Significance
Typically, comparative developmental studies focus on the question whether key genes are conserved over large evolutionary distances. How the gene regulatory network of a whole developmental process evolves is virtually unknown. This requires a functional comparison of homologous processes in species closely related enough to find similarities and distant enough to see functional changes. We approach this question by genetically and molecularly comparing trichome development in Arabidopsis thaliana with that in Arabis alpina. We show which steps are regulated similarly and which are different. A closer analysis of trichome patterning revealed fascinating changes in the gene regulatory network; in particular, the finding that overexpression of one key regulator leads to opposite phenotypes in the two species suggested fundamental changes.
Keywords: Arabisalpina, trichomes, genetic analysis
Abstract
The genetic and molecular analysis of trichome development in Arabidopsis thaliana has generated a detailed knowledge about the underlying regulatory genes and networks. However, how rapidly these mechanisms diverge during evolution is unknown. To address this problem, we used an unbiased forward genetic approach to identify most genes involved in trichome development in the related crucifer species Arabis alpina. In general, we found most trichome mutant classes known in A. thaliana. We identified orthologous genes of the relevant A. thaliana genes by sequence similarity and synteny and sequenced candidate genes in the A. alpina mutants. While in most cases we found a highly similar gene-phenotype relationship as known from Arabidopsis, there were also striking differences in the regulation of trichome patterning, differentiation, and morphogenesis. Our analysis of trichome patterning suggests that the formation of two classes of trichomes is regulated differentially by the homeodomain transcription factor AaGL2. Moreover, we show that overexpression of the GL3 basic helix–loop–helix transcription factor in A. alpina leads to the opposite phenotype as described in A. thaliana. Mathematical modeling helps to explain how this nonintuitive behavior can be explained by different ratios of GL3 and GL1 in the two species.
Evolutionary genetics has been widely used to study how molecular changes of gene functions or regulatory networks lead to phenotypic differences (1–3). To understand the source of molecular variation that ultimately results in phenotypic changes, microevolutionary approaches (4) and comparative studies of closely related species have been used (1, 3). These approaches enable an understanding of functional changes of genes in developmental processes. In this work we study the evolution of leaf trichome development. Leaf trichome development is best studied in Arabidopsis thaliana. The characterization of individual mutants in closely related species suggests that trichome development has a common basis in crucifer (5, 6). However, within the crucifer family, trichomes exhibit a wide range of trichome density and morphology phenotypes that correlate well with the phylogenic tree (7). It is therefore conceivable that the basic regulatory machinery for trichome development is conserved in the crucifer family but exhibits significant variation. We therefore reasoned that functional comparison of trichome development of A. thaliana with that of another evolutionarily distant crucifer species should enable us to recognize functional differences or diversifications. Toward this end, we chose Arabis alpina as a second genetic model for trichome development. A. alpina is diverged about 26–40 million years from A. thaliana and is in a distinct clade in the Brassicaceae family (8–10). These species might therefore be distant enough to find differences in the regulation of trichome development, while being closely enough related to compare orthologous gene functions. To approach this problem, we decided to first perform a genetic screen aiming to identify gene functions based on mutant phenotypes and then to compare the spectrum of phenotypes to that known in A. thaliana. In A. thaliana, trichomes develop on young leaves in regular patterns and are separated by protodermal cells (11). Incipient trichomes proceed through several rounds of DNA replication in the absence of cell divisions (endoreduplication) and emerge from the leaf surface (11). Typically three or four branches are formed in a regular arrangement before the cell expands to its final size (11). Genetic screens and subsequent molecular analyses have revealed most genes relevant for the initiation and development of trichomes (12–14). Based on mutant phenotypes, various developmental steps have been defined (11). The first step is the selection of individual epidermal cells among others to adopt a trichome cell fate, which involves positive and negative regulators of trichome development. They are considered to generate a trichome pattern through intercellular interactions (15, 16). The initiation of trichome differentiation is a separate step and requires the GLABRA2 gene (17). Various genes are important for the proper endoreduplication driven cell enlargement (12, 18–20). Trichome branching is governed by several genes that regulate either the number or the position of branches (21, 22). Regular cell expansion is defined by a group of mutants that are characterized by grossly distorted trichomes (23). Finally, maturation of trichomes as recognized by the formation of small papilla on their outer surface is dependent on genes of the glassy group (11, 24).
Leaf trichome development in A. alpina is reminiscent to that in A. thaliana (6). In contrast to A. thaliana, two types of trichomes are formed on A. alpina leaves. One class of small trichomes is regularly distributed and a second class of much larger trichomes is interspersed with regular distances to each other (6). Leaf trichome development is readily accessible to genetic approaches (6) and the molecular analysis is facilitated by the availability of a fully sequenced genome (10, 25) and because A. alpina can be transformed by Agrobacterium-mediated floral dip (26).
In this study, we systematically screened an ethyl methanesulfonate (EMS)-mutagenized population of A. alpina plants for trichome mutants. We identified mutants affecting most steps of trichome development, as previously reported for A. thaliana. Orthologous genes in A. alpina were identified based on sequence similarity and synteny and candidate genes were sequenced in A. alpina mutants. While most gene-phenotype relations in A. alpina were the same as in A. thaliana, we found some notable differences in the regulation of trichome patterning, differentiation, and morphogenesis.
Results
Trichome Development in A. alpina.
