Significance
Mimicry among bumble bees has driven them to diversify and converge in their color patterns, making them a replicate-rich system for connecting genes to traits. Here, we discover that mimetic color variation in a bumble bee is driven by changes in Hox gene expression. Hox genes are master regulators of numerous segment-specific morphologies and thus are some of the most conserved developmental genes across animals. In these bees, the posterior Hox gene Abd-B is upregulated in a more anterior location to impart phenotypic change. This homeotic shift happens late in development, when nonspecific effects are minimized, thus availing these genes for color pattern diversification. Similar mimetic color patterns were inferred to use different mutations, suggesting diverse routes to mimicry.
Keywords: Bombus, Müllerian mimicry, evo–devo, Abd-B, Hox
Abstract
Natural phenotypic radiations, with their high diversity and convergence, are well-suited for informing how genomic changes translate to natural phenotypic variation. New genomic tools enable discovery in such traditionally nonmodel systems. Here, we characterize the genomic basis of color pattern variation in bumble bees (Hymenoptera, Apidae, Bombus), a group that has undergone extensive convergence of setal color patterns as a result of Müllerian mimicry. In western North America, multiple species converge on local mimicry patterns through parallel shifts of midabdominal segments from red to black. Using genome-wide association, we establish that a cis-regulatory locus between the abdominal fate-determining Hox genes, abd-A and Abd-B, controls the red–black color switch in a western species, Bombus melanopygus. Gene expression analysis reveals distinct shifts in Abd-B aligned with the duration of setal pigmentation at the pupal–adult transition. This results in atypical anterior Abd-B expression, a late developmental homeotic shift. Changing expression of Hox genes can have widespread effects, given their important role across segmental phenotypes; however, the late timing reduces this pleiotropy, making Hox genes suitable targets. Analysis of this locus across mimics and relatives reveals that other species follow independent genetic routes to obtain the same phenotypes.
Understanding how genomic change drives phenotypic evolution is a central goal of evolutionary biology. Such genotype-to-phenotype connections enable us to decipher the genetic code and together provide predictable trends, such as whether evolution tends to be mediated by few large-effect or many small-effect loci (1–3), targets the same genes and mutations for the same phenotypes (4–7), and modifies genes in cis- or trans- (8, 9), and whether certain types of genes are evolutionary hot spots (8, 10, 11). Adaptive traits that evolve independently in two or more lineages are especially suited for understanding these trends as they provide natural replicates (11).
Müllerian mimicry, in which two or more distasteful species share similar aposematic signals to avoid predation, presents exceptional examples of evolutionary convergence and thus potential for evolutionary genetics (12–15). Numerous insights have been gained, for example, in studying the genetic basis of wing pattern convergence in mimicry complexes of Heliconius butterflies. These studies have revealed that butterfly wing patterns are generated by cis-regulatory modifications to a few major developmental genes co-opted for novel function in the wing (16–20) and that diversity and convergence in phenotypes can involve different regulatory modifications to these same loci (21–23). Targeting of these genes has also revealed microevolutionary processes, showing that mimetic phenotypes can be differentially partitioned across populations and species through processes of ancestral allele sorting (24) and adaptive introgression (21, 25).
Bumble bees exhibit a similarly striking level of color pattern diversity partially as a result of Müllerian mimicry. The ∼260 species in the genus (26) exhibit over 400 color patterns, making most bumble bee species polymorphic (27). Bumble bee color patterns are bright and contrasting, an aposematic signal of their toxic sting (28, 29). Color is imparted by dense, setal pile coating the cuticle and is highly modular, tending to change color—switching between white, black, red-orange, brown, or yellow—by segmental sclerites of the head, thorax (mesosoma), and abdomen (metasoma) (27, 30). Many of the conceivable combinations of color across these segments have been achieved (refs. 30 and 31 and Fig. 1A). Although multiple ecological factors may play a role in bumble bee color diversity, Müllerian mimicry is believed to drive much of their polymorphism (27). Color patterns have been documented to converge geographically onto over 24 global mimicry complexes, and species that span multiple mimicry complexes tend to converge onto each local pattern (27, 32, 33). This system, thus, has potential for revealing evolutionary developmental and genetic processes: it can inform processes driving sclerite identity and has abundant replicates across global mimicry complexes to assess whether similar mechanisms generate the same phenotypes and how these variants sort across populations and species.
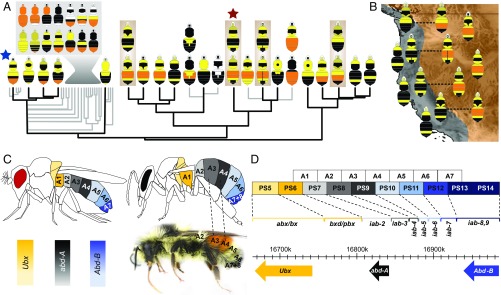
Fig. 1.
An evolutionary framework for color pattern evolution in bumble bees. (A) Phylogeny of one subgroup of bumble bees, Pyrobombus (phylogenetic relationships data from ref. 31; 19% of all bumble bee species), that contains the study taxon B. melanopygus (red star), with the primary color patterns for those clades mapped. This emphasizes the diversity and modularity of coloration, the most common and ancestral bumble bee pattern (blue star), and four comimetic species that change their color in parallel (brown boxes). Gray box, Primary patterns from the indicated clade; gray branches, non-North American taxa. (B) Map conveying mimetic patterns of western North America showing species in each mimicry zone from A, with polymorphic species indicated with dotted lines. Additional species outside of Pyrobombus also mimic these patterns. (C) Spatial distribution of the BX-C Hox genes in D. melanogaster (Left), expected expression for male bumble bees (Right) applying the Drosophila model, and its translation to B. melanopygus. The color polymorphism in B. melanopygus takes place at A3 (metasomal T2) and A4 (metasomal T3), corresponding to segments controlled by abd-A. Color intensity in diagrams reflects relative amount of expression of the dominant gene. (D) Protein coding sequences of BX-C Hox genes (bottom) and infraabdominal (iab) expression domains (bracketed) in Drosophila melanogaster. BX-C gene expression is controlled by colinear regulatory domains, with each parasegment (PS) controlled by a different iab region (bracketed). Boxes are colored by the gene and corresponding segments of the adult fly that they regulate (C). Figure drawn from data in ref. 40.
To begin to uncover the genomic drivers of the mimetic color pattern radiation in these bumble bees, we examine color pattern variation in mimicry complexes of western North America. Two major mimicry complexes, the Pacific Coastal and the Rocky Mountain complexes (34) occur in this region and are converged upon by multiple monomorphic (∼10 spp.) and polymorphic bumble bee species (∼7 spp.). Most polymorphic species spanning these zones converge onto each local complex simply by changing their midabdominal coloration from black (Pacific) to orange-red (Rocky Mountain) (Fig. 1 A and B). In this study, we decipher the genetic basis of this red–black color switch in Bombus melanopygus, a species that exhibits a discrete red/black dimorphism (Fig. 1A). Previous genetic crosses and population genetic work on this species revealed that its color variation is likely driven by a single two-allele Mendelian locus of unknown identity, with red dominant, and that there is no population structure between color forms in the hybrid zone (35, 36). This combination of traits is ideal for targeting the locus driving this trait.
