Significance
Sinking particles composed of both organic and inorganic material feed the deep-sea ecosystem and contribute centrally to ocean carbon sequestration. Despite their importance, little is known about the biological composition of sinking particles reaching the deep sea. Time-series analyses of sinking particles unexpectedly revealed bacterial assemblages that were simple and homogeneous over time. Particle-associated eukaryote assemblages, however, were more variable and complex. Several modes of export were observed, including summer inputs from the surface, more stochastic export of surface-derived protists and animals, and contributions from midwater animals and deep-sea bacteria. In summary, sinking particles exhibited temporally variable, heterogeneous biological sources and activities that reflected their important roles in the downward transport and transformation of organic matter in the deep sea.
Keywords: deep sea, marine microbes, particulate organic matter, marine carbon cycle, piezophile
Abstract
Sinking particles are a critical conduit for the export of organic material from surface waters to the deep ocean. Despite their importance in oceanic carbon cycling and export, little is known about the biotic composition, origins, and variability of sinking particles reaching abyssal depths. Here, we analyzed particle-associated nucleic acids captured and preserved in sediment traps at 4,000-m depth in the North Pacific Subtropical Gyre. Over the 9-month time-series, Bacteria dominated both the rRNA-gene and rRNA pools, followed by eukaryotes (protists and animals) and trace amounts of Archaea. Deep-sea piezophile-like Gammaproteobacteria, along with Epsilonproteobacteria, comprised >80% of the bacterial inventory. Protists (mostly Rhizaria, Syndinales, and ciliates) and metazoa (predominantly pelagic mollusks and cnidarians) were the most common sinking particle-associated eukaryotes. Some near-surface water-derived eukaryotes, especially Foraminifera, Radiolaria, and pteropods, varied greatly in their abundance patterns, presumably due to sporadic export events. The dominance of piezophile-like Gammaproteobacteria and Epsilonproteobacteria, along with the prevalence of their nitrogen cycling-associated gene transcripts, suggested a central role for these bacteria in the mineralization and biogeochemical transformation of sinking particulate organic matter in the deep ocean. Our data also reflected several different modes of particle export dynamics, including summer export, more stochastic inputs from the upper water column by protists and pteropods, and contributions from sinking mid- and deep-water organisms. In total, our observations revealed the variable and heterogeneous biological origins and microbial activities of sinking particles that connect their downward transport, transformation, and degradation to deep-sea biogeochemical processes.
Photosynthetic primary production in marine surface waters is delivered to the abyss via sinking particles, fueling food webs, and biogeochemical cycles in the deep ocean (1). Sinking particulate organic matter (POM) is the primary conduit for connecting surface water productivity to the deep sea, with lesser contributions from downward diffusion of dissolved organic carbon and macrozooplankton vertical migration (2). The solar energy-driven “biological pump” also contributes to the export and sequestration of carbon dioxide to the deep sea.
Microbes associated with sinking particles may influence carbon export efficiency by facilitating aggregation/disaggregation activities, POM degradation, and trophic transfer of POM-associated organic carbon and energy (3–6). The taxonomic composition of marine particle-associated microbes has been examined on a variety of different particle types using different methodologies (7–16). Recent studies have focused on near-surface water samples that are derived from single time points, and most have used filter fractionation techniques to capture particle-associated microbes. A few deep-sea studies have examined microbial diversity on filter-fractionated deep-sea particles as well (17–20), that sample both suspended as well as slowly sinking POM. Because filtration methods can be biased by the volume of water filtered (21), also collect suspended particles, and may under-sample larger, more rapidly sinking particles, it remains unclear how well they represent microbial communities on sinking POM in the deep sea. Sediment-trap sampling approaches have the potential to overcome some of these difficulties because they selectively capture sinking particles.
The Hawaii Ocean Time-series Station ALOHA is an open-ocean study site (22° 45′ N, 158° W) where long-term ecosystem variability and associated microbial biogeochemistry have been intensively monitored in the North Pacific Subtropical Gyre for over 30 y (22). One component of this effort has included time-series studies of particle export to the deep sea (at 2,800 m and 4,000 m) for over 25 y (23). In combination with surface primary production and euphotic zone export measurements, these efforts have revealed the variability of organic matter export to the deep sea at Station ALOHA, and its quantitative biogeochemical significance. In particular, strong seasonality in particulate organic carbon (POC) flux to the seafloor was observed with values ranging from 98.6 µmol C⋅m−2⋅d−1 in winter, to 282.7 µmol C⋅m−2⋅d−1 in the summer (23). This approximately threefold increase in in summertime particle flux, referred to as the “summer export pulse” (SEP), is believed to be fueled by blooms associated with symbiotic nitrogen-fixing cyanobacteria-diatom phytoplankton assemblages (23). However, for unknown reasons, in some years (including the 2014 samples reported here) the average summer and winter particle fluxes are not significantly different (23). Beyond these details, and despite the extensive and informative biogeochemical time-series datasets available, little is known about the biological origins and processes occurring on sinking POM reaching abyssal depths at Station ALOHA.
To better characterize the biological composition and microbial dynamics associated with sinking particles at abyssal depths in the North Pacific Subtropical Gyre, we collected and preserved sinking particles in moored sequencing sediment traps deployed at 4,000-m depth (800 m above the seafloor) at Station ALOHA, over a period of 9 mo in 2014. A total of 21 samples taken over sequential 12-d intervals were recovered. After trap recovery, DNA and RNA were extracted from sinking deep-sea particles. Metagenomic, metatranscriptomic, and rRNA amplicon libraries were prepared for all samples to characterize particle-associated taxa, genomes, gene contents, and gene transcripts. Our observations and analyses provide insight into the biological sources, processes, and temporal dynamics associated with sinking POM reaching abyssal depths in the oligotrophic ocean.
Results
Sinking Particle Flux at Station ALOHA in 2014.
Although increased particle export to the deep sea in summer months is typical for Station ALOHA (23), during 2014 particle flux was near the low end of historical values (SI Appendix, Fig. S1). Specifically, from March to November 2014, particulate carbon flux ranged between 104.3 and 219.9 µmol C⋅m−2⋅d−1 and did not exceed 232.2 µmol C⋅m−2⋅d−1, the annual mean between 1992 to 2004 at Station ALOHA (23) (SI Appendix, Fig. S1A). Similarly, particulate nitrogen and particulate phosphorus (PP) fluxes were low compared with previously reported values (23) (SI Appendix, Fig. S1 B and C). A pronounced SEP that is more typical of Station ALOHA was not observed during the 2014 time series.
Biological Composition of Sinking Particles at Abyssal Depths.