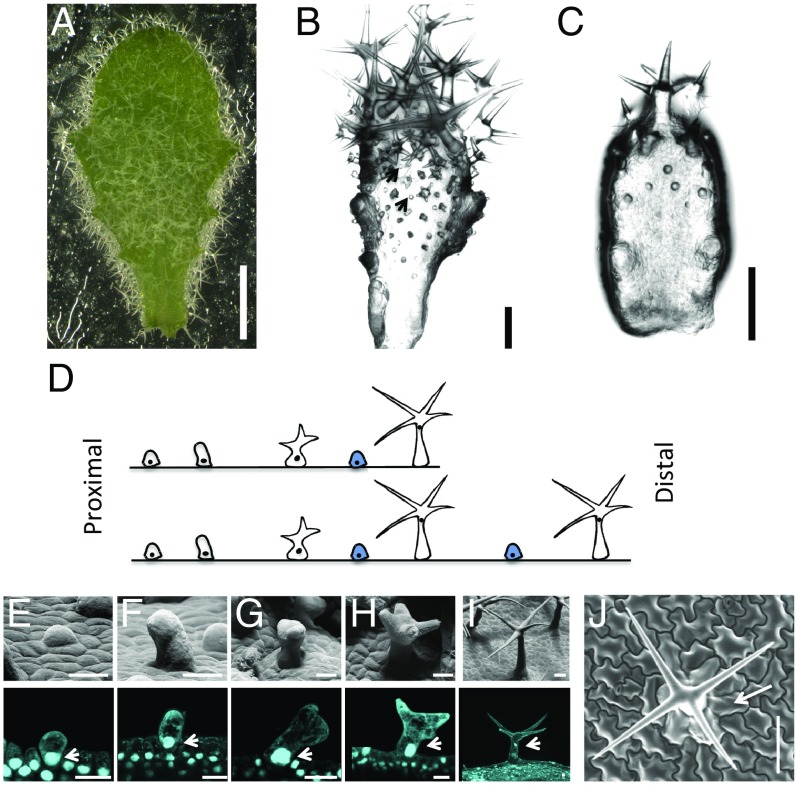
Trichomes in A. alpina densely cover the surface of adult leaves during vegetative development (Fig. 1A). They are initiated at the base of young leaves and in more distal regions between already existing trichomes (ref. 6 and Fig. 1 B–D). At the leaf base, incipient trichomes are typically formed four to five cells away from each other. Subsequently, trichomes are spatially separated due to expansion and cell divisions of the intervening epidermal cells. In more distal regions, new trichomes are formed between more advanced trichome stages. These are typically formed in a similar distance as between incipient trichomes at the leaf base (typically four to five cells).
Fig. 1.
Trichome development on A. alpina rosette leaves. (A) Mature leaf; trichomes densely cover the whole surface. (B) In slightly older stages, incipient trichomes are found between older trichomes (arrowheads). (C) On very young leaves, incipient trichomes are found at the leaf base and advanced developmental stages in more distal regions. (D) Schematic representation of the trichome distribution along the proximal-distal axis on a very young leaf as shown in C, and an older leaf as shown in B. Blue-colored trichomes are intercalating between already existing ones. (E–J) Scanning electron micrographs and optical section of DAPI-stained trichomes at different developmental stages. (E) Incipient trichomes beginning to expand. (F) Unbranched trichomes. (G) Two-branched trichomes. (H) Four-branched trichomes. (I) Mature trichomes. (J) Top view of a mature trichome. Note that the immediately adjacent cells are shaped like pavement cells. n = 20. (Scale bars: A, B, and E–I: 10 μm; C and D: 20 μm; and J: 100 μm.)
Similar as in A. thaliana (11, 27), trichome development proceeds through a series of characteristic stages (Fig. 1 E–I). Incipient trichomes begin to expand and grow perpendicular to the leaf surface (Fig. 1 E and F). Subsequently, trichomes typically undergo three branching events (Fig. 1 G and H), followed by extensive elongation growth. During maturation, numerous papillae are formed on the trichome surface. It is noteworthy that the cells immediately next to a mature trichome are shaped like normal epidermal pavement cells (Fig. 1J). This suggests that in contrast to A. thaliana (28), these cells do not differentiate into socket cells. Cell growth in A. thaliana occurs concomitant with an increase in nuclear size and DNA content due to several rounds of endoreduplication, leading to an average DNA content of 32C in mature trichome cells (18, 29). We used DAPI staining to study the nuclear size in different stages of trichome development in A. alpina. Incipient unbranched trichomes already show increased nuclear sizes (Fig. 1 E and F). A further increase in nuclear size was observed during branch formation (Fig. 1 G and H). Similar to A. thaliana (11, 21), the nucleus moves up to the branch points at this stage. We determined the relative DNA content of nuclei in mature trichomes by comparing the DAPI fluorescence in trichomes with that in stomata. In A. thaliana, stomata have a DNA content of 2C (29) and were therefore used as a reference to judge the C-value of trichome cells (11). Using this strategy, we found an average C-value of 64C in A. alpina trichomes (SI Appendix, Fig. S1A). When analyzing the population of the larger trichome class separately from the smaller trichome class, we found a clear separation such that the smaller trichome class has on average 16C and 32C and the larger trichome class 64–128C (SI Appendix, Fig. S1B). This suggests that the small trichomes undergo on average four endoreduplication rounds and the larger trichomes five or six cycles.
