Significance
Despite its prominent role in structural biology, NMR spectroscopy is hampered by extremely poor sensitivity. In this work, we develop a laser-assisted methodology, LC-photo-CIDNP, to enhance NMR sensitivity for the detection of amino acids and proteins in solution down to nanomolar concentration. Our method is significant because it enables detection of nanomolar concentrations of aromatic amino acids (Trp and Tyr), either in their free state or within proteins, in only a few seconds. Hence, the LC-photo-CIDNP approach renders data collection thousands of times faster than laser-free NMR experiments performed on higher-field spectrometers. Further, LC-photo-CIDNP is readily applicable to both 1D and 2D data collection mode. Rapid detection of proteins in solution at nanomolar concentration by NMR spectroscopy is unprecedented.
Keywords: hyperpolarization, photo-CIDNP, NMR, proteins, amino acids
Abstract
Solution-state NMR typically requires 100 μM to 1 mM samples. This limitation prevents applications to mass-limited and aggregation-prone target molecules. Photochemically induced dynamic nuclear polarization was adapted to data collection on low-concentration samples by radiofrequency gating, enabling rapid 1D NMR spectral acquisition on aromatic amino acids and proteins bearing aromatic residues at nanomolar concentration, i.e., a full order of magnitude below other hyperpolarization techniques in liquids. Both backbone H1-C13 and side-chain resonances were enhanced, enabling secondary and tertiary structure analysis of proteins with remarkable spectral editing, via the 13C PREPRINT pulse sequence. Laser-enhanced 2D NMR spectra of 5 μM proteins at 600 MHz display 30-fold better S/N than conventional 2D data collected at 900 MHz. Sensitivity enhancements achieved with this technology, denoted as low-concentration photo-CIDNP (LC-photo-CIDNP), depend only weakly on laser intensity, highlighting the opportunity of safer and more cost-effective hypersensitive NMR applications employing low-power laser sources.
NMR is an atomic-resolution noninvasive method to probe molecular structure and dynamics. This technique is, however, inherently insensitive due to the unfavorable distribution of nuclear spin states at the near-ambient temperature used in most applications. Methods implemented over the years to overcome the low sensitivity of NMR in liquids include the use of high applied magnetic fields, data acquisition in the time domain followed by Fourier transform, fast data collection schemes, and cryogenic probes (1–3). More recently, nuclear-spin hyperpolarization including Overhauser dynamic nuclear polarization (4, 5), optical pumping (6–9), parahydrogen-induced polarization (10, 11), signal amplification by reversible exchange (12), and dissolution dynamic nuclear polarization (D-DNP) (13–17) have displayed significant potential (3).
Despite the above technological advances, typical liquid-state NMR experiments employing hyperpolarization still require very expensive instrumentation; harsh hyperpolarization conditions; long polarization times; and last but not least, ≥50–100 μM sample concentration. In addition, NMR data collection of dilute biomolecules in physiologically relevant milieux is often unfeasible. While some of the above challenges may in principle be overcome by concentrating NMR samples and employing probes accommodating small sample volumes, this process is often unfeasible due to limited amounts of available material or to undesirable aggregation. In summary, there is a compelling need to further enhance the sensitivity of solution-state NMR spectroscopy.
Photochemically induced dynamic nuclear polarization (photo-CIDNP) is a spin-selective technique involving the transient generation of radical pairs (Fig. 1A). This methodology has been traditionally employed to gauge macromolecular solvent-exposure (18–20). More recently, photo-CIDNP has been combined with heteronuclear correlation spectroscopy (21, 22) and heavy atom-containing additives (23) to achieve spectral-editing and large sensitivity gains. The main recent advances, which led to the emergence of photo-CIDNP as a nuclear hyperpolarization technique in solution, are listed below.
Fig. 1.
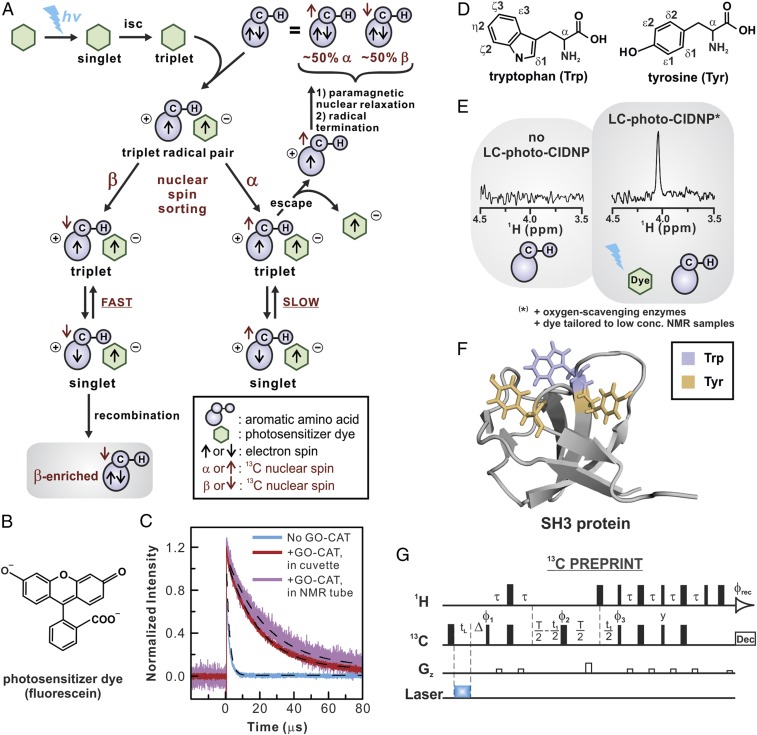
Basic aspects of hypersensitive NMR spectroscopy via low-concentration photochemically induced dynamic nuclear polarization (LC-photo-CIDNP). (A) Cartoon representation of photo-CIDNP radical pair mechanism. The symbol isc denotes inter-system crossing. (B) Structure of fluorescein, the photo-CIDNP dye. (C) Transient electronic absorption spectrum of fluorescein, illustrating the triplet–triplet absorption in the absence and presence of the GO-CAT oxygen-scavenging system. Black dashed lines denote single-exponential fits (see also SI Appendix, section 2.7). (D) Structure of tryptophan (Trp) and tyrosine (Tyr), two representative photo-CIDNP–active amino acids. (E) Dark (laser-off) and light (laser-on) 1H-detected 13C photo-CIDNP acquired with a room temperature probe (600 MHz) on 1 µM Trp. (F) Structure of the SH3 protein (56), highlighting its Trp and Tyr. (G) The 13C PREPRINT pulse sequence, which enables 1H-detected 13C photo-CIDNP NMR of low-concentration samples (LC-photo-CIDNP) in the presence of a cryogenic probe. The parameter tL denotes the laser irradiation time (488 nm, Ar ion laser, 200 ms). The τ delay was set to 1/4 JCH (1.7 ms). T is the constant chemical shift evolution time in the indirect dimension, which was set to 13.3 and 26.7 ms for side-chain and Cα carbons, respectively. The parameter denotes the perturbation-recovery delay (150 ms) that follows laser irradiation. The phase cycling was ; .
First, 13C steady-state photo-CIDNP (22) proved to more effectively boost NMR signal-to-noise (S/N) than 1H or 15N approaches. Second, fluorescein (Fig. 1B) was found to be a much more efficient photo-sensitizer than the traditionally employed flavin mononucleotide (FMN) at low sample concentration (24). Third, data collection in the presence of catalytic amounts of the oxygen-scavenging enzymes glucose oxidase (GO) and catalase (CAT) (25) enhanced S/N by largely mitigating quenching of the photoexcited triple state of fluorescein (9, 26) (Fig. 1C). The above three innovations led to the rapid detection of small molecules in solution at low micromolar concentration (24).