The setae (“hairs”) of bumble bees are single-cellular cuticular structures extending from a single epithelial layer underlying the cuticle. Black and red setae differ primarily in relative amount of melanin, with black setae containing some combination of both eumelanin and pheomelanin and red containing exclusively pheomelanin, a pigment until recently not thought to occur in insects (37). In Drosophila, multiple enzymes in the melanin pathway have been targeted to drive melanic variation in body cuticle, each time through cis-regulatory mutations in the pigment enzyme that presumably alters binding of upstream transcription factors (38). Segment-specific variation, such as sex-specific variation in posterior melanization (39), is regulated by altering the ability of segment-specific genes to bind. Considering this and the segmental fashion of the bumble bee color pattern, our initial hypothesis was that melanic pattern variation in these bees likely involves a cis-regulatory modification in an unknown pheomelanin enzyme, which alters its ability to bind upstream segmental identity genes, such as the Hox genes. Given the midabdominal location of the phenotype, the most likely Hox gene affected would be abdominal-A (abd-A) (ref. 40 and Fig. 1 C and D).
In the current study, we employ a comparative approach, combining genome-wide association analysis, subsequent population-level genotyping to refine the locus, and gene expression analysis to characterize the genomic locus driving adaptive mimetic color variation in B. melanopygus. We examine the refined locus across sister lineages and comimics to determine whether comimetic species have evolved their patterns using similar mechanisms. The results establish the bumble bees as a system fruitful for further evolutionary genetics research.
Results and Discussion
Genetic Locus Driving Color Pattern Variation.
Genome-wide association analysis between 21 red and black B. melanopygus males sampled from across the hybrid zone resulted in a single fixed ∼50-kb peak of association in an intergenic domain between Hox genes abd-A and Abd-B of the bithorax complex (BX-C) (Fig. 2). In insects, the BX-C cluster involves three segmental Hox genes, abd-A, Abd-B, and Ubx, which specify posterior segmental phenotypes (Fig. 1C). Further fine-scale Sanger sequencing of 133 individuals across this block narrowed the associated region to a 4-kb, zero-recombinant interval closest to and ∼50 kb downstream from the Abd-B transcript. In Drosophila, the intergenic region between abd-A and Abd-B (the iab region) is the primary regulatory region directing expression of both adb-A and Abd-B. The iab region is divided into several noncoding domains (iab-2 to -9) that direct the segmental expression of these genes in a collinear fashion, with the regions closest to abd-A directing the most anterior segmental expression of this gene (iab-2) and those closest to Abd-B directing the most posterior Abd-B expression (iab-9) (Fig. 1 C and D) (41–43). Only a small proportion of the B. melanopygus abd-A/Abd-B region can be reliably aligned to Drosophila, including the iab-4/8 micro-RNA (44) and the protein-coding sequences of the two Hox genes (SI Appendix, Fig. S1). This is not unexpected, as the iab regions exhibit 12% sequence homology even across Drosophila species (45). The relative location of the iab-4 miRNA in this intergenic region, however, appears to be conserved across insect lineages, including B. melanopygus (46–48), suggesting some level of conservation in relative locations of regulatory domains. If in bumble bees iab regions were to function in the same collinear manner as in Drosophila, the relative location of the fixed color locus in B. melanopygus would most likely fall in iab-6/7, which in Drosophila would direct the expression of primarily Abd-B in the most posterior tergites (A6 and A7; bumble bee “tail”) (49) (Fig. 1 C and D and SI Appendix, Fig. S1).
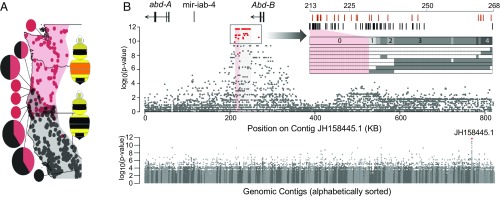
Fig. 2.
The color locus is revealed using a genome-wide association study (GWAS). (A) Map of localities from which GWAS samples were collected. The dots on the map indicate the overall distribution of each color form from specimen data (34). The pie charts indicate sites where samples were collected for this analysis with the size and portion of these charts reflecting the number of samples per locality (n = 1–5). (B) Whole-genome association (Bottom) showing that sites of fixed association (indicated in red) are confined to a single contig (contigs in alternating gray). The association within this contig (above) demonstrate that fixation (red dots) is confined to a ∼50-kb block 3′ of Abd-B (highlighted gray). Highlighted in red is the region that remains fixed with additional genotyping, further highlighted with the Inset SNP plot. The rows at the Bottom of the Inset plot represent ordered SNP/indel variants of recombinant individuals, with gray indicating SNP/indels belonging to the opposite phenotype in genomic samples and which thus break fixation. The number of individuals breaking fixation is indicated above these lines. At the Top, SNPs (black) and indels (red) and their position are indicated, and fixed sites are aligned with the respective sites in the plot below using highlighting.
BX-C Gene Expression Across Bumble Bee Development.
The location of the color locus between abd-A/Abd-B suggests that this phenotype is likely driven by shifts in expression of one of these Hox genes. Holometabolous insects undergo two major developmental transitions: (i) in the embryo to build larvae, and (ii) during late larval and pupal stages to build adult structures. The majority of developmental work focuses on how these Hox genes operate to establish segmental identity in embryos; however, Hox genes are likely to operate throughout development (50, 51). All three BX-C genes were shown to be expressed in the developing adult abdominal epidermis in Drosophila pupae in a spatial pattern similar to the embryo, although with more discrete domains (52). In bumble bees, setae develop in early white to pink-eyed pupal stages and are pigmented in the late pupal stages; thus, the relevant gene expression patterns for color should occur in pupae (53). To test how Hox genes fluctuate across metamorphosis in bumble bees and determine their potential to play a role in pupal stages, we performed qPCR in the model bumble bee Bombus impatiens at different developmental stages including whole embryos and larvae, and on metasomal terga of pupal and early adult stages (callow). mRNA of the three BX-C genes were detected across all developmental stages, even into adulthood (Fig. 3A). The level of mRNA expression, however, fluctuated, with peak levels of mRNA appearing at the major periods of metamorphosis, including in embryos and during the larval–pupal transition. This correlation with developmental change suggests that Hox genes continue to be players as fate-determining selector genes across development rather than relying on triggered effector genes downstream to regulate these phenotypes.
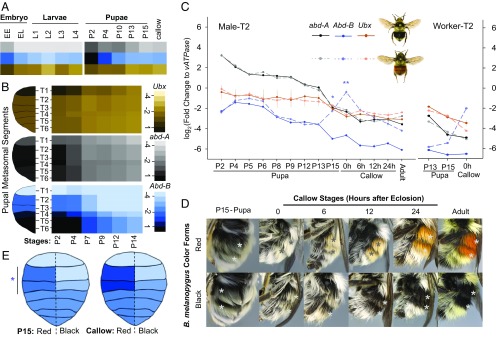
Fig. 3.