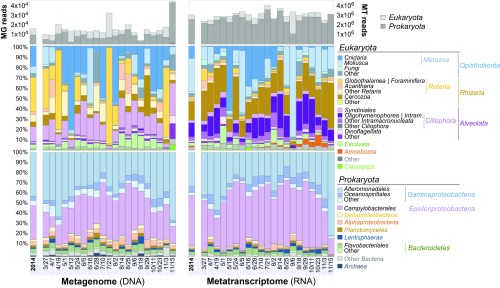
To characterize taxa associated with abyssal sinking particles, we analyzed small subunit ribosomal RNA (SSU rRNA) gene sequences associated with DNA extracted from sinking particles, as well as SSU rRNAs extracted directly from bulk particle RNA using several different approaches (metagenomic, metatranscriptomic, and amplicon libraries). Summed over all 21 samples collected from March to November 2014, the majority of SSU rRNA genes in DNA-based metagenomic libraries, as well as rRNAs from RNA-based metatranscriptomic libraries, were derived from Bacteria and Eukarya, with minimal contributions from Archaea (Datasets S1–S6) (24, 25). The number of annotated SSU rRNA genes recovered from metagenomes for each sample ranged between 4,145 (September 18) and 45,215 (November 15), with an average over the 9-mo period of 16,364 per each 12-d sampling period. Annotated SSU rRNAs from metatranscriptomes ranged in number between 2,366,569 (March 27) and 3,978,938 (July 21), with an average of 3,022,365 per 12-d sampling period over the 9-mo deployment. In metagenomic DNA libraries, 59.4% of the annotated SSU rRNA genes originated from Bacteria, followed by 40% Eukaryote and 0.6% Archaea rRNA genes (Fig. 1 and Table 1). In the RNA-based metatranscriptome libraries, 83.9% of the annotated rRNAs originated from Bacteria, followed by 16% Eukaryote and 0.1% Archaea rRNAs.
Fig. 1.
Time-series abundance profiles of organisms associated with sinking particles in the deep sea. The diversity of eukaryotic (Middle) and prokaryotic (Bottom) SSU rRNA genes from metagenome libraries (Left) and rRNAs from metatranscriptome libraries (Right) are shown. The total number of reads mapping to eukaryotic 18S rRNA genes (light gray) and prokaryotic 16S rRNA genes (dark gray) are displayed on the Top. Each date references the endpoint of one individual 12-d sampling period.
Table 1.
Average taxon representation estimated from SSU rRNA and rRNA gene counts in deep-sea sediment trap metatranscriptomes and metagenomes
| Domain | Taxa | %MG* reads | %MT reads | Rank MG | Rank MT |
| Prokaryota | Gammaproteobacteria | Alteromonadales | 25.04 | 26.94 | 1 | 2 |
| Prokaryota | Epsilonproteobacteria | Campylobacterales | 19.25 | 40.47 | 2 | 1 |
| Eukaryota | Rhizaria | Foraminifera | Globothalamea | 13.24 | 0.35 | 3 | 20 |
| Eukaryota | Opisthonkonta | Metazoa | Cnidaria | 7.92 | 1.92 | 4 | 9 |
| Eukaryota | Alveolata | Dinoflagellata | Syndiniales | 5.55 | 0.41 | 5 | 19 |
| Prokaryota | Misc. Gammaproteobacteria | 2.94 | 2.27 | 6 | 6 |
| Eukaryota | Rhizaria | Misc Radiolaria | 2.75 | 0.18 | 7 | 26 |
| Prokaryota | Gammaproteobacteria | Oceanospirillales | 2.56 | 5.31 | 8 | 3 |
| Eukaryota | Opisthonkonta | Metazoa | Mollusca | 2.54 | 5.14 | 9 | 4 |
| Prokaryota | Alphaproteobacteria | 2.36 | 1.49 | 10 | 11 |
| Prokaryota | Flavobacteriales | 1.94 | 2.22 | 11 | 7 |
| Eukaryota | Rhizaria | Cercozoa | 1.91 | 2.91 | 12 | 5 |
| Prokaryota | Misc. Bacteria | 1.70 | 1.26 | 13 | 12 |
| Eukaryota | Excavata | 1.52 | 0.46 | 14 | 18 |
| Eukaryota | Misc. Opisthokonta | 1.37 | 0.54 | 15 | 17 |
| Prokaryota | Deltaproteobacteria | 1.24 | 0.99 | 16 | 13 |
| Eukaryota | Rhizaria | Radiolaria | Acantharia | 1.13 | 0.18 | 17 | 25 |
| Prokaryota | Planctomycetes | 0.97 | 0.72 | 18 | 14 |
| Prokaryota | Lentisphaerae | 0.77 | 1.53 | 19 | 10 |
| Prokaryota | Archaea | 0.57 | 0.08 | 20 | 29 |
| Eukaryota | Misc. Eukaryota | 0.44 | 0.35 | 21 | 21 |
| Eukaryota | Alveolata | Misc Dinoflagellata | 0.43 | 0.21 | 22 | 24 |
| Prokaryota | Misc. Bacteroidetes | 0.42 | 0.62 | 23 | 15 |
| Eukaryota | Misc. Alveolata | 0.40 | 0.13 | 24 | 27 |
| Eukaryota | Alveolata | Ciliophora | Misc Intramacronucleata | 0.31 | 0.56 | 25 | 16 |
| Eukaryota | Chloroplast | 0.30 | 0.12 | 26 | 28 |
| Eukaryota | Opisthonkonta | Fungi | 0.27 | 0.25 | 27 | 22 |
| Eukaryota | Alveolata | Ciliophora | Intramacronucleata | Oligohymenophorea | 0.11 | 2.16 | 28 | 8 |
| Eukaryota | Misc. Rhizaria | 0.03 | 0.00 | 29 | 31 |
| Eukaryota | Alveolata | Misc. Ciliophora | 0.01 | 0.02 | 30 | 30 |
| Eukaryota | Amoebozoa | 0.01 | 0.23 | 31 | 23 |
Top 10 rankings are indicated in bold. Misc. indicates additional subtaxa that fall within the indicated taxon ranking.
Values indicate percentage taxon representation relative to all annotated taxa, averaged across all time points for each taxon.
Across the 9-mo sampling period, the sinking particle-associated microbial assemblage was dominated in both the metagenome and the metatranscriptome by two main bacterial groups: the gammaproteobacterial order Alteromonadales and the epsilonproteobacterial order Campylobacterales (Fig. 1 and Table 1). Alteromonadales comprised 25% of SSU rRNA genes in DNA and 26.9% SSU rRNAs in RNA. Campylobacterales represented 19.3% and 40.5% of particle-associated SSU rRNA genes and SSU rRNAs, respectively (Table 1). Together, these two bacterial groups represented 45% of all annotated SSU RNA genes in the metagenomes, and 67% SSU rRNAs of all annotated rRNAs in the metatranscriptomes. This trend was consistent within individual samples across the entire 2014 time series in the metagenome, metatranscriptome (Fig. 1), and PCR-based rRNA amplicon libraries (SI Appendix, Fig. S2).
Among bacterial SSU rRNAs and rRNA genes, the Epsilonproteobacteria genus Arcobacter (Campylobacterales) was most highly represented, followed by the Gammaproteobacteria genera Colwellia, Moritella, Shewanella (Alteromonadales), and Neptunomonas (Oceanospirillales) (Table 2). In many of the time-series samples, Colwellia species closely related to known piezophiles were the most abundant Gammaproteobacteria present on sinking POM. In the late November sampling interval, however (November 15, 2014) (Fig. 1), where bacterial SSU rRNA genes were particularly abundant in DNA (95.7%), other Alteromonadales dominated. Specifically, Shewanella and Moritella species closely related to known psychrophilic and piezophilic bacterial isolates were the most abundant (+39.3% and +32.7%, above their 9-mo averages, respectively) (Fig. 1 and Table 2). In the metatranscriptome, bacterial SSU rRNAs exhibited similar trends as were observed in the metagenomic rRNA gene pool, as did the PCR-generated bacterial SSU rRNA amplicon sequences (SI Appendix, Fig. S2). The concordance between metagenomic, metatranscriptomic and amplicon results for bacteria was striking (Fig. 1 and SI Appendix, Fig. S2).
