Significance
Epithelial cells perform critical protective, secretory, absorptive, and sensory functions, for which they require plasma membrane polarization into apical and basolateral domains. Impaired polarity causes cancer and developmental and degenerative disorders. Research on fundamental polarity mechanisms has been hindered by the paucity of model proteins and by the use of overexpression systems. Here, we introduce a high-throughput surface proteomics approach based on domain-selective biotinylation and quantitative mass spectrometry that provides candidate proteins to study polarity under normal expression levels. Using this approach, we described that clathrin adaptors mediate apical and basolateral distribution of surface proteins, expanding the traditional notion that clathrin adaptors mediate only basolateral polarity. Our results establish quantitative surface proteomics as a powerful tool to study epithelial polarity.
Keywords: epithelial polarity, apical basolateral polarity, SILAC mass spectrometry, AP-1A, AP-1B
Abstract
The current model of polarized plasma membrane protein sorting in epithelial cells has been largely generated on the basis of experiments characterizing the polarized distribution of a relatively small number of overexpressed model proteins under various experimental conditions. Thus, the possibility exists that alternative roles of various types of sorting machinery may have been underestimated or missed. Here, we utilize domain-selective surface biotinylation combined with stable isotope labeling with amino acids in cell culture (SILAC) and mass spectrometry to quantitatively define large populations of apical and basolateral surface proteins in Madin-Darby canine kidney (MDCK) cells. We identified 313 plasma membrane proteins, of which 38% were apical, 51% were basolateral, and 11% were nonpolar. Silencing of clathrin adaptor proteins (AP) AP-1A, AP-1B, or both caused redistribution of basolateral proteins as expected but also, of a large population of apical proteins. Consistent with their previously reported ability to compensate for one another, the strongest loss of polarity was observed when we silenced AP-1A and AP-1B simultaneously. We found stronger evidence of compensation in the apical pathway compared with the basolateral pathway. Surprisingly, we also found subgroups of proteins that were affected after silencing just one adaptor, indicating previously unrecognized independent roles for AP-1A and AP-1B. While AP-1B silencing mainly affected basolateral polarity, AP-1A silencing seemed to cause comparable loss of apical and basolateral polarity. Our results uncover previously overlooked roles of AP-1 in polarized distribution of apical and basolateral proteins and introduce surface proteomics as a method to examine mechanisms of polarization with a depth not possible until now.
Epithelial cells perform key vectorial functions in secretion and absorption that depend on the accurate localization of transporters, channels, and receptors to apical and basolateral surface domains. Polarized sorting of plasma membrane proteins is believed to occur via recognition of apical and basolateral signals by the intracellular sorting machinery (1, 2). The current model postulates that apical and basolateral sorting is mediated by separate machineries. While apical sorting signals are poorly understood, they seem to involve glycans and other determinants that engage lectins, lipid rafts, and cytoplasmic motors (3). On the other hand, basolateral sorting is believed to be mediated by discrete tyrosine or dileucine motifs, which engage clathrin (4) and the clathrin adaptor protein 1 (AP-1). AP-1 is a heterotetramer of different β-, γ-, μ-, and σ-subunits. The μ-subunit has two isoforms (μ1A and μ1B), which define the tissue-specific expression of the AP-1 complex, being AP-1A ubiquitous and AP-1B specific of epithelia (5).
Our understanding of the current epithelial sorting model and the role of AP-1 in polarized protein distribution in epithelial cells still has many gaps. (i) Some epithelia lack μ1B, including liver (5), retinal pigment epithelium (6), kidney proximal tubule (7), and choroid plexus (8), and they still manage to direct proteins to the basolateral domain. (ii) Do AP-1A and AP-1B complement each other or have independent roles? Studies in Madin-Darby canine kidney (MDCK) cells show that, while loss of μ1B decreases polarization of several basolateral proteins (9–13), the combined silencing of μ1A plus μ1B has a more dramatic effect (14–16), indicating some degree of compensation. However, to our knowledge, a totally independent role of AP-1A in basolateral protein distribution has not been demonstrated. (iii) Do apical proteins also rely on AP-1 for polarized trafficking? In recent years, the role of AP-1 in determining the localization of basolateral proteins exclusively has been expanded to also include apical proteins in Caenorhabditis elegans (17, 18) and mice (19) and glycosylphosphatidylinositol (GPI)-anchored proteins in MDCK cells (20).
Many important aspects of the polarity mechanisms in epithelial cells remain unsolved, because conclusions have been drawn largely by observing the behavior of a very small set of representative proteins, which are often overexpressed via transfection. This approach poses the risk of overlooking the behavior of a less well-characterized repertoire of endogenous proteins under more physiological conditions. Mass spectrometry-based proteomics techniques have the potential of overcoming these limitations. The surface proteome of MDCK cells may encompass hundreds of proteins (21). Hence, a quantitative global surface proteomics approach would be highly advantageous to accurately measure the polarized distribution of surface proteins and its changes in response to deletion of potential components of the polarity machinery. Here, we applied domain-selective biotinylation of MDCK cells, a technique originally introduced by our laboratory (22), in combination with stable isotope labeling with amino acids in cell culture (SILAC) (23) and mass spectrometry to quantitatively define apical and basolateral plasma membrane protein populations in MDCK cells. This approach was recently performed successfully in nonpolarized cells to characterize the role of the retromer in plasma membrane protein recycling (24) and to define a newly identified retriever complex (25). Adapting this approach to polarized epithelial cells, here we identified previously overlooked roles of AP-1A and AP-1B in the polarized distribution of apical and basolateral proteins.
Results
Polarized Surface Proteome of MDCK Cells.
