Significance
IgM antibodies protect mammals against humoral microbial infection and mediate clearance of cellular debris. IgM activates the immune complement system only after binding to cell-surface antigens. Here we report the in situ 3D structures of surface-antigen–bound IgM antibodies in complex with both C1 and C4b complement components. The data indicate the structural arrangement of pentameric and hexameric IgM upon antigen binding, exposing the C1q-binding sites with both adopting hexagonal symmetry. The structures reveal the entire C1qr2s2 complex and elucidate several protein–protein interactions with C4b and IgM. Based on the structural data, we hypothesize a C1q-transmitted surface trigger that activates C1, leading to C4 cleavage and C4b deposition on membranes.
Keywords: complement, IgM, C1, cryoelectron tomography, subtomogram averaging
Abstract
Antigen binding by serum Ig-M (IgM) protects against microbial infections and helps to prevent autoimmunity, but causes life-threatening diseases when mistargeted. How antigen-bound IgM activates complement-immune responses remains unclear. We present cryoelectron tomography structures of IgM, C1, and C4b complexes formed on antigen-bearing lipid membranes by normal human serum at 4 °C. The IgM-C1-C4b complexes revealed C4b product release as the temperature-limiting step in complement activation. Both IgM hexamers and pentamers adopted hexagonal, dome-shaped structures with Fab pairs, dimerized by hinge domains, bound to surface antigens that support a platform of Fc regions. C1 binds IgM through widely spread C1q-collagen helices, with C1r proteases pointing outward and C1s bending downward and interacting with surface-attached C4b, which further interacts with the adjacent IgM-Fab2 and globular C1q-recognition unit. Based on these data, we present mechanistic models for antibody-mediated, C1q-transmitted activation of C1 and for C4b deposition, while further conformational rearrangements are required to form C3 convertases.
Secreted IgM complexes in human serum occur as (disulfide-linked) pentamers and hexamers of monomeric antibody complexes (SI Appendix, Fig. S1A). They are the first antibodies to recognize antigens on newly invading microbes (1–3). Weak, but multivalent, binding affinities concur with polyreactivity of naturally occurring IgM antibodies that recognize various danger and damage signals on microbes and host cells (1). Natural IgM-induced complement opsonization of apoptotic host cells yields clearance by phagocytosis, without causing inflammatory responses (4, 5). Deficiency in either IgM or the early complement components C1, C4, or C2 causes failure in silent clearance and is strongly linked to autoimmune diseases, such as systemic lupus erythematosus (6, 7). However, complement activation by IgM autoantibodies against, for example, gangliosides on peripheral nerves causes neuropathies and Guillain-Barré syndrome (8). Although IgM antibodies and IgM-mediated complement activation play important roles in immune responses and tissue homeostasis, the underlying molecular processes of IgM immune-complex formation and complement activation have remained largely unclear. Hexameric or pentameric IgM complexes adopt large stellate structures in solution that undergo a marked conformational change into a staple-like arrangement upon binding to surface-exposed antigens (9, 10). While fivefold and sixfold structural models of soluble IgM pentamers and hexamers have been proposed (11–13), the overall organization of domains in intact IgM pentamers and hexamers, and their transitions from solution to complement-activating surface-bound states, have remained elusive.
C1 consists of a multimeric C1q-recognition molecule and four associated proteases C1r2s2 (14) (SI Appendix, Fig. S1B). C1q is a hexamer of the heterotrimeric chains A, B, and C, forming a bundle of six (heterotrimeric) collagen helices with six (heterotrimeric) globular C1q (gC1q) recognition units at the C termini and an N-terminal stalk of 18 peptide chains with disulfide cross-links between A-B and C-C chains, respectively, within and between collagen helices (15). C1r and C1s are homologous serine proteases, each consisting of six domains (SI Appendix, Fig. S1C). The N-terminal (CUB1-EGF-CUB2) domains of C1r and C1s bind C1q-lysine residues through Ca2+-binding sites in the CUB1 and CUB2 domains of C1r and CUB1 of C1s (16, 17), yielding a heterotetrameric C1r2s2 CUB1-EGF-CUB2 platform positioned between the C1q-collagen helices (17–19). Binding of C1q to surface-bound immune complexes activates proenzymes C1r and C1s that are associated with C1q. Subsequently, activated C1 proteases cleave the complement component C4 into C4b (SI Appendix, Fig. S1D), which propagates the classical pathway of complement (14) (SI Appendix, Fig. S1E). Previous studies revealed that IgG antibodies oligomerize on surfaces to activate complement through forming Fc platforms that present C1q-binding sites separated by 9–18 nm (20–22). However, disorder in C1-IgG1 complexes limited the resolution of previous EM studies, hindering complete modeling.
Results
Imaging IgM-C1 Complexes on Liposomes.
We generated a synthetic peptide mimotope of CD52 (23) conjugated to cholesterol and formed liposomes with 1-mol% CD52-cholesterol haptens, encapsulating self-quenching sulforhodamine B to allow spectroscopic measurement of membrane-attack-complex pore formation through release of the fluorophore (24). In the presence of monoclonal rat anti-CD52 IgM (25) and normal human serum (NHS), rapid lysis of antigenic liposomes was observed (SI Appendix, Fig. S2A). No lysis was detected over long time periods (up to 1 h) at 4 °C (26), while complement activity was rapidly reconstituted after raising the temperature of the sample to 21 °C (SI Appendix, Fig. S2B).
