Significance
NAD+, a key compound in cellular redox metabolism, can also be incorporated into RNA as the first nucleotide and serve as an alternative RNA cap. NAD+-capped RNAs have been found in bacteria, yeast, and human cells, but whether they exist in plants is not known. Here, we show that thousands of transcripts from the Arabidopsis transcriptome contain the NAD+ cap. These transcripts are derived primarily from protein-coding genes in nuclear and mitochondrial, but not chloroplast, genomes. NAD+-capped transcripts from nuclear genes are spliced, polyadenylated, and preferentially associated with actively translating polysomes. Our findings indicate that NAD+ can serve as a functional mRNA cap in plants and implicate crosstalk between cellular redox status and gene expression through RNA NAD+ capping.
Keywords: NAD+ cap, m7G cap, NAD captureSeq, polysome, translation
Abstract
As the most common RNA cap in eukaryotes, the 7-methylguanosine (m7G) cap impacts nearly all processes that a messenger RNA undergoes, such as splicing, polyadenylation, nuclear export, translation, and degradation. The metabolite and redox agent, nicotinamide adenine diphosphate (NAD+), can be used as an initiating nucleotide in RNA synthesis to result in NAD+-capped RNAs. Such RNAs have been identified in bacteria, yeast, and human cells, but it is not known whether they exist in plant transcriptomes. The functions of the NAD+ cap in RNA metabolism or translation are still poorly understood. Here, through NAD captureSeq, we show that NAD+-capped RNAs are widespread in Arabidopsis thaliana. NAD+-capped RNAs are predominantly messenger RNAs encoded by the nuclear and mitochondrial genomes, but not the chloroplast genome. NAD+-capped transcripts from the nuclear genome appear to be spliced and polyadenylated. Furthermore, although NAD+-capped transcripts constitute a small proportion of the total transcript pool from any gene, they are enriched in the polysomal fraction and associate with translating ribosomes. Our findings implicate the existence of as yet unknown mechanisms whereby the RNA NAD+ cap interfaces with RNA metabolic processes as well as translation initiation. More importantly, our findings suggest that cellular metabolic and/or redox states may influence, or be regulated by, mRNA NAD+ capping.
All eukaryotic messenger RNAs (mRNAs) contain the evolutionarily conserved cap structure—a 7-methylguanosine (m7G) moiety linked to the initiating nucleotide of the transcript via an inverted 5′–5′ triphosphate bridge (1). This structure is added cotranscriptionally and exerts profound effects on the fate of mRNAs. The cap protects mRNAs from 5′ to 3′ exonucleolytic degradation and recruits protein factors for splicing, polyadenylation, nuclear export, and translation (2). In particular, the methyl group linked to the N7 amine of guanosine in the m7G cap ensures RNA recognition by eukaryotic translation initiation factor 4E for cap-dependent translation initiation (3). Cap-independent translation can occur via the recognition of other features in mRNAs, such as internal ribosome entry sites or m6A modification (4).
The nucleotide-containing metabolite, nicotinamide adenine diphosphate (NAD+), has emerged as a new RNA cap in diverse organisms (5–9). The existence of the NAD+ cap was first discovered in Escherichia coli RNA by liquid chromatography mass spectrometry analyses (5). Subsequently, through combined chemoenzymatic capture and next-generation sequencing (NAD captureSeq), NAD+-capped transcripts were globally identified in E. coli, Bacillus subtilis, Saccharomyces cerevisiae, and human cells (6–9). Among the identified NAD+-capped RNAs (sometimes simplified as NAD-RNAs), specific regulatory small RNAs (sRNAs) are particularly enriched in E. coli (6). In S. cerevisiae, a small subset of nuclear genes encoding the translational machinery and mitochondrial genes preferentially give rise to NAD+-capped RNAs (8). In contrast to E. coli and S. cerevisiae, NAD+-capped RNAs are widespread in the transcriptome of human cells and are mainly composed of mRNAs and small nucleolar RNAs (snoRNAs) (9). Despite the well-known, essential roles of the m7G cap in pre-mRNA splicing and polyadenylation, NAD+-capped mRNAs were found to be spliced and polyadenylated in human cells (8, 9). While the m7G cap is added to the first nucleotide of an RNA after transcription initiation, NAD+ is thought to be incorporated by RNA polymerase (RNAP) as the initiating nucleotide (10, 11). Upon the transfection of in vitro-synthesized, m7G-capped or NAD+-capped RNAs into human cells, the NAD+-capped RNAs had a shorter half-life and were not translated, which led to the conclusion that the NAD+ cap fails to confer RNA stability or translatability (9).
While NAD+-capped RNAs are found in bacteria, yeast, and human cells, it is not known whether they exist in plants, which are multicellular and photosynthetic eukaryotes containing 2 types of organelles with their own genomes, namely mitochondria and chloroplasts. Here, we report the identification of NAD+-capped RNAs in the model plant Arabidopsis thaliana. NAD captureSeq performed with seedling and inflorescence samples revealed a large number of NAD+-capped RNAs. Intriguingly, NAD+-capped RNAs are predominantly mRNAs from nuclear and mitochondrial genes, but not chloroplast genes. Nuclear encoded, NAD+-capped mRNAs are spliced and polyadenylated. Furthermore, we show that NAD+-capped mRNAs are enriched in the polysomal fraction and associate with translating ribosomes. This implies the existence of as yet unknown mechanisms of translation initiation on NAD+-capped mRNAs.
Results
NAD+-Capped RNAs Are Present in A. thaliana.
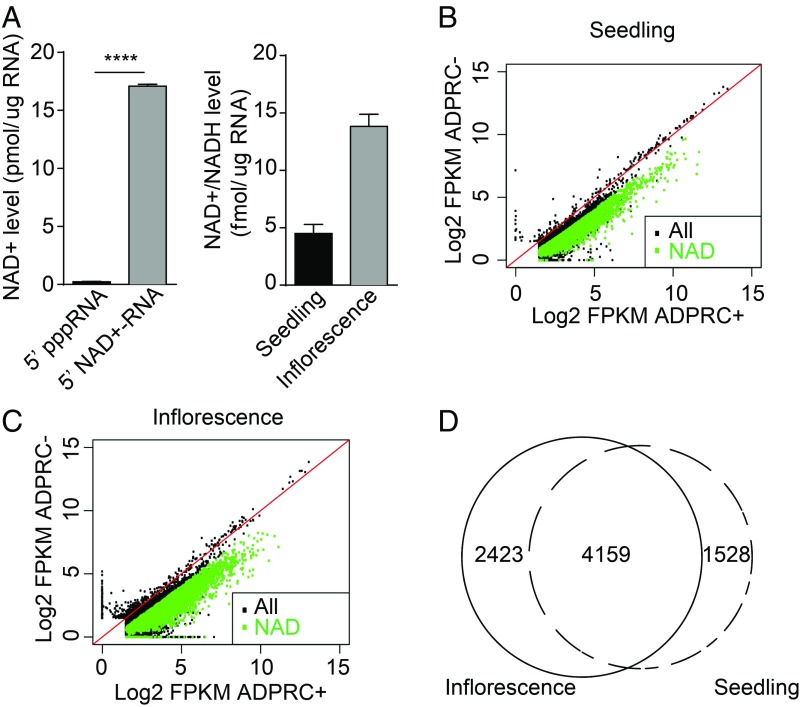
To determine if NAD+-capped RNAs exist in plants, we analyzed total Arabidopsis RNAs with the NAD-capQ approach wherein the amount of NAD+/NADH in cellular total RNAs was measured by a 2-step procedure consisting of enzymatic treatment of RNA with nuclease P1 to release NAD+/NADH, followed by quantitation of the released NAD+/NADH by a colorimetric assay (12). In vitro-transcribed 5′ triphosphate RNA (pppRNA) and 5′ NAD+-RNA served as a negative and a positive control, respectively. As shown in Fig. 1A, the signal intensity of colorimetric products from the NAD+-RNA was drastically higher than that from the pppRNA. We measured the levels of NAD+/NADH released from total RNAs of 2 samples: seedlings (which consisted largely of leaves) and inflorescences (which consisted largely of flowers). Results showed that the NAD+/NADH level in RNA from seedlings was around 5 fmol/μg RNA, and that in inflorescences was approximately 3-fold higher (Fig. 1A), revealing the existence of Arabidopsis NAD+/NADH-capped RNAs and their differing levels between 2 tissue/organ types.
