Significance
Overexpression of human homologue of mouse double minute 2 (HDM2) is associated with aggressive cancer malignancy. It is therefore critical to understand cellular mechanisms for the control of HDM2 abundance. By identifying Skp1-Cullin 1-FBXO22-ROC1 (SCFFBXO22) as an E3 ubiquitin ligase for HDM2, our work has significantly improved knowledge of the cellular regulatory network required for the maintenance of HDM2 protein stability. The results of tumor cell invasion, mouse 4T1 breast tumor model study, and human breast cancer tissue expression analyses suggest a role for SCFFBXO22 in the inhibition of breast cancer metastasis by down-regulating HDM2.
Keywords: HDM2 abundance, E3 SCF-FBXO22, breast cancer metastasis
Abstract
Human homolog of mouse double minute 2 (HDM2) is an oncogene frequently overexpressed in cancers with poor prognosis, but mechanisms of controlling its abundance remain elusive. In an unbiased biochemical search, we discovered Skp1-Cullin 1-FBXO22-ROC1 (SCFFBXO22) as the most dominating HDM2 E3 ubiquitin ligase from human proteome. The results of protein decay rate analysis, ubiquitination, siRNA-mediated silencing, and coimmunoprecipitation experiments support a hypothesis that FBXO22 targets cellular HDM2 for ubiquitin-dependent degradation. In human breast cancer cells, FBXO22 knockdown (KD) increased cell invasiveness, which was driven by elevated levels of HDM2. Moreover, mouse 4T1 breast tumor model studies revealed that FBXO22 KD led to a significant increase of breast tumor cell metastasis to the lung. Finally, low FBXO22 expression is correlated with worse survival and high HDM2 expression in human breast cancer. Altogether, these findings suggest that SCFFBXO22 targets HDM2 for degradation and possesses inhibitory effects against breast cancer tumor cell invasion and metastasis.
Mouse double minute 2 (MDM2; human homolog, HDM2) is defined as a RING-type E3 ubiquitin ligase with a prominent role in tumorigenesis, acting both as an oncogene and a tumor suppressor (1). MDM2/HDM2 is best understood for its oncogenic role, which is executed predominantly via a mechanism that utilizes the intrinsic E3 ligase activity to target the p53 tumor suppressor for ubiquitin-dependent degradation (2, 3). However, accumulating evidence suggests p53-independent mechanisms that contribute to MDM2/HDM2’s oncogene function as well (4).
As noted by a large body of clinical studies, HDM2 amplification and overexpression are common to a variety of cancers that often have a poor prognosis (4–6). Elevated levels of HDM2 are regarded as a significant risk factor in distant metastasis (7). Although, mechanistically, such phenomena can be attributed to the ability of HDM2 to drive p53 degradation (5), emerging studies have revealed additional contributing mechanisms involving previously underappreciated activity by MDM2/HDM2 to promote tumor cell invasiveness (8). In this regard, MDM2 was shown to act as an E3 ligase to target epithelial marker E-cadherin for ubiquitin-dependent degradation (8). However, a subsequent study suggests that, in renal cell carcinoma, MDM2 can promote cell motility and invasiveness without the function of MDM2 RING finger domain (9). Another mechanism may involve matrix metalloproteinase (MMP) capable of degrading membrane-associated extracellular proteins, a process critically implicated for metastasis (10). Intriguingly, MDM2 overexpression was found to transcriptionally up-regulate the expression of MMP-9 (11), although the precise mechanism remains to be elucidated.
The compelling association between HDM2 overexpression and aggressive cancer malignancy argues for a potentially critical importance concerning cellular mechanisms for the control of HDM2 abundance. On this note, earlier work employed ectopic expression methods, suggesting that MDM2 regulates its own levels by RING domain-mediated autoubiquitination (12, 13). However, subsequent mouse genetic experiments suggested alternative mechanisms. By using knock-in technique, a mouse line was created to express a mutant form of MDM2 without a functional RING domain (MDM2C462A/C462A) (14). Fibroblasts derived from the MDM2C462A/C462A mouse, however, exhibit an ability to drive proteasomal degradation of the mutant MDM2 at a rate indistinguishable from that seen in cells expressing the wild-type MDM2. In a follow-up study, the MDM2Y487A/Y487A mouse was generated, expressing a form of MDM2 with impaired E3 activity, but retaining its ability to heterodimerize with MDMX (15). Still, the MDM2Y487A/Y487A mouse shows a rate of degradation of the mutant MDM2 nearly identical to the wild-type. Collectively, these findings strongly suggest that the cellular control of MDM2/HDM2 stability requires mechanisms mediated by extrinsic E3 ubiquitin ligase(s).
To this end, the present study launched an unbiased search for cellular HDM2 E3 ligase activities using chromatographic methods. This search identified Skp1-Cullin 1-FBXO22-ROC1 (SCFFBXO22) as the most dominating HDM2 E3 ligase from the HeLa cell proteome. SCF represents a well-characterized class of modular E3 complexes, in which Cullin 1 (CUL1) acts as a scaffold that anchors the Skp1 adaptor protein at the N terminus and the ROC1/RBX1 RING finger protein at the C-terminal half (16, 17). An F-box protein, such as FBXO22, belongs to a family of F-box–containing proteins (∼70 in humans) (18, 19) and assembles into the SCF E3 complex through affinity interactions predominantly with Skp1. The resulting full SCF complex, such as SCFFBXO22, utilizes the F-box protein subunit to bind to a protein substrate, which is positioned for ubiquitin transfer in reactions facilitated by the coordinated actions of the ROC1 RING protein and the ROC1-bound E2 conjugating enzyme (20). Intriguingly, previous studies have implicated roles for SCFβTrCP and SCFFBXO31 in stress/DNA damage-dependent ubiquitination and degradation of MDM2 (21–24).
The present work provides evidence suggesting that SCFFBXO22 targets HDM2 for degradation and that this previously unrecognized proteolytic regulation may have inhibitory effects against breast cancer metastasis.
Results
Identification of SCFFBXO22 as an E3 Ubiquitin Ligase for HDM2 in Vitro.
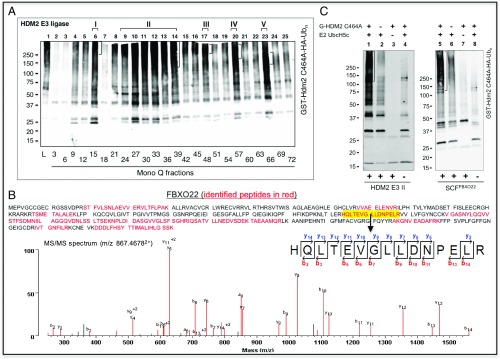
To systematically search for E3 ubiquitin ligase activities capable of directing HDM2 ubiquitination, we developed an unbiased screen of extrinsic E3 enzymes present in human proteome. In this protocol, fractions of HeLa cell extracts derived from chromatographic separation were subjected to ubiquitination reaction using GST-HDM2 C464A as a substrate. The purpose of using this mutant form of HDM2 is twofold. First, MDM2/HDM2 is known to catalyze autoubiquitination in vitro that is mediated by E3’s RING finger domain (12). Such in vitro autoubiquitination activity would severely interfere with our campaign of searching for extrinsic HDM2 E3 activity. The use of GST-HDM2 C464A avoids this obstacle because this mutant was shown previously to impair the RING finger function and hence eliminate HDM2 autoubiquitination in vitro (12). Second, MDM2 C462A (equivalent to HDM2 C464A) was shown to be a substrate of the ubiquitin–proteasome system in vivo (14), suggesting that this mutant can still act as an ubiquitination substrate targeted by an extrinsic E3 enzyme in search. To facilitate E3 identification, HA-tagged ubiquitin was used for this reaction. Following modification, GST-HDM2 C464A and modified substrate were purified through glutathione matrix. Ubiquitin-modified HDM2 was visualized by immunoblots analysis using anti-HA antibody following separation of protein products by denaturing gel electrophoresis. The results revealed the presence of five HDM2 E3 peak activities marked as I–V, with HDM2 E3 II appearing as the most dominating activity (Fig. 1A).
Fig. 1.
