Significance
The 5′ end of a eukaryotic messenger RNA generally contains an 7-methylguanosine (m7G) cap, which has an essential role in regulating gene expression. Recent discoveries of RNAs with a noncanonical NAD+ moiety indicate the existence of a previously unknown mechanism for controlling gene expression. We have developed a method termed NAD tagSeq for the accurate identification and quantification of NAD+-capped RNAs and for revealing the complete sequences of NAD-RNAs using single-molecule RNA sequencing. Using this method, we found that NAD-RNAs in Arabidopsis were mostly derived from protein-coding genes and that they have essentially the same overall sequence structures as the canonical m7G-RNAs. The identification of NAD-RNAs and their sequence structures facilitates the elucidation of their possible molecular and physiological functions.
Keywords: NAD+ cap, RNA cap, NAD tagSeq, Arabidopsis, Oxford Nanopore sequencing
Abstract
The 5′ end of a eukaryotic mRNA transcript generally has a 7-methylguanosine (m7G) cap that protects mRNA from degradation and mediates almost all other aspects of gene expression. Some RNAs in Escherichia coli, yeast, and mammals were recently found to contain an NAD+ cap. Here, we report the development of the method NAD tagSeq for transcriptome-wide identification and quantification of NAD+-capped RNAs (NAD-RNAs). The method uses an enzymatic reaction and then a click chemistry reaction to label NAD-RNAs with a synthetic RNA tag. The tagged RNA molecules can be enriched and directly sequenced using the Oxford Nanopore sequencing technology. NAD tagSeq can allow more accurate identification and quantification of NAD-RNAs, as well as reveal the sequences of whole NAD-RNA transcripts using single-molecule RNA sequencing. Using NAD tagSeq, we found that NAD-RNAs in Arabidopsis were produced by at least several thousand genes, most of which are protein-coding genes, with the majority of these transcripts coming from <200 genes. For some Arabidopsis genes, over 5% of their transcripts were NAD capped. Gene ontology terms overrepresented in the 2,000 genes that produced the highest numbers of NAD-RNAs are related to photosynthesis, protein synthesis, and responses to cytokinin and stresses. The NAD-RNAs in Arabidopsis generally have the same overall sequence structures as the canonical m7G-capped mRNAs, although most of them appear to have a shorter 5′ untranslated region (5′ UTR). The identification and quantification of NAD-RNAs and revelation of their sequence features can provide essential steps toward understanding the functions of NAD-RNAs.
In eukaryotic cells, a messenger RNA (mRNA) molecule typically comprises a 7-methylguanylate (m7G) cap at its 5′ end, which is added through an unusual 5′ to 5′ triphosphate linkage. This cap protects the mRNA from decay by 5′ to 3′ exonucleases, as well as plays an essential role in almost all aspects of gene expression by acting as a unique identifier when recruiting proteins for transcription, pre-mRNA splicing, polyadenylation, nuclear transport, and protein synthesis initiation (for reviews, see refs. 1–3).
It was once thought that prokaryotic RNAs lack a 5′ cap. Several years ago, an NAD+ moiety was found to be covalently linked to the 5′ end of some RNAs in Escherichia coli (4), although the identities of these NAD+-capped RNAs (NAD-RNAs) were unknown. A method termed NAD captureSeq was developed for the identification of NAD-RNAs (5, 6). In this method, the nicotinamide moiety of NAD in NAD-RNAs is replaced by an alkyne through a reaction catalyzed by adenosine diphosphate (ADP)-ribosyl cyclase (ADPRC). The resulting “clickable” products react with biotin azide through an azide–alkyne cycloaddition reaction. Biotinylated RNAs can then be captured by a streptavidin resin. The enriched RNAs can be used to produce a cDNA library for identification and quantification of NAD-RNAs using second-generation sequencing technology. Using this method, 29 mRNAs and 15 noncoding RNAs (ncRNAs) were found to be NAD capped in E. coli (5). The NAD captureSeq method was recently used to identify at least 20 NAD-RNAs in yeast and a much wider range of NAD-RNAs in mammals (7, 8).
NAD-RNAs can be synthesized when RNA polymerase incorporates NAD as the first nucleotide, in place of adenosine triphosphate (ATP), during transcription initiation (9, 10), although NAD capping might also occur posttranscriptionally (5, 7). However, the molecular and physiological functions of NAD-RNAs remain unclear.
The NAD captureSeq method (5, 6) has some shortcomings. Nonspecific binding of RNAs to the streptavidin resin might lead to false positives and reduced sensitivity. RNA fragmentation introduced during the click chemistry reaction and sequencing library construction can cause loss of information concerning the overall sequence structures of NAD-RNAs. Recently, another method, termed CapZyme-seq, was used to identify NAD-RNAs and other noncanonical initiating nucleotides (11). In this method, RNA extracts are treated with an NAD-RNA decapping enzyme. The decapped RNAs can then be ligated to a linker to make a library for sequencing. One shortcoming of CapZyme-seq is that an NAD-RNA decapping enzyme might have substrate specificity such that the method might identify only a subset of NAD-RNAs. In addition, partially degraded RNAs might ligate to the linker, producing false positives.