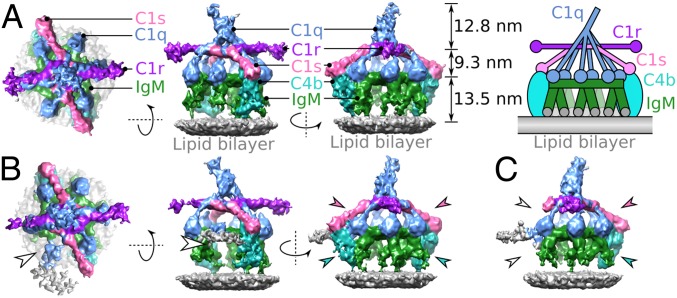
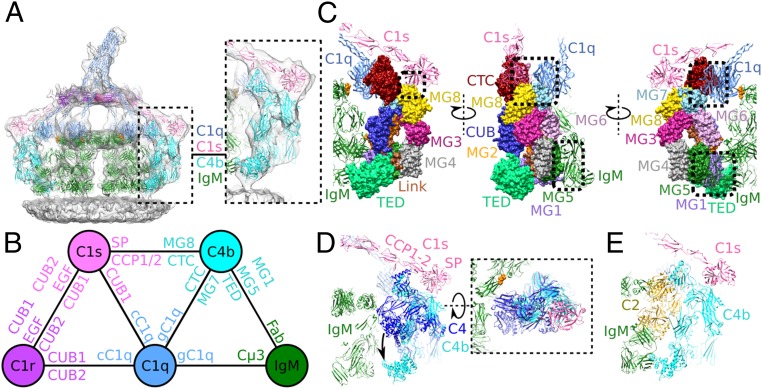
Next, we used phase-plate cryoelectron tomography to image CD52-mimotope–bearing liposomes in the presence of anti-CD52 IgM and NHS, as a source of (nonpurified) complement proteins. Cryotomograms of samples kept for 20 min at 4 °C before vitrification showed clear C1 binding (SI Appendix, Fig. S2C), while cryotomograms of samples subsequently warmed to 21 °C for 20 min showed C1 binding, extensive opsonization, and pore formation (SI Appendix, Fig. S2D). Subvolumes of IgM-C1 complexes were selected from 54 phase-plate tomograms, collected from samples incubated at 4 °C. Averaging and classification of 1,522 particles yielded separate classes for pentameric and hexameric IgM bound to C1 at ∼26-Å resolution (Fig. 1 and SI Appendix, Fig. S3). Surface-bound IgM pentamers and hexamers bind C1 at 4 °C in a far more structurally homogeneous manner than observed for C1-IgG hexamers (22). Density was present for IgM, C1q, and the four C1r and C1 proteases and, to our surprise, for C4b bound to IgM-C1 (Fig. 1).
Fig. 1.
IgM-C1-C4b complexes imaged by phase-plate cryoelectron tomography. (A) Top and two side views of the hexameric IgM-C1-C4b2 density map. Shown are assemblies containing C1q (blue), two molecules of C1r (purple), two molecules of C1s (pink), hexameric IgM (green), and two molecules of C4b (cyan) formed on liposome bilayers (gray). (Right) Relative heights of components are indicated along with a simplified schematic of the complex. (B) Density map of the pentameric IgM-C1-C4b2 complex indicating two molecules of C4b (cyan arrowheads) and two C1s arms (pink arrowheads). Weak density is observed for the C1q leg above empty space and noisy density of pentameric IgM (white arrowhead). (C) Density map of the pentameric IgM-C1-C4b1 complex, showing a single molecule of C4b (cyan arrowhead) and the associated C1s CCP1/2-SP domains (pink arrowhead). On the opposite side disordered noisy density (gray) and increased flexibility of C1s due to the absent C4b molecule are apparent as missing density (white arrowheads).
Surface-Bound IgM Pentamers and Hexamers.
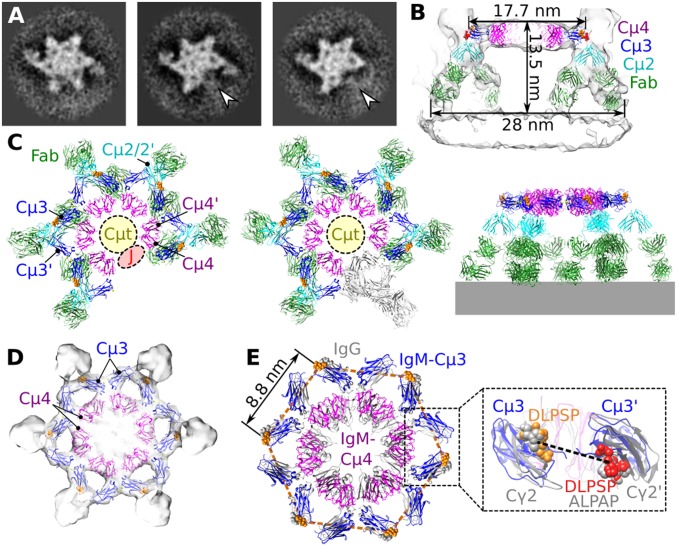
Class-averaged densities revealed that IgM pentamers and hexamers, bound to surface antigens and in complex with C1, adopt isomorphic, C6-symmetric dome-shape structures of ∼13.5-nm height and ∼28-nm width (Fig. 2 A–C). The IgM pentamer thus is structurally equivalent to an IgM hexamer, with its five protomers separated by ∼60° and one replaced by a J chain (Fig. 2C).
Fig. 2.
Pentameric and hexameric IgM complexes form dome-shape structures. (A) Tomographic slices through the Fc platforms of hexameric IgM-C1-C4b2 (Left), pentameric IgM-C1-C4b2 (Middle), and pentameric IgM-C1-C4b1 (Right) showing 60° spacing of protomers. The site of the missing IgM protomer in the pentameric Fc platforms is indicated (white arrowheads). (B) Side view of the hexameric IgM complex fitted into the density, showing binding of both Fab2 arms (green) to the antigenic surface, dimeric Cµ2/2′-hinge domains (cyan), and the Fc (Cµ3–4/3′-4′) platform (Cµ3 and Cµ4 domains are colored blue and purple, respectively). DLPSP residues (432–436) are colored orange (top face) and red (bottom face). (C) Models of pentameric IgM (Left) and hexameric IgM from the side (Middle) and top (Right). The region for the 18 Cµt peptide extensions is indicated by yellow circles, the positions of the DLPSP residues mediating C1q binding are shown in orange, and the putative site for the J-chain is shown as a red ellipse in the pentameric complex. A single protomer in hexameric IgM is colored gray. (D) Fc platform comprising Cµ3 and Cµ4 domains fitted into the C6-symmetrised density map. (E) Top view of the hexameric IgM model (colored) overlaying the hexameric IgG Fc platform (gray). The distance between adjacent DLPSP motifs (orange) is 8.8 nm (orange hexagon). (Inset) A side view of a Fc region indicating the 31-Å Cα distance between Pro434-Pro434’ (dashed line) in IgM (blue) with DLPSP (orange and red, as described above) and hexamerized IgG1 (gray) aligned to the homologous ALPAP residues (gray spheres).
