et al. Homeostatic and injury-induced microglia behavior in the aging brain. Aging Cell 13, 60–69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaughan DW & Peters A Neuroglial cells in the cerebral cortex of rats from young adulthood to old age: an electron microscope study. J Neurocytol. 3, 405–429 (1974). [DOI] [PubMed] [Google Scholar]
- 13.Tremblay M-È, Zettel ML, Ison JR, Allen PD, & Majewska AK Effects of aging and sensory loss on glial cells in mouse visual and auditory cortices. Glia 60, 541–558 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sierra A, Gottfried-Blackmore AC, McEwen BS, & Bulloch K Microglia derived from aging mice exhibit an altered inflammatory profile. Glia 55, 412–424 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Hickman SE, Allison EK, & Khoury, El J Microglial dysfunction and defective amyloid clearance pathways in aging Alzheimer’s disease mice. J Neurosci. 28, 8354–8360 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickman SE et al. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 16, 1896–1905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grabert K et al. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat Neurosci 19, 504–516 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgens DW et al. Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat. Commun. 8, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haney MS et al. Identification of phagocytosis regulators using magnetic genome-wide CRISPR screens. Nat. Genet. 50, 1716–1727 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohlen CJ, Bennett FC, & Bennett ML Isolation and Culture of Microglia. Current Protocols in Immunology 339, e70–21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bohlen CJ et al. Diverse requirements for microglial survival, specification, and function revealed by defined-medium cultures. Neuron 94, 759–773 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butovsky O et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgens DW, Deans RM, Li A, & Bassik MC Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 34, 634–636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macauley MS, Crocker PR, & Paulson JC Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 14, 653–666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitschke L, Carsetti R, Ocker B, Köhler G & Lamers MC CD22 is a negative regulator of B-cell receptor signalling. Curr. Biol. 7, 133–143 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Li Y-Q, Sun L, & Li J Macropinocytosis-dependent endocytosis of Japanese flounder IgM+ B cells and its regulation by CD22. Fish and Shellfish Immunology 84, 138–147 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Linnartz-Gerlach B, Kopatz J, & Neumann H Siglec functions of microglia. Glycobiology 24, 794–799 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Griciuc A, et al. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 78, 631–643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett ML et al. (2016). New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA 113, E1738–E1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Chen K, Sloan SA et al. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. J Neurosci. 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller J et al. CD22 ligand-binding and signaling domains reciprocally regulate B-cell Ca2+ signaling. Proc. Natl. Acad. Sci. U.S.A. 110, 12402–12407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hudak JE, Canham SM, & Bertozzi CR Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat. Chem. Biol. 10, 69–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ereño-Orbea J et al. Molecular basis of human CD22 function and therapeutic targeting. Nature Communications 8, 764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahlgren KN, Manelli AM, Stine WB, Baker LK, Krafft GA, & LaDu MJ Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J. Biol. Chem. 277, 32046–32053 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Deczkowska A, Keren-Shaul H, Weiner A, Colonna M, Schwartz M, and Amit I Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 173, 1073–1081 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Deczkowska A et al. Mef2C restrains microglial inflammatory response and is lost in brain ageing in an IFN-I-dependent manner. Nature Communications 8, 717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y et al. TREM2 Lipid Sensing Sustains the Microglial Response in an Alzheimer’s Disease Model. Cell 160, 1061–1071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marciniak E et al. The Chemokine MIP-1alpha/CCL3 impairs mouse hippocampal synaptic transmission, plasticity and memory. Sci Rep 5, 15862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cole AJ, Saffen DW, Baraban JM & Worley PF Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature 340, 474–476 (1989). [DOI] [PubMed] [Google Scholar]
- 40.Sheng M, McFadden G & Greenberg ME Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron 4, 571–582 (1990). [DOI] [PubMed] [Google Scholar]
- 41.Gosselin D et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavin Y et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett FC et al. A combination of ontogeny and CNS environment establishes microglial identity. Neuron 98, 1170–1183.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101, 207–223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kress BT et al. Impairment of paravascular clearance pathways in the aging brain. Ann Neurol. 76, 845–861 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Da Mesquita S et al. Functional aspects of meningeal lymphatics in ageing and Alzheimer’s disease. Nature 560, 185–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bradshaw EM et al. CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat. Neurosci. 16, 848–850 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siddiqui SS et al. The Alzheimer’s disease-protective CD33 splice variant mediates adaptive loss of function via diversion to an intracellular pool. J. Biol. Chem. 292, 15312–15320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kleinberger G et al. TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci. Transl. Med. 6, 243ra86 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Huang K-L et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat. Neurosci. 20, 1052–1061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedman BA et al. Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer’s disease not evident in mouse models. Cell Reports 22, 832–847 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Olah M et al. A single cell-based atlas of human microglial states reveals associations with neurological disorders and histopathological features of the aging brain. bioRxiv 343780 (2018). [Google Scholar]
- 53.Zhang Y, Sloan SA, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89, 37–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lambert J-C et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nature Genetics 45, 1452–1458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Funikov SY et al. FUS(1–359) transgenic mice as a model of ALS: pathophysiological and molecular aspects of the proteinopathy. Neurogenetics 19, 189–204 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Cougnoux A et al. Microglia activation in Niemann–Pick disease, type C1 is amendable to therapeutic intervention. Hum. Mol. Genet. 27, 2076–2089 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao H, Woods EC, Vukojicic P, & Bertozzi CR Precision glycocalyx editing as a strategy for cancer immunotherapy. Proc Natl Acad Sci USA 113, 10304–10309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larocca JN & Norton WT Isolation of myelin. Curr. Protoc. Cell Biol. Chapter 3, 25 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Stine WB, Jungbauer L, Yu C, & LaDu MJ Preparing Synthetic Aβ in Different Aggregation States In Biological Microarrays, Khademhosseini A, Suh K-Y, and Zourob M, eds. (Totowa, NJ: Humana Press; ), pp. 13–32 (2010). [Google Scholar]
- 60.Polinski NK et al. Best Practices for Generating and Using Alpha-Synuclein Pre-Formed Fibrils to Model Parkinson’s Disease in Rodents. Journal of Parkinson’s disease 8, 303–322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolf A, Bauer B, Abner EL, Ashkenazy-Frolinger T, & Hartz AMS A Comprehensive Behavioral Test Battery to Assess Learning and Memory in 129S6/Tg2576 Mice. PLoS ONE 11, e0147733–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castellano JM et al. Human umbilical cord plasma proteins revitalize hippocampal function in aged mice. Nature 544, 488–492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq datasets have been deposited online in the Gene Expression Omnibus (GEO) under accession numbers GSE127542 and GSE127543.