Significance
Acute myeloid leukemia is a disease that lacks effective therapies, especially in postremission patients. Immunotherapies directed against a lineage-specific antigen (LSA), such as CD33 has demonstrated on-target effects on AML cells but is limited by toxicities because normal myeloid cells and hematopoietic progenitors also express CD33. Here we show that genetically ablating CD33 in human stem/progenitor cells, using Cas9/guide RNA mediated strategies, enables immunotherapy against leukemias using anti-CD33 CAR-T or antibody therapy. We model a postremission human marrow with minimal residual leukemic disease in mice and show effective clearance of acute myeloid leukemia and the reconstitution of the CD33-deleted human graft, enabling future clinical studies. This study presents an approach to treat myeloid leukemias and could be extended to other cancers and other antigens.
Keywords: acute myeloid leukemia, chimeric antigen receptor, CRISPR/Cas9, transplantation, CD33
Abstract
Antigen-directed immunotherapies for acute myeloid leukemia (AML), such as chimeric antigen receptor T cells (CAR-Ts) or antibody-drug conjugates (ADCs), are associated with severe toxicities due to the lack of unique targetable antigens that can distinguish leukemic cells from normal myeloid cells or myeloid progenitors. Here, we present an approach to treat AML by targeting the lineage-specific myeloid antigen CD33. Our approach combines CD33-targeted CAR-T cells, or the ADC Gemtuzumab Ozogamicin with the transplantation of hematopoietic stem cells that have been engineered to ablate CD33 expression using genomic engineering methods. We show highly efficient genetic ablation of CD33 antigen using CRISPR/Cas9 technology in human stem/progenitor cells (HSPC) and provide evidence that the deletion of CD33 in HSPC doesn’t impair their ability to engraft and to repopulate a functional multilineage hematopoietic system in vivo. Whole-genome sequencing and RNA sequencing analysis revealed no detectable off-target mutagenesis and no loss of functional p53 pathways. Using a human AML cell line (HL-60), we modeled a postremission marrow with minimal residual disease and showed that the transplantation of CD33-ablated HSPCs with CD33-targeted immunotherapy leads to leukemia clearance, without myelosuppression, as demonstrated by the engraftment and recovery of multilineage descendants of CD33-ablated HSPCs. Our study thus contributes to the advancement of targeted immunotherapy and could be replicated in other malignancies.
Acute myeloid leukemia (AML) is a hematopoietic stem cell (HSC) disorder characterized by clonal expansion of functionally abnormal immature myeloid progenitors (1). The current approach for the treatment of AML includes induction of remission and postremission therapy that dates from the 1980s (2). Complete remission is achieved in most cases, but most patients relapse within weeks or months. Postremission therapies may include allogeneic HSC transplantation (HSCT), which can be curative in 30–50% of patients, but many patients relapse. Hence, AML represents a disease with a huge unmet need.
Because the success of HSCT in AML patients depends on donor immune cells attacking and killing the host leukemia cells (a phenomenon termed “graft-versus-leukemia”), immunological therapy may represent a powerful mode to treat AML. However, standard immune checkpoint inhibition has generally not proven successful in AML. An alternative is to use chimeric antigen receptor T (CAR-T) cell therapy, or antibody-drug conjugates (ADCs) directed against the AML cells. However, both approaches require antigens that are uniquely or preferentially expressed on AML cells (3). In the absence of such AML-specific antigens, these immunotherapies rapidly reach their toxicity threshold (4, 5) and can no longer be used because of the on-target killing of normal, non-AML myeloid cells (e.g., neutrophils, or hematopoietic progenitors that express the antigen) that are necessary for survival. Here, we show that by genetically editing and thereby deleting a target (CD33) on normal blood stem cells, we can create a platform that allows maximal delivery of ADC and CAR-T therapy, either singly or in combination, in AML. This approach does not compromise normal myeloid numbers (because they lack the antigen and are therefore resistant to the immune therapy), enables maximal immunological treatment, and thus represents a potentially novel means to treat AML that deserves clinical evaluation in humans.
Our studies began with the observation of the success of CD19-directed CAR-T cell therapy for B cell malignancies. In the CAR-T therapy approach, T cells are genetically modified to express chimeric antigen receptors, which are generated by fusing one or more signaling domain to a single-chain variable fragment of a monoclonal antibody (mAb) with a high affinity for an antigen uniquely and abundantly expressed on cancer cells (6). Patients treated with anti-CD19 CAR-T showed antiacute lymphoblastic leukemia responses, but also prolonged B cell aplasia, due to the killing of both normal and tumor B cells (7–9). In humans, B cell ablation could be tolerated by virtue of Ig intravenous infusions.
Despite the success of CAR-T therapy in B cell-related malignancies, this approach could not be replicated in other hematological malignancies, including AML, due to lack of unique targetable cell surface antigens. An ideal antigen candidate for a CAR-T therapy should be unique to cancer cells and not expressed on normal cells of the same lineage type or elsewhere in the body. CD19, a B cell lineage-specific marker, shares some of these “ideal” properties: it is lineage-specific and predominantly expressed on malignant cells but also on normal B cells but, as described above, B cell aplasia can be managed with Ig supplements. For cancers, where such an ideal antigen is absent, we propose an approach combining the targeting of an antigen that is lineage-specific and overexpressed by malignant cells with the transplantation of genetically engineered stem cells lacking that lineage-specific antigen (LSA).
CD33, a transmembrane protein with two extracellular Ig-like domains, is a member of the sialic acid-binding Ig-like lectins (SIGLEC). CD33 makes an attractive target for immunotherapy in myeloid malignancies for several reasons. CD33 is constantly expressed on both normal and malignant myeloid cells and its expression is found in >90% adult and childhood AML blasts and on leukemia stem cells (10). Both in preclinical and clinical studies, anti-CD33 therapy using mAbs, such as Lintuzumab (SGN33) and Gemtuzumab Ozogamicin (GO), have shown a good response in a subset of AML (11, 12). Preclinical studies, both in vitro and in vivo (AML xenograft), using CD33 CAR-T (CART33) (13) showed good response by eliminating leukemia and improving survival. Despite such good response in preclinical and clinical AML studies, anti-CD33 therapies were not in routine use in AML patients until recently for several reasons. In preclinical models, hematopoietic toxicities, including cytopenia and reduction in myeloid progenitors, were observed (14). Myelosuppression was associated with severe impairment of immune and clotting functions due to severe neutropenia and thrombocytopenia, respectively. The most recent National Comprehensive Cancer Network guidelines have reintroduced GO as a single agent or in combination for AML patients.
Although its function is poorly characterized, CD33 knockout mice develop normally without any apparent hematological defects (15), suggesting CD33 is either dispensable for hematopoietic functions or is functionally redundant. Furthermore, we note the existence of rare deletion (dbSNP ID rs201074739) in CD33 (with an allele frequency of 0.014) that results in a frameshift in exon 3 in humans; homozygotes carrying two copies of this allele would be expected to be null for CD33, thereby providing further evidence for the functional redundancy of this molecule in humans. Based on the above information, we designed an approach to treat AML (Fig. 1A). In this proof-of-concept preclinical study, we provide evidence that genetically ablating a myeloid LSA, such as CD33 in human stem/progenitor cells (HSPC), enables immune therapy against leukemias using anti-CD33 CAR-T or antibody therapy. We model a postremission marrow with minimal residual disease and CD33 ablated HSPCs, and show that anti-CD33 therapy leads to the elimination of myeloid leukemia for at least 12 wk, and the concomitant engraftment and growth of CD33 deleted hematopoietic cells transplanted into the same host. We note that this approach could be extended to other cancers and other antigens that have similar properties (i.e., the antigens that are expressed on the cancer cells, but have minor or dispensable functions in normal cells). Our study thus contributes to the advancement of targeted immunotherapy and could be broadened to other malignancies.
