Significance
G protein-coupled receptors (GPCRs) regulate a wide variety of important cellular processes and are targeted by a large fraction of approved drugs. GPCRs signal by activating heterotrimeric G proteins and must couple to a select subset of G proteins to produce appropriate intracellular responses. It is not known how GPCRs select G proteins, but it is generally accepted that the Gα subunit C terminus is the primary G protein determinant of coupling selectivity. We systematically studied coupling of GPCRs to four families of G proteins and chimeras with C terminal regions that were exchanged between families. We uncovered rules for coupling selectivity and found that different GPCRs can recognize different features of the same G protein for selective coupling.
Keywords: GPCR, G protein-coupled receptor, G protein selectivity, ternary complex
Abstract
G protein-coupled receptors (GPCRs) activate four families of heterotrimeric G proteins, and individual receptors must select a subset of G proteins to produce appropriate cellular responses. Although the precise mechanisms of coupling selectivity are uncertain, the Gα subunit C terminus is widely believed to be the primary determinant recognized by cognate receptors. Here, we directly assess coupling between 14 representative GPCRs and 16 Gα subunits, including one wild-type Gα subunit from each of the four families and 12 chimeras with exchanged C termini. We use a sensitive bioluminescence resonance energy transfer (BRET) assay that provides control over both ligand and nucleotide binding, and allows direct comparison across G protein families. We find that the Gs- and Gq-coupled receptors we studied are relatively promiscuous and always couple to some extent to Gi1 heterotrimers. In contrast, Gi-coupled receptors are more selective. Our results with Gα subunit chimeras show that the Gα C terminus is important for coupling selectivity, but no more so than the Gα subunit core. The relative importance of the Gα subunit core and C terminus is highly variable and, for some receptors, the Gα core is more important for selective coupling than the C terminus. Our results suggest general rules for GPCR-G protein coupling and demonstrate that the critical G protein determinants of selectivity vary widely, even for different receptors that couple to the same G protein.
G protein coupled receptors (GPCRs) exert many of their physiological effects by coupling to and activating heterotrimeric G proteins. The 16 human Gα subunit genes are classified into four families (Gs, Gi/o, Gq/11, and G12/13), and members of each family interact with different effector molecules to produce distinct cellular responses (1, 2). Individual cells generally express many different G proteins from multiple families, and, therefore, GPCRs must be able to selectively activate subsets of G proteins. Many receptors show a preference for G proteins from just one of the four families, although promiscuous coupling to G proteins from multiple families is also not uncommon (3, 4). The mechanisms of selective GPCR-G protein coupling are not completely understood, and the structural determinants of selectivity have not been determined with precision. Analysis of GPCR primary sequences has not revealed simple conserved motifs for coupling to different G protein families (5), and analysis of GPCR–G protein complex structures has only begun to uncover features that may be important for coupling selectivity (6–12). From the standpoint of the G protein, it has long been appreciated that the Gα carboxy (C) terminus is critical for activation by receptors and also a key determinant of selectivity (13–17). The distal part of the C-terminal alpha helix (helix 5; H5) is enveloped by GPCR transmembrane (TM) domains during coupling (6), and several studies have shown that mutations within this region can promote coupling of receptors to noncognate heterotrimers (17–21). Although other Gα regions have been shown to be important for recognition by individual GPCRs (22–25), the Gα C terminus is widely considered to be the most important structural determinant of coupling selectivity.
Here, we test this idea by measuring coupling of GPCRs to four representative Gα subunits (Gαs, Gαi1, Gαq, and Gα12) and 12 Gα chimeras with the C terminus of one representative and the Gα core region of another. We monitor coupling using a sensitive energy transfer assay that allows comparison of all GPCR–G protein combinations directly at the coupling step. Our results demonstrate unexpectedly promiscuous coupling of many GPCRs to wild-type G proteins and support the notion that the Gα C terminus is one important determinant of coupling selectivity for many receptors. However, our results also show that the Gα C terminus is a minor contributor to selective coupling of several other receptors. Taken together our findings suggest that GPCRs recognize widely distributed structural features of G proteins and are consistent with the suggestion that different GPCRs recognize different conserved selectivity determinants of a given G protein (5).
Results
Promiscuous Coupling to Wild-Type G Proteins.
