Abstract
In a screen for human kinases that regulate Xenopus laevis embryogenesis, we identified Nagk and other components of the UDP-GlcNAc glycosylation salvage pathway as regulators of anteroposterior patterning and Wnt signaling. We find that the salvage pathway does not affect other major embryonic signaling pathways (Fgf, TGFβ, Notch, or Shh), thereby demonstrating specificity for Wnt signaling. We show that the role of the salvage pathway in Wnt signaling is evolutionarily conserved in zebrafish and Drosophila. Finally, we show that GlcNAc is essential for the growth of intestinal enteroids, which are highly dependent on Wnt signaling for growth and maintenance. We propose that the Wnt pathway is sensitive to alterations in the glycosylation state of a cell and acts as a nutritional sensor in order to couple growth/proliferation with its metabolic status. We also propose that the clinical manifestations observed in congenital disorders of glycosylation (CDG) in humans may be due, in part, to their effects on Wnt signaling during development.
Keywords: Nagk, UDP-GlcNAc salvage pathway, Wnt signaling, development
Summary Statement
Nagk regulates Wnt signaling and anterior-posterior patterning in the early embryo via the UDP-GlcNAc salvage pathway.
Introduction (Section 1)
Section 1.1
N-acetyl-D-glucosamine (GlcNAc) moieties are key building blocks of N- and O-glycans, glycolipids, glycosaminoglycans, and the glycosyl phosphatidylinositol anchor of membrane-bound glycoproteins (Freeze et al., 2015). Roughly half of all amino sugars from endocytosed glycans are recycled, and approximately 80% of the GlcNAc salvaged from degraded glycoproteins is converted into uridine diphosphate N-acetylglucosamine (UDP-GlcNAc), the substrate utilized in the first step in the assembly of dolichol-linked oligosaccharide intermediates required for N-linked glycosylation (Freeze et al., 2015). In a screen for regulators of early development in Xenopus laevis, we identified N-acetylglucosamine kinase (Nagk) as a regulator of anterior-posterior patterning. Nagk is a key enzyme in the production of UDP-GlcNAc and converts GlcNAc into GlcNAc 6-phosphate (GlcNAc-6-P), the first step in the UDP-GlcNAc salvage pathway. We show that Nagk and other components of this salvage pathway mediate their developmental effects through the Wnt pathway. We demonstrate that this latter activity is selective and conserved in zebrafish, Drosophila, and intestinal enteroids.
Results (Section 2)
Section 2.1 Genome-scale kinase screen identifies Nagk as a regulator of development in Xenopus embryos
To identify kinases that regulate vertebrate development, an arrayed plasmid DNA library of human kinases available from the Harvard Institute of Proteomics FLEXGene human kinase collection was subjected to a series of PCR reactions that resulted in the addition of a T7 promoter and an SV40 polyadenylation signal to the 5’ and 3’ region of the open reading frame, respectively (Turner and Weintraub, 1994) (Fig S1). The final PCR products were then purified and subjected to in vitro transcription to generate capped mRNAs (transcribed PCR products with poly (A) tails or TPATs) for injection into Xenopus laevis embryos (Fig. S1). The addition of the SV40 polyadenylation signal from pCS2+ was critical for robust expression of mRNAs injected into Xenopus embryos (Fig. S2). Although TPATs generated from unpurified final PCR products were expressed when injected into Xenopus embryos, we observed more consistent results using purified final PCR products (Fig. S2).
Section 2.1.1
For screening, 29 pools of 8 TPATs encoding human kinases were injected into the dorsal blastomeres of 2–4 cell stage Xenopus embryos. Embryos were then analyzed for developmental defects at post-neurula stages (Keller, 1991). mRNAs from pools of TPATs that perturbed development were then injected individually to identify the kinase mediating the observed phenotype. Several kinases were identified that perturbed early development. Among these were casein kinase 1 epsilon and delta, known regulators of Wnt signaling (Peters et al., 1999; Sakanaka et al., 1999; Swiatek et al., 2004), thereby validating our approach. Of the kinases not previously characterized as regulators of early vertebrate development, N-Acetylglucosamine kinase (Nagk) exhibited the most penetrant phenotype, and we proceeded with its further characterization.
Section 2.2 Nagk regulates primary axis formation in Xenopus embryos
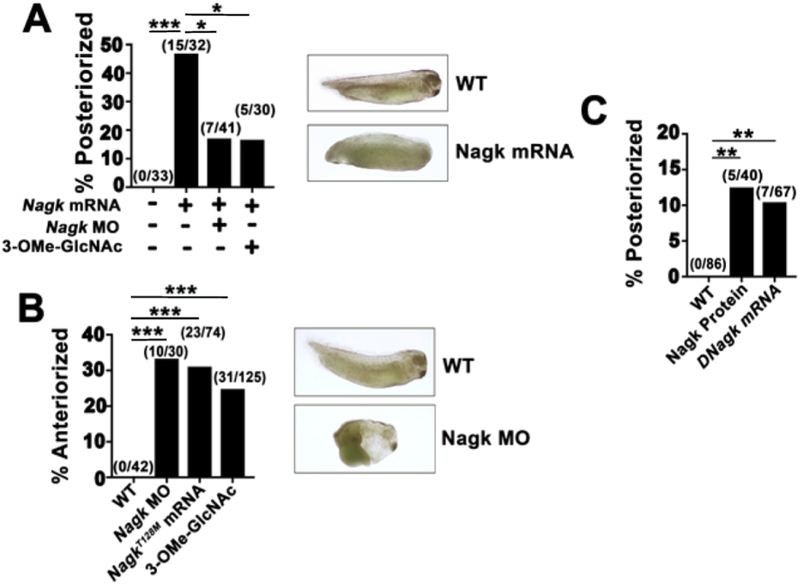
Nagk is the first enzyme in the salvage pathway and converts free, cytoplasmic GlcNAc generated from degradative cellular pathways into UDP-GlcNAc, which is then transferred onto oligosaccharide chains that are incorporated into glycosylated proteins and glycosaminoglycans (Hinderlich et al., 2000) (Fig. S3). Injection of Nagk mRNA into Xenopus embryos resulted in posteriorized embryos with reduced anterior trunk and head structures (Fig. 1A). Conversely, downregulating Xenopus Nagk by morpholino oligonucleotide (MO) injections resulted in anteriorized embryos (Fig. 1B and S4C). Previous studies of a related sugar kinase, Glucokinase, indicated that a mutation in the ATP binding region (T228M) resulted in a kinase that acted in a dominant-negative manner (Mahalingam et al., 1999). We generated the corresponding mutation in Nagk (NagkT128M) and showed that, similar to Nagk MO, injections of NagkT128M mRNA anteriorized embryos (Fig. 1B, S4A). A small molecule competitive inhibitor of Nagk, 3-O-methyl N-acetylglucosamine (3-OMe-GlcNAc) (Blume et al., 2008; Miwa et al., 1994; Zeitler et al., 1992), also resulted in anteriorization of embryos (Fig. 1B, S4A). As predicted, co-injections of either Nagk MO or 3-OMe-GlcNAc suppressed the effects of Nagk mRNA (Fig. 1A). Finally, we show that injecting recombinant Nagk protein or mRNA of the Drosophila melanogaster orthologue of Nagk (CG6218; DNagk) (Fig. 1C, S4A) posteriorized embryos. These data provide strong evidence that Nagk plays an evolutionary role in primary axis formation in Xenopus embryos.
Fig. 1.