Sixfold averaging of the top platform of the hexameric dome-shaped structure yielded a map at 20-Å resolution (Fig. 2D and SI Appendix, Fig. S3). Independent fitting of two monomeric Cµ3 domains and one dimeric Cµ4/4′ unit (12) yielded a ring of Fc units, similar to that of oligomerized IgG1 molecules (20, 22) (Fig. 2 D and E). The GlcNAc2Man8-glycosylated C-terminal µ-chain tail (Cµt) extensions (27) accounted for the central density of ∼40-nm3 volume (Fig. 2C). The observed arrangement agrees with cross-linking of Cµ3 Cys414 residues between neighboring Fc units (28) and between Cys575 residues of the Cµt extensions, needed for IgM oligomerization (29) (SI Appendix, Fig. S4A).
Below the IgM-Fc platform the density bent down toward the membrane (Fig. 2B). Density directly below the Fc platform fitted crystal structures of a Cµ2/2′ dimer (12) at angles of ∼100° with respect to the Fc platform (Fig. 2 B and C). Below the Cµ2/2′-hinge domains, the densities extended into pairs of Fab arms forming an inner and outer ring of surface-bound Fabs. Fitting Fab structures (30) into the density indicated that all (10 or 12) antigen-binding regions are bound to surface antigens (Fig. 2B and SI Appendix, Fig. S4B). The lower resolution of the Fab densities close to the membrane perhaps indicates increasing flexibility with increasing distance from the Cµ2 hinge region. Compared with sixfold symmetric positions, the Fab2 arms adjacent to C4b densities were shifted up to ∼2.5 nm, indicating steric rearrangements upon C4 binding or C4b production (SI Appendix, Fig. S4C).
Multivalent surface binding by IgM induces conformational changes that expose the critical DLPSP (Asp-Leu-Pro-Ser-Pro) residues 432–436 for C1 binding (31) (SI Appendix, Fig. S5). In the observed IgM-C1 complexes, these residues in the Cµ3 domains were positioned on the periphery of the Fc platform spaced 8.8–17.5 nm apart (Fig. 2E), similar to the homologous ALPAP (Ala-Leu-Pro-Ala-Pro) residues of IgG1, which were 8.9–17.9 nm apart in the IgG1-C1 complexes (22). Furthermore, the IgM-Fc regions displayed “open” configurations as seen in IgG1-C1 complexes (22), as characterized by the 31-Å distances between Pro434 residues in Cµ3 and Cµ3′ of one Fc (Fig. 2E), yielding alternating upward and downward-oriented DLPSP peptides on Fc-Cµ3 and Cµ3′ domains, respectively.
C1q Binding to IgM.
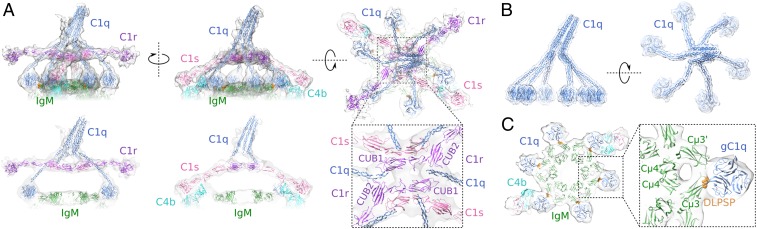
The ∼27-Å resolution density map of C1 in complex with hexameric IgM revealed density for all components of the C1 (C1qr2s2) complex (Fig. 3A and SI Appendix, Fig. S6 A–F). The density was consistent with six C1q-collagen helices that extend down, with a right-handed supercoil, from a tilted stalk to gC1q modules that are bound to the hexameric IgM-Fc platform (Fig. 3B), and with four C1r and C1s proteases that create a platform between the C1q-collagen helices with four protruding arms, two of which point straight out parallel to the membrane and two of which bend down toward the densities for C4b (Figs. 1A and 3A). Densities at ∼26-Å resolution of pentameric IgM-C1 complexes revealed very similar C1 structures, with five C1q legs bound to the IgM-Fc platform and a sixth leg hovering above the missing element (Fig. 1 B and C).
Fig. 3.
C1 binding to hexameric IgM platforms. (A) Model of C1qr2s2 in density (truncated below the IgM Fc platform) bound to hexameric IgM showing the orientations of C1r and C1s. (Below) Central slices through the model. The Inset displays the CUB1 and CUB2 domains of the two antiparallel C1r molecules binding to C1q-collagen helices. (B) C1q has six collagen legs spiraling down to hexagonally arranged globular head domains. (C) Binding of heterotrimeric gC1q units to DLPSP residues (orange) of the FG loop of Fc Cµ3 (SI Appendix, Fig. S6).