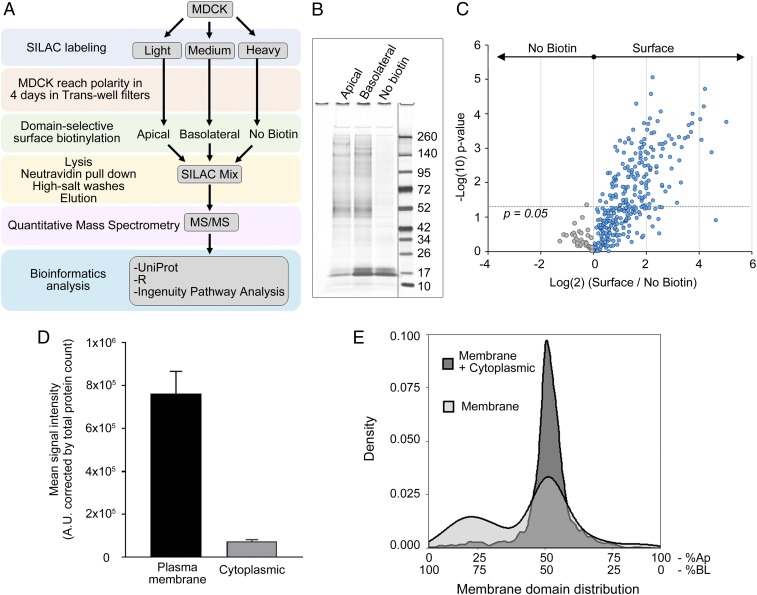
To study the polarized surface proteome of MDCK cells, we followed the approach depicted in Fig. 1A. We cultured MDCK cells in SILAC medium and allowed them to polarize for 4 d. Then, we performed surface biotinylation on the apical or basolateral surface domains and analyzed the surface samples by mass spectrometry. We included a control sample without biotinylation to account for the background signal (Fig. 1B). We detected the majority of the signal at the surface protein fraction (Fig. 1 B and C), indicating very little background. We identified a total of 1,737 proteins in three or more replicates in six separate experiments. Of these proteins, 313 were transmembrane, GPI-anchored, or peripheral extracellular proteins that had a plasma membrane annotation in the UniProt knowledgebase; the rest (∼80% of the proteins) were annotated as cytoplasmic. However, despite the higher abundance of cytoplasmic proteins, quantitative analysis showed that they contributed less than 10% of the total peptide signal in the mass spectrometry (Fig. 1D). It is reasonable to expect a certain degree of intracellular biotinylation due to endogenous biotinylation and to biotin leakage through the plasma membrane combined with the higher abundance of cytoplasmic proteins. Any nonspecific biotinylation is expected to occur indiscriminately from the apical or basolateral domain. Therefore, when we analyzed the percentage frequency (density) distribution of proteins annotated as cytoplasmic or plasma membrane, we observed that most of the signal from cytoplasmic proteins is not polarized (Fig. 1E), ruling out any bias that may be associated with residual nonspecific biotinylation. Altogether, these results highlight the suitability of the surface biotinylation method.
Fig. 1.
Polarized surface proteome in MDCK cells via surface biotinylation followed by SILAC. (A) Flow chart illustrating the procedure for obtaining a polarized surface proteome in MDCK cells. (B) Representative silver-stained gel showing apical and basolateral proteins retrieved after surface biotinylation and background from nonbiotinylated samples. Dividing line indicates noncontiguous gel lanes. (C) SILAC ratios of surface vs. nonbiotinylated proteins. Each data point represents a single protein. Gray datapoints represent proteins from the no biotin sample. Blue datapoints are proteins from the biotinylated sample. Dotted line indicates the −log(10) of a P value = 0.05. (D) Quantification of signal obtained from proteins annotated as plasma membrane or cytoplasmic proteins corrected by total number of proteins in each fraction. (E) Density plot showing percentage frequency of proteins annotated as plasma membrane and cytoplasmic (dark gray) and membrane proteins only (light gray) according to their polarized distribution at the apical and basolateral membrane domains. Ap, apical; BL, basolateral.
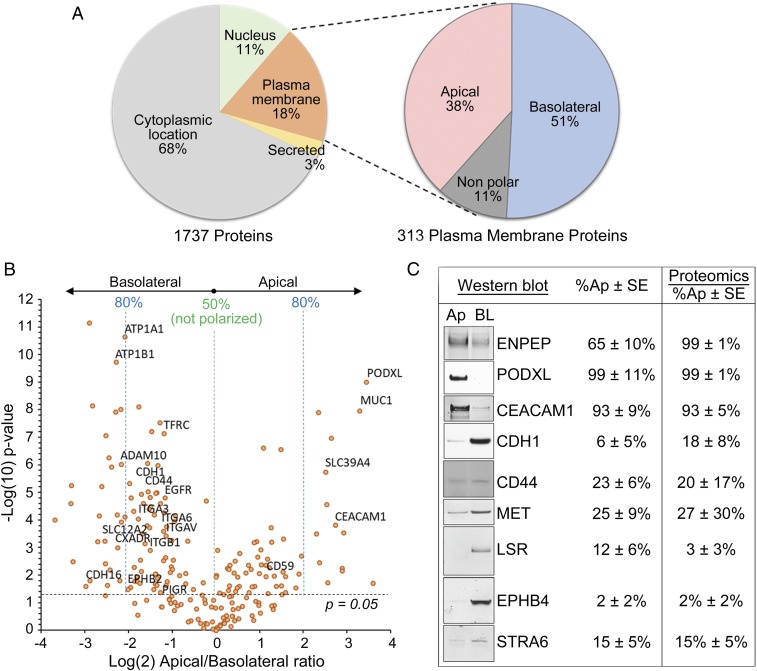
Because we wished to study the surface proteome of MDCK cells, we focused our analysis on plasma membrane proteins only. The quantitative nature of our approach allowed us to determine the relative proportion of each protein at the apical or basolateral domain. Fig. 2A shows the location annotation for all 1,737 proteins detected in MDCK cells (Fig. 2A, Left) and the polarized surface distribution of the subset of 313 proteins annotated as plasma membrane proteins (Fig. 2A, Right). Among these, 38% were apical, 51% were basolateral, and 11% were nonpolar, defined as being enriched by more than 60% in the respective membrane domain. The apical/basolateral ratio is easily visualized in the volcano plot in Fig. 2B, where we indicated the relative enrichment in each membrane domain and highlighted proteins of previously known distribution. Our proteomics data are in good agreement with the published literature. To further validate our data, we studied a subset of surface proteins by Western blot (Fig. 2C), and we found their polarity assessed with this procedure to be consistent with the proteomics results.