Xenopus embryos are posteriorized by Nagk overexpression and anteriorized by Nagk downregulation, respectively. (A) Injection of Nagk mRNA (1.5 ng) posteriorizes Xenopus embryos, and can be suppressed by coinjecting Nagk MO (1 pg) or 3-OMe-GlcNAc (125 pmol). (Right) Representative embryo posteriorized upon injection of NAGK mRNA is shown. (B) Injection of Nagk MO (1 pg), NAGKT128M mRNA (1.5 ng), or 3-OMe-GlcNAc (125 pmol) anteriorizes Xenopus embryos. (Right) Representative anteriorized embryo injected with Nagk MO. (C) Nagk protein (20 pg) and Drosophila Nagk (DNagk) mRNA (1.5 ng) posteriorizes Xenopus embryos (A–C) Aggregated from n ≥ 3 replicates. Embryos were scored for anteriorization or posteriorization according to the dorsal-anterior index (DAI) as previously described (Kao and Elinson, 1988). DAI of ventralized embryos ranged from 4 to 2, whereas dorsalized embryos ranged from 6 to 8. Absolute numbers are indicated above bars. Significance was calculated using Fisher's exact test with Bonferroni correction. **ρ < 0.00334, ***ρ < 0.000334, and *ρ < 0.0253.
Section 2.3 Disruption of the UDP-GlcNAc salvage pathway lead to defects in axiation
We tested whether the effects of Nagk activity on primary axis formation is due to its role in glycosylation. We found that soaking embryos in N-acetyl-D-glucosamine, the first substrate in the biosynthetic pathway that produces N-acetylglucosamine-6-P (GlcNAc-6-P), posteriorized Xenopus embryos (Fig. S5A and C). As Wnt/β-catenin signal transduction is essential for anteroposterior patterning, this finding suggests that the role of Nagk in anteroposterior patterning may occur through Wnt/β-catenin signaling.
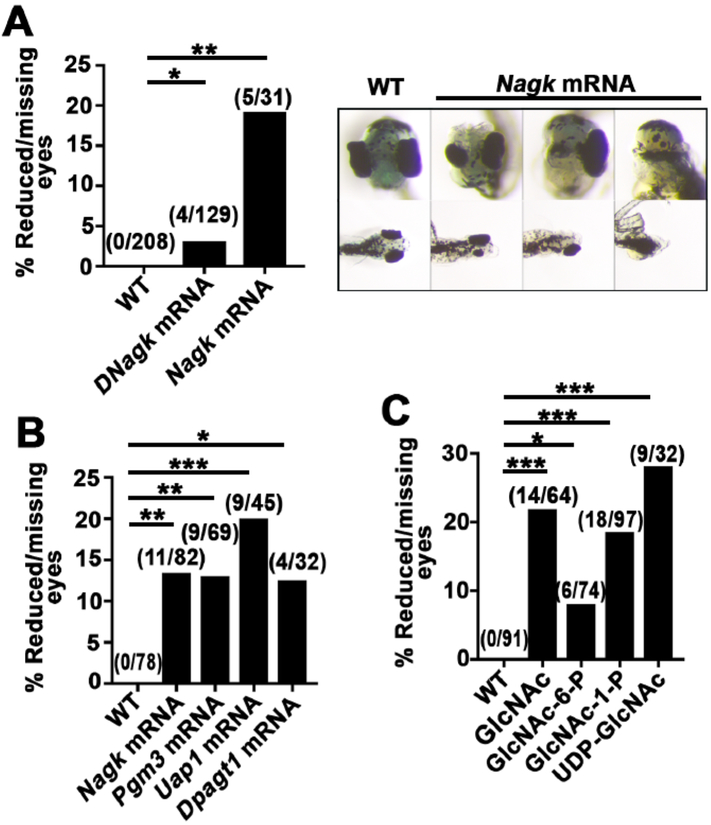
In the UDP-GlcNAc salvage pathway, Nagk phosphorylates GlcNAc to GlcNAc-6-P, which is converted to GlcNAc-1-P by phosphoglucomutase 3 (Pgm3, Fig. S4B) (Berger et al., 2002). GlcNAc-1-P is then converted to UDP-GlcNAc by UDP-NAcetylglucosamine Pyrophosphorylase 1 (Uap1, Fig. S4B) (Berger et al., 2002). To test the effects of these other UDP-GlcNAc salvage enzymes on Xenopus axis formation, we injected individually Pgm3 or Uap1 mRNAs and found that they, like Nagk, posteriorized Xenopus embryos (Fig. 2A, S4B). Conversely, injection of Pgm3 or Uap1 MOs anteriorized embryos (Fig. 2B, S4C). Furthermore, the posteriorizing effects of Pgm3 or Uap1 mRNA injections were suppressed by co-injection of MOs of other enzymes in the pathway (Fig. S6A–C). We next assessed whether the sugar intermediates of the UDP-GlcNAc salvage pathway would affect axiation of Xenopus embryos and found that their injection resulted in posteriorized embryos (Fig. 2C, S4D). Furthermore, we found that the effects of inhibiting the salvage pathway could be suppressed by co-injection of the downstream sugars (Fig. S7). Conversely, the phenotypes produced by injected salvage pathway sugars could be suppressed by injecting MOs of downstream enzymes in the pathway (Fig. S8). These results provide strong evidence that the UDP-GlcNAc salvage pathway regulates anterior-posterior patterning in the early Xenopus embryo.
Fig. 2.
FThe anterior-posterior patterning defect caused by altered Nagk activity in Xenopus embryos is phenocopied upon disruption of other UDP-GlcNAc salvage pathway components. (A) Overexpression of enzymes from the UDP-GlcNAc glycosylation salvage pathway posteriorizes Xenopus embryos. (B) In contrast, downregulating enzymes of the salvage pathway by MO anteriorizes Xenopus embryos. (C) Injection of the sugars from the UDP-GlcNAc salvage pathway, similar to overexpression of their enzymes, results in posteriorized embryos. (D) Injection of mRNA encoding Ngly1, which removes Nlinked glycans from glycoproteins, posteriorizes Xenopus embryos. 1.5 ng of mRNA, 1 pg MO, or 125 pmol compounds were used per injection. Results are aggregates of n ≥ 3 replicates. Absolute numbers are indicated above bars. Significance was calculated using Fisher's exact test with Bonferroni correction. (A–C) *ρ < 0.0127, **ρ < 0.00250, ***ρ < 0.000250. (D) **ρ < 0.01.
Section 2.4 Perturbation of the N-linked glycosylation pathway disrupts normal axiation in Xenopus embryos
To determine whether the salvage pathway alters anteroposterior patterning via N-linked glycosylation, we tested whether perturbation of the rate-limiting enzyme in the N-linked glycosylation pathway, dolichol phosphate GlcNac-1-phosphate transferase 1(Dpagt1), disrupted primary body axis formation. Dpagt1 transfers the first sugar (GlcNAc) onto dolichol in the endoplasmic reticulum to initiate the synthesis of N-linked glycoproteins (Fig. S3) (Lehrman et al., 1988). Similar to our observations with salvage pathway components, injection of Dpagt1 mRNA or MO posteriorized embryos and anteriorized embryos, respectively (Fig. 2A,B and S4E). The posteriorized phenotype of Dpagt1 mRNA could be rescued by co-injection of salvage pathway MOs (Fig. S6D). Consistent with these results, soaking embryos in tunicamycin, an inhibitor of Dpagt1, anteriorized Xenopus embryos (Fig. S5B and C) (Heifetz A et al., 1979; Lehrman et al., 1988). Finally, injection of N-glycanase 1 (Ngly1) mRNA, which removes N-linked glycans from glycoproteins, anteriorized embryos (Fig. 2D, S4F). These results provide strong evidence that the anteroposterior patterning phenotypes we observed upon altering the UDP-GlcNAc salvage pathway are due to its effects on protein N-glycosylation.
Section 2.5 The UDP-GlcNAc salvage pathway selectively regulates Wnt signaling
To test whether altering the UDP-GlcNAc salvage pathway specifically affects the Wnt pathway, we assessed the expression of target genes of other signaling pathways known to control early embryonic patterning in Xenopus (at the appropriate time for which they have been shown to be upregulated) using quantitative real time PCR (qRTPCR). Overexpression or knockdown of Dpagt1 or other UDP-GlcNac salvage pathway components significantly altered expression of the Wnt reporter genes, Xnr3 and chordin (Fig. S9A and S10). In contrast, the expression of target genes for the Fgf (Dusp6), TGFβ (Tbxt), Notch (Hes1), or Shh (Ptch1) pathways were not affected (Fig. S9B and S10) (Gu et al., 2012; Mir et al., 2008; Moriishi et al., 2005; Nishimoto and Nishida, 2007; Wilson et al., 1997; Zheng et al., 2017). These results suggest that during early embryonic development the UDP-GlcNAc salvage pathway specifically regulates Wnt signaling.