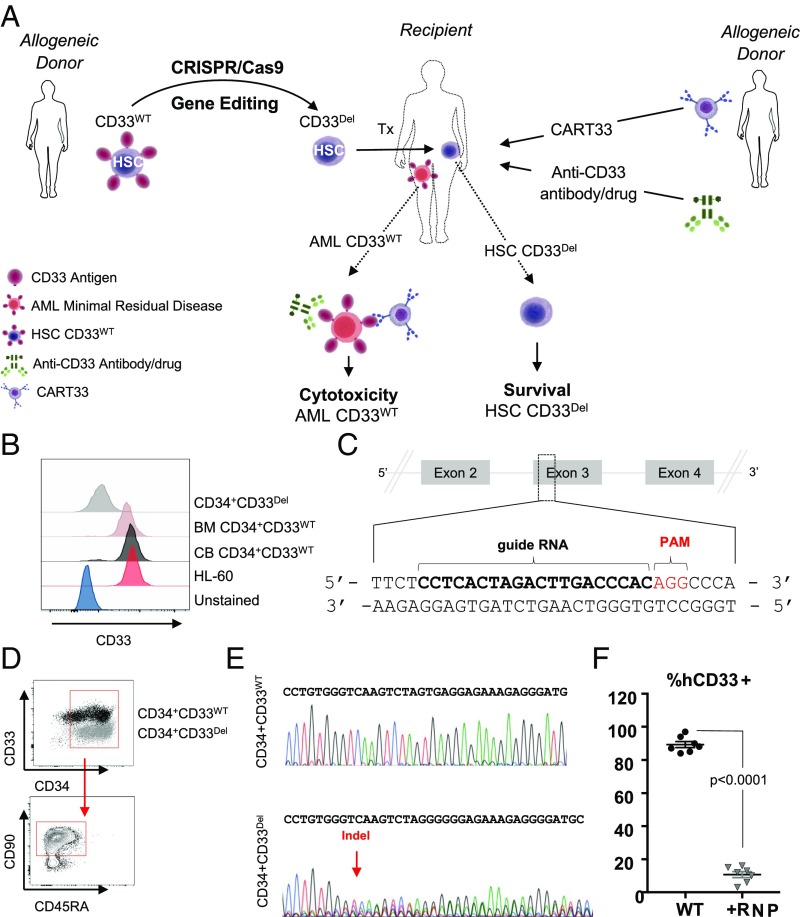
Fig. 1.
(A) Approach: stem cells, either mobilized or CB obtained from a donor, will be genetically manipulated to ablate CD33 expression using gene-editing technology, such as CRISPR/Cas9, and transplanted to relapsed patients eligible for HSCT. Subsequent to transplantation, T cells from an allogeneic donor will be genetically manipulated, using a viral delivery system, to express chimeric antigen receptors targeting CD33 and infused in the recipient. Alternatively, patients can receive ADC (GO) either alone or in combination with CAR-T. (B–F) CD33 expression and its ablation in human cells. (B) Expression of CD33 in human AML cell line HL-60, in human primary CD34+CD33WT cells from BM and CB and human primary CD34+CD33Del after CRISPR/Cas9-mediated ablation. (C) Schematic representation of CD33 genomic locus showing exons 2–4 and location and sequence of sgRNA (in bold, PAM in red) targeting CD33. (D) Surface expression of CD33 by flow cytometry after electroporation in CD34+CD33WT cells and CD34+CD33Del. All cells maintain their stem cells phenotype as assessed by CD90 expression. (E) Chromatogram of Sanger sequencing showing a region surrounding the DNA double-strand break site, (Upper) CD34+CD33WT cells and (Lower) CD34+CD33Del. (F) Five to 7 d after electroporation, CD34+ cultured cells show consistent deletion of CD33 compared with control (15 independent donors).
Results
CRISPR/Cas9-Mediated Genetic Ablation of CD33 Antigen.
We used CD33 expressing HL-60 to model myeloid leukemia and primary CD34+ cells, either from cord blood (CB) or from adult bone marrow (BM), as the donor HSPC. Surface expression of CD33 was confirmed in both HL-60 cells and in CD34+ cells using flow cytometry (Fig. 1B), as previously described (16–19). CRISPR/Cas9, a recently developed versatile RNA-guided DNA editing technology (17), was used to genetically edit genomic loci of CD33 to ablate its expression. Guides were designed to target exon 3 genomic loci (Fig. 1C and SI Appendix, Fig. S1) because it is common to all CD33 transcripts and has little to no similarity with Siglec family pseudogenes of which CD33 is a member. We tested plasmid, lentivirus, and ribonucleoprotein (RNP) -based delivery systems to check the efficiency of several guides in cell lines and primary cells, and found the RNP system in combination with chemically modified guides to be the most efficient in primary cells (Fig. 1 B and D). At the optimal conditions, we found the loss of CD33 expression in greater than 80% of CD34+ HSPC, henceforth referred as CD33Del (Fig. 1 B and D), measured on a flow cytometer using the anti-CD33 clone HIM34, which recognizes an epitope located in the C2 domain common to all CD33 isoforms. This reduction in CD33 expression was accompanied by the presence of insertions/deletions (indels) at the expected cut site of Cas9 on DNA as measured by Sanger sequencing of DNA (Fig. 1E, Lower chromatogram). Two other single-guide RNAs (sgRNAs), also targeting exon 3, were tested and displayed the same efficiency (SI Appendix, Fig. S1 A and B). As expected, electroporation of Cas9 alone in the absence of sgRNA did not induce any indels at the target site (Fig. 1E, Upper chromatogram). CD33Del cells retained high expression of CD34 and CD90 (Fig. 1D, Lower). We saw consistently high deletion efficiency in several independent experiments (Fig. 1F). At baseline, on average, 85% of CD34+ cells showed CD33 expression. After Cas9-mediated deletion, on average 10% showed CD33 expression, 5–7 d post-RNP electroporation. In addition, we also used B cells that lacked CD33 expression as a negative control to confirm loss of expression after Cas9-mediated deletion (SI Appendix, Fig. S1C). The remaining 10% of CD33 expression observed is likely from cells that are unedited (wild type for both alleles) or partially edited (only one allele with indels).
CD34+ CD33Del HSPCs Show Engraftment and Multilineage Differentiation in Vivo.
Because the central element of our approach is to transplant CD33 gene-edited stem cells (CD33Del) as a platform for CART33 and ADC delivery (GO), it is important to test the ability of CD33Del cells to engraft and contribute to myelopoiesis and lymphopoiesis. We tested both BM- and CB-derived CD34+ cells (Fig. 2). CD33Del HSPCs were injected in sublethally irradiated NSG-SGM3 (NSGS) mice via tail vein injection and BM and blood were analyzed (Fig. 2A). In mice injected with BM-derived cells, peripheral blood analysis at 7 wk posttransplant revealed the presence of human CD45+ cells, and mature cells of myeloid (CD14+ cells) and lymphoid (CD19+ cells) origin (Fig. 2B). The analysis of BM aspirate at 15 wk (SI Appendix, Fig. S2A) and whole BM from sacked mice at 21 wk (Fig. 2C), summarized in SI Appendix, Fig. S2B, showed chimerism with a sustained contribution of human CD45+ cells over time. In all of the tissues examined, there was multilineage engraftment with the presence of progenitors and mature cells of both myeloid (monocytes) and lymphoid (B cells) origin. All cells remained CD33− (SI Appendix, Fig. S2 A and C). No significant differences in multilineage engraftment of CD33Del cells were observed compared with wild-type cells.