Fig. 2.
Apical and basolateral proteome of MDCK. (A) Subcellular localization of all 1,737 proteins identified by mass spectrometry (Left), of which 313 were plasma membrane proteins and were classified as apical, basolateral, or not polarized based on the SILAC ratios (Right). (B) Volcano plot of log(2) SILAC ratios for apical vs. basolateral proteins in MDCK cells (horizontal axis) against −log(10) P value (vertical axis). Each data point represents a single protein. Dotted line indicates the −log(10) of a P value = 0.05. Labels indicate proteins of previously known polarized distribution. (C) Validation of a subset of proteins via surface biotinylation followed by Western blot (Left) compared with the percentage detected at the apical and basolateral membranes in proteomics (Right). Ap, apical; BL, basolateral.
Functional classification of the surface proteins identified by global proteomics showed distinct groups at the apical and basolateral membranes (Table 1). Certain proteins, like adhesion molecules, tight junction proteins, and membrane-bound proteases, were enriched at the basolateral domain, while GPI-anchored proteins and proteins involved in purine metabolism among others were enriched apically. Surface receptors, ion channels, and transporters populate both membranes in comparable proportions (a complete list is in Dataset S1).
Table 1.
Apical and basolateral distribution of surface proteins in MDCK
| Name | Gene identification | Peptides | Sequence coverage (%) | n/6 | % Apical | % Basolateral | P value |
| Basal cell adhesion molecule | BCAM | 28 | 62.9 | 6 | 11 | 89 | 1.16293E-08 |
| Plexin A1 | PLXNA1 | 24 | 9.9 | 6 | 15 | 85 | 4.61961E-06 |
| Plexin B1 | PLXNB1 | 27 | 17.4 | 4 | 12 | 88 | 0.023047413 |
| Semaphorin 4D | SEMA4D | 19 | 7.9 | 4 | 24 | 76 | 0.149175744 |
| Integrin subunit-α6 | ITGA6 | 48 | 47.8 | 6 | 22 | 78 | 0.000253324 |
| Integrin subunit-β1 | ITGB1 | 28 | 42.2 | 6 | 24 | 76 | 0.001229654 |
| Integrin subunit-αV | ITGAV | 61 | 58 | 6 | 25 | 75 | 0.000361996 |
| Integrin subunit-α2 | ITGA2 | 42 | 43.9 | 6 | 29 | 71 | 0.022472422 |
| Insulin-like growth factor 1 receptor | IGF1R | 42 | 37.8 | 6 | 40 | 60 | 0.163886842 |
| Intercellular adhesion molecule 1 | ICAM1 | 9 | 19.5 | 5 | 96 | 4 | 0.017956289 |
| Piezo-type mechanosensitive ion channel component 1 | PIEZO1 | 19 | 8.2 | 5 | 18 | 82 | 0.006293186 |
| Transmembrane and coiled-coil domains 1 | TMCO1 | 2 | 12.2 | 4 | 74 | 26 | 0.170385728 |
| Chloride intracellular channel 1 | CLIC1 | 5 | 27.4 | 6 | 89 | 11 | 0.234170658 |
| Solute carrier family 9 member A1 | SLC9A1 | 9 | 8.7 | 4 | 5 | 95 | 0.016457311 |
| Anoctamin 6 | ANO6 | 25 | 26.6 | 6 | 16 | 84 | 4.74988E-05 |
| Leucine-rich repeat containing 8 VRAC subunit A | LRRC8A | 11 | 12.5 | 4 | 29 | 71 | 0.139840212 |
| Cell migration-inducing hyaluronidase 2 | TMEM2 | 8 | 5.1 | 6 | 27 | 73 | 0.004854648 |
| Pyrophosphatase (inorganic) 1 | PPA1 | 5 | 33.1 | 6 | 86 | 14 | 0.005309926 |
| 2′,3′-Cyclic nucleotide 3′ phosphodiesterase | CNP | 20 | 46.4 | 6 | 87 | 13 | 0.005965224 |
| Prenylcysteine oxidase 1 | PCYOX1 | 8 | 6.5 | 4 | 100 | 0 | 0.01011956 |
| Endonuclease domain containing 1 | ENDOD1 | 7 | 33.7 | 6 | 83 | 17 | 0.004053892 |
| Lamin B receptor | LBR | 13 | 29.1 | 6 | 71 | 29 | 0.096990048 |
| Cadherin 16 | CDH16 | 26 | 46.7 | 4 | 8 | 92 | 0.024358355 |
| Ectonucleoside triphosphate diphosphohydrolase 3 | ENTPD3 | 17 | 34 | 6 | 72 | 28 | 2.3237E-07 |
| Paraoxonase 2 | PON2 | 4 | 18.1 | 6 | 77 | 23 | 0.001226897 |
| Tripartite motif-containing 25 | TRIM25 | 14 | 27.9 | 6 | 70 | 30 | 0.004216133 |