Expression pattern of the bithorax complex (BX-C) homeotic genes during bumble bee development. (A) Relative expression of BX-C genes across B. impatiens developmental stages, including embryo, larvae (whole body), pupa, and early adult (metasomal tergites). (B) Spatial and temporal distribution of the BX-C genes in metasomal tergal segments of B. melanopygus pupae (worker, red form) from stage P2 to state P14. Shades for both (A and B) represent log2 relative fold change to vATPase. Right lane, scales for relative expression level. (C) Temporal expression pattern of the BX-C genes at the dimorphic metasomal tergal segment 2 (T2) of male B. melanopygus. Note that, in the callow adult stage, the relative abundance of Abd-B is higher than abdA and Ubx. *P < 0.01, **P < 0.001, Mann–Whitney U test. Abd-B expression was marginally significantly higher with <2× difference in the red form at P5E (P = 0.04) and P6 (P = 0.03); however, this difference is only seen in T2 but not T3 (SI Appendix, Fig. S2). A similar homeotic shift of Abd-B in the red form is also observed in polymorphic segments of workers (Right) (P = 0.057 at P15; no statistics applied at 0 h as only one sample is in black form). (D) Setal color development in late pupal and early adult stages of B. melanopygus. The stars indicate polymorphic segments. For P15 pupae, pupal cuticle was removed to reveal the developing adult setae underneath. The red and black setae are distinguishable in males starting from 0-h callow adult. Color continues to intensify in the first 24 h posteclosion. (E) Abd-B expression across abdominal tergites of B. melanopygus at stage P15 and in the newly eclosed callow adult showing up-regulation of Abd-B only in T2 and T3 for both of these stages, and only in the red form (segments on the Left side), not the other segments and the black form.
We further examined the location and timing of expression of each of the BX-C genes in B. melanopygus by metasomal segment. In pupal metasomal tergites of B. melanopygus, Abd-B is expressed primarily in metasomal segments T4–6 (Fig. 3B) [abdominal segments A5–7 (Fig. 1D)], abd-A in metasomal segments T1–6 (Fig. 3B) [abdominal segments A2–7 (Fig. 1D)], with highest levels in T3–4, and Ubx primarily in metasomal segment T1 (Fig. 3B) [abdominal segment A2 (Fig. 1D)]. The pupal expression pattern of the BX-C genes in bumble bees is thus consistent with their conserved spatial pattern in embryos across insects and other arthropods (i.e., Ubx anteriorly, abd-A medially, Abd-B posteriorly; Fig. 1 C and D) (40, 46, 51, 54). Consistently across segments, the expression of each BX-C gene was highest at the earliest stages of pupal development and declined thereafter (Fig. 3B).
BX-C Gene Expression Between Color Phenotypes.
To determine whether setal color variation in B. melanopygus is associated with differential expression of BX-C genes during adult setal development, we performed qPCR on dissected pupal epidermal tissues of the dimorphic second and third metasomal terga (T2 and T3), run separately, on male pupae of the red and black phenotypes. As BX-C genes were determined to be expressed throughout early pupal to adult stages, we analyzed differential expression patterns across pupal–adult development to avoid missing any critical stages of gene regulation. Nine pupal and four callow adult stages, representing fine increments of pupal–adult development, were sampled with staging based on morphological criteria (53). In both red and black bees, all three BX-C genes exhibit the highest levels of expression at the early pupal stage, with abd-A expression being higher than Ubx and Abd-B (Fig. 3C), as is expected given the typical dominance of abd-A in midabdominal segments. The expression of all three genes gradually decreases during pupal development, while their relative levels remain the same. Dramatic differences, however, were seen at the late pupal stages where a significant up-regulation of Abd-B was observed only in the red form in both dimorphic metasomal terga (T2 and T3), while abd-A and Ubx expression continued to decrease [T2 (Fig. 3C); T3 (SI Appendix, Fig. S2)]. In the red form, mRNA levels of Abd-B increase starting between stages P13 and P15 and peak with a 10–20× expression difference (P < 0.001) from the black form at the time of eclosion (0-h callow adult), returning to low background levels by 24 h posteclosion (Fig. 3C). Similar differentiation of Abd-B expression between the two color forms was also observed among the female worker caste (Fig. 3C). This Abd-B up-regulation was confined only to the dimorphic second and third metasomal segments, and not to the first segment or the more posterior segments (Fig. 3E), supporting its role in generating the dimorphic differences observed in these segments. This is further supported by the timing of the differentiation, which is well aligned with the timing of differential setal pigmentation (Fig. 3D). Pigments for both red and black setae start to deposit as early as the late-P12/P13 pupal stage (∼6 d after pupation in workers) as a pale greyish orange color; however, the differences in color between black and red setae are not macroscopically distinguishable until around stage P15 in queens and 0-h callow adults in male and workers (∼8 d after pupation), when black setae exhibit a grayer tinge, while the red setae become more orange (Fig. 3D). Both red and black setae attain most of their color within the first 6 h posteclosion and complete pigmentation by 12–24 h (Fig. 3D) The expression of this gene concordant with the expression of the phenotype is in contrast to regulators of pigmentation in butterflies, which are turned on to premap the wing pattern early in development before wing pigmentation actually takes place (16, 55). The function of Abd-B is similar to its mode of action during embryonic development, where it controls onsite segment identification, and is typical for a selector gene (56, 57).
During this up-regulation, Abd-B mRNA has higher expression than that of the other BX-C genes, including abd-A (Fig. 3C). In Drosophila, Abd-B is considered to exhibit dominance over abd-A, leading to repression of abd-A translation and Abd-B segmental identity where it occurs (58). In other cases, combinatorial effects rather than complete dominance interactions have been observed (59, 60). The increase of Abd-B over abd-A expression suggests that regardless of the dominance mode, this uptick likely generated a homeotic shift late in development of the T2 and T3 segments from midabdominal to more posterior-abdominal identity.
These results suggest that shifting the expression of the major developmental gene Abd-B likely was the initial trigger driving a shift in melanin gene expression in B. melanopygus. Previous work examining genes driving natural and laboratory-generated differences in melanization in Drosophila support mutations occurring primarily in cis-regulatory regions of pigment enzymes (38). These studies have served as examples of how the genome tends to be targeted by changing downstream targets in cis-, where these mutations have reduced pleiotropic consequences (38, 61). Although they are not typically the evolutionary target, upstream players like Hox genes have been found to play a role in differential melanization in Drosophila (61). For example, sex-specific variation in the expression of developmental gene bric-a-brac impacts the ability of Abd-B to trigger tissue-specific differences in melanization by sex (39, 62). Abd-B is not known in this case to change its expression to mediate natural adaptive color pattern variation, but rather directs the posterior location of the dimorphism, acting as it typically does as a segment-specific selector. Shifts in the Hox genes themselves could be occurring in Drosophila, as minor effect mutations that could drive differences in abd-A and Abd-B expression have been linked to sex-specific pigment variation in some species (63).