Section 2.6 Perturbation of the UDP-GlcNAc salvage pathway induces a Wnt phenotype in zebrafish
To provide further evidence for the role of the UDP-GlcNAc salvage pathway in regulating Wnt signaling and to determine whether its role on anteroposterior patterning is evolutionarily conserved, we tested the effects of perturbing the salvage pathway in the zebrafish Danio rerio. Wnt signaling patterns the anteroposterior neuraxis in zebrafish (Chan et al., 2009; Hikasa and Sokol, 2013). The predominant phenotype of perturbing Wnt signaling in the early developing zebrafish embryo differs from that of perturbing Wnt signaling in the early Xenopus embryo. Ectopic activation of the Wnt/β-catenin pathway blocks eye formation in zebrafish embryos, and zebrafish with activating Wnt/β-catenin pathway mutations produce embryos with defects in anterior structures (Cavodeassi et al., 2005; Kim et al., 2000; van de Water et al., 2001). Consistent with ectopic activation of Wnt signaling, we found that injection of zebrafish embryos with mRNAs encoding salvage enzymes disrupt eye formation (Fig. 3A–B). Injection of sugars of the salvage pathway also inhibited eye formation in zebrafish embryos (Fig. 3C). Conversely, injection of NAGKT128M mRNA or 3-O-Methyl-N-acetyl-D-glucosamine (3-OMe-GlcNAc) gave rise to cyclopic embryos (Fig. 4A). The cyclopic phenotype of NAGKT128M mRNA injected embryos could be rescued by co-injection of intermediate sugars of the salvage pathway (Fig. 4B). Injection of Ngly1 also gave rise to cyclopic embryos, phenocopying inhibition of the salvage pathway (Fig. 4C). These results suggest that the effects of perturbing the UDP-GlcNAc salvage pathway on embryonic development are conserved in Xenopus and Danio rerio and that the observed phenotypes are likely due to altered Wnt signaling.
Fig. 3.
Increasing levels of salvage pathway enzymes, sugars, or Dpagt1 inhibits eye formation in zebrafish. (A) (Left) Injection of DNagk or Nagk mRNAs inhibits eye formation in zebrafish. (Right) Representative images (5 days post fertilization) of disrupted eye formation in zebrafish injected with Nagk mRNA. Observed phenotypes ranged from reduced eye size to missing one or both eyes. Reduced eyes were defined as less than half the size of a wild-type eye. (B) Injection of mRNAs encoding Nagk, Pgm3, Uap1, or Dpagt1 inhibits eye formation. (C) Injection of GlcNAc, GlcNAc-6-P, GlcNAc-1-P, or UDP-GlcNAc inhibits eye formation. Results are aggregates of n ≥ 3 replicates. Injections were performed with 0.5 ng mRNA or 25 pmol sugar. Significance was calculated using Fisher's exact test with Bonferroni correction. Absolute numbers are indicated above bars. (A) *ρ < 0.0253 and **ρ < 0.00501. (B–C) *ρ < 0.0127, **ρ < 0.00251, and *** ρ < 0.000251.
Fig. 4.
Inhibition of Nagk and N-glycosylation in zebrafish induces cyclocephaly. (A) (Left) Injection of NagkT128M mRNA or 3-OMe-GlcNAc results in cyclopic zebrafish. (Right) Representative images of cyclopic zebrafish (5 days post fertilization) injected with NagkT128M mRNA. Animals were defined as cyclopic as previously described (Marlow et al., 1998). *ρ < 0.0253. (B) Injection of NagkT128M mRNA induces cyclopia, which is rescued by co-injection with GlcNAc, GlcNAc-6-P, GlcNAc-1-P, or UDPGlcNAc. *ρ < 0.0127. (C) Injection of Ngly1 mRNA also induces cyclopia. *ρ < 0.05. Results are aggregates of n ≥ 3 replicates. Injections were performed with 0.5 ng mRNA or 25 pmol sugar. Absolute numbers are indicated above bars. Significance was calculated using Fisher's exact test with Bonferroni correction.
Section 2.7 Salvage pathway perturbation results in convergent extension defects in zebrafish
In zebrafish, disruption of eye formation leading to cyclopia is indicative of perturbation in Wnt signaling. Furthermore, inhibition of certain Wnts, including Wnt5b (Pipetail) and Wnt11 (Silberblick), leads to defects in convergent extension (Goudevenou et al., 2011; Heisenberg et al., 2000; Rauch et al., 1997; Stoick-Cooper et al., 2007). In these studies, convergent extension malformations were observed in association with and independent of eye malformations. We found that perturbation of any of the salvage pathway components similarly caused convergent extension defects in zebrafish (Fig. S11). These results suggest the UDP-GlcNAc salvage pathway also regulates convergent extension through Wnt signaling in the early embryo.
Section 2. 8 DNagk and DPgm3 promote Wingless/Wnt signaling in Drosophila
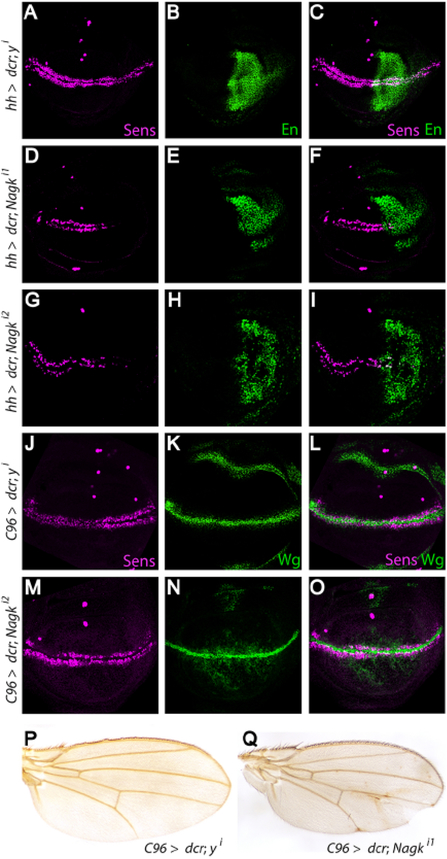
To determine whether DNagk also promotes Wnt/Wingless (Wg) signaling in Drosophila, we tested the effects of its knockdown in vivo. Wg signaling controls wing development by directing patterning at the dorsoventral boundary of the larval wing imaginal disc, the precursor of the adult wing. Inhibition of Wg signaling at this boundary results in the loss of marginal bristles and aberrantly notched wings (Diaz-Benjumea and Cohen, 1995; Struhl and Basler, 1993). We performed RNAi-mediated knockdown of DNagk in the posterior region of third instar larval wing imaginal discs using the hedgehog (hh)-Gal4 driver. In wild-type wing discs, the Wg target gene senseless (sens) is expressed along the entire dorsoventral boundary (Figure 5A–C, J–O) (Nolo et al., 2000). In contrast, DNagk knockdown in the posterior wing disc resulted in a reduction of sens expression in this region. To rule out RNAi off-target effects, we repeated these experiments with a second DNagk RNAi construct and also observed reduction of sens expression in the posterior region (Figure 5G–I). By comparison, knockdown of the control gene yellow (y) had no effect on sens expression (Figure 5A–C). Moreover, RNAi-mediated knockdown of DNagk using C96-Gal4, which drives expression in a broad stripe that overlaps the dorsoventral wing disc boundary (Gustafson and Boulianne, 1996), also resulted in decreased sens expression in the larval wing disc and in the formation of notches at the margin of adult wings (Figure 5Q), whereas RNAi-mediated knockdown of the control gene y neither decreased sens expression (Figure 5J–L) nor altered the adult wing margin (Figure 5P). Furthermore, the expression of wg was not decreased by DNagk knockdown (Figure 5M–O), indicating that DNagk is important for Wg signal transduction but not Wg expression.