C1 caps the top of the dome-shape structures by gC1q binding to all (five or six) exposed DLPSP sites of the IgM-Fc platforms (Fig. 3C). Even though the Fc platforms of both pentameric and hexameric IgM and hexamerized IgG1 (22) have comparable widths (17.4 and 17.9 nm, respectively), the C1q-collagen helices are spread markedly wider apart in IgM-C1 complexes than in IgG1-C1, with their collagen ends separated by 24 nm instead of 19 nm (SI Appendix, Fig. S6 G and H). A kink in the collagen-gC1q connection allows edge-on positioning of the gC1q modules, enabling residues ArgB150 and PheB178 of the C1q (22) to interact with the DLPSP-containing FG loop of Fc Cµ3, and ArgB108 with the BC loop of Cµ3′ (32) (SI Appendix, Fig. S6I). Overall, the wider structure of C1q brings the platform of C1r-C1s protease in C1-IgM ∼1.5 nm closer to the antibody-Fc platform than in C1-IgG1.
IgM-Mediated C1 Activation.
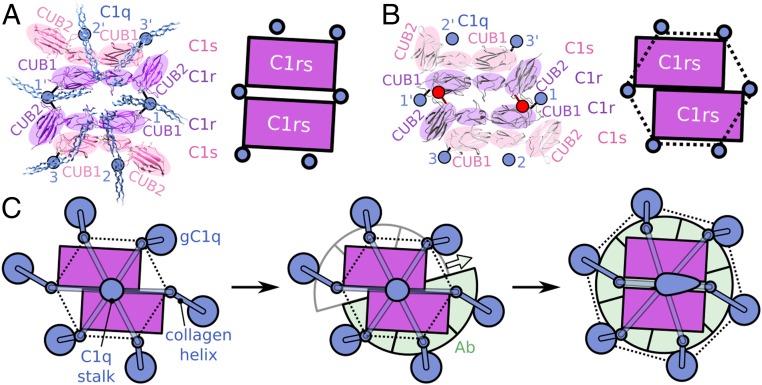
Density of the protease platform and its four protruding arms accounted for all six (CUB1-EGF-CUB2-CCP1-CCP2-SP) domains of two C1r and two C1s molecules (Fig. 3A). The C1r2s2-protease platform between the C1q-collagen arms is formed by two antiparallel dimers, each consisting of antiparallel arranged N-terminal domains of C1r and C1s, analogous to previous models (17–19). The density revealed the two outermost molecules bending down toward the C4b density (Fig. 3A). Since it is C1s that binds and cleaves C4, this implies a C1s-C1r-C1r-C1s stacking of the CUB1-EGF-CUB2 domains, which is consistent with the heterotetrameric arrangement of C1r and C1s CUB1-EGF-CUB2 fragments found in the asymmetric unit of a recent crystal structure (33). Remodeling of C1r2s2 CUB1-EGF-CUB1 in the density of C1-IgG1 (22) improved the fit and thus provides additional evidence for the model (SI Appendix, Fig. S7A). In these models of IgM-C1 and IgG1-C1, each stable C1rs dimer binds three collagen helices through their binding sites on C1r CUB1 and CUB2 and C1s CUB1 (Fig. 4A), while the two C1r molecules in the middle bind the same two central C1q-collagen helices, resulting in a head-to-tail C1r CUB1-EGF-CUB2 arrangement and antiparallel binding of the two C1rs heterodimers.
Fig. 4.
Comparing C1r2s2 in the IgM-C1-C4b complex and the isolated crystal structure suggests a mechanism for C1 activation. (A) Interactions of C1q-collagen helices with C1r and C1s observed in IgM-C1; structures of CUB1-EGF-CUB2 domains in C1r and C1s with positions of C1q indicated by blue circles with specific binding to collagen-binding sites indicated by connecting black lines. Schematic shows the C1rs cores with positions of collagen arms. (B) Crystal structure (Protein Data Bank code 6F1C) (33) and schematic representations of C1rs heterodimers with projected collagen locations added (blue circles), as deduced from C1-IgM; the two obscured collagen positions are indicated by red circles and lines. (C) Schematic representation of C1 conformational rearrangement upon binding to antibody-Fc platforms; with C1q (blue), C1rs heterodimers (purple), and antibodies (green). Empty sites for antibody binding are indicated in gray; C1q positions at the height of the proteases by small blue circles; gC1q headpieces by large blue circles, connecting C1q collagen helices by blue bars; and upright and tilted C1q stalks as a blue circle or ovoid. Dotted lines highlight the hexagonal arrangements. The sliding motion of two halves of the complex is indicated by the colored arrow. See also Movie S1.
C1r-C1r interactions are mediated by two central C1q-collagen helices in IgM-C1 and IgG1-C1 (Figs. 3A and 4A), whereas in the C1r2s2 CUB1-EGF-CUB2 crystal structure (33) the domains of the two C1r molecules interact directly with each other (Fig. 4B). The two observed arrangements differ predominantly by a single sliding motion of the C1rs heterodimers, with stable cores consisting of CUB1-EGF-CUB2 of C1r and CUB1-EGF of C1s. Each C1rs heterodimer is associated with three collagen helices that are invariant with respect to the translation and remain accessible, while the “collagen-bridging” C1r-CUB2 sites are obstructed for collagen binding in the C1r2s2 CUB1-EGF-CUB2 crystal structure (Fig. 4 A and B). Moreover, projection of collagen helices onto the crystal structure suggests an apparent hexagonal configuration for C1q binding (Fig. 4B). In contrast, in IgM-C1 and IgG1-C1 (22) we observed a skewed configuration of C1q collagens bound to C1r and C1s (Fig. 4A), while in these complexes gC1q units were bound to sixfold symmetric Fc platforms. Considering the stiffness of collagens (34), a single sliding motion of two halves of the C1 complex may convert nonskewed and skewed hexagonal arrangements of C1r2s2-bound collagen and gC1q units, respectively, in unbound C1 to vice versa arrangements in antibody-bound C1 (Fig. 4C and Movie S1).
C4b Bound to IgM-C1 Complexes.
