Abstract
Mononegaviruses are promising tools as oncolytic vectors and transgene delivery vectors for gene therapy and regenerative medicine. By using the Magnet proteins, which reversibly heterodimerize upon blue light illumination, photocontrollable mononegaviruses (measles and rabies viruses) were generated. The Magnet proteins were inserted into the flexible domain of viral polymerase, and viruses showed strong replication and oncolytic activities only when the viral polymerases were activated by blue light illumination.
Keywords: mononegavirus, measles virus, rabies virus, vector, oncolytic
The order Mononegavirales includes the Paramyxoviridae, Pneumoviridae, Bornaviridae, Filoviridae, and Rhabdoviridae families. Certain mononegaviruses are used for oncolytic viral therapy (1, 2). Oncolytic virotherapy is a new therapeutic option for cancer treatment using replication-competent viruses that preferentially target, replicate in, and kill cancer cells by inducing tumor-specific immune responses. They are also used as nonintegrating transgene delivery vectors to generate inducible pluripotent stem (iPS) cells (3). However, the designed removal of transgenes is still ideal to establish fully reprogrammed iPS cells. The control of viral replication also reduces the risk of adverse effects by oncolytic vectors. Here, we demonstrate a system for the spatiotemporal control of mononegavirus gene expression and replication.
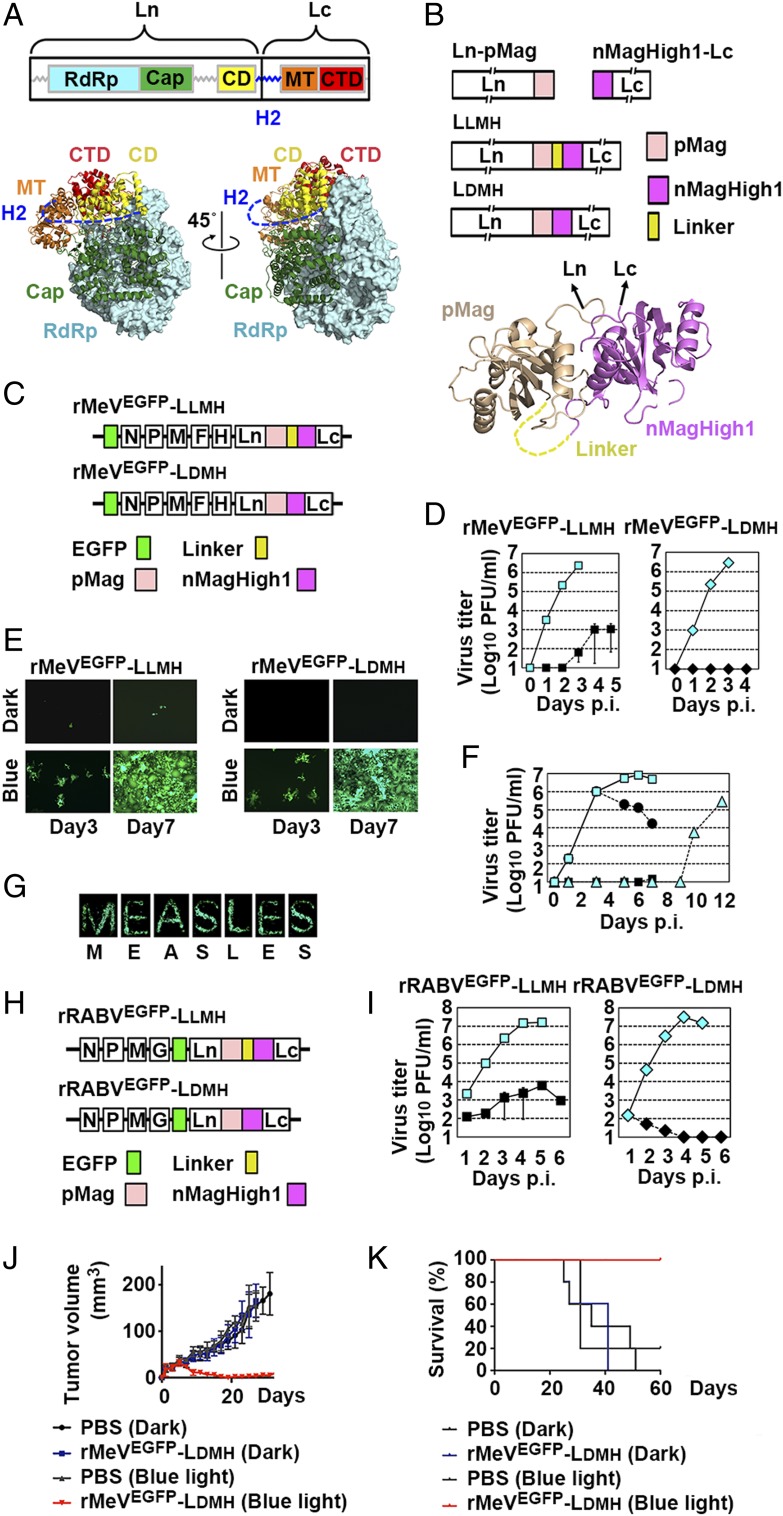
The mononegavirus genome forms a ribonucleoprotein complex with the nucleocapsid protein and the RNA-dependent RNA polymerase composed of the large (L) protein and the cofactor proteins. The L protein exhibits the major catalytic activities for RNA synthesis, such as nucleotide polymerization, mRNA capping, and polyadenylation. Measles virus (MeV) belongs to the genus Morbillivirus, family Paramyxoviridae. The MeV polymerase L protein has a highly variable linker region, called the hinge 2 (H2) region, between the connector domain (CD) and the methyltransferase (MT) domain (4) (Fig. 1A). We attempted to optically regulate the L protein polymerase activity by adding a photocontrollable domain at the H2 region, for which we used the Magnet system (5). The Magnet system consists of paired photoswitchable proteins—positive Magnet (pMag) and negative Magnet (nMag or nMagHigh1)—which heterodimerize upon blue light irradiation (5) (Fig. 1B).