Fig. 5.
DNagk promotes expression of the Wg target gene sens in Drosophila. (A-I) RNAi-mediated knockdown of the y control or Nagk in the posterior compartment of third instar wing imaginal discs using the hh-Gal4 driver. The region of hh-Gal4 activity is indicated by expression of Engrailed (En) (green in B, C, E, F, H, I). Knockdown of y (A–C) does not affect sens expression (magenta, A and C), whereas knockdown of Nagk using two independent RNAi lines (D–G) reduces sens expression in the posterior compartment of the discs (D, F, G and I). (J–Q) RNAi-mediated knockdown of Nagk with the C96-Gal4 driver (M–O, Q) reduces sens expression in larval imaginal discs (magenta, M and O) and results in formation of aberrant adult wing notches (Q), whereas RNAi-mediated knockdown of the control y affects neither sens expression (J) nor formation of the wing margin (P). 33% (n = 60) of adult males expressing the Nagk RNAi display wing notches compared to 0% (n = 36) of males expressing the control y RNAi. wg expression is not reduced upon knockdown of Nagk (green, N). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To determine whether DPgm3 also promotes Wg signaling, we tested the effects of its knockdown in third instar larval imaginal discs. RNAi-mediated knockdown of DPgm3 in the posterior wing disc using the hh-Gal4 driver resulted in a severe reduction of sens expression (Figure S12A–F). To rule out RNAi off-target effects, we tested a second line for RNAi-mediated DPgm3 knockdown and observed similarly decreased sens expression (Figure S12G–I). In contrast, RNAi-mediated knockdown of the control gene y resulted in no reduction in sens expression (Figure S12A–C). Additionally, RNAi-mediated knockdown of DPgm3 with the C96-Gal4 driver resulted in notches at the adult wing margin (Figure S12Q), whereas knockdown of the y control had no effect. Moreover, DPgm3 knockdown did not result in decreased Wg levels, indicating that DPgm3 is important for Wg signaling but not Wg expression (Figure S12J–L). Together, these results indicate that both DNagk and DPgm3 promote Wg signal transduction. Thus, regulation of Wnt signaling by the UDP-GlcNAc salvage pathway is conserved in Xenopus, zebrafish, and Drosophila.
Section 2.9 The growth of intestinal enteroids is significantly altered by changes in the GlcNAc pool
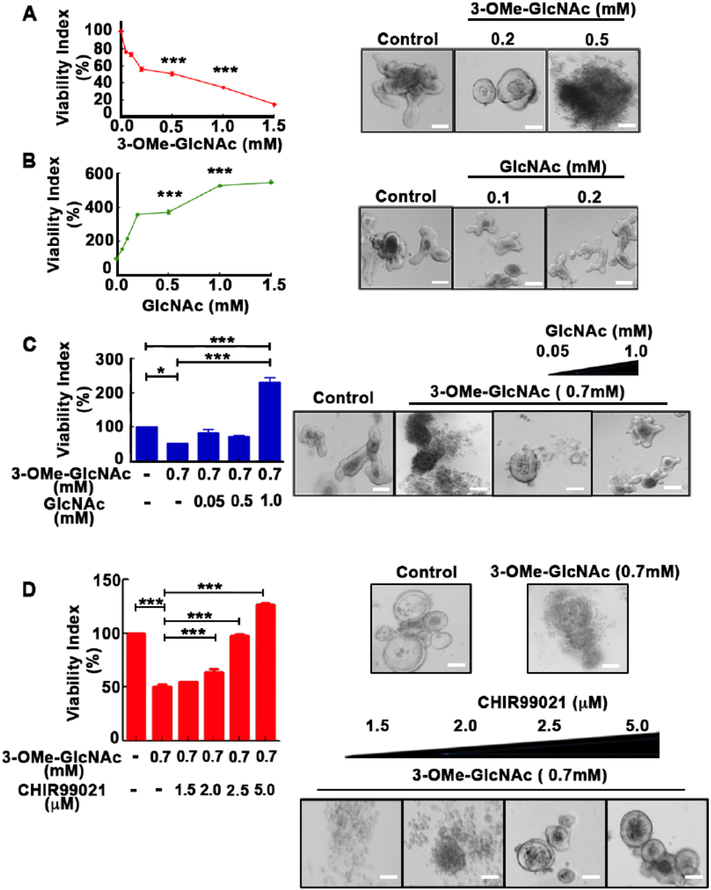
We next asked whether the UDP-GlcNAc salvage pathway also plays a role in regulating Wnt signaling in mammalian cells. Mini-organ or organoid cultures represent a powerful ex vivo system to model mammalian development. We tested whether altering the GlcNAc pool by addition of the Nagk substrate, GlcNAc, or the Nagk inhibitor, 3-OMe-GlcNAc, affected the growth of intestinal crypt-derived enteroids, which require the establishment of a Wnt-dependent stem cell niche for growth and maintenance (Sato et al., 2009). Surprisingly, we found that the viability of intestinal organoids was promoted several fold by the addition of GlcNAc and inhibited by the addition of 3-OMe-GlcNAc (Fig. 6A and B). The inhibitory effect of 3-OMe-GlcNAc could be reverse with the addition of GlcNAc (Fig 6C). Significantly, the Wnt activator, CHIR99021, could also reverse the effect of 3-OMe-GlcNAc on organoid viability (Fig 6D). These studies are consistent with a role for the UDP-GlcNAc salvage pathway in regulating a Wnt-dependent process in mammalian cells.
Fig. 6.
Viability of Wnt-dependent intestinal enteroids is sensitive to changes in the UDP-GlcNAc salvage pathway. Wnt-dependent intestinal enteroids grown in the presence of increasing concentrations of (A) 3-OMe-GlcNAc or (B) GlcNAc demonstrate decreased and increased viability, respectively. (C) The inhibitory effect of 3-OMe-GlcNAc could be reversed by addition of GlcNAc and (D) the GSK3 inhibitor, CHIR99021, which activates the Wnt pathway by stabilizing β-catenin. (A–D) Graphs (Left) with representative images from three independent replicates (Right). Viability Index is relative to no drug control (100%). Scale bars, 200 μm. Statistical analyses were performed using the GraphPad Prism version 5 software tool (La Jolla, CA, USA). Data shown are mean values ± SD (n = 3) determined by one-way analysis of variance (ANOVA) followed by Bonferroni multiple comparison testing. *p < 0.05 and ***p < 0.001.
Section 2.10 Regulation of Wnt signaling by the salvage pathway occurs at the level of the Wnt ligand and/or receptors
Wnt signaling is normally not detectable in undifferentiated ectodermal explants (animal caps) of early Xenopus embryos. To determine the level of the Wnt pathway at which the UDP-GlcNAc salvage pathway and Dpagt1 act, we tested whether pathway activation by different Wnt components could be enhanced or inhibited by overexpressing or downregulating salvage pathway components (and Dpagt1), respectively. We focused on Wnt ligand and receptor components because they are known to be glycosylated. We found that co-injecting Xwnt8, Wnt8-FZ5 (a constitutively active form of the Wnt receptor Frizzled (Fz)) (Holmen et al., 2002), or the Wnt co-receptor low-density lipoprotein receptor-related protein 6 (Lrp6) with mRNA encoding salvage pathway enzymes or Dpagt1 increased expression of the Wnt target genes, Xnr3, and chordin (Fig. 7A–C, S13 A–C). Conversely, co-injecting MOs of Xwnt8, Wnt8-FZ5, or Lrp6 with mRNA encoding salvage pathway enzymes or Dpagt1 decreased expression of Xnr3 and chordin (Fig. 7A–C, S13A–C). Dsh, a cytoplasmic nonglycosylated Wnt component, is thought to function at the level of the plasma membrane to regulate activation of Wnt receptors (Saito-Diaz et al., 2013). We found that co-injecting mRNA encoding or MO directed against Dsh with salvage mRNA encoding pathway enzymes or Dpagt1 did not alter the expression of Xnr3 or chordin (Fig. 7D, S13D). These data strongly suggest that the UDP-GlcNAc salvage pathway controls Wnt signaling at the ligand-receptor level in the early embryo.