| Lipolysis stimulated lipoprotein receptor | LSR | 20 | 29.3 | 6 | 3 | 97 | 5.49804E-06 |
| Signal transducer and activator of transcription 3 | STAT3 | 18 | 35.3 | 5 | 42 | 58 | 0.169979355 |
| Protein tyrosine phosphatase, receptor type M | PTPRM | 26 | 28.3 | 4 | 22 | 78 | 0.078938671 |
| Protein tyrosine phosphatase, receptor type F | PTPRF | 79 | 64.5 | 6 | 28 | 72 | 6.96458E-05 |
| Calpain 2 | CAPN2 | 40 | 64.9 | 6 | 64 | 36 | 0.134192804 |
| Transmembrane serine protease 4 | TMPRSS4 | 6 | 18.6 | 6 | 13 | 87 | 1.52653E-05 |
| ADAM metallopeptidase domain 17 | ADAM17 | 13 | 21.9 | 6 | 7 | 93 | 9.63391E-05 |
| ADAM metallopeptidase domain 10 | ADAM10 | 20 | 36.7 | 6 | 13 | 87 | 9.53067E-07 |
| Suppression of tumorigenicity 14 | ST14 | 7 | 11 | 5 | 23 | 77 | 0.026355564 |
| Carboxypeptidase M | Cpm | 5 | 12.2 | 5 | 74 | 26 | 0.024747236 |
| Glutamyl aminopeptidase | ENPEP | 30 | 22.6 | 6 | 100 | 0 | 2.70449E-05 |
| Xenotropic and polytropic retrovirus receptor 1 | XPR1 | 7 | 11.2 | 6 | 14 | 86 | 8.78517E-05 |
| Cadherin EGF LAG seven-pass G-type receptor 2 | CELSR2 | 25 | 12.3 | 6 | 19 | 81 | 0.005019663 |
| Adhesion G protein-coupled receptor G1 | ADGRG1 | 4 | 2.5 | 4 | 24 | 76 | 0.13836571 |
| Frizzled class receptor 6 | FZD6 | 8 | 8.6 | 5 | 36 | 64 | 0.128016301 |
| G protein-coupled receptor class C group 5 member A | GPRC5A | 4 | 10.1 | 6 | 87 | 13 | 0.000616634 |
| ATP binding cassette subfamily E member 1 | ABCE1 | 29 | 52 | 6 | 61 | 39 | 0.027707251 |
| ATP binding cassette subfamily F member 1 | ABCF1 | 12 | 21.2 | 6 | 69 | 31 | 0.080170041 |
| Solute carrier family 35 member A1 | SLC35A1 | 4 | 10.7 | 6 | 82 | 18 | 0.02315915 |
| Solute carrier family 44 member 2 | SLC44A2 | 20 | 28.6 | 6 | 8 | 92 | 5.95465E-06 |
| Solute carrier family 39 member 10 | SLC39A10 | 4 | 9.9 | 6 | 26 | 74 | 2.85037E-08 |
| Solute carrier family 4 member 2 | SLC4A2 | 37 | 11.2 | 5 | 8 | 92 | 0.002769117 |
| ATPase Na+/K+-transporting subunit β1 | ATP1B1 | 19 | 46.9 | 6 | 12 | 88 | 1.82896E-10 |
| ATPase Na+/K+-transporting subunit-α1 | ATP1A1 | 71 | 56.7 | 6 | 16 | 84 | 2.21093E-11 |
| Scavenger receptor class B member 1 | SCARB1 | 11 | 22.2 | 6 | 25 | 75 | 0.076726321 |
| ATPase Na+/K+-transporting subunit-β3 | ATP1B3 | 7 | 53.8 | 4 | 25 | 75 | 0.102593542 |
| Hephaestin | HEPH | 23 | 25 | 6 | 26 | 74 | 0.019696101 |
| Polymeric Ig receptor | PIGR | 10 | 18.8 | 6 | 27 | 73 | 0.041687344 |
| Transferrin receptor | TFRC | 32 | 45.3 | 6 | 27 | 73 | 7.07717E-08 |
| Folate receptor-β | FOLR2 | 4 | 23.5 | 6 | 86 | 14 | 0.000183835 |
| Carcinoembryonic antigen-related cell adhesion molecule 1 | CEACAM1 | 16 | 29 | 6 | 93 | 7 | 0.000146391 |
| EPH receptor B4 | EPHB4 | 16 | 22.4 | 6 | 1 | 99 | 0.000591366 |
| EPH receptor A1 | EPHA1 | 17 | 25.3 | 5 | 8 | 92 | 0.003683916 |
| Erb-b2 receptor tyrosine kinase 2 | ERBB2 | 38 | 41.2 | 6 | 13 | 87 | 2.46446E-05 |
| Epidermal growth factor receptor | EGFR | 29 | 31.9 | 6 | 18 | 82 | 2.48095E-05 |
| MET protooncogene, receptor tyrosine kinase | MET | 38 | 36.8 | 6 | 27 | 73 | 0.111350932 |
| Ephrin B1 | EFNB1 | 7 | 37.2 | 6 | 28 | 72 | 0.012405291 |
| Podocalyxin | PODXL | 13 | 24.7 | 6 | 99 | 1 | 9.85873E-10 |
The complete table is in Dataset S1.
Altogether, these results indicate that the combination of domain-specific surface biotinylation and quantitative SILAC-based proteomics provides a robust assessment of the steady-state global surface proteome in polarized MDCK cells.
AP-1A and AP-1B Maintain the Polarized Proteome of MDCK Cells.