Fig. 1.
NAD+-capped RNAs are present in plant tissues/organs. (A) Quantitation of NAD+/NADH levels in RNA. (A, Left) NAD+/NADH concentrations in negative (5′ pppRNA) and positive (5′ NAD+-RNA) controls that were generated through in vitro transcription. (A, Right) NAD+/NADH concentrations in cellular RNAs from 12-d-old seedlings and inflorescences. Concentrations of NAD+/NADH were calculated by dividing the detected value of NAD+/NADH by the amount of RNAs analyzed. Three independent replicates were performed. Error bars represent SD. ****P < 0.0001 (Student’s t test). (B and C) Detection of NAD+-capped RNAs from total RNAs by NAD captureSeq. Read counts in ADPRC-treated (ADPRC+) and mock-treated (ADPRC−) samples from seedlings (B) and inflorescences (C) were plotted. Green dots indicate NAD+-capped RNAs; black dots indicate non–NAD+-capped RNAs. “All” represents all expressed genes; “NAD” represents genes producing NAD+-capped RNAs. (D) Venn diagram showing the degree of overlap in genes producing NAD+-capped RNAs between seedling and inflorescence samples. Numbers indicate the number of genes that give rise to NAD+-capped RNAs.
Given that NAD+/NADH was detected in Arabidopsis RNA, we then performed NAD captureSeq, which specifically captures NAD+-capped RNAs (rather than NADH-capped RNAs) (6) to globally identify NAD+-capped RNA species. Our NAD captureSeq (workflow depicted in SI Appendix, Fig. S1A) was adapted from the original NAD captureSeq approach developed for bacterial RNAs (13). Total RNAs from seedlings and inflorescences were either treated or not with adenosine diphosphate (ADP)-ribosyl cyclase (ADPRC), which catalyzes the transglycosylation reaction of NAD+. The “clickable” transglycosylation products were subsequently biotinylated by a copper-catalyzed azide–alkyne cycloaddition (CuAAC) reaction. This procedure was first performed with in vitro-synthesized 5′ pppRNA and 5′ NAD+-RNA; detection of biotinylated RNA by streptavidin confirmed that this method specifically biotinylated NAD+-RNA (SI Appendix, Fig. S1B). Total RNAs from Arabidopsis were similarly treated, and the biotinylated RNAs were captured by streptavidin-conjugated beads and subjected to alkaline hydrolysis followed by library construction. Random primers were used in the reverse-transcription (RT) step during library construction to include RNAs that were not polyadenylated. We will refer to this modified procedure still as NAD captureSeq.
Three separate experiments with seedlings and 2 with inflorescences gave highly reproducible results (SI Appendix, Fig. S1C) (14). The ADPRC− samples, which underwent all treatments described above except that ADPRC was left out, probably represented RNAs that were nonspecifically attached to the beads, as the abundance of transcripts in ADPRC− samples was largely proportional to that in total RNAs without treatment (SI Appendix, Fig. S2A). The distribution of reads along gene bodies differed between the ADPRC− and ADPRC+ samples. The former had even distribution of reads along gene bodies as normally seen in RNA sequencing (RNA-seq), while the latter showed an enrichment of reads toward the 5′ ends of genes (SI Appendix, Fig. S2B). This pattern was also seen in NAD captureSeq from yeast (8) and was likely due to random RNA cleavage caused by Cu2+ that was present in the CuAAC reaction followed by enrichment for 5′ NAD+ ends of RNAs through streptavidin capture. In both seedlings and inflorescences, RNA levels from most genes were higher in the ADPRC+ samples than in the ADPRC− samples, suggesting that most genes gave rise to NAD+-capped RNAs (Fig. 1 B and C). A total of 5,687 and 6,582 NAD+-capped RNAs from seedlings and inflorescences, respectively (Datasets S1 and S2), were identified as those that passed the filter of fragments per kilobase per million (FPKM) fold change in ADPRC+ vs. ADPRC− of >2 and a false discovery rate (FDR) of <0.05 in the biological replicates. A total of 4,159 NAD+-capped RNAs were common between seedlings and inflorescences (Fig. 1D); thus, a large set of genes produced NAD+-capped RNAs in the 2 tissue/organ types. Note that the identification of NAD+-capped RNAs in NAD captureSeq relies on the ratio of ADPRC+ to ADPRC−. Given that RNA abundance in the ADPRC− samples largely reflected RNA abundance in total RNAs, the ADPRC+/ADPRC− ratio reflected the proportion of NAD+-capped transcripts in the total transcript pool for any gene, as opposed to the absolute abundance of NAD+-capped transcripts. For simplicity, we refer to NAD+-capped RNAs as NAD-RNAs hereafter.
Nuclear and Mitochondrial Genes Produce NAD-RNAs.
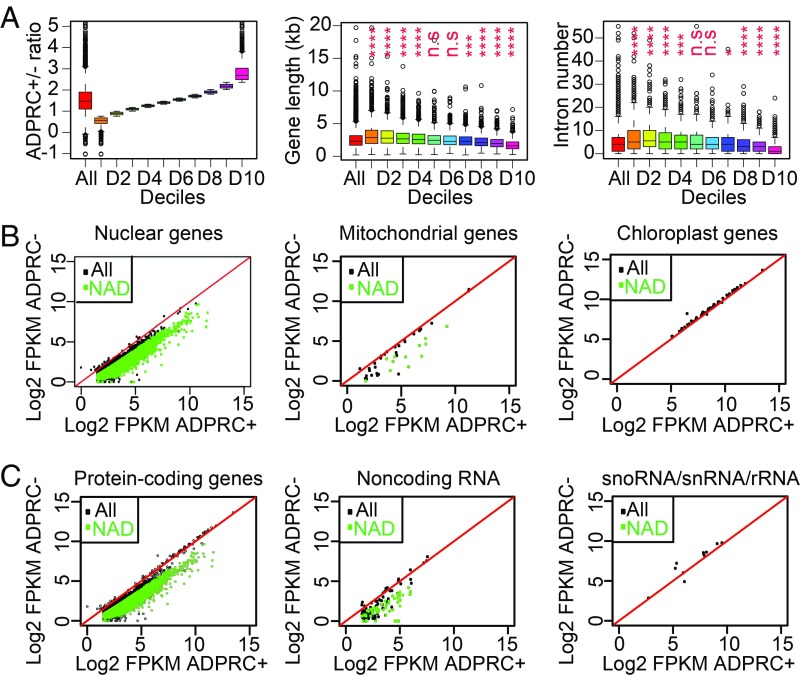
To characterize the features of genes that produce NAD-RNAs, we equally divided the 5,687 genes found to give rise to NAD-RNAs in seedlings into 10 parts (deciles) based on the FPKM fold change of ADPRC+ versus ADPRC− samples, and then compared the lengths and intron numbers of genes in the 10 deciles. It was apparent that shorter genes tended to give a higher proportion of NAD-RNAs (Fig. 2A). It is most likely that shorter genes also tended to have fewer introns; thus, it was not surprising that genes with fewer introns also tended to produce a higher proportion of NAD-RNAs (Fig. 2A).