Fig. 7.
The UDP-GlcNAC salvage pathway acts at the level of the Wnt ligand and/or receptors. (A–C, Left) Activation of the Wnt pathway by Xwnt8, Wnt8-Fz5, and Lrp6 is inhibited by co-injection with MOs against salvage pathway enzymes as indicated by expression of the Wnt target gene, Xnr3. (A–C; Right) Conversely, co-injection with mRNAs encoding pathway enzymes enhanced Xnr3 expression. (D) In contrast, activation of the Wnt pathway by Dsh was unaffected by inhibition (Left) or overexpression (Right) of salvage pathway enzymes. Data shown are representative of n ≥ 3 biological replicates using n = 3 technical replicates. Each replicate was a pool of n = 5 animal caps. Each dorsal blastomere was injected or co-injected with 1 ng mRNA or 1 pg MO. Expression was normalized to Odc. Graphs display fold change calculated using the 2−ΔΔCt method. Significance was calculated using t-tests with equal variance with Bonferroni correction on the ΔCt values. *ρ < 0.0127, **ρ < 0.00251, ***ρ < 0.000251.
Discussion (Section 3)
Section 3.1
In the current study, we found that increased and decreased activity of the UDPGlcNAc salvage pathway phenocopied Wnt activation and inhibition, respectively. Furthermore, we showed that these effects are evolutionarily conserved in Xenopus, zebrafish, Drosophila, and mouse intestinal enteroids. Perhaps it is not suprising that alterations in glycosylation would impact Wnt signaling as it has been previously shown that glycosylation is important for the maturation of Wnt ligands and receptors (MacDonald and He, 2012). Interestingly, glycosylation does not appear to play a role in Wnt-Frizzled interaction (Janda et al., 2012). In the case of LRP6, conserved N-glycan chains have been shown to be important for modulation the conformation of the ectodomain and, potentially, Wnt signaling (Matoba et al., 2017). Various heparin sulfate proteoglycans have been shown to play important roles in Wnt signaling, primarily by facilitating ligand-receptor interactions (Filmus et al., 2008; Pataki et al., 2015; Wodarz and Nusse, 1998). Because of our demonstration that perturbing the UDP-GlcNAc pathway alters pathway activation by Frizzled and LRP6, we favor a model in which changes in the UDP-GlcNAc pool affect the Wnt receptors themselves (either via their maturation or via their structural conformation) and, consequently, downstream signaling (Fig. S14).
Section 3.2
Most surprisingly are our findings that changes in the UDP-GlcNAc salvage pathway have such a profound affect on Wnt signaling, but not on other critical embryonic signaling pathways (e.g., Fgf, TGFβ, Notch, or Shh) during development. Cellular levels of UDP-GlcNAc reflect changes in the intracellular levels of glucose, amino acids, fatty acids, and nucleotides, and they has been proposed to act as a sensor of nutrient availability (Cheatham, 2004; Rossetti, 2000). Because the Wnt pathway is a major growth pathway in humans, one evolutionarily conserved role of the Wnt pathway may be to couple the overall available metabolic resources of the cell to growth and proliferation.
Section 3.3
Although beyond the scope of the present study, a major question is how alterations in the UDP-GlcNAc pool affect signaling by Wnt receptors. It is possible that the effects are due to global changes on the glycosylation state of Wnt receptors proteins. We believe this is unlikely given that other signaling pathways would likely to be affected as well. Thus, we favor a model in which one or more critical glycosylation event(s) on the Wnt receptor may be altered in response to the changing intracellular pool of UDP-GlcNAc (Fig. S14). Identifying the relevant Wnt receptor glycosylation event(s) altered by changes in UDP-GlcNAc levels will be the focus of future studies.
Section 3.4
Finally, partial loss-of-function mutations in glycosylation cause a group of human diseases termed congenital disorders of glycosylation (CDGs) (Freeze, 2006). The biological and molecular defects by which mutations in CDGs cause their symptoms are not known. While some symptoms of CDGs differ, depending on the particular mutated gene, many CDGs have similar symptoms. Recent evidence suggests that CDGs may result from mutations in Pgm3 (Pgm3-CDG) that cause alterations in the free pool of UDP-GlcNAc and its utilization in glycosylation (Pacheco-Cuellar et al., 2017; Stray-Pedersen et al., 2014). Patients with Pgm3-CDG mutations present with immunodeficiency, short stature, brachydactyly, dysmorphic facial features, and intellectual disability (Pacheco-Cuellar et al., 2017; Stray-Pedersen et al., 2014). Interestingly, all of these phenotypes have been attributed to developmental defects resulting from mutations in Wnt pathway components (Brugmann et al., 2007; Wang et al., 2011; White et al., 2018). Thus, it is possible that disruption in Wnt signaling during development may contribute significantly to the phenotypes associated with N-glycosylation mutations, and patients with CDGs may benefit from therapeutics that target the Wnt pathway. Future studies in model systems such as Xenopus, zebrafish, and Drosophila will be helpful in testing these possibilities at the molecular and phenotypic level.
Methods (Section 4)
Section 4.1 Kinase screen
We obtained 232 cDNAs from the Harvard Institute of Proteomics FLEXGene human kinase cDNA collection (pDNR-dual complete set). Primers designed to facilitate in vitro transcription were used to generate PCR constructs. Human kinase coding regions were amplified using a primer set containing flanking plasmid sequence. The 5’ oligonucleotide contained a T7 promoter sequence and the 3’ oligonucleotide was designed to overlap with the 5’ oligonucleotide sequence of the CS2 poly(A) fragment: Forward: 5’ GGCCCGCGCGCCAAACGAATGGTC 3’
Reverse: 5’CCAAGCCTTCTAATACGACTCACTATAGGGAGACAGTGAGCGAGGAAGCGGCCG C 3’.
The pCS2 poly(A) fragment was amplified in a separate PCR reaction using the following primers:
Forward: 5’ GACCATTCGTTTGGCGCGCGGGCCTGAGATCCAGACATGATAAGATAC 3’
Reverse: 5’ GAATTAAAAAACCTCCCACACCTCCCCCTGAACCTG 3’
Both DNA fragments were stitched together in a third PCR reaction to produce a single fragment containing a 5’ human kinase coding region and a 3’ poly(A) tail. mRNAs were generated using the MEGAscript T7 Transcription Kit (Ambion). Pools of 8 human kinase mRNAs were injected equatorially into dorsal blastomeres of four-cell stage Xenopus laevis embryos. Pools were assessed for perturbation of development. Positive pools were further characterized by single mRNA injections.
Section 4.2 DNA constructs, mRNA, and protein
Kinases were subcloned into pCS2 plasmids, and these constructs were used for all further experiments. A kinase-dead Nagk mutant was generated by introducing a threonine to methionine substitution at amino acid 128 (NagkT128M) by site-directed mutagenesis. cDNAs encoding Pgm3, Uap1, Dpagt1 and Ngyl1 were obtained from Open Biosystems, and Drosophila CG6218/DNagk cDNA was obtained from the Drosophila Genomics Resource. mRNAs were generated using the mMessage Machine SP6 transcription kit (Ambion). Nagk was subcloned into the pMAL vector (New England Biolabs), and recombinant MBP-tagged Nagk was expressed and purified according to manufacturer’s protocol.
Section 4.3 Sugars and morpholinos
3-OMe-GlcNAc, UDP-GlcNAc, and GlcNAc-6-P were obtained from Cayman Chemical Company. GlcNAc and GlcNac-1-P were obtained from Sigma-Aldrich. Morpholinos were designed using J-Strain 9.2 and acquired from Gene Tools, LLC. The following MO sequences were used: Nagk 5’ CCCCCATACACAGCAGCCATCTC 3’, Pgm3 5’ ATTCAGCACTGCTTCCATCTTCATC 3’, Uap1 5’ TGACGAACAACTGCCACATCCATAC 3’, and Dpagt1 5’ CCGGCATGTTTGCCAATAGTTTACG 3’.