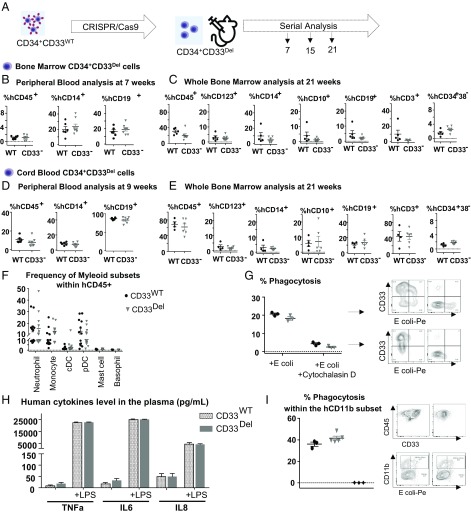
Fig. 2.
Deletion of CD33 does not impair engraftment, hematopoietic repopulation, and function in NSGS mice. (A) Schematic of experimental design. (B and C) BM-derived CD34+ cells engraftment and repopulation: (B) Peripheral blood (7 wk) and (C) whole BM (21 wk) posttransplant analyzed for cells of various lineages, as indicated. CD34+CD33Del cells show the same engraftment (CD45+) as control cells as well as comparable percentage of mature myeloid and lymphoid cells. BM CD34+CD33Del cells show comparable percentage of myeloid (progenitor CD123+, mature CD14+), and lymphoid (progenitor CD10+, mature CD19+) T cells (CD3+) and stem cells CD34+38−. (D and E) CB-derived CD34+ cell engraftment and repopulation: (D) Peripheral blood (9 wk) and (E) BM (21 wk) posttransplant analyzed for cells of various lineages, as indicated. CD34+CD33Del cells show same engraftment (CD45+) as control cells, as well as comparable percentage of mature myeloid and lymphoid cells. BM CD34+CD33Del cells show comparable percentage of myeloid (progenitor CD123+, mature CD14+) and lymphoid (progenitor CD10+, mature CD19+), T cells (CD3+), and stem cells CD34+38−. Data were analyzed using unpaired t test and no significant differences were found in all of the groups examined (P > 0.05). All data are represented as mean ± SEM (two independent experiments, two donors). (F–I) In vitro and in vivo functional assays. (F) CD34+CD33Del show comparable development of myeloid lineage than CD34+CD33WT in NSGS mice. Frequencies of neutrophils, monocytes, cDC, pDC, mast cells, and basophils in the BM aspirates of NSGS mice injected with CB CD34+CD33WT or CD34+CD33Del cells. (Control n = 12, CD34+CD33Del n = 13). (G) In vitro E. coli bioparticles phagocytosis assay of in vitro CD33WT or CD33Del differentiated monocytes. CD33Del monocytes show similar phagocytosis capacity (two independent experiments, two donors). (H) Response to LPS-induced Toll-like receptor activation is similar in NSGS mice injected with CD34+CD33WT or CD34+CD33Del cells. Analysis of plasma cytokines level at 0 and 4h30 after intraperitoneal injection of 15 μg LPS (Control n = 12, CD34+CD33Del n = 13). (I) Peritoneal cavity analysis 2 h after intravenous injection of E. coli bioparticles (Control n = 3 CD34+CD33Del n = 5), untreated mice (♦). Mouse and syringe images designed by Freepik and Kiranshastry from Flaticon.
In parallel, we followed a similar strategy with CB-derived CD34+ cells and obtained similar results. We saw multilineage engraftment in peripheral blood at 9 wk (Fig. 2D), in BM aspirate at 16 wk (SI Appendix, Fig. S2C), and in whole BM at 21 wk posttransplant (Fig. 2E). The above results suggest that CD33 is not necessary for the engraftment of either the CB or BM CD34+ cells or for the sustained repopulation of a complete human hematopoietic system in animal models.
CD33Del Cells Are Proficient in Myeloid Differentiation and Function.
Because the goal of our approach is its translation to the clinic, we assessed the functional capacity of myeloid CD33Del cells in vitro and in vivo. First, we analyzed the diversity of myeloid lineage in CD34+CD33del compared with CD34+CD33WT humanized mice and found no noticeable difference among the myeloid subsets (Fig. 2F). We then tested the ability of monocyte differentiated CD34+CD33WT and CD34+CD33Del to phagocytose Escherichia coli bioparticles in vitro (Fig. 2G). No significant difference was noticed. We also analyzed the LPS-induced cytokine production by monocytes/macrophages in NSGS mice transplanted with CD34+CD33WT or CD34+CD33Del cells and saw that plasma levels of TNF-α, IL-6, and IL-8 after induction were comparable (Fig. 2H). Finally, to assess phagocytic function of CD33-deleted cells in vivo, we analyzed the peritoneal cavity of humanized mice, 2 h after intraperitoneal injection of E. coli bioparticles. Flow cytometry analysis showed similar phagocytic uptake by the hCD45+hCD11b+hCD14−hCD16− subset in both CD34+CD33WT and CD34+CD33Del humanized mice (Fig. 2I). All of these findings show intact function of CD33Del myeloid cells.
No Detectable Off-Target Mutagenesis or Loss of Functional p53 Pathways in Gene-Edited Cells.
Previous studies have shown varying degree of off-target mutagenesis with Cas9 depending upon the cell type and guide RNA sequence used. To evaluate whether the two guides used in this study introduce indels at off-target sites in HSP cells, we assessed for indels in whole-genome sequencing data of human CB CD34+ HSP cells electroporated with Cas9/sgRNA RNP complex (CD33Del) and compared with the cells electroporated with Cas9 protein only (CD33WT). We obtained over 629 million passed filter reads with a base quality of over Q30 in over 93% of the reads (SI Appendix, Table S1). The mean coverage depth was over 26×. To identify single-nucleotide variants and small indels, the reads were aligned to the human hg38 reference genome.
A summary of variants detected in both samples is presented in SI Appendix, Table S1. We saw a robust on-target activity with indels in >90% reads aligning to the expected cut sites, chr19:51225811 and chr19:51225846 (Fig. 3A). Importantly, all indels were located within expected cut sites of the two sgRNA used. Few small indels that were observed outside of the targeted region in the entire CD33 locus were not unique to CD33Del electroporated cells and were also present in cells electroporated with Cas9 only. We next examined our data for indels in predicted off-target sites that showed a high degree of similarity, with up to four mismatches with the sgRNAs used (Fig. 3B and SI Appendix, Tables S2 and S3). Again, we did not find any indels in all of the off-target loci examined (SI Appendix, Tables S2 and S3). We also looked for indels in TP53 loci and did not find any indels that were unique to CD33Del cells.
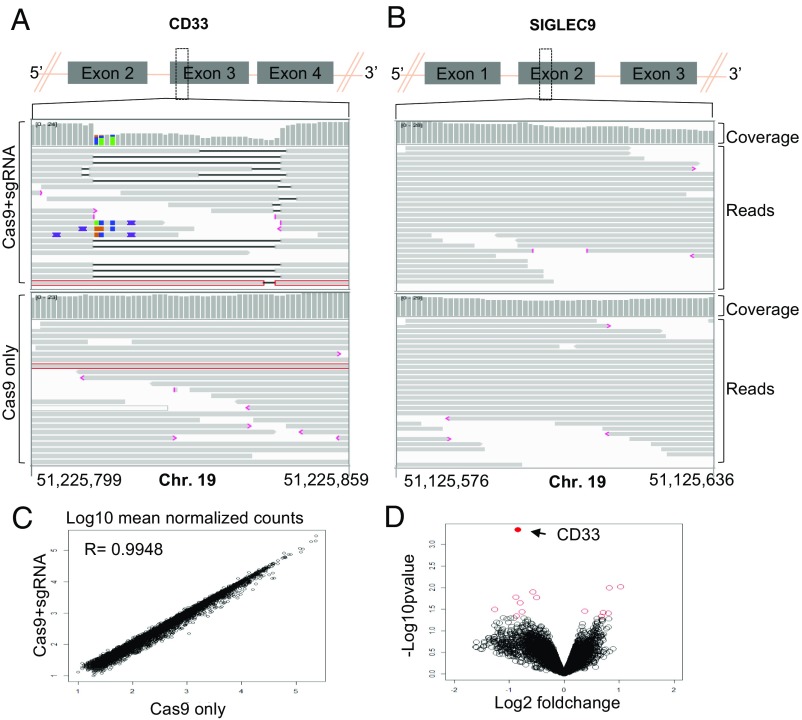
Fig. 3.