Isolation of Trichome Mutants in A. alpina.
We took a forward genetic approach to dissect trichome development into functional steps. Toward this end, seeds of five M1 plants from two independent EMS populations were pooled and about 48 M2 plants from each pool were screened for trichome phenotypes. The first screen was done in an A. alpina Pajares population representing 4165 M1 plants (26). The second screen was done in the A. alpina flowering time pep1-1 background that does not have an obvious trichome phenotype compared with Pajares (26, 30–32). Here we screened an M2 population derived from 6800 M1 plants. Trichome mutant phenotypes were confirmed in the M3 generation. Our screens yielded 49 trichome mutants. The mutant spectrum of patterning and morphogenesis mutants was similar to that in A. thaliana (11) though we did not find higher trichome density mutants and glassy mutants that are otherwise normal in shape (SI Appendix, Table S1).
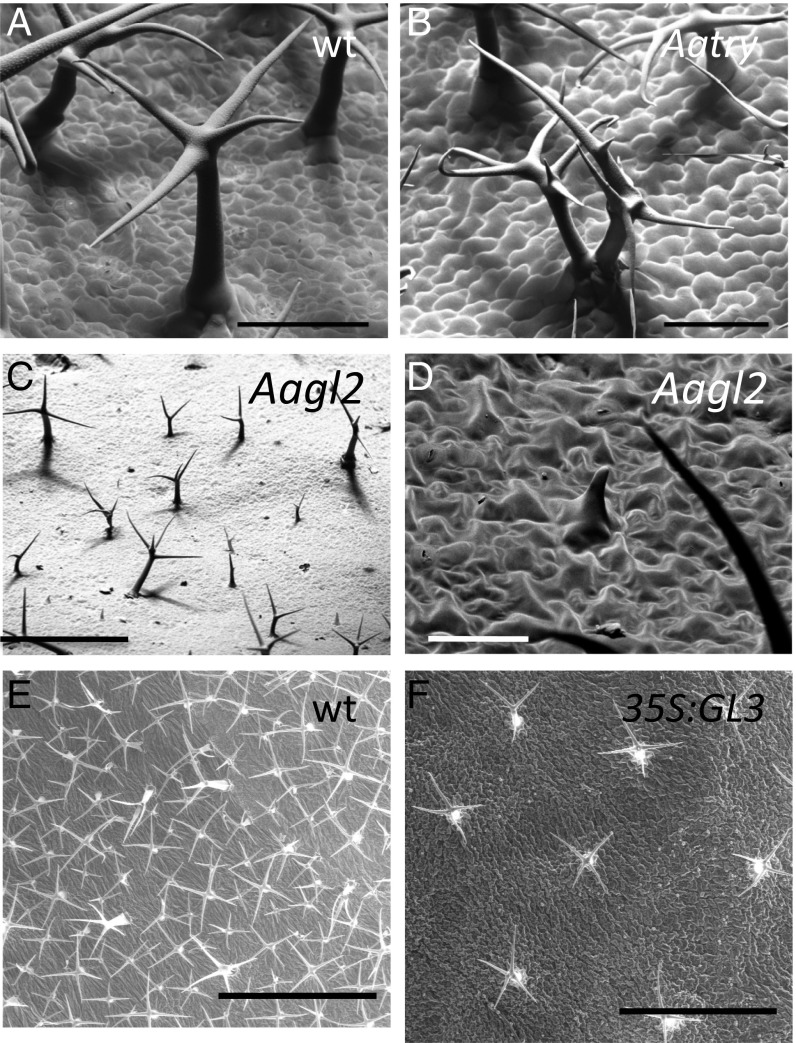
We identified 13 mutant lines in which trichome initiation and/or their distribution is affected. Eleven mutants show no trichomes on the leaf blade of 4-wk-old rosette leaves and variable numbers of trichomes at the leaf margin. Two lines were already reported to be Aattg1 alleles (6). We excluded the presence of very small and aborted trichomes by analyzing the leaf surface at a high magnification. The second class of patterning mutants comprises two lines in which trichomes occur in clusters with the individual trichomes being larger and over-branched (Fig. 2 A and B). Trichome clusters contained two to three trichomes immediately next to each other. We also found trichome clusters on young leaves indicating that the corresponding gene is important to single out one trichome cell during trichome initiation.
Fig. 2.
Phenotypes of patterning mutants. (A) Scanning electron microscope (SEM) picture of a mature wild-type trichome. (B) SEM picture of a mature Aatry mutant trichome. (C) SEM picture of an Aagl2 mutant leaf showing the distribution of large wild-type shaped trichomes and small aborted trichomes. (D) SEM picture of an aborted Aagl2 mutant trichome. (E) Wild-type A. alpina (Pajares) leaf with large and small trichomes. (F) 35S:AaGL3 with large trichomes. (Scale bars: A and B: 100 μm; C: 500 μm; D: 50 μm; E and F: 1 mm.)