Identification of SCFFBXO22 as HDM2 E3 II. (A) HeLa cell proteome contains at least five peaks of HDM2 E3 ligase activity. Aliquots of the indicated Mono-Q fraction (1 µL) were assayed for E3 activity to support the ubiquitination of GST-HDM2 C464A, which is described in detail in SI Appendix, Methods. Lane 1 is the reaction with HeLa cell extract (10 µg). Note that flow-through to Mono-Q was found to contain little E3 activity. (B) Identification of FBXO22 by MS/MS analysis. MS identified 20 unique peptides that cover 44.4% of the FBXO22 sequence (Top). (Bottom) Representative MS/MS spectrum showing a tryptic FBXO22 peptide (m/z 867.46782+) with the sequence identified as HQLTEVGLLDNPELR. (C) SCFFBXO22 supports the ubiquitination of HDM2. HDM2 E3 II (Superose 6 fraction 27, 0.66 µg) or SCFFBXO22 (0.3 pmol; purity shown in SI Appendix, Fig. S5) was assayed for E3 activity to support the ubiquitination of GST-HDM2 C464A.
GST alone yielded barely detectable ubiquitination signal under these conditions (SI Appendix, Fig. S1, lane 4), suggesting specific attachment of ubiquitin chains to HDM2 C464A. Note that HeLa lysates plus E2 UbcH5c supported ubiquitination of GST-HDM2 C464A in the absence of added E1 (SI Appendix, Fig. S1, lane 7), suggesting that HeLa lysates contain E1 at levels sufficient for supporting ubiquitination under the conditions used. In addition, a significant amount of GST-HDM2 C464A ubiquitination products appear to be smaller in size than the full-length GST-HDM2 C464A (Fig. 1A). This finding is most likely a result of the heterogeneous size of purified GST-HDM2 C464A, which appears to be highly sensitive to nonspecific proteolysis that occurs during its expression in bacteria.
To purify HDM2 E3 II, we developed a procedure as diagrammed in SI Appendix, Fig. S2A. As shown in SI Appendix, Fig. S2B, gel filtration by Superose 6 and density sedimentation by glycerol gradient determined the Stokes radius and S value of HDM2 E3 II: 5.833 nm and 6.7S, respectively. These values were applied to the formula of Siegel and Monty (25), yielding HDM2 E3 II’s apparent molecular weight: 164 kDa (SI Appendix, Fig. S2C).
To determine the identity of HDM2 E3 II, the Superpose 6 fraction containing the peak E3 activity was separated by SDS/PAGE, and individually excised protein-containing bands, as visualized in SI Appendix, Fig. S3A, were subjected to mass spectrometry (MS). The results revealed multiple E3 peptides but dominated with those corresponding to CUL1, ROC1/Rbx1, Skp1, and FBXO22 (Fig. 1B and SI Appendix, Fig. S3B), which together form an E3 complex known as SCFFBXO22 (26). In addition, the theoretical molecular weight of SCFFBXO22 is 165 kDa, which matches the experimentally determined molecular weight for HDM2 E3 II (SI Appendix, Fig. S2C). These findings suggest that SCFFBXO22 was a candidate for HDM2 E3 II.
Several lines of evidence supported the hypothesis that SCFFBXO22 is HDM2 E3 II. First, fractionation experiments showed that HDM2 E3 II peaked in Superose 6 fractions 26 and 27 (SI Appendix, Fig. S4, Top), precisely coincidental with the migration of protein peaks corresponding to CUL1, Skp1, and FBXO22, as revealed by immunoblot analyses (SI Appendix, Fig. S4, Bottom). In contrast, the peak of FBXO7, another F-box protein identified by MS (SI Appendix, Fig. S3B), was found enriched in fractions 29–31 that are separated from HDM2 E3 II. FBXO3 and FBXL15, two additional F-box proteins identified by MS (SI Appendix, Fig. S3B), were not detected in Superose 6 fractions. Thus, SCFFBXO22 and HDM2 E3 II are comigrated. Second, SCFFBXO22 was overexpressed and purified by using the baculovirus/insect cell system (SI Appendix, Fig. S5; see ref. 27). When subjected to ubiquitination assays, the reconstituted SCFFBXO22 exhibited HDM2 E3 activity at levels similar to those observed for HDM2 E3 II (Fig. 1C). Thus, SCFFBXO22 is capable of supporting the ubiquitination of HDM2 in vitro.
Evidence for FBXO22 in Targeting Cellular HDM2 for Degradation.
Given that HDM2 overexpression is associated with aggressive cancer malignancy (4–6), we sought to explore the impact of FBXO22 on HDM2 protein stability and biological consequences using triple-negative breast cancer cell lines MDA-MB-231 and BT-549. These cell lines are characterized by lacking expression of estrogen receptor, progesterone receptor, and HER2 (28). In addition, MDA-MB-231 and BT-549 express mutant forms of p53 R280K or R249S (29), respectively, both of which are located in p53’s core DNA-binding domain and are expected to impact DNA binding directly or indirectly (30).
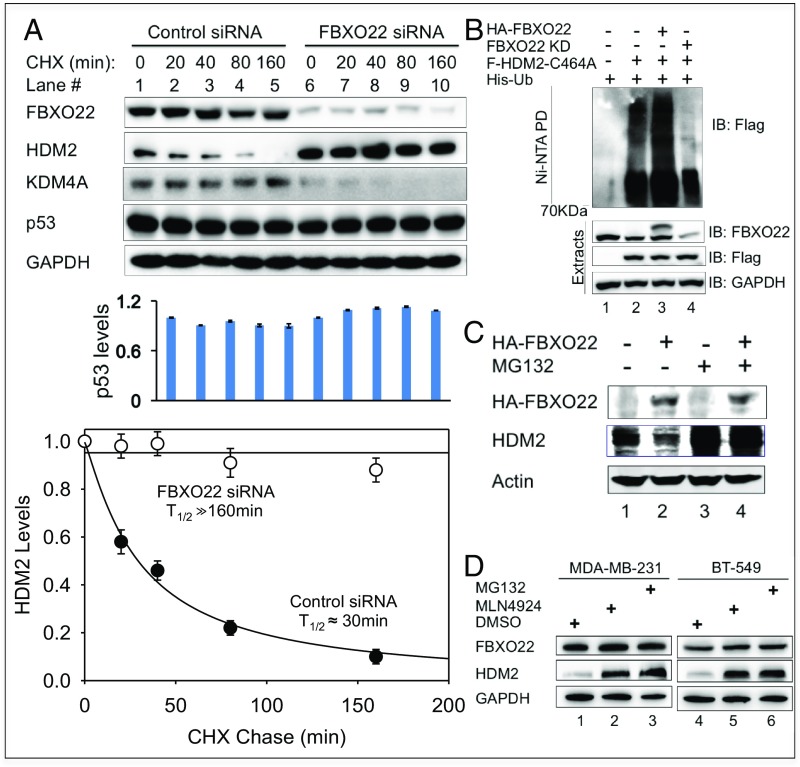
Protein stability is best characterized by its decay rate, known as half-life (t1/2), which is typically determined by chase experiments using cycloheximide that blocks protein synthesis. As shown (Fig. 2A, lanes 1–5 and graph), HDM2’s t1/2 in MDA-MB-231 cells treated with control small interference RNA (siRNA) was ∼30 min, similar to the rate determined previously for HDM2 in a range of tumor cell lines (21, 24). However, FBXO22 knockdown (KD) by a pool of two siRNAs elevated HDM2 levels (Fig. 2A, compare lanes 1 and 6). Importantly, this treatment drastically slowed HDM2’s decay rate, increasing its t1/2 from ∼30 min to far greater than 160 min (Fig. 2A, compare lanes 1–5 and 6–10; graph). Similar effects were observed in MDA-MB-231 cells treated with either siRNA alone (SI Appendix, Fig. S6A), strongly suggesting that the decreased rate of HDM2 degradation by FBXO22 KD is not caused by an off-target effect of one of the siRNAs used. Note that, under the conditions used, FBXO22 KD increased HDM2 mRNA only slightly (∼20%) in MDA-MB-231 cells as measured by real-time PCR experiments (SI Appendix, Fig. S6B).
Fig. 2.
FBXO22 targets cellular HDM2 for degradation. (A) Depletion of FBXO22 results in prolonged t1/2 of HDM2 in MDA-MB-231 cells. Cycloheximide assays were performed to measure HDM2 decay rates in cells treated with control or FBXO22 KD siRNA. Graphs represent an average of three independent experiments (biological replicates), with error bars indicating SD. Graphing and calculation of t1/2 were done by using the scientific data analysis and graphing software SigmaPlot. (B) FBXO22 is critical for cellular ubiquitination of HDM2. MDA-MB-231 cells were transfected with the His-ubiquitin expression vector and various combinations of HA-FBXO22 and Flag-HDM2 C464A in the presence or absence of anti-FBXO22 siRNA as indicated. At 48 h posttransfection, cells were treated with MG132 (10 μM) for 6 h to block protein degradation. His-ubiquitin–modified proteins were isolated and purified under denaturing conditions as described in SI Appendix, Methods. Anti-Flag Western detects Flag-HDM2 C464A modified by His-ubiquitin. (C) Forced expression of FBXO22 decreased the protein level of HDM2 in a manner that depends on proteasomal activity. HeLa cells were transfected with a vector expressing HA-FBXO22, and, before lysis, the cells were treated with MG132 (10 μM) for 6 h. The levels of the indicated proteins were determined by immunoblot analysis. (D) Stabilization of HDM2 by MLN4924. MDA-MB-231 or BT-549 cells were treated with DMSO, MLN4924 (3 µM), or MG132 (10 µM) for 24 h, followed by immunoblot analysis.