(A and B) IGV screenshot of genomic region of CD33 (A) and SIGLEC9 (B) genes surrounding the guides in Cas9+sgRNA (Top) and Cas9 only (Bottom) cells as indicated in the left. The gray bars in the coverage track (indicated on right) show the depth of the reads displayed at each locus. Generally, the coverage should be uniform and hence the bar height should be same but deletions results in dip in the height. The reads track shows all of the reads (gray boxes) mapped in this region. The deletions are represented by a solid black line and insertions with purple boxes. Reads with red border are those without a mapped mate. One read in each group was without a mapped mate. Mismatch bases are colored in green, blue, brown, and red for nucleotides A, C, G, and T, respectively. SIGLEC9 genomic region was selected as representative region to show absence of indels at off-target site because (i) it belongs to SIGLEC family with homology to CD33, and (ii) it has the highest homology within 10 bp of the expected cut site compared with any other guide (SI Appendix, Table S2). Chromosome coordinates at the bottom are based on hg38. (C) Scatter plot showing correlation between log10 mean normalized counts, normalized using the DEseq2 method, between CD33 edited cells and control cells. (D) Volcano plot showing log2 fold-change and −log10 P value for genes analyzed using the edgeR method; genes that were significantly differentially expressed (P < 0.05) are shown as red open circles and the CD33 gene is represented by a filled red circle and indicated by a left arrow (four donors).
To assess whether the loss of CD33 expression causes changes in expression of other genes, we compared gene-expression profiles of CD33 deleted (n = 5) and control (n = 5) CD34+ cells obtained from four different donors. A gene-expression profile for each sample was obtained using RNA sequencing and comparison between groups was made using edgeR. Comparable gene-expression profiles were observed between two groups with a Pearson correlation coefficient of 0.9948 (Fig. 3C) and no significant differences were observed based on P-adjusted value (SI Appendix, Table S4). Fourteen genes were found to be significantly different based on P value, with the most significant difference being the down-regulation of CD33 in the CD33Del samples compared with the controls (Fig. 3D and SI Appendix, Table S4). These results confirm that the absence of CD33 in CD34+ cells in vitro does not grossly affect downstream gene’s expression. Among the 13 genes that showed changed expression based on P value, there was no enrichment for any one pathway or cellular process. Of note, gene-expression signatures did not suggest that the TP53 pathway, or other DNA damage pathways, that could compromise HSC function or diminish their long-term potential, had been activated. We therefore conclude that CD33 ablation in CB cells and adult HSCs using the gene-editing technologies described here does not appear to compromise their future function.
We also manually inspected the data for indels in reads mapping to exon 3 of CD33 and all coding exons of the TP53 transcript in RNA-sequencing data using an integrated genomic viewer (IGV). As expected, there were indels in >95% reads in CD33 exon 3 (SI Appendix, Fig. S3). We found few reads with indels in TP53 but they were within repeat sequences and were also present in control samples, suggesting sequencing artifacts or their presence before editing. Taken together, these data suggest that CRISPR/Cas9-mediated genomic editing at the CD33 locus using the guides we used in this study results in no detectable off-target indels in our stem cell system. A more comprehensive analysis using deep sequencing and other approaches recently developed to study off-target effects may be required to study rare events.
Expression of CD33 Specific CARs in T Cells.
CARs are classified into different generations based on the number of costimulatory domains. We have designed a second-generation CAR (Fig. 4A), including a single-chain variable region of anti-CD33 (clone My96) paired with a CD28 transmembrane domain, a 4-1BB (CD137) costimulatory domain, and a CD3 ζ-chain from a CD3 T cell receptor as an intracellular domain. The CAR cDNA was cloned into the pHIV-zsGreen lentiviral vector under the control of an EF1-α promoter, enabling bicistronic expression with zsGreen. Peripheral blood obtained from normal donors was fractionated to obtain peripheral blood mononuclear cells (PBMCs), and transduced with lentiviral particles carrying either vector only or the CAR construct (Fig. 4B). We noticed that the transduction efficiency of CD4 and CD8 cells within the PBMCs was not equal, as we observed higher transduction of CD4 compared with CD8 cells, and similar observations were also made in other studies (20). Given the unequal transduction of CD4 and CD8 cells in PBMCs, and to obtain a more defined composition of CD4 and CD8 cells, we transduced purified CD4 and CD8 cells separately and sorted these cells based on GFP expression from a downstream IRES element and mixed them in an equimolar ratio. We confirmed the CAR expression by measuring the surface expression and binding of CAR to purified biotinylated CD33 protein conjugated to a streptavidin fluorochrome. We saw a robust expression of the CAR and its binding with the CD33 molecule (Fig. 4B).
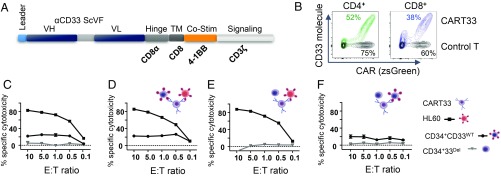
Fig. 4.
CD33 deletion protects CD34+ cells from CART33 cytotoxicity in vitro. (A) Schematic of CART33 construct. (B) Contour plot showing CAR expression in human primary T cells after lentiviral transduction with control (black) or CART33 (green or blue) virus. Percentage transduction in each group is specified next to the plots. CD4+ and CD8+ cells were transduced independently and comixed 1:1 before experiment. (C–F) Cytotoxicity assays. (C) CART33 cells or control T were incubated with HL-60 or CD34+CD33WT or CD34+CD33Del and cytotoxicity assessed by flow cytometry. (D–F) Triple culture cytotoxicity assay. CART33 cells or control T cells were coincubated with (D) HL-60 and CD34+CD33WT or (E) with HL-60 and CD34+CD33Del or (F) with CD34+CD33WT and CD34+CD33Del cells (four independent experiments).
CAR-Expressing T Cells Show CD33-Dependent Cytotoxicity in Vitro.
We first evaluated the cytotoxicity of CART33 cells over targets with variable CD33 expression. The high killing of CD33 myeloid leukemia cells HL-60 was confirmed, and lower killing of CD33WT stem cells, which express CD33 at a reduced level, was observed. Notably, the absence of CD33 expression (due to Cas9/sgRNA-mediated deletion) protected CD34+ cells from killing as no cytotoxicity of CART33 was observed when incubated with CD33Del CD34+ cells. This first experiment (Fig. 4C) also confirmed a correlation between CART33 cytotoxicity level and CD33 expression level on target cells: that is, that CART33 cytotoxicity is proportional to the expression level of CD33 on target cells.
We then devised a triple culture assay to assess CART33 cell killing when coincubated with targets of variable CD33 expression. When CD33WT HL-60 cells and CD34+CD33WT cells were coincubated with CART33, we saw correlated cytotoxicity level of CART33 cells over both cell types (Fig. 4D), but when CD33WT HL-60 cells and CD34+CD33Del cells were coincubated with CART33 cells, only HL-60 cells were killed (Fig. 4E). Similarly, the coincubation of all three cell types—CD34+CD33WT, CD34+CD33Del, and CART33 cells (Fig. 4F)—displayed only killing of CD34+CD33WT at a level proportional to its CD33 expression. Additionally, we tested CART33 cytotoxicity over another acute myeloid cell line, KG1, and found similar CD33-dependent cytotoxicity of CART33 in vitro (SI Appendix, Fig. S4).