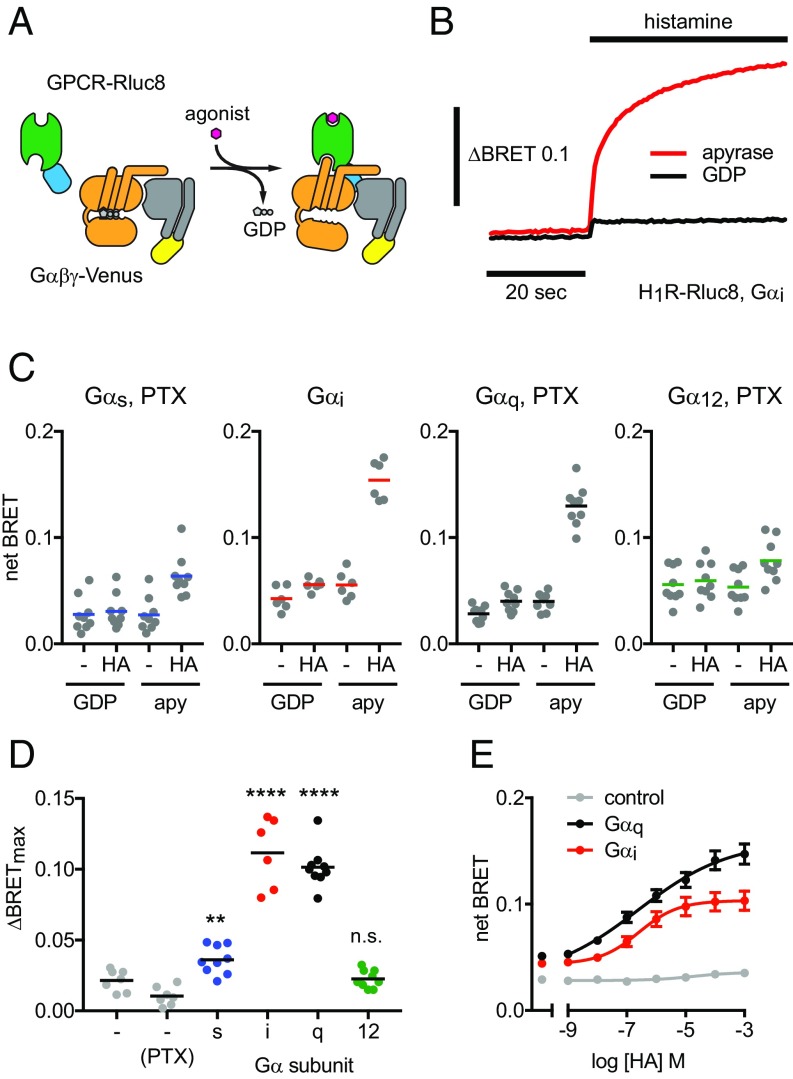
Coupling of a GPCR to a heterotrimeric G protein can be defined as the allosteric interaction between the ligand binding site of the receptor and the nucleotide binding site of the associated Gα subunit, such that the agonist-bound state of the receptor promotes the nucleotide-free (empty) state of the G protein and vice versa (26, 27) (Fig. 1A). To quantify coupling we monitored agonist-dependent and nucleotide-sensitive association of GPCRs with different heterotrimers using bioluminescence resonance energy transfer (BRET) (28). To interfere as little as possible with receptor–G protein interactions, we positioned BRET donor and acceptor labels far from regions identified in functional and structural studies as important for coupling. A luciferase was fused to the GPCR C terminus, and complementary fragments of the fluorescent protein Venus were fused to Gβ1 and Gγ2 subunits. These components were transfected together with unlabeled Gα subunits into CRISPR/Cas9-edited HEK293 cells deficient in either Gαs, Gαq, and Gα12 subunit families (three-family knockouts; 3GKO) or all four Gα families (four-family knockouts; 4GKO) (29). When 3GKO cells were used, the S1 subunit of pertussis toxin (PTX) was coexpressed to prevent coupling to endogenous Gαi/o subunits, except when the overexpressed Gα subunit was itself sensitive to PTX. Cells were permeabilized with digitonin and either supplemented with GDP or treated with apyrase to remove residual nucleotides.
Fig. 1.
Coupling to wild-type heterotrimers in permeabilized, nucleotide-depleted cells. (A) Receptors were fused to Renilla luciferase (Rluc8), and Gβγ dimers were fused to Venus. (B) Time course of BRET between H1R-Rluc8 and Gαi1βγ-Venus in response to 100 μM histamine (HA) in permeabilized cells in the presence (GDP) and absence (apyrase) of nucleotides. (C and D) H1R couples to Gs, Gi1, and Gq heterotrimers; **P = 0.0011, ****P < 0.0001, n.s. not significant; one-way ANOVA, Sidak’s multiple comparisons. Pertussis toxin S1 subunit (PTX) was coexpressed in experiments with Gαs, Gαq, and Gα12. (E) Similar potency of histamine-induced coupling to Gi1 (EC50 = 258 nM; n = 4) and Gq (EC50 = 245 nM; n = 7) heterotrimers.
Complexes between agonist-bound GPCRs and G proteins are short-lived when guanine nucleotides are present at concentrations similar to those found in the cytosol of intact cells and are greatly stabilized when guanine nucleotides are absent (30). Accordingly, nucleotide depletion significantly enhanced the magnitude of agonist-induced BRET between receptors and G proteins (Fig. 1B), and maximal BRET was observed when agonist was present and GDP was absent (Fig. 1C). As a simple index of coupling, we defined ΔBRETmax as the difference between the BRET observed when receptor–G protein complexes are least stable (agonist absent, GDP present) and that observed when receptor–G protein complexes are most stable (agonist present, GDP absent). This experimental system allows direct comparison of receptor coupling to heterotrimers with Gα subunits from different families and is sufficiently sensitive to detect secondary coupling to nonpreferred G proteins. Using this assay, we found that receptors generally coupled to the same G proteins as indicated by other methods and as annotated in the IUPHAR/BPS Guide to Pharmacology (3, 31). However, many receptors coupled to wild-type G proteins more promiscuously than expected. For example, we found that H1 histamine receptors (H1R), which are generally classified as Gq-coupled (3), couple strongly to both Gq and Gi heterotrimers, couple weakly to Gs heterotrimers, and do not couple significantly to G12 heterotrimers (Fig. 1D). The EC50 values for H1R coupling to Gq and Gi were comparable (Fig. 1E), suggesting that activation of Gi heterotrimers by this receptor may have physiological significance, as some studies have indicated (32). We carried out similar experiments on a panel of 19 receptors (SI Appendix, Table S1), 17 of which are classified in the IUPHAR/BPS Guide to Pharmacology (3) as coupling primarily to Gs, Gi, or Gq proteins, and two of which are unclassified orphan receptors that transduce signals through G12/13 proteins (33, 34). We found that promiscuous coupling to more than one wild-type G protein was a universal feature of the Gs-, Gq- and G12-coupled receptors that we studied. In contrast, the Gi-coupled receptors that we studied were much more selective for Gi heterotrimers (Fig. 2 and SI Appendix, Fig. S1). From the perspective of the G protein, we found that Gi heterotrimers coupled to all of the receptors that we studied. It should be noted that the assay we used for these studies reflects allosteric coupling between agonist and nucleotide binding sites, but this is only a minimum requirement for agonist-dependent G protein signaling. Additional factors may be required for efficient G protein activation and signaling under physiological conditions in cells (see below).