The maps revealed additional densities adjacent to the IgM-Fab arms that corresponded to C4b (35) with the thioester-containing domain (TED) positioned next to the inner Fab and its thioester moiety oriented toward the membrane, consistent with covalent attachment to the surface (Fig. 5A and SI Appendix, Fig. S1D). Particle classification resulted in two populations of structures of pentameric IgM-C1, with either one or two C4b molecules present, and only one for hexameric IgM-C1 with two C4b molecules bound (Fig. 1). In all three cases, the C1q stalk was tilted toward the side of the single-bound C4b or toward the C4b with the strongest density in classes where two C4b complexes were present (SI Appendix, Fig. S8). There was no density for the C1s CCP1-CCP2-SP arm above the C4b-absent site (Fig. 1C and SI Appendix, Fig. S8), indicating that the C1s arm by itself is flexible without the C4b-C1s interactions, consistent with the disorder observed in the C1-IgG1 complex (22).
Fig. 5.
C4b binding to IgM-C1 complexes. (A) The full model in density of the hexameric IgM-C1-C4b2 complex, colored as in Fig. 1. (Inset) The C4b density fit. (B) Protein interaction map showing the domains mediating binding between the protein complexes. (C) Interactions of C4b (surface model) with IgM, C1q, and C1s (ribbon models) are indicated by dashed boxes. C4b colored as in SI Appendix, Fig. S1. (D) Hypothetical IgM-C1-C4 model. The TED domain is structurally unhindered and free to rearrange upon cleavage to C4b. (E) Hypothetical IgM-C1-C4bC2 model showing steric clashes with both the Fc and Fab regions of IgM.
Modeling C4b (35) into the density maps indicated that C4b interacts with multiple components of C1 and IgM (Fig. 5 B and C). Specific electrostatic interactions needed for C1s-C4 binding (SI Appendix, Fig. S9A) (36) were also shown to be between CCP1-CCP2 and the C-terminal C345c (CTC) domain of C4 in the structure of the C1s homolog MASP2 CCP1-CCP2-SP complexed with C4 (37, 38) but were not present between C1r and C4b (SI Appendix, Fig. S9B). These are possibly maintained and supported by additional contacts between C1s SP and macroglobulin (MG) domain 8 of C4b (Fig. 5C). Furthermore, gC1q contacts the helices on top of the C4b CTC, C4b-core domains MG1 and MG5 contact the adjacent outer IgM-Fab arm, while the newly formed, negatively charged (35), α′-chain N-terminal region (bound onto MG7) exhibits potential electrostatic interactions with gC1q (Fig. 5C). At the other side, C4b TED contacts the variable region of the inner Fab arm. Interactions of C4b with both the inner and outer arms of the adjacent Fab2 are likely responsible for the induced structural changes that widen the Fab-Fab distance from 6 to 8 nm and distort the sixfold symmetry of IgM (SI Appendix, Fig. S4C).
Discussion
Activation of the complement cascade at 4 °C leads to the formation of IgM-C1-C4b complexes on the membrane surface, as observed by cryo-EM tomography of samples containing IgM, human serum, and antigen-bearing liposomes (Fig. 1 and SI Appendix, Fig. S2). Density reconstruction up to ∼26–27 Å resolution of liposome-associated IgM complexes revealed that IgM pentamers and hexamers adopt dome-shaped structures on the membrane (Fig. 2), which are highly reminiscent of images of the previously described staple conformation (9). The IgM dome-shaped structures are formed by (five or six) antibody protomers creating a platform of Fc regions on top, comprising Cµ3-Cµ4/Cµ3′-Cµ4′ Fc domains, and Cµt extensions filling the central hole. The Fc platforms are supported by dimeric Cµ2/2′ domains and Fab2 regions that yield an inner and outer ring of bound Fabs with their antigen-binding regions oriented toward the membrane, consistent with binding the short CD52 peptide mimotopes (Fig. 2). The pair-wise surface binding of IgM-Fab2 arms differs from that of surface-bound structures of IgG1-C1, where only one arm of each Fab pair binds surface antigens (20, 22) (SI Appendix, Fig. S4D). Most likely, the dimeric Cµ2/Cµ2′ domains in IgM restrain the two connected Fab arms to adopt correlated orientations, whereas independently flexible hinge regions in IgG1 leave the orientations of the two Fab arms largely unrestrained, yielding the observed difference in surface binding. Analogous to oligomerization of surface-bound IgG, gradual surface binding of adjacent Fab2 arms bends the hinges preceding the Fc segments orienting connected Cµ2/Cµ2′-Fab2 toward the membrane. This stellate to dome-shaped conformational change upon membrane binding exposes the Fc-Cµ3 FG loops that contain the DLPSP residues critical for C1 binding (31) on the periphery of the IgM-Fc platform (SI Appendix, Fig. S5).
Both IgM hexamers and pentamers bind and activate C1 through adopting (respectively, complete and incomplete) hexagonal arrangements, irrespective of the substitution of one antibody protomer by a J chain (Fig. 1). In solution, however, IgM pentamers do not adopt hexagonal symmetry (39), implying that C1 binding directs IgM pentamers to adopt hexagonal symmetry. The C1-binding and activating Fc platforms generated by surface-bound IgM pentamers and hexamers are remarkably similar to those generated by surface-bound and oligomerized IgG1 molecules. Both expose C1q-binding sites on the periphery of the hexagonal platforms at 8.8- to 17.5-nm distances in IgM (Fig. 2E) and 8.9–17.9 nm in hexamerized IgG1 (22). However, C1 binds IgM through spreading its C1q helices far more widely than when binding IgG1 complexes, with angles between the adjacent collagen helices of ∼98° instead of ∼70°, respectively (SI Appendix, Fig. S6G). As a consequence, the IgM-C1q bound C1r2s2 protease platform is lowered by ∼1.5 nm with respect to the membrane surface, which compensates partially for the increase in height of ∼2.5 nm due to the presence of Cµ2/Cµ2′ domains in IgM (SI Appendix, Fig. S6 G and H). Overall, pentameric and hexameric IgM form highly similar complement-activating complexes on membranes that appear structurally more homogeneous and ordered than surface-bound and hexamerized IgG1-C1 complexes.