Anti-CD33 Immunotherapy Show CD33-Dependent Leukemia Clearance in a Cell Line-Derived Xenograft (CDX) Mouse Model.
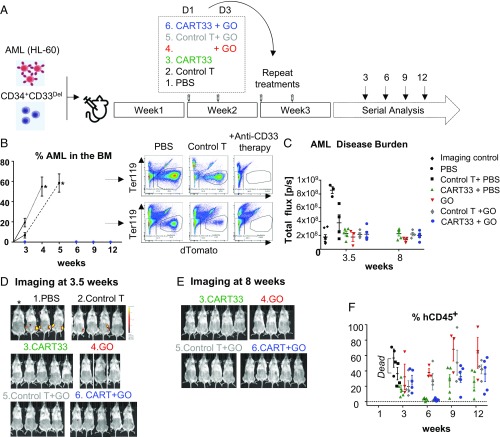
For our in vivo experiments, we designed a strategy that we felt would best represent the human therapeutic setting in the context of minimal residual disease (Fig. 5A). In this model, we first initiated leukemia by injecting 500,000 HL-60 cells in sublethally irradiated mice and simultaneously injected 500,000 CD34+CD33Del cells, to mimic an AML relapsing model. Our preliminary experiments suggested a 1-wk period was sufficient to enable homing and engraftment of AML cells, and cause 100% of mice to become leukemic after an additional 2 wk. This, to the best of our ability, recapitulates the postremission BM where AML cells may still remain, although be clinically undetectable. One week after coinjection of leukemia and CD34+ stem cells, mice were divided into various groups and treated with agents as described (Fig. 5A). Then leukemia burden and CD34+ cells engraftment was monitored over time using imaging and flow cytometry (Fig. 5 B–F and SI Appendix, Fig. S5). By week 3, we saw high tumor burden in the BM aspirate of PBS and control CAR-T cell (i.e., T cells transduced with the vector alone, lacking the CAR-T constructs) -treated mice, and by weeks 3 and 4 all of the mice in these two groups died of their disease (Fig. 5B and SI Appendix, Fig. S5). Two control T-treated mice had relatively low leukemia burden at 3 wk but progressed to a very high leukemia burden in BM at death (SI Appendix, Fig. S5A). In contrast, over the course of 12 wk, no CD33WT leukemia cells were found in BM aspirate (Fig. 5B and SI Appendix, Fig. S5B) or on imaging at 3.5 wk (Fig. 5 C and D and SI Appendix, Fig. S5C) or 8 wk (Fig. 5 C and E and SI Appendix, Fig. S5D) of mice treated with CART33 cells, with the anti-CD33 ADC GO or with a combination of GO and CART33. These results suggest that both CART33 and GO are potent agents against CD33-expressing leukemia in our model.
Fig. 5.
Therapy model: CD34+CD33Del cells resist CD33-targeted immunotherapy. (A) Schematic of experimental design: 5 × 105 HL-60 and 5 × 105 CD34+CD33Del were injected in NSGS mice on day 0. One week after, mice were treated with PBS or allogeneic CART33 or control T cells. Three days after a new group received GO only, while allogeneic CART33 and control T cells-injected mice received GO or PBS. Treatment was repeated on week 3. Leukemia progression and CD34+CD33Del engraftment were then monitored by serial BM aspiration. (B) Monitoring of leukemia burden in BM aspirates. Leukemia burden in the whole BM of control groups mice at the time of death are shown with an asterisk (*). Leukemia cells were gated on Ter119−dtomato+. (C) Leukemia burden measure via epifluorescence quantification of images shown in D and SI Appendix, Fig. S5C at 3.5 wk, and E and SI Appendix, Fig. S5D at 8 wk. One mouse representative of each treatment is shown in C and E. See SI Appendix, Fig. S5 for full imaging panel. Background was removed with untreated mouse (*Imaging control). (F) CART33 or GO leukemia clearance doesn’t impair engraftment of CD34+CD33Del cells overtime (% hCD45+ cells), as shown by flow cytometry of BM aspirates. CD34+-injected derived human cells were gated on Ter119−dtomato−, Ly5−/H2kd−human CD45+CART− (three independent experiments with GO, two independent experiments with CART33). Mouse and syringe images designed by Freepik and Kiranshastry from Flaticon.
CD34+CD33Del HSPC Show Multilineage Engraftment and Differentiation in the Therapy Model.
Simultaneously, we monitored mice for multilineage engraftment of CD34+CD33Del cells in our therapy model, described above. We observed engraftment as demonstrated by the presence of human CD45+ cells that are CAR-T and CD33− in BM aspirate of all of the groups (Fig. 5F), suggesting that CD34+CD33Del cells can also engraft in our therapy model. Interestingly, we saw a dip in the percentage of CD33Del human CD45+ cells at the week 6 time point relative to the initial point of week 3 in the CART33-treated groups (CART33+PBS and CART33+GO), but these levels were recovered to their initial levels by week 9 and maintained until the last point of week 12. The relatively low engraftment and the subsequent dip in two groups of mice likely reflect treatment-related stress (including the antileukemia response by CAR-T cells in the BM), which was more pronounced in the CAR-T group and persisted longer than the GO group. Alternatively, the use of different CD34+ and T cell donors might have triggered an allo-response that could explain the repopulation delay observed in mice injected with CART33 cells. Notably, this phenomenon was reversible as engraftment levels recovered over 8 wk.
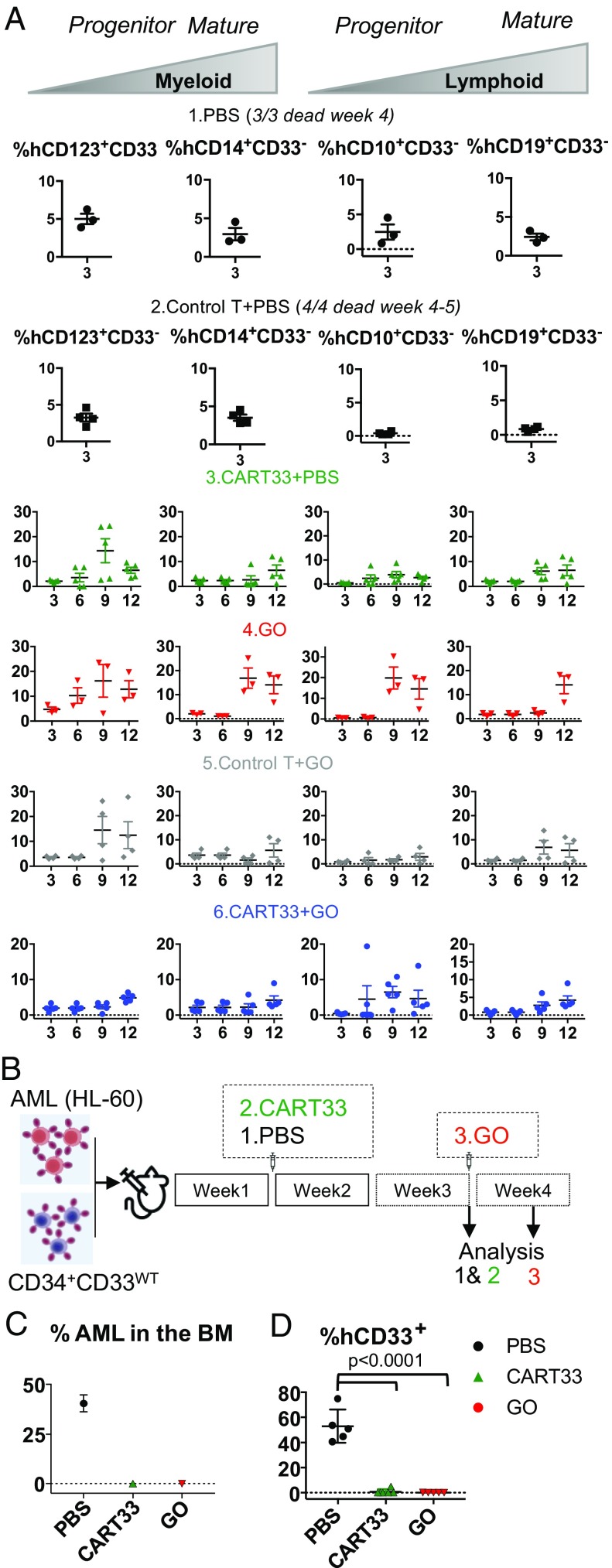
We next looked at the multipotential nature of the engrafted cells by analyzing myelopoiesis and lymphopoiesis (Fig. 6A). We found CD33− myeloid and lymphoid progenitors, as well as mature myeloid and lymphoid cells at all time points analyzed (Fig. 6A). While a full hematopoietic system repopulation was observed over time in all treated mice, we noticed a lymphoid repopulation delay in mice injected with CAR-T cells. This could be the result of an allo response, as lymphoid progenitor cells are known to be more sensitive to an allo-specific effect.