Table 2.
Average Gammaproteobacteria and Epsilonproteobacteria genus representation in deep-sea sediment trap metagenomes and metatranscriptomes estimated from rRNA genes and rRNAs
| Class | Genus | %MG* reads | %MT reads | Rank MG | Rank MT |
| Epsilonbacteria | Arcobacter | 38.260 | 53.538 | 1 | 1 |
| Gammaproteobacteria | Colwellia | 20.121 | 18.850 | 2 | 2 |
| Gammaproteobacteria | Moritella | 15.285 | 8.649 | 3 | 3 |
| Gammaproteobacteria | Shewanella | 9.644 | 1.972 | 4 | 5 |
| Gammaproteobacteria | Neptunomonas | 2.414 | 3.327 | 5 | 4 |
| Gammaproteobacteria | Psychrobium | 1.879 | 1.414 | 6 | 6 |
| Gammaproteobacteria | Pseudoalteromonas | 1.081 | 1.344 | 7 | 7 |
| Gammaproteobacteria | Other | 1.073 | 0.908 | 8 | 9 |
| Gammaproteobacteria | Vibrio | 0.799 | 0.323 | 9 | 17 |
| Gammaproteobacteria | Thalassotalea | 0.669 | 0.589 | 10 | 12 |
| Gammaproteobacteria | Psychromonas | 0.478 | 0.568 | 11 | 14 |
| Gammaproteobacteria | Other Colwelliaceae | 0.467 | 0.635 | 12 | 11 |
| Gammaproteobacteria | Other Pseudoalteromonadaceae | 0.396 | 1.263 | 13 | 8 |
| Gammaproteobacteria | Photobacterium | 0.359 | 0.125 | 14 | 26 |
| Gammaproteobacteria | Other Spongiibacteraceae | 0.319 | 0.230 | 15 | 19 |
| Gammaproteobacteria | Oleispira | 0.310 | 0.586 | 16 | 13 |
| Gammaproteobacteria | Kangiella | 0.299 | 0.543 | 17 | 15 |
| Gammaproteobacteria | Other Vibrionaceae | 0.277 | 0.057 | 18 | 40 |
| Gammaproteobacteria | Other Halieaceae | 0.267 | 0.085 | 19 | 34 |
| Epsilonbacteria | Sulfurovum | 0.253 | 0.274 | 20 | 18 |
| Gammaproteobacteria | Haliea | 0.244 | 0.115 | 21 | 29 |
| Gammaproteobacteria | Pseudomonas | 0.241 | 0.129 | 22 | 23 |
| Gammaproteobacteria | Halioglobus | 0.230 | 0.172 | 23 | 21 |
| Gammaproteobacteria | Halomonas | 0.204 | 0.193 | 24 | 20 |
| Gammaproteobacteria | Oleiphilus | 0.196 | 0.859 | 25 | 10 |
Top 10 rankings are indicated in bold, and only the top 25 rankings are shown.
Values indicate average genus representation as a percent of all Gammaproteobacteria and Epsilonproteobacteria across all time points for each taxon. Genera are ordered according to their abundance in metagenomes.
Unlike the bacteria, which were dominated by just a few major groups in both RNA and DNA pools, particle-associated eukaryotes were more variable in their representation across the time series, within and between the metagenomic, metatranscriptomic and amplicon libraries. In the metagenomic DNA, eukaryotic rRNA genes were most highly represented by the supergroup Rhizaria (47.4%, mostly Foraminifera), followed by the supergroup Opisthokonta (30.1% mostly Cnidaria), and next the supergroup Alveolata (16.9%, mostly Syndiniales) (Fig. 1). In contrast, eukaryotic rRNAs in the RNA pool were most highly represented by Opisthokonta (48.7% mostly Mollusca), followed by Rhizaria (22.5%, mostly Cercozoa) and Alveolates (21.7%, mostly Oligohymenophorea), and lesser amounts from Fungi and other eukaryotic groups (Fig. 1). In the metagenomic DNA, the most highly represented eukaryote groups ranked from third to fifth most abundant of all annotated SSU rRNAs (Table 1) were Foraminifera (class Globothalamea), Cnidaria (mostly siphonophores and Anthomedusae), and Syndiniales comprising 13.2%, 7.9%, and 5.6% of all SSU rRNA genes recovered, respectively (Table 1). In contrast, the most abundant eukaryotic SSU rRNAs in the metatranscriptome were from metazoa (Mollusca, mostly pteropods) followed by the protists (Cercozoa), ranking fourth and fifth, representing 5.3% and 1.9% of all annotated SSU rRNAs, respectively (Table 1). Additionally, Ciliophora (mostly Oligohymenophorea) were highly represented in metatranscriptomic rRNAs, especially relative to their proportions in the metagenomic DNA. Other metazoa present in lesser amounts appeared consistent with pelagic and deep-sea origins included tunicates (mostly appendicularians and salps), polychaetes, and crustacea (ostracods and copepods).
While Bacteria dominated the rRNA pool on deep-sea sinking particles throughout the time series (Fig. 1 and Table 1), shifts in abundance of some eukaryotic groups over the time series were observed during some sampling intervals, but never concomitantly in both DNA and RNA (Fig. 1). Eukaryote SSU rRNA genes dominated the metagenome with either Foraminifera, (especially the Globorotalia genus) or Cnidaria (mostly an unknown genus of Siphonophorae) in a few samples (Fig. 1). Eukaryotic rRNAs in the metatranscriptome were more abundant than those of bacteria in only one sample (September 6, 2014) (Fig. 1). In this sample, mollusks sharing highest rRNA similarity to the pteropod genus Diacria, represented 87.1% of the total rRNA pool.
Eukaryotic SSU rRNA amplicons prepared from RNA-derived cDNA exhibited abundance patterns generally consistent with those found in the metatranscriptome, including consistent and elevated proportions of Cercozoa and Ciliophora (SI Appendix, Fig. S3). In the DNA samples, however, eukaryote SSU rRNA amplicon yields were very low, and groups such as Excavata and Foraminifera were not well represented. The complexity of these samples, along with potential PCR biases and inhibition, in addition to variable SSU rRNA gene copy numbers among different eukaryotic groups, likely accounts for inconsistencies for eukaryotes observed between DNA-based SSU rRNA amplicon results and metagenomic SSU rRNA gene abundances.
Genomic Characteristics of Sinking, Deep-Sea Particle-Associated Bacteria.