Fig. 1.
Replication of photocontrollable rMeVs and rRABVs in cultured cells. (A) The schema of mononegavirus L proteins (Upper) and the three-dimensional (3D) structure of vesicular stomatitis virus L protein (8) (Lower). CTD, C-terminal domain; RdRp, RNA-dependent RNA polymerase. (B) The schema of photocontrollable L proteins (Upper) and the 3D structure of fungal photoreceptor Vivid (9) (Lower), from which Magnet proteins were generated (5). The light-state dimer is shown. (C) The genome structure of rMeVs. (D) Replication kinetics. Vero/hSLAM cells were infected with rMeV [multiplicity of infection (MOI) = 0.01] and illuminated by blue light (cyan) or kept in the dark (black). (E) EGFP fluorescence by rMeVs. Vero/hSLAM cells were infected with rMeV and illuminated by blue light or kept in the dark. (Magnification: 50×.) (F) Vero/hSLAM cells were infected with rMeVEGFP-LLMH (MOI = 0.01) and illuminated by blue light (cyan squares) or kept in the dark (black squares). Certain cells were illuminated by blue light for the first 3 d and kept in the dark thereafter (black circles) or kept in the dark for the first 6 d and illuminated after 7 d p.i. (cyan triangles). (G) Monolayers of Vero/hSLAM cells were infected with rMeVEGFP-LDMH (MOI = 0.01). The bottom of the culture dish was covered with black vinyl tape with letter-shaped slits, and the cells were illuminated continuously by blue light from below. At 5 d p.i., the vinyl tape was removed and EGFP fluorescence was observed under a fluorescence microscope. (Magnification: 10×.) (H) The genome structure of rRABVs. (I) Replication kinetics. BHK cells were infected with rRABVs (MOI = 0.01) and illuminated by blue light (cyan) or kept in the dark (black). (J and K) Balb-c nu/nu mice bearing MDM-MB-468 cell tumors were intratumorally injected with rMeVEGFP-LDMH or PBS at days 1, 3, 5, 7, and 9. Mice were illuminated by blue light for 12 h/d or kept in the dark throughout the day. (J) Mean tumor volumes (n = 5). (K) Kaplan–Meier survival curve (n = 5).