We identified one mutant that appeared glabrous on the first glance; however, on closer inspection, revealed two classes of trichomes: extremely small and aborted trichomes and wild-type-like small trichomes (Fig. 2C). The small trichomes were reminiscent of young unbranched trichomes. Their shape, however, differed from young wild-type trichomes in that their cell body exhibited a puzzle-piece-like form that is characteristic of epidermal pavement cells (Fig. 2D). This suggests that trichome initiation was not affected and that trichomes have a mixed trichome/pavement fate. The wild-type-like trichomes were regularly distributed on the leaf surface at large distances with typically one or two small trichome cells in between (Fig. 2C). This phenotype suggests the presence of two types of genetically distinct trichomes in A. alpina that have not been observed in A. thaliana.
In addition, we isolated a number of mutants affecting the morphogenesis of trichomes. One class comprising 24 mutants exhibited defects in branch number or their spatial arrangement. A second class showed distorted and twisted trichomes (SI Appendix, Figs. S2 and S3 and Table S2)
Identification of Trichome Genes in A. alpina.
Given that A. alpina and A. thaliana are crucifers that diverged only about 26–40 million years ago (8, 9), we assumed that the majority of the mutant phenotypes found in our screen are caused by mutations in the genes orthologous to the respective A. thaliana genes. In a first step, we searched for putative orthologous genes in the A. alpina genome based on sequence similarity and synteny (33). By these criteria, we identified orthologs of the selected Arabidopsis genes in the A. alpina genome (SI Appendix, Fig. S4 and Table S3).
Identification of Mutant-Specific Alleles.
Under the assumption that mutations in A. alpina trichome genes lead to the same phenotype as in A. thaliana, one would expect to find relevant mutations in the respective genes in the trichome mutants. We therefore sequenced candidate genes in selected trichome mutants focusing on the unambiguous phenotypes, similar as done before with AaTTG1 (6). As summarized in SI Appendix, Table S4 we found 23 mutant-specific alleles.
The analysis of the 11 glabrous mutants revealed three lines with mutations in the AaTTG1 gene, as reported previously (6). In the remaining eight mutants, we discovered relevant mutations in the AaGL3 gene (SI Appendix, Fig. S6). Mutations included changes leading to acceptor splice site, premature STOP codons, or amino acid exchanges. These findings were unexpected, as Atgl3 mutants have trichomes. Atgl3 Ategl3 double mutants are completely glabrous, indicating that GL3 and EGL3 act in a partially redundant manner in A. thaliana (34). Sequencing the AaEGL3 gene in four gl3 alleles revealed no mutations (SI Appendix, Table S5). Thus, in A. alpina, AaGL3 possesses the full basic helix–loop–helix (bHLH) function in trichome patterning and AaEGL3 does not appear to be functionally relevant in this context. However, rescue experiments show that the AaEGL3 protein can rescue the Atgl3 Ategl3 Arabidopsis mutant efficiently when expressed under the 35S promoter, indicating that the protein is fully functional in this context (SI Appendix, Fig. S7). We also sequenced the AaGL1 gene in all glabrous mutants. None displayed mutations in the AaGL1 gene (SI Appendix, Table S5).
Two patterning mutants exhibited trichome clusters reminiscent to try mutants in A. thaliana. Sequence analysis revealed a premature STOP codon in one mutant and a Leucine to Phenylalanine change in a conserved region in the other. Both mutations are expected to lead to severe defects in protein function, indicating that the two mutants are Aatry alleles (SI Appendix, Fig. S6).
The differentiation mutant displaying large and fairly normal trichomes plus small underdeveloped trichomes has no clear counterpart in A. thaliana. However, the population of underdeveloped trichomes shares similarities with gl2 trichomes in A. thaliana. When sequencing the AaGL2 gene in this line, we found a deletion of one base pair in exon 4 leading to a premature STOP codon at amino acid position 348 (SI Appendix, Fig. S6).
The molecular analysis of the morphogenesis mutants also revealed mutations in several known trichome morphogenesis genes (SI Appendix, Fig. S6).
Analysis of the GL3 Function in A. alpina.
Our finding that mutations in GL3 result in a strong trichome phenotype in A. alpina, whereas in A. thaliana an additional mutation in EGL3 is required for a glabrous phenotype, suggests a functional divergence of GL3 and EGL3 between the two species. In A. thaliana, it was reported that Atgl3 single mutants exhibit a weak trichome phenotype but none of the other ttg1 traits. The Atgl3 Ategl3 double mutants strongly enhance the trichome phenotype, show ectopic root hairs, lack anthocyanin, and show a weak defect in seed coat mucilage production (34). Seed color is normal in Atgl3 Ategl3. Together this suggests that AtGL3 and AtEGL3 act redundant in four TTG1 traits. As Aagl3 single mutants in A. alpina display the same trichome phenotype as the Atgl3 Ategl3 mutant in A. thaliana, we determined whether representative Aagl3 alleles of A. alpina, Aagl3-1 and Aagl3-2, show additional phenotypes. Seed color was normal in both Aagl3 alleles (SI Appendix, Fig. S8 A–C). The analysis of root hair formation revealed hairy roots (SI Appendix, Fig. S8 D–F). A closer inspection showed that all files including the N-files produced almost only root hairs. Thus, similar as observed for trichomes, Aagl3 mutants produce the full Aattg1 root hair phenotype suggesting that EGL3 has also no relevant function in root hair patterning. In contrast, seed coat mucilage production and anthocyanin production in seedlings grown on 3% sucrose was unaffected in both alleles (SI Appendix, Fig. S8 G–L).