The 2014 sediment-trap time-series metagenome assemblies were binned into 16 draft bacterial metagenomic-assembled genomes (MAGs) (SI Appendix, Table S1). No eukaryotic MAGs were recovered, because eukaryote genome sizes, lower coding density, and repetitive sequences precluded their efficient recovery in our samples. The 16 bacterial MAGs were interrelated by their average nucleotide identity (ANI). The bacterial classes recovered in MAGs mirrored dominant bacterial groups identified in SSU rRNA analyses (SI Appendix, Table S1). Gammaproteobacteria MAGs included several Colwelliaceae members (four of eight of the gammaproteobacterial MAGs). Moritellaceae, Shewanellaceae, and Oceanospirillaceae represented the other families of Gammaproteobacteria recovered in MAGs. A total of five MAG bins of Epsilonproteobacteria were recovered, all belonging to the order Campylobacterales. The other bacterial MAG bins were associated with Alphaproteobacteria (Rhodobacteraceae) and Flavobacteriia (Flavobacteriaceae).
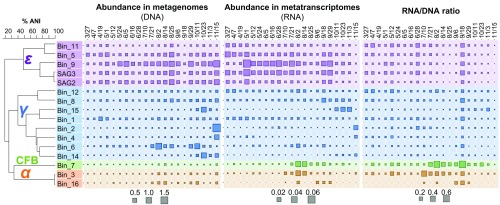
Similar to SSU rRNA analyses, metagenome and metatranscriptome read mapping to Campylobacterales MAGs (bins 5, 9, 11) (SI Appendix, Table S1) indicated that they were abundant and persistent throughout the time-series sampling (Fig. 2). Also similar to rRNA analyses, the representation of Colwelliaceae (bins 1, 6, 14, 15) and Shewanellaceae (bin 2) genomes was more sporadic over the sampling period. Fewer metatranscriptomic reads were recruited for the Gammaproteobacteria than for Epsilonproteobacteria MAGs. MAGs belonging to the Alphaproteobacteria (bins 3, 16) and Flavobacteria (bin 7) exhibited a much lower read coverage in the deep-sea POM DNA, but were relatively more highly represented in the RNA pool, especially in summer months (Fig. 2). Comparisons of the Alphaproteobacteria and Flavobacteria MAG bins to Station ALOHA metagenomic times-series samples (26) suggested that these Flavobacteria were most prevalent in surface waters (SI Appendix, Fig. S4). Although a strong SEP (23) was not observed in our 2014 samples, these data suggest potential export of Alphaproteobacteria and Flavobacteria from surface waters, possibly as epiphytes on sinking phytoplankton or within rapidly sinking fecal pellets.
Fig. 2.
Time-resolved abundance profiles of bacterial genomes associated with sinking particles in the deep-sea. Relative read coverage of MAG bins in the metagenome libraries (Left) and metatranscriptome libraries (Center), and their ratios (Right) are shown. The MAG bins are colored coded according to their taxonomic affiliation and ANI clustering ordinates (far Left). α, Alphaproteobacteria (orange); CFB, Bacteroidetes (green); ε, Epsilonproteobacteria (purple); γ, Gammaproteobacteria (blue).
Transcriptional Characteristics of Sinking, Deep-Sea Particle-Associated Bacteria.
Consistent with rRNA results, ∼70% of all annotated Gammaproteobacteria transcripts from the metatranscriptome samples shared highest similarity to reference genes from psychophilic or piezophilic deep-sea bacterial isolates (including Moritella sp. PE36, Shewanella benthica, Colwellia piezophila, Colwellia sp. TT2012, and Colwellia sp. MT41). Among the Epsilonproteobacteria, 80% of all annotated transcripts shared highest similarity to Arcobacter species, also consistent with taxonomic information derived from rRNA analyses.
Functional annotations of transcripts were determined by a homology search against the eggNOG database (27), and divided into four broad functional classifications: (i) Information Storage and Processing, (ii) Cellular Processing and Signaling, (iii) Metabolism, and (iv) Poorly Characterized. Of those reads mapping to orthologous groups involved in metabolism, 38% were associated with energy production and conversion. Nine of 10 of these most abundant transcripts were most similar Epsilonproteobacteria homologs, and fell into two general categories: short-chain organic acid metabolism and nitrate reduction.
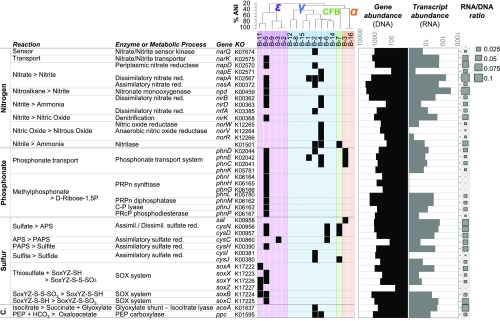
To further characterize bacterial genes and transcripts in the sinking POM, we examined functional gene categories that appeared abundant in the metatrascriptome pool in bacterial draft metagenome-assembled genomes (MAGs). MAG bin 5 affiliated with the epsilonproteobacterium genus Arcobacter (SI Appendix, Table S1), and MAG bins 4 and 2, affiliated with Gammaproteobacteria of the Alteromonadales (Moritellaceae and Shewanella, respectively), encoded genes for a complete or near complete glycolysis or pentose phosphate pathway, as well as genes of the TCA cycle. None of the MAGs contained key enzymes involved in CO2 fixation, such as RuBisCO or ATP-isocitrate lyase (reductive TCA) or carbon-monoxide dehydrogenase (reductive acetyl-CoA pathway), albeit these represent only partial draft genomes. However, Moritellaceae and Shewanella MAGs did encode the ppc gene for PEP-carboxylase, involved in the anapluerotic fixation of carbon dioxide and the depletion of phosphoenolpyruvate (PEP) in the TCA cycle (Fig. 3). The Shewanella MAG also contained the glyoxylate shunt-associated aceA gene for isocitrate lyase (Fig. 3). Its relatively high abundance in the POM DNA and RNA suggests that fatty acid or other simple carbon compound utilization may be important for sinking-particle–associated Alteromonadales.
Fig. 3.
Abundance profiles of functional genes from bacterial genomes associated with sinking particles in the deep sea. Metabolic pathway reconstructions in MAGs for nitrogen, phosphonate, sulfur metabolisms, and carbon fixation. Key genes and enzymes are identified by their KEGG Ortholog number, gene name, and Enzyme Commission (EC) number (when applicable); presence in a MAG is displayed in black. Gene (Left) and transcript (Right) abundances are displayed in the barchart, and ratio between them indicated by the bubble plot to the right. MAG bins are colored according their taxonomic affiliation and ANI clustering ordinates (Top). α, Alphaproteobacteria (orange); CFB, Bacteroidetes (green); ε, Epsilonproteobacteria (purple); γ, Gammaproteobacteria (blue). C., carbon; red., reduction.