Fig. 6.
(A) CD34+CD33Del cells resist CD33-targeted immunotherapy and contribute to myelopoiesis and lymphopoiesis. Left two panels in each condition is monitoring overtime of the repopulation of myeloid progenitors, and Right two panels shows lymphoid progenitors and mature cells in BM aspirates. No significant differences were observed between different treatment groups at all time points analyzed. (B–D) CD34+CD33WT cells are sensitive to CD33-targeted immunotherapy. (B) A schematic of experimental design: 5 × 105 CD34+CD33WT alone or in combination with 5 × 105 HL-60 were injected in NSGS mice on day 0. One week after, mice were treated with PBS or allogeneic CART33 cells. Leukemia progression and CD34+CD33WT engraftment were then monitored by bone marrow aspiration at week 3 for CART33. The same day a group of mice was injected with GO and analyzed 4 d after. (C and D) BM aspirates show complete elimination of CD33WT leukemia cells (C) and CD33WT primary cells (D) in mice treated with CART33 or GO compared with PBS alone. Significant difference was observed between CART33 and GO compared with PBS. CD34+ injected derived human cells were gated on Ter119−dtomato−, Ly5−/H2kd− human CD45+CART−. All data are represented as mean ± SEM (two independent experiments, two donors). Mouse and syringe images designed by Freepik and Kiranshastry from Flaticon.
Concomitantly, to demonstrate the specificity of CART33 and GO toward CD34+CD33WT primary HSPCs, sublethally irradiated NSGS mice were coinjected with 500,000 HL-60 cells and 500,000 CD34+CD33WT cells (Fig. 6B). One week after, a group of mice was treated with CART33 cells and BM aspirated on week 3, the same day another group was injected with GO and their BM aspirate analyzed 4 d after. As observed in our therapy model, we confirmed full leukemia clearance following treatments (Fig. 6C). Additionally, we saw full ablation of CD33WT cells (Fig. 6D). Therefore, CD33WT cells remain sensitive to GO and CART33 therapies, while CD33 ablated cells are insensitive.
Discussion
The success of any antigen-dependent immune therapy using agents like CAR-T or mAbs is dependent on the presence of a unique antigen on the cancer cell surface and not on normal cells or other cells in the body. Unfortunately, such antigens are rare in cancers. We reasoned that by ablating LSA using genomic engineering methods in stem cells, we could generate stem/progenitor cells that are resistant to antigen-dependent immune therapy, thereby enabling maximal immunotherapy. After demonstrating that such antigen-depleted cells are functionally similar to the wild-type cells, they can be used to supplant the diseased cells. Careful selection of an LSA that is dispensable to the normal function of that lineage is key to this approach. In an alternative approach, if an LSA is indispensable, instead of ablating the expression of the LSA completely, one can use gene-editing technology to modify the epitope recognized by the antigen-dependent immune therapy agent on LSA while maintaining the LSA function (we term this approach “functionally redundant epitope switching,” or FRES, and will test this in the future studies).
In this proof-of-concept study, we show that combining stem cells lacking a lineage antigen, CD33, with allogeneic engineered T cells or an ADC, we can enable leukemia ablation and full hematopoietic repopulation. We used AML, a disease with unmet need in the area of therapy, and demonstrate that such an approach is feasible. Because CD33 is an LSA and targeting of CD33 in AML, using either CAR-T or CD33 mAbs, results in severe myelosuppression and lympho-depletion due to the elimination of stem/progenitor cells, as well as cells of myeloid lineage, our proposed approach for treating AML is to rebuild the hematopoietic system with cells lacking CD33. We used a CRISPR-based approach to disrupt CD33 expression in donor stem cells, either CB or BM CD34+ cells, to render them “resistant” to CAR-T cell attack.
Recently, two groups made similar observations and independently reported the approach described in this study (21, 22). Our data strengthen the observations made by these studies and also add insights using complementary approaches. Unlike the Kim et al. study (21), in which the mice were first injected with CD33 gene-edited CD34+ cells to allow for complete engraftment before the leukemia introduction and treatment, our approach more closely mimics the situation of AML relapse with minimal residual disease because we coinject leukemia cells and gene-edited stem cells, followed by CAR-T or ADC therapy. Furthermore, by stringently selecting CD33 guide RNAs with high on-target and low off-target activity, we observe efficient ablation of CD33 expression in HSCs with no off-target indels observed in other genes, thus enabling confidence in the safety of this approach in human studies (21). Indeed, we did not find any indels within any of the Siglec family of genes and pseudogenes examined. Kim et al. observed off-target activity in SIGLEC22P, including a deletion of a 4-kb fragment, most likely due to 100% homology with SIGLEC22P of the sgRNA designed for CD33 (21). Our choice of location within exon 3, and our confirmation of the absence of homology of the chosen sgRNA with other genes, may have enabled the specificity of CD33-only ablation. The absence of indels or other genomic rearrangements in TP53 gene (as analyzed using whole-genome sequencing), and the absence of any deregulated genes related to the p53 pathway or p53 itself, suggest that the approach that we have developed might provide efficient genome-editing with high specificity without compromising HSC function. Finally, the use of Cas9 RNP (which is only present transiently in the HSCs during their ex vivo manipulation), as opposed to virally mediated expression of Cas9 that is constitutive and continues in the HSCs in vivo, avoids future issues with preexisting immunity against Cas9, which is found to be present in >50% of the population (23, 24).
Furthermore, in contrast to Kim et al. (21), our approach involves allo-BM transplant with CD33-edited HSCs (from CB or adult BM) followed by ADC treatment or CAR-T treatment with T cells derived from the allogeneic donor. We feel that this approach is more practical in the clinical setting. Patients with hematologic malignancies who have been heavily pretreated with cytotoxic chemotherapies often produce poor autologous T cell yields, limiting the efficiency and effectiveness of autologous CAR-T. We circumvent this problem by using allo-BM transplantation and allo-T cells, where yield and quality is not an issue. More importantly, by using ADC (rather than CAR-T) to target the disease, we show that humoral therapy can act in concert with, or as an alternative to, CAR-T cells, further expanding the approach to antileukemia therapy using humoral approaches.
We also note that the GO drug (comprised of the anti-CD33 antibody clone P67.6) recognizes an epitope located in exon 2. Two isoforms of CD33 are found in humans. The more common isoform is the full-length protein including exon 2 that is sensitive to GO; the less common is an isoform that lacks exon 2. Around 30% of the population carries a homozygote SNP (T/T) resulting in the exclusive expression of the less common CD33 variant that lacks exon 2. This population could also be considered a potential pool of HSCT donors in combination with the targeted CD33 immunotherapy described in this work, thereby eliminating the need for Cas9-directed ablation. However, this might limit the donor pool drastically, making the approach practically unfeasible in human studies. In this regard, Humbert et al. (22) used a CRISPR/Cas9 approach to target flanking introns using two different sgRNAs to delete exon 2. It is unclear whether selectively removing the V-domain has any advantage over disruption of the entire CD33, as we and others did not observe engraftment or functional defects in both mice and monkeys (Rhesus macaques) by removing all of CD33 (21). Additionally, the use of multiple guides multiplies potential off-targets and the efficiency of the guides will be likely limited by the least-efficient guide in the pool.