We attempted to generate recombinant MeVs (rMeVs) accommodating the photocontrollable L protein and enhanced green fluorescent protein (EGFP) using the method reported previously (6). The MeV L protein was split into the N-terminal half region before position1708/9 (Ln) and C-terminal half region after position1708/9 (Lc). The position1708/9 was located at the site corresponding to amino acid positions 1708 and 1709 in the H2 region. pMag was fused to the Ln C terminus (Ln-pMag), while nMagHigh1 was fused to the Lc N terminus (nMagHigh1-Lc) (Fig. 1B). Ln-pMag and nMagHigh1-Lc were connected by a 26-amino acid flexible linker (Linker), generating Linker MagHigh L protein (LLMH) (Fig. 1B). Ln-pMag and nMagHigh1-Lc were also connected directly, generating Direct MagHigh L protein (LDMH) (Fig. 1B). It was expected that the CD in Ln and the MT domain in Lc get closer upon blue light illumination (the Magnet dimerization). rMeVs possessing LLMH or LDMH (rMeVEGFP-LLMH or rMeVEGFP-LDMH, respectively) were successfully generated under blue light (470 ± 20 nm) (Fig. 1C). They replicated efficiently under blue light illumination (Fig. 1D). Notably, rMeVEGFP-LLMH showed low but detectable levels of replication capacity in the dark, while rMeVEGFP-LDMH showed none (Fig. 1D). The replication of rMeVEGFP-LLMH was accelerated when infected cells, which were initially kept in the dark, were illuminated by blue light after a 7-d incubation period (Fig. 1F). Conversely, the viral titers decreased when the illumination with blue light was stopped 3 d postinfection (p.i.) (Fig. 1F). When monolayers of rMeVEGFP-LDMH–infected cells were illuminated by blue light through letter-shaped slits at the bottom of the culture dishes, EGFP-fluorescent letters emerged on the monolayers (Fig. 1G).
We investigated whether the method used for MeV is applicable to rabies virus (RABV), a member of the genus Lyssavirus, family Rhabdoviridae. Since RABV and vesicular stomatitis virus, another member of Rhabdoviridae family, are used as tracers of neural circuits, this control method of virus gene expression and replication would be greatly beneficial for neuroscientists. The RABV reverse genetics system reported previously (7) was used. Successful insertion of Magnet proteins was achieved for the position corresponding to the amino acid position between 1623 and 1625. Rescued recombinant RABVs (rRABVs) were termed rRABVEGFP-LLMH and rRABVEGFP-LDMH, respectively (Fig. 1H). Their replication was accelerated greatly by blue light illumination (Fig. 1I). rRABVEGFP-LDMH showed a strong switching-off effect under dark conditions (Fig. 1I).
The utility of this system as an oncolytic vector was assessed in vivo. Balb-c nu/nu mice bearing MDM-MB-468 cell tumors were treated intratumorally with rMeVEGFP-LDMH. Mice were kept in the dark throughout the day or under the blue light for 12 h/d. Tumors in the phosphate-buffered saline (PBS) injection control group grew aggressively (Fig. 1K). Treatment with rMeVEGFP-LDMH resulted in a substantial reduction in tumor growth under the blue light (Fig. 1K). Only rMeVEGFP-LDMH–treated mice under the blue light survived (Fig. 1L).
This report presents photocontrollable viral vectors using mononegaviruses. The benefits of mononegaviruses in cancer treatment, gene therapy, and regenerative medicine have been comprehensively demonstrated (1–3). We believe that this control strategy of mononegaviruses would enhance the clinical utility of mononegaviruses for state-of-the-art medical treatments.
Methods
Construction of the Full-Genome MeV and RABV Plasmids.