Deep sediment trap Arcobacter, Moritellaceae, and Shewanella MAGs all encoded gene suites involved in nitrate reduction (nap and nir genes) (Fig. 3). Transcripts for these genes were among the most abundant in the metatranscriptome, resulting in a concomitant high ratio of transcripts to genes (Fig. 3). The Arcobacter MAG encoded genes for nitrate transport (narK), as well as genes involved in reduction of nitrate, nitrite, and nitroalkanes (napDA, nasA, nirBD, nitronate monooxygenase). This combination of genes was not found within any single Alteromonadales MAG, and neither was nitronate monooxygenase. Some Alteromonadales MAGs did encode the gene for nitrilase (for converting nitrile to ammonium). The nirK gene associated with nitrite reduction to nitrous oxide was highly represented in the metagenome with high levels of transcripts in the RNA pool. Although no norBC genes were identified, the norRVW genes that can replace them in anoxic conditions were found in an Alteromonas MAG. The absence of some of these genes may be due to the incomplete nature of MAGs, because some genes missing in MAGs were detected in aggregate metatranscriptome reads (e.g., nirS, nor, nosZ). In total, the data suggest that both assimilatory and respiratory dissimilatory nitrate reduction to nitrite, nitric oxide, nitrous oxide, and ammonia may occur on sinking POM-associated low-oxygen microhabitats.
Genes involved in assimilatory sulfate reduction (cys genes) from sulfate to sulfite (but not sulfide) were present in Arcobacter, as well as a complete system for thiosulfate oxidation (the SOX system) (Fig. 3). The genes cysND and cysC (converting sulfate to adenosine 5′-phosphosulfate, APS to PAPS, respectively), and cysIJ (sulfite to sulfide) were also present in a few of the Alteromonadales MAGs, but none encoded the SOX system. One Arcobacter MAG also encoded a complete phosphonate utilization pathway (phn genes) (Fig. 3), suggesting that organic phosphate sources may be utilized in the mineralization of deep-sea particles.
Discussion
Describing the nature and variability of sinking POM is important for characterizing deep-sea ecosystem dynamics and biogeochemistry, as well as for understanding the role of the biological pump in carbon export and sequestration. A number of previous studies have focused on the characterization of suspended particle-associated microbes using serial filtration techniques, but there are few direct observations of microbes associated with sinking particles reaching the seafloor. Although extensive and informative biogeochemical time-series datasets are available, surprisingly little is known about the biological nature and variability of sinking POM that reaches the seafloor in the oligotrophic ocean. To address these challenges, we combined time-resolved, deep-sea sediment-trap collections with nucleic acid preservation and analytical techniques, to better assess sinking POM-associated biological origins and dynamics across all three domains of life.
A relatively simple community of bacteria dominated both the RNA and DNA fractions recovered from sinking particles collected at 4,000 m, with a more complex and variable assemblage of eukaryotes being the next most abundant. Among eukaryotes were diverse metazoa (most notably cnidaria and mollusks, with smaller contributions from tunicates, polychaetes, crustacea, and other groups) and protists (Alveolates, most notably dinoflagellates, syndinids, and ciliates; and Rhizaria, most notably foraminiferans, radiolarians and cercozoans). Archaeal rRNAs were scarce on sinking particles at 4,000 m, representing only 0.6% of the total rRNAs detected on average.
A surprisingly low bacterial diversity, dominated by Epsilonproteobacteria (Arcobacter and Sulfurovum) and Gammaproteobacteria (Colwellia, Moritella, and Shewanella) (Table 2) was confirmed in all datasets (metagenome, metatranscriptome, and amplicon libraries) that was not anticipated from previous studies. Surveys of free-living, deep-sea planktonic bacteria collected at Station ALOHA and elsewhere have typically found oligotrophic bacterial groups, like SAR11, SAR202, SAR406, SAR324, as well as copiotrophic genera like Alteromonas, Pseudoalteromomas, Halomonas, and Alcanivorax (17, 28, 29). The sinking POM microbes captured by the sediment traps reported here also appear very different from previous reports of particle-associated microbes collected in the water column using filter fractionation techniques (19, 20, 28). In particular, one report (20) recently claimed that the most abundant prokaryotes on sinking particles in the deep ocean are those that are also present in surface waters. Our data suggest a different scenario, and we postulate the lack of congruence between our observations and prior reports (20) is due mainly to fundamental differences between the sediment trap samples reported here, versus previously reported filtered water column samples. On balance, some of the less abundant bacterial taxa we observed on sinking POM were consistent with previous bathy- and abyssopelagic studies, where >3.0-μm fractions were enriched in Bdellovibrio, Planctomyces, Firmicutes, and Bacteroidetes (19, 28). It seems clear, however, that sediment traps sample very different populations compared with seawater filter fractionation techniques, and that trap-collected material is likely more relevant when considering export processes associated with sinking POM and its associated microbial community dynamics and activities.
The deep-sea heterotrophic bacteria we found on the sinking particles varied little over the time series, and their high transcript representation suggested their persistence and activity on deep-sea sinking particles. The most abundant bacterial rRNA genes in particle-associated DNA also corresponded to those most abundant in the metatranscriptome, indicating that dominant Epsilonproteobacteria and Gammaproteobacteria were active in situ at deep-sea temperatures and pressures. Our results are consistent with a previous study that documented piezophilic nutrient responses on deep-sea sinking particles incubated at in situ pressures and temperatures (30). The prevalence of piezophile-like Gammaproteobacteria on sinking particles supports an important role for them in the degradation and turnover of sinking organic matter at abyssal depths. The combined activities of these heterotrophic bacteria, along with protistan assemblages that we observed, likely facilitate in situ degradation and turnover of both surface-derived phytoplankton detritus (23), as well as dead metazoan-tissues that comprised much of the deep-sea sinking POM. Additionally, given the well-known ability of piezophilic bacteria to biosynthesize essential polyunsaturated fatty acids, their abundance and activity on deep-sea particles supports their hypothesized role as important in situ sources of essential nutrients in the deep sea (31–33).
Considering their consistently large overall contribution to the RNA transcript pool, our evidence suggests that Epsilonproteobacteria were active on sinking particles at the in situ temperatures and pressures of the deep sea. Arcobacter species are often found as eukaryotic symbionts, epibionts, or pathogens (34–38), and are commonly observed in association with marine invertebrates, including sponges, polychaetes, cnidaria, molluscs, ascidians, and vestimentiferans. Arcobacter species can also form syntrophic symbioses with protists driven by interspecies hydrogen transfer (38). Deep-sea particle-associated Epsilonproteobacteria potentially were symbiotically associated with the protists (ciliates, rhizaria) or animals (cnidaria, mollusks) also present in the sinking POM. Indeed, previous reports have noted the presence of Arcobacter-like Epsilonproteobacteria in 500-m sediment traps (but not shallower), and those trap-associated Arcobacter encoded genes known to be induced in epibionts by their protistan hosts (12, 38, 39).