Hox genes play diverse roles in driving the fate of all phenotypes in their respective segments; thus, changes in these genes can have highly pleiotropic consequences. Likely because of the deleterious effects of such mutations, the relative location of expression in these genes during embryogenesis is conserved across insects and other arthropods (51). In the case of bumble bees, the genetic changes in the Abd-B cis-regulatory region are not associated with any other apparent phenotype that would suggest segmental homeosis. This may be because the shift of Abd-B generated by these genetic changes occurs at the end of development, when the only marked changes in morphology involve melanization and sclerotization. Shifting Abd-B expression at these stages may thus switch setal pigmentation with minimal pleiotropic impacts on other tissues. These consequences can be even further reduced if up-regulation of Abd-B was cell-specific, as insect setae are single-celled structures (64). If negative pleiotropic consequences can be avoided, upstream regulators are excellent targets for evolution and diversification, as these genes have already established numerous developmental genetic connections to impart change (8, 65).
Late developmental spatial shifts in expression of Hox and other upstream regulators can be an important mechanism for driving phenotypic change (57, 66). For example, in arthropods, localized shifts in Hox genes in later stages are responsible for variation in limb morphologies. Shifts in Scr expression influence male sex comb morphology in Drosophila forelimbs (67) and the formation of crustacean maxillipeds (68). Similarly, regional up-regulation of Ubx along insect hindlimbs drives growth and setal loss (69, 70), such as that observed in the expanded corbicula pollen basket in honey bees and bumble bees (71). Such variation during late development could be driven by Hox genes evolving to respond to downstream components of regulatory networks, further enabling developmental micromanagement (57, 67).
This leaves open the question as to how a homeotic shift generates an on–off switch for pigmentation in bumble bees. Many genes throughout the genome pick up binding sites for Hox genes to direct their segment-specific expression. At one point, cis-regulatory binding of Abd-B may have been picked up by a gene in the melanin pathway to turn on red pigmentation where it occurs in the posterior of the abdomen. The most common pattern across bumble bees is a yellow bee with a red-tipped abdomen (ref. 27 and Fig. 1A), an aposematic signal to frame its venomous sting. This is most likely the ancestral bumble bee pattern (e.g., it is common in the early diverging Mendacibombus) and is common throughout the Old World where bumble bees originated (31). The location of ferruginous stripes more anteriorly in bumble bees is a derived condition primarily found in the North American Pyrobombus, belonging to B. melanopygus and several of its close relatives (Fig. 1A). A more anterior expression of red color could have thus been achieved merely by relocating Abd-B expression from posterior to anterior segments, a mutation that, once generated, may have no other fitness consequences due to the loss of pleiotropy in these tissues late in development. Co-option of established gene networks in new locations to generate new phenotypes (heterotopy) is an emerging source of evolutionary novelties. For example, Drosophila wing spots were generated by the wingless gene being co-opted to turn on pigmentation in its original location of expression, followed by wingless shifting expression to multiple additional parts of the wing to generate spots (72). Similarly, the optix gene is involved in eye formation and pigmentation in insects and has been co-opted in butterflies to turn on the same pigments in the wing (19, 73). As Hox genes are major genes influencing the location of phenotypes, these genes could help promote rapid changes in color pattern.
Concerted Mutations Operate in the Regulatory Domain.
Beyond the 4-kb locus, several recombinant red-form individuals were found bearing black haplotypes in the remaining block that is fixed in genomic samples (Fig. 2B and SI Appendix, Fig. S3). Although appearing primarily red by eye, detailed color analysis revealed that individuals with these recombinant haplotypes exhibit various levels of penetrance of the black phenotype, with more black setae mixed among the red. This suggests that, although the narrowed 4-kb fixed region contains sequence variants able to cause a nearly full color switch, there are variants within the interval outside this region that contribute to the phenotype. This explains why the fixed region occurs in a large block and lends support to adaptive phenotypes being driven by multiple linked SNPs that have evolved in concert as opposed to single mutations, as has been shown in regulatory regions of Drosophila (74–76). The red form was previously determined to have complete dominance, retaining red color in heterozygous individuals (35). With our sequencing of the color locus, we were able to identify and compare homozygous and heterozygous females, and found that heterozygotes exhibited black hair penetrance similar to the recombinants (SI Appendix, Fig. S3), suggesting that gene expression is reduced in a similar manner for both. This indicates that dominance is partially incomplete.
The Color Locus and Abd-B Regulation.
The function of the mutations in this color locus in driving Abd-B differential regulation remains unknown. The location of the color locus relative to known segmental regulatory domains in Drosophila suggests it may normally regulate more posterior segments than it does, in line with the idea that this region may have first evolved to regulate color in more posterior phenotypes and have been subsequently mutated to generate an anterior shift. Laboratory-generated iab cis-regulatory mutants in Drosophila can generate such an effect (77, 78), with homeotic shifts in segmental identity generated, for example, by altering insulators demarcating iab segmental regulatory regions (79, 80). Furthermore, in Drosophila, the entire region between abd-A and Abd-B is transcribed and spliced into a massive nc-RNA (iab-8-ncRNA) (41, 42, 49). This could be another source of Abd-B regulation (81). Further research on this regulatory locus would enable a more complete understanding of the regulatory mechanisms driving the Hox toolkit.
Implication of the Color Locus in Comimetic Species.
To test whether the same genetic mechanism underlies parallel color variation displayed by comimetic species and decipher the ancestry of these phenotypes, we compared sequences of the fixed color locus from four dimorphic species displaying similar black and red color variation in metasomal segments T2 and T3 as well as in six monomorphic sister lineages bearing either red or black phenotypes in these segments. None of the fixed SNPs and indels for B. melanopygus in an 18.5-kb genomic region including our fixed interval show the same association with color variation in other species, even those species most closely related to B. melanopygus (e.g., B. sylvicola) (Fig. 4). This supports the independent evolution of at least one of the two phenotypes from the ancestor in B. melanopygus. Mimetic color variation in these western North American mimicry complexes thus involves multiple, independent genetic mutations, rather than sorting of ancestral variation, even among close relatives with similar phenotypes. This lends support to the potential for mutation as opposed to alternative allele sorting in contributing to phenotypic diversity (82–84). This contrasts with Heliconius butterflies, where close relatives obtained their patterns from a common ancestral mutation, either through introgression (25) or ancestral polymorphisms (24). Although the same SNPs are not implicated, it is possible the same gene may be involved in these other species, but using different genetic targets. This was observed, for example, in Heliconius butterflies H. erato and H. melpomene, where comimicking forms were generated by the independent targeting of different parts of the cis-regulatory region of same gene, optix (23, 85, 86).
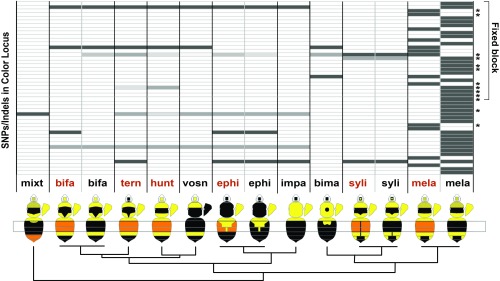
Fig. 4.