Section 4.4 Animal care
All vertebrate animals in this study (Xenopus and zebrafish) were treated in accordance with Vanderbilt’s Institutional Animal Care and Use Committee.
Section 4.5 Xenopus embryo injections
Xenopus embryos were in vitro fertilized, dejellied, cultured, and injected as previously described (Peng, 1991). Staging was as previously described (Nieuwkoop and Faber, 1994). Embryos were injected equatorially in both dorsal blastomeres at the 4-cell stage and allowed to develop to stage 35 before phenotyping. For soaking embryos, stage 3 embryos were transferred to 2.5% (w/v) GlcNAc or Tunicamycin, suspended, and allowed to develop to stage 35 at room temperature.
Section 4.6 Zebrafish embryo injections
Wild-type (AB) zygotes (1 cell) were injected (1 nl) in the single cell. Embryos were raised in egg water (0.03% Instant Ocean) + 0.01 mg/L methylene blue at 28.5°C at a density of 50 or fewer embryos per 100 × 20 mm petri dish. Embryos were phenotyped at 5 days post fertilization. Embryos were fixed overnight in 4% paraformaldehyde in phosphate-buffered saline (PBS) overnight and stored in PBS at 4°C until imaged. Embryos with severe and non-specific edema were excluded from analysis.
Section 4.7 Generation of cDNA and quantitative-PCR
Total RNA was collected from whole embryos at stage 10.5, 13, or 16, and animal caps were collected from stage 10.5 embryos. Samples were homogenized in 1 ml RNA Stat-60 (Amsbio) with a disposable pestle and extracted with chloroform. cDNAs were synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCR was performed using the GoTaq® qPCR Master Mix (Promega) on a CFX96 qPCR machine (Bio-Rad). All qPCR reactions were performed in triplicate. mRNA levels were normalized to the house keeping gene ornithine decarboxylase (Odc). Fold changes were calculated using 2−ΔΔct as described (Rao et al., 2013). The following previously described qPCR primers were used (Batut et al., 2005; Jin et al., 2016; Miyazaki et al., 2012; Sun et al., 2015; Swain et al., 2005):
- Odc:
-
Forward: 5’ GTCAATGATGGAGTGTATGGATC 3’Reversec: 5’ TCCATTCCGCTCTCCTGAGCAC 3’.
-
- Hes1:
-
Forward: 5’ AAAGTCCTCCAAGCCCATC 3’Reverse: 5’ CCGGGAGCTATCTTTCTTGAG 3’.
-
- Dusp6:
-
Forward: 5’ GTGACACCAAACTTGCCTAATC 3’Reverse: 5’ CGGGCTTCATCTATAAACGAGAT 3’.
-
- Tbxt:
-
Forward: 5’-GGATCGTTATCACCTCTG-3’Reverse: 5’-GTGTAGTCTGTAGCAGCA-3’.
-
- Ptch1:
-
Forward: 5’-GGACAAGAATCGCAGAGCTG-3’Reverse: 5’-GGATGCTCAGGGAACCTTAC-3’.
-
- Chordin:
-
Forward: 5’-AACTGCCAGGACTGGATGGT-3’Reverse: 5’-GGCAGGATTTAGAGTTGCTTC-3’.
-
- Xnr3:
-
Forward: 5’-CTTCTGCACTAGATTCTG-3’Reverse: 5’-CAGCTTCTGGCCAAGACT-3’.
-
Section 4.8 Microscopy
Bright field images were obtained using a Stemi 2000-CS microscope (Zeiss, Oberkochen, Germany) with an Olympus DP72 camera. Fluorescent images were obtained using a Nikon Eclipse 80i microscope with a Cool SNAP ES camera (Photometrics, Tucson, USA). Images were analyzed in Fiji or Photoshop.
Section 4.9 Drosophila stocks and crosses
DNagk RNAi lines (Nagki1: Bloomington Drosophila Stock Center (BDSC) #28386 and Nagki2: Vienna Drosophila Resource Center (VDRC) #108069) and y (BDSC #64527) were expressed, in addition to UAS-Dcr-2 (BDSC #24648), in third instar larval wing imaginal discs using the hh-Gal4 (Tanimoto et al., 2000) or C96-Gal4 (BDSC #25757) drivers. Pgm3: Pgm3i1 (VDRC #31298) and Pgm3i2 (VDRC #105398) RNAi lines were similarly expressed in third instar wing imaginal discs. Crosses with C96-Gal4 were reared at 29°C, whereas crosses with hh-Gal4 were reared at 25°C.
Section 4.10 Drosophila immunohistochemistry
Third instar wing imaginal discs were dissected on ice in PBS and fixed in 4% paraformaldehyde in PBS for 20 minutes. The discs were then washed with 0.1% Triton X-100 in PBS followed by a 1 hour incubation in PBS with 0.5% Triton X-100 and 10% BSA. Wing discs were incubated with guinea pig anti-Senseless (1:2000) (Nolo et al., 2000), mouse anti-Wg (4D4, Developmental Studies Hybridoma bank [DSHB], 1:20), and mouse anti-Engrailed (4D9, DSHB, 1:20) at 4°C overnight in PBS with 0.1% Triton X-100. Discs were subsequently incubated with secondary antibodies (anti-guinea pig and anti-mouse Alexa Fluor 488 and 555 conjugates, ThermoFisher Scientific, 1:500) for 2 hours at room temperature. The discs were stained in 2 μg/ml DAPI and mounted in Prolong Gold (Invitrogen). All fluorescent images were obtained using a Nikon A1RSi confocal microscope.
Section 4.11 Enteroid culture
Enteroids were prepared as previously described (Li et al., 2017). Briefly, jejunum from ~8-week-old mice was obtained and washed with PBS followed by incubation for 20 min with cold PBS containing 1.5 mM dithiothreitol and 30 mM EDTA. Tissue was incubated with warm PBS containing 15 mM EDTA for 6 min, vigorously shaken to release intestinal crypts, and centrifuged. The resulting crypt pellet was washed with 30X volume of basal media (Dulbecco’s modified Eagle media/F12 plus 2 mM GlutaMAX, 10 mM Hepes, penicillin/streptomycin (100 U/ml), and 1X N2 and 1X B27 supplements). Purified crypts were filtered with a 100 μm cell strainer and embedded in Matrigel (Corning). Isolated enteroids were grown in IntestiCult Organoid Growth Medium (Mouse) (Stemcell Technologies) in the absence or presence of drugs. The media was replenished every other day for 10 days and cell viability determined using the Cell Titer Glo assay (Promega). CHIR99021 was purchased from Stemcell Technologies.
Section 4.12 Statistics
All statistical analyses were performed in R v3.1.0. Fisher’s exact test and multiple T-test (two tailed, equal variance) were used as indicated in figure legends. Post hoc analysis of Fisher’s exact test and multiple T-test was performed by Bonferroni correction.
Supplementary Material
Highlights:
The UDP-GlcNAc glycosylation salvage pathway regulates axiation in Xenopus embryos
Disruption of Xenopus axiation by the salvage pathway is mediated by Wnt signaling
The salvage pathway regulates Wnt but not Fgf, TGFβ, Notch, or Shh signaling
Regulation of Wnt signaling by the salvage pathway is evolutionarily conserved
Acknowledgments (Section 5)
We would like to thank Emilio Tahinci for Xenopus embryo injections, Emily Crispi for her assistance in preparing reagents and animal care, and Laura Lee for reading the manuscript. This work was funded by NIH grants R01CA105038, R01GM122222, R01121421, and the Norris Cotton Cancer Center to Y.A.; R35GM122516 and CTSA award (UL1TR000445) from the National Center for Advancing Translational Sciences to E.L.; R01CA219189 to D.J.R.; RO1EY024354 to J.G.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests (Section 6)
E.L. and D.J.R. are co-founders of StemSynergy Therapeutics Inc., a company that seeks to develop inhibitors of major signaling pathways (including the Wnt pathway) for the treatment of cancer.