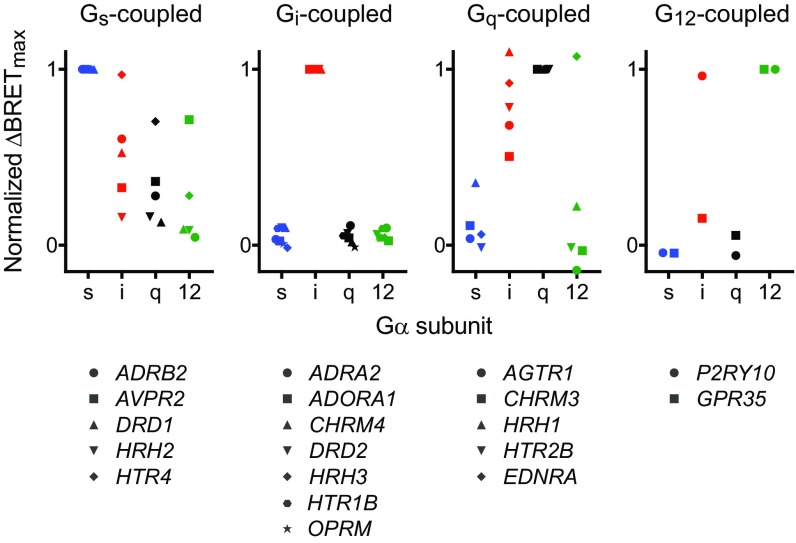
Fig. 2.
Gi-coupled receptors are relatively selective. Normalized ΔBRETmax for a panel of Gs-, Gi-, Gq-, and G12-coupled receptors. Each value represents the mean ΔBRETmax (normalized to the value obtained using the presumed cognate G protein) from n = 3–6 independent experiments. Individual data points for each receptor are shown in SI Appendix, Fig. S1.
Coupling to G Protein C-Terminal Chimeras.
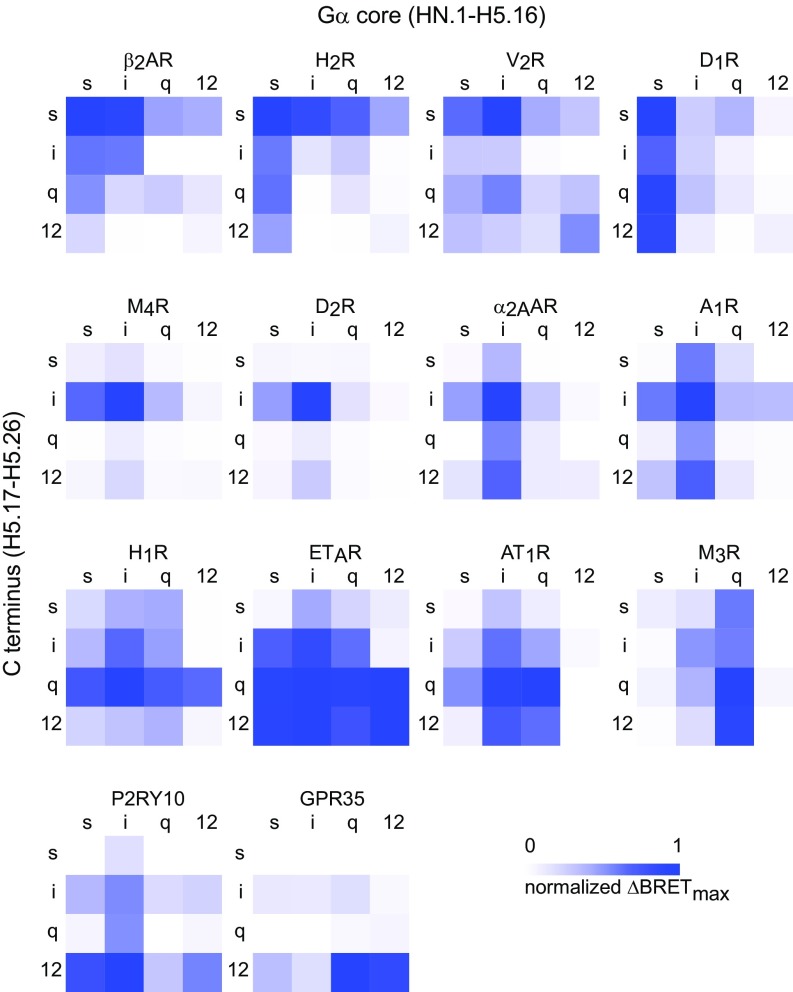
To assess the importance of the Gα C terminus for coupling selectivity, we constructed 12 Gα chimeras consisting of the main Gα subunit core [HN.1-H5.16 using the common G protein numbering (CGN) system] of Gαs (long isoform), Gαi1, Gαq, or Gα12 and the final 10 amino acids (H5.17–26) of each of the others. This portion of the Gα C terminus is deeply embedded in the TM domains of GPCRs in all receptor–G protein complex structures and has been called the interface module of the C-terminal alpha helix (helix 5 or H5) (35). Previous mutagenesis studies have primarily targeted residues within this region to alter coupling selectivity (36). This region is also unstructured in G protein crystal structures (35), suggesting that it is not involved in folding or stability of the Gα subunit core and, therefore, can be exchanged without compromising the structural integrity of heterotrimers. We confirmed the ability of Gα chimeras to form heterotrimers by monitoring sequestration of overexpressed Gβγ dimers (37), and all expressed at comparable levels as indicated by immunoprecipitation (SI Appendix, Fig. S2). Conditions were optimized to provide roughly equivalent stoichiometry for each receptor–G protein pair, with the latter in excess. For simplicity, we refer to heterotrimers that incorporate these chimeras as Gsi, Gsq, Gs12. We generated full coupling profiles (4 wild-type Gα subunits and 12 chimeras) for 14 GPCRs and plotted the results as ΔBRETmax heat maps (Fig. 3). Similar heat maps were constructed for GDP-sensitive BRET in the absence of agonist (ΔBRETGDP; SI Appendix, Fig. S3) and agonist-induced BRET in the presence of GDP (ΔBRETag; SI Appendix, Fig. S4).