Fig. 2.
NAD+-capped transcripts are derived from nuclear and mitochondrial genes. (A) Correlation between ADPRC+/ADPRC− ratio and gene length or intron number. Genes were separated into 10 groups based on their ADPRC+/ADPRC− ratios (Left), and either gene length (Middle) or intron number (Right) was determined for each group. “All” denotes all genes. Each decile was compared with all genes for differences in gene length or intron number. n.s., not significant; *P < 0.05; ***P < 0.001; ****P < 0.0001 (Mann–Whitney–Wilcoxon test). (B and C) Comparison of read counts between ADPRC+ and ADPRC− samples for nuclear, mitochondrial, and chloroplast genes (B), as well as protein-coding genes, noncoding transcripts, and snoRNA/snRNA/rRNA (C) in NAD captureSeq. Green dots indicate NAD-RNAs; black dots indicate non–NAD-RNAs. “All” represents all expressed genes; “NAD” represents genes producing NAD-RNAs.
Most NAD-RNAs were from the nuclear genome (Fig. 2B). Similar to human and yeast (8, 9) mitochondrial transcripts, many such transcripts were also NAD+ capped in Arabidopsis (Fig. 2B). Intriguingly, no transcripts from the chloroplast genome were found to be NAD+ capped (Fig. 2B). Among the nuclear and mitochondrial NAD-RNAs, most of them were from protein-coding genes, but a small subset was from noncoding genes (Fig. 2C). Most snoRNAs and small nuclear RNAs (snRNAs) were not represented in our libraries, and the ones that were present, as well as ribosomal RNAs (rRNAs), did not show an enrichment in the ADPRC+ samples compared with the ADPRC− samples (Fig. 2C).
To explore the potential biological processes that NAD+-capped RNAs may be involved in, we performed GO enrichment analysis for 1,000 genes with the highest ADPRC+/ADPRC− ratio (SI Appendix, Fig. S3). The enriched GO terms were responses to various stimuli, including oxidative stress and other cellular processes such as photosynthesis and redox homeostasis, as reflected by the nuclear genes AT5G58250 (Low Chlorophyll Accumulation A/Hypothetical Chloroplast ORF 54, LCAA/YCF54) and AT5G38420 (Rubisco small subunit 2B, RBCS2B) and the mitochondrial genes ATMG00160 (Cytochrome oxidase 2, COX2) and ATMG00516 (NADH dehydrogenase), as well as many other nuclear and mitochondrial genes. This suggests that RNA NAD+ capping may serve to regulate gene expression in response to stresses.
NAD-RNAs Are Predominantly mRNAs.
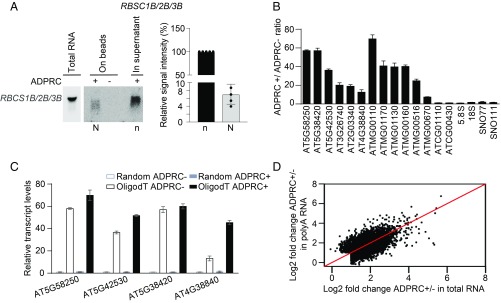
To validate the presence of NAD-RNAs for individual genes, RNA gel blot assay was performed against the Rubisco small subunit (RBCS) genes, which were found to produce NAD-RNAs in NAD captureSeq. Since the RBCS1B, -2B, and -3B genes all produced NAD-RNAs, and they share a high degree of sequence similarity so that the probe could not distinguish them, the gel blot signals reflected NAD-RNAs from all 3 genes (Fig. 3A). The lack of signal in the negative control (ADPRC−) confirmed the specificity of the capture scheme for NAD-RNAs. RNA gel blot assay had the added advantage of determining the relative abundance of NAD-RNAs vs. non–NAD-RNAs from the same genes (i.e., RNAs on beads vs. in supernatant after NAD capture; Fig. 3A). Based on gel blot assays in 5 biological replicates, the level of NAD-RNAs was estimated to be ∼5% of total transcripts from the RBS1B/2B/3B genes (Fig. 3A). This level is comparable to the estimated proportions of NAD-RNAs in human cells (9).
Fig. 3.
NAD-RNAs are poly(A) RNAs. (A) RNA gel blot assay to detect NAD-RNAs and non–NAD-RNAs derived from RBCS1B/2B/3B genes. NAD-RNAs (labeled as “N”) were isolated by NAD capture with streptavidin beads, while RNAs in the supernatant were considered as non–NAD-RNAs (indicated as “n”). One-fifth of non–NAD-RNAs in the supernatant were resolved in an agarose gel together with all captured NAD-RNAs. The intensities of bands were measured by ImageJ. The bar plot on the Right shows the relative signal intensities (N vs. n) from 5 independent replicates (after correcting for differences in loading). Error bars represent SD. Dots indicate the values of the 5 replicates. (B) Quantification of transcript levels in ADPRC+ vs. ADPRC− samples from seedlings by qRT-PCR. From left to right are 6 nuclear genes, 6 mitochondrial genes, 2 chloroplast genes, 2 rRNAs, and 2 snoRNAs. (C) Detection of NAD+-capped poly(A) RNAs by qRT-PCR. After NAD capture from total RNAs, NAD-RNAs were eluted from beads, reverse transcribed by either oligo (dT) (“OligodT ADPRC+/−”) or random primers (“Random ADPRC+/−”), and then detected with gene-specific primers. For B and C, qRT-PCR values were normalized to those of the ADPRC− samples, which were arbitrarily set to 1. Two biological replicates were performed, each with 3 technical replicates. Error bars represent SD. (D) Comparison of signal intensity (ADPRC+/ADPRC− ratio) in NAD captureSeq from total RNAs and that from poly(A) RNAs.
To verify the production of NAD-RNAs from more genes, we performed qRT-PCR on ADPRC+ and ADPRC− samples for 12 different nuclear and mitochondrial protein-coding genes. Transcripts from all these genes showed higher levels in the ADPRC+ sample than in the ADPRC− sample, confirming that NAD-RNAs were generated from these genes (Fig. 3B). Conversely, transcripts from 2 chloroplast genes and the tested snoRNAs and rRNAs were similar in abundance in ADPRC+ and ADPRC− samples (Fig. 3B). Thus, NAD+-capping varies among intracellular organelles and RNA classes.
As most of the NAD-RNAs identified via NAD captureSeq were from protein-coding genes, we wondered whether the transcripts were spliced and polyadenylated. Visual inspection of genome-mapped reads in NAD captureSeq revealed that NAD-RNAs were properly spliced (SI Appendix, Fig. S4). However, the polyadenylation status could not be inferred from NAD captureSeq, as total RNAs were used for NAD capture and random primers were used for RT. We performed qRT-PCR on individual RNAs to determine whether NAD-RNAs harbored a poly(A) tail. Total RNAs were subjected to NAD capture, and the captured NAD-RNAs were divided into 2 parts: One was reverse transcribed with random primers while the other underwent oligo (dT)-primed RT. NAD-RNAs from 4 genes were found in both randomly primed and oligo (dT)-primed cDNA, suggesting that these NAD+-capped transcripts possessed a poly(A) tail (Fig. 3C).