In this work, we show that photo-CIDNP hyperpolarization can be extended from free amino acids (Fig. 1 D and E) to folded and unfolded proteins (Fig. 1F) and can be combined with cryogenic probe technology via the 13C PREPRINT sequence (Fig. 1G) and low-power lasers to achieve large sensitivity gains. Rapid NMR data collection of small and large biomolecules in solution is now possible within the nanomolar concentration regime.
This approach, which is tailored to dilute samples in solution, is denoted here as low-concentration photo-CIDNP (LC-photo-CIDNP).
Results and Discussion
LC-Photo-CIDNP in the Presence of a Cryogenic Probe: Challenges and Solutions.
In this section, we use the term cryogenic probes to denote solution-state NMR probes where the sample temperature is not cryogenic. These probes ought not to be confused with solid-state NMR probes, which typically cool the sample and coils but often not the preamplifier.
NMR cryogenic probes for data collection in liquids enable increases in sensitivity by threefold to fourfold relative to room-temperature probes (27–29) by supercooling the spectrometer components used for detection, hence reducing the observed noise induced by random currents (27–30). In cryogenic probes, the temperature of receiver coil and preamplifier is independent of sample temperature. Hence, the latter can typically be set within a wide range of values (ca. −40 to +120 °C). The ratio of the current induced by a signal to the current induced by noise (US/UN) can be expressed as (31)
| [1] |
where Tc, Ta, and Ts are the temperatures of the coil, preamplifier, and sample, respectively, and Rc and Rs are the resistance of the coil in the absence and presence of the NMR sample, respectively. Eq. 1 shows that lowering Tc and Ta leads to S/N enhancements by decreasing the noise level.
The 1H-detected 13C photo-CIDNP, on the other hand, increases the NMR signal by perturbing the Boltzmann 13C nuclear-spin distribution via transient enhancement of nuclear spin polarization (22).
Hence, combining photo-CIDNP with cryogenic probe technology offers unique opportunities to synergistically improve S/N in NMR spectroscopy. However, performing photo-CIDNP in the presence of a cryogenic probe poses some challenges.
In protonated solvents, cryogenic probes are more susceptible to radiation damping than room-temperature probes (32). This effect often gives rise to analog-to-digital-converter (ADC) overflow. We observed that the likelihood of ADC overflow in the presence of a cryogenic probe is higher under laser-on (i.e., light) than laser-off (i.e., dark) conditions.
To prevent this drawback, we started by optimizing gradient-pulse lengths and strengths and offset frequencies. ADC overflow was mitigated by this procedure but not eliminated. Next, we reasoned that ADC overflow upon laser irradiation may be due to a small degree of sample heating or some other type of moderate, transient, and reversible perturbation, which in turn may cause the residual solvent resonance to undergo a slight chemical shift. As a result, the pulse sequence no longer perfectly aligns the solvent magnetization along +z by the end of the pulse sequence (i.e., immediately before free-induction decay acquisition). This effect must be due to either a fast and reversible temperature variation or to nonthermally related phenomena, given that we were not able to detect any apparent temperature variations via the thermocouple sensor of the spectrometer.
We reasoned that a few dummy scans (≥2) at the beginning of the laser-on experiments could help reach a probe steady state. This simple procedure, in combination with careful optimization of gradient strength/length and frequency offset, enabled an improvement in the ADC overflow challenge. However, the problem was still occasionally present.
We were able to fully and reproducibly eliminate ADC overflow challenges by inserting a crucial 150–250 ms delay immediately after laser irradiation (Fig. 1G) in the 13C PRINT pulse scheme (22). This delay serves the effective purpose of gating most of the radiofrequency pulses of the sequence, so that they are only applied after the probehead has fully reestablished its regular status. To highlight this small but essential modification, the resulting pulse sequence is denoted as 13C PREPRINT (i.e., perturbation-recovery photo-CIDNP-INEPT). The 13C PREPRINT NMR pulse sequence is used in all of the LC-photo-CIDNP experiments discussed in this work.
Combining Laser-Enhanced NMR with a Cryogenic Probe Leads to Unprecedented Sensitivity Gains.
All of the LC-photo-CIDNP experiments shown in this work employed fluorescein as a photosensitizer. This dye is more photostable and yields much higher nuclear spin polarization at low sample concentration than the more commonly used FMN (24). In addition, we employed catalytic amounts of the commercially available oxygen-scavenging enzymes CAT and GO (25), which mitigate the deleterious effect of singlet oxygen generated upon laser irradiation (33). Electronic triplet–triplet transient absorption shows that the presence of these additives increases the triplet lifetime of the photoexcited fluorescein dye (Fig. 1C).
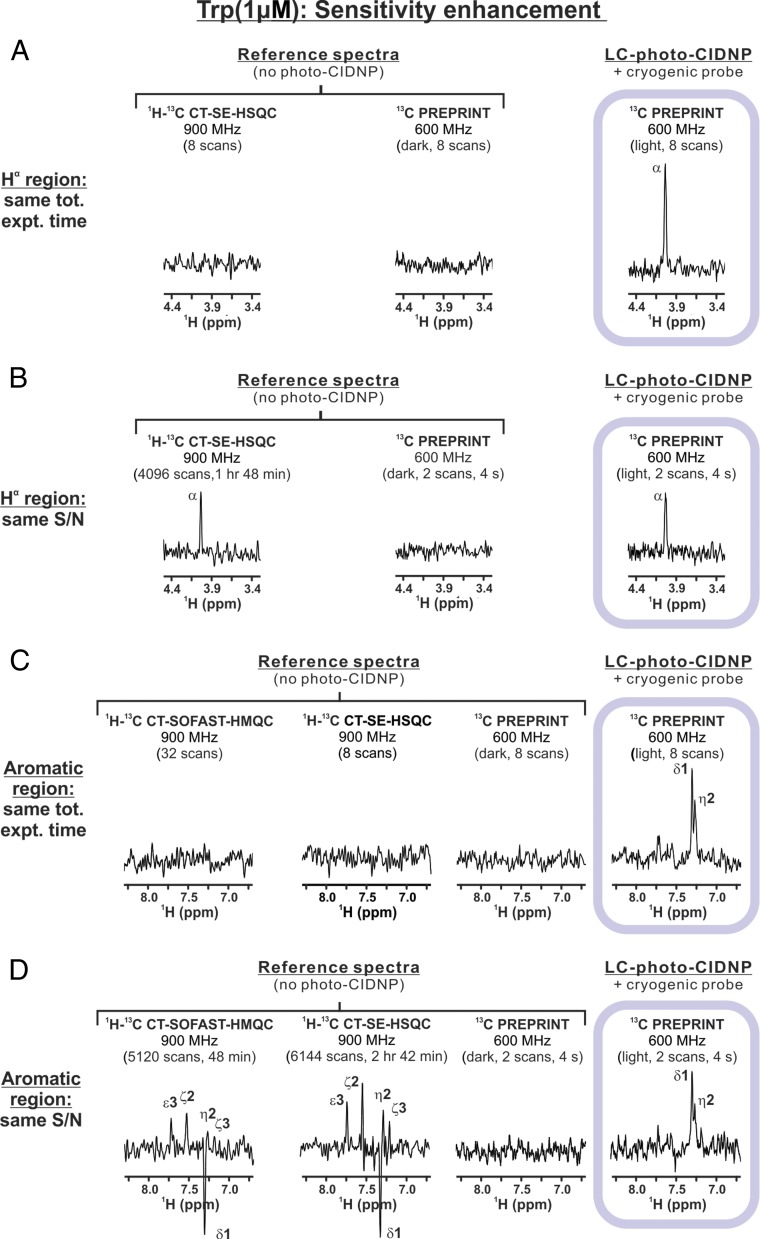
The 1H-detected 13C LC-photo-CIDNP experiments focusing on the 1Hα region were carried out with the 13C PREPRINT pulse sequence on a 600-MHz spectrometer equipped with a cryogenic probe. An excellent S/N was achieved on a 1-μM Trp sample in only 16 s (Fig. 2A). Reference data acquired either on the same 600-MHz spectrometer under dark (i.e., laser-off) conditions or with the pulse-field-gradient sensitivity-enhanced constant-time 1H,13C HSQC (1H,13C CT-SE-HSQC) pulse sequence (34) at 900 MHz, yielded only noise (Fig. 2A).
Fig. 2.
Increase in NMR sensitivity achieved upon carrying out 1H-detected 13C LC-photo-CIDNP with a cryogenic probe. All data were collected on 1 μM Trp on spectrometers equipped with cryogenic probes. The 1D 13C PREPRINT LC-photo-CIDNP spectra were acquired on a 600-MHz spectrometer in the presence of submicromolar amounts of oxygen-depleting enzymes and the fluorescein photosensitizer (Materials and Methods). Data were compared with corresponding 1D 13C PREPRINT experiments collected under dark (i.e., laser-off) conditions and with non–laser-enhanced 1D 1H-13C CT-SE-HSQC or 1H-13C CT-SOFAST-HMQC experiments acquired on a 900-MHz spectrometer. Vertical intensities were normalized to display approximately the same noise levels. (A) Hα spectral region. Data were collected with the same number of scans and had an identical total experiment time (excluding the dummy scans: 16 for 1H-13C CT-SOFAST-HMQC and 2 for the other experiments). (B) Hα spectral region. Data collection and processing parameters were kept consistent in all experiments except that the non–photo-CIDNP experiments comprised a larger number of transients. (C) Aromatic spectral region. Data collection and processing were carried out as in A. (D) Aromatic spectral region. Data collection and processing were carried out as in B.
We then collected similar 1H,13C CT-SE-HSQC reference data until the Trp-Hα resonance intensity reached the same values as LC-photo-CIDNP. Under these conditions, the S/N achievable via LC-photo-CIDNP in two scans (4 ± 0.5 s) can only be matched upon collection of ca. 4,096 scans (1 h and 48 min) with 1H,13C CT-SE-HSQC (Fig. 2B). Hence, remarkably, laser-enhanced data collection leads to a 1,620-fold savings in time, equivalent to a ca. 40-fold gain in S/N.
The above time savings apply even though the LC-photo-CIDNP time per scan is longer by about 0.35 s than the duration of a single scan in the 1H,13C CT-SE-HSQC reference sequence, due to the laser irradiation and perturbation-recovery delays.
Similar results were achieved in the aromatic region of the Trp spectrum (Fig. 2 C and D). A S/N similar to that of 2-scan LC-photo-CIDNP was only achieved when 5,120 (ca. 48 min) and 6,144 scans (ca. 2 h 42 min) were acquired with the reference 1H,13C CT-SOFAST HMQC and 1H,13C CT-SE-HSQC pulse sequences, respectively (Fig. 2D).
We then measured the enhancement in sensitivity [defined as (S/N)t per unit time; Materials and Methods] yielded by LC-photo-CIDNP (on a 600-MHz spectrometer) relative to the non–laser-enhanced reference experiments. We obtained a 30-fold sensitivity enhancement for aromatic protons at 600 MHz relative to 1H-13C SOFAST-HMQC at 900 MHz. In addition, we achieved a 50- and 40-fold sensitivity enhancement for aromatic and Hα protons, respectively, relative to 1H-13C CT-SE-HSQC at 900 MHz. The latter values highlight the remarkable practical utility of our 1H-detected 13C PREPRINT experiment, which yields 1,600- to 2,500-fold savings in data collection time relative to the most widely used non–laser-enhanced high-sensitivity NMR experiments performed at higher field.
Reference experiments acquired at the same applied field as LC-photo-CIDNP (600 MHz) are expected to compare even more favorably, with a 55-fold sensitivity enhancement for aromatic protons relative to 1H-13C SOFAST-HMQC, and a 92- and 73-fold sensitivity enhancement for aromatic and Hα protons, respectively, relative to 1H-13C CT-SE-HSQC.
We assessed the LC-photo-CIDNP enhancement (defined in SI Appendix, section 2.2) yielded by 13C PREPRINT and found it to be 1,800 for 13C, a remarkably high value. We also determined the actual 13C percent polarization is 2.2% (defined in SI Appendix, section 2.3). Despite the moderate numerical value of the latter quantity, the significant 1,800-fold increase relative to the thermal 13C percent polarization (i.e., 0.0012%) and the dramatic savings in data collection time fully support the utility of this technology.
Nanomolar Concentrations of Trp Are Readily Detected.
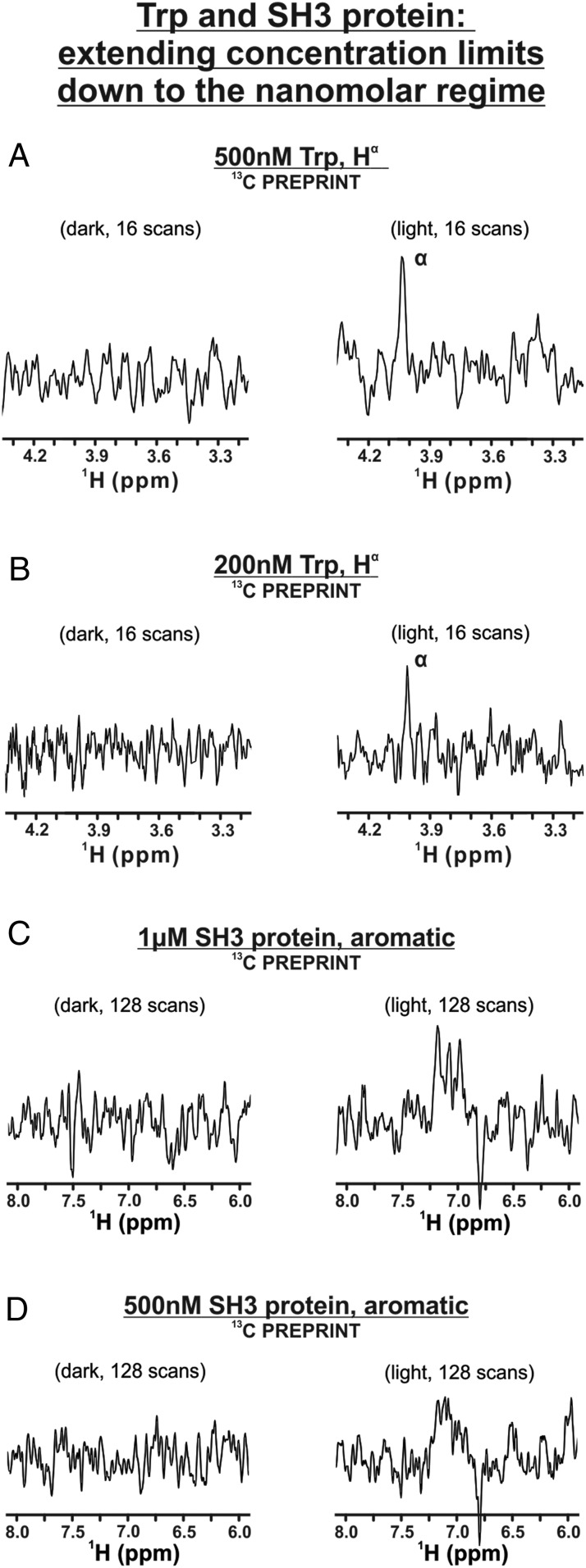
Using the cryogenic probe in conjunction with 13C-PREPRINT, we collected data on 500 nM aqueous Trp with only 16 scans under light conditions (Fig. 3A). We further reduced the concentration down to 200 nM Trp and were able to collect a clear signal with 16 scans (Fig. 3B). To the best of our knowledge, this is the lowest sample concentration detected via solution-state NMR, to date.
Fig. 3.
LC-photo-CIDNP of amino acids and proteins at nanomolar concentration in the presence of a cryogenic probe. The 13C PREPRINT data acquired on (A) 500 nM and (B) 200 nM Trp and on (C) 1 µM and (D) 500 nM SH3 protein.
While the light (i.e., laser-on) spectrum of Fig. 3B only required 16 scans, it is estimated (1–3) that a spectrum of comparable S/N would require 20,670 scans and a 1,378-fold longer data collection time, in the absence of photo-CIDNP hyperpolarization, at 600 MHz. Hence, it is clear that LC-photo-CINPD NMR greatly facilitates 1D data collection.
1D LC-Photo-CIDNP Enables Efficient Data Collection on Proteins at 1 Micromolar Concentration.
We investigated LC-photo-CIDNP on the N-terminal SH3 domain of the Drosophila melanogaster adaptor protein drk, denoted here as SH3 (Fig. 1F). The slow chemical-shift exchange regime between the SH3 native and unfolded states enables visualization of both unfolded- and folded-state resonances (35, 36). SH3 consists of 59 amino acids including one Trp (W36) and two Tyr (Y37 and Y52) and populates the folded and unfolded states in a ca. 3:2 ratio at room temperature (37).
We first performed 1H,13C 1D LC-photo-CIDNP on SH3 at 1 μM concentration (Fig. 3C). A concentration as low as 500 nM was also detected with adequate signal-to-noise (Fig. 3D). Collection of more transients than in the case of Trp was necessary. This result can be rationalized upon considering the larger surface area and added geometrical constraints of proteins relative to free amino acids, leading to an expected decrease in collision efficiency. Alternative non–photo-CIDNP reaction pathways (e.g., reductive electron transfer) may also play a role in photo-CIDNP of proteins (38). Despite the above caveat, the ability to detect proteins at 500 nM concentration and site-specific resolution in solution under mild, physiologically relevant conditions is unprecedented and simple to achieve with our experimental setup.
The 13C photo-CIDNP in the presence of the fluorescein dye only enhances Trp and Tyr (24). The intrinsic phase of the photo-CIDNP Trp aromatic protons (absorptive) is opposite to that of Tyr (emissive) (24). Hence, despite the fact that the aromatic side-chain resonances are not resolvable and assignments are not currently available in Fig. 3 C and D, amino acid types can be readily discriminated due to their opposite phase.
LC-photo-CIDNP enhancement requires solvent exposure of the target nuclei, so that intermolecular protein–dye collisions can occur. The photo-CIDNP–active Tyr and Trp are the most polar aromatic amino acids, and a significant percent of these residues, namely, 76.9% of Trp and 86.8% of Tyr, is known to be solvent-exposed in the folded state of proteins (39). It follows that LC-photo-CIDNP is applicable to a variety of protein types and states, including the majority of Trp and Tyr residues of unfolded and intrinsically disordered proteins (IDPs), as well as protein folded states.
2D LC-Photo-CIDNP Is a Powerful Tool to Collect High-Resolution Data on Proteins.
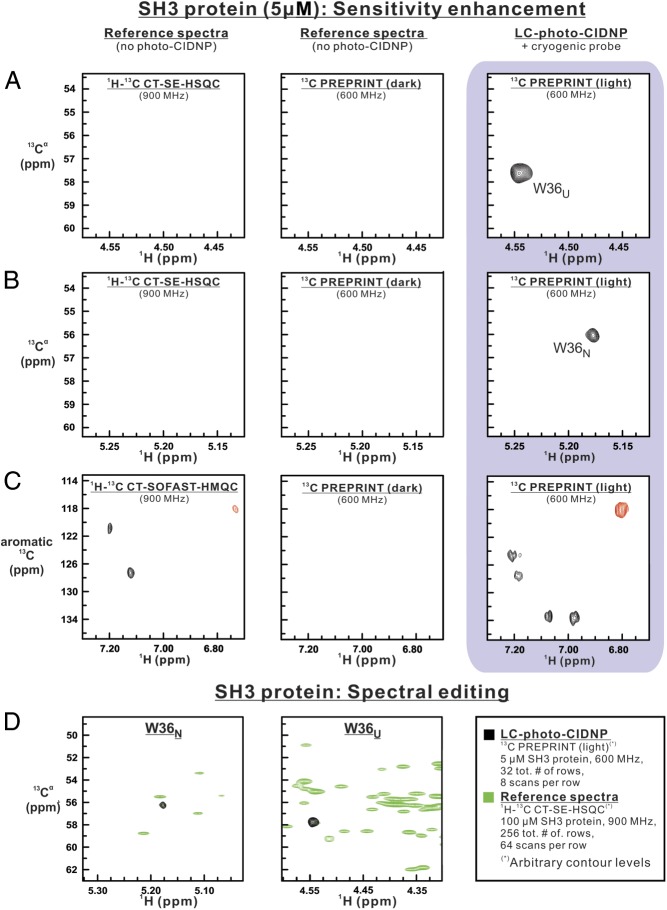
We performed 2D LC-photo-CIDNP experiments on the SH3 model protein at 5 μM concentration on a 600-MHz spectrometer (Fig. 4 A–C). Data collection was completed in less than 11 min. We also acquired reference 1H-13C CT-SE-HSQC and 1H-13C CT-SOFAST HMQC data on a 900-MHz spectrometer, to provide a stringent comparison at even higher magnetic field.
Fig. 4.
The 2D 13C-PREPRINT analysis of the SH3 protein (5 μM), highlighting the benefits of laser-enhanced NMR. (A) Upfield and (B) downfield Hα-Cα regions of 2D heteronuclear correlation spectra of the SH3 protein. The 2D 13C PREPRINT spectrum (16 complex points in indirect dimension, eight scans per row) under light (i.e., laser-on) conditions is shown along with reference spectra (1H-13C CT-SE-HSQC and dark 13C PREPRINT, i.e., under laser-off conditions) collected with identical number of scans per row, recycle delays, sweep widths, and number of points in both dimensions. To highlight the power of photo-CIDNP, LC-photo-CIDNP and 1H-13C CT-SE-HSQC data were collected at 600 and 900 MHz, respectively. (C) The 2D spectra analogous to A and B, except that the focus is on the 1H-13C aromatic region. In this case, 1H-13C CT-SOFAST-HMQC was employed as a reference experiment. To achieve an identical total experiment time, the 1H-13C CT-SOFAST-HMQC experiment included 36 scans per row. (D) Overlay of 13C PREPRINT 2D spectrum of the SH3 protein (5 μM; black) and the 2D 1H-13C CT-SE-HSQC reference spectrum (100 μM; green). The overlay highlights the dramatic spectral-editing capabilities of 13C PREPRINT, enabling the rapid selective detection of photo-CIDNP-active resonances of complex macromolecules.
Intense resonances were observed in the Hα region of the LC-photo-CIDNP light spectrum, while no signals were observed in the dark and the reference spectra (Fig. 4 A and B and SI Appendix, Fig. S8), which comprised the same number of scans and row in the indirect dimension. Our results demonstrate the applicability of 2D LC-photo-CIDNP to proteins in the low-micromolar concentration regime, which is unprecedented in solution.
The ability to gather high-resolution data on 1Hα-13Cα pairs, highlighted in Fig. 4 A and B, is particularly powerful. The 1Hα-13Cα pairs enable assessing protein backbone secondary structure at unprecedented sensitivity via the 13Cα secondary chemical shift (40) and SSP (41) parameters.
The resonances in the Hα regions are due to the folded and unfolded states of SH3’s single Trp (W36; Fig. 4 A and B). None of the Tyr resonances were observed at 5 μM, but weak signals due to Y37 and Y52 could be identified in 2D spectra at 20 μM (SI Appendix, Fig. S9).
Five aromatic side-chain SH3 resonances were observed under LC-photo-CIDNP light conditions (Fig. 4C). As noted in the previous section, even though assignments for these resonances are not known, LC-photo-CIDNP enables rapid amino acid type identification. Given that free-Tyr aromatic resonances have emissive H-detected 13C polarization (24), we tentatively assigned the aromatic resonance at 6.78 ppm (1H) and 117 ppm (13C) to Tyr.
The 1H,13C CT-SOFAST HMQC reference spectrum shows some resonances corresponding to the highest-intensity signals in the aromatic region. The same resonances were not observed in the LC-photo-CIDNP experiment, which specifically highlights only Trp and Tyr aromatic side chains.
One major advantage of 2D LC-photo-CIDNP is its spectral editing capability. This feature is particularly advantageous in the case of complex data sets, as shown in Fig. 4D. Resolution of Hα-Cα resonance pairs is often impossible in two dimensions. LC-photo-CIDNP circumvents the problem by enabling selective visualization of resonances arising from Trp and Tyr. This dramatic spectral simplification enables the rapid identification of the secondary structure and side-chain identity of aromatic residues at extremely high sensitivity.
We conclude that liquid-state LC-photo-CIDNP in the presence of a cryogenic probe is a powerful tool to increase NMR sensitivity for the detection of proteins in a spectrally edited setting.
LC-Photo-CIDNP Depends only Weakly on Laser Power, at Extremely Low Sample Concentration.
Earlier theoretical and experimental work (25, 38) showed that at high and moderate sample concentration (i.e., >5 μM), photo-CIDNP enhancement is proportional to the steady-state concentration of triplet photoexcited dye [T1DSS]. On the other hand, the scenario is significantly different at lower sample concentration (e.g., 500 nM).
To properly illustrate the dependence of photo-CIDNP hyperpolarization on [T1DSS] at different sample concentrations, we derived appropriate analytical expressions, as shown in SI Appendix, section 1.1. Importantly, one of these relations enables predicting that photo-CIDNP enhancement is expected to depend only weakly on [T1DSS], at submicromolar sample concentration (SI Appendix, section 1.1 and Eq. S8).
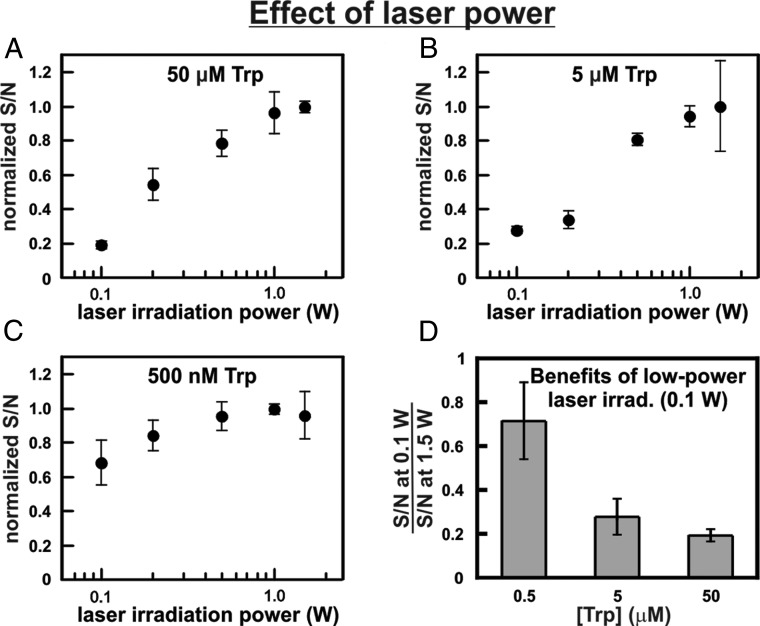
To experimentally probe the above predictions, we investigated the laser power dependence of LC-photo-CIDNP at a moderate (5 and 50 μM) and extremely low (500 nM) Trp concentration, keeping in mind that [T1DSS] is directly proportional to laser power via the laser excitation rate constant (42, 43). As shown in Fig. 5 A and B, at 50 and 5 μM Trp concentration, photo-CIDNP intensities depend significantly on laser power. In contrast, only a weak laser power dependence is observed at 500 nM sample concentration (Fig. 5C). As summarized in Fig. 5D, at the lowest tested sample concentration (500 nM), the ratio between S/N at high (1.5 W) and low (0.1 W) laser power is much higher than the corresponding ratio at higher sample concentrations (5 and 50 μM). These experimental results are fully consistent with our theoretical predictions (SI Appendix, section 1.1).
Fig. 5.
Laser power dependence of photo-CIDNP intensities in the presence of a cryogenic probe. Photo-CIDNP data at variable laser power were collected on the Trp amino acid in 90% H2O at variable Trp concentration, namely, (A) 50 μΜ, (B) 5 μM, and (C) 500 nM. At each Trp concentration, the S/N was normalized relative to the highest S/N within the series. (D) Block diagram illustrating the fractional S/N achieved at low laser power (0.1 W, 0.2 s irradiation time) at various Trp concentrations, relative to high-power irradiation (1.5 W, 0.2 s irradiation). This plot shows that low power irradiation is particularly effective at low (ca. submicromolar) sample concentration.
The results shown in Fig. 5 lead the way to future LC-photo-CIDNP applications with low-power lasers in the submicromolar sample concentration range. Low-power (50–500 mW) lasers are inexpensive and do not typically need air or water cooling. Hence, they offer the added advantage of increased affordability, portability, and safety.
Salt Effects Are Predictable.
High ionic strength leads to losses in NMR sensitivity in the presence of cryogenic probes (31, 44) due to an increased noise level (SI Appendix, Fig. S3A). High ionic strength is also expected to reduce photo-CIDNP enhancements when data are collected with room-temperature probes (45, 46). Upon performing LC- photo-CIDNP with a cryogenic probe, we found that up to ∼100 mM salt, the ionic strength dependence is indistinguishable in dark and light experiments, indicating that the observed moderate S/N losses are solely due to the cryogenic probe (SI Appendix, Fig. S3B). At higher salt concentration, up to 500 mM NaCl, however, additional losses are experienced under laser-on (light) conditions (SI Appendix, Fig. S3B). These losses can be entirely accounted for by theoretical predictions (SI Appendix, section 1.2 and Fig. S3C). Hence, LC-photo-CIDNP in the presence of a cryogenic probe under high-ionic strength condition is less advantageous yet still effective, and the more moderate enhancements are entirely predictable.
Effect of Long-Term Laser Irradiation.
As illustrated in SI Appendix, Fig. S2A, a progressive decrease in 13C Trp resonance intensity is observed in successive sets of 8-scan photo-CIDNP experiments acquired on the same sample with high-power laser irradiation (1.5 W). As a result of this effect, the cumulative S/N of Trp does not significantly increase after the initial 16 scans (SI Appendix, Fig. S2B). On the other hand, inspection of 1D SE-HSQC spectra collected before and after the above 80-scan photo-CIDNP experiment (SI Appendix, Fig. S2C) shows that the Trp population has decreased by only ca. 50%.
Therefore, given that the intensity losses due to both dye and sample concentration changes (see photo-CIDNP experiment in SI Appendix, Fig. S2A) are much larger than the losses due to Trp alone (SI Appendix, Fig. S2C), we conclude that the progressive decrease in photo-CIDNP enhancement is dominated by irreversible processes acting mainly upon the fluorescein photosensitizer and not upon the molecule of interest (i.e., Trp). The above result is fully consistent with previous findings in the Cavagnero group (24).
It is worth noticing that while prolonged laser irradiation is generally undesirable, it induces chemical modifications leading to products that are spectroscopically unobservable. Hence, no losses in spectral quality are observed. As a consequence, prolonged laser irradiation at high power only leads to more moderate levels of enhancements than expected, as data collection progresses. Conveniently, the sample of interest can undergo several photo-CIDNP cycles (i.e., several scans can be collected) before any significant extent of photo-induced substrate modifications take over.
The above applies to data collected at high laser power (1.5 W) and relatively high sample concentration (10 μM Trp; SI Appendix, Fig. S2). On the other hand, at low sample concentration (e.g., 500 nM), Fig. 5C shows that collecting photo-CIDNP data at low laser power leads to S/N comparable to data collected at high power. Hence, low-power lasers, which are far less expensive and considerably safer, are to be preferred for photo-CIDNP data collection at small sample concentration. On the other hand, it remains to be determined whether photo-induced degradative processes at low laser power and low sample concentration are less prominent than the effects shown in SI Appendix, Fig. S2. Collecting data on photodegradation at low sample concentration is nontrivial, given the long necessary experimental times. Future plans include carrying out additional systematic studies involving long-term data collection of reference experiments. In this way we will be able to quantitatively address the extent of photo-induced degradation at low sample concentration.
Laser-Assisted NMR Can Be Performed in Cell-Like Media.
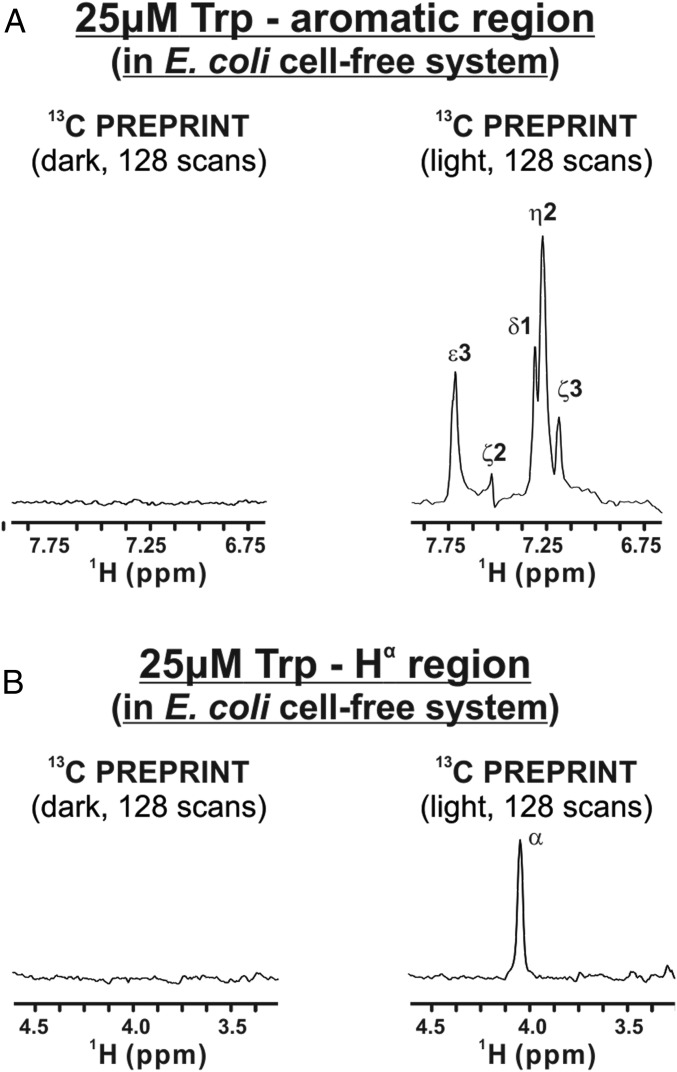
Finally, we carried out LC-photo-CIDNP experiments on a diluted cell-free extract to demonstrate the versatility of LC-photo-CIDNP in complex, physiologically relevant environments (Fig. 6 and SI Appendix, section 2.6). We were able to observe dramatic photo-CIDNP enhancements, relative to laser-off (dark) conditions. This result shows that the LC-photo-CIDNP technology is successful in complex media, and it is not limited to pure biomolecules in buffer.
Fig. 6.
The 1D 13C PREPRINT photo-CIDNP spectra of Trp in an E. coli cell-free system. Dark (i.e., laser-off) and light (i.e., laser-on) NMR data on 15N-13C-Trp (25 μM) are shown in the 1H (A) aromatic and (B) aliphatic region of a diluted cell-free solution prepared as described in SI Appendix, section 2.5, with data collected and processed as described in SI Appendix, section 2.6.
This result is far from trivial, given that nonspecific binding of photosensitizer dye to cell-free components, traces of heavy metals (23), and the photo-CIDNP competition effect (38) could have potentially prevented detection of hyperpolarization in this cell-like medium.
Comparisons with Other Hyperpolarization Techniques in Liquids.
In this work, we show that LC-photo-CIDNP in combination with a cryogenic probe enables fast data collection on 500 nM proteins in solution. This is, to the best of our knowledge, the lowest concentration achieved by high-resolution NMR spectroscopy in liquids, even relative to other hyperpolarization techniques. For instance, Ragavan et al. reported detection of 15 and 20 μM proteins via D-DNP (47, 48). This is a considerably higher concentration than the one achieved in the present study. D-DNP requires harsh freeze–thaw cycles and can therefore only be reliably employed on IDPs or small molecules. Another recently introduced method, employing diamond-based magnetometers bearing nitrogen vacancies carrying stable electron-spin centers and employing optical/quantum-logic detection, yielded a remarkable single-molecule sensitivity on protein nuclear spins in solution (49). However, the digital resolution provided by this pioneering approach at the present stage of implementation is only of the order of kHz. This value is far beyond the resolution required by most studies on protein structure and dynamics.
Here we were also able to detect free Trp at 200 nM concentration with regular 5-mm NMR tubes, corresponding to 26 ng. This value greatly surpasses the sensitivity reported on amino acids so far. For instance, Mompean et al. (50) detected 1 μg N-acetyltyrosine via photo-CIDNP in combination with microfluidics. Other small molecules, however, were quantified in solution (by magnetic resonance methods) at smaller total mass than reported here. For instance, p-fluorophenol and water were detected at 90 pg (50) and ca. 1 ng (51) levels, respectively.
However, the methods employed in these investigations demand extremely small volumes coupled with very large sample concentrations. On the other hand, the latter conditions are frequently not applicable to biological studies in aqueous solution, which often involve samples with limited solubility. This is often the case in biophysics, biotechnology, and clinical research on small biomarkers and proteins.
Conclusions
High NMR sensitivity is crucial when samples of interest are only available at millimolar to nanomolar concentration. Here we present a method to address this need: the 13C PREPRINT pulse sequence was developed to carry out LC-photo-CIDNP on high-field spectrometers equipped with cryogenic probes. We achieved increases in NMR S/N by a factor of ca. 30–90 relative to reference non–laser-assisted experiments, even though the latter were performed at significantly higher magnetic field (Figs. 2 and 3 and SI Appendix, Fig. S4). In addition, we showed that LC-photo-CIDNP can be performed on proteins at 500 nM to 5 μM concentration in only a few seconds. Further, this technique yields dramatic S/N enhancements relative to dark spectra in complex physiologically relevant environments including a bacterial cell-free system (Fig. 6). Finally, LC-photo-CIDNP intensity depends only weakly on triplet-state concentration. This finding, which is consistent with a theoretical treatment of LC-photo-CIDNP hyperpolarization at extremely low sample concentration, suggests future cost-effective and safer LC-photo-CIDNP implementations employing low-power lasers.
Materials and Methods
Materials.
The experiments described in this work employed 13C,15N-Trp (Cambridge Isotopes) as a small-molecule model system and fluorescein (Sigma-Aldrich) as a photosensitizer. Aspergillus niger GO and bovine liver CAT were purchased as freeze-dried powders from Sigma-Aldrich. GO (Enzyme commission classification code EC 1.1.3.4) and CAT (EC 232-577-1) had a nominal specific activity of 155,000 and 14,000 units per milligram, respectively.
Oxygen-Scavenging Enzyme Stock Solutions.
GO and CAT enzyme stock solutions were generated by separately dissolving solid GO and CAT in 30 mM potassium phosphate buffer (pH 7.2). Enzyme concentrations were assessed by electronic absorption spectrophotometry via the 267,200 M−1 cm−1 at 280 nm (52) and 912,500 M−1 cm−1 at 276 nm (52) extinction coefficients, respectively. Stock-solution aliquots were flash-frozen, stored at −80 °C, and used only once. GO and CAT concentrations were optimized for each independently generated stock solution. The same GO and CAT stock solutions were employed in all experiments.
SH3 Protein Preparation.
The 13C, 15N-labeled SH3 protein was prepared and purified according to known procedures (37). Protein concentration was assessed by electronic absorption spectrophotometry using the known extinction coefficient 8,480 M−1 cm−1 at 280 nm (37). Protein stock solutions were prepared in aqueous Tris buffer (50 mM Tris, 5 mM MgCl2, 50 mM KCl, pH 7.2), then aliquoted, flash-frozen, and stored at −80 °C.
NMR Sample Preparation.
All Trp NMR samples were in 10 mM phosphate buffer (pH 7.0) and 10% D2O. All SH3-protein NMR samples were in 16 mM phosphate buffer (pH 7.2 ± 0.1) and 10% D2O. All photo-CIDNP experiments contained 0.20 μM GO, 0.20 μM CAT and 2.5 μM fluorescein. In addition, 2.5 mM glucose was added to initiate oxygen depletion (25), and the sample was immediately incubated at room temperature for ca. 20 min before the beginning of photo-CIDNP data collection.
Photo-CIDNP Experiments.
Photo-CIDNP data were collected at 24 °C on an Avance III HD 600-MHz NMR spectrometer (Bruker Biospin Corp.) equipped with a 1H{19F/13C/15N} triple resonance (TCI-F) cryogenic probe fitted with a z-gradient. An argon-ion laser (INNOVA Sabre DBW 25/7; Coherent) in single-line mode (488 nm) was employed. Laser light was focused into an optical fiber via a convex lens (Thorlabs) and a fiber-coupler (F-91-C1-T; Newport Corporation). The optical fiber was inserted into a glass coaxial insert (WGS-5BL; Wilmad-Labglass) as previously described (22, 42, 43). Laser power was assessed with a power meter at the tip of the coaxial insert, and the coaxial insert was inserted into a 5-mm NMR tube (Wilmad-Labglass). The bottom tip of the insert was vertically adjusted to be 5 mm above the top region of the receiver coil.
The 13C PREPRINT pulse sequence, which is a perturbation-recovered version of 13C-PRINT (22), was used in all of the LC-photo-CIDNP experiments. The 1D version of this sequence has the 13C chemical-shift evolution time t1 set to zero. The 13C PREPRINT experiments were run separately for the aromatic and Cα spectral regions, with a constant time T of 13.3 and 26.6 ms, respectively, to properly optimize S/N, after taking into account typical values of 1-bond homonuclear 13C–13C scalar coupling constants for aromatic-side-chain and sp3 carbons. The 1H sweepwidth was 10,000 Hz. Unless otherwise stated, a laser power of 1.5 W was employed. Laser irradiation times were empirically optimized for optimum signal (after 8 and 128 transients for Trp and SH3, respectively) and were set to 0.2 and 0.3 s for Trp and SH3 protein, respectively. The recovery delay Δ following the laser pulse was set to 150 ms. The recycle delay was set to 1.5 s. 2D experiments were run and processed in echo–antiecho mode (53). Chemical shift referencing was carried out with external 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS).
The 1D time-domain data comprised 3,998 total points. Free-induction decays were apodized with exponential multiplication (5-Hz linebroadening) and zero-filled to 4,096 complex points. The 1D data were processed with the MestreNova software package (v. 10.0; Mestrelab Research).
The 2D experiments comprised 5,998 total points in the direct dimension. An unshifted Gaussian was applied to both dimensions, and data were zero-filled 2× and 5× in the direct and indirect dimensions, respectively. The 2D data were processed with NMRPipe (v. 9.0.0-b108) (54), and visualized with the NMRDraw and NMRView J (v. 2009.015.15.35) (55) software packages.
1H-13C CT-SE-HSQC and 1H-13C CT-SOFAST-HMQC Reference Experiments.
The 1H-13C CT-SE-HSQC and 1H-13C CT-SOFAST-HMQC reference experiments were run at 24 °C on an Avance III HD 900 MHz (21.2 T) spectrometer equipped with a 1H {13C,15N} cryogenic probe. The 2D 1H-13C CT-SE-HSQC data were collected/processed in echo–antiecho mode (53), and 2D 1H-13C CT-SOFAST-HMQC data were acquired/processed in States-TPPI mode. The number of complex rows of each 2D experiment is listed in the relevant figure legends. All other acquisition parameters were identical to 13C PREPRINT except that in 1H-13C CT-SOFAST-HMQC 1,024 total points were collected in the 1H dimension, and the recycle delay was empirically optimized as described in SI Appendix, section 1.4, and set to 0.48 s. All processing parameters were the same as in 13C PREPRINT.
Supplementary Material
Acknowledgments
We thank Collin Goebel and Matt Dalphin for gifts of the SH3 protein and Escherichia coli cell-free systems, respectively. We are also thankful to Clemens Anklin, Daniel Mathieu, Bernhard Brutscher, and Heike Hofstetter for helpful discussions. This work was funded by the NIH (Grants R21AI088551, R01GM125995, S10RR13866-01, and S10OD012245) and by the University of Wisconsin–Madison (Bridge Funds from the College of Letters & Science). M.F.M. and O.Y. were recipients of NIH Molecular Biophysics predoctoral training fellowships. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH Grant P41GM103399 (NIGMS), old N P41RR002301. Equipment was purchased with funds from the University of Wisconsin–Madison, the NIH (P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, and S10RR029220), the NSF (DMB-8415048, OIA-9977486, and BIR-9214394), and the US Department of Agriculture.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820573116/-/DCSupplemental.