Allelic variants in the color locus of B. melanopygus comimics and close relatives. No fixed SNP or indel are similarly differentiated by color in other species. At Bottom are species related to B. melanopygus (Right, “mela”) that vary in the red/black color of metasomal segments T2 and T3 (segments indicated with a gray box) and their phylogenetic positions (Fig. 1). Species names and the respective color of the specimens are abbreviated above each picture. A map of the character state of each ordered SNP or indel (starred) across an 18.5-kb window that includes the narrowed fixed block (Fig. 2) is shown above. SNPs that are derived are shaded dark gray and ancestral alleles are white. Medium gray shading indicates alternative SNPs and indels to those in B. melanopygus.
Most likely, the independently evolved phenotype is a black form from the red form. This is inferred because the red form tends to contain the same conserved ancestral sequences as the other taxa with most of the functional SNPs and indels derived in the black form (Fig. 4). The black form exhibits higher linkage disequilibrium (LD) and reduced nucleotide diversity across the color locus contig, suggesting that the region is under selection and more so in this color form (SI Appendix, Fig. S5). If black is derived, this seems counter to gene expression patterns, as red, not black, up-regulates Abd-B. Further gene expression analyses across bumble bee species are needed to examine when Abd-B up-regulation originated to determine whether this is a gain in the red form or a loss in the black form.
Conclusion and Future Directions.
Our study demonstrates that evolution can target well-connected, highly conserved, pleiotropic developmental genes to generate phenotypic variation. Modifying regional selector genes, such as Hox genes, in cis- allows time-specific changes in expression, thus reducing pleiotropic consequences and making these genes suitable targets for driving region-specific variation in phenotypes. Targeting positional genes, such as Hox genes, may enable the diversification of segmental phenotypes seen in this lineage. Bumble bee coloration is a new system for evolutionary genetics. With this discovery, we can begin to explore how segmental variation is regulated and the multitude of ways evolution has achieved the same phenotypes. As a nonmodel group, functional genetics is more challenging, but the comparative potential provided by the extent of interspecific and intraspecific variation still enables strong candidates to be identified and can inform evolutionary processes. Studying abdominal Hox gene expression during pupal development across species will be important for understanding the evolutionary events leading to this phenotypic transition. Furthermore, few natural adaptive loci have been found in cis-regulatory regions of Hox genes. Deciphering how this regulatory region works in this system will help inform how Hox genes operate and evolve in insects beyond Drosophila.
Materials and Methods
Genomic Analysis.
We sequenced genomes of red (n = 11) and black (n = 10) males of B. melanopygus from multiple localities throughout their morphological transition zone (Fig. 1 and SI Appendix, Table S1) including one individual per form outside this zone. Males are haploid, thus enabling improved alignment, assembly (87), and avoidance of dominance effects. DNA extraction was performed on thoracic muscle tissue using Qiagen DNeasy or EZNA (Omega Bio-tek) DNA extraction kits with RNaseA treatment. Samples were barcoded, and prepared for Illumina sequencing using Illumina TruSeq DNA Nano kits. Whole-genome sequencing was performed 5–10 samples/lane on an Illumina HiSeq 2500 sequencer in Rapid Run mode to generate 150-bp paired-end read libraries at the Penn State Huck Genomics Core Facility. Trimming was applied using Trimmomatic, version 0.38 (88), to remove low-quality bases, clip adaptors, and discard short-length sequences (<36 bp). Reads were then aligned to the published genome assembly (89) of closely related species Bombus impatiens (GCA_000188105.2, BIMP_2.0) [common ancestor with B. melanopygus ∼10 mya (31)], using BWA-mem algorithm (90) of BWA, version 0.7.17 (91), aligner. Alignment postprocessing was conducted in SAMtools 1.8 (92) and Picard 1.119 (broadinstitute.github.io/picard/index.html). Multisample variant calling was performed using GATK Unified Genotyper (version 3.6-0) (93, 94) with parameters set for haploidy. After filtration of variable sites with substantial missing data (>25%) with VCFtools 0.1.15 (95), a genotype–phenotype association analysis between case-control phenotypes (black and red) and the SNP dataset was performed in PLINK 1.9 (96) using Fisher’s exact test.
To identify fixed insertions and deletions in the color locus, de novo genome assemblies of B. melanopygus including two for each color form were performed using MIRA assembler, version 4.9.6 (97). Indels were manually identified from assembled contigs and checked for fixation using IGV (98). Summary statistics for genomic samples are available in SI Appendix, Table S1. Genomic sequencing data are available under NCBI BioProject accession number PRJNA526235 (99). Additional data are available in the Dryad Digital Repository (100).
Genotyping.
To narrow the locus regulating the phenotype, genotyping was performed on 80 red and 53 black B. melanopygus, including both males (n = 108) and females (n = 10 queens, 15 workers). These were sampled mostly from within the transition zone in Oregon and California, but also included areas where exclusively red or black morphotypes occurred in California, Oregon, Washington, and Utah (SI Appendix, Table S2). Individuals were subsampled for SNPs and indels across the fixed 50-kb interval to look for potential recombinants, which were then sequenced across all SNPs and indels to better define the window of fixation. DNA was extracted with Qiagen DNeasy or EZNA DNA kits, purified with ExoSAP-It (Thermo Scientific), and PCR amplified (HotStart Mastermix; NEB) with custom-designed primers (SI Appendix, Table S3). Sanger sequencing was performed at the Penn State Huck Genomics Facility. SNPs were called from chromatograms with heterozygosity determined from double peaks. A subset of black males and females, red homozygous males and females, all recombinants, and red heterozygous females were phenotyped under the microscope to determine the extent of mixture of black hairs in red (SI Appendix, Fig. S3).
To examine whether the fixed SNPs in B. melanopygus are implicated in seemingly parallel color variation in closely related species, we sequenced all SNPs and indels in the narrowed fixed interval as well as an additional 4 kb flanking this from red and black T2/T3 polymorphic and monomorphic sister lineages, including nearly all of the closest relatives. SNPs were called as above and designated as ancestral or derived using parsimony principles. Data are available from the Dryad Digital Repository (100).
qPCR.
Developmental stages of B. impatiens (Koppert Biological Systems) were used to assess general expression of Ubx, abd-A, and Abd-B across bumble bee development (experiment I; Fig. 3A). Sampling included early (no visible larval segments) and late (visible larval segments) embryos, four larval stages determined based on head capsule width (101), pupae from five stages (53), and a young adult callow, including a single specimen per stage. Embryos and larvae were sampled whole, while pupae and callow adults included dissected (1× PBS) abdominal tergite epidermis with fat body and muscle tissues removed.