REFERENCES: (Section 7)
- Batut J, Vandel L, Leclerc C, Daguzan C, Moreau M, Neant I, 2005. The Ca2+-induced methyltransferase xPRMT1b controls neural fate in amphibian embryo. Proc Natl Acad Sci U S A 102, 15128–15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M, Chen H, Reutter W, Hinderlich S, 2002. Structure and function of N-acetylglucosamine kinase. Identification of two active site cysteines. European journal of biochemistry /FEBS 269, 4212–4218. [DOI] [PubMed] [Google Scholar]
- Blume A, Berger M, Benie AJ, Peters T, Hinderlich S, 2008. Characterization of ligand binding to N-acetylglucosamine kinase studied by STD NMR. Biochemistry 47, 13138–13146. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Goodnough LH, Gregorieff A, Leucht P, ten Berge D, Fuerer C, Clevers H, Nusse R, Helms JA, 2007. Wnt signaling mediates regional specification in the vertebrate face. Development 134, 3283–3295. [DOI] [PubMed] [Google Scholar]
- Cavodeassi F, Carreira-Barbosa F, Young RM, Concha ML, Allende ML, Houart C, Tada M, Wilson SW, 2005. Early stages of zebrafish eye formation require the coordinated activity of Wnt11, Fz5, and the Wnt/beta-catenin pathway. Neuron 47, 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TM, Longabaugh W, Bolouri H, Chen HL, Tseng WF, Chao CH, Jang TH, Lin YI, Hung SC, Wang HD, Yuh CH, 2009. Developmental gene regulatory networks in the zebrafish embryo. Biochim Biophys Acta 1789, 279–298. [DOI] [PubMed] [Google Scholar]
- Cheatham B, 2004. Enough is enough: nutrient sensors and insulin resistance. Endocrinology 145, 2115–2117. [DOI] [PubMed] [Google Scholar]
- Diaz-Benjumea FJ, Cohen SM, 1995. Serrate signals through Notch to establish a Wingless-dependent organizer at the dorsal/ventral compartment boundary of the Drosophila wing. Development 121, 4215–4225. [DOI] [PubMed] [Google Scholar]
- Filmus J, Capurro M, Rast J, 2008. Glypicans. Genome Biol 9, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze HH, 2006. Genetic defects in the human glycome. Nature reviews 7, 537–551. [DOI] [PubMed] [Google Scholar]
- Freeze HH, Hart GW, Schnaar RL, 2015. Glycosylation Precursors, in: rd Varki, Cummings A, Esko RD, Stanley JD, Hart P, Aebi GW, Darvill M, Kinoshita AG, Packer T, Prestegard NH, Schnaar JH, Seeberger RL, P H (Eds.), Essentials of Glycobiology, Third Edition ed Cold Spring Harbor, Cold Spring Harbor; (NY: ), pp. 51–63. [Google Scholar]
- Goudevenou K, Martin P, Yeh YJ, Jones P, Sablitzky F, 2011. Def6 is required for convergent extension movements during zebrafish gastrulation downstream of Wnt5b signaling. PLoS One 6, e26548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Shi HJ, Gao L, Zhang HY, Tao QH, 2012. Maternal Mga is required for Wnt signaling and organizer formation in the early Xenopus embryo. Acta Bioch Bioph Sin 44, 939–947. [DOI] [PubMed] [Google Scholar]
- Gustafson K, Boulianne GL, 1996. Distinct expression patterns detected within individual tissues by the GAL4 enhancer trap technique. Genome 39, 174–182. [DOI] [PubMed] [Google Scholar]
- Heifetz A, Keenan RW, AD E, 1979. Mechanism of action of tunicamycin on the UDP-GlcNAc:dolichyl-phosphate Glc-NAc-1 -phosphate transferase. Biochemistry 18, 2186–2192. [DOI] [PubMed] [Google Scholar]
- Heisenberg CP, Tada M, Rauch GJ, Saude L, Concha ML, Geisler R, Stemple DL, Smith JC, Wilson SW, 2000. Silberblick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature 405, 76–81. [DOI] [PubMed] [Google Scholar]
- Hikasa H, Sokol SY, 2013. hiWnt signaling in vertebrate axis specification. Cold Spring Harb Perspect Biol 5, a007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderlich S, Berger M, Schwarzkopf M, Effertz K, Reutter W, 2000. Molecular cloning and characterization of murine and human N-acetylglucosamine kinase. European journal of biochemistry / FEBS 267, 3301–3308. [DOI] [PubMed] [Google Scholar]
- Holmen SL, Salic A, Zylstra CR, Kirschner MW, Williams BO, 2002. A novel set of Wnt-Frizzled fusion proteins identifies receptor components that activate beta -catenin-dependent signaling. J Biol Chem 277, 34727–34735. [DOI] [PubMed] [Google Scholar]
- Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC, 2012. Structural basis of Wnt recognition by Frizzled. Science 337, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Schwend T, Fu J, Bao Z, Liang J, Zhao H, Mei W, Yang J, 2016. Members of the Rusc protein family interact with Sufu and inhibit vertebrate Hedgehog signaling. Development 143, 3944–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao KR, Elinson RP, 1988. The entire mesodermal mantle behaves as Spemann’s organizer in dorsoanterior enhanced Xenopus laevis embryos. Dev Biol 127, 64–77. [DOI] [PubMed] [Google Scholar]
- Keller R, 1991. Early embryonic development of Xenopus laevis. Methods Cell Biol 36, 61–113. [DOI] [PubMed] [Google Scholar]
- Kim C, Oda T, Itoh M, Jiang D, Artinger K, Chandrasekharappa S, Driever W, Chitnis A, 2000. Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature 407, 913–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman MA, Zhu XY, Khounlo S, 1988. Amplification and molecular cloning of the hamster tunicamycin-sensitive N-acetylglucosamine-1 -phosphate transferase gene. The hamster and yeast enzymes share a common peptide sequence. J Biol Chem 263, 19796–19803. [PubMed] [Google Scholar]
- MacDonald BT, He X, 2012. Frizzled and LRP5/6 receptors for Wnt/beta-catenin signaling. Cold Spring Harb Perspect Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam B, Cuesta-Munoz A, Davis EA, Matschinsky FM, Harrison RW, Weber IT, 1999. Structural model of human glucokinase in complex with glucose and ATP: implications for the mutants that cause hypo- and hyperglycemia. Diabetes 48, 1698–1705. [DOI] [PubMed] [Google Scholar]
- Marlow F, Zwartkruis F, Malicki J, Neuhauss SC, Abbas L, Weaver M, Driever W, Solnica-Krezel L, 1998. Functional interactions of genes mediating convergent extension, knypek and trilobite, during the partitioning of the eye primordium in zebrafish. Dev Biol 203, 382–399. [DOI] [PubMed] [Google Scholar]
- Matoba K, Mihara E, Tamura-Kawakami K, Miyazaki N, Maeda S, Hirai H, Thompson S, Iwasaki K, Takagi J, 2017. Conformational Freedom of the LRP6 Ectodomain Is Regulated by N-glycosylation and the Binding of the Wnt Antagonist Dkk1. Cell Rep 18, 32–40. [DOI] [PubMed] [Google Scholar]
- Mir A, Kofron M, Heasman J, Mogle M, Lang S, Birsoy B, Wylie C, 2008. Long-and short-range signals control the dynamic expression of an animal hemisphere-specific gene in Xenopus. Dev Biol 315, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa I, Mita Y, Murata T, Okuda J, Sugiura M, Hamada Y, Chiba T, 1994. Utility of 3-O-methyl-N-acetyl-D-glucosamine, an N-acetylglucosamine kinase inhibitor, for accurate assay of glucokinase in pancreatic islets and liver. Enzyme & protein 48, 135–142. [DOI] [PubMed] [Google Scholar]