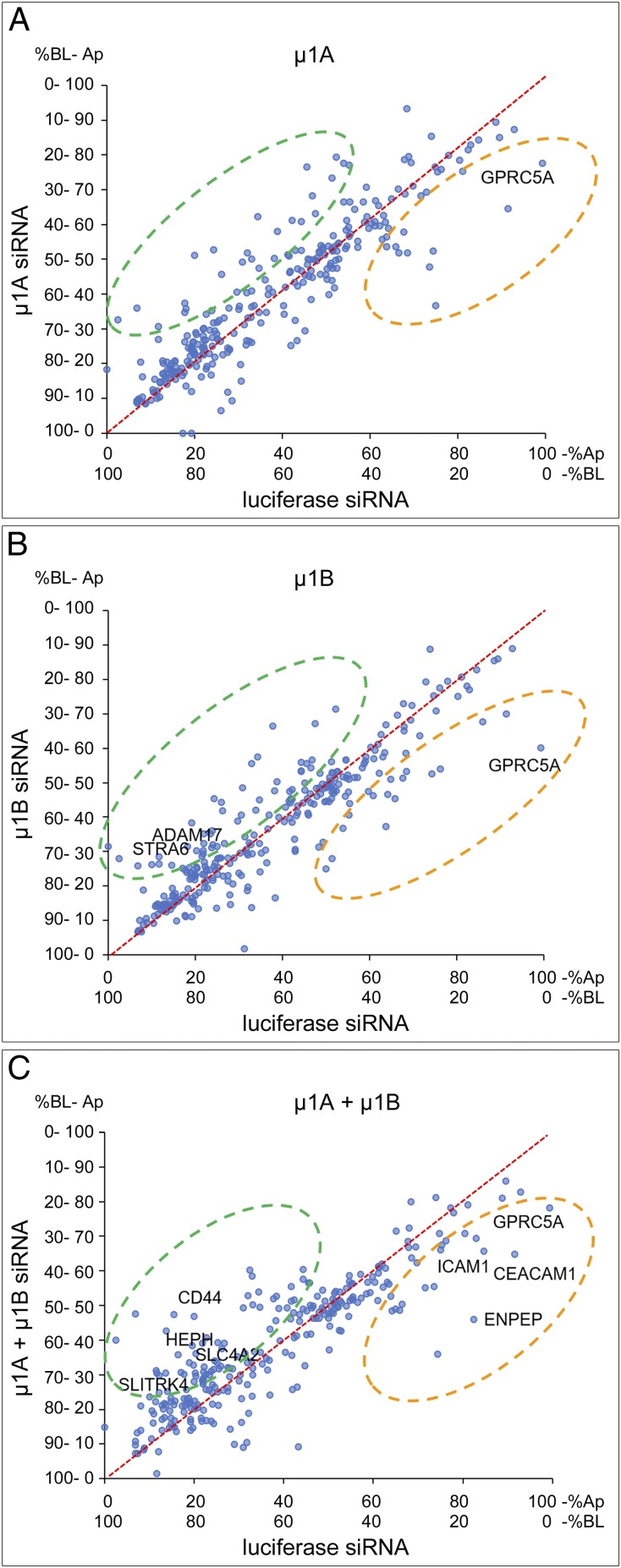
To study how AP-1A and AP-1B contribute to generating the polarized surface proteome of MDCK cells, we silenced μ1A and μ1B independently or simultaneously using small interfering RNA (siRNA) and measured the resulting changes in distribution of apical and basolateral plasma membrane proteins. To rule out artifacts due to biotin leakage through compromised tight junctions during gene silencing, we measured transepithelial resistance as indicative of functional tight junctions. In agreement with a previous report showing selective apical and basolateral surface biotinylation after AP-1 silencing (15), we did not detect any statistical difference in transepithelial resistance among silencing treatments (luciferase siRNA = 238 ± 5 Ω⋅cm2; μ1A-siRNA = 240 ± 3 Ω⋅cm2; μ1B-siRNA = 232 ± 4 Ω⋅cm2; μ1A + μ1B-siRNA = 231 ± 8 Ω⋅cm2; ANOVA: P = 0.1805), indicating that AP-1 silencing does not compromise the epithelial monolayer. Plots in Fig. 3 represent the relative polarity of all detected plasma membrane proteins in control MDCK cells (luciferase siRNA) compared with each of the silencing conditions (siRNA for μ1A, μ1B, or both). We performed a linear regression analysis to detect changes in protein polarity as deviations from the straight line that represents no change. In all cases, we observed basolateral proteins with decreased polarity as expected, but surprisingly, we also observed apical proteins with decreased polarity, suggesting that AP-1 may also determine apical protein localization. In addition, a large number of surface proteins did not have their polarity affected, indicating that a separate mechanism, mediated by a different adaptor or a clathrin-independent polarization mechanism, operates in epithelia.
Fig. 3.
Silencing of the μ1A or μ1B subunits of the AP-1 complex in MDCK cells produces shifts in the polarity of surface proteins. Linear regressions obtained by comparing the apical distribution of control proteins (luciferase siRNA) with (A) μ1A siRNA, (B) μ1B siRNA, and (C) double silencing of μ1A and μ1B simultaneously (μ1A + μ1B). The fitted line indicating the region of no change is represented in red and crosses the origin at x = 0 and y = 0. Protein shifts resulting from loss of apical polarity fall in the quadrant indicated in orange. Protein shifts resulting from loss of basolateral polarity fall in the quadrant indicated in green. A select set of proteins (indicated by name) was identified to verify the results by surface biotinylation and Western blot after silencing of μ1A, μ1B, or both in MDCK cells. Ap, apical; BL, basolateral.
From these data, we conclude that there is a previously underestimated role of AP-1 in apical protein localization in MDCK cells and that a nonidentified trafficking mechanism independent of AP-1 may operate in apical-basolateral distribution of plasma membrane proteins.
Endogenous Markers to Study AP-1A– and AP-1B–Mediated Polarized Protein Distribution in MDCK Cells.