In this study, the deletion of CD33 in human CD34+ BM and CB did not result in any noticeable side effects. More than 21 wk after transplantation, NSGS mice have not presented any abnormal phenotypes. The absence of an observable CD33 deletion-linked phenotype might be explained by functional redundancy or compensation among Siglecs members.
Despite an increasing number of clinical trials involving modified immune cells, few have resulted in improved outcomes for patients, mainly because of the on-target/off-tissue toxicity on normal tissues. While CAR-T is a recent approach whose long-term effects are less well established, the use of mAbs and ADCs is routine in cancer care and generally safe. We show that combining CART cells and ADCs, such as GO, with engineered stem cells protects normal tissues from on-target/off-tissue toxicity and can lead to full remission and full hematopoietic reconstitution in an animal model. Despite proven benefits, virtually all GO-treated patients experience substantial depletion of normal myeloid lineage cells, which can lead to potentially lethal febrile neutropenias and abnormal bleeding problems due to GO-induced myelosuppression (25). These severe adverse events limit the use of GO to brief exposures during induction chemotherapy and essentially prohibit its chronic use. Our research also suggests that the combination of lower GO doses with or without CART33 infusion could be a fundamentally new approach to treat AML patients. Finally, the recent reapproval of GO, and the resurgence of current clinical trials of novel CD33-directed reagents (including new anti-CD33 CAR-T, anti-CD33 ADCs, and CD33 bispecific T cell engagers, or BiTEs) render this approach transferable to the clinic in the near future.
We have also designed an easily usable pipeline to test new potential targets that share the properties that make CD33 an attractable target: that is, a functionally redundant lineage marker that is strictly expressed by hematopoietic cells and also expressed by the cancer cells (e.g., CD123, CLL-1 or CD244). This antigen is rendered “cancer specific” by CRISPR-mediated ablation of the antigen from HSCs (17). Our strategy could also be replicated in solid tumors where a functional organoid might be generated from embryonic or induced pluripotent stem cells that are edited to ablate expression, or where the primary organ has already been removed (i.e., to target a normal prostate lineage antigen in a patient after radical prostatectomy). Finally, we propose the possibility of “epitope modification” of a tissue-specific antigen using DNA-based editing methods. “Epitope modification” might allow a protein to retain its function, but switch a small antigenic determinant. In this strategy, the stem cells retain a functional protein, but no longer possess the binding site for the immune therapy, while the cancer cells, carrying the unmodified protein, remain uniquely sensitive to immune therapies.
Materials and Methods
CRISPR/Cas9-Mediated CD33 Genomic Targeting.
The TrueCut Cas9 protein V2 was purchased from Invitrogen. The chemically modified sgRNA targeting CD33 were designed using Synthego CRISPR Gene KO design tool and purchased from Synthego. Three micrograms of TrueCut Cas9 protein and 1.5 μg sgRNA for 200,000 CD34+ cells were mixed in P3 buffer (Lonza, Amaxa P3 Primary Cell 4D-Nucleofector Kit) and incubated 10 min at 37 °C. The cells were then washed with PBS, resuspended in P3 buffer, mixed with the Cas9/sgRNA RNP complex, and then electroporated with the 4D-Nucleofector (program DZ100). After electroporation, cells were cultured at 37 °C until analysis or maintained 48–72 h in vitro and then intravenously injected (5 × 105–1 × 106 per mouse) into sublethally irradiated NSGS mice (The Jackson Laboratory).
Whole-Genome and RNA Sequencing.
After electroporation with Cas9 only or RNP complex Cas9/sgRNA, CD34+ cells were kept in vitro for 10 d and their DNA or RNA isolated as followed. DNA was purified with a QIAAmp DNA mini kit, following the manufacturer’s protocol, then eluted with 30 μL and DNA concentration measured using Nanodrop and Qubit dsDNA BR assay. RNA was purified with a miRNeasy micro kit, following the manufacturer’s protocol, then eluted with 18 μL. Nanodrop and Bioanalyzer Pico chip assays were performed to measure concentration and quality.
For whole-genome sequencing, we used an NEBNext Ultra II DNA Library Prep Kit for Illumina, clustering, and sequencing reagents. Briefly, the genomic DNA was fragmented by acoustic shearing, cleaned up, and end repaired. Adapters were ligated and DNA libraries were made. The DNA libraries were also quantified by real-time PCR (Applied Biosystems), clustered on two lanes of a flowcell, and loaded on the Illumina HiSeq instrument according to the manufacturer’s instructions. The samples were sequenced using a 2× 150 paired-end (PE) configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS) on the HiSeq instrument. DNA sequences were processed using Illumina HiSeq Analysis Software v2.1 (HAS 2.1) using default parameters.
For RNA sequencing, cDNA synthesis and amplification were performed using SMART-Seq v4 Ultra Low Input Kit for Sequencing (Clontech). The sequencing library was prepared using Nextera XT (Illumina). The samples were sequenced using a 2 × 150 PE configuration. After investigating the quality of the raw data, the trimmed reads were mapped to the Homo sapiens reference genome available on ENSEMBL using the STAR aligner v.2.5.2b. BAM files were generated as a result of this step. Unique gene hit counts were calculated by using feature Counts from the Subread package v1.5.2. Only unique reads that fell within exon regions were counted. After extraction of gene hit counts, the gene hit counts table was used for downstream differential expression analysis using the edgeR package within the SARTools package (26). Genes were considered significantly differentially expressed if the P value is <0.05.
Flow Cytometry and Sorting.
In vitro.
After transduction, CAR-T cells were expanded up to 15 d then sorted for GFP+ using the Bio-Rad S3e sorter (dead cells were excluded using propidium iodide) and comixed 1:1 for in vitro and in vivo experiments. CAR expression and their ability to recognize and bind CD33 was assessed by incubating CAR-T cells with biotinylated human CD33 protein (ACRO biosystem) for 20 min at 4 °C and then stained with fluorochrome-conjugated streptavidin.
Human CD34+ stem cells were analyzed 5–7 d after electroporation using the following antibodies from BioLegend: hCD34-PerCp/Cy5.5 and hCD33-FITC.
In vivo.
Engraftment and repopulation of the hematopoietic system over time was assessed by analysis of peripheral blood, BM aspiration, whole BM (from sacked mice) using the consequent antibodies from BioLegend or BD Biosciences: Ter119-PeCy5, Ly5/H2kD-BV711, hCD45-BV510, hCD3-Pacific Blue, hCD123-BV605, hCD33-APC, hCD14-APC/Cy7, hCD10-BUV395, hCD19-BV650, hCD34-BV421, hCD90-PeCy7, hCD38-BUV661, and hCD45RA-BUV737. CAR-T cells stably express fluorescent protein zsGreen, leukemic cells stably express dTomato, and dead cells were excluded using propidium iodide. Leukemia cells were gated on Ter119−dtomato+. CD34+-injected derived human cells were gated on Ter119−dtomato−, Ly5−/H2kd−human CD45+CART−. Myeloid lineage development was assessed by analysis of BM aspiration using the following antibodies from BioLegend or BD Biosciences: Ter119-Pecy5, hCD19-Pecy5, hCD3-Pecy5, and propidium iodide for dead cells were assigned as Dump Channel. Cells were then gated on Ly5-APC−, hCD45-PerCp/Cy5.5+. Within hCD45-PerCp/Cy5.5+ population, neutrophils were gated as SSC-Ahigh, hCD15-AF700+; monocytes were gated as HLA-DR-BV510+, hCD14-APC/Cy7+, hBDCA1/3-BV711−; cDC were gated as HLA-DR-BV510+, hCD14-APC/Cy7−, hBDCA1/3-BV711+, hCD11c-PeCy7+; pDC were gated as HLA-DR-BV510+, hCD123-BV421+, BDCA2-PeDazzle594+; mast cells were gated as HLA-DR-BV510−, hCD117-BV650+, hCD203c-BV605+; basophils were gated as HLA-DR-BV510−, hCD117-BV650−, hCD203c-BV605+. Cells from the peritoneal cavity were first incubated 10 min at room temperature with Human TruStain FcX and True-Stain Monocyte blocker (BioLegend) and then analyzed using antibodies from BioLegend: Ter119-Pecy5, hCD19-Pecy5, hCD3-Pecy5, and propidium iodide for dead cells were assigned as Dump Channel. Cells were then gated on Ly5/H2kD-BV711−, hCD45-BV510+. Then identification of phagocytic cells was determined by plotting Ecoli-Pe over hCD14-APC/Cy7 or hCD11b-Alexa488, or hCD16-PeCy7.