References
- 1.Lee J. H., Okuno Y., Cavagnero S., Sensitivity enhancement in solution NMR: Emerging ideas and new frontiers. J. Magn. Reson. 241, 18–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernst R. R., Bodenhausen G., Wokaun A., Principles of Nuclear Magnetic Resonance in One and Two Dimensions (Oxford University Press, Oxford, 1987). [Google Scholar]
- 3.Ardenkjaer-Larsen J. H., et al. , Facing and overcoming sensitivity challenges in biomolecular NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 54, 9162–9185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carver T. R., Slichter C. P., Polarization of nuclear spins in metals. Phys. Rev. 92, 212–213 (1953). [Google Scholar]
- 5.Kaminker I., Barnes R., Han S., Overhauser dynamic nuclear polarization studies on local water dynamics. Methods Enzymol. 564, 457–483 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Nikolaou P., et al. , Near-unity nuclear polarization with an open-source 129Xe hyperpolarizer for NMR and MRI. Proc. Natl. Acad. Sci. U.S.A. 110, 14150–14155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosen M., Chupp T., Coulter K., Welsh R., Swanson S., Polarized 129Xe optical pumping/spin exchange and delivery system for magnetic resonance spectroscopy and imaging studies. Rev. Sci. Instrum. 70, 1546–1552 (1999). [Google Scholar]
- 8.Willmering M. M., et al. , Probing the magnetic field dependence of the light hole transition in GaAs/AlGaAs quantum wells using optically pumped NMR. Phys. Rev. B 97, 075303 (2018). [Google Scholar]
- 9.Navon G., et al. , Enhancement of solution NMR and MRI with laser-polarized xenon. Science 271, 1848–1851 (1996). [Google Scholar]
- 10.Green R. A., et al. , The theory and practice of hyperpolarization in magnetic resonance using parahydrogen. Prog. Nucl. Magn. Reson. Spectrosc. 67, 1–48 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Bowers C. R., Weitekamp D. P., Parahydrogen and synthesis allow dramatically enhanced nuclear alignment. J. Am. Chem. Soc. 109, 5541–5542 (1987). [Google Scholar]
- 12.Rayner P. J., Duckett S. B., Signal amplification by reversible exchange (SABRE): From discovery to diagnosis. Angew. Chem. Int. Ed. Engl. 57, 6742–6753 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Ardenkjaer-Larsen J. H., et al. , Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 100, 10158–10163 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bornet A., et al. , Boosting dissolution dynamic nuclear polarization by cross polarization. J. Phys. Chem. Lett. 4, 111–114 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Szekely O., Olsen G. L., Felli I. C., Frydman L., High-resolution 2D NMR of disordered proteins enhanced by hyperpolarized water. Anal. Chem. 90, 6169–6177 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Kurzbach D., et al. , Investigation of intrinsically disordered proteins through exchange with hyperpolarized water. Angew. Chem. Int. Ed. Engl. 56, 389–392 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Zhang G., Hilty C., Applications of dissolution dynamic nuclear polarization in chemistry and biochemistry. Magn. Reson. Chem. 56, 566–582 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Bargon J., Fischer H., Johnsen U., Nuclear magnetic resonance emission lines during fast radical reactions. I. Recording methods and examples. Z. Naturforsch. A 22, 1551–1555 (1967). [Google Scholar]
- 19.Ward H. R., Lawler R. G., Nuclear magnetic resonance emission and enhanced absorption in rapid organometallic reactions. J. Am. Chem. Soc. 89, 5518–5519 (1967). [Google Scholar]
- 20.Hore P. J., Broadhurst R. W., Photo-CIDNP of biopolymers. Prog. Nucl. Magn. Reson. Spectrosc. 24, 345–402 (1993). [Google Scholar]
- 21.Lyon C. E., Jones J. A., Redfield C., Dobson C. M., Hore P. J., Two-dimensional 15N—1H photo-CIDNP as a surface probe of native and partially structured proteins. J. Am. Chem. Soc. 121, 6505–6506 (1999). [Google Scholar]
- 22.Lee J. H., Sekhar A., Cavagnero S., 1H-Detected 13C photo-CIDNP as a sensitivity enhancement tool in solution NMR. J. Am. Chem. Soc. 133, 8062–8065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuno Y., Cavagnero S., Effect of heavy atoms on photochemically induced dynamic nuclear polarization in liquids. J. Magn. Reson. 286, 172–187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okuno Y., Cavagnero S., Fluorescein: A photo-CIDNP sensitizer enabling hypersensitive NMR data collection in liquids at low micromolar concentration. J. Phys. Chem. B 120, 715–723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J. H., Cavagnero S., A novel tri-enzyme system in combination with laser-driven NMR enables efficient nuclear polarization of biomolecules in solution. J. Phys. Chem. B 117, 6069–6081 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song L., Hennink E. J., Young I. T., Tanke H. J., Photobleaching kinetics of fluorescein in quantitative fluorescence microscopy. Biophys. J. 68, 2588–2600 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill H. D., Improved sensitivity of NMR spectroscopy probes by use of high-temperature superconductive detection coils. ITAS 7, 3750–3755 (1997). [Google Scholar]
- 28.Styles P., Soffe N. F., Scott C. A., An improved cryogenically cooled probe for high-resolution NMR. J. Magn. Reson. 84, 376–378 (1989). [Google Scholar]
- 29.Styles P., et al. , A high-resolution NMR probe in which the coil and preamplifier are cooled with liquid helium. J. Magn. Reson. 60, 397–404 (1984). [DOI] [PubMed] [Google Scholar]
- 30.Kovacs H., Moskau D., Spraul M., Cryogenically cooled probes—A leap in NMR technology. Prog. Nucl. Magn. Reson. Spectrosc. 46, 131–155 (2005). [Google Scholar]
- 31.Kelly A. E., Ou H. D., Withers R., Dötsch V., Low-conductivity buffers for high-sensitivity NMR measurements. J. Am. Chem. Soc. 124, 12013–12019 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Krishnan V. V., Murali N., Radiation damping in modern NMR experiments: Progress and challenges. Prog. Nucl. Magn. Reson. Spectrosc. 68, 41–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindqvist L., A flash photolysis study of fluorescein. Ark. Kemi 16, 79–138 (1960). [Google Scholar]
- 34.Vuister G. W., Bax A., Resolution enhancement and spectral editing of uniformly 13C-enriched proteins by homonuclear broadband 13C decoupling. J. Magn. Reson. 98, 428–435 (1992). [Google Scholar]
- 35.Zhang O., Kay L. E., Olivier J. P., Forman-Kay J. D., Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR 4, 845–858 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Zhang O., Forman-Kay J. D., Structural characterization of folded and unfolded states of an SH3 domain in equilibrium in aqueous buffer. Biochemistry 34, 6784–6794 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Lee J. H., et al. , Heterogeneous binding of the SH3 client protein to the DnaK molecular chaperone. Proc. Natl. Acad. Sci. U.S.A. 112, E4206–E4215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okuno Y., Cavagnero S., Photochemically induced dynamic nuclear polarization: Basic principles and applications. Emagres 6, 283–313 (2017). [Google Scholar]
- 39.Richards F. M., Areas, volumes, packing and protein structure. Annu. Rev. Biophys. Bioeng. 6, 151–176 (1977). [DOI] [PubMed] [Google Scholar]
- 40.Wishart D. S., Sykes B. D., Chemical shifts as a tool for structure determination. Methods Enzymol. 239, 363–392 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Marsh J. A., Singh V. K., Jia Z., Forman-Kay J. D., Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: Implications for fibrillation. Protein Sci. 15, 2795–2804 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daviso E., et al. , A 10,000-fold nuclear hyperpolarization of a membrane protein in the liquid phase via a solid-state mechanism. J. Am. Chem. Soc. 133, 16754–16757 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Zysmilich M. G., McDermott A., Photochemically induced dynamic nuclear polarization in the solid-state 15N spectra of reaction centers from photosynthetic bacteria rhodobacter sphaeroides. J. Am. Chem. Soc. 116, 8362–8363 (1994). [Google Scholar]
- 44.Biswas Z., Merkley N., Syvitski R. T., Biomolecular sample considerations essential for optimal performance from cryogenic probes. Metabolomics 10, 607–615 (2014). [Google Scholar]
- 45.Pedersen J. B., Freed J. H., A hydrodynamic effect on chemically-induced dynamic spin polarization. J. Chem. Phys. 62, 1790–1795 (1975). [Google Scholar]
- 46.Freed J. H., Pedersen J. B., The theory of chemically induced dynamic spin polarization. Adv. Magn. Opt. Reson. 8, 1–84 (1976). [Google Scholar]
- 47.Ragavan M., Chen H. Y., Sekar G., Hilty C., Solution NMR of polypeptides hyperpolarized by dynamic nuclear polarization. Anal. Chem. 83, 6054–6059 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Ragavan M., Iconaru L. I., Park C. G., Kriwacki R. W., Hilty C., Real-time analysis of folding upon binding of a disordered protein by using dissolution DNP NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 56, 7070–7073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lovchinsky I., et al. , Nuclear magnetic resonance detection and spectroscopy of single proteins using quantum logic. Science 351, 836–841 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Mompeán M., et al. , Pushing nuclear magnetic resonance sensitivity limits with microfluidics and photo-chemically induced dynamic nuclear polarization. Nat. Commun. 9, 108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glenn D. R., et al. , High-resolution magnetic resonance spectroscopy using a solid-state spin sensor. Nature 555, 351–354 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Fasman G. D., Sober H. A., Handbook of Biochemistry and Molecular Biology (CRC Press, Cleveland, OH, 1977). [Google Scholar]
- 53.Kay L., Keifer P., Saarinen T., Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114, 10663–10665 (1992). [Google Scholar]
- 54.Delaglio F., et al. , NMRPIPE - A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 55.Johnson B. A., Blevins R. A., NMRVIEW - A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994). [DOI] [PubMed] [Google Scholar]
- 56.Tollinger M., Skrynnikov N. R., Mulder F. A. A., Forman-Kay J. D., Kay L. E., Slow dynamics in folded and unfolded states of an SH3 domain. J. Am. Chem. Soc. 123, 11341–11352 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.