All of the full-genome MeV plasmids in this study were generated using p(+)MV-IC-EGFP-M/P64S/E89K (6) as the backbone plasmid. To generate the LLMH construct, coding sequences for tandem-linked pMag, Linker, nMagHigh1, and HA tag [Linker MagHigh (LMH)] were inserted into the L gene at the site corresponding to amino acid positions 1708 and 1709 (position1708/9) (the two asparagine residues at positions 1708 and 1709 were removed). The LDMH construct contained tandem linked coding sequences for pMag, nMag, and HA tag [Direct MagHigh (DMH)] at the position1708/9. All RABV plasmids in this study were generated using p3.0-GFP (7) as the backbone plasmid. The LMH or DMH coding sequence was inserted into the RABV L gene at the site corresponding to the amino acid position between 1623 and 1625 (position 2) (a lysine residue at position 1624 was removed), generating the full-length RABV plasmids encoding photocontrollable L proteins.
Intratumor Treatment with Photocontrollable MeV.
The breast cancer MDM-MB-468 cell line was obtained from American Type Culture Collection. Balb-c nu/nu mice, 5 to 6 wk of age (Charles River Laboratories International, Inc.), were injected subcutaneously in the ventral area with 5 × 106 MDM-MB-468 cells to produce tumors. The tumor dimensions and body weight of the mice were measured every other day. After a tumor developed to over 2 mm in diameter, rMeVEGFP-LDMH was injected intratumorally. Each tumor was infected with 5 × 105 plaque-forming units per injection at days 1, 3, 5, 7, and 9, while control mice received PBS (Nacalai Tesque). Infected mice were kept in the dark continuously or illuminated by ISL-150X150-HBB (CCS Inc.) from outside of the mouse cage with blue light for 12 h/d. Mice were killed when tumor reached over 200 mm3. Animal experiments ware approved by the Institutional Animal Care and Use Committee (Permission no. PA17-47) and carried out according to the University of Tokyo animal experimentation regulations.
Acknowledgments
The reverse genetics system for the RABV was kindly provided by Drs. Mutsuyo Takayama-Ito and Masayuki Saijo. BHK/T7 cells and Vero/hSLAM cells were kindly provided by Drs. Naoto Ito and Yusuke Yanagi, respectively.
Footnotes
Conflict of interest statement: K.T. is the principal investigator of Project Division of ALA Advanced Medical Research, financially supported by grants from neopharma Japan Co., Ltd., Shinnihonseiyaku Co., Ltd., and Takara Bio Inc. The remaining authors have no conflicts of interest to declare.
References
- 1.Matveeva O. V., Kochneva G. V., Zainutdinov S. S., Ilyinskaya G. V., Chumakov P. M., [Oncolytic paramyxoviruses: Mechanism of action, preclinical and clinical studies]. Mol. Biol. (Mosk.) 52, 360–379 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Melzer M. K., Lopez-Martinez A., Altomonte J., Oncolytic vesicular stomatitis virus as a viro-immunotherapy: Defeating cancer with a “hammer” and “anvil.” Biomedicines 5, E8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haridhasapavalan K. K., et al. , An insight into non-integrative gene delivery approaches to generate transgene-free induced pluripotent stem cells. Gene 686, 146–159 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Duprex W. P., Collins F. M., Rima B. K., Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame. J. Virol. 76, 7322–7328 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawano F., Suzuki H., Furuya A., Sato M., Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat. Commun. 6, 6256 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Tahara M., Takeda M., Yanagi Y., Contributions of matrix and large protein genes of the measles virus Edmonston strain to growth in cultured cells as revealed by recombinant viruses. J. Virol. 79, 15218–15225 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khawplod P., et al. , A novel rapid fluorescent focus inhibition test for rabies virus using a recombinant rabies virus visualizing a green fluorescent protein. J. Virol. Methods 125, 35–40 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Liang B., et al. , Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162, 314–327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaidya A. T., Chen C. H., Dunlap J. C., Loros J. J., Crane B. R., Structure of a light-activated LOV protein dimer that regulates transcription. Sci. Signal. 4, ra50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]