Fig. 3.
Coupling to heterotrimers with C-terminal Gα subunit chimeras. Heat maps of ΔBRETmax (normalized to the largest value) for Gs-coupled (Top), Gi-coupled (second row), Gq-coupled (third row), and G12-coupled (Bottom) receptors. Maps are arranged from left to right depending on coupling that is determined more (e.g., H1R) or less (e.g., D1R) by the Gα C terminus (H5.17–H5.26). Each value represents the mean normalized ΔBRETmax from n = 3–6 independent experiments.
In agreement with our results using wild-type Gα subunits, we found that Gs-, Gq- and G12-coupled receptors coupled more promiscuously to G protein chimeras than Gi-coupled receptors. The Gs-, Gq- and G12-coupled receptors that we studied always coupled well to several chimeras in addition to wild-type G proteins, sometimes with ΔBRETmax values that approached or even exceeded that of the cognate wild-type G protein (Fig. 3). The Gi-coupled receptors that we studied were more selective. For example, the D2 dopamine receptor (D2R) failed to couple to any Gα chimera with a ΔBRETmax value that reached half of that observed with wild-type Gαi1. We also observed that the Gα12 core was often nonpermissive for coupling, particularly for Gi- and Gq-coupled receptors. This was sometimes partially overcome when a cognate C terminus was present, e.g., for A1R and H1R. By comparison, the Gα12 C terminus was more accommodating to most receptors (Fig. 3). This suggests that many receptors reject Gα12 core regions rather than the C terminus to select against G12 heterotrimers. The most promiscuous receptor that we studied was the endothelin A (ETAR) receptor, which coupled well to Gαi1, Gαq, Gα12, and eight of the nine chimeras bearing the C termini of these subunits. This receptor coupled poorly to Gαs and chimeras bearing the Gαs C terminus, suggesting that selection against Gs coupling for this receptor is determined primarily by the C terminus.
We anticipated that GPCRs would couple poorly to chimeras bearing noncognate C termini, i.e., adding a noncognate C terminus to a cognate Gα core would impair coupling. Indeed, this was observed for all 42 receptor-chimera pairs that we studied. However, the extent of impairment was highly variable, ranging from a near complete loss of coupling to almost no loss (SI Appendix, Fig. S5A). Gi-coupled receptors, in particular the M4 acetylcholine receptor (M4R) and D2R, were the most sensitive to noncognate C termini. Conversely, we expected that GPCRs would generally couple well to chimeras bearing cognate C termini, i.e., noncognate Gα core regions would only modestly impair coupling. However, we found that a noncognate Gα core was often as detrimental to coupling as a noncognate C terminus (SI Appendix, Fig. S5B). Although adding a cognate C terminus to a noncognate Gα core usually enhanced coupling, the gain of coupling was again highly variable and rarely a complete “switch.” For most of the receptors that we studied, coupling selectivity was determined both by the Gα subunit core and the C terminus. A clear C terminus-dominant coupling pattern was observed for only a few receptors, the most obvious examples being H1R, P2RY10, and GPR35 (Fig. 3). Other receptors, such as H2R and A1R, showed a more balanced influence of the cognate C terminus and G protein core. These receptors coupled equally well to chimeras bearing either the cognate C terminus or the cognate G protein core, although coupling to these chimeras was never as efficient as coupling to cognate wild-type G proteins. Unexpectedly, D1R and M3R showed coupling that was determined almost entirely by the G protein core and was virtually insensitive to the Gα C terminus (Fig. 3). Full concentration–response curves for these receptors confirmed the patterns observed with single saturating concentrations of agonists. Specifically, for the D1R and M3R agonist potency was greater for chimeras with cognate G protein cores, whereas for the H1R agonist potency was greater for chimeras with cognate C termini (SI Appendix, Fig. S6 and Table S2).
Efficient coupling to G proteins with noncognate Gα C termini was unexpected, and suggested either that the C terminus contributes little to the stability of some receptor–G protein complexes or, alternatively, that some receptors can interact strongly with both cognate and noncognate C termini. To better understand the basis of efficient coupling to Gα subunits with noncognate C termini, we performed atomistic molecular dynamics (MD) simulations of D1R complexes with Gs, Gsi, Gsq, and Gs12. These simulations revealed that in all cases, the C terminus contributed a substantial portion (at least one-third) of the total Gs protein interaction energy with D1R (SI Appendix, Fig. S7B) and, therefore, was critical for the overall stability of the complex. However, this receptor accommodated noncognate C termini with small energetic penalties (SI Appendix, Fig. S7), which corresponds to the observation that the C terminus was not critical for D1R selectivity. These simulations also identified several regions of the Gαs core that contribute to the interaction and may also contribute to coupling selectivity for this receptor, including HN, the hns1, s2s3, and h4s6 linkers and N-terminal residues of H5 (SI Appendix, Fig. S7A).