The densities for C1r and C1s proteases in the IgM-C1-C4b complexes reveal that the heterotetramer of C1r and C1s binds C1q in an antiparallel stacked arrangement with C1r molecules on the inside and C1s on the outside, thus reversing previous C1r-C1s-C1s-C1r stacking models into C1s-C1r-C1r-C1s (17, 18). In IgM-C1-C4b, densities of the two “outside” arms are oriented down to interact with C4b (Fig. 3A). Since C1s, and not C1r, cleaves C4 to produce C4b, we conclude that C1r proteases are positioned on the inside and C1s on the outside, yielding an antiparallel stacking of C1s-C1r-C1r-C1s. This same stacking arrangement was also observed in a recent crystal structure (33) and is confirmed by an improved fit after remodeling of C1-IgG1 density from single-particle data (SI Appendix, Fig. S7) (22). In the new C1s-C1r-C1r-C1s model, the collagen-binding sites in the CUB1 and CUB2 domains of the two central antiparallel C1r molecules bind the two C1q-collagen helices in the middle (Figs. 3A and 4A). The collagen-binding sites in the CUB1 domains of the two C1s molecules each bind C1q collagen helices positioned on the outside. Variable orientations of C1s CUB2 (19, 22, 33) (SI Appendix, Fig. S7B) reflect the absence of specific collagen-binding interactions with the remaining C1q-collagen helix on either side (17). The absence of collagen binding to C1s CUB2 likely provides the molecular complex an increased flexibility for its C1s CUB2-CCP1-CCP2-SP arms, accounting for diverse arrangements needed in order for C1s to be cleaved by C1r and to reach and, subsequently, cleave its substrates C4 and C4bC2 (SI Appendix, Figs. S9 and S10).
Comparing our model of IgM-C1 with the crystal structure of C1r2s2 CUB1-EGF-CUB2 (33) led us to hypothesize that the configuration observed in the crystal structure may reflect the C1r2s2-C1q arrangement as occurs in (unbound) C1 in solution, yielding a mechanism for surface-triggered activation of C1 upon binding to surfaced-bound IgM and oligomerized IgG complexes (Fig. 4). Projection of the C1q-collagen helices at accessible sites in the crystal-structure configuration suggests a near-hexagonal arrangement of C1q collagen bound to the C1rs protease dimers (Fig. 4B). In contrast, a skewed arrangement of C1q helices is observed when bound to C1r and C1s in the IgM-C1 and IgG1-C1 complexes, while the gC1q units in these complexes are restrained to a hexagonal configuration due to binding the hexagonal Fc platforms (Figs. 3C and 4A). We hypothesize that C1 functions as two halves, each formed by a stable C1rs heterodimer that associates with three adjacent C1q-collagen helices (with the outer two C1q-collagen helices bound to the CUB1 domains of C1r and C1s, while the third collagen helix in the middle is not bound). Binding to hexagonal Fc platforms will induce the gC1q units of the two halves to adopt hexagonal symmetry (Movie S1). Transmission of this gC1q rearrangement through the short, and thereby relatively stiff (34), C1q-collagen helices skews the arrangement of the collagens at the height of the proteases, which consequently induces a shift of C1rs dimers and tilts the C1q stalk along the sliding direction (Fig. 4C and Movie S1). We previously considered compaction of C1q-collagen helices as a trigger for activation (22). The wide spreading of C1q-collagen helices in IgM-C1 excludes such a simple model (SI Appendix, Fig. S6 G and H). In agreement with C1q compaction, however, our current hypothesis specifies a single sliding motion that transforms a skewed arrangement of gC1q into a hexagonal one upon gC1q binding to Fc platforms, which compacts C1q in one direction and induces rearrangement of the C1r2s2 protease platform.
At 4 °C, C4 cleavage by IgM-C1 on liposomes leads to formation of IgM-C1-C4b complexes that block C4 turnover and C4b deposition. Superposing C4 (37) onto C4b in the IgM-C1-C4b structures (Fig. 5D) suggests that binding of C4 initially involves interactions through C4-substrate binding to C1s CCP1-CCP2-SP (37) supported by interactions that latch C4 CTC onto gC1q (Fig. 5 B and C). Cleavage of C4 into C4b induces large structural rearrangements that expose the thioester moiety in C4b TED for covalent attachment to the surface (Fig. 5A). In addition, a conformational switch of the C4b CTC-anchor region (residues 1,579–1,593) from α-helix to β-hairpin [as previously observed in structures of C3 and C4 (35, 37, 40)] allows the MG1–6 core of C4b to align with the outer Fab arm. The final structure of IgM-C1-C4b is in agreement with covalent attachment of C4b to the membrane and IgM-C1 binding of C4b through C4b interactions with gC1q, CCP2-SP of C1s, and both Fab arms (Fig. 5 B and C). Displacement of the Fab arms by C4b (SI Appendix, Fig. S4C), yielding the final IgM-C1-C4b structures, implies mobility or flexibility of the epitopes, which is facilitated by membrane fluidity, but may be restricted in the case of antigens on solid surfaces.