Bacterial transcripts associated with aerobic respiration via the TCA cycle and anaerobic nitrogen cycling by Epsilonproteobacteria were abundant on sinking POM bacterial assemblages. Transcripts of genes associated with denitrification, sulfur oxidation, and dissimilatory nitrate reduction to ammonium (DNRA) pathways from both Epsilonproteobacteria and Gammaproteobacteria were also well represented. Denitrification and DNRA are anaerobic processes, yet it seems unlikely that rates of aerobic respiration on sinking particles would be sufficient to counter act O2 diffusion, because the water column at Station ALOHA is comparatively well oxygenated from the surface to the seafloor (2). There are several possible explanations for the observed high levels of gene representation and transcript expression of putative anaerobic respiratory genes. For example, in coastal marine sediments denitrification has been shown to occur simultaneously with aerobic respiration in well-oxygenated habitats (40). Another possibility is that the expression of anaerobic respiratory genes on deep-sinking particles reflects the symbiotic association of facultatively anaerobic Gamma- and Epsilonproteobacteria, with midwater or deep-sea animals (or protists), where oxygen-limiting microhabitats might occur in body cavities, tissues, or intracellularly. Indeed, nitrogen cycling dynamics in invertebrate–bacterial symbiotic systems studied under in situ deep-sea conditions has been shown to involve DNRA, which supplies reduced nitrogen nutrients to both symbiotic partners (36, 37).
Eukaryotes comprised a considerable fraction of the DNA and RNA in sinking particles collected and preserved at 4,000 m, but taxonomic representation between the two datasets was markedly different. The variable representation in DNA and RNA among different eukaryotic groups was likely due to several different factors, including their origins and activity in situ at 4,000 m, relative time since death, or relative sinking rates. Rhizaria, particularly Foraminifera and Radiolaria, were abundant in the DNA, but much less so in the RNA, suggesting they represent nonliving biomass sinking from surface waters above. Along with other recent studies (41–43), our observations support the hypothesis that Rhizaria (Foraminifera and Radiolaria) are important contributors to organic matter flux at abyssal depths. Additionally, pteropods from the upper water column, along with mid- and deep-water cnidaria, also appeared to contribute significantly to sinking POM fluxes. Taken together, our results imply that in addition to fast sinking POM, considerable trophic “reworking” of phytoplankton-derived POM is likely mediated by midwater and the abyssal animals and their associated deep-sea bacterial and protistan microbiomes.
Connectivity between the surface and deep ocean is mediated by several different and variable export modalities in space and time. Previous studies at Station ALOHA (23) have documented an SEP that occurs in late summer, but not in every year, for unknown reasons. The SEP at Station ALOHA results in elevated flux of carbon, nitrogen, phosphorus, and biogenic silica to the deep sea that are associated with blooms of diatoms harboring symbiotic nitrogen-fixing cyanobacteria. The observations we report here appear to be capturing several different modes of POM export at Station ALOHA, including: (i) sporadic inputs of surface-water–derived Rhizaria, in particular foraminifera and radiolaria; (ii) sporadic inputs of animal biomass presumably derived from the upper- and midwater column, especially from pelagic molluscs and pteropods; (iii) colonization of sinking particles and mineralization by piezophilic Gammaproteobacteria, Epsilonproteobacteria, and ciliates; and (iv) summer export, as indicated by elevated levels of surface-water–derived bacterial groups (e.g., Flavobacteria and Alphaproteobacteria). Although a large SEP event was not observed in our 2014 samples (SI Appendix, Fig. S1) (23), more extended time-series sampling may provide deeper insight into heterogeneous export modalities and their variability that mediate POM flux to the deep sea.
Conclusions
The precise biological composition of sinking particles in the deep sea is largely unknown. This study provides relevant information on the biological composition and microbial activities of deep-sea sinking POM. The bacterial assemblages on sinking particles captured in the abyss were dominated by deep-sea Epsilonproteobacteria and piezophile-like Gammaproteobacteria present either as gut microbiota, epibionts, or endosymbionts of animals and protists (living or dead) associated with sinking particles. Several different protistan groups were abundant components of the eukaryotic assemblage, and likely acted as active grazers or parasites within other particle-associated organisms. Others, like surface-water–derived foraminifera and radiolarians, apparently arrive at the seafloor in large numbers more sporadically on fast-sinking particles. Animal biomass, derived in particular from siphonophores [presumably originating from mid- and deep-waters (44)], and pteropods [from the upper mesopelagic (45, 46)], were abundant in the deep-sea sinking POM. This highlights the important role of sinking pelagic animal biomass in fueling bacterial, protistan, and fungal heterotrophic activities in the deep sea. While POM-associated bacterial assemblages were generally homogeneous in their taxonomic representation and presumptive activities over our 9-mo study, sinking eukaryotic components were more variable, reflecting their different and more variable surface, midwater, and deep-sea origins. In total, these data are consistent with hypotheses proposing that a considerable amount of sinking POM degradation may be due to its consumption by mid- and deep-water animals, and their associated deep-sea adapted microbiota (3).
Materials and Methods
Sample Collection.
For each trap, a total of 21 samples were collected, each capturing 12 d of accumulated sediment. In the first trap, sinking particles were collected in buffered formalin brine for analyses of particulate carbon, nitrogen, and phosphorus according to the Hawaii Ocean Time-series, as described previously (23). In a second trap, sinking particles were collected in a nucleic acid preservative solution, as previously described (12), using the same sampling times as formalin-fixed traps. Particulate material was collected by centrifugation, and subsequently stored at −80 °C (see SI Appendix text for further details on sample processing and analyses).
RNA and DNA Extraction and Purification.
DNA extractions and purifications were performed using the MoBio PowerBiofilm DNA Isolation kit (MoBio #2400050) following the manufacturer’s protocol. RNA extractions were performed as previously described (47–50). A secondary RNA column clean-up step was performed using the MoBio PowerClean Pro RNA Clean-Up Kit (MoBio #1399750) after initial mirVana (ThermoFisher Scientific #AM1560) RNA purification.
Small Subunit rRNA Amplicon Sequencing and Analyses.
Bacterial and archaeal rRNA amplicon libraries were prepared by PCR amplification of the V4 hypervariable region of the SSU rRNA using barcoded 515F (5′GTGYCAGCMGCCGCGGTAA3′) and 806R (5′GGACTACNVGGGTWTCTAAT3′) primers, as previously described (51), with minor modifications (52, 53). Sequences were then demultiplexed using QIIME2 (51). Error modeling and correction was performed using DADA2 (51, 54). Finally, taxonomy was assigned using the SILVA v123 database (55).
Eukaryote rRNAs were amplified and sequenced using previously described methods (56). For RNA samples extracted total RNA was reverse transcribed into cDNA (Bio-Rad iScript). The full protocol used to create eukaryotic SSU rRNA metabarcoding libraries can be found online at protocols.io: dx.doi.org/10.17504/protocols.io.hdmb246. Amplicon sequence libraries were created using PCR primers for the V4 hypervariable region of the 18S rRNA gene from the DNA and RNA extracts. The PCR primers were: forward (5′-CCAGCASCYGCGGTAATTCC-3′) and reverse (5′-ACTTTCGTTCTTGATYRA-3′), targeting the V4 hypervariable region (57).
Metagenomic and Metatranscriptomic Library Preparation.
Metagenomic libraries were prepared using an automated NeoPrep instrument with TruSeq Nano DNA library preparation kit (Illumina #NP1011001), using an input of 25 ng of sheared genomic DNA. Libraries were sequenced either using a 150-bp paired-end NextSeq500/550 High Output v2 reagent kit (Illumina #FC4042004).
Metatranscriptomic libraries were prepared with 0.5–50 ng of total RNA using the ScriptSeq v2 RNA-Seq kit (Illumina #SSV21124). Unique single-plex barcodes were annealed onto cDNA fragments during the PCR enrichment for Illumina sequencing primers with 12 cycles following the manufacturer’s guidelines. Libraries were normalized to 4-nM final DNA concentration, pooled in equal volumes, and sequenced using an Illumina NextSeq 500 system with a V2 high output 300 cycle reagent kit. The PhiX quality control (Illumina) reagent was added to an estimated final contribution of 5% of the total estimated sequence density.
DNA and RNA Sequence Analyses and Workflow.
The metatranscriptomic paired-end reads were prepared for analysis with two quality-control workflows resulting in 257 million high-quality metatranscriptomic and 1.05 billion cleaned metagenomic sequences. The taxonomic affiliations of the SSU rRNA sequences from the metatranscriptomic and metagenomic libraries, as well as SSU rRNA amplicon sequences, were assigned by sequence identities to the databases using a 97% similarity threshold. SSU rRNA sequences were compared with the SILVA SSU NR99 database Release 123_1 (55) and the Protist Ribosomal Reference (PR2) database release 4.5 (58).
Protein-coding genes and resulting proteins were predicted in cleaned sequences not containing rRNA using Prodigal 2.60 (parameters: -p meta -c) (parameters: -p meta -c) (59). Protein sequences were taxonomically and functionally annotated using homology search by LAST 756 (parameters: -b 1 -x 15 -y 7 -z 25) (60) against the RefSeq database release 75 (61), and functionally annotated by profile search by HMMER 3.1b1 (http://hmmer.org/) against eggNOG database release 4.5 (27), respectively.
Contig Assessment and Quality Control of MAGs.
For MAG assemblies, the quality of reads from the 21 metagenomes were filtered using two passes of BBDuk software as implemented in BBTools 36.32 (https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/) to remove Illumina adapters, known Illumina artifacts, phiX, and reads with extreme GC values (parameters for first pass: “ktrim=r k=23 mink=11 hdist=1 tbo tpe tbo tpe”, second pass: “k=27 hdist=1 qtrim=rl trimq=17 cardinality=t mingc=0.05 maxgc=0.95”).
Contigs were taxonomically assigned by predicting genes with Prodigal 2.60 (parameters: -p meta -c) (59) and comparing predicted genes to RefSeq database release 79 using LAST 756 (parameters: -F 15 -b 1 -x 15 -y 7 -z 25) (60). The 2,733 assemblies identified as bacterial were imported into Anvi’o 2.1.0 (61, 62). In parallel, the taxonomic affiliation and the completion of MAGs were also assessed by a multiple marker genes approach using CheckM (63). Based on the length, number of predicted genes, and ANI clustering, 16 bins were selected for further analysis.
MAG Distributions and Key-Functional Genes.
Genes and corresponding proteins from MAGs were predicted using Prodigal v.2.6.3 (parameters: -p meta). The abundance and number of transcripts for each MAG gene was retrieved from that of the closest relative in the gene catalog as identified by homology search. The abundance and transcript representation of MAG genes were assessed by mapping quality controlled metagenomic and metatranscriptomic reads against them. MAGs were annotated by protein sequence homology search against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (64) using LAST 756 (60).
Deep-Trap Gene Catalog.
In addition to the MEGAHIT assembly of all samples, cleaned reads from each sample were also assembled with Meta-3.9.0 (parameters: “-meta -k 21,33,55,77,99,127”) (65). A nonredundant gene catalog was built from the 21 individual sample assemblies and the combined megahit assembly.
Supplementary Material
Acknowledgments
We thank Tara Clemente, Blake Watkins, and Eric Grabowski for their expert efforts in sediment trap deployment, recovery, and biogeochemical analysis; the Captain and crew of the R/V Kilo Moana for able assistance at sea; Lisa Y. Mesrop for assistance with eukaryotic rRNA amplicon sequencing and analyses; and Paul Den Uyl for assisting in metagenomic library preparation and sequencing. This research was supported by a Grant 329108 from the Simons Foundation (to E.F.D., D.M.K., and D.A.C.), and Gordon and Betty Moore Foundation GBMF 3777 (to E.F.D.) and GBMF 3794 (to D.M.K.). This work is a contribution of the Simons Collaboration on Ocean Processes and Ecology and the Daniel K. Inouye Center for Microbial Oceanography: Research and Education.
Footnotes
The authors declare no conflict of interest.
Data deposition: The metagenome and metatranscriptome and prokaryote small subunit ribosomal RNA amplicon sequence datasets reported in this study have been deposited in the NCBI Sequence Read Archive (project no. PRJNA482655); the eukaryotic small subunit ribosomal RNA amplicon sequences have been deposited in the NCBI Sequence Read Archive (project no. PRJNA393049).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903080116/-/DCSupplemental.
References
- 1.Volk T., Hoffert M. I., “Ocean carbon pumps: analysis of relative strengths and efficiencies in ocean-driven atmospheric CO2 changes” in The Carbon Cycle and Atmospheric CO2: Natural Variations Archean to Present, Sundquist E., Broecker W., Eds. (Geophysical Monograph Series, American Geophysical Union, Washington, DC, 1985), pp. 99–110. [Google Scholar]
- 2.Karl D. M., Church M. J., Ecosystem structure and dynamics in the North Pacific Subtropical Gyre: New views of an old ocean. Ecosystems 20, 433–457 (2017). [Google Scholar]
- 3.Karl D. M., Knauer G. A., Martin J. H., Downward flux of particulate organic matter in the ocean: A particle decomposition paradox. Nature 332, 438–441 (1988). [Google Scholar]
- 4.Smith D. C., Simon M., Alldredge A. L., Azam F., Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359, 139–142 (1992). [Google Scholar]
- 5.Steinberg D. K., et al. , Bacterial vs. zooplankton control of sinking particle flux in the ocean’s twilight zone. Limnol. Oceanogr. 53, 1327–1338 (2008). [Google Scholar]
- 6.Grabowski E., Letelier R. M., Laws E. A., Karl D. M., Coupling carbon and energy fluxes in the North Pacific Subtropical Gyre. Nat. Commun. 10, 1895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong E. F., Franks D. G., Alldredge A. L., Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38, 924–934 (1993). [Google Scholar]
- 8.Bidle K. D., Fletcher M., Comparison of free-living and particle-associated bacterial communities in the chesapeake bay by stable low-molecular-weight RNA analysis. Appl. Environ. Microbiol. 61, 944–952 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon M., Grossart H.-P., Schweitzer B., Ploug H., Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28, 175–211 (2002). [Google Scholar]
- 10.Amacher J., Neuer S., Anderson I., Massana R., Molecular approach to determine contributions of the protist community to particle flux. Deep Sea Res. Part I 56, 2206–2215 (2009). [Google Scholar]
- 11.Ganesh S., Parris D. J., DeLong E. F., Stewart F. J., Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J. 8, 187–211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontanez K. M., Eppley J. M., Samo T. J., Karl D. M., DeLong E. F., Microbial community structure and function on sinking particles in the North Pacific Subtropical Gyre. Front. Microbiol. 6, 469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rieck A., Herlemann D. P., Jürgens K., Grossart H. P., Particle-associated differ from free-living bacteria in surface waters of the Baltic Sea. Front. Microbiol. 6, 1297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.López-Pérez M., Kimes N. E., Haro-Moreno J. M., Rodriguez-Valera F., Not all particles are equal: The selective enrichment of particle-associated bacteria from the Mediterranean Sea. Front. Microbiol. 7, 996 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farnelid H., et al. , Diverse diazotrophs are present on sinking particles in the North Pacific Subtropical Gyre. ISME J. 13, 170–182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez-Rodriguez A., et al. , High contribution of Rhizaria (Radiolaria) to vertical export in the California Current Ecosystem revealed by DNA metabarcoding. ISME J. 13, 964–976 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salazar G., et al. , Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J. 10, 596–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bochdansky A. B., Clouse M. A., Herndl G. J., Eukaryotic microbes, principally fungi and labyrinthulomycetes, dominate biomass on bathypelagic marine snow. ISME J. 11, 362–373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peoples L. M., et al. , Vertically distinct microbial communities in the Mariana and Kermadec trenches. PLoS One 13, e0195102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mestre M., et al. , Sinking particles promote vertical connectivity in the ocean microbiome. Proc. Natl. Acad. Sci. U.S.A. 115, E6799–E6807 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padilla C. C., et al. , Standard filtration practices may significantly distort planktonic microbial diversity estimates. Front. Microbiol. 6, 547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karl D. M., Lukas R., The Hawaii Ocean Time-series (HOT) program: Background, rationale and field implementation. Deep Sea Res. Part II 43, 129–156 (1996). [Google Scholar]
- 23.Karl D. M., Church M. J., Dore J. E., Letelier R. M., Mahaffey C., Predictable and efficient carbon sequestration in the North Pacific Ocean supported by symbiotic nitrogen fixation. Proc. Natl. Acad. Sci. U.S.A. 109, 1842–1849 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boeuf D., et al. , Data from “Biological composition and microbial dynamics of sinking particulate organic matter at abyssal depths in the oligotrophic open ocean”. NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA482655. Deposited 31 January 2019. [DOI] [PMC free article] [PubMed]
- 25.Boeuf D., et al. , Data from “Biological composition and microbial dynamics of sinking particulate organic matter at abyssal depths in the oligotrophic open ocean”. NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA393049. Deposited 4 July 2017. [DOI] [PMC free article] [PubMed]
- 26.Mende D. R., et al. , Environmental drivers of a microbial genomic transition zone in the ocean’s interior. Nat. Microbiol. 2, 1367–1373 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Huerta-Cepas J., et al. , eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu R., et al. , Depth-resolved distribution of particle-attached and free-living bacterial communities in the water column of the New Britain Trench. Front. Microbiol. 9, 625 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pham V. D., Konstantinidis K. T., Palden T., DeLong E. F., Phylogenetic analyses of ribosomal DNA-containing bacterioplankton genome fragments from a 4000 m vertical profile in the North Pacific Subtropical Gyre. Environ. Microbiol. 10, 2313–2330 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Deming J. W., Colwell R. R., Observations of barophilic microbial activity in samples of sediment and intercepted particulates from the demerara abyssal plain. Appl. Environ. Microbiol. 50, 1002–1006 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delong E. F., Yayanos A. A., Biochemical function and ecological significance of novel bacterial lipids in deep-sea procaryotes. Appl. Environ. Microbiol. 51, 730–737 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shulse C. N., Allen E. E., Diversity and distribution of microbial long-chain fatty acid biosynthetic genes in the marine environment. Environ. Microbiol. 13, 684–695 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Shulse C. N., Allen E. E., Widespread occurrence of secondary lipid biosynthesis potential in microbial lineages. PLoS One 6, e20146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller W. G., et al. , The complete genome sequence and analysis of the epsilonproteobacterium Arcobacter butzleri. PLoS One 2, e1358 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fera M. T., et al. , Detection of Arcobacter spp. in the coastal environment of the Mediterranean Sea. Appl. Environ. Microbiol. 70, 1271–1276 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girguis P. R., et al. , Fate of nitrate acquired by the tubeworm Riftia pachyptila. Appl. Environ. Microbiol. 66, 2783–2790 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders J. G., Beinart R. A., Stewart F. J., Delong E. F., Girguis P. R., Metatranscriptomics reveal differences in in situ energy and nitrogen metabolism among hydrothermal vent snail symbionts. ISME J. 7, 1556–1567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamann E., et al. , Environmental Breviatea harbour mutualistic Arcobacter epibionts. Nature 534, 254–258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelve E. A., Fontanez K. M., DeLong E. F., Bacterial succession on sinking particles in the ocean’s interior. Front. Microbiol. 8, 2269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchant H. K., et al. , Denitrifying community in coastal sediments performs aerobic and anaerobic respiration simultaneously. ISME J. 11, 1799–1812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guidi L., et al. ; Tara Oceans coordinators , Plankton networks driving carbon export in the oligotrophic ocean. Nature 532, 465–470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biard T., et al. , In situ imaging reveals the biomass of giant protists in the global ocean. Nature 532, 504–507 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Biard T., et al. , Biogeography and diversity of Collodaria (Radiolaria) in the global ocean. ISME J. 11, 1331–1344 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mapstone G. M., Global diversity and review of Siphonophorae (Cnidaria: Hydrozoa). PLoS One 9, e87737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Betzer P. R., et al. , The oceanic carbonate system: A reassessment of biogenic controls. Science 226, 1074–1077 (1984). [DOI] [PubMed] [Google Scholar]
- 46.Burridge A. K., et al. , Diversity and abundance of pteropods and heteropods along a latitudinal gradient across the Atlantic Ocean. Prog. Oceanogr. 158, 213–223 (2017). [Google Scholar]
- 47.Ottesen E. A., et al. , Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc. Natl. Acad. Sci. U.S.A. 110, E488–E497 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ottesen E. A., et al. , Ocean microbes. Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science 345, 207–212 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Aylward F. O., et al. , Microbial community transcriptional networks are conserved in three domains at ocean basin scales. Proc. Natl. Acad. Sci. U.S.A. 112, 5443–5448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aylward F. O., et al. , Diel cycling and long-term persistence of viruses in the ocean’s euphotic zone. Proc. Natl. Acad. Sci. U.S.A. 114, 11446–11451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caporaso J. G., et al. , QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Apprill A., McNally S., Parsons R., Weber L., Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137 (2015). [Google Scholar]
- 53.Parada A. E., Needham D. M., Fuhrman J. A., Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Callahan B. J., et al. , DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quast C., et al. , The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu S. K., et al. , Estimating protistan diversity using high-throughput sequencing. J. Eukaryot. Microbiol. 62, 688–693 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Stoeck T., et al. , Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19 (suppl. 1), 21–31 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Guillou L., et al. , The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyatt D., et al. , Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kiełbasa S. M., Wan R., Sato K., Horton P., Frith M. C., Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487–493 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Leary N. A., et al. , Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eren A. M., et al. , Anvi’o: An advanced analysis and visualization platform for ‘omics data. PeerJ 3, e1319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K., KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bankevich A., et al. , SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.