We then compared Ubx, abd-A, and Abd-B gene expression by tergite throughout pupal development in B. melanopygus black color form workers (experiment II; Fig. 3B). Colonies of B. melanopygus were started from queens from California and Oregon. Abdominal tergites were dissected as above, from reared worker pupae of both red and black forms across six developmental stages (P2, P4, P7, P9, P12, and P14) (53) collecting four tergal regions: metasomal segment T1, T2+3, T4, and T5+6 with three to five replicates per condition.
To compare gene expression between red and black metasomal tergites T2 and T3 (experiment III; Fig. 3C), segments were dissected from male pupae and callow adults of both B. melanopygus color forms from nine pupal stages (SI Appendix, Fig. S4) covering time points fairly evenly across pupal development (∼12 h/stage) and four callow adult stages (0, 6, 12, and 24 h posteclosion). Tergites of the polymorphic metasomal segment T2 and T3 were sampled separately. The same tissues were also collected from P13 and P15 pupae and 0-h callows of workers for comparison. For stage P15 male pupae and freshly eclosed male (0-h) callow adults, we also collected tergites from metasomal T1 and metasomal T4–7 (pooled) for comparison (Fig. 3E). Three to seven replicates/pupal stage were used. Pupae and callow samples were collected from five black queen and nine red queen colonies.
All bees were reared at 29 °C and 60 ± 5% relative humidity in the dark. Samples were flash frozen and stored at −80 °C. RNA was extracted using an RNAqueous total RNA kit (Invitrogen) for experiment I, RNAqueous Micro RNA isolation kits (Invitrogen) for experiment II, and the Direct-zol RNA MicroPrep (Zymo Research) kits for experiment III. RNA extractions, purification, and subsequent DNase treatment were performed following manufacturer recommendations, with purity and concentration of RNA confirmed using a Nanodrop (Thermo Scientific).
First-strand cDNA was synthesized from 500 ng of total RNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCRs were performed in triplicate using PowerSYBR Green PCR master mix (Applied Biosystems) on an ABI 7900 real-time PCR detection system. Primers and conditions for abd-A, Abd-B, and Ubx are provided in SI Appendix, Table S4. Stability analysis on five different housekeeping genes was performed in RefFinder (95), leading to the selection of vATPase. A standard curve (5× dilution series) per gene was used to quantify sample expression, followed by normalization with vATPase (target gene/vATPase) (96). Mann–Whitney U statistical tests were performed to test differences in expression between color forms by stage and gene. Data are available from the Dryad Digital Repository (100).
Population Genetics Analysis.
Nucleotide diversity (π) and FST across the contig containing the color locus was assessed using custom Python scripts (https://github.com/simonhmartin/genomics_general). LD decay analysis was carried out using PopLDdecay (102).
Sequence Conservation Analysis.
Comparison of sequence conservation was performed across the abd-A/Abd-B intergenic region between B. impatiens and B. melanopygus red and black forms, bumble bee Bombus terrestris, Drosophila, and corbiculate bees Apis mellifera and Melipona quadrifasciata, using mVISTA tools (103, 104) and LAGAN (105) sequence alignment.
Supplementary Material
Acknowledgments
We thank Guillaume Ghisbain, Rebecca Sommers, Andy Deans, and Michelle Duennes for assisting in sample collection; Rebecca Sommers and Claudia Rosales for assistance in DNA extraction; Kaustubh Adhikari for assistance in Manhattan plot R custom script design; Mark Rebeiz for training in gene expression approaches; and Sydney Cameron and Jeff Lozier for intellectual guidance. We thank Penn State Genomics Core Facility for providing sequencing support. Computations for this research were performed on the Pennsylvania State University’s Institute for CyberScience Advanced CyberInfrastructure. This project was funded by a National Science Foundation Division of Environmental Biology CAREER Grant 1453473 (to H.M.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw sequencing reads for genomic samples reported in this paper have been deposited in the National Center for Biotechnology Information BioProject database, https://www.ncbi.nlm.nih.gov/bioproject (accession no. PRJNA526235). The bioinformatic analysis protocols, codes, scripts, associated analysis files, and qPCR CT value data reported in this paper have been deposited in the Dryad Digital Repository: https://doi.org/10.5061/dryad.3v6f25v.
See Commentary on page 11573.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900365116/-/DCSupplemental.
References
- 1.Rockman MV. (2012) The QTN program and the alleles that matter for evolution: All that’s gold does not glitter. Evolution 66:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Remington DL. (2015) Alleles versus mutations: Understanding the evolution of genetic architecture requires a molecular perspective on allelic origins. Evolution 69:3025–3038. [DOI] [PubMed] [Google Scholar]
- 3.Dittmar EL, Oakley CG, Conner JK, Gould BA, Schemske DW (2016) Factors influencing the effect size distribution of adaptive substitutions. Proc Biol Sci 283:20153065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenblum EB, Parent CE, Brandt EE (2014) The molecular basis of phenotypic convergence. Annu Rev Ecol Evol Syst 45:203–226. [Google Scholar]
- 5.Manceau M, Domingues VS, Linnen CR, Rosenblum EB, Hoekstra HE (2010) Convergence in pigmentation at multiple levels: Mutations, genes and function. Philos Trans R Soc Lond B Biol Sci 365:2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elmer KR, Meyer A (2011) Adaptation in the age of ecological genomics: Insights from parallelism and convergence. Trends Ecol Evol 26:298–306. [DOI] [PubMed] [Google Scholar]
- 7.Conte GL, Arnegard ME, Peichel CL, Schluter D (2012) The probability of genetic parallelism and convergence in natural populations. Proc Biol Sci 279:5039–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stern DL, Orgogozo V (2009) Is genetic evolution predictable? Science 323:746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoekstra HE, Coyne JA (2007) The locus of evolution: Evo devo and the genetics of adaptation. Evolution 61:995–1016. [DOI] [PubMed] [Google Scholar]
- 10.Papa R, Martin A, Reed RD (2008) Genomic hotspots of adaptation in butterfly wing pattern evolution. Curr Opin Genet Dev 18:559–564. [DOI] [PubMed] [Google Scholar]
- 11.Martin A, Orgogozo V (2013) The loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution 67:1235–1250. [DOI] [PubMed] [Google Scholar]
- 12.Wilson JS, Williams KA, Forister ML, von Dohlen CD, Pitts JP (2012) Repeated evolution in overlapping mimicry rings among North American velvet ants. Nat Commun 3:1272. [DOI] [PubMed] [Google Scholar]
- 13.Turner JRG. (1976) Adaptive radiation and convergence in subdivisions of the butterfly genus Heliconius (Lepidoptera: Nymphalidae). Zool J Linn Soc 58:297–308. [Google Scholar]
- 14.Santos JC, Coloma LA, Cannatella DC (2003) Multiple, recurring origins of aposematism and diet specialization in poison frogs. Proc Natl Acad Sci USA 100:12792–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marek PE, Bond JE (2009) A Müllerian mimicry ring in Appalachian millipedes. Proc Natl Acad Sci USA 106:9755–9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reed RD, et al. (2011) optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333:1137–1141. [DOI] [PubMed] [Google Scholar]
- 17.Nadeau NJ. (2016) Genes controlling mimetic colour pattern variation in butterflies. Curr Opin Insect Sci 17:24–31. [DOI] [PubMed] [Google Scholar]
- 18.Mazo-Vargas A, et al. (2017) Macroevolutionary shifts of WntA function potentiate butterfly wing-pattern diversity. Proc Natl Acad Sci USA 114:10701–10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin A, et al. (2014) Multiple recent co-options of Optix associated with novel traits in adaptive butterfly wing radiations. Evodevo 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallant JR, et al. (2014) Ancient homology underlies adaptive mimetic diversity across butterflies. Nat Commun 5:4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heliconius Genome Consortium (2012) Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Belleghem SM, et al. (2017) Complex modular architecture around a simple toolkit of wing pattern genes. Nat Ecol Evol 1:0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Supple MA, et al. (2013) Genomic architecture of adaptive color pattern divergence and convergence in Heliconius butterflies. Genome Res 23:1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hines HM, et al. (2011) Wing patterning gene redefines the mimetic history of Heliconius butterflies. Proc Natl Acad Sci USA 108:19666–19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pardo-Diaz C, et al. (2012) Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genet 8:e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams PH. (1998) An annotated checklist of bumble bees with an analysis of patterns of description (Hymenoptera: Apidae, Bombini). Bull Nat Hist Mus Entomol Ser 67:79–152. [Google Scholar]
- 27.Williams PH. (2007) The distribution of bumblebee colour patterns worldwide: Possible significance for thermoregulation, crypsis, and warning mimicry. Biol J Linn Soc Lond 92:97–118. [Google Scholar]
- 28.Brower LP, Brower JVZ, Westcott PW (1960) Experimental studies of mimicry. 5. The reactions of toads (Bufo terrestris) to bumblebees (Bombus americanorum) and their robberfly mimics (Mallophora bomboides), with a discussion of aggressive mimicry. Am Nat 94:343–355. [Google Scholar]
- 29.Evans DL, Waldbauer GP (1982) Behavior of adult and naive birds when presented with a bumblebee and its mimic. Z Tierpsychol 59:247–259. [Google Scholar]
- 30.Rapti Z, Duennes MA, Cameron SA (2014) Defining the colour pattern phenotype in bumble bees (Bombus): A new model for evo devo. Biol J Linn Soc Lond 113:384–404. [Google Scholar]
- 31.Hines HM. (2008) Historical biogeography, divergence times, and diversification patterns of bumble bees (Hymenoptera: Apidae: Bombus). Syst Biol 57:58–75. [DOI] [PubMed] [Google Scholar]
- 32.Hines HM, Williams PH (2012) Mimetic colour pattern evolution in the highly polymorphic Bombus trifasciatus (Hymenoptera: Apidae) species complex and its comimics. Zool J Linn Soc 166:805–826. [Google Scholar]
- 33.Plowright RC, Owen RE (1980) The evolutionary significance of bumble bee color patterns: A mimetic interpretation. Evolution 34:622–637. [DOI] [PubMed] [Google Scholar]
- 34.Ezray BD, Wham DC, Hill C, Hines HM (2019) Mullerian mimicry in bumble bees is a transient continuum. bioRxiv:10.1101/513275. Preprint, posted January 8, 2019. [DOI] [PMC free article] [PubMed]
- 35.Owen RE, Plowright RC (1980) Abdominal pile color dimorphism in the bumble bee, Bombus melanopygus. J Hered 71:241–247. [Google Scholar]
- 36.Owen RE, Whidden TL, Plowright RC (2010) Genetic and morphometric evidence for the conspecific status of the bumble bees, Bombus melanopygus and Bombus edwardsii. J Insect Sci 10:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hines HM, Witkowski P, Wilson JS, Wakamatsu K (2017) Melanic variation underlies aposematic color variation in two hymenopteran mimicry systems. PLoS One 12:e0182135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kronforst MR, et al. (2012) Unraveling the thread of nature’s tapestry: The genetics of diversity and convergence in animal pigmentation. Pigment Cell Melanoma Res 25:411–433. [DOI] [PubMed] [Google Scholar]
- 39.Jeong S, Rokas A, Carroll SB (2006) Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell 125:1387–1399. [DOI] [PubMed] [Google Scholar]
- 40.Kyrchanova O, et al. (2015) The boundary paradox in the Bithorax complex. Mech Dev 138:122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akbari OS, Bousum A, Bae E, Drewell RA (2006) Unraveling cis-regulatory mechanisms at the abdominal-A and Abdominal-B genes in the Drosophila bithorax complex. Dev Biol 293:294–304. [DOI] [PubMed] [Google Scholar]
- 42.Bae E, Calhoun VC, Levine M, Lewis EB, Drewell RA (2002) Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc Natl Acad Sci USA 99:16847–16852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maeda RK, Karch F (2009) The bithorax complex of Drosophila an exceptional Hox cluster. Curr Top Dev Biol 88:1–33. [DOI] [PubMed] [Google Scholar]
- 44.Garaulet DL, Lai EC (2015) Hox miRNA regulation within the Drosophila bithorax complex: Patterning behavior. Mech Dev 138:151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho MC, et al. (2009) Functional evolution of cis-regulatory modules at a homeotic gene in Drosophila. PLoS Genet 5:e1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dearden PK, et al. (2006) Patterns of conservation and change in honey bee developmental genes. Genome Res 16:1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hui JHL, et al. (2013) Structure, evolution and function of the bi-directionally transcribed iab-4/iab-8 microRNA locus in arthropods. Nucleic Acids Res 41:3352–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miura S, Nozawa M, Nei M (2011) Evolutionary changes of the target sites of two microRNAs encoded in the Hox gene cluster of Drosophila and other insect species. Genome Biol Evol 3:129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maeda RK, Karch F (2006) The ABC of the BX-C: The bithorax complex explained. Development 133:1413–1422. [DOI] [PubMed] [Google Scholar]
- 50.Hueber SD, Lohmann I (2008) Shaping segments: Hox gene function in the genomic age. BioEssays 30:965–979. [DOI] [PubMed] [Google Scholar]
- 51.Hughes CL, Kaufman TC (2002) Hox genes and the evolution of the arthropod body plan. Evol Dev 4:459–499. [DOI] [PubMed] [Google Scholar]
- 52.Kopp A, Duncan I (2002) Anteroposterior patterning in adult abdominal segments of Drosophila. Dev Biol 242:15–30. [DOI] [PubMed] [Google Scholar]
- 53.Tian L, Hines HM (2018) Morphological characterization and staging of bumble bee pupae. PeerJ 6:e6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akam M, et al. (1994) The evolving role of hox genes in arthropods. Development 120(Suppl):209–215. [PubMed] [Google Scholar]
- 55.Martin A, et al. (2012) Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc Natl Acad Sci USA 109:12632–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akam M. (1998) Hox genes: From master genes to micromanagers. Curr Biol 8:R676–R678. [DOI] [PubMed] [Google Scholar]
- 57.Akam M. (1998) Hox genes, homeosis and the evolution of segment identity: No need for hopeless monsters. Int J Dev Biol 42:445–451. [PubMed] [Google Scholar]
- 58.Gehring WJ, Kloter U, Suga H (2009) Evolution of the Hox gene complex from an evolutionary ground state. Curr Top Dev Biol 88:35–61. [DOI] [PubMed] [Google Scholar]