Cleavage of C4 by IgM-C1 leads to C4b opsonization and, next, formation of C3 convertases leading to opsonization by C3b. The state captured here, IgM-C1-C4b at 4 °C, indicates C4b product release as a temperature-limited step in the complement cascade. Moreover, docking of proenzyme C2 onto C4b in IgM-C1-C4b [based on the homologous structure of C3bB in complex with its protease factor D (41) (Fig. 5E)] results in steric clashes, indicating that formation of the C4b-C2 proconvertase complex also requires conformational changes that apparently do not occur at 4 °C. The observation of stable IgM-C1-C4b complexes at 4 °C suggests the presence of (weak) interactions that facilitate, on the one hand, C4b release and C4 turnover yielding C4b deposition, and, on the other hand, the retention of C4b close to C1 for C2 binding and cleavage of C4bC2 by the resident C1s. Previous cryo-EM tomograms of IgG-C1 complexes, formed on liposomes at ambient temperature when using C2-depleted sera, showed potential individual occurrences of related IgG-C1-C4b complexes (42) (SI Appendix, Fig. S9C). Notably, the height difference between surface-bound IgM and hexamerized IgG1 is in part compensated by the height difference due to the two different modes of C1 binding to IgM and IgG1 hexamers (SI Appendix, Fig. S6H). Evidently, higher temperatures allow conformational changes to occur and expose C4b for C2 binding, thus generating C4bC2 complexes. Based on the homologous C3bB in complex with its protease factor D (41), a reorientation of C1s SP-catalytic domain would be needed after C4 cleavage to subsequently cleave C4bC2 and generate a C4b2a complex or C3 convertase (SI Appendix, Fig. S9D).
In conclusion, we used a mimic of the short CD52 antigen and a cognate CDC-inducing monoclonal IgM antibody to obtain stable and homogeneous complexes of IgM-C1-C4b formed at 4 °C on liposomal membranes. The resulting structures of IgM pentamers and hexamers in complex with C1 and C4b reflect geometries of antibodies bound close to lipid bilayers for binding, activation, and activity of C1, producing C4b that is deposited directly onto the targeted surface. However, C1 reacts in vivo upon binding to a range of variable patterns presented on diverse surfaces, either by direct C1 binding or via antibodies or other mediators of inflammation (see, e.g., ref. 43). These insights enable design of further experiments to determine effects of important parameters, such as epitope location and orientation, antigen density and diffusion of antigens, and type of mediator (44–46). Moreover, diverse conditions encountered in vivo may influence the packing and stability of C1-C4b complexes, which in turn may affect the relative rates of C4b deposition and C3 convertase (C4b2a) formation. How and whether these effects contribute to distinguish self from nonself, and how complement processes host cells versus microbes, remains to be established.
Materials and Methods
Synthesis of CD52 and Complement Activation Fluorescence Assay.
A CD52 mimotope with the amino acid sequence TSSPSAD was synthesized by solid-phase peptide synthesis and used to form antigenic liposomes as described in SI Appendix. Complement activity was assessed as described in SI Appendix.
Cryoelectron Tomography and Subtomogram Averaging.
Samples were prepared for cryo-EM and imaged as described in SI Appendix. Tomograms were reconstructed (SI Appendix) before particles of IgM-C1 complexes were picked, aligned, and classified using multireference alignment as described in SI Appendix.
Model Building.
Models were constructed as described in SI Appendix. The structures used for the final models are summarized in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
We thank Julio Ortiz and the NeCEN, (Leiden, The Netherlands) for help with data collection and Stuart Howes for help with Dynamo. This research was supported by European Research Council Grant 759517 (to T.H.S.); The Netherlands Organization for Scientific Research Grants 722.015.006 (to A.L.B.), 714.013.002 (to P.G. and A.J.K.), and 024.002.009 (to P.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.R.B. is a guest editor invited by the Editorial Board.
Data deposition: Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank, www.ebi.ac.uk/pdbe/emdb [accession codes: EMD-4878 (Hexameric-IgM-C1-C4b2), EMD-4945 (pentameric-IgM-C1-C4b2), and EMD-4943 (pentameric-IgM-C1-C4b1)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901841116/-/DCSupplemental.
References
- 1.Boes M., Role of natural and immune IgM antibodies in immune responses. Mol. Immunol. 37, 1141–1149 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Frank M. M., Humphrey J. H., The subunits in rabbit anti-Forssman IgM antibody. J. Exp. Med. 127, 967–982 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mäkelä O., Ruoslahti E., Seppälä I. J., Affinity of IgM and IgG antibodies. Immunochemistry 7, 917–932 (1970). [DOI] [PubMed] [Google Scholar]
- 4.Zhou Z. H., et al. , Polyreactive antibodies plus complement enhance the phagocytosis of cells made apoptotic by UV-light or HIV. Sci. Rep. 3, 2271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quartier P., Potter P. K., Ehrenstein M. R., Walport M. J., Botto M., Predominant role of IgM-dependent activation of the classical pathway in the clearance of dying cells by murine bone marrow-derived macrophages in vitro. Eur. J. Immunol. 35, 252–260 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Lintner K. E., et al. , Early components of the complement classical activation pathway in human systemic autoimmune diseases. Front. Immunol. 7, 36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott D., Botto M., The paradoxical roles of C1q and C3 in autoimmunity. Immunobiology 221, 719–725 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Willison H. J., Jacobs B. C., van Doorn P. A., Guillain-Barré syndrome. Lancet 388, 717–727 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Feinstein A., Munn E. A., Conformation of the free and antigen-bound IgM antibody molecules. Nature 224, 1307–1309 (1969). [DOI] [PubMed] [Google Scholar]
- 10.Burton D. R., Is IgM-like dislocation a common feature of antibody function? Immunol. Today 7, 165–167 (1986). [DOI] [PubMed] [Google Scholar]
- 11.Czajkowsky D. M., Shao Z., The human IgM pentamer is a mushroom-shaped molecule with a flexural bias. Proc. Natl. Acad. Sci. U.S.A. 106, 14960–14965 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller R., et al. , High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation. Proc. Natl. Acad. Sci. U.S.A. 110, 10183–10188 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perkins S. J., Nealis A. S., Sutton B. J., Feinstein A., Solution structure of human and mouse immunoglobulin M by synchrotron X-ray scattering and molecular graphics modelling. A possible mechanism for complement activation. J. Mol. Biol. 221, 1345–1366 (1991). [DOI] [PubMed] [Google Scholar]