Here, we report the development of a high-throughput method termed NAD tagSeq for the accurate identification and quantification of NAD-RNAs. Our study found evidence that NAD-RNAs in Arabidopsis are produced from at least several thousand genes. For some genes, over 5% of their transcripts were NAD capped. We found that Arabidopsis NAD-RNAs have essentially the same sequence structures as the m7G-capped RNAs (m7G-RNAs).
Results
Arabidopsis Produces NAD-RNAs.
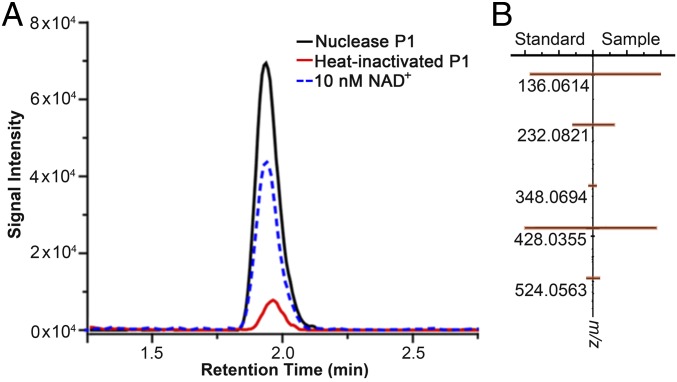
To determine whether Arabidopsis produces NAD-RNAs, we digested total Arabidopsis RNA with nuclease P1 to release single nucleotides and small molecules conjugated to RNA molecules. The digest was separated by high-performance liquid chromatography (HPLC), and the fraction corresponding to the retention time of an NAD+ standard was collected and analyzed by liquid chromatography mass spectrometry (LC-MS) for identification. Both the retention time and the product ions of the fraction from the P1 digest matched those of the NAD+ standard (Fig. 1), indicating that some RNA molecules from Arabidopsis contain the NAD+ moiety.
Fig. 1.
Detection of NAD+ in total RNA extract from Arabidopsis seedling. Total RNAs were digested with P1. The digest was separated by HPLC, and the fraction containing NAD+ was collected and analyzed by LC-MS. (A) Representative LC-MS chromatograph of the NAD+ standard, the NAD+ fraction from the P1-digested RNAs, and the control sample (the RNA sample treated with heat-inactivated P1). The experiment was repeated 3 times with similar results. (B) Product ions from NAD+ of the P1 digest were identical to those from the NAD+ standard.
Development of the NAD tagSeq Method for Transcriptome-Wide Identification of NAD-RNAs.
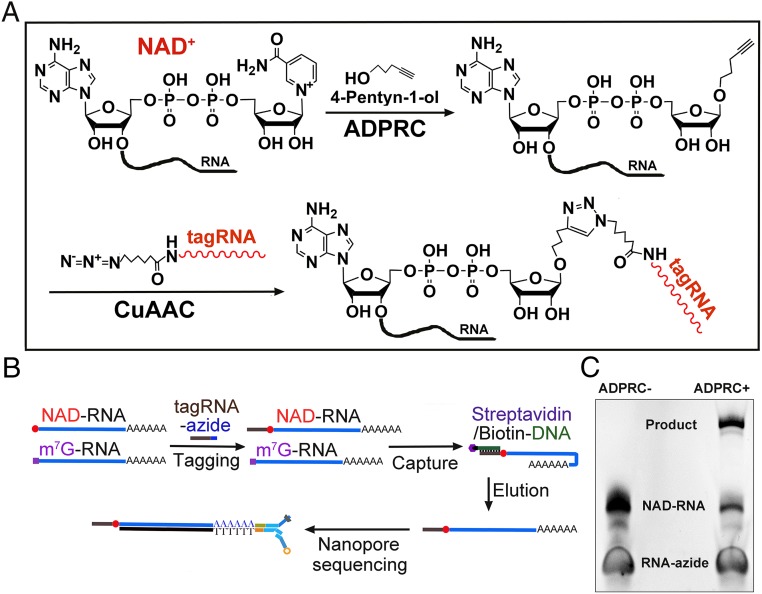
We developed the method NAD tagSeq for transcriptome-wide identification and quantification of NAD-RNAs (Fig. 2). The first step in NAD tagSeq is the same as that in NAD captureSeq (5, 6): RNA samples were treated with an alkyne (4-pentyn-1-ol) in the presence of ADPRC to replace the nicotinamide of NAD-RNA with the alkyne. In the second step, the clickable products were reacted with a synthetic RNA (tagRNA) that has an azide group at its 3′ end. Copper-catalyzed azide–alkyne cycloaddition (CuAAC) was used to link the original NAD-RNA to the tagRNA (Fig. 2A). The tagged RNA molecules can be enriched by hybridization to a DNA probe that is complementary to the tagRNA sequence (Fig. 2B). The enriched RNA sample can be directly sequenced using the Oxford Nanopore single RNA molecule sequencing platform. Sequence reads that contained the tagRNA sequence were deemed to be NAD-RNAs. A parallel experiment without ADPRC served as the negative control.
Fig. 2.
The NAD tagSeq method. (A) Diagram illustrating the reactions for tagging NAD-RNA with a synthetic RNA. In the presence of ADPRC, the alkyne (4-pentyn-1-ol) replaces the nicotinamide of NAD-RNA, resulting in NAD-RNA being functionalized with an alkyne group. In the second step, through the CuAAC reaction, the alkyne-functionalized NAD-RNA is ligated to a synthetic RNA (tagRNA) with an azide group at its 3′ end. (B) Workflow for identification of NAD-RNAs from poly(A)-containing RNAs. Poly(A)-enriched RNAs are tagged with the tagRNA through the reactions shown in A. Tagged NAD-RNAs are enriched by hybridization to the 5′ biotin-DNA probe attached to the streptavidin beads. The eluted RNAs are subjected to Oxford Nanopore library preparation and sequencing. (C) Tagging of a 38-nt NAD-RNA with a synthetic 25-nt RNA-azide. The NAD-RNAs were reacted with 4-pentyn-1-ol in the presence or absence of ADPRC and then with the RNA-azide through CuAAC, resulting in its ligation with the RNA tag. No such product was detected in the absence of ADPRC. The RNAs were resolved on a denaturing polyacrylamide gel.
To test the feasibility of the tagging process, we synthesized an NAD-RNA containing 38 nucleotides (nt) and a 25-nt RNA-azide molecule, which we refer to as tagRNA. After the synthetic NAD-RNAs were reacted with 4-pentyn-1-ol in the presence of ADPRC and then with the tagRNA, the 2 RNA molecules were found to be ligated (Fig. 2C). In the ADPRC negative control, no such ligation product was detected.
The typical experimental workflow of the NAD tagSeq method shown in Fig. 2B can be varied for the generation of different types of information about cellular NAD-RNAs. For example, to identify and quantify NAD-RNAs among poly(A)-containing RNAs, a poly(A) enrichment step could be carried out before the tagging step. To identify total NAD-RNAs, including those without a native poly(A) tail, polyadenylation of total RNA would have to be performed, since RNAs without a poly(A) tail cannot be sequenced by the current Oxford Nanopore sequencing method. Including a step in which tagged NAD-RNAs are enriched by hybridization to a cDNA probe would increase the sequencing coverage of NAD-RNAs. If the enrichment step is not implemented, both tagged and untagged RNAs could be sequenced, allowing comparison of the relative abundance of NAD-RNAs and their total transcripts, although this approach would reduce the sequencing depth of NAD-RNAs. Different tagRNAs might be used as barcodes for multiplex sequencing. We used a 40-nt RNA-azide molecule (SI Appendix, Supplementary Methods) as the tagRNA in this study.
NAD-RNAs in Arabidopsis Are Mostly Produced from Protein-Coding Nuclear Genes.
We carried out transcriptome-wide identification of NAD-RNAs in Arabidopsis. To determine which types of RNAs might be NAD capped, total RNA samples from 12-d-old Arabidopsis seedlings were subjected to the tagging process. A polyadenylation step was carried out using a poly(A) polymerase after the tagging step. Oligo (dT) beads were then used to increase the proportion of poly(A)-containing RNAs and remove excess free tagRNAs. Tagged RNAs were enriched by hybridization to the DNA probe. The eluted RNAs were directly sequenced by nanopore RNA sequencing (12). We used 1 nanopore flow cell to sequence each sample. For the negative control, the RNA samples were subjected to the same treatment but without ADPRC. This experiment is referred as the “total RNA tagging experiment.”
We obtained a total of 13,839 sequencing reads from the ADPRC+ sample and 4,634 reads from the ADPRC− sample. The most abundant reads were rRNA molecules, suggesting that a poly(A) tail was added to a large portion of RNAs which lacked a native poly(A) tail. From the ADPRC+ sample, 917 reads were found to be NAD-RNAs. These reads mapped to 192 Arabidopsis genes (Dataset S1), including 188 protein-coding genes and 4 nonprotein-coding genes. From the ADPRC− sample, 2 reads were found to contain tagRNA, suggesting the presence of a low level of noise from the use of this method. The most abundant NAD-RNAs were from AT5G38410, which encodes a Rubisco small subunit. The NAD-RNA reads from nonprotein-coding genes included 2 ncRNA genes: 1 long noncoding (lnc) gene, and 1 read from a chloroplast rRNA gene. Among the NAD-RNAs from protein-coding genes, one was from a mitochondrial gene, ATMG01360, which encodes cytochrome c oxidase subunit 1, and the others from nuclear genes.
The sequencing depth for both NAD-RNA reads and total reads was low when the total RNA samples were used. Nevertheless, the results reported above indicate that NAD-RNAs in Arabidopsis are produced primarily from protein-coding nuclear genes. The sequencing coverage was considerably improved when poly(A) RNAs were subjected to the NAD tagSeq analysis (see below).
NAD-RNAs Generally Contain a Poly(A) Tail and Are Produced from at Least Several Thousand Protein-Coding Genes.
The NAD-RNAs from the protein-coding nuclear genes (and possibly also the noncoding nuclear genes) identified in the total RNA tagging experiment are probably transcribed by RNA polymerase II. Because RNAs transcribed by RNA polymerase II are generally polyadenylated, we reasoned that most NAD-RNAs are also polyadenylated.
In the next experiment, poly(A)-containing RNAs were enriched and subjected to the tagging process. This experiment is referred to as the “poly(A) RNA tagging experiment.” Three biological replicates were included for both ADPRC+ and ADPRC− samples. Each pair of ADPRC+ and ADPRC− samples was processed in parallel, and 3 pairs of the samples processed on different days were treated as 3 replicates.
For each ADPRC+ sample, an average of 1.4 million sequence reads were generated, while an average of 0.79 million reads were generated from each ADPRC− sample. From 3 ADPRC+ samples, we identified a total of 710,098 NAD-RNA reads, which were derived from over 10,000 different genes (Dataset S2). Over 95% of the NAD-RNAs identified in the total RNA tagging experiment were also identified in the poly(A) RNA tagging experiment. The above results further indicated that most Arabidopsis NAD-RNAs contain a poly(A) tail.
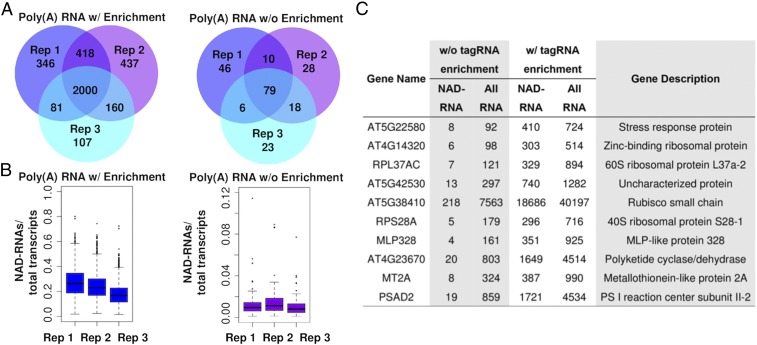
In the negative control, 4,623 reads from the 3 replicates were found to contain the tagRNA sequence. The low level of noise might have been due to nonspecific ligation of tagRNA to non–NAD-RNAs, probably during the click chemistry reaction. Dataset S2 lists the NAD-RNAs identified from each sample, including those from the negative control. Among all of the NAD-RNA reads, over 60% were produced from fewer than 200 genes. We calculated transcripts per million reads (TPMs) for each sample to normalize the read counts. NAD-RNAs from 6,486 genes had the TPM value differing by 3 or more in the comparison between the ADPRC+ sample and the ADPRC− sample. For a large majority of these genes, no NAD-RNA read was detected from the ADPRC− samples (Dataset S2). These genes were considered as high-confident NAD-RNA–producing genes identified from this analysis. Among them, NAD-RNAs from 2,000 genes were found to have the TPM value differing by 7 or more in the comparison between the ADPRC+ sample and the ADPRC− sample in all 3 replicates (Fig. 3A and Dataset S3). These top 2,000 NAD-RNA–producing genes include 1,980 protein-coding genes, 12 ncRNA genes, and 8 lncRNA genes.
Fig. 3.
NAD-RNAs and their relative abundance compared with total transcripts. (A) The numbers of NAD-RNAs identified from the 3 replicates (Rep 1, 2, and 3) in the poly(A) RNA tagging experiment with (w/) (Left) or without (w/o) (Right) the tagged RNA enrichment step. Only the NAD-RNAs with a normalized TPM value differing by 7 or more in the comparison between the ADPRC+ sample and ADPRC− sample were selected for the analysis. (B) Percentage of NAD-RNA reads in the total transcript reads from the same genes detected from the poly(A) RNA tagging experiment with (Left) or without (Right) the tagged RNAs enrichment step. (C) Ten Arabidopsis genes with the highest ratios of NAD-RNA reads to total RNA reads. Shown are the counts of NAD-RNA reads and total transcript reads from the 2 poly(A) RNA tagging experiments with or without the enrichment step of tagged RNAs.
More than 5% of Transcripts from Some Arabidopsis Genes Were NAD Capped.
To determine the ratio of NAD-RNAs to total transcripts from the same gene, we carried out an experiment similar to the poly(A) RNA tagging experiment, except that the hybridization-based enrichment step for tagRNA-linked RNAs was excluded so that all transcripts from the same genes could be counted.
From this experiment, we obtained an average of 0.66 million sequence reads from each of 3 ADPRC+ samples, and 0.57 million sequence reads from 1 ADPRC− sample. From the 3 ADPRC+ samples, we identified a total of 10,123 NAD-RNA reads from 2,370 genes (Dataset S4). In the ADPRC− sample, a total of 9 reads contained the tagRNA sequence, with no more than 1 read from any individual gene, again indicating a very low level of background noise. As the background noise was very low, we included just 1 ADPRC− sample in the experiment.
We selected the top 210 NAD-RNAs from this experiment with a TPM value differing by at least 7 between the ADPRC+ sample and the ADPRC− sample. The ratio of the NAD-RNA counts to the total transcript counts from the same gene was calculated. We selected these relatively high abundant NAD-RNAs for calculation of the ratio to reduce randomness arisen from the NAD-RNAs with a lower read number. These 210 NAD-RNAs were also identified in the previous poly(A) RNA tagging experiment, which included the tagged RNA enrichment step, and a high correlation was observed in the NAD-RNA counts for individual genes between the previous experiment and this experiment (SI Appendix, Fig. S1), indicating that the tagRNA enrichment step was not noticeably biased toward specific transcripts. For these 210 genes, ∼1% of their total transcripts were found to be NAD-RNA reads (Fig. 3B). However, for some genes, over 5% of their transcripts were NAD-RNA reads. The 10 genes with the highest ratios of NAD-RNA counts to total transcript counts are listed in Fig. 3C. The actual abundance of the NAD-RNAs is likely to be much higher, because not all NAD-RNAs are expected to have been tagged, and some NAD-RNA molecules could not be sequenced because their poly(A) tails were missing due to fragmentation occurring during the tagging process. With the tagged RNA enrichment step, ∼20% of the total transcripts were NAD-RNA reads (Fig. 3B), indicating a 20-fold enrichment in tagged RNAs.
High Abundant NAD-RNAs Were Mostly from Highly Expressed Genes.
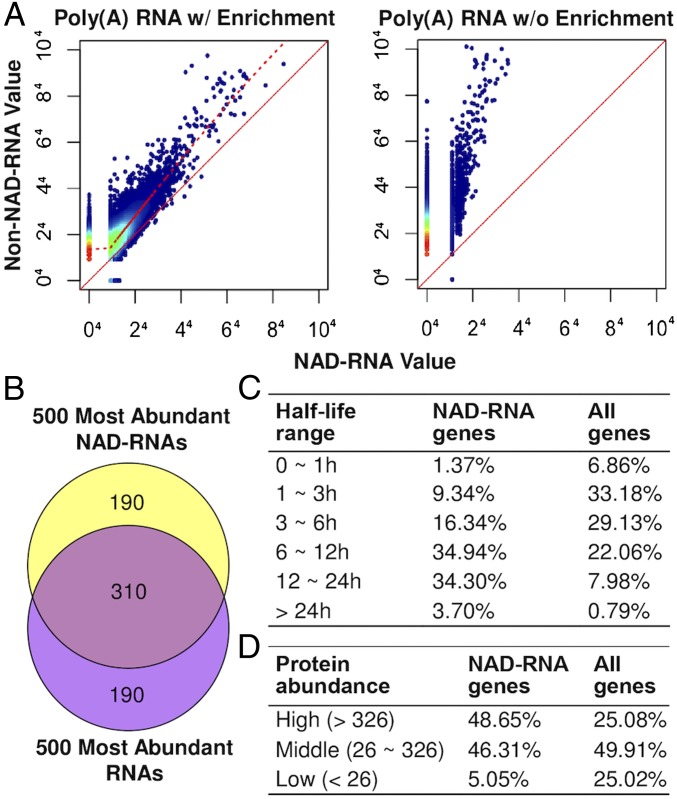
In general, the genes that produced relatively high abundant NAD-RNAs also had relatively high counts of total transcripts, although not all highly expressed genes produced NAD-RNAs (Fig. 4 A and B).
Fig. 4.
NAD-RNA–producing genes tend to be highly expressed. (A) Scatter plots showing abundance of NAD-RNAs and non–NAD-RNAs from the same genes in the poly(A) RNA tagging experiments with (w/) (Left) or without (w/o) (Right) enrichment of tagged RNAs. The different colors of the dots indicate transcript density: red (highest) > yellow > green > blue (lowest). The RNA levels shown were normalized TPM and under the fourth root to better represent genes with no NAD-RNA read. The solid red line is the identity line; the dashed red line represents the locally weighted regression line, which indicates a high positive correlation between NAD-RNA read values and non–NAD-RNA read values. (B) Overlap between the 500 most abundant NAD-RNAs and the 500 most abundant total transcripts detected in the poly(A) RNA tagging experiment. (C) Comparison of transcript half-lives between the 2,000 NAD-RNA–producing genes and all genes. The half-lives of transcripts were based on ref. 13. (D) Comparison of abundance of proteins from NAD-RNA–producing genes and all genes. The protein abundances were based on normalized spectral counts from the proteomic data from ref. 14.
Using the data from Narsai et al. (13) on transcriptome-wide analysis of mRNA decay rates in Arabidopsis as a reference, it was found that transcripts from over 69% of the top 2,000 NAD-RNA–producing genes had a half-life of >6 h (Fig. 4C). In contrast, 30.8% of all transcripts analyzed by Narsai et al. (13) had a half-life of >6 h. Based on the proteomic data for Arabidopsis seedlings reported by Motohashi et al. (14), a total of 1,647 proteins detected in the proteomic study were grouped into high-abundance (0 to 25% quantile), middle-abundance (25 to 75% quantile), and low-abundance (75 to 100% quantile) proteins according to their normalized spectral counts. Proteins from 555 of the 2,000 NAD-RNA–producing genes were detected in the proteomic analysis. Among them, 49.7% are in the high-abundance category and 46.3% in the middle-abundance group (Fig. 4D). The above analyses indicate that the NAD-RNA–producing genes tend to produce mRNAs that are relatively more stable and proteins that are relatively more abundant than the non–NAD-RNA–producing genes.
Gene ontology (GO) enrichment analysis showed that GO terms annotated to the 2,000 NAD-RNA–producing genes were enriched for photosynthesis, translational processes, responses to various stresses (oxidative stress and biotic and abiotic stresses), and response to cytokinin (SI Appendix, Fig. S2).
NAD-RNAs Share Similar Primary Sequence Structures to Those of m7G-RNAs.
The length of the detected NAD-RNA reads ranged from 128 to 5,021 nt, not including the poly(A) tail or the tag RNA sequence, whereas the length of all detected reads ranged from 101 to 12,134 nt (SI Appendix, Fig. S3). As the current nanopore sequencing method is not effective for sequencing small transcripts, it is possible that some small RNAs (for example, smaller than 100 nt) might have eluded identification by this method.
One of the main advantages of NAD tagSeq is that it allows the determination of the entire sequences of individual NAD-RNA transcripts using nanopore single-molecule RNA sequencing. However, a small number of bases downstream or upstream of the junction between the tagRNA and the NAD cap, which does not resemble typical nucleotides (Fig. 2A), might be miscalled or not be called, making it difficult to determine the exact 5′ end of an NAD-RNA transcript.
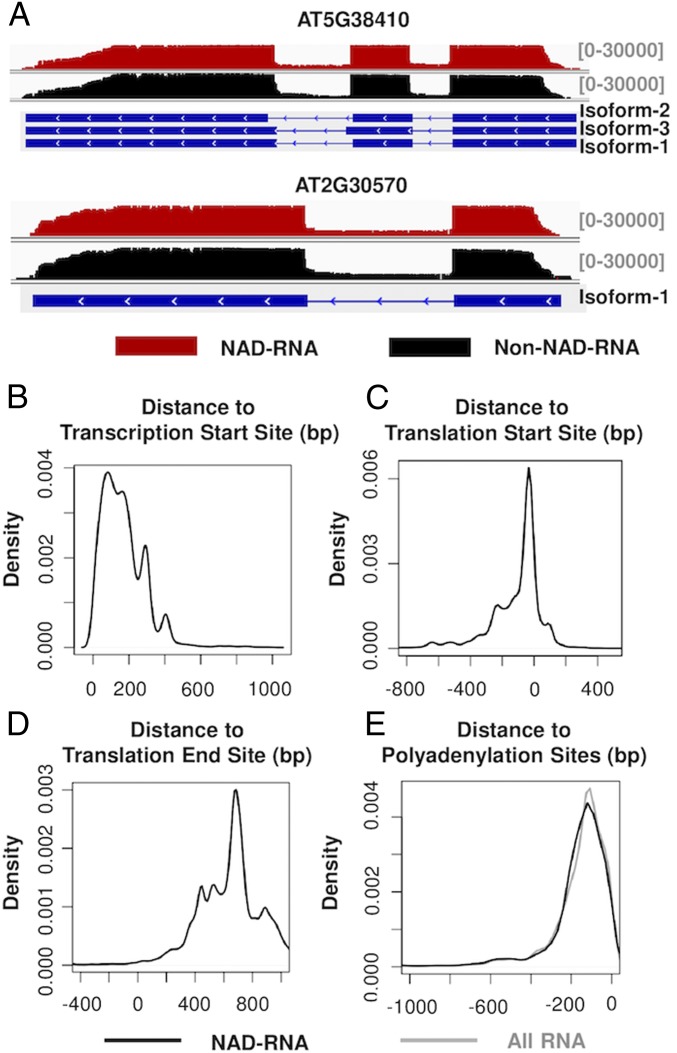
We compared the primary sequences of 710,098 NAD-RNA reads identified by the poly(A) RNA tagging experiment with the annotated sequences of the m7G-capped transcripts in the Arabidopsis genome database. It was found that the NAD-RNAs were generally spliced in the same manner as their m7G counterparts, as exemplified by the 2 genes with the highest abundance of NAD-RNAs (Fig. 5A). Only a very small portion of the NAD-RNA reads were aberrantly spliced. Among the 710,098 NAD-RNA reads, 171 reads from 99 protein-coding genes were found to be unspliced (SI Appendix, Fig. S4), as shown for one of these genes (SI Appendix, Fig. S4A), indicating that they were pre-mRNAs. Although the number of unspliced NAD-RNA reads was very small, the presence of the NAD cap in the pre-mRNAs supports the conception that the NAD cap could be installed during transcription. In addition to the 171 unspliced reads, 93 NAD-RNA reads had at least 1 intron retained, while the remaining intron or introns were spliced.
Fig. 5.
Sequence organization of NAD-RNAs. (A) Alignment of NAD-RNA reads and other RNA reads to the genomic sequences of 2 genes. Aligned sequencing reads were visualized using the Integrative Genomics Viewer, with the density of reads denoted above the genes. For the organization of the genes, boxes represent exons, and the lines represent introns. (B) Locations of 5′ ends of the NAD-RNAs relative to annotated TSSs. The reads in the positive x axis had the identified 5′ ends located downstream of the annotated TSSs. (C) Locations of 5′ ends of the NAD-RNAs relative to annotated translation start sites. The reads in the negative x axis had their 5′ ends located upstream of translation start sites. (D) Locations of the 3′ ends of the NAD-RNA reads relative to annotated translation end sites. The reads in the positive x axis contained a 3′ UTR. (E) Distribution of 3′ ends of the NAD-RNA reads (black) and all RNA reads (gray) relative to annotated polyadenylation sites.
We compared the detected 5′ ends of the NAD-RNA reads with the annotated transcription start sites (TSSs) from the corresponding genes. The detected 5′ ends of a large majority of NAD-RNAs were located around 30 to 400 bases downstream of the annotated TSS (Fig. 5B). Although some bases at the 5′ ends of NAD-RNAs might not be called by nanopore sequencing, this result indicates that a majority of NAD-RNAs might have a shorter 5′ UTR than m7G-RNAs. However, most NAD-RNA reads included the translation start sites (Fig. 5C) and translation end sites (Fig. 5D). Comparison of the 3′ ends of the NAD-RNA reads with those of non–NAD-RNA reads showed that the distributions of their polyadenylation sites were highly similar (Fig. 5E), suggesting that the NAD cap did not have an obvious effect on the selection of polyadenylation sites.
Discussion
Eukaryotic mRNAs have long been known to contain the m7G cap, which not only protects mRNA from degradation by 5′ to 3′ exonucleases but also functions in almost every other step of gene expression (15). The occurrence of the noncanonical NAD cap shows that RNA capping and decapping is more complex than previously assumed.
Identifying NAD-RNAs and revealing their structural features are essential steps toward understanding the functions of the NAD cap. The NAD tagSeq method allows more accurate identification and quantification of NAD-RNAs than previous methods. Furthermore, it can reveal the sequences of whole NAD-RNA transcripts. Using NAD tagSeq, we found that NAD-RNAs in Arabidopsis were produced from at least several thousand genes, mostly protein-coding nuclear genes.
Most of the NAD-RNAs in Arabidopsis appear to have a 5′ end mapped to a region downstream of the annotated TSS, raising the possibility that an alternative promoter might be used for transcriptional initiation for some NAD-RNAs. Other than their shorter 5′ UTR, the NAD-RNAs have the same nucleotide sequences as their m7G-RNA counterparts and usually include all coding sequences, suggesting that they could be translated. This notion is supported by the findings reported in PNAS by Wang et al. (16) that Arabidopsis NAD-RNAs are preferentially associated with the polysomal fraction under active translation. For m7G-RNAs, the cap recruits the nuclear cap binding complex, which mediates pre-mRNA splicing, polyadenylation, nuclear transport, and translation initiation (15). The way in which NAD-mRNAs can be processed like m7G-mRNAs and transported to the cytosol for translation remains to be determined. As NAD-mRNAs generally share the same sequences as m7G-mRNAs and have a shorter 5′ UTR, it also raises the possibility that for some NAD-mRNAs, the NAD cap could be added posttranscriptionally, perhaps following decapping of m7G-mRNAs.
The mechanisms by which NAD-RNAs regulate gene expression also remain unclear. Among the top 2,000 NAD-RNA–producing genes, 1,712 were also identified using NAD captureSeq by Wang et al. (16) (Dataset S3). It appears that transcripts from these 2,000 NAD-RNA–producing genes are more stable than the average over all transcripts. In addition, proteins from these genes are apparently relatively abundant, suggesting that NAD-RNAs might enhance the expression of the corresponding genes. These 2,000 NAD-RNA–producing genes are highly enriched in the GO terms related to photosynthesis, translational processes, and responses to cytokinin and stresses (oxidative stress and other stresses). NAD-RNAs could be involved in cellular energy and redox homeostasis. It is plausible that NAD-RNA levels might be modulated by cellular NAD levels or the NAD/NADH ratio, both of which are affected by environmental conditions and cellular energy status. NAD-RNAs could act as a signal to mediate cellular responses to environmental and intracellular cues. The NAD tagSeq method provides a very useful tool for comparing cellular NAD-RNA profiles under different environmental conditions.
Materials and Methods
Plant Growth Conditions.
Arabidopsis Col-0 seeds were germinated on solid half-strength Murashige and Skoog medium with 1% sucrose. Seedlings were grown on plates in a growth room at 23 °C under a 16-h light/8-h dark cycle using a light unit (photons) of 100 μmol⋅m−2⋅s−1 provided by cool white fluorescent lights.
Detection of the NAD+ Moiety in NAD-RNAs by MS.
Total RNAs were digested by nuclease P1 (Sigma). The retention time of the standard NAD+ was determined on an HPLC system. Fractions corresponding to the retention time of NAD+ were collected and applied to a LC-MS system. The method is described in more detail in SI Appendix, Supplementary Methods.
In Vitro Transcription for NAD-RNA Synthesis.
The template for the 38-nt NAD-RNA was generated by annealing a DNA oligo containing the T7 class II promoter (ɸ2.5) with its complementary sequence (SI Appendix, Supplementary Methods). Transcription was performed in a 400-μL reaction using the T7 RNA polymerase under the condition described in SI Appendix, Supplementary Methods.
RNA Extraction and Poly(A) RNA Isolation.
Seedlings for RNA extraction were harvested during 11:00 AM to 12:00 PM. Total RNA was extracted using the TransZol reagent (TransGen Biotech), and contaminated genomic DNA was removed with DNase l (New England Biolabs) following the manufacturers’ instructions. Poly(A) RNAs were isolated from total RNAs using Oligo d(T)25 Magnetic Beads (New England Biolabs) following the manufacturer’s instructions. RNA concentration was determined using a Qubit fluorometer (Invitrogen).
ADPRC Catalyzed Reaction.
This reaction was conducted following the method used for NAD captureSeq. (6). The procedure is described in more detail in SI Appendix, Supplementary Methods.
CuAAC.
RNA samples purified after the ADPRC reaction were incubated in 100-μL reactions containing 50 mM Hepes (pH 7.0), 5 mM MgCl2, 5 μM azide-RNA oligo (SI Appendix, Supplementary Methods), 1 mM CuSO4, 0.5 mM tris(3-hydroxypropyltriazolylmethyl)amine (THPTA), and 2 mM sodium ascorbate at 24 °C for 30 min with gentle vortex mixing. The RNAs were then purified using an RNA clean-up kit (Zymo Research).