- 59.Foronda D, Estrada B, de Navas L, Sánchez-Herrero E (2006) Requirement of Abdominal-A and Abdominal-B in the developing genitalia of Drosophila breaks the posterior downregulation rule. Development 133:117–127. [DOI] [PubMed] [Google Scholar]
- 60.Martin A, et al. (2016) CRISPR/Cas9 mutagenesis reveals versatile roles of hox genes in crustacean limb specification and evolution. Curr Biol 26:14–26. [DOI] [PubMed] [Google Scholar]
- 61.Massey JH, Wittkopp PJ (2016) The genetic basis of pigmentation differences within and between Drosophila species. Curr Top Dev Biol 119:27–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kopp A, Duncan I, Godt D, Carroll SB (2000) Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 408:553–559. [DOI] [PubMed] [Google Scholar]
- 63.Signor SA, Liu Y, Rebeiz M, Kopp A (2016) Genetic convergence in the evolution of male-specific color patterns in Drosophila. Curr Biol 26:2423–2433. [DOI] [PubMed] [Google Scholar]
- 64.Keil TA. (1997) Comparative morphogenesis of sensilla: A review. Int J Insect Morphol Embryol 26:151–160. [Google Scholar]
- 65.Stern DL, Frankel N (2013) The structure and evolution of cis-regulatory regions: The shavenbaby story. Philos Trans R Soc Lond B Biol Sci 368:20130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Atallah J, et al. (2014) Sex-specific repression of dachshund is required for Drosophila sex comb development. Dev Biol 386:440–447. [DOI] [PubMed] [Google Scholar]
- 67.Eksi SE, Barmina O, McCallough CL, Kopp A, Orenic TV (2018) A Distalless-responsive enhancer of the Hox gene Sex combs reduced is required for segment- and sex-specific sensory organ development in Drosophila. PLoS Genet 14:e1007320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abzhanov A, Kaufman TC (1999) Novel regulation of the homeotic gene Scr associated with a crustacean leg-to-maxilliped appendage transformation. Development 126:1121–1128. [DOI] [PubMed] [Google Scholar]
- 69.Stern DL. (1998) A role of Ultrabithorax in morphological differences between Drosophila species. Nature 396:463–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mahfooz N, Turchyn N, Mihajlovic M, Hrycaj S, Popadić A (2007) Ubx regulates differential enlargement and diversification of insect hind legs. PLoS One 2:e866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Medved V, Huang ZY, Popadić A (2014) Ubx promotes corbicular development in Apis mellifera. Biol Lett 10:20131021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Werner T, Koshikawa S, Williams TM, Carroll SB (2010) Generation of a novel wing colour pattern by the Wingless morphogen. Nature 464:1143–1148. [DOI] [PubMed] [Google Scholar]
- 73.Monteiro A. (2012) Gene regulatory networks reused to build novel traits: Co-option of an eye-related gene regulatory network in eye-like organs and red wing patches on insect wings is suggested by optix expression. BioEssays 34:181–186. [DOI] [PubMed] [Google Scholar]
- 74.Bickel RD, Kopp A, Nuzhdin SV (2011) Composite effects of polymorphisms near multiple regulatory elements create a major-effect QTL. PLoS Genet 7:e1001275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McGregor AP, et al. (2007) Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448:587–590. [DOI] [PubMed] [Google Scholar]
- 76.Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB (2009) Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326:1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Celniker SE, Sharma S, Keelan DJ, Lewis EB (1990) The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J 9:4277–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sánchez-Herrero E. (1991) Control of the expression of the bithorax complex genes abdominal-A and abdominal-B by cis-regulatory regions in Drosophila embryos. Development 111:437–449. [DOI] [PubMed] [Google Scholar]
- 79.Galloni M, Gyurkovics H, Schedl P, Karch F (1993) The bluetail transposon: Evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J 12:1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F (1997) In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124:1809–1820. [DOI] [PubMed] [Google Scholar]
- 81.Gummalla M, et al. (2012) abd-A regulation by the iab-8 noncoding RNA. PLoS Genet 8:e1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hedrick PW. (2013) Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol Ecol 22:4606–4618. [DOI] [PubMed] [Google Scholar]
- 83.Nei M. (2013) Mutation-Driven Evolution (Oxford Univ Press, Oxford: ). [Google Scholar]
- 84.Jones FC, et al. ; Broad Institute Genome Sequencing Platform; Whole Genome Assembly Team (2012) The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baxter SW, et al. (2008) Convergent evolution in the genetic basis of Müllerian mimicry in Heliconius butterflies. Genetics 180:1567–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wallbank RW, et al. (2016) Evolutionary novelty in a butterfly wing pattern through enhancer shuffling. PLoS Biol 14:e1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iwasaki Y, et al. (2016) Effective de novo assembly of fish genome using haploid larvae. Gene 576:644–649. [DOI] [PubMed] [Google Scholar]
- 88.Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sadd BM, et al. (2015) The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol 16:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H. (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2. Preprint, posted May 26, 2013.
- 91.Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H, et al. ; 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.DePristo MA, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McKenna A, et al. (2010) The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Danecek P, et al. ; 1000 Genomes Project Analysis Group (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Purcell S, et al. (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chevreux B, Wetter T, Suhai S (1999) Genome sequence assembly using trace signals and additional sequence information. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics, ed Wingender E. Wingender E (German Conference on Bioinformatics, Hannover, Germany: ), Vol 99, pp 45–56. [Google Scholar]
- 98.Robinson JT, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rahman SR, et al. (2019) Data from “A homeotic shift late in development drives mimetic color variation in a bumble bee.” National Center for Biotechnology Information Bioproject. Available at https://www.ncbi.nlm.nih.gov/bioproject/526235. Deposited March 6, 2019.
- 100.Rahman SR, et al. (2019) Data from “A homeotic shift late in development drives mimetic color variation in a bumble bee.” Dryad Digital Repository. Available at https://doi:10.5061/dryad.3v6f25v. Deposited April 14, 2019.
- 101.Cnaani J, Schmid-Hempel R, Schmidt JO (2002) Colony development, larval development and worker reproduction in Bombus impatiens Cresson. Insectes Soc 49:164–170. [Google Scholar]
- 102.Zhang C, Dong S-S, Xu J-Y, He W-M, Yang T-L (October 15, 2018) PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics, 10.1093/bioinformatics/bty875. [DOI] [PubMed] [Google Scholar]
- 103.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: Computational tools for comparative genomics. Nucleic Acids Res 32:W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mayor C, et al. (2000) VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046–1047. [DOI] [PubMed] [Google Scholar]
- 105.Brudno M, et al. ; NISC Comparative Sequencing Program (2003) LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res 13:721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.