Next, we sought to determine whether NAD-RNAs were polyadenylated at the genomic scale. We first isolated poly(A) RNAs from total RNAs and subjected poly(A) RNAs to NAD captureSeq. Two biological replicates gave highly reproducible results (SI Appendix, Fig. S5A) (14). As observed in NAD captureSeq with total RNAs, numerous transcripts were found to have higher FPKM counts in the ADPRC+ samples than in the ADPRC− samples in the poly(A) RNA NAD captureSeq (SI Appendix, Fig. S5B), suggesting that polyadenylated RNAs harbored the NAD+ cap. A total of 6,089 transcripts were identified as NAD-RNAs with the filter of FPKM fold change in ADPRC+ vs. ADPRC− of >2 and an FDR of <0.05 in 2 biological replicates (Dataset S3). The NAD+-capped transcripts identified from poly(A) and total RNA libraries largely overlapped (SI Appendix, Fig. S5C). For the population of NAD+-capped transcripts identified in either total RNA or poly(A) RNA libraries, we examined the signal intensities (FPKM fold change in ADPRC+ vs. ADPRC− samples) in the 2 types of libraries. A linear relationship was observed for the 2 types of libraries (Fig. 3D). These data indicated that NAD-RNAs were polyadenylated.
NAD-RNAs Are Enriched in the Polysomal Fraction.
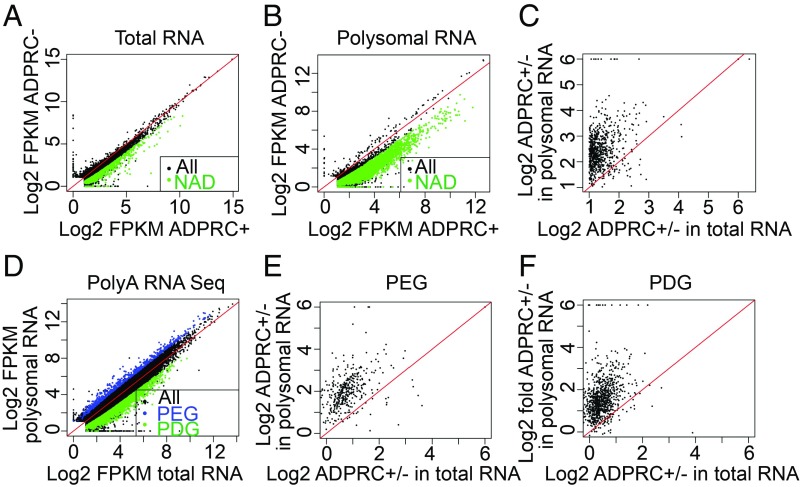
Since the m7G cap is a major determinant of the translatability of an mRNA in eukaryotes (15), and most NAD+-capped transcripts identified above are from protein-coding genes, we thus asked whether the NAD-RNAs are translated. Eukaryotic mRNA initiation begins with the recruitment of the small ribosomal subunit (40S) to the mRNA via the recognition of the 5′ m7G cap (16). The poly(A) tail also enhances translation initiation (17). The coupling of the 40S subunit with a 60S subunit at the start codon gives an 80S ribosome, and the elongation phase starts (18). An mRNA undergoing translation usually associates with multiple ribosomes to form polyribosome complexes (polysomes), and the number of ribosomes per transcript reflects the efficiency of translation (19). To ascertain whether NAD-RNAs are associated with polysomes, we isolated total cellular ribosomes and then fractionated ribosomes by sucrose gradient centrifugation to separate the heavy fraction containing polysomes from the light fraction containing monosomes and ribosomal subunits (SI Appendix, Fig. S6A). We refer to the heavy and light fractions as polysomal and nonpolysomal fractions, respectively. RNAs associated with the polysomal fraction were isolated and subjected to NAD captureSeq, and total RNAs from the same tissue samples were included for comparison (SI Appendix, Fig. S6B) (14). With the same cutoff as before, NAD-RNAs were identified not only in total RNA (Fig. 4A and Dataset S4) but also in polysomal RNA (Fig. 4B and Dataset S5), suggesting that NAD-RNAs associated with polysomes. We next compared the NAD+ signal intensity (FPKM ratios of ADPRC+ vs. ADPRC− samples) in total RNA and polysomal RNA NAD captureSeq for the population of NAD-RNAs found in either total RNA or polysomal RNA samples. Surprisingly, polysomal RNAs had markedly higher signal intensity compared with total RNAs (Fig. 4C).
Fig. 4.
NAD-RNAs are enriched on polysomes. (A and B) Scatter plots showing transcript abundance in ADPRC+ vs. ADPRC− samples in NAD captureSeq from total RNAs (A) and polysomal RNAs (B). Green dots indicate NAD-RNAs; black dots indicate non–NAD-RNAs. “All” represents all expressed genes; “NAD” represents genes producing NAD-RNAs. (C) Comparison of signal/noise ratios (ADPRC+/ADPRC− ratios) between total RNA and polysomal RNA NAD captureSeq. Genes that were found to produce NAD-RNAs in either total RNAs or polysomal RNAs were included. (D) Comparison of mRNA abundance between total RNA and polysomal RNA samples by mRNA-seq. Blue dots represent PEGs, and green dots show PDGs. (E and F) Comparison of signal/noise ratios (ADPRC+/ADPRC− ratios) between total RNA and polysomal RNA NAD captureSeq for PEGs (E) and PDGs (F).
The higher ADPRC+/ADPRC− ratio suggested that, in the total pool of transcripts from a gene, the polysomal fraction had a higher proportion of NAD+-capped transcripts. This would implicate that NAD+-capped transcripts are preferentially polysome associated relative to m7G-capped transcripts (illustrated in SI Appendix, Fig. S7A). To rule out the scenario that the enhanced detection of NAD-RNAs from a gene in the polysomal fraction was due to higher levels of polysome loading of the total transcript pool from this gene (SI Appendix, Fig. S7B), we first globally determined polysome loading status of transcripts by performing poly(A) mRNA sequencing for polysomal RNAs and total RNAs. A total of 3,132 genes were defined as polysome-depleted genes (PDGs), as their transcripts had higher abundance in total RNAs than polysomal RNAs (fold change of FPKM in total RNAs vs. polysomal RNAs of >2; FDR of <0.05; Fig. 4D, green dots). A total of 638 genes had higher RNA levels in polysomal RNAs than in total RNAs (fold change of FPKM in polysomal RNAs vs. total RNAs of >2; FDR of <0.05) and were designated as polysome-enriched genes (PEGs, indicated by blue dots in Fig. 4D). Next, we examined the NAD+ signal intensities of the 2 groups of genes in polysomal RNAs and total RNAs. For both PEGs and PDGs, an enrichment of NAD-RNAs in the polysomal fraction was seen (Fig. 4 E and F), suggesting that this enrichment was independent of the abundance of transcripts in the polysomal fraction.
NAD-RNAs Are on Actively Translating Polysomes.