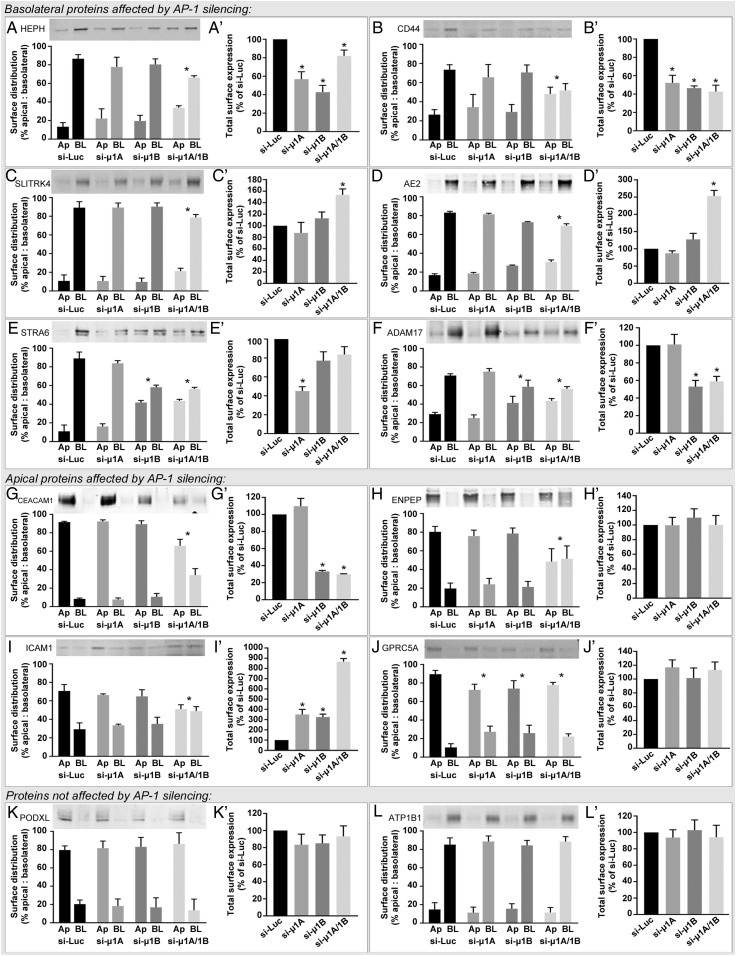
To confirm the roles of AP-1A and AP-1B in polarized protein distribution that we observed by proteomics, we silenced μ1A and μ1B and measured the polarized localization of a select group of endogenous proteins by surface biotinylation and Western blot (Fig. 4). We found a high degree of agreement between our proteomics data and the subset assessed by Western blot. We observed that some basolateral proteins, including Hephaestin (HEPH), CD44, SLIT and NTRK Like Family Member 4 (SLITRK4), and Anion Exchanger 2 (AE2), required the presence of both adaptors, since basolateral polarity was decreased only after simultaneous silencing of μ1A and μ1B (Fig. 4 A–D). Some basolateral proteins, like the Retinol-Binding Protein Receptor STRA6, and the membrane protease ADAM17 were only dependent on AP-1B (Fig. 4 E and F). Conversely, apical proteins, like gp114 (CEACAM1), the aminopeptidase ENPEP, the intercellular adhesion molecule 1 (ICAM1), and the G protein-coupled receptor C5A (GPRC5A) also required AP-1 for apical localization (Fig. 4 G–J). In this last set, CEACAM1, ENPEP, and ICAM1 had reduced apical polarity after simultaneous silencing of μ1A and μ1B. However, individual silencing of either μ1A or μ1B was enough to decrease apical polarity of GPRC5A. This suggests that, while AP-1A and AP-1B are individually sufficient for apical localization of GPRC5A, they require the presence of each other to determine the distribution of other apical proteins, like CEACAM1, ENPEP, and ICAM1. Finally, two other proteins that we tested, the apical Podocalyxin (PODXL) and the basolateral β1-subunit of the Na/K ATPase (ATP1B1), had no major change in their polarized distribution after silencing either of the adaptors alone or together (Fig. 4 K and L) (similar to the proteomics results), indicating that some apical and basolateral proteins rely on an AP-1–independent mechanism(s) for polarized localization.
Fig. 4.
Selected apical and basolateral proteins affected by silencing the μ1A or μ1B subunits of the AP-1 complex in MDCK cells as measured by surface biotinylation followed by Western blot. The examples provided illustrate basolateral proteins that had reduced polarity after silencing AP-1 subunits (A–F), apical proteins that had reduced polarity after silencing μ1A and μ1B simultaneously (G–I) or individually (J), and an apical (K) and a basolateral protein (L) that were not affected by silencing of μ1A or μ1B. For each protein, we calculated the total surface levels as indicated in A′–L′. Bars represent mean percentage surface distribution ± SD. n = 3–4. Ap, apical; BL, basolateral. *P < 0.05 vs. control (si-Luc) one-way ANOVA with Dunnett’s correction for multiple comparisons against control.
Altogether, we report here a set of endogenous basolateral and apical proteins previously unknown to be dependent on AP-1 for apical-basolateral distribution and describe various polarity patterns that AP-1 supports. The reporter proteins that we identified will be useful to study not only roles of AP-1 in basolateral polarity but also, the role of AP-1 in apical polarized trafficking that we describe here. As we also uncovered reporter proteins that depend differently for their polarized distribution from individual adaptors or both, it will be possible in the future to dissect more precisely the specific functions of AP-1A and AP-1B. Last but not least, using endogenous proteins to study AP-1 function has the appeal of eliminating the confounding effects resulting from overexpression of exogenous proteins.
Independent and Compensatory Roles of AP-1A and AP-1B in Polarized Distribution Analyzed by Empirical Bayes Method.