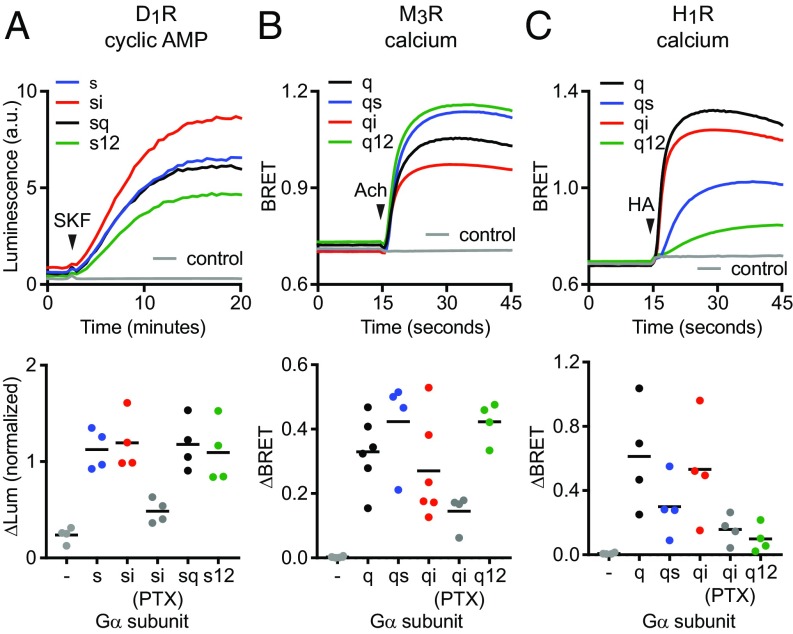
We then asked if receptors that interacted well with nucleotide-free G proteins bearing noncognate Gα C termini in our direct BRET coupling assay and MD simulations could also efficiently activate these G proteins and produce downstream signals. We found that wild-type Gs, Gsi, Gsq, and Gs12 all effectively restored potent activation of adenylate cyclase in response to D1R activation in cells lacking endogenous Gαs subunits (Fig. 4A and SI Appendix, Fig. S8), whereas our assays did not detect signals mediated by wild-type Gi1, Gq, or G12 in these cells (SI Appendix, Fig. S9). Likewise, we found that wild-type Gq, Gqi, Gqs, and Gq12 all fully restored calcium release from intracellular stores in response to M3R activation in cells lacking endogenous Gαq subunits (Fig. 4B and SI Appendix, Fig. S8), whereas our assays did not detect signals mediated by wild-type Gs, Gi1, or G12 in these cells (SI Appendix, Fig. S9). Notably, both D1R and M3R coupled significantly to Gi1 heterotrimers in our direct BRET assay, with ΔBRETmax values that were approximately half that of the respective cognate heterotrimers (Fig. 2) but failed to inhibit cAMP accumulation through Gi1 (SI Appendix, Fig. S9). This suggests that receptors may couple to heterotrimers, as defined by formation of an agonist- and nucleotide-sensitive complex, and yet fail to activate a given signaling pathway even when overexpressed. In contrast to what we observed with D1R and M3R, G protein chimeras with noncognate C termini only partially restored calcium release from intracellular stores in response to H1R activation. The rank order of restoration by chimeras was Gqi > Gqs > Gq12 (Fig. 4C), which was consistent with the rank order of H1R coupling to wild-type Gα subunits (Fig. 1). These results confirm that some GPCRs can efficiently couple to and activate heterotrimers with a variety of noncognate Gα C termini. Taken together with our direct coupling BRET results, these findings indicate a highly variable influence of the Gα subunit C terminus on coupling selectivity for different GPCRs.
Fig. 4.
Activation of Gα subunit chimeras with noncognate C termini. (A) Gsi, Gsq, and Gs12 chimeras fully restored cAMP accumulation in response to D1R stimulation (10 μM SKF 81297) in cells lacking endogenous Gαs family subunits (n = 4). (B) Gqs, Gqi, and Gq12 chimeras fully restored calcium release in response to M3R stimulation (100 μM acetylcholine) in cells lacking endogenous Gαq family subunits (n = 6). (C) Gqi but not Gqs and Gq12 chimeras fully restored calcium release in response to H1R stimulation (100 μM histamine) in cells lacking endogenous Gαq subunits (n = 4). Traces are averages of several experiments, and data points represent independent experiments. Pertussis toxin S1 subunit (PTX) was coexpressed as indicated.
Discussion
In this study, we used energy transfer between GPCRs and heterotrimeric G proteins to monitor allosteric coupling between agonist and nucleotide binding. This approach allowed us to directly compare coupling of several receptors to multiple G protein families without relying on downstream signals such as second messenger production. The use of G protein-deficient cell lines restricted coupling to defined wild-type and chimeric Gα subunits, and permeabilization allowed us to control occupancy of both agonist and nucleotide binding sites.
Our results revealed that many receptors couple more promiscuously than expected. For example, every GPCR that we tested coupled to Gi1 to some extent, and every Gs-coupled receptor that we tested also coupled somewhat to Gq. As a general rule, it appears that receptors that couple primarily to Gi heterotrimers are more selective than receptors that couple primarily to Gs and Gq heterotrimers. These trends are not readily apparent in annotated databases of coupling selectivity (3, 4). We suspect that the sensitivity of our assay, which detects stable complexes between agonist-occupied receptors and nucleotide-free G proteins, allowed us to detect secondary coupling (particularly to Gi1) that might easily be overlooked in functional studies. Indeed, our results with D1R and M3R coupling to Gi1 suggest that this method can detect interactions that are too inefficient to lead to physiologically significant G protein activation in cells. However, it is also possible that weak secondary coupling interactions have physiological significance under certain circumstances, as previously demonstrated for β2AR activation of Gi heterotrimers (38, 39). We also suspect that a single assay that reports coupling to G proteins directly from all four families is likely to produce a more accurate profile of subtype selectivity than comparison across families with multiple assays based on second messenger accumulation and/or gene expression.
Our results with wild-type Gα subunits are consistent with recent computational and structural studies, which have suggested that the outward displacement of transmembrane domain 6 (TM6) is restricted in active Gi-coupled receptors (8, 10–12, 40, 41). This produces a relatively small pocket in the cytoplasmic surface of the receptor that can only accommodate the relatively small C termini of Gi/o family Gα subunits. This mechanism predicts stringent rejection of Gs and Gq heterotrimers by Gi-coupled receptors, but no similar barrier to promiscuous Gi activation, as we observed. However, our results with Gα chimeras also show that some Gi-coupled receptors (e.g., α2AR and A1R) can tolerate noncognate C termini to some extent and, therefore, suggest that the Gαi core region also partly determines selectivity for these receptors.
Overall our results support the well-established role of the Gα subunit C terminus as a key determinant of receptor-G protein coupling selectivity. However, our findings also emphasize that other selectivity determinants exist that are equally important or more important for G protein recognition by many receptors. This is consistent with several previous reports of selectivity determinants that lie outside of the distal C terminus (22–25) and suggests that recognition of several spatially distributed regions of Gα subunits is likely to be a general property of GPCRs. Remarkably, a few receptors virtually ignored the distal C terminus for the purposes of coupling selectivity. This demonstrates that the broad functional diversity of GPCRs extends to the mechanism of receptor-G protein coupling selectivity and provides direct evidence to support the prediction that different receptors will recognize different conserved features of a particular G protein family (5).
Materials and Methods
Materials.
Trypsin, Dulbecco's phospate-buffered saline (DPBS), phosphate-buffered saline (PBS), Hanks' balanced salt solution (HBSS), fetal bovine serum (FBS), Minimal Essential Medium (MEM), Dulbecco's Modified Eagle Medium (DMEM), penicillin/streptomycin, and l-glutamine were from GIBCO (ThermoFisher Scientific). Some receptor ligands, d-luciferin, and forskolin were purchased from Cayman Chemical. The remaining receptor ligands, digitonin, apyrase, protease inhibitor, and GDP were purchased from MilliporeSigma. All detergents were purchased from Anaspec. PEI MAX was purchased from Polysciences Inc.
Plasmids.
Several different GPCR-luciferase constructs were made by appending either the Renilla luciferase variant Rluc8 or NanoLuc (Nluc) directly to the receptor C terminus either by QuikChange PCR or by subcloning into pRluc8-N1 or pNluc-N1 vectors. The V2R-Rluc8 plasmid was received as a gift from Kevin Pfleger (Harry Perkins Institute of Medical Research, Nedlands, WA, Australia). In some cases, a short GGSG linker was inserted between the GPCR and luciferase. Untagged GPCR sequences were obtained either from the cDNA Resource Center, https://www.cdna.org/home.php?cat=0 (Bloomsburg University), as a gift from Jonathan Javitch (Columbia University, New York, NY) or as a gift from Bryan Roth (University of North Carolina, Chapel Hill, NC; PRESTO-Tango Kit: no. 1000000068, Addgene, Watertown, MA). Plasmids encoding Gα subunits were purchased from the cDNA Resource Center. To generate chimeric Gα subunits and Gαs Q227L10delct, we used the PCR, reverse primers incorporating alternative C-terminal sequences, and wild-type Gα templates to amplify full-length Gα subunit sequences that were ligated into pcDNA3.1(+) using KpnI and XhoI. To generate p115RhoGEF-Rluc8, the sequence encoding p115RhoGEF was amplified from p115RhoGEF-GFP and ligated into a Rluc8-N1 vector with BglII and AgeI. All plasmid constructs were verified by Sanger sequencing. A plasmid encoding the S1 subunit of pertussis toxin (PTX-S1) was kindly provided by Stephen R. Ikeda (National Institute on Alcohol Abuse and Alcoholism, Rockville, MD). Plasmids encoding masGRKct-Rluc8, Venus-Kras, Venus-1–155-Gγ2, and Venus-155–239-Gβ1 have been described previously (37, 42). The Glosensor-22F cAMP plasmid (E2301) was obtained from Promega, p115RhoGEF-GFP was received as a gift from Phil Wedegaertner (Thomas Jefferson College, Philadelphia, PA), and the pT7-CalfluxVTN plasmid (43) was a gift from Carl Johnson (Vanderbilt University, Nashville, TN; Addgene plasmid 83926).
Cell Culture and Transfection.
HEK 293 cells (American Type Culture Collection) were propagated in plastic flasks and on six-well plates according to the supplier’s protocol. HEK 293 cells with targeted deletion of GNAS and GNAL (GSKO), HEK 293 cells with additional targeted deletion of GNAS, GNAL, GNAQ, GNA11, GNA12, and GNA13 that are G protein three family knockouts (3GKO), and HEK 293 cells with additional targeted deletions to the 3GKO cells of GNAI1, GNAI2, GNAI3, GNAT1, GNAT2, GNAZ, and GNAO1 that are G protein four family knockouts (4GKO) were derived, authenticated, and propagated as previously described (29, 44). Cells were transiently transfected in growth medium using linear polyethyleneimine MAX (PEI MAX; MW 40,000) at an N/P ratio of 20 and were used for experiments 12–48 h later. Up to 3.0 μg of plasmid DNA was transfected in each well of a six-well plate.
BRET and luminescence assays.
Validation of Gα subunit ability to form heterotrimers: HEK 293 cells were transiently transfected with masGRKct-Rluc8, Venus-1–155-Gγ2, Venus-155–239-Gβ1, pcDNA3.1(+), and a Gα subunit in a (1:1:1:5:0) ratio or a (1:1:1:4:1) ratio or a (1:1:1:0:5) ratio. After a 24-h incubation, cells were washed twice with 1× DPBS, harvested by trituration, and transferred to opaque black 96-well plates.
Measurement of coupling between receptor and G protein in nucleotide-depleted cells.
3GKO or 4GKO cells were transiently transfected with a GPCR-Rluc8 and Gα subunit pair, Venus-1-155-Gγ2, Venus-155-239-Gβ1, and pcDNA3.1(+) or PTX-S1 in a (1:3:1:1:1) ratio. Experiments with Gαi C termini were conducted in 4GKO cells for Gαi cognate receptors and in 3GKO cells for all other receptors. Experiments with Gαi C termini were conducted without PTX-S1, all other Gα subunits were cotransfected with PTX-S1. After a 48-h incubation, cells were washed twice with permeabilization buffer (potassium permeabilization solution; KPS) containing 140 mM KCl, 10 mM NaCl, 1 mM MgCl2, 0.1 mM KEGTA, 20 mM NaHEPES (pH 7.2), harvested by trituration, permeabilized in KPS buffer containing 10 μg·mL−1 high purity digitonin, and transferred to opaque black 96-well plate. Measurements were made from permeabilized cells supplemented either with 0.5 mM GDP or 2 U·mL−1 apyrase, in both cases with or without agonist (SI Appendix, Table S1).
Data processing.
Net BRET was calculated as the raw BRET ratio minus the same ratio measured from cells expressing only the BRET donor. Heatmaps represent ΔBRETmax (Fig. 3; BRET in the presence of agonist and apyrase minus BRET in the presence of GDP alone), ΔBRETGDP (SI Appendix, Fig. S2; BRET in the presence of apyrase alone minus BRET in the presence of GDP alone), and ΔBRETag (SI Appendix, Fig. S3; BRET in the presence of agonist and GDP minus BRET in the presence of GDP alone). Background ΔBRET (presumably due to endogenous Gα subunits remaining in 3GKO and 4GKO cells) measured from control cells not expressing exogenous Gα subunits was routinely subtracted. Control cells expressed PTX-S1 for experiments with non-Gαi C termini. Normalized ΔBRET values were obtained by dividing the ΔBRET observed for each chimera in a heatmap by the highest value observed for each receptor. For the Glosensor assays with Gs, Gsi, Gsq, and Gs12, vehicle-subtracted luminescence changes in response to agonist were normalized to vehicle-subtracted luminescence changes in response to forskolin. For the Glosensor assay with Gi1 the percent change in luminescence was given as agonist-induced change in luminescence over baseline luminescence. For the Calflux assay ΔBRETHA and ΔBRETAch were calculated from the kinetic experiments by subtracting the average of the time points before agonist addition from the average of the final five time points after agonist addition.
Detailed methods related to second messenger assays, BRET and luminescence measurements, Gα subunit immunoprecipitation and structural modeling and molecular dynamics simulations can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Steve Ikeda, Jonathan Javitch, Kevin Pfleger, Philip Wedegaertner, Carl Johnson, and Bryan Roth for providing plasmid DNA used in this study. This work was supported by National Institutes of Health Grants GM130142 (to N.A.L.) and GM117923 (to N.V.) and Ruth L. Kirschstein National Research Service Award Individual Fellowship GM131672 (to N.O.). A.I. was funded by the Advanced Research & Development programs PRIME JP17gm5910013 and LEAP JP17gm0010004 from Japan Agency for Medical Research and Development, and a grant-in-aid for scientific research KAKENHI 17K08264 from the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905993116/-/DCSupplemental.
References
- 1.Pierce K. L., Premont R. T., Lefkowitz R. J., Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3, 639–650 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Simon M. I., Strathmann M. P., Gautam N., Diversity of G proteins in signal transduction. Science 252, 802–808 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Alexander S. P. H., et al. , G protein-coupled receptors. IUPHAR/BPS Guide to PHARMACOLOGY, http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=694. Accessed 13 November 2018.
- 4.Pándy-Szekeres G., et al. , GPCRdb in 2018: Adding GPCR structure models and ligands. Nucleic Acids Res., 46, D440–D446 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flock T., et al. , Selectivity determinants of GPCR-G-protein binding. Nature 545, 317–322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasmussen S. G. F., et al. , Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter B., Tate C. G., Engineering a minimal G protein to facilitate crystallisation of G protein-coupled receptors in their active conformation. Protein Eng. Des. Sel. 29, 583–594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang Y., et al. , Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature 558, 553–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai C.-J., et al. , Crystal structure of rhodopsin in complex with a mini-G o sheds light on the principles of G protein selectivity. Sci. Adv. 4, eaat7052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Draper-Joyce C. J., et al. , Structure of the adenosine-bound human adenosine A1 receptor-Gi complex. Nature 558, 559–563 (2018). [DOI] [PubMed] [Google Scholar]
- 11.García-Nafría J., Nehmé R., Edwards P. C., Tate C. G., Cryo-EM structure of the serotonin 5-HT1B receptor coupled to heterotrimeric Go. Nature 558, 620–623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koehl A., et al. , Structure of the µ-opioid receptor-Gi protein complex. Nature 558, 547–552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan K. A., et al. , Identification of receptor contact site involved in receptor-G protein coupling. Nature 330, 758–760 (1987). [DOI] [PubMed] [Google Scholar]
- 14.West R. E. Jr, Moss J., Vaughan M., Liu T., Liu T. Y., Pertussis toxin-catalyzed ADP-ribosylation of transducin. Cysteine 347 is the ADP-ribose acceptor site. J. Biol. Chem. 260, 14428–14430 (1985). [PubMed] [Google Scholar]
- 15.Simonds W. F., Goldsmith P. K., Codina J., Unson C. G., Spiegel A. M., Gi2 mediates alpha 2-adrenergic inhibition of adenylyl cyclase in platelet membranes: In situ identification with G alpha C-terminal antibodies. Proc. Natl. Acad. Sci. U.S.A. 86, 7809–7813 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamm H. E., et al. , Site of G protein binding to rhodopsin mapped with synthetic peptides from the alpha subunit. Science 241, 832–835 (1988). [DOI] [PubMed] [Google Scholar]
- 17.Conklin B. R., Farfel Z., Lustig K. D., Julius D., Bourne H. R., Substitution of three amino acids switches receptor specificity of Gq α to that of Gi α. Nature 363, 274–276 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Conklin B. R., et al. , Carboxyl-terminal mutations of Gq alpha and Gs alpha that alter the fidelity of receptor activation. Mol. Pharmacol. 50, 885–890 (1996). [PubMed] [Google Scholar]
- 19.Liu J., Conklin B. R., Blin N., Yun J., Wess J., Identification of a receptor/G-protein contact site critical for signaling specificity and G-protein activation. Proc. Natl. Acad. Sci. U.S.A. 92, 11642–11646 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostenis E., Conklin B. R., Wess J., Molecular basis of receptor/G protein coupling selectivity studied by coexpression of wild type and mutant m2 muscarinic receptors with mutant G α(q) subunits. Biochemistry 36, 1487–1495 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Semack A., Sandhu M., Malik R. U., Vaidehi N., Sivaramakrishnan S., Structural elements in the Gαs and Gαq C termini that mediate selective G protein-coupled receptor (GPCR) signaling. J. Biol. Chem. 291, 17929–17940 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kostenis E., Degtyarev M. Y., Conklin B. R., Wess J., The N-terminal extension of Galphaq is critical for constraining the selectivity of receptor coupling. J. Biol. Chem. 272, 19107–19110 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Grishina G., Berlot C. H., A surface-exposed region of G(salpha) in which substitutions decrease receptor-mediated activation and increase receptor affinity. Mol. Pharmacol. 57, 1081–1092 (2000). [PubMed] [Google Scholar]
- 24.Blahos J., et al. , A novel site on the Galpha -protein that recognizes heptahelical receptors. J. Biol. Chem. 276, 3262–3269 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Lee C. H., Katz A., Simon M. I., Multiple regions of G alpha 16 contribute to the specificity of activation by the C5a receptor. Mol. Pharmacol. 47, 218–223 (1995). [PubMed] [Google Scholar]
- 26.De Lean A., Stadel J. M., Lefkowitz R. J., A ternary complex model explains the agonist-specific binding properties of the adenylate cyclase-coupled beta-adrenergic receptor. J. Biol. Chem. 255, 7108–7117 (1980). [PubMed] [Google Scholar]
- 27.Maguire M. E., Van Arsdale P. M., Gilman A. G., An agonist-specific effect of guanine nucleotides on binding to the beta adrenergic receptor. Mol. Pharmacol. 12, 335–339 (1976). [PubMed] [Google Scholar]
- 28.Galés C., et al. , Real-time monitoring of receptor and G-protein interactions in living cells. Nat. Methods 2, 177–184 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Grundmann M., et al. , Lack of beta-arrestin signaling in the absence of active G proteins. Nat. Commun. 9, 341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao X. J., et al. , The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc. Natl. Acad. Sci. U.S.A. 106, 9501–9506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander S. P., et al. ; CGTP Collaborators , The concise guide to pharmacology 2017/18: G protein-coupled receptors. Br. J. Pharmacol. 174 (suppl. 1), S17–S129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seifert R., Höer A., Offermanns S., Buschauer A., Schunack W., Histamine increases cytosolic Ca2+ in dibutyryl-cAMP-differentiated HL-60 cells via H1 receptors and is an incomplete secretagogue. Mol. Pharmacol. 42, 227–234 (1992). [PubMed] [Google Scholar]
- 33.Mackenzie A. E., Milligan G., The emerging pharmacology and function of GPR35 in the nervous system. Neuropharmacology 113, 661–671 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Mackenzie A. E., et al. , Receptor selectivity between the G proteins Gα12 and Gα13 is defined by a single leucine-to-isoleucine variation. FASEB J. 33, 5005–5017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flock T., et al. , Universal allosteric mechanism for Gα activation by GPCRs. Nature 524, 173–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wess J., Molecular basis of receptor/G-protein-coupling selectivity. Pharmacol. Ther. 80, 231–264 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Hollins B., Kuravi S., Digby G. J., Lambert N. A., The c-terminus of GRK3 indicates rapid dissociation of G protein heterotrimers. Cell. Signal. 21, 1015–1021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao R. P., Ji X., Lakatta E. G., Functional coupling of the beta 2-adrenoceptor to a pertussis toxin-sensitive G protein in cardiac myocytes. Mol. Pharmacol. 47, 322–329 (1995). [PubMed] [Google Scholar]
- 39.Zhu W.-Z., et al. , Dual modulation of cell survival and cell death by beta(2)-adrenergic signaling in adult mouse cardiac myocytes. Proc. Natl. Acad. Sci. U.S.A. 98, 1607–1612 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose A. S., et al. , Position of transmembrane helix 6 determines receptor G protein coupling specificity. J. Am. Chem. Soc. 136, 11244–11247 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Van Eps N., et al. , Gi- and Gs-coupled GPCRs show different modes of G-protein binding. Proc. Natl. Acad. Sci. U.S.A. 115, 2383–2388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lan T.-H., Liu Q., Li C., Wu G., Lambert N. A., Sensitive and high resolution localization and tracking of membrane proteins in live cells with BRET. Traffic 13, 1450–1456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J., et al. , Coupling optogenetic stimulation with NanoLuc-based luminescence (BRET) Ca++ sensing. Nat. Commun. 7, 13268 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stallaert W., et al. , Purinergic receptor transactivation by the β2-adrenergic receptor increases intracellular Ca2+ in nonexcitable cells. Mol. Pharmacol. 91, 533–544 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.