- Miyazaki A, Ishii K, Yamashita S, Nejigane S, Matsukawa S, Ito Y, Onuma Y, Asashima M, Michiue T, 2012. mNanog possesses dorsal mesoderm-inducing ability by modulating both BMP and Activin/nodal signaling in Xenopus ectodermal cells. PLoS One 7, e46630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriishi T, Shibata Y, Tsukazaki T, Yamaguchi A, 2005. Expression profile of Xenopus banded hedgehog, a homolog of mouse Indian hedgehog, is related to the late development of endochondral ossification in Xenopus laevis. Biochem Biophys Res Commun 328, 867–873. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J, 1994. Normal Table of Xenopus laevis (Daudin). Routledge, New York, NY. [Google Scholar]
- Nishimoto S, Nishida E, 2007. Fibroblast growth factor 13 is essential for neural differentiation in xenopus early embryonic development. Journal of Biological Chemistry 282, 24255–24261. [DOI] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ, 2000. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102, 349–362. [DOI] [PubMed] [Google Scholar]
- Pacheco-Cuellar G, Gauthier J, Desilets V, Lachance C, Lemire-Girard M, Rypens F, Le Deist F, Decaluwe H, Duval M, Bouron-Dal Soglio D, Kokta V, Haddad E, Campeau PM, 2017. A Novel PGM3 Mutation Is Associated With a Severe Phenotype of Bone Marrow Failure, Severe Combined Immunodeficiency, Skeletal Dysplasia, and Congenital Malformations. J Bone Miner Res 32, 1853–1859. [DOI] [PubMed] [Google Scholar]
- Pataki CA, Couchman JR, Brabek J, 2015. Wnt Signaling Cascades and the Roles of Syndecan Proteoglycans. J Histochem Cytochem 63, 465–480. [DOI] [PubMed] [Google Scholar]
- Peng HB, 1991. Xenopus laevis: Practical uses in cell and molecular biology. Solutions and protocols. Methods Cell Biol 36, 657–662. [PubMed] [Google Scholar]
- Peters JM, McKay RM, McKay JP, Graff JM, 1999. Casein kinase I transduces Wnt signals. Nature 401, 345–350. [DOI] [PubMed] [Google Scholar]
- Rao X, Huang X, Zhou Z, Lin X, 2013. An improvement of the 2A(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath 3, 71–85. [PMC free article] [PubMed] [Google Scholar]
- Rauch GJ, Hammerschmidt M, Blader P, Schauerte KE, Strahle U, Ingham PW, McMahon AP, Haffter P, 1997. WNT5 is required for tail formation in the zebrafish embryo. Cold Spring Harb Sym 62, 227–234. [PubMed] [Google Scholar]
- Rossetti L, 2000. Perspective: Hexosamines and nutrient sensing. Endocrinology 141, 1922–1925. [DOI] [PubMed] [Google Scholar]
- Saito-Diaz K, Chen TW, Wang X, Thorne CA, Wallace HA, Page-McCaw A, Lee E, 2013. The way Wnt works: components and mechanism. Growth Factors 31, 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakanaka C, Leong P, Xu L, Harrison SD, Williams LT, 1999. Casein kinase Is in the Wnt pathway: Regulation of p-catenin function. Proceedings of the National Academy of Sciences of the United States of America 99, 12548–12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H, 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. [DOI] [PubMed] [Google Scholar]
- Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT, 2007. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134, 479–489. [DOI] [PubMed] [Google Scholar]
- Stray-Pedersen A, Backe PH, Sorte HS, Morkrid L, Chokshi NY, Erichsen HC, Gambin T, Elgstoen KB, Bjoras M, Wlodarski MW, Kruger M, Jhangiani SN, Muzny DM, Patel A, Raymond KM, Sasa GS, Krance RA, Martinez CA, Abraham SM, Speckmann C, Ehl S, Hall P, Forbes LR, Merckoll E, Westvik J, Nishimura G, Rustad CF, Abrahamsen TG, Ronnestad A, Osnes LT, Egeland T, Rodningen OK, Beck CR, Baylor-Johns Hopkins Center for Mendelian, G., Boerwinkle EA, Gibbs RA, Lupski JR, Orange JS, Lausch E, Hanson IC, 2014. PGM3 mutations cause a congenital disorder of glycosylation with severe immunodeficiency and skeletal dysplasia. Am J Hum Genet 95, 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Basler K, 1993. Organizing activity of wingless protein in Drosophila. Cell 72, 527–540. [DOI] [PubMed] [Google Scholar]
- Sun G, Hu Z, Min Z, Yan X, Guan Z, Su H, Fu Y, Ma X, Chen YG, Zhang MQ, Tao Q, Wu W, 2015. Small C-terminal Domain Phosphatase 3 Dephosphorylates the Linker Sites of Receptor-regulated Smads (R-Smads) to Ensure Transforming Growth Factor beta (TGFbeta)-mediated Germ Layer Induction in Xenopus Embryos. J Biol Chem 290, 17239–17249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain RK, Katoh M, Medina A, Steinbeisser H, 2005. Xenopus frizzled-4S, a splicing variant of Xfz4 is a context-dependent activator and inhibitor of Wnt/beta-catenin signaling. Cell Commun Signal 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek W, Tsai IC, Klimowski L, Pepler A, Barnette J, Yost HJ, Virshup DM, 2004. Regulation of casein kinase I epsilon activity by Wnt signaling. J Biol Chem 279, 13011–13017. [DOI] [PubMed] [Google Scholar]
- Tanimoto H, Itoh S, ten Dijke P, Tabata T, 2000. Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs. Mol Cell 5, 59–71. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H, 1994. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes & Development 8, 1434–1447. [DOI] [PubMed] [Google Scholar]
- van de Water S, van de Wetering M, Joore J, Esseling J, Bink R, Clevers H, Zivkovic D, 2001. Ectopic Wnt signal determines the eyeless phenotype of zebrafish masterblind mutant. Development 128, 3877–3888. [DOI] [PubMed] [Google Scholar]
- Wang B, Sinha T, Jiao K, Serra R, Wang J, 2011. Disruption of PCP signaling causes limb morphogenesis and skeletal defects and may underlie Robinow syndrome and brachydactyly type B. Hum Mol Genet 20, 271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JJ, Mazzeu JF, Coban-Akdemir Z, Bayram Y, Bahrambeigi V, Hoischen A, van Bon BWM, Gezdirici A, Gulec EY, Ramond F, Touraine R, Thevenon J, Shinawi M, Beaver E, Heeley J, Hoover-Fong J, Durmaz CD, Karabulut HG, Marzioglu-Ozdemir E, Cayir A, Duz MB, Seven M, Price S, Ferreira BM, Vianna-Morgante AM, Ellard S, Parrish A, Stals K, Flores-Daboub J, Jhangiani SN, Gibbs RA, Baylor-Hopkins Center for Mendelian, G., Brunner HG, Sutton VR, Lupski JR, Carvalho CMB, 2018. WNT Signaling Perturbations Underlie the Genetic Heterogeneity of Robinow Syndrome. Am J Hum Genet 102, 27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PA, Lagna G, Suzuki A, Hemmati-Brivanlou A, 1997. Concentration-dependent patterning of the Xenopus ectoderm by BMP4 and its signal transducer Smad1. Development 124, 3177–3184. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Nusse R, 1998. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol 14, 59–88. [DOI] [PubMed] [Google Scholar]
- Zeitler R, Giannis A, Danneschewski S, Henk E, Henk T, Bauer C, Reutter W, Sandhoff K, 1992. Inhibition of N-acetylglucosamine kinase and N-acetylmannosamine kinase by 3-O-methyl-N-acetyl-D-glucosamine in vitro. European journal of biochemistry / FEBS 204, 1165–1168. [DOI] [PubMed] [Google Scholar]
- Zheng X, Narayanan S, Zheng X, Luecke-Johansson S, Gradin K, Catrina SB, Poellinger L, Pereira TS, 2017. A Notch-independent mechanism contributes to the induction of Hes1 gene expression in response to hypoxia in P19 cells. Exp Cell Res 358, 129–139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.