It was previously shown that SCFFBXO22 has multiple cellular targets, including Histone Demethylase KDM4A (26) and p53 in methylated forms (31). For comparison, the effects of FBXO22 KD in the stability of p53 or KDM4A in MDA-MB-231 cells were examined by the cycloheximide chase assay as well. As shown (Fig. 2A, lanes 1–5 and bar graph), p53 remained unaltered over the testing time period of 160 min, an observation consistent with a previously reported finding demonstrating that p53 was highly stable in MDA-MB-231 cells (32). FBXO22 KD resulted in only slight increase of p53 (Fig. 2A, compare lanes 1–5 and 6–10; bar graph). On the contrary, the level of KDM4A in MDA-MB-231 cells appeared low and decayed slowly (Fig. 2A, lanes 1–5). For reasons not understood, FBXO22 KD reproducibly decreased the expression of KDM4A (Fig. 2A, compare lanes 1–5 and 6–10). Despite overall low expression of KDM4A that precludes accurate quantification, it is evident that no stabilization of KDM4A was observed in MDA-MB-231 cells depleted of FBXO22. Taken together, these results support the hypothesis that FBXO22 specifically targets cellular HDM2 for degradation in MDA-MB-231 cells. Note that FBXO22 appeared stable (Fig. 2A, lanes 1–5), suggesting that this protein may differ from other F-box family proteins that are short-lived as a result of autocatalytic mechanisms (33, 34).
Similar observations were made with BT-549 cells (SI Appendix, Fig. S7), revealing that FBXO22 KD (i) slowed HDM2 degradation, (ii) produced no changes on the protein levels of p53 (which was highly stable), and (iii) lowered the expression of KDM4A. Real-time PCR experiments showed that FBXO22 KD increased HDM2 mRNA only slightly (∼20%) in BT-549 cells (SI Appendix, Fig. S6B). Note that, under the conditions used, in comparison with MDA-MB-231 (Fig. 2A), BT-549 cells exhibited a slower, but measurable, degradation rate of HDM2 with a t1/2 of ∼245 min (SI Appendix, Fig. S7). Although the reasons for this discrepancy are unclear, it is evident that FBXO22 KD caused near-complete HDM2 stabilization for a period of 240 min (SI Appendix, Fig. S7, graph).
To evaluate cellular ubiquitination, MDA-MB-231 cells were forced to express His-tagged ubiquitin. The impact of FBXO22 overexpression and/or FBXO22 KD was determined by transfection as specified. His-ubiquitin chains attached to Flag-HDM2-C464A were isolated by nickel pull-down (Ni-NTA) under denaturing conditions followed by immunoblot (anti-Flag) analyses. Anti-Flag signal detected in the absence of Flag-HDM2-C464A denoted background/noise levels (Fig. 2B, lane 1). Expression of Flag-HDM2-C464A significantly increased anti-Flag signal (Fig. 2B, compare lanes 1 and 2), which represented ubiquitination signal specific to Flag-HDM2-C464A. Forced expression of HA-FBXO22 increased the total levels of FBXO22 by approximately twofold (Fig. 2B, lane 3, see anti-FBXO22 blot). Elevated FBXO22 resulted in a marked increase of the Flag-HDM2-C464A–specific ubiquitination signal (Fig. 2B, compare lanes 2 and 3). On the contrary, FBXO22 KD reduced HDM2-C464A–specific ubiquitination signal (Fig. 2B, compare lanes 2 and 4). Thus, in line with the results of in vitro experiments (Fig. 1C), FBXO22 targeted cellular HDM2 for ubiquitination.
In addition to the FBXO22 KD approach, overexpression of HA-FBXO22 was found to decrease the level of HDM2 in HeLa cells (Fig. 2C, lanes 1 and 2), in keeping with the role for FBXO22 in targeting HDM2 for degradation. Importantly, the effect of overexpression of FBXO22 on HDM2 reduction was reversed by the treatment of cells with proteasome inhibitor MG132 (Fig. 2C, lanes 3 and 4), suggesting that the FBXO22-driven down-regulation of HDM2 requires the 26S proteasomal activity. Moreover, treatment of MDA-MB-231 or BT-549 cells with MLN4924, an inhibitor of Nedd8 E1 enzyme that blocks E3 SCF and related Cullin-RING ligases (35), resulted in elevation of HDM2 protein levels to the same extent as those with the MG132 treatment (Fig. 2D). These findings are consistent with a role for E3 SCF in the control of HDM2 stability. In all, these results support the hypothesis that SCFFBXO22 targets cellular HDM2 for proteolytic degradation.
In further support of the role of FBXO22 in targeting HDM2 for degradation, FBXO22 KD was found to correlate with accumulation of HDM2 in an additional panel of transformed cell lines that included HeLa (SI Appendix, Fig. S8A), HCT116 p53−/−, HCT116 p53+/+, and MCF-7 cells (SI Appendix, Fig. S8B). Because similar HDM2 accumulation effects by FBXO22 KD were observed in HCT116 p53−/− or HCT116 p53+/+ cells (SI Appendix, Fig. S8B, lanes 1–4), it appears that the observed FBXO22 KD-induced accumulation of HDM2 is a p53-independent effect.
Because MDA-MB-231 and BT-549 cells contain p53 mutations (29) that might impact its stability control by FBXO22, we tested the effects of FBXO22 KD on HCT116 and MCF7 cells that carry a wild-type p53 gene. However, the results showed no alteration in p53 abundance in these cells depleted of FBXO22 (SI Appendix, Fig. S8C). Given that the previously reported degradation effects of FBXO22 on p53 were observed in retinal pigment epithelial (RPE) cells and mouse embryonic fibroblasts, it remains to be determined whether such effects were mediated by cell type/stage-specific signaling that impacts p53 methylation, which is required for degradation by FBXO22 (31). In addition, it is important to bear in mind that the lack of effects on p53 levels by FBXO22 KD in cell lines tested in this study does not mean inaction of SCFFBXO22 in targeting p53. This is because SCFFBXO22 targets both p53 (31) and HDM2 (this work) and this dual activity would have opposite effects on p53 abundance. It can be envisioned that, even though FBXO22 KD acts to increase p53 and HDM2, enhanced HDM2 may offset any p53 increase, resulting in the “no changes in p53 abundance” phenotype observed. Regardless of the precise mechanism, because MDA-MB-231 or BT-549 cells carry mutant p53 gene and showed no alteration in p53 abundance in response to FBXO22 KD, they provided excellent experimental models to examine the biological effects of targeted degradation of HDM2 by FBXO22 independently of the wild-type p53 function (as described later).
Finally, we examined the impact of FBXO22 KD in HDM2 levels during progression of the cell cycle. It was previously shown that FBXO22 KD led to cell cycle arrest at G1 phase (31). It was therefore possible that the observed HDM2 accumulation in FBXO22 KD cells was caused by an indirect cell cycle effect. FBXO22 KD cells progressed more slowly than control in MDA-MB-231 or BT-549 cells, with significantly more G1 populations at all time points examined (SI Appendix, Fig. S9 A and B, Top). This finding is in keeping with a previously reported effect of FBXO22 deletion in causing G1 arrest (31). However, HDM2 levels did not change significantly in MDA-MB-231 or BT-549 cells (SI Appendix, Fig. S9 A and B, Middle, lanes 1–6), which differed from HeLa cells, in which HDM2 was shown to vary during the cell cycle (21). Nevertheless, at all time points examined, FBXO22 KD in MDA-MB-231 or BT-549 cells increased HDM2 levels by approximately twofold (SI Appendix, Fig. S9 A and B, Middle, compare lanes 1–6 and 7–12; Bottom, bar graph for quantification). These data suggest an ability of FBXO22 to target HDM2 for degradation in a manner independent of cell cycle progression.
Contributions of Multiple E3 Ligases to the Control of HDM2 Stability.
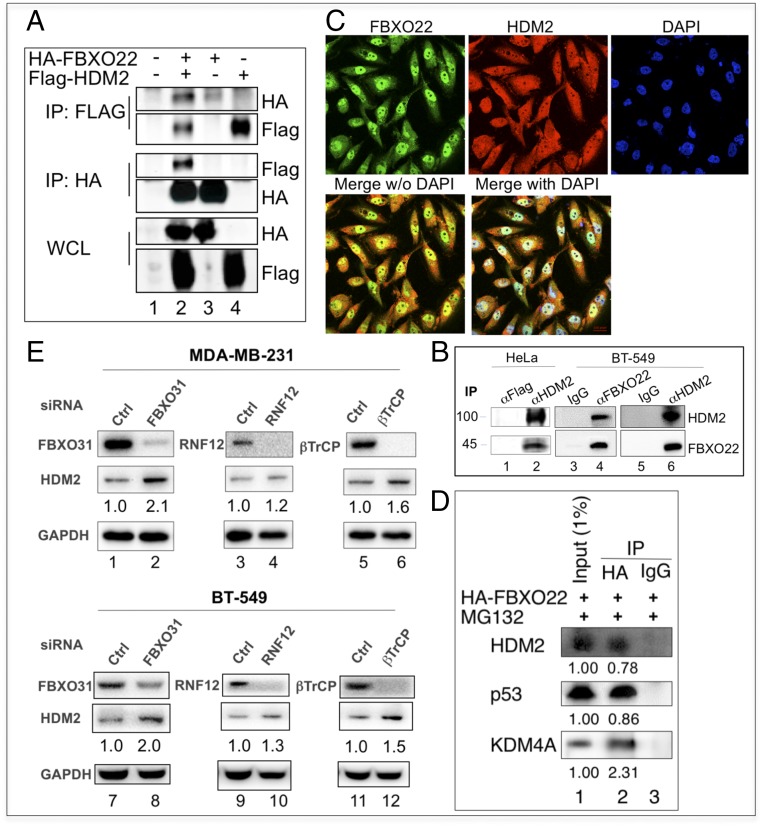
We examined the interactions between FBXO22 and HDM2, as well as influences by previously determined FBXO22 additional targets and HDM2 E3s. To probe FBXO22–HDM2 interactions, reciprocal coimmunoprecipitation experiments were carried out, and the results showed that the recombinant forms of FBXO22 and HDM2 interacted in HEK293T cells (Fig. 3A). In addition, FBXO22 and HDM2 interacted at the endogenous levels as revealed by immunoprecipitation experiments with extracts from HeLa or BT-549 cells (Fig. 3B). The subcellular location of FBXO22 and HDM2 in MDA-MB-231 cells was determined by immunofluorescence (Fig. 3C). FBXO22 was predominantly located to nucleus, although cytoplasmic signals were clearly visible. HDM2 was both nuclear and cytoplasmic perhaps because of constant nucleocytoplasmic shuttling (36). Merged staining revealed colocalization between fractions of FBXO22 and HDM2. These results are consistent with the hypothesis that FBXO22 targets cellular HDM2 for degradation.
Fig. 3.
Contributions of multiple E3 ligases to the control of HDM2 stability. (A and B) FBXO22 interacts with HDM2 in cells. (A) HEK293T cells were transfected with expression vectors as indicated. Extract proteins (3 mg) were used for immunoprecipitation. (B) HeLa or BT-549 cells were treated with MG132 (10 μM) for 6 h before lysis. Extract proteins (10 mg) were used for immunoprecipitation. (C) Immunofluorescence. MDA-MB-231 cells were used for immunofluorescence analysis as described in SI Appendix, Methods. (D) FBXO22 interacts with HDM2, p53, and KDM4A. MDA-MB-231 cells were transfected with a vector expressing HA-FBXO22, and the cells were treated with MG132 (10 μM) for 6 h before lysis. Extract proteins (5 mg) were used for immunoprecipitation as described in SI Appendix, Methods. (E) Effects of various E3 KDs in a panel of tumor cell lines. Immunoblot analysis of the relative protein level of FBXO22 and HDM2 in indicated cell lines treated with siRNA against various E3s. HDM2 quantification is shown below each blot image.
To compare the ability of FBXO22 to interact with HDM2 or other previously reported targets including p53 and KDM4A, extracts from MDA-MB-231 cells expressing HA-FBXO22 were immunoprecipitated by using anti-HA antibodies followed by immunoblot analysis. The results confirmed the ability of FBXO22 to interact with HDM2, p53, or KDM4A (Fig. 3D). Quantification of the bound target protein against input revealed >2% of KDM4A recovered in the HA-FBXO22 immunoprecipitates. p53 and HDM2 bound to HA-FBXO22 with similar efficiency at ∼0.8%.
Finally, we tested and compared the effects of several previously reported HDM2 E3s that include FBXO31 (24), RNF12 (37), or βTrCP (21). As shown (Fig. 3E), depletion of any of these E3s (or E3 components) led to an increase of HDM2 in MDA-MB-231 or BT-549 cells, albeit at varying degrees. On average, the observed ratio of HDM2 in FBXO22 KD vs. control in MDA-MB-231 and BT-549 cells is ∼2:1 (SI Appendix, Fig. S9). By comparison, the average ratios of HDM2 in E3 KD vs. control in MDA-MB-231 and BT-549 cells are 2:1, 1.6:1, or 1.3:1 for FBXO31 KD, βTrCP KD, or RNF12 KD, respectively (Fig. 3E). These results suggest that multiple E3 ligases contributed to the control of HDM2 stability. At least in MDA-MB-231 and BT-549 breast cancer cells, it appears that FBXO22 and FBXO31 play a more dominant role in directing HDM2 for degradation.
FBXO22 Inhibits Migration and Invasion of Human Breast Cancer Cells in Vitro.
MDA-MB-231 and BT-549 belong to the mesenchymal-like subtypes characterized by high levels of invasiveness (28). The ability of these cancer cells to migrate or invade can be quantified by Boyden chamber assay. The cell mobility can be measured by penetration through a membrane inserted between upper and lower chambers. On the contrary, the cell invasiveness can be detected by penetration through extracellular matrix-loaded membrane, which requires protein degradation by metalloproteinases.
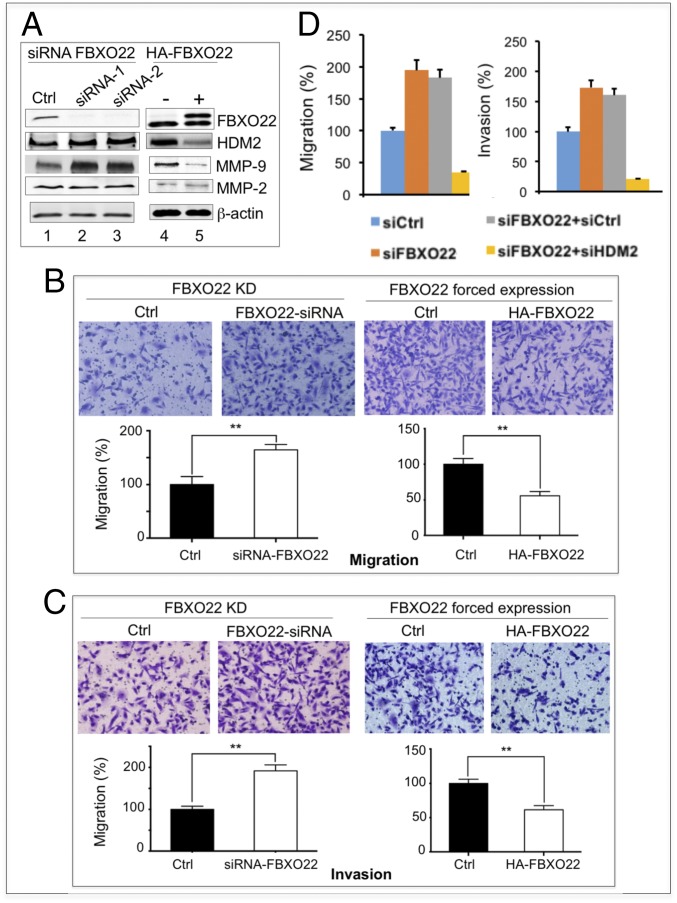
We determined the effects of FBXO22 on cell migration and invasion in MDA-MB-231 cells by silencing FBXO22 using siRNA. As revealed by immunoblot analyses (Fig. 4A, lanes 1–3), FBXO22 KD was correlated with increase of HDM2 and MMP-9. No significant effect on MMP-2 was observed. Under this condition, FBXO22 KD increased the ability of MDA-MB-231 cells to migrate (Fig. 4B, Left) and invade (Fig. 4C, Left) by approximately twofold. On the contrary, modest overexpression of FBXO22 by no more than twofold decreased HDM2 and MMP-9 (Fig. 4A, lanes 4–5). As a consequence, the ability of MDA-MB-231 cells to migrate (Fig. 4B, Right) or invade (Fig. 4C, Right) was reduced by approximately twofold. Nearly identical observations were made with BT-549 cells (SI Appendix, Fig. S10). Note that, under the conditions used, FBXO22 KD decreased proliferation and clonogenic survival of MDA-MB-231 or BT-549 cells only slightly (SI Appendix, Fig. S11). Thus, FBXO22 appears to predominantly inhibit the invasive potential in at least a subset of breast cancer cell lines.
Fig. 4.
FBXO22 inhibits migration and invasion of breast cancer cells. MDA-MB-231 cells were treated with siRNA against FBXO22 or transfected with vector that expresses HA-FBXO22. (A) Immunoblot analyses confirmed FBXO22 depletion or overexpression. The levels of HDM2, MMP-9, and MMP-2 are also shown. The siRNA-treated or HA-FBXO22–expressing cells were examined for changes in migration (B) or invasion (C). Data are shown as means ± SD (three biological replicates; **P < 0.01). (D) Effects of FBXO22/HDM2 double KD on cell migration and invasion. FBXO22/HDM2 double KD suppresses the ability of FBXO22 KD cells to promote cell migration and invasion in MBA-MD-231 cells. Quantification is shown. Data are shown as means ± SD (three biological replicates). Western blots and images of cell migration or invasion are shown in SI Appendix, Fig. S12.
To confirm that the FBXO22 KD-driven effects on cell migration/invasion was caused by the accumulation of HDM2, we attempted to rescue the FBXO22 KD effect by simultaneous depletion of HDM2 (double-KD approach) in MDA-MB-231 or BT-549 cells. Immunoblot analysis confirmed that, whereas FBXO22 KD increased HDM2 (SI Appendix, Fig. S12A, lanes 2 and 6), FBXO22/HDM2 double KD diminished such increase and restored the HDM2 level to that observed in the control cells (SI Appendix, Fig. S12A, lanes 1 and 4 and lanes 5 and 8). In agreement with previous observations (Fig. 4 A–C and SI Appendix, Fig. S10), FBXO22 KD caused increase of migration or invasion by approximately twofold in MDA-MB-231 (Fig. 4D) and BT-549 cells (SI Appendix, Fig. S12). However, FBXO22/HDM2 double KD inhibited migration or invasion in MDA-MB-231 (Fig. 4D) or BT-549 cells (SI Appendix, Fig. S12) to levels even lower than those seen in the control cells. Thus, HDM2 KD was able to rescue the phenotype on cell migration/invasion caused by FBXO22 KD. These findings strongly suggest that it was the increased level of HDM2 (as a result of FBXO22 KD) that promotes cell migration and invasion.
MMP-9 is a transcriptional target of MDM2/HDM2 (11). To further determine whether the increased levels of MMP-9 in FBXO22 KD cells are enzymatically active, we performed gelatin zymography assay. As shown in SI Appendix, Fig. S13A, MMP-9 activity (but not MMP-2 activity) was increased in MDA-MB-231 or BT-549 cells depleted of FBXO22, coincident with the elevated MMP-9 protein levels (Fig. 4A and SI Appendix, Fig. S10A). Conversely, forced expression of HA-FBXO22 led to decreased MMP-9 activity in the same breast cancer cell lines tested (SI Appendix, Fig. S13B). In addition, previous studies showed that HDM2/MDM2 acted as an E3 to target E-cadherin for degradation (8). It was observed that E-cadherin was decreased in MDA-MB-231 cells depleted of FBXO22 (SI Appendix, Fig. S13C), perhaps as a result of HDM2 accumulation. Taken together, these findings suggest that FBXO22 is a negative regulator of breast cancer cell invasion in a manner that is correlated with its targeting activity on HDM2.
FBXO22 Down-Regulation Enhances Breast Cancer Cell Metastasis in Vivo.
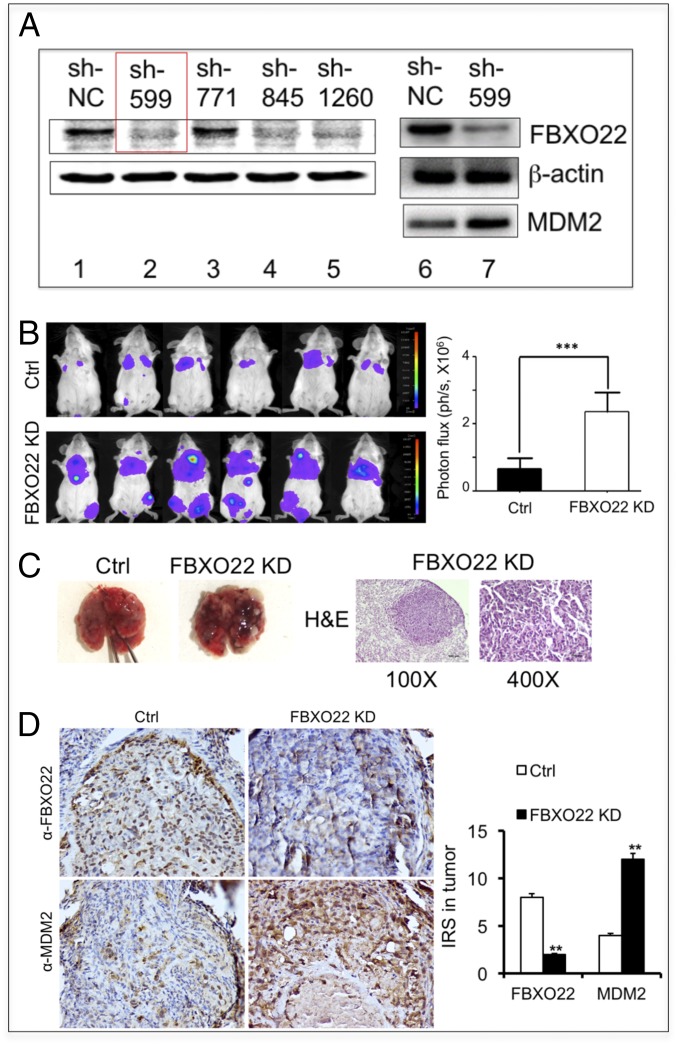
We next examined the role of FBXO22 in breast cancer metastasis using the mouse 4T1 breast tumor model. The 4T1 mammary carcinoma is p53-null (38) and is a transplantable tumor cell line that is highly tumorigenic and invasive. 4T1 tumor cells can spontaneously metastasize from the primary tumor site to multiple distant sites, including lung. To this end, we created lentivirus expressing shRNA that targets FBXO22, or control shRNA, thereby generating 4T1-luc-FBXO22 KD or 4T1-luc-Ctrl cell lines, respectively. A luciferase reporter gene (luc) is included to allow imaging the transplanted 4T1 tumor cells. Fig. 5A shows the results of immunoblot analysis that confirmed depletion of FBXO22 by three of four shRNAs used (sh-599, sh-845, and sh-1260; Fig. 5A, lanes 1–5). As expected, the MDM2 level was increased in 4T1 cells infected with the virus expressing shRNA sh-599 that targets FBXO22 (Fig. 5A, lanes 6–7).
Fig. 5.
FBXO22 down-regulation enhances breast cancer cell metastasis in vivo. (A) Immunoblot analysis confirmed depletion of FBXO22 in the 4T1-luc-FBXO22 KD cell lines (constructed as described in SI Appendix, Methods). (B) FBXO22 down-regulation enhances breast cancer cell metastasis. Tail-vein metastasis assay was performed as described in SI Appendix, Methods. (B) Images of luminescent signals expressed by the transplanted 4T1 cells carrying control shRNA or shRNA-599 against FBXO22. There is a significant increase of luminescent signals in the lung area of mice injected with the 4T1-luc-FBXO22 KD cells in comparison with control. Bar graphs show quantification of results, expressed as means ± SD (n = 6 mice for each group; ***P < 0.001). (C) Close-up examination of the metastatic nodules in the lung. (Left) Representative images of metastatic nodules from the control or FBXO22 KD group. (Right) H&E staining sections of the lung derived from the FBXO22 KD mouse. Detailed procedures are described in SI Appendix, Methods. Original magnifications are ×100 and ×400. (D) Immunohistochemistry shows low FBXO22 expression, but high HDM2 expression, in lung tumors of the FBXO22 KD group.
The BALB/c nude mice were injected via tail vein with 4T1-luc-FBXO22 KD (sh-599) and 4T1-luc-Ctrl cells, respectively. Bioluminescent imaging was performed regularly to monitor tumor burden in vivo, followed by end-of-study necropsy and histopathological analysis of the lungs. At 1 mo postinjection, the 4T1-luc-FBXO22 KD and control groups displayed luminescent signal in the lung area (Fig. 5B), indicative of metastasis of 4T1 cells that express the luciferase reporter gene. However, the level of luminescent signals observed in the 4T1-luc-FBXO22 KD group were significantly (threefold) higher than those seen in the control group (Fig. 5B). Moreover, visual inspection of the lungs retrieved from the experimental mice detected significantly higher number of metastatic nodules in the 4T1-luc-FBXO22 KD group than in the control (Fig. 5C, Left). Close-up examination of the 4T1-luc-FBXO22 KD lung nodules by hematoxylin and eosin (H&E) stain confirmed tumor cell masses (Fig. 5C, Right). Finally, immunohistochemistry analysis of the lungs derived from mice showed significant decrease of FBXO22, but elevated levels of HDM2, in tumor samples (Fig. 5D). These results revealed correlation between enhanced metastasis with down-regulation of FBXO22 and increased levels of HDM2. In all, these findings suggest that FBXO22 down-regulation significantly enhances breast cancer cell metastasis.
Low FBXO22 Expression Is Correlated with Worse Survival in Human Breast Cancer.
To explore the potential role of FBXO22 in human breast cancer, we examined the relationship between FBXO22 expression and survival rates as well as other clinicopathologic parameters in breast cancer patients. For this purpose, we performed immunohistochemistry experiments to profile FBXO22 expression across an entire tissue microarray (TMA) that contains a total of 410 human primary breast carcinoma samples (SI Appendix, Methods). The resulting FBXO22 staining intensity was quantified by immunoreactive score (IRS). Samples with IRS in the range of 0–3 or 4–12 were classified as low or high expression of FBXO22, respectively. SI Appendix, Table S1 shows the relationship between FBXO22 expression and clinicopathologic parameters. Overall, low or high FBXO22 expression accounted for 44.9% or 55.1% of the 410 tumor samples analyzed, respectively. FBXO22 signal was dramatically decreased in histology grade III in comparison with histology grades I and II (P = 0.011, χ2 test). In addition, low FBXO22 expression was significantly correlated with lymph node metastasis (P = 0.001, χ2 test). In contrast, there was no significant correlation between FBXO22 expression and other clinicopathologic variables, including patient age, tumor size, ER status, PR status, HER2 status, or p53 status.
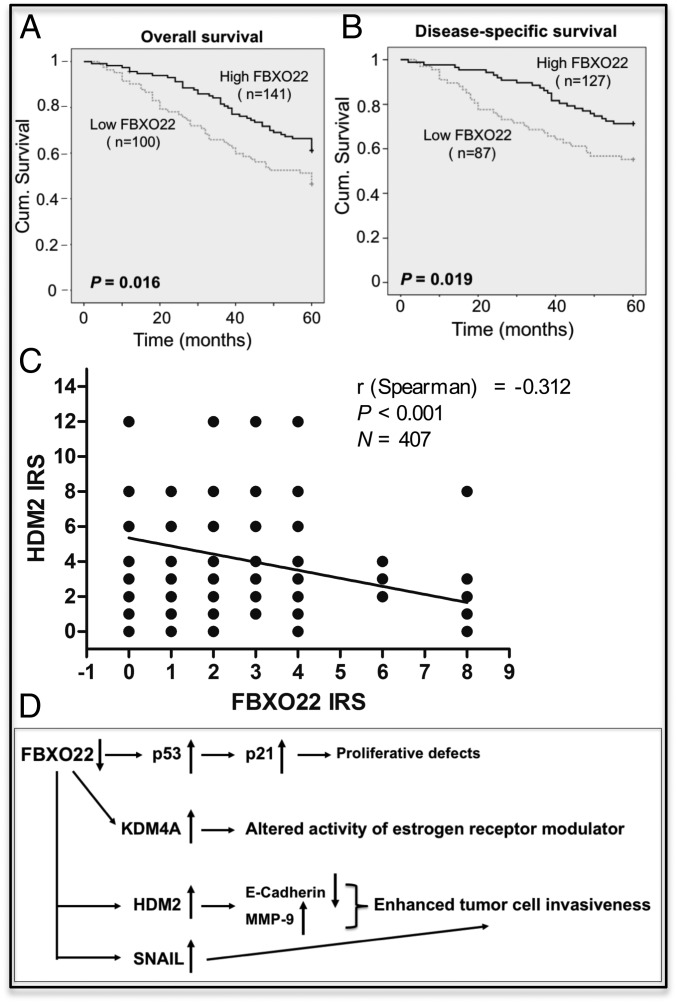
Fig. 6 A and B show the Kaplan–Meier survival curves that were constructed based on the data on FBXO22 staining with human breast cancer samples. Note that, of the 410 patient specimens collected, 241 samples have available clinical follow-up data for 5 y. As shown in Fig. 6A, the 5-y overall cumulative survival rate decreased from 61.4% in patients with high FBXO22 expression to 46.3% in those with low FBXO22 expression. In addition, the 5-y disease-specific cumulative survival rate decreased from 71.3% in patients with high FBXO22 expression to 55.2% in those with low FBXO22 expression (Fig. 6B). Thus, low FBXO22 expression appears to correlate with worse overall and disease-specific patient survival rates.
Fig. 6.
(A and B) Low FBXO22 expression is correlated with worse breast cancer survival. FBXO22 expression was examined in a total of 410 human breast carcinoma tissues, and statistical analysis of the relationship between FBXO22 expression and survival was performed as described in SI Appendix, Methods. Kaplan–Meier curves are shown to correlate low FBXO22 expression with worse 5-y overall (A) and disease-specific (B) survival in breast cancer patients. Cum., cumulative. (C) Low FBXO22 expression is correlated with high HDM2 expression in human breast cancer. A total of 407 human breast cancer samples were analyzed for expression levels of FBXO22 and HDM2, which were quantified and expressed by IRS as described in SI Appendix, Methods. Graph shows inverse correlation between expression of FBXO22 and HDM2 in 407 human breast cancer samples. (D) Summary of proposed SCFFBXO22 biological roles. A diagram is presented to summarize the biological roles and pathways impacted by SCFFBXO22 based on data shown in this work and in the literature. The Discussion includes a detailed description.
Moreover, univariate Cox regression analyses revealed that FBXO22 expression was an independent prognostic marker for breast cancer patient overall survival (hazard ratio, 0.604; 95% CI, 0.398–0.918; P = 0.018; SI Appendix, Table S2) and disease-specific survival (hazard ratio, 0.536; 95% CI, 0.315–0.912; P = 0.021; SI Appendix, Table S2). In multivariate Cox regression analysis, we found that FBXO22 expression was also an independent prognostic marker for 5-y overall survival (hazard ratio, 0.596; 95% CI, 0.373–0.952; P = 0.030; SI Appendix, Table S3) and disease-specific survival (hazard ratio, 0.495; 95% CI, 0.327–0.681; P = 0.018; SI Appendix, Table S3). Thus, low FBXO22 expression is associated with poor prognosis, suggesting this F-box protein as a prognostic marker for breast cancer.
To determine the relationship between the expression of FBXO22 and HDM2 in human breast cancer, 407 breast cancer tissue samples were analyzed by immunohistochemistry. The results were quantified, and the graph showed inverse correlation between FBXO22 and HDM2, with a negative coefficient value of −0.312 (Fig. 6C).
Discussion
SCFFBXO22 Acts as an E3 Ubiquitin Ligase for HDM2.
This study presents multiple lines of biochemical evidence strongly suggesting that SCFFBXO22 acts as an E3 ligase for HDM2. First, unbiased chromatographic studies coupled with MS have identified SCFFBXO22 as the most dominating E3 ligase activity from the HeLa cell proteome that directs ubiquitination of the unmodified form of HDM2 (Fig. 1 A and B and SI Appendix, Figs. S2–S4). Second, purified recombinant SCFFBXO22 supports HDM2 ubiquitination in vitro (Fig. 1C), and FBXO22 positively contributes to HDM2 cellular ubiquitination (Fig. 2B). Third, immunoprecipitation experiments revealed that recombinant and endogenous forms of FBXO22 and HDM2 interact in cells (Fig. 3 A and B). Fourth, silencing FBXO22 expression by siRNA prolonged the half-life of HDM2 in MDA-MB-231 and BT-549 breast cancer cells (Fig. 2A and SI Appendix, Fig. S6A and S7). This increase of HDM2 driven by FBXO22 depletion was observed in all stages of the cell cycle (SI Appendix, Fig. S9). Fifth, FBXO22 overexpression decreases HDM2 abundance in a proteasomal-dependent manner (Fig. 2C). Sixth, blocking E3 SCF and Cullin-RING ligase activity by inhibitor MLN4924 causes accumulation of HDM2 (Fig. 2D). Finally, FBXO22 KD did not cause significant change in HDM2 mRNA (SI Appendix, Fig. S6B), but increased HDM2 protein levels in HCT116 cells with or without p53 (SI Appendix, Fig. S8B). These findings eliminate the possibility that the observed HDM2 increase is a result of elevated gene expression driven by p53. Taken together, SCFFBXO22 and HDM2 meet the criteria for the E3–substrate relationship.
SCFFBXO22 has multiple cellular targets, including Histone Demethylase KDM4A (26), p53 in methylated forms (31), KLF transcription factor 4 (39), and SNAIL (40). Indeed, results of immunoprecipitation experiments confirm the ability of FBXO22 to bind KDM4A, p53, and HDM2 with comparable efficiency (Fig. 3D). However, in four cancer cell lines tested (MDA-MB-231, BT-549, HCT116, and MCF7), FBXO22 KD did not result in significant changes in p53 abundance (Fig. 2A and SI Appendix, Figs. S7A, S8C, and S9). The lack of effect on p53 abundance by FBXO22 KD could be the result of balanced acts between SCFFBXO22, HDM2, and p53. Conceivably, enhanced HDM2 may offset any p53 increase in cells depleted of FBXO22. Additionally, given the requirement of methylation on p53 for FBXO22-mediated targeting (31), the effect on p53 abundance by FBXO22 KD may be strongly influenced by cell types or developmental stages that might vary significantly in cellular p53 methylation.
In addition to SCFFBXO22 (this work), three MDM2/HDM2 E3 ligases have been reported. SCFβTrCP was shown to act as an E3 to mediate ubiquitin-dependent degradation of MDM2 in response to DNA damage and in a manner that requires phosphorylation of MDM2 by casein kinase I (21). SCFFBXO31 was found to interact with the phosphorylated from of MDM2 by ATM and to direct its degradation in response to genotoxic stress (24). Finally, RNF12 was identified as a MDM2-interacting protein and was shown to direct ubiquitin-dependent degradation of MDM2 (37). In agreement with these reports, we observed that depletion of FBXO31, βTrCP, or RNF12 led to an increase of HDM2 in MDA-MB-231 or BT-549 cells, albeit to varying degrees (Fig. 3E). Notably, depletion of SCFβTrCP, SCFFBXO31, or RNF12 results in MDM2/HDM2 increase that diminishes p53 level and activity (21, 24, 37). In contrast, at least in a subset of cancer cells, FBXO22 KD elevates HDM2 without affecting the p53 level (Fig. 2A and SI Appendix, Figs. S7A, S8C, and S9). This unique property of FBXO22 provides an opportunity to investigate the ability of MDM2/HDM2 overexpression to promote tumorigenesis via p53-independent mechanisms (Discussion and Figs. 4 and 5). Moreover, whereas SCFβTrCP and SCFFBXO31 act to target HDM2/MDM2 for degradation in response to DNA damage (21, 24), SCFFBXO22 is capable of mediating the ubiquitination of the unmodified form of HDM2 (Fig. 1C) and supporting cellular HDM2 ubiquitination (Fig. 2B) and degradation (Fig. 2C) in the absence of DNA damage agents.
Despite efforts in the identification of extrinsic E3s to target HDM2/MDM2 (refs. 21, 24 and 37; this work), it remains to be determined how HDM2/MDM2 mediates specific interactions with various E3s. Future in-depth degron mapping is critical to understand molecular recognition and to explore therapeutic potential by altering HDM2 stability in a manner that benefits human health.
Role of FBXO22 in Inhibiting Breast Cancer Metastasis.
This work has revealed a role for FBXO22 in suppressing breast cancer metastasis. We observed in at least two human breast cancer cell lines that, whereas KD of FBXO22 increased the cell’s ability to migrate and invade, overexpression decreased these invasive potentials (Fig. 4 and SI Appendix, Fig. S10). Double-KD experiments confirmed the role of HDM2 as a dominant factor that drives cell migration and invasion (Fig. 4D and SI Appendix, Fig. S12). Importantly, the results in mouse 4T1 breast tumor model studies revealed that KD of FBXO22 led to a significant increase of breast tumor cell metastasis to the lung (Fig. 5). Finally, statistical studies showed that low FBXO22 expression is correlated with worse 5-y survival (Fig. 6 A and B and SI Appendix, Table S1) and high HDM2 expression (Fig. 6C) in human breast cancer. FBXO22 expression was an independent prognostic marker for this disease (SI Appendix, Tables S2 and S3). While the present manuscript was in preparation, a study by Sun et al. (40) was published that showed that FBXO22 suppresses cell mobility and invasiveness in vitro and metastatic lung colonization in vivo. These findings on metastasis suppression by FBXO22 are consistent with observations presented in the present work. In addition, the correlation of low FBXO22 expression with poor prognosis in human breast cancer patients revealed in the present work (Fig. 6 A and B and SI Appendix, Tables S1 and S3) was supported by recent studies from Sun et al. (40) and Johmura et al. (41).
HDM2 overexpression has been linked to tumor invasiveness with a poor patient prognosis (4–6). In this context, Yang et al. (8) have shown that MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. In addition, correlations were observed between HDM2 overexpression and enhanced MMP-9 protein in clinical samples (42). Chen et al. (11) have provided evidence demonstrating that overexpression of MDM2 increased MMP-9 expression primarily at the transcriptional levels by mechanisms yet to be determined. In breast cancer cells examined in the present work, altered FBXO22 abundance by siRNA or overexpression was shown to change the protein levels of HDM2 in an inverse manner (Fig. 4A). Note that the FBXO22-driven changes of HDM2 were accompanied by alterations in the protein levels of MMP-9 and E-cadherin in a manner that is consistent with them being HDM2’s transcriptional and proteolytic targets, respectively (Fig. 4A and SI Appendix, Figs. S10A and S13C). These results suggest that enhanced MMP-9 and decreased E-cadherin, as a result of HDM2 increase caused by FBXO22 KD, are the possible factors that mediate the observed increase in cell migration and invasion (Fig. 4 and SI Appendix, Fig. S10).
The FBXO22-knockout mouse has been created by CRISPR-cas9 technique and exhibits severe growth defects (31). Marked accumulation of p53, p21, and MDM2 was found in multiple FBXO22−/− mouse tissues, including liver, thymus, spleen, heart, kidney, and brain (31). However, the accumulation of MDM2 can be contributed to by p53-driven transcriptional effect and loss of FBXO22 targeting activity (this work). In addition, the potential role in cancer metastasis has not been examined in FBXO22−/− mice. It should be noted that Johmura et al. (31) have observed proliferative defects in immortalized retina pigmented epithelial cells depleted of FBXO22 and in FBXO22−/− mouse epithelial fibroblasts. Similar growth defects were reported in human liver cancer HepG2 cells treated with shRNA against FBXO22 (39). In addition, overexpression of FBXO22 accelerated cell proliferation and colony formation in a number of breast cancer cells (40). However, we observed only small inhibitory effects on growth and clonogenic survival in cancer lines MDA-MB-231 and BT-549 by silencing FBXO22 (SI Appendix, Fig. S11). Instead, these treated cells showed a marked increase in cell migration and invasion (Fig. 4 and SI Appendix, Fig. S10). Sun et al. (40) suggest that FBXO22 plays a dual role in mammary tumorigenesis and metastasis, with its effect on metastasis, but not primary cancer, as the major determinant of the mortality of breast cancer patients.
Thus, it appears that FBXO22 has multiple biological functions as a result of its ability to target many cellular proteins. Fig. 6D summarizes these activities and provides a perspective. FBXO22 clearly has a role in growth control because the FBXO22−/− mouse showed severe proliferative defects (31). It is suggested that the loss of FBXO22 leads to p53 increase and hence p21 accumulation, resulting in G1 arrest and proliferative inhibition. However, in at least a subset of breast cancer cells, high levels of HDM2, as a result of FBXO22 KD, promote cell invasion (Fig. 4 and SI Appendix, Fig. S10). This effect is possibly mediated by enhanced activity of MMP-9 (Fig. 4A and SI Appendix, Fig. S10A and S13A) and E-cadherin reduction (SI Appendix, Fig. S13C). In addition, FBXO22-targeted degradation of SNAIL (40) may contribute to the regulation of tumor cell invasion as well. The increased invasiveness likely contributes to elevated levels of tumor metastasis observed in the mouse 4T1 breast tumor model study (Fig. 5). Finally, the effects by FBXO22 on breast cancer could be mediated by targeted degradation of KDM4A (26), which was shown to modulate estrogen receptor modulator activity (41).
Previous mouse genetics experiments strongly suggest that MDM2 overexpression can promote tumorigenesis via p53-independent mechanisms because elevated levels of MDM2 increased sarcomas in p53-null background (43). In addition, Cordon-Cardo et al. (44) have noted a significant proportion of tumor samples containing overexpressed HDM2 and p53 mutations. Intriguingly, the present study detected correlations between low FBXO22 expression and worse breast cancer survival and poor prognosis (Fig. 6 A and B and SI Appendix, Tables S1–S3). Moreover, low FBXO22 expression is correlated with high HDM2 expression in human breast cancer (Fig. 6C). It thus raises a question of whether FBXO22 abundance can be developed into a predictive biomarker for the prognosis of a subset of human tumors characterized by HDM2 overexpression. Additionally, increasing targeted degradation of HDM2 by SCFFBXO22 might be a viable strategy for down-regulating HDM2. It remains to be determined how FBXO22 is down-regulated in human breast cancers. Future work is required to assess whether this low expression is the consequence of transcriptional, translational, and/or posttranslational effects.
Methods
SI Appendix, Methods describes in detail the experimental procedures used in this study, including DNA plasmids, protein substrate purification, cell lines and animals used, isolation and identification of HDM2 E3 peak II, in vitro ubiquitination, siRNA transfection, DNA transfection and extract preparation, immunoprecipitation, immunoblot, cycloheximide chase, in vivo ubiquitination, immunofluorescence, cell migration and invasion, lentiviral shRNA stable cell line, tail-vein assay of metastasis, immunohistochemistry, patient specimens and TMA, TMA immunohistochemistry, and statistical analysis.
Supplementary Material
Acknowledgments
We thank Dr. Yanping Zhang of the University of North Carolina for his strong support of this work including project initiation, reagents, and advice; and Dr. Serge Fuchs for reagents and advice on ubiquitination analysis. This work was supported by National Natural Science Foundation of China Grant 81572710; Jiangsu Province (China) Grant “Project of Invigorating Health Care through Science, Technology and Education”; National Natural Science Foundation of China Grants 81672845 and 8187304 (to J.B.); and NIH Grants 5R01GM074830-07 (to L.H.), GM61051 (to Z-Q.P.), and GM122751 (to Z-Q.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820990116/-/DCSupplemental.
References
- 1.Manfredi J. J., The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 24, 1580–1589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haupt Y., Maya R., Kazaz A., Oren M., Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Honda R., Tanaka H., Yasuda H., Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Bohlman S., Manfredi J. J., p53-independent effects of Mdm2. Subcell. Biochem. 85, 235–246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leach F. S., et al. , p53 mutation and MDM2 amplification in human soft tissue sarcomas. Cancer Res. 53(10, Suppl)2231–2234 (1993). [PubMed] [Google Scholar]
- 6.Momand J., Jung D., Wilczynski S., Niland J., The MDM2 gene amplification database. Nucleic Acids Res. 26, 3453–3459 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathew R., et al. , Alterations in p53 and pRb pathways and their prognostic significance in oesophageal cancer. Eur. J. Cancer 38, 832–841 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Yang J.-Y., et al. , MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol. Cell. Biol. 26, 7269–7282 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polański R., et al. , MDM2 promotes cell motility and invasiveness through a RING-finger independent mechanism. FEBS Lett. 584, 4695–4702 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Stamenkovic I., Matrix metalloproteinases in tumor invasion and metastasis. Semin. Cancer Biol. 10, 415–433 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Chen X., et al. , MDM2 promotes invasion and metastasis in invasive ductal breast carcinoma by inducing matrix metalloproteinase-9. PLoS One 8, e78794 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., Weissman A. M., Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275, 8945–8951 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Honda R., Yasuda H., Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene 19, 1473–1476 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Itahana K., et al. , Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell 12, 355–366 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Tollini L. A., Jin A., Park J., Zhang Y., Regulation of p53 by Mdm2 E3 ligase function is dispensable in embryogenesis and development, but essential in response to DNA damage. Cancer Cell 26, 235–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petroski M. D., Deshaies R. J., Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Sarikas A., Hartmann T., Pan Z. Q., The cullin protein family. Genome Biol. 12, 220 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skaar J. R., D’Angiolella V., Pagan J. K., Pagano M., SnapShot: F box proteins II. Cell 137, 1358.e1–1358.e2 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Skaar J. R., Pagan J. K., Pagano M., SnapShot: F box proteins I. Cell 137, 1160–1160.e1 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Duda D. M., et al. , Structural regulation of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 21, 257–264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inuzuka H., et al. , Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell 18, 147–159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., et al. , DNA damage-induced activation of ATM promotes β-TRCP-mediated Mdm2 ubiquitination and destruction. Oncotarget 3, 1026–1035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt M. L., Calvisi D. F., Clark G. J., NORE1A regulates MDM2 via β-TrCP. Cancers (Basel) 8, E39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malonia S. K., Dutta P., Santra M. K., Green M. R., F-box protein FBXO31 directs degradation of MDM2 to facilitate p53-mediated growth arrest following genotoxic stress. Proc. Natl. Acad. Sci. USA 112, 8632–8637 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegel L. M., Monty K. J., Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta 112, 346–362 (1966). [DOI] [PubMed] [Google Scholar]
- 26.Tan M. K., Lim H. J., Harper J. W., SCF(FBXO22) regulates histone H3 lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation. Mol. Cell. Biol. 31, 3687–3699 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu K., et al. , DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J. Biol. Chem. 278, 28882–28891 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Lehmann B. D., et al. , Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Runnebaum I. B., Nagarajan M., Bowman M., Soto D., Sukumar S., Mutations in p53 as potential molecular markers for human breast cancer. Proc. Natl. Acad. Sci. USA 88, 10657–10661 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joerger A. C., Fersht A. R., Structure-function-rescue: The diverse nature of common p53 cancer mutants. Oncogene 26, 2226–2242 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Johmura Y., et al. , SCF(Fbxo22)-KDM4A targets methylated p53 for degradation and regulates senescence. Nat. Commun. 7, 10574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hui L., Zheng Y., Yan Y., Bargonetti J., Foster D. A., Mutant p53 in MDA-MB-231 breast cancer cells is stabilized by elevated phospholipase D activity and contributes to survival signals generated by phospholipase D. Oncogene 25, 7305–7310 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Zhou P., Howley P. M., Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell 2, 571–580 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Galan J. M., Peter M., Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA 96, 9124–9129 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soucy T. A., et al. , An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Roth J., Dobbelstein M., Freedman D. A., Shenk T., Levine A. J., Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 17, 554–564 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao K., et al. , RNF12 promotes p53-dependent cell growth suppression and apoptosis by targeting MDM2 for destruction. Cancer Lett. 375, 133–141 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Aslakson C. J., Miller F. R., Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 52, 1399–1405 (1992). [PubMed] [Google Scholar]
- 39.Tian X., et al. , F-box protein FBXO22 mediates polyubiquitination and degradation of KLF4 to promote hepatocellular carcinoma progression. Oncotarget 6, 22767–22775 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun R., et al. , FBXO22 possesses both protumorigenic and antimetastatic roles in breast cancer progression. Cancer Res. 78, 5274–5286 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Johmura Y., et al. , Fbxo22-mediated KDM4B degradation determines selective estrogen receptor modulator activity in breast cancer. J. Clin. Invest. 128, 5603–5619 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozdemir E., et al. , Strong correlation of basement membrane degradation with p53 inactivation and/or MDM2 overexpression in superficial urothelial carcinomas. J. Urol. 158, 206–211 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Jones S. N., Hancock A. R., Vogel H., Donehower L. A., Bradley A., Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc. Natl. Acad. Sci. USA 95, 15608–15612 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cordon-Cardo C., et al. , Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 54, 794–799 (1994). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.