Poly(A) Tailing.
After total RNA samples were tagged through the ADPRC reaction and CuAAC, the RNA samples were incubated in a 1-mL reaction containing 50 mM Tris⋅HCl pH 7.9, 250 mM NaCl, 10 mM MgCl2, 2 mM ATP, and 5,000 U E. coli poly(A) polymerase (New England Biolabs) at 37 °C for 30 min. The reaction was stopped, and RNA was purified using an RNA clean-up kit (Zymo Research).
Removal of Free tagRNAs.
Unreacted azide-RNAs were separated from poly(A) RNAs by using 3.0 mg Oligo d(T)25 Magnetic Beads (New England Biolabs) according to the manufacturer’s instructions. The buffers are described in SI Appendix, Supplementary Methods.
Enrichment of tagRNA-Linked NAD-RNA.
The DNA-biotin probe for capturing tagRNAs (see SI Appendix, Supplementary Methods for its sequence) was immobilized on streptavidin magnetic beads (New England Biolabs). tagRNA-linked RNAs were captured by hybridization with the probe DNA. Following wash with wash buffer and low-salt buffer, the hybridized RNAs were eluted and purified. The buffers are described in SI Appendix, Supplementary Methods. RNA concentration was determined using a Qubit fluorometer (Invitrogen).
Nanopore RNA Sequencing.
For each RNA sample, 100 to 500 ng was used to prepare a library using the Nanopore Direct RNA Sequencing Kit following the manufacturer’s instructions (Oxford Nanopore Technologies). Each library was loaded onto a flow cell (R9.4) and sequenced on the sequencing devices MinION or GridION. Base calling was conducted using the Albacore or Guppy software (Oxford Nanopore Technologies).
Processing and analysis of sequencing reads.
Reads were aligned to the reference genome and transcriptome of Arabidopsis thaliana Ensembl 41 separately using minimap 2 (v2.12) (17). For reads with multiple alignments, only alignments with the best mapping quality scores were kept. Reads with or without the tagRNA sequence were differentiated using a Python script (see SI Appendix, Supplementary Methods for details). Counts of reads from each gene were normalized as TPM.
Correlation, Functional Analysis, and Visualization.
Pairwise Pearson correlation values were calculated by using the stats package in R 3.5.1 (https://www.R-project.org/). GO analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (18), and terms with a P value after Bonferroni correction of <0.05 were considered to be significant. The Integrative Genomics Viewer (19) was used to visualize the NAD-RNAs and non–NAD-RNAs.
Raw Data.
Raw fastq data have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus repository (accession no. GSE127755).
Supplementary Material
Acknowledgments
We thank Drs. Runsheng Li (Department of Biology, Hong Kong Baptist University), Daogang Guan (School of Chinese Medicine, Hong Kong Baptist University), and Thomas Bray (Oxford Nanopore Technologies) for helpful discussions. This work was supported by Research Grants Council of Hong Kong (General Research Fund Grant nos. 262212, 12100415, 12100018, and AoE/M-403/16 to Y.X.) and by Hong Kong Baptist University (Grant nos. RC-ICRS/16-17/04 and SDF15-10120-P04 to Y.X.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Raw fastq data have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus repository, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE127755).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903683116/-/DCSupplemental.
References
- 1.Meyer K. D., Jaffrey S. R., The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapp L. D., Lorsch J. R., The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 73, 657–704 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Ramanathan A., Robb G. B., Chan S. H., mRNA capping: Biological functions and applications. Nucleic Acids Res. 44, 7511–7526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y. G., Kowtoniuk W. E., Agarwal I., Shen Y., Liu D. R., LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 5, 879–881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cahová H., Winz M. L., Höfer K., Nübel G., Jäschke A., NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519, 374–377 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Winz M. L., et al. , Capture and sequencing of NAD-capped RNA sequences with NAD captureSeq. Nat. Protoc. 12, 122–149 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Jiao X., et al. , 5′ End nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-mediated deNADding. Cell 168, 1015–1027.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walters R. W., et al. , Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 114, 480–485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird J. G., et al. , The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 535, 444–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frindert J., et al. , Identification, biosynthesis, and decapping of NAD-capped RNAs in B. subtilis. Cell Rep. 24, 1890–1901.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Vvedenskaya I. O., et al. , CapZyme-seq comprehensively defines promoter-sequence determinants for RNA 5′ capping with NAD. Mol. Cell 70, 553–564.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Workman R. E., et al. , Nanopore native RNA sequencing of a human poly(A) transcriptome. 10.1101/459529 (9 November 2018). [DOI] [PMC free article] [PubMed]
- 13.Narsai R., et al. , Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19, 3418–3436 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motohashi R., Rödiger A., Agne B., Baerenfaller K., Baginsky S., Common and specific protein accumulation patterns in different albino/pale-green mutants reveals regulon organization at the proteome level. Plant Physiol. 160, 2189–2201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topisirovic I., Svitkin Y. V., Sonenberg N., Shatkin A. J., Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA 2, 277–298 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Wang Y., et al. , NAD+-capped RNAs are widespread in the Arabidopsis transcriptome and can probably be translated. Proc. Natl. Acad. Sci. U.S.A. 116, 12094–12102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dennis G., Jr, et al. , DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3 (2003). [PubMed] [Google Scholar]
- 19.Robinson J. T., et al. , Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.