To support the idea that the observed mutant phenotypes of Aagl3-1 and Aagl3-2 alleles are caused by mutations in the GL3 gene, we performed a complementation test. F1 plants showed a glabrous trichome phenotype and hairy roots (SI Appendix, Fig. S9), indicating that the causal mutations are allelic.
To better understand the function of AaGL3 in A. alpina, we generated transgenic plants expressing the AaGL3 gene under the 35S promoter. In A. thaliana, overexpression of AtGL3 leads to extra trichome formation (35) and reduced root hair numbers (36) supporting the idea that AtGL3 promotes trichome formation and nonroot hair formation. We recovered two plants in which we found 30-fold and 10-fold higher expression levels of AaGL3 compared with wild type in qRT-PCR experiments (SI Appendix, Tables S6–S8). In the next generation, single plants were analyzed with respect to the trichome and root hair phenotypes (SI Appendix, Tables S9–S11). In four individual plants, we confirmed the overexpression of AaGL3 by qRT-PCR (SI Appendix, Tables S7 and S8). We also found reduced root hair formation (SI Appendix, Fig. S10) further supporting that AaGL3 promotes nonroot hair fate also in A. alpina. Trichome density was determined on the third leaf when they reached a length of 0.6–1 cm. Trichome density was reduced to 10% of that found in wild type (SI Appendix, Tables S9–S11). This is in sharp contrast to A. thaliana where overexpression of AtGL3 causes extra trichome formation (35). The reduced trichome density was also observed on older leaves. Trichomes were fairly similar in size and regularly distributed (Fig. 2 E and F).
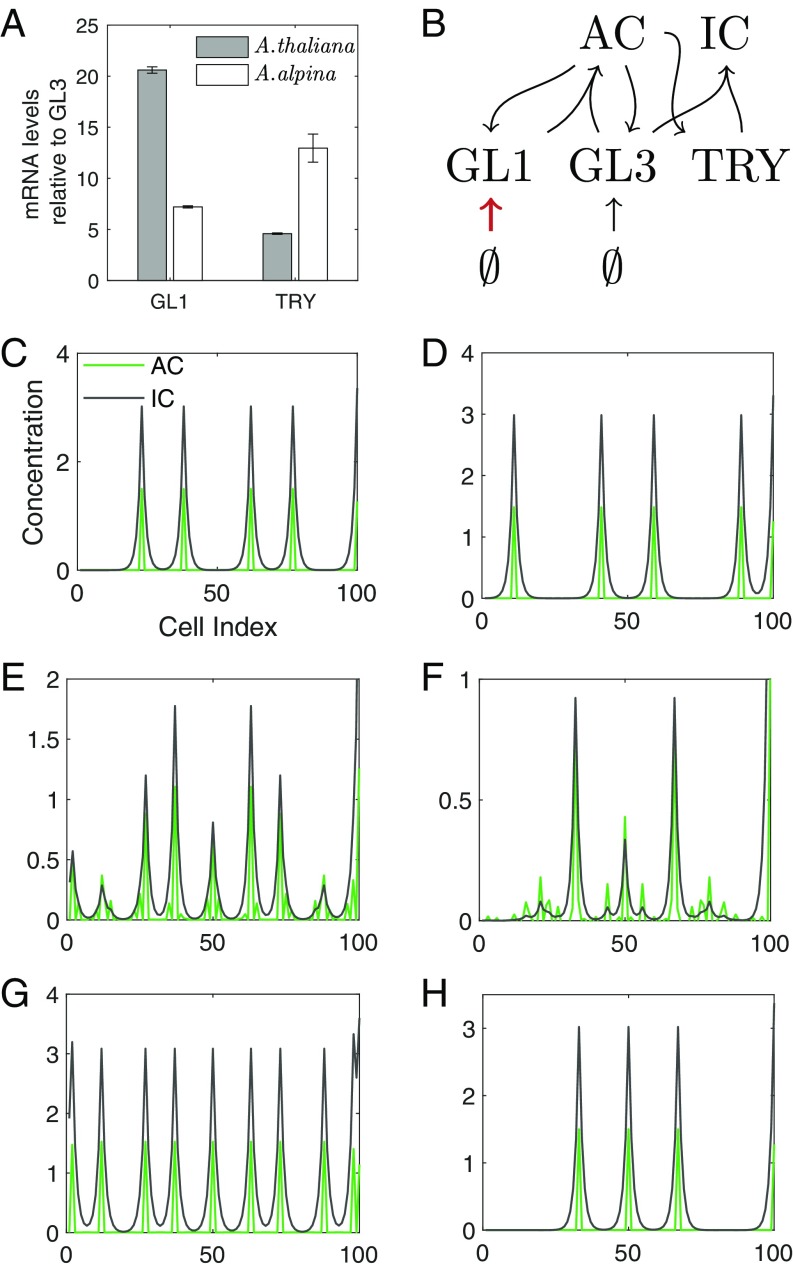
We reasoned that this nonintuitive difference in the response to GL3 overexpression might be correlated with differences in the relative expression levels of the core patterning genes GL1, GL3, and TRY. To test this assumption, we compared their expression between Arabidopsis and Arabis in young leaves corresponding to stages shown in Fig. 1B by qPCR analysis. When comparing the relative expression levels of GL1 with that of GL3, we found a large difference (Fig. 3A). Compared with Arabidopsis, the relative levels of GL1 levels were strongly reduced. To explore whether reduced GL1 levels relative to GL3 are sufficient to explain the different responses to GL3 overexpression in Arabis, we used a modeling approach. We use a previously published model that includes GL1 and GL3 that together form the activator complex (AC) (37). The AC activates TRY, which in turn forms the inhibitor complex (IC) together with GL3 (Fig. 3B) (34). Using this model, we screened for parameter sets that lead to increased trichome numbers upon a simulated overexpression of GL3 [A. thaliana situation (Fig. 3 C, E, and G)]. These sets were screened for parameter sets in which the expression of GL1 was decreased and overexpression of GL3 leads to less trichomes [A. alpina situation (Fig. 3 D, F, and H)]. Our results show that the relative expression difference of GL1 compared with GL3 can explain the different response to the overexpression of GL3 in the two species. A closer analysis of the model revealed that in both, the Arabidopsis and the Arabis situation, overexpression of GL3 leads to an increase of the inhibitor and activator activity; however, in the Arabis situation, the relative increase of the inhibitor activity is more pronounced around the activator peaks leading to an ∼3 times broader repression domain (SI Appendix, Fig. S11). This is consistent with the qPCR results in which the relative levels of TRY are higher in Arabis. As a result, in the Arabidopsis situation, overexpression of GL3 effectively leads to less repression and therefore a higher density. By contrast, in the Arabis situation, only the strongest peaks survive and the lower peaks are suppressed over time by the broader TRY domain. This is reminiscent to presence of the large trichomes and the lack of small trichomes in 35S:GL3 Arabis plants.
Fig. 3.
Modeling A. thaliana and A. alpina wild-type and 35S:GL3 phenotypes. (A) mRNA levels determined by qPCR of GL1 and TRY relative to GL3 for A. thaliana (gray bars) and A. alpina (white bars). Note that in Arabis we find relatively low levels of GL1 and relatively higher levels of TRY compared with Arabidopsis. (B) Schematic network of the model. The GL1 basal production parameter is decreased (red arrow) to reproduce A. alpina phenotypes. Simulation examples of (C) A. thaliana wild-type and (D) A. alpina wild-type on a 1D grid discretized into 100 cells (x-axis). Concentrations of activating complex (AC) and inactive complex (IC) are indicated by green and black lines, respectively. E and G show the development of 35S:GL3 patterns for A. thaliana conditions over time, where (E) is intermediate and (G) is the final state. F and H show the development of 35S:GL3 patterns for A. alpina conditions over time, where F is intermediate and (H) is the final state.
Discussion
In this study, we set out to systematically identify all genes involved in trichome development in A. alpina by a forward genetic mutagenesis screen to enable a comparison with the genetic inventory in A. thaliana. We reasoned that the evolutionary distance between the two species is close enough to compare orthologous processes and genes, yet distant enough to expect differences. Our phenotypic comparison supports this hypothesis as trichome patterning and morphogenesis in A. alpina is almost indistinguishable from that in A. thaliana except for the presence of two differently sized trichome classes and the absence of morphologically distinct accessory cells. We therefore expected genetic and molecular differences explaining the two classes of trichomes. For all other patterning and morphogenetic aspects, we assumed not to find major differences in the function of the involved genes.
How Well Does the Mutant Spectrum Reflect the Gene Inventory?
EMS mutagenesis screens can be considered to induce random mutations in the genome and as a consequence the frequency of deleterious mutations should be similar in all genes. It is therefore in principle possible to judge the saturation of the screen by the allele frequency. Our sequence analysis of candidate genes in the respective mutants revealed on average 2.6 mutations for each considered gene. Although, as in other studies, the distribution of alleles is very asymmetric, the allele frequency lies in a similar range as in other systematic genetic screens (38). In this context, it is noteworthy that we also found several alleles of fairly small genes such as TRY. This suggests that we have identified a fairly representative set of mutants.
Trichome Patterning and Differentiation: Differences and Similarities Between A. alpina and A. thaliana.
In A. thaliana, trichome initiation is governed by the WD40 gene AtTTG1, the MYB gene AtGL1, and the redundantly acting bHLH genes AtGL3 and AtEGL3 (12, 39). Orthologous genes of these regulators are also present in the A. alpina genome as judged by sequence similarity and synteny. It was therefore surprising that we found no gl1 mutant in our screen though this may be due to statistical reasons. Our attempts to create transgenic lines suppressing AaGL1 expression failed as we could not recover any transgenic plants. All glabrous mutants carried either relevant mutations in AaTTG1 (6) or in AaGL3. The latter is striking, as this indicates that Aagl3 mutants are completely devoid of trichomes. This is in sharp contrast to A. thaliana, where only the additional loss of AtEGL3 results in a glabrous phenotype (34). Our result indicates that the AaEGL3 gene in A. alpina has no function in trichome patterning. Since AaEGL3 can efficiently rescue the A. thaliana Atgl3 Ategl3 mutant when expressed from a heterologous promoter, it is likely that the functional change is due to differences in the regulation of expression. Also the function of AaGL3 in trichome patterning of A. alpina appears to be different as judged by our overexpression data. While overexpression of AtGL3 in A. thaliana Columbia causes a higher trichome density, 10–30 fold overexpression of AaGL3 in A. alpina leads to a reduction of trichome density. On first glance this is difficult to understand. One possibility is that in A. alpina overexpression of AaGL3 counteracts intercalating trichome formation. In support of this, we found only similar sized trichomes on older leaves. However, as both trichome size classes depend on AaGL3 and AaTTG1, it is difficult to envision why only one patterning event should be sensitive to AaGL3 overexpression. In principle it could be due to changes in the gene regulatory network structure or additional regulators. A simpler explanation would explain the difference by changes of the parameters in the Arabidopsis network. The latter was suggested to us by the finding that the relative expression levels of GL1 and GL3 differ between the two species. Mathematical modeling revealed how a reduction of GL1 relative to GL3 can explain the different responses to GL3 overexpression. It is important to note that this mathematical explanation of the phenotypes is already possible with a simple model capturing the protein-protein interactions and that this conclusion is therefore also valid for any models extended by additional genes and/or interactions.
Trichome patterning in A. thaliana involves also seven redundantly acting inhibitor genes including AtTRY, AtCPC, AtETC1, AtETC2, AtETC3, AtTCL1, and AtTCL2. Among these, AtTRY and AtCPC are the most relevant as judged by the single mutant phenotypes, though they show qualitatively different phenotypes. While Attry mutants exhibit trichome clusters, Atcpc mutants show a higher trichome density. It is not surprising that we did not find Aacpc mutants in our screens as wild-type A. alpina plants display a very dense trichome pattern and higher densities can easily be missed under screening conditions. We did, however, find two Aatry alleles. Both show trichome clusters and larger, more-branched trichomes. The latter is interesting as it indicates that the dual function of the TRY gene in trichome patterning and cell size/branching is evolutionary conserved and therefore possibly functionally relevant.
In A. thaliana, trichome differentiation is regulated by the AtGL2 gene (17). This is suggested by the Atgl2 mutant trichome phenotype: Trichomes are smaller, less branched, and in extreme cases, trichomes have a puzzle-piece-like shape with a little bump suggesting that trichomes are initiated but lost their trichome fate. The Arabis Aagl2 mutant found in this study differs in that only the small trichomes but not the large ones are affected. As our attempts to verify the phenotype by microRNA suppression or rescue of the Aagl2 mutant failed because we could not recover any transformants the phenotype needs to be interpreted with caution. Within this limits one possible explanation is that AaGL2 is not necessary for the proper differentiation of large trichomes implying that small and large trichomes are genetically distinct. An alternative explanation is that increased size can compensate the requirement for AaGL2. This scenario is supported by the previous finding that Atgl2 A. thaliana mutants can be rescued in genetic situations in which the DNA content and thereby trichome size is increased (40).
Perspective.
Our forward genetic approach in A. alpina has enabled us to recognize the developmental processes and the underlying genes that are similar to that in A. thaliana, as well as those that are different. Based on this work it will be possible to unravel the molecular basis of the evolutionary differences of trichome development between Arabidopsis and Arabis.
Materials and Methods
Plant Material and Growth Conditions.
All A. alpina mutants were isolated from EMS-mutagenized A. alpina Pajares and pep1-1 populations (26). A. alpina seeds on soil were stratified in darkness at 4 °C for 5 d and then placed in growth chambers under long day (LD; 16 h light, 8 h darkness) conditions at 21 °C. Pajares required 12 wk of vernalization to flower whereas pep1-1 flowered in ∼80 d without vernalization.
Sequence and Synteny Analysis.
A. alpina gene sequences were analyzed with the CLC DNA Workbench 5.6.1 (CLC bio, Aarhus, Denmark) by comparison with the coding sequences of the relevant A. thaliana genes obtained from TAIR 10 (www.arabidopsis.org). National Center for Biotechnology Information Blastn (2.2.28; https://blast.ncbi.nlm.nih.gov/Blast.cgi) (41) along with the assembled A. alpina genome was used to confirm the synteny of the selected genes using conserved order and appearance of the neighboring genes. For sequence analysis, primers were designed outside the coding sequence of a given A. alpina gene to sequence it in wild type and mutants. NCBI’s conserved domain database (CDD; https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to search for conserved domains within the protein sequence (42). The Net2Gene server was used to predict splicing sites (http://www.cbs.dtu.dk/services/NetGene2/) (43).
Constructs and Stable Plant Transformation.
The binary vector pAMPAT-CaMV35S-GW was used to create pAMPAT- CaMV35S:AaGL3 and pAMPAT- CaMV35S:AaEGL3 using the Gateway system with PCR-amplified AaGL3 and AaEGL3 coding sequences, respectively (SI Appendix, Table S12). The constructs were introduced in the A. thaliana gl3 egl3 double mutant (34) and in the Arabis alpina pep1-1 background (26) by Agrobacterium-mediated transformation (strain GV3101-pMP90RK) using floral dip (44). qPCR analysis was done with established reference primers (SI Appendix and ref. 45).
Supplementary Material
Acknowledgments
We would like to thank Siegfried Werth for taking pictures from whole leaves and seeds and Hans-Peter Bollhagen and Dr. Frank Nitsche for scanning electron microscope analysis. This work was supported by a Deutsche Forschungsgemeinschaft grant (to M.H.), the SFB680 (to M.H.) and an International Max Planck Research School fellowship (to D.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819440116/-/DCSupplemental.
References
- 1.Simpson P., Evolution of development in closely related species of flies and worms. Nat. Rev. Genet. 3, 907–917 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Purugganan M. D., The molecular evolution of development. Bioessays 20, 700–711 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Fernández-Mazuecos M., Glover B. J., The evo-devo of plant speciation. Nat. Ecol. Evol. 1, 110 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Nunes M. D. S., Arif S., Schlötterer C., McGregor A. P., A perspective on micro-evo-devo: Progress and potential. Genetics 195, 625–634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nayidu N. K., et al. , Comparison of five major trichome regulatory genes in Brassica villosa with orthologues within the Brassicaceae. PLoS One 9, e95877 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chopra D., et al. , Analysis of TTG1 function in Arabis alpina. BMC Plant Biol. 14, 16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beilstein M. A., Al-Shehbaz I. A., Kellogg E. A., Brassicaceae phylogeny and trichome evolution. Am. J. Bot. 93, 607–619 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Koch M. A., et al. , Three times out of Asia Minor: The phylogeography of Arabis alpina L. (Brassicaceae). Mol. Ecol. 15, 825–839 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Beilstein M. A., Nagalingum N. S., Clements M. D., Manchester S. R., Mathews S., Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 18724–18728 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willing E. M., et al. , Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat. Plants 1, 14023 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Hülskamp M., Misŕa S., Jürgens G., Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Hülskamp M., Plant trichomes: A model for cell differentiation. Nat. Rev. Mol. Cell Biol. 5, 471–480 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Tominaga-Wada R., Ishida T., Wada T., New insights into the mechanism of development of Arabidopsis root hairs and trichomes. Int. Rev. Cell Mol. Biol. 286, 67–106 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Marks M. D., Esch J., Herman P., Sivakumaran S., Oppenheimer D., A model for cell-type determination and differentiation in plants. Symp. Soc. Exp. Biol. 45, 77–87 (1991). [PubMed] [Google Scholar]
- 15.Pesch M., Hülskamp M., One, two, three...models for trichome patterning in Arabidopsis? Curr. Opin. Plant Biol. 12, 587–592 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Grebe M., The patterning of epidermal hairs in Arabidopsis–Updated. Curr. Opin. Plant Biol. 15, 31–37 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Rerie W. G., Feldmann K. A., Marks M. D., The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8, 1388–1399 (1994). [DOI] [PubMed] [Google Scholar]
- 18.Traas J., Hülskamp M., Gendreau E., Höfte H., Endoreduplication and development: Rule without dividing? Curr. Opin. Plant Biol. 1, 498–503 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Lee H. O., Davidson J. M., Duronio R. J., Endoreplication: Polyploidy with purpose. Genes Dev. 23, 2461–2477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guimil S., Dunand C., Cell growth and differentiation in Arabidopsis epidermal cells. J. Exp. Bot. 58, 3829–3840 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Folkers U., Berger J., Hülskamp M., Cell morphogenesis of trichomes in Arabidopsis: Differential control of primary and secondary branching by branch initiation regulators and cell growth. Development 124, 3779–3786 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Zhang X., Grey P. H., Krishnakumar S., Oppenheimer D. G., The IRREGULAR TRICHOME BRANCH loci regulate trichome elongation in Arabidopsis. Plant Cell Physiol. 46, 1549–1560 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Szymanski D. B. Breaking the WAVE complex: The point of Arabidopsis trichomes. Curr. Opin. Plant Biol. 8, 103–112 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Fornero C., Suo B., Zahde M., Juveland K., Kirik V., Papillae formation on trichome cell walls requires the function of the mediator complex subunit Med25. Plant Mol. Biol. 95, 389–398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiao W. B., et al. , Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Res. 27, 778–786 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang R., et al. , PEP1 regulates perennial flowering in Arabis alpina. Nature 459, 423–427 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Szymanski D. B., Jilk R. A., Pollock S. M., Marks M. D., Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125, 1161–1171 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Valdivia E. R., et al. , DVL genes play a role in the coordination of socket cell recruitment and differentiation. J. Exp. Bot. 63, 1405–1412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melaragno J. E., Mehrotra B., Coleman A. W., Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5, 1661–1668 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nordström K. J., et al. , Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nat. Biotechnol. 31, 325–330 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Albani M. C., et al. , PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering. PLoS Genet. 8, e1003130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergonzi S., et al. , Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340, 1094–1097 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Lyons E., et al. , Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 148, 1772–1781 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang F., Gonzalez A., Zhao M., Payne C. T., Lloyd A., A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Payne C. T., Zhang F., Lloyd A. M., GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernhardt C., et al. , The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130, 6431–6439 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Digiuni S., . A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol. Syst. Biol. 4, 217 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollock D. D., Larkin J. C., Estimating the degree of saturation in mutant screens. Genetics 168, 489–502 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishida T., Kurata T., Okada K., Wada T., A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59, 365–386 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Bramsiepe J., et al. , Endoreplication controls cell fate maintenance. PLoS Genet. 6, e1000996 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchler-Bauer A., et al. , CDD: A Conserved Domain Database for protein classification. Nucleic Acids Res. 33, D192–D196 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hebsgaard S. M., et al. , Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 24, 3439–3452 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Stephan L., Tilmes V., Hülskamp M., Selection and validation of reference genes for quantitative Real-Time PCR in Arabis alpina. PLoS One 14, e0211172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.