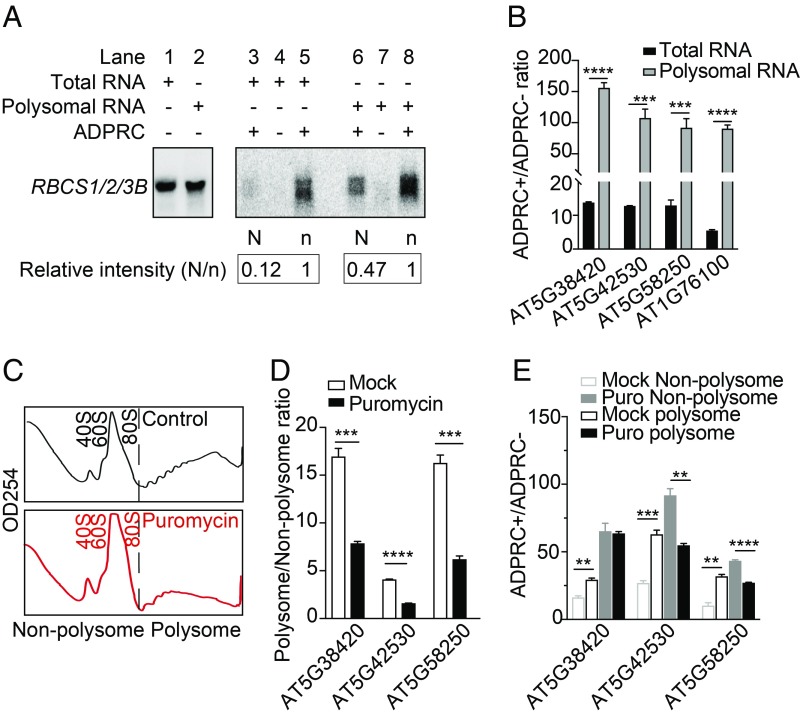
The sequencing results above strongly indicated that polysomes had a higher proportion of NAD-RNAs from the transcript pool of any gene. To further confirm this, we turned to RNA gel blot assays to directly compare the abundance of RBCS1B/2B/3B RNAs in various fractions. NAD capture was performed for both total RNAs and polysomal RNAs, and then RNAs on streptavidin beads (NAD-RNAs) and in the supernatant (non–NAD-RNAs) were recovered and subjected to gel blot analysis. Relative to the levels of non–NAD-RNAs in total extract and polysomes (arbitrarily set as 1), the abundance of NAD-RNAs in total extract was 0.12, while in the polysome fraction, it increased to 0.47 (Fig. 5A). This confirmed the preferential association of NAD+-capped RBCS1B/2B/3B RNAs with polysomes. We next conducted qRT-PCR to compare the abundance of NAD-RNAs between total extract and polysomes for 4 other genes (Fig. 5B). In each case, an enrichment of NAD-RNAs in the polysomal fraction was observed, indicating that NAD-RNAs were associated with polysomes and may undergo translation in plant cells.
Fig. 5.
Puromycin treatment leads to the dissociation of NAD-RNAs from polysomes. (A) RNA gel blot assays showing the enrichment of NAD-RNAs in the polysomal fraction. NAD capture was performed with total RNAs or polysomal RNAs, and NAD-RNAs (on beads; labeled as “N”) and non–NAD-RNAs (in supernatant; labeled as “n”; one-fifth of supernatant RNAs were loaded) were resolved in a gel. Hybridization was performed with a probe common to RBCS1B/2B/3B genes. Signal intensity of the bands was measured by ImageJ. Levels of non–NAD-RNAs in total extract and polysomes were arbitrarily set as 1, and the relative abundance of NAD-RNAs is indicated by the numbers. The numbers reflected the N/n ratio without correction for the differences in loading. Note that the “ADPRC−” samples (lanes 4 and 7) had little background, which demonstrated the specificity of NAD capture. Two biological replicates were performed and gave similar results. (B) Quantification of transcript levels in ADPRC+ vs. ADPRC− samples by qRT-PCR. NAD capture was performed with total or polysomal RNAs. qRT-PCR values were normalized to those of the ADPRC− samples, which were arbitrarily set to 1. Two biological replicates, each with 3 technical replicates, were performed. Error bars represent SD. ***P < 0.001; ****P < 0.0001 (Student’s t test). (C) Profile of ribosomes on continuous sucrose gradients. Ribosomes were isolated from lysates treated (Bottom) or not (Upper) with puromycin and separated by sucrose gradient centrifugation. Absorbance at 254 nm is shown for various fractions (from left to right: the lighter to the heavier fractions). The fractions to the left of the black dashed line were combined as the nonpolysomal fraction; the fractions to the right were combined as the polysomal fraction. (D) Quantification of transcript levels in the polysomal vs. nonpolysomal fractions with or without puromycin treatment. Three genes were examined by qRT-PCR, which showed that the puromycin treatment led to the dissociation of their mRNAs from polysomes. qRT-PCR values were normalized to those of the nonpolysomal fractions, which were arbitrarily set to 1. Two biological replicates, each with 3 technical replicates, were performed. Error bars represent SD. ***P < 0.001; ****P < 0.0001 (Student’s t test). (E) Puromycin treatment led to the dissociation of NAD-RNAs from polysomes. RNAs from the polysomal and nonpolysomal fractions in C were subjected to NAD capture followed by qRT-PCR to measure transcript levels in ADPRC+ vs. ADPRC− samples for 3 genes. qRT-PCR values were normalized to those of the ADPRC− samples, which were arbitrarily set to 1. Two biological replicates, each with 3 technical replicates, were performed. Error bars represent SD. **P < 0.01; ***P < 0.001; ****P < 0.0001 (Student’s t test).
However, it still remained a formal possibility that NAD-RNAs were bound by nontranslating polysomes. We took advantage of a well-known protein synthesis inhibitor, puromycin, to interrogate whether NAD-RNAs were associated with translating ribosomes. As the structure of puromycin is analogous to aminoacyl-transfer RNAs (tRNAs), it enters the A site of translating ribosomes and causes premature translation termination (20). In bacterial cultures, the addition of puromycin leads to the breakdown of polyribosomes with a concomitant accumulation of 70S monosomes (21). We treated plant lysates with or without puromycin, followed by ribosome isolation. The profiles of ribosomes on sucrose gradients revealed puromycin-induced polysome dissociation and accumulation of 80S monosomes (Fig. 5C). We then detected poly(A) RNAs from 3 genes in the nonpolysomal fraction (combination of 40S and 60S ribosomal subunits and 80S ribosomes) and in the polysomal fraction of mock- and puromycin-treated lysates. The abundance of RNAs from these genes was significantly higher in the polysomal than in the nonpolysomal fraction in untreated lysates, but the fold change was greatly reduced upon puromycin treatment (Fig. 5D), consistent with puromycin-induced polysome dissociation. To elucidate whether NAD-RNAs were associated with active polysomes, NAD-RNAs were captured from both polysomal and nonpolysomal fractions from mock- and puromycin-treated lysates and then detected by qRT-PCR. Without puromycin treatment, NAD-RNAs from the 3 genes were enriched in the polysomal fraction relative to the nonpolysomal fraction (Fig. 5E). Puromycin treatment abolished the polysomal enrichment or even resulted in an enrichment in the nonpolysomal fraction (Fig. 5E), suggesting that the NAD-RNAs were on actively translating polysomes in vivo. We also took advantage of a published ribo-seq dataset (Gene Expression Omnibus database, accession no. GSE50597, no stress group) (22) in seedlings to examine the translation efficiency of genes with high levels of NAD-RNAs. Translation efficiency was calculated as the ratio between ribosome protected fragment counts and mRNA levels (FPKM from RNA-seq). The 1,000 genes with the highest ADPRC+/ADPRC− ratio had higher translation efficiency than 1,000 randomly selected genes (SI Appendix, Fig. S8). Thus, genes that generate higher levels of NAD-RNAs tend to be more efficiently translated. As NAD-RNAs only constitute a small portion of the total transcripts from these genes, the higher translation efficiency of these genes is not conclusive evidence for the preferential translation of NAD-RNAs, but it is consistent with our observation that NAD-RNAs are preferentially polysome associated.
Discussion
Our studies demonstrate that thousands of genes in the nuclear and mitochondrial genomes in Arabidopsis produce NAD-RNAs. Using a different approach, Zhang et al. (23) in PNAS also discovered thousands of NAD-RNAs from Arabidopsis. Between the 6,089 NAD-RNA species identified in our poly(A) RNA NAD captureSeq and the 2,000 NAD-RNAs found in their poly(A) RNA NAD tagSeq, 1,712 were in common (Dataset S6). Thus, we conclude that NAD-RNAs are widespread in the Arabidopsis transcriptome in that many genes give rise to NAD-RNAs. The high degree of overlap between the 2 independent studies also suggests that the NAD+-capped transcriptome is not random: Although a large number of genes produce NAD-RNAs, some genes have a higher propensity than others to generate NAD-RNAs.
Our findings of the widespread presence of NAD-RNAs are consistent with findings from human cells (9) and, together with discoveries from bacteria and yeast (6, 8), indicate that NAD+ capping of RNA is an ancient and conserved RNA modification. Unlike the m7G cap, the NAD+ cap can be directly incorporated into transcripts as the initiating nucleotide by bacterial RNAP, eukaryotic nuclear RNAP II, and mitochondrial RNAP (mtRNAP) (7, 10, 11, 24). Intriguingly, transcripts from chloroplasts were not found to be NAD+ capped in our NAD captureSeq. Angiosperm chloroplasts employ 2 kinds of RNAPs for transcription: a nuclear-encoded, mtRNAP-like, single subunit RNAP (NEP) and a chloroplast-encoded, multisubunit RNAP (PEP) (25). A functional chloroplast depends on the coordinated action of these 2 RNAPs; many chloroplast genes have both PEP and NEP promoters, and both PEP and NEP are essential for chloroplast transcription (26). The absence of chloroplast NAD-RNAs could be due to the inability of NEP and PEP to incorporate NAD+. Another potential reason for the absence of chloroplast NAD-RNAs is the low concentration of free NAD+ in chloroplasts. The levels of NAD-RNAs are highly correlated with those of cellular free NAD+ (12). NAD+ is essential for metabolic energy production, and the largest cellular NAD+/NADH pool is believed to be located in mitochondria (2 mM in mitochondria vs. 0.6 mM in the cytosol) (27). On the other hand, chloroplasts have a high level of phosphorylated NADH, which serves as the terminal electron acceptor in the photosynthetic electron transport chain (28). Thus, the uneven distribution of NAD-RNAs among organelles may be due to differences in free NAD+ levels in the cellular compartments. Finally, it is worth noting that the lack of detection of chloroplast NAD-RNAs via NAD captureSeq cannot rule out the existence of such RNAs in chloroplasts. The identification of NAD-RNAs in this approach depends on a high signal (ADPRC+) to noise (ADPRC−) ratio. We note that the chloroplast transcripts had high FPKM values in the ADPRC− samples (Fig. 2B), suggesting that they might have been biotinylated in the absence of ADPRC treatment, perhaps due to some unknown modifications. A similar argument goes for rRNAs, for which no NAD+ capping was detected.
The m7G cap provides a platform to form cap-binding complexes, which then recruit other factors to promote splicing, polyadenylation, and translation (2). It is not known whether or how the NAD+ cap interfaces with various RNA processing and translational machineries. The NAD+-capped transcripts detected in this study and in the Zhang et al. (23) study were properly spliced and polyadenylated, which is consistent with observations in human HEK293T cells (9). Thus, NAD+ may serve as a functional cap to promote splicing and polyadenylation with as yet unknown mechanisms. A previous study reported the lack of translation capacity of an NAD-RNA, which was assessed by transfection of an in vitro-synthesized, NAD+-capped luciferase mRNA into HEK293T cells (9). On the contrary, our studies implicate that NAD-RNAs are not only translated but preferentially translated compared with m7G-capped RNAs. The polysomal fraction has a higher proportion of NAD-RNAs in the total transcript pool for RBCS genes and other genes, as determined by gel blot assays, qRT-PCR, and NAD captureSeq. Furthermore, treatment of cell lysates with puromycin led to the release of NAD-RNAs from polysomes, implying that NAD-RNAs are on actively translating polysomes. The difference in results between these 2 studies could be attributed to different mechanisms in the 2 species (Arabidopsis vs. human) or to different sources of NAD-RNAs evaluated. The in vitro-synthesized NAD-luciferase transcripts transfected into human cells may not undergo proper co- and posttranscriptional processing as do endogenous transcripts, such as splicing, protein binding, nuclear export, etc., and may not reflect the fate of endogenous NAD-RNAs.
Our findings that NAD-RNAs are likely translated raise many questions that await investigation. Can the NAD+ cap recruit a cap-binding complex, which further recruits translation initiation factors and the 40S ribosomal subunit? How do NAD+-capped mRNAs undergo translation initiation? Future investigations will focus on these and other questions regarding this newly identified RNA cap. More importantly, as NAD+ is a metabolite that reflects cellular energy and redox conditions, the existence and translatability of NAD-RNAs in plants suggest that NAD+ capping may be a mechanism whereby cellular energy and redox status impacts the transcriptome and translatome, which may in turn contribute to energy and redox homeostasis.
Materials and Methods
Plant Materials and Growth Conditions.
A. thaliana Col-0 seeds were stratified at 4 °C for 3 d in the dark. Sterilized seeds were dispersed onto plates containing 1/2 Murashige and Skoog medium, 1% (wt/vol) sucrose, and 0.7% (wt/vol) agar and grown for 12 d in growth rooms with 16 h light/8 h dark cycles at 22 °C. For this study, 12-d-old seedlings or unopen flower buds were collected.
In Vitro Transcription.
A DNA template with a T7 ɸ2.5 promoter at the 5′ end and an adenosine at the transcription start site, but without any adenosine in the rest of the sequence (9), was prepared as follows. First, a single-stranded DNA with the above features was synthesized. Next, PCR was performed to generate a double-stranded DNA (dsDNA) template for in vitro transcription (primer sequences in SI Appendix, Table S1). In vitro transcription reactions contained 5 µg of dsDNA templates; 1X T7 polymerase buffer (New England Biolabs); 10 mM cytidine triphosphate (CTP), guanosine triphosphate (GTP), and uridine triphosphate (UTP); 10 mM NAD+ (for 5′ NAD+-RNA) or 10 mM ATP (for 5′ pppRNA); and T7 RNAP (New England Biolabs) and were allowed to proceed at 37 °C overnight. RNAs were treated with DNase I (Roche) at 1 U/μg at 37 °C for 30 min, extracted by acid phenol and chloroform, and then precipitated with ethanol. Unincorporated nucleotides were removed using Micro Bio-Spin P-30 Gel Columns (Bio-Rad).
Quantification of NAD Content in RNA via NAD-capQ.
For the measurement of NAD+/NADH level in plant RNAs from seedlings and inflorescences, the NAD-capQ method was used (12). An in vitro-transcribed, NAD-RNA was included as a positive control. Briefly, total RNAs from seedlings and inflorescences were extracted with the TRIzol reagent. Free cellular NAD+/NADH was removed by incubating the extracted RNAs with 10 mM Tris⋅HCl (pH 7.5) and 2 M urea, followed by RNA precipitation in the presence of 2 M ammonium acetate. The in vitro-transcribed RNA (1 µg) and total RNAs from seedlings (800 µg) and inflorescences (300 µg) were digested into nucleotides by nuclease P1 (1 U/50 µg RNA). Next, the amount of NAD+/NADH was measured by a colorimetric assay according to the manufacturer’s instructions (NAD+/NADH Quantitation Kit, Sigma). The same RNA samples without nuclease P1 digestion served as negative controls for the background NAD+/NADH levels.
NAD captureSeq.
Total RNAs were extracted from 12-d-old seedlings and inflorescences by the TRIzol reagent and treated with DNase I (Roche; 1 U/μg RNA at 37 °C for 60 min). Isolation of NAD-RNAs was according to the original capture protocol (6, 13), with minor modifications as follows: 100-μg portions of total or polysomal RNAs were incubated in 100-μL reactions containing 0.85 μM ADPRC (or left out as a control), 10% (vol/vol) 4-pentyn-1-ol (Sigma), 50 mM Hepes, and 5 mM MgCl2 (pH 7) at 37 °C for 30 min. The reactions were stopped by low-pH phenol/chloroform extraction. The RNAs were precipitated and then treated with 250 μM biotin-PEG3-azide, 1 mM fresh CuSO4, 0.5 mM Tris (3-hydroxypropyltriazolylmethyl)amine (THPTA; Sigma), 2 mM sodium ascorbate, 50 mM Hepes, and 5 mM MgCl2 (pH 7) at 25 °C for 30 min. After acid phenol/chloroform extraction and precipitation, the RNAs were incubated with 25 μL of MyOne Streptavidin C1 beads (preblocked with bovine serum albumin. and E. coli tRNA for 20 min) for 1 h in 100 μL of immobilization buffer [1 M NaCl, 10 mM Hepes (pH 7), and 5 mM ethylenediaminetetraacetic acid]. Beads were then washed 4 times with streptavidin wash buffer [8 mM urea, 50 mM Tris⋅HCl (pH 7.4), and 0.25% Triton X-100] and an additional 3 times with nuclease-free H2O. NEBNext Ultra Directional RNA Library Prep Kit was used for subsequent RNA library preparation. First Strand Synthesis Reaction Buffer (containing zinc ions to fragment RNA) was mixed with the random primers to make the RT mixture. The beads with bound NAD-RNAs were suspended with the RT mixture and incubated at 94 °C for 6 min to (i) elute the RNAs from beads, (ii) fragment the RNAs, and (iii) denature the RNAs. Next, RNase Inhibitor and reverse transcriptase were added to initiate first-strand cDNA synthesis. The remaining steps were performed according to the manufacturer’s instructions.
For poly(A) mRNA NAD captureSeq, ∼4 μg of poly(A) RNAs were isolated from 500 μg of total RNAs using oligo (dT) magnetic beads (New England Biolabs). These RNAs were made up to 100 μg by adding 96 μg of E. coli tRNA (to mitigate RNA loss during the NAD capture process). The 100-μg RNA mixture was subjected to the same NAD capture and RNA library preparation procedures described above. The same batch of RNAs was also used for RNA-seq library preparation using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (E7420; New England Biolabs).
The libraries were sequenced with the 150-nucleotide paired-end mode with Illumina HiSeqX Ten. The sequences were deposited at the National Center for Biotechnology Information Gene Expression Omnibus database under the accession number GSE127002.
Analysis of NAD captureSeq Libraries.
NAD captureSeq reads were collapsed into nonredundant reads and mapped back to the TAIR10 genome with Araport 11 annotation using STAR aligner with a maximum of 8 mismatches per pair-end read being allowed (29). Transcript levels were quantified using cuffdiff (30), and those with FPKM higher than 1 were retained for further analysis. NAD-RNAs were defined as ones with 2-fold higher levels in ADPRC+ samples than in ADPRC− samples at a maximum FDR of 0.05.
The gene body coverage of NAD captureSeq reads was calculated through RSeQC (31). Briefly, all transcripts were scaled to 100 nucleotides, and the number of reads covering each nucleotide position was counted and normalized based on the position with the highest number of reads.
Analysis of RNA-Seq and Polysomal RNA-Seq.
All RNA-seq reads were collapsed into nonredundant reads and mapped back to the TAIR10 genome with Araport 11 annotation using STAR aligner with a maximum of 8 mismatches per paired-end read being allowed (29). Transcript levels were quantified using cuffdiff (30), and those with FPKM higher than 1 were retained for further analysis. PEGs were defined as genes having at least 2-fold higher expression in RNA-seq from the polysomal fraction than in RNA-seq from total extracts at a maximum FDR of 0.05. PDGs were defined as genes with at least 2-fold higher expression in RNA-seq from total extracts than in RNA-seq from the polysomal fraction at a maximum FDR of 0.05.
Clustering Analysis of NAD captureSeq Libraries and RNA-Seq Libraries.
The 10,000 top-varying transcripts were used for hierarchical clustering analysis, and samples were grouped based on the correlation distance.
Validation of NAD-RNAs by qRT-PCR and Gel Blot Assays.
Captured NAD-RNAs were eluted from magnetic MyOne Streptavidin C1 beads by incubation in diethyl pyrocarbonate (DEPC) water at 70 °C for 10 min, and reverse transcribed into cDNA in 20-μL reactions containing 1 μL of random hexamer (50 ng/μL; Invitrogen), 1 μL of SuperScript III Reverse Transcriptase (200 U/μL; Invitrogen), 1 μL of deoxynucleotide (10 mM), 1 μL of 0.1 M dithiothreitol (DTT), 4 μL of 5X RT buffer, and 1 μL of RiboLock RNase Inhibitor (40 U/μL; Thermo Fisher). The reactions were kept at 25 °C for 10 min followed by 60 min at 50 °C, and were quenched at 70 °C for 15 min.
Real-time PCR was performed with gene-specific primers (SI Appendix, Table S1) using iTaq Supermix (Bio-Rad) on the Bio-Rad CFX96 Real-Time PCR System. RNA levels were computed by the comparative Ct method. For RNA gel blot assays, captured NAD-RNAs were released from magnetic MyOne Streptavidin C1 beads by the addition of denaturing RNA loading buffer (Thermo Fisher) and incubation at 70 °C for 10 min. The purified NAD-RNAs from beads and non–NAD-RNAs from the supernatant were resolved in 2% denaturing agarose gels and transferred to nylon membranes by the capillary method. A probe corresponding to RBCS1B/2B/3B genes was generated by PCR amplification from genomic DNA using gene-specific primers (SI Appendix, Table S1) and labeled with [a-32P]-dCTP using the DecaLabel DNA Labeling Kit (Thermo Fisher). Hybridization was performed as described (32). Signals were detected and quantified using the Typhoon phosphorimaging system.
Isolation of Polysomes and Puromycin Treatment.
Total cellular polysomes were isolated as previously described (33). Briefly, seedlings (1 g) were ground in liquid nitrogen and the powder was resuspended in 8 mL of polysome extraction buffer [0.2 M Tris⋅HCl (pH 8.5), 0.1 M KCl, 70 mM Mg(C2H3O2)2, 50 mM ethylene glycol-bis (β-aminoethyl ether)-N, N, N′, N′-tetraacetic acid (EGTA), 0.25 M sucrose, 10 mM DTT, 50 μg/mL cycloheximide, 50 μg/mL chloramphenicol, and 2.5 U/mL SUPERase⋅In]. The slurry was filtered through 2 layers of miracloth at 16,000 × g for 15 min at 4 °C twice. The supernatant was loaded onto an 8-mL 1.75 M sucrose cushion, and ribosomes were pelleted by centrifugation at 170,000 × g for 3 h at 4 °C in a Type70 Ti rotor (Beckman Coulter) and resuspended in 400 μL of resuspension buffer [0.2 M Tris⋅HCl (pH 9.0), 0.2 M KCl, 0.025 M EGTA, 0.035 M MgCl2, 5 mM DTT, 50 μg/mL cycloheximide, and 50 μg/mL chloramphenicol].
For separation of nonpolysomal and polysomal fractions, the resuspended ribosomes were applied onto a 15 to 60% sucrose step gradient and centrifuged at 100,000 × g in an SW55 Ti rotor (Beckman Coulter) for 1.5 h. Fractions were collected from the top to the bottom of the gradient with continuous monitoring of 254-nm absorbance. Nonpolysomal (40S, 60S, and 80S) and polysomal fractions were pooled separately for RNA extraction.
For puromycin treatment, total plant lysates were prepared in polysome extraction buffer (with the exclusion of cycloheximide and chloramphenicol). Puromycin was added to 0.25 mg/mL or not added (mock control). After 30 min incubation at room temperature, the lysate was loaded on a sucrose cushion for ribosome isolation, followed by the separation of nonpolysomal and polysomal fractions as described above.
For determination of the polysome association of transcripts under mock and puromycin treatment (Fig. 5D), we isolated ribosomes from the same amount of seedlings (1 g); the ribosomes were mock or puromycin treated. The ribosomes were fractionated, and nonpolysomal and polysomal fractions were collected. RNAs isolated from the 2 fractions were dissolved to the same volume (20 μL) with DEPC water, and then 3 μL of each sample corresponding to different amounts of RNA (nonpolysomal mock sample: 3 μg; polysomal mock sample: 6 μg; nonpolysomal puromycin sample: 4 μg; polysomal puromycin sample: 4 μg) was subjected to RT to generate cDNA. Although the RNA amount used for RT from each sample was different, the RNAs were proportionally derived from each fraction. qRT-PCR was then performed with the same dilution of the cDNA. Thus, the nonpolysomal and polysomal qRT-PCR values could be directly compared.
Supplementary Material
Acknowledgments
We thank Mr. Hailei Zhang for discussions. This work was supported by the NIH (GM061146) and the National Institute of Food and Agriculture (CA-R-BPS-5084-H). Y.W. and C.Y. were supported by postdoctoral fellowships from Shenzhen University. B.L. was supported by an NIH T32 training grant (T32 ES018827).
Footnotes
The authors declare no conflict of interest.
Data deposition: Genomics datasets have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE127002).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903682116/-/DCSupplemental.
References
- 1.Shatkin A. J., Capping of eucaryotic mRNAs. Cell 9, 645–653 (1976). [DOI] [PubMed] [Google Scholar]
- 2.Ramanathan A., Robb G. B., Chan S. H., mRNA capping: Biological functions and applications. Nucleic Acids Res. 44, 7511–7526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filipowicz W., et al. , A protein binding the methylated 5′-terminal sequence, m7GpppN, of eukaryotic messenger RNA. Proc. Natl. Acad. Sci. U.S.A. 73, 1559–1563 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shatsky I. N., Terenin I. M., Smirnova V. V., Andreev D. E., Cap-independent translation: What’s in a name? Trends Biochem. Sci. 43, 882–895 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Chen Y. G., Kowtoniuk W. E., Agarwal I., Shen Y., Liu D. R., LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 5, 879–881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cahová H., Winz M. L., Höfer K., Nübel G., Jäschke A., NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519, 374–377 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Frindert J., et al. , Identification, biosynthesis, and decapping of NAD-capped RNAs in B. subtilis. Cell Rep 24, 1890–1901.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Walters R. W., et al. , Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 114, 480–485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao X., et al. , 5′ end nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-mediated deNADding. Cell 168, 1015–1027.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malygin A. G., Shemyakin M. F., Adenosine, NAD and FAD can initiate template-dependent RNA synthesis catalyzed by Escherichia coli RNA polymerase. FEBS Lett. 102, 51–54 (1979). [DOI] [PubMed] [Google Scholar]
- 11.Bird J. G., et al. , The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 535, 444–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grudzien-Nogalska E., Bird J. G., Nickels B. E., Kiledjian M., “NAD-capQ” detection and quantitation of NAD caps. RNA 24, 1418–1425 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winz M. L., et al. , Capture and sequencing of NAD-capped RNA sequences with NAD captureSeq. Nat. Protoc. 12, 122–149 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Wang Y., et al. , NAD captureSeq in Arabidopsis. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE127002 (26 February 2019).
- 15.Filipowicz W., Functions of the 5,-terminal m7G cap in eukaryotic mRNA. FEBS Lett. 96, 1–11 (1978). [DOI] [PubMed] [Google Scholar]
- 16.Hinnebusch A. G., Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75, 434–467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munroe D., Jacobson A., mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol. Cell. Biol. 10, 3441–3455 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey-Serres J., Selective translation of cytoplasmic mRNAs in plants. Trends Plant Sci. 4, 142–148 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Kawaguchi R., Girke T., Bray E. A., Bailey-Serres J., Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 38, 823–839 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Yarmolinsky M. B., Haba G. L., Inhibition by puromycin of amino acid incorporation into protein. Proc. Natl. Acad. Sci. U.S.A. 45, 1721–1729 (1959). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohler R. E., Ron E. Z., Davis B. D., Significance of the free 70 s ribosomes in Escherichia coli extracts. J. Mol. Biol. 36, 71–82 (1968). [DOI] [PubMed] [Google Scholar]
- 22.Juntawong P., Girke T., Bazin J., Bailey-Serres J., Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, E203–E212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H., et al. , NAD tagSeq reveals that NAD+-capped RNAs are mostly produced from a large number of protein-coding genes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 116, 12072–12077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bird J. G., et al. , Highly efficient 5′ capping of mitochondrial RNA with NAD+ and NADH by yeast and human mitochondrial RNA polymerase. eLife 7, e42179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray M. W., Lang B. F., Transcription in chloroplasts and mitochondria: A tale of two polymerases. Trends Microbiol. 6, 1–3 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Börner T., Aleynikova A. Y., Zubo Y. O., Kusnetsov V. V., Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim. Biophys. Acta 1847, 761–769 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Igamberdiev A. U., Gardeström P., Regulation of NAD- and NADP-dependent isocitrate dehydrogenases by reduction levels of pyridine nucleotides in mitochondria and cytosol of pea leaves. Biochim. Biophys. Acta 1606, 117–125 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Heber U. W., Santarius K. A., Compartmentation and reduction of pyridine nucleotides in relation to photosynthesis. Biochim. Biophys. Acta 109, 390–408 (1965). [DOI] [PubMed] [Google Scholar]
- 29.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C., et al. , Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L., Wang S., Li W., RSeQC: Quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Liu J., He Y., Amasino R., Chen X., siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev. 18, 2873–2878 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mustroph A., et al. , Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 18843–18848 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.