All data were acquired with the Bio-Rad ZE5 flow cytometry analyzer in regular or high-throughput mode and analysis was performed using FlowJo 10.4.2. Concomitantly, leukemia progression was also assessed by fluorescent imaging using the PerkinElmer IVIS Spectrum Optical Imaging System. Images were acquired and analyzed with Living Image 4.4 Optical Imaging Analysis Software.
In Vivo Experiments.
NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ (NSG-SGM3) mice (The Jackson Laboratory) were conditioned with sublethal (1.2 Gy) total-body irradiation. Human CD34+CD33Del BM or CB stem cells (5 × 105–1 × 106), along with 5 × 105 dTomato− HL-60 cells were injected intravenously into the mice within 8–24 h after total-body irradiation. One to 2 wk later, mice were treated with 2–3 × 106 anti-CD33 or control CAR-T cells (premixed CD4:CD8 = 1:1), or 6 μg of GO (Gemtuzumab Ozagamicin) or PBS intravenously injected.
All experiments were performed under protocols approved by the Institutional Animal Care and Use Committee of Columbia University.
Supplementary Material
Acknowledgments
Flow cytometry and cell-sorting experiments were performed using instrumentation maintained by the Columbia Stem Cell Initiative Flow Cytometry core facility directed by Michael Kissner. DNA and RNA from CD34+ cells were purified and quality-controlled by Molecular Pathology Shared Resource. Mice imaging was performed using instrumentation maintained by the Cancer Center Small Animal Imaging Shared Resource with NIH Grant #P30 CA013696 (National Cancer Institute). This work was supported by a grant from Vor Biopharma and PureTech Health.
Footnotes
Conflict of interest statement: This study was funded by a grant from Vor Biopharma and PureTech Health, which has launched a company called Vor Biopharma. Columbia University owns equity in Vor Biopharma and has licensed technology that is the subject of this study to Vor Biopharma. F.B., A.M.A., and S.M. are coinventors on issued and pending patent applications licensed to Vor Biopharma. S.M. has equity ownership and is on the Scientific Advisory Board of Vor Biopharma. A.R. received funding from PureTech Health.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819992116/-/DCSupplemental.
References
- 1.O’Donnell M. R., et al. , Acute myeloid leukemia, version 3.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 15, 926–957 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Rai K. R., et al. , Treatment of acute myelocytic leukemia: A study by cancer and leukemia group B. Blood 58, 1203–1212 (1981). [PubMed] [Google Scholar]
- 3.Alcantara M., Tesio M., June C. H., Houot R., CAR T-cells for T-cell malignancies: Challenges in distinguishing between therapeutic, normal, and neoplastic T-cells. Leukemia 32, 2307–2315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan R. A., et al. , Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol. Ther. 18, 843–851 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamers C. H., et al. , Treatment of metastatic renal cell carcinoma with CAIX CAR-engineered T cells: Clinical evaluation and management of on-target toxicity. Mol. Ther. 21, 904–912 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dotti G., Gottschalk S., Savoldo B., Brenner M. K., Design and development of therapies using chimeric antigen receptor-expressing T cells. Immunol. Rev. 257, 107–126 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davila M. L., et al. , Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci. Transl. Med. 6, 224ra25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalos M., et al. , T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci. Transl. Med. 3, 95ra73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porter D. L., Levine B. L., Kalos M., Bagg A., June C. H., Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 365, 725–733 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laing A. A., Harrison C. J., Gibson B. E. S., Keeshan K., Unlocking the potential of anti-CD33 therapy in adult and childhood acute myeloid leukemia. Exp. Hematol. 54, 40–50 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Kung Sutherland M. S., et al. , SGN-CD33A: A novel CD33-targeting antibody-drug conjugate using a pyrrolobenzodiazepine dimer is active in models of drug-resistant AML. Blood 122, 1455–1463 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Jurcic J. G. What happened to anti-CD33 therapy for acute myeloid leukemia? Curr. Hematol. Malig. Rep. 7, 65–73 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Kenderian S. S., et al. , CD33-specific chimeric antigen receptor T cells exhibit potent preclinical activity against human acute myeloid leukemia. Leukemia 29, 1637–1647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill S., et al. , Preclinical targeting of human acute myeloid leukemia and myeloablation using chimeric antigen receptor-modified T cells. Blood 123, 2343–2354 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brinkman-Van der Linden E. C., et al. , CD33/Siglec-3 binding specificity, expression pattern, and consequences of gene deletion in mice. Mol. Cell. Biol. 23, 4199–4206 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taussig D. C., et al. , Hematopoietic stem cells express multiple myeloid markers: Implications for the origin and targeted therapy of acute myeloid leukemia. Blood 106, 4086–4092 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haubner S., et al. , Coexpression profile of leukemic stem cell markers for combinatorial targeted therapy in AML. Leukemia 33, 64–74 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wisniewski D., Affer M., Willshire J., Clarkson B., Further phenotypic characterization of the primitive lineage- CD34+CD38-CD90+CD45RA- hematopoietic stem cell/progenitor cell sub-population isolated from cord blood, mobilized peripheral blood and patients with chronic myelogenous leukemia. Blood Cancer J. 1, e36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krupka C., et al. , CD33 target validation and sustained depletion of AML blasts in long-term cultures by the bispecific T-cell-engaging antibody AMG 330. Blood 123, 356–365 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Blaeschke F., et al. , Induction of a central memory and stem cell memory phenotype in functionally active CD4+ and CD8+ CAR T cells produced in an automated good manufacturing practice system for the treatment of CD19+ acute lymphoblastic leukemia. Cancer Immunol. Immunother. 67, 1053–1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim M. Y., et al. , Genetic inactivation of CD33 in hematopoietic stem cells to enable CAR T cell immunotherapy for acute myeloid leukemia. Cell 173, 1439–1453.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humbert O., et al. , Engineering resistance to CD33-targeted immunotherapy in normal hematopoiesis by CRISPR/Cas9-deletion of CD33 exon 2. Leukemia 33, 762–808 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Charlesworth C. T., et al. , Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 25, 249–254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crudele J. M., Chamberlain J. S., Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat. Commun. 9, 3497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amadori S., et al. , Gemtuzumab Ozogamicin versus best supportive care in older patients with newly diagnosed acute myeloid leukemia unsuitable for intensive chemotherapy: Results of the randomized phase III EORTC-GIMEMA AML-19 trial. J. Clin. Oncol. 34, 972–979 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Varet H., Brillet-Guéguen L., Coppée J. Y., Dillies M. A., SARTools: A DESeq2- and edgeR-based R pipeline for comprehensive differential analysis of RNA-seq data. PLoS One 11, e0157022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.