- 14.Merle N. S., Church S. E., Fremeaux-Bacchi V., Roumenina L. T., Complement system Part I: Molecular mechanisms of activation and regulation. Front. Immunol. 6, 262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid K. B., Porter R. R., Subunit composition and structure of subcomponent C1q of the first component of human complement. Biochem. J. 155, 19–23 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bally I., et al. , Expression of recombinant human complement C1q allows identification of the C1r/C1s-binding sites. Proc. Natl. Acad. Sci. U.S.A. 110, 8650–8655 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bally I., et al. , Identification of the C1q-binding sites of human C1r and C1s: A refined three-dimensional model of the C1 complex of complement. J. Biol. Chem. 284, 19340–19348 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips A. E., et al. , Analogous interactions in initiating complexes of the classical and lectin pathways of complement. J. Immunol. 182, 7708–7717 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venkatraman Girija U., et al. , Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation. Proc. Natl. Acad. Sci. U.S.A. 110, 13916–13920 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diebolder C. A., et al. , Complement is activated by IgG hexamers assembled at the cell surface. Science 343, 1260–1263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jong R. N., et al. , A novel platform for the potentiation of therapeutic antibodies based on antigen-dependent formation of IgG hexamers at the cell surface. PLoS Biol. 14, e1002344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ugurlar D., et al. , Structures of C1-IgG1 provide insights into how danger pattern recognition activates complement. Science 359, 794–797 (2018). [DOI] [PubMed] [Google Scholar]
- 23.James L. C., Hale G., Waldmann H., Bloomer A. C., 1.9 A structure of the therapeutic antibody CAMPATH-1H fab in complex with a synthetic peptide antigen. J. Mol. Biol. 289, 293–301 (1999). Correction in: J. Mol. Biol. 292, 1161 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Sharp T. H., Koster A. J., Gros P., Heterogeneous MAC initiator and pore structures in a lipid bilayer by phase-plate cryo-electron tomography. Cell Rep. 15, 1–8 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Hale G., et al. , Removal of T cells from bone marrow for transplantation: A monoclonal antilymphocyte antibody that fixes human complement. Blood 62, 873–882 (1983). [PubMed] [Google Scholar]
- 26.Borsos T., Rapp H. J., Walz U. L., Action of the first component of complement. Activitation of C′1 to C′1a in the hemolytic system. J. Immunol. 92, 108–117 (1964). [PubMed] [Google Scholar]
- 27.Arnold J. N., et al. , Human serum IgM glycosylation: Identification of glycoforms that can bind to mannan-binding lectin. J. Biol. Chem. 280, 29080–29087 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Wiersma E. J., Shulman M. J., Assembly of IgM. Role of disulfide bonding and noncovalent interactions. J. Immunol. 154, 5265–5272 (1995). [PubMed] [Google Scholar]
- 29.Yoo E. M., et al. , Structural requirements for polymeric immunoglobulin assembly and association with J chain. J. Biol. Chem. 274, 33771–33777 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Graille M., et al. , Crystal structure of a Staphylococcus aureus protein A domain complexed with the fab fragment of a human IgM antibody: Structural basis for recognition of B-cell receptors and superantigen activity. Proc. Natl. Acad. Sci. U.S.A. 97, 5399–5404 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arya S., et al. , Mapping of amino acid residues in the C mu 3 domain of mouse IgM important in macromolecular assembly and complement-dependent cytolysis. J. Immunol. 152, 1206–1212 (1994). [PubMed] [Google Scholar]
- 32.Gadjeva M. G., et al. , Interaction of human C1q with IgG and IgM: Revisited. Biochemistry 47, 13093–13102 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Almitairi J. O. M., et al. , Structure of the C1r-C1s interaction of the C1 complex of complement activation. Proc. Natl. Acad. Sci. U.S.A. 115, 768–773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghavanloo E., Persistence length of collagen molecules based on nonlocal viscoelastic model. J. Biol. Phys. 43, 525–534 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mortensen S., et al. , Structural basis for the function of complement component C4 within the classical and lectin pathways of complement. J. Immunol. 194, 5488–5496 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Rossi V., Bally I., Thielens N. M., Esser A. F., Arlaud G. J., Baculovirus-mediated expression of truncated modular fragments from the catalytic region of human complement serine protease C1s. Evidence for the involvement of both complement control protein modules in the recognition of the C4 protein substrate. J. Biol. Chem. 273, 1232–1239 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Kidmose R. T., et al. , Structural basis for activation of the complement system by component C4 cleavage. Proc. Natl. Acad. Sci. U.S.A. 109, 15425–15430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry A. J., et al. , A molecular switch governs the interaction between the human complement protease C1s and its substrate, complement C4. J. Biol. Chem. 288, 15821–15829 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiramoto E., et al. , The IgM pentamer is an asymmetric pentagon with an open groove that binds the AIM protein. Sci. Adv. 4, eaau1199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forneris F., et al. , Regulators of complement activity mediate inhibitory mechanisms through a common C3b-binding mode. EMBO J. 35, 1133–1149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forneris F., et al. , Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science 330, 1816–1820 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharp T. H., Faas F. G. A., Koster A. J., Gros P., Imaging complement by phase-plate cryo-electron tomography from initiation to pore formation. J. Struct. Biol. 197, 155–162 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Richards D. B., et al. , Therapeutic clearance of amyloid by antibodies to serum amyloid P component. N. Engl. J. Med. 373, 1106–1114 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Pawluczkowycz A. W., et al. , Binding of submaximal C1q promotes complement-dependent cytotoxicity (CDC) of B cells opsonized with anti-CD20 mAbs ofatumumab (OFA) or rituximab (RTX): Considerably higher levels of CDC are induced by OFA than by RTX. J. Immunol. 183, 749–758 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Cleary K. L. S., Chan H. T. C., James S., Glennie M. J., Cragg M. S., Antibody distance from the cell membrane regulates antibody effector mechanisms. J. Immunol. 198, 3999–4011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cragg M. S., et al. , Complement-mediated lysis by anti-CD20 mAb correlates with segregation into lipid rafts. Blood 101, 1045–1052 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.