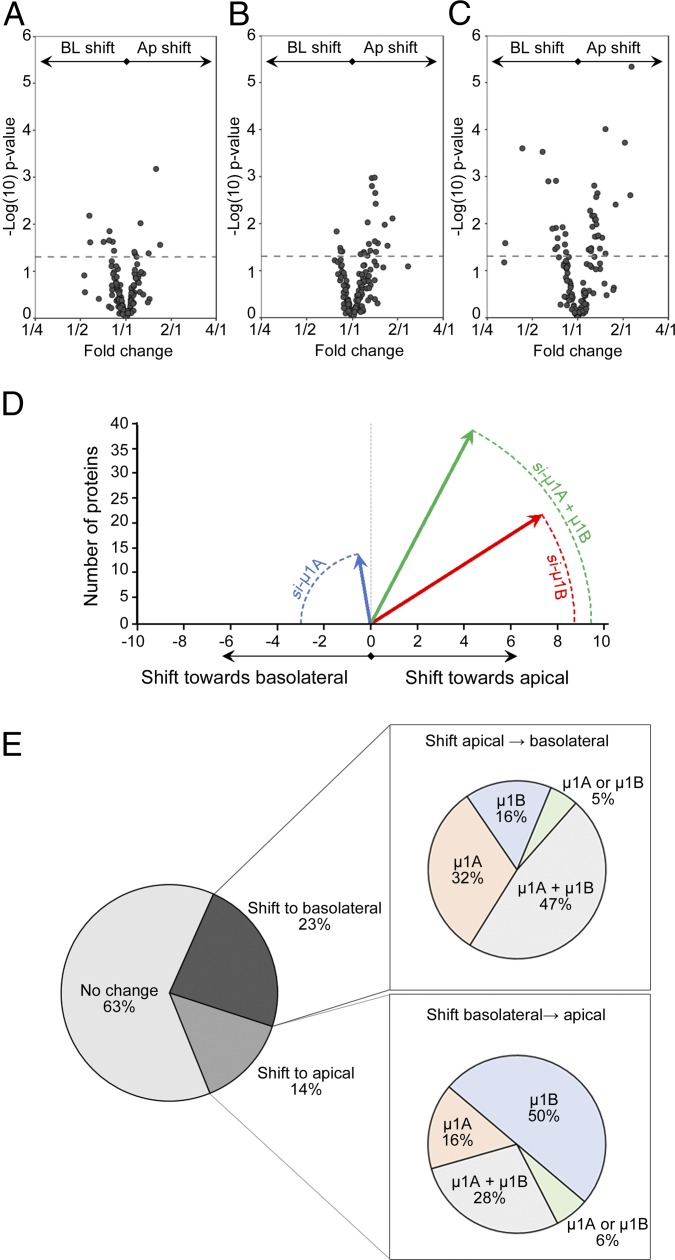
Our data so far suggest that AP-1A and AP-1B may have independent roles in protein polarity, and they also compensate for each other in some cases. The compensatory mechanism is evident during simultaneous silencing of μ1A and μ1B, which had a larger effect than each individual silencing. The proteomics approach that we introduce here provides an excellent tool to analyze the global effects of silencing a component of the polarization machinery. Accordingly, statistical analysis was conducted through the limma package in R, wherein an empirical Bayes procedure was implemented to shrink protein sample variances toward mean protein sample variance (26, 27). Thereafter, differential protein expression was determined through moderated t tests between experimental conditions. We applied this kind of analysis to detect global patterns when silencing μ1A and μ1B in MDCK cells. To further reduce variability, we also minimized potential batch effects by performing a set of triplicate experiments in parallel, thus minimizing variation associated with sample processing. Fig. 5 summarizes the magnitude of changes toward the apical or basolateral directions when we silenced μ1A or μ1B. While similar numbers of proteins experienced an apical or basolateral shift during μ1A silencing (Fig. 5A), more proteins shifted toward the apical membrane when we silenced μ1B (Fig. 5B). These shifts in both directions involved more proteins when we silenced μ1A and μ1B simultaneously (Fig. 5C). To assess the magnitude of changes in each direction, we constructed vectors to quantify the global effect of silencing by compounding the count, the direction, and the degree of the shift for all proteins affected beyond the significance threshold computed by the empirical Bayes approach. The vectors obtained in this way are summarized in Fig. 5D. We observed that μ1A silencing had the smaller effect in magnitude, but the shift leaned toward the basolateral direction. Silencing μ1B had the largest effect but in the opposite direction toward the apical membrane. Simultaneous silencing of μ1A and μ1B had the largest effect and still leaned toward an apical shift, most likely due to the relatively larger contribution of μ1B. This analysis indicates that AP-1A and AP-1B may have independent roles. Since this was previously unrecognized, we analyzed directly this possibility by comparing the effects of silencing μ1A and μ1B on the polarized distribution of all proteins detected. We observed a significant bias toward the basolateral direction after silencing μ1A and toward the apical direction when silencing μ1B (μ1A: 43.8% toward apical, 56.2% toward basolateral; μ1B: 59.9% toward apical, 40.1% toward basolateral; P < 0.05, Fisher’s exact test). Altogether, these data indicate that μ1A and μ1B are independent and that μ1A had a predominant influence in maintaining polarity of both apical and basolateral proteins, while μ1B predominated in maintenance of basolateral polarity.
Fig. 5.
Global patterns of major shifts in polarity after silencing AP-1A, AP-1B, or both. Volcano plots showing the magnitude of (A) the shifts toward the apical and basolateral sides when silencing μ1A, (B) the shifts when silencing μ1B, and (C) the shifts with the double silencing. Dotted lines represent P = 0.05. Data points above the line are statistically significant. (D) Quantification of the magnitude of shifts toward the apical (positive values) or basolateral (negative values) domains after silencing μ1A (blue), μ1B, (red), or both (green). The lengths of the vectors (radii) are directly proportional to the compounded contribution of the number of proteins affected and the intensity of the shift. (E) Percentages of proteins not affected by silencing of AP-1 subunits or affected by experiencing a shift toward the basolateral or apical sides. Subcharts indicate the proportions of proteins that experienced a shift toward the apical or basolateral side after silencing of the μ-subunits as indicated. Ap, apical; BL, basolateral.
These separate roles of AP-1A and AP-1B are evident when looking at the proportions of proteins affected by the single and double silencing (Fig. 5E). Of all of the proteins that experienced a shift in the apical-to-basolateral direction, the largest number of proteins was included in the double-silencing group. Individual μ1A or μ1B silencing affected a smaller proportion of proteins, but the predominant shift was due to silencing of μ1A. In contrast, among the proteins that experienced a shift in the basolateral-to-apical direction, the largest group was due to silencing of μ1B.
Altogether, our analysis indicates that μ1B silencing favored shifts in the basolateral-to-apical direction, while μ1A silencing had an independent effect that was comparable in both directions. This may indicate physiological differences between the two adaptors. While AP-1B function may be more relevant to basolateral polarity, AP-1A provides a mechanism for some proteins to achieve apical distribution. In certain cases, however, the two adaptors seem to cooperate in the polarized distribution of specific proteins.
Discussion
With the advent of high-throughput mass spectrometry-based techniques, the study of membrane surface proteins has advanced toward discovery approaches and a more comprehensive characterization of surface proteomes (28). However, the study of surface proteins presents unique challenges, such as their relative low abundance compared with intracellular proteins, solubility issues, and contamination with intracellular proteins. These limitations can be minimized to some extent by enriching the samples via membrane fractionation or biochemical methods as discussed elsewhere (29). In this work, we used a domain-specific surface biotinylation approach followed by quantitative mass spectrometry to characterize the polarized surface proteome of epithelial cells. These techniques are readily available in the toolkit of most laboratories, making it attractive for routine studies. However, it is important to pinpoint the limitations of our approach. One of them is inherent to mass spectrometry, which fails to detect proteins that might be underrepresented in the total protein pool. An example of this may be the LDL receptor, previously used as a model protein in epithelial polarity via overexpression methods (9, 12, 15). However, it lies below the limit of detection in our dataset of endogenous MDCK proteins. The N-hydroxysulfosuccinimide (NHS)-SS-Biotin reagent requires access to primary amines, therefore underestimating proteins with few or no exposed extracellular lysines or arginines. Alternative techniques used in proteomics target glycosylated proteins either by aminooxybiotinylation (30) or by cell surface capturing technique (31). These methods apply to the detection of glycosylated proteins and may be advantageous in minimizing intracellular labeling. However, the membrane-impermeant NHS-SS-Biotin reagent that we used here has been previously shown to have negligible intracellular labeling (32). In this work, we show that the signal ratio from plasma membrane/cytoplasmic proteins with NHS-SS-Biotin is at least 10:1 (Fig. 1), which indicates that intracellular biotinylation may not be a great concern. We propose that future applications taking advantage of combinations of surface-labeling methods as performed in nonpolarized cells (33) will expand the repertoire of surface proteins detected in epithelia.
To minimize confounding signals from cytoplasmic proteins, we considered only proteins to which a plasma membrane annotation could be assigned based on inclusion criteria from the UniProt knowledgebase. It is possible that, in so doing, we underestimated the number of plasma membrane proteins due to incomplete annotations, as the database is constantly updated based on new data. We expect this to improve as progress is made in efforts toward more complete “surfaceomes” (34, 35).
To our knowledge, few attempts have been made to apply proteomics to study polarity in MDCK cells, mainly focused on secreted (36) and GPI-anchored proteins (37). A previous work by Mathias et al. (21) used an approach consisting of isolation of apical and basolateral membranes followed by label-free mass spectrometry. This technique highly enriches membrane proteins, but intracellular proteins may still be overrepresented, as many of the highest-abundance proteins detected are cytoplasmic. Our approach selectively labels surface proteins and in addition, is quantitative, since we used SILAC to measure the relative distribution of each protein at the apical and basolateral membranes. This allowed us to determine the polarity of proteins even if there is a minimal presence in the opposite membrane. This is advantageous when performing measurements of polarity after different treatments or gene silencing, like we conducted here.
To study the role of AP-1A and AP-1B in polarity, we silenced μ1A and μ1B in MDCK cells, because there are several open questions regarding these adaptors. The general notion is that these two adaptors complement each other (15, 16). We found evidence of this cooperation, since many proteins required double silencing of μ1A and μ1B to have their polarity affected. This effect was more pronounced for a larger set of proteins that experienced a shift in the apical-to-basolateral direction, suggesting higher overlapping function in the apical pathway. This is intriguing, because the cooperation between AP-1A and AP-1B has only been recognized before in the basolateral pathway, although it is still debated whether they share the same subcellular location (16) or they act at sequential trafficking compartments (15). However, while any one of these scenarios entails overlap between AP-1A and AP-1B function, our current data indicate that each adaptor also has an independent trafficking role. We draw this conclusion from subsets of apical and basolateral proteins that had their polarity affected by silencing only one of the adaptors but not the other one and vice versa. Future research should address the complex trafficking behavior of AP-1 variants. An important point in our study is that we identified endogenous cargo proteins that showed altered polarity when AP-1 subunits are silenced. These cargo proteins expand the repertoire of model proteins to study the role of AP-1 in polarized trafficking and are expressed at physiological levels, thus minimizing confounding effects associated with overexpression (38, 39). In addition, because not all plasma membrane proteins were depolarized by AP-1 silencing, which is consistent with previous reports (15, 20), the decreased polarity of apical or basolateral proteins is likely not the result of a global indirect effect on epithelial polarity.
The role of the AP-1 adaptor as mediator of basolateral polarity has been extensively studied and involves recognition of well-defined sorting signals in basolateral cargo proteins (9–15). However, previous evidence indicates that AP-1 may also play a role in the distribution of apical proteins. For example, in μ1B knockout mice, the apical localization of sucrase and villin was disrupted in intestinal cells (19). More recently, the apical localization of GPI-anchored proteins has been shown to depend on AP-1 in MDCK cells (20). Here, we show that AP-1 also mediates the apical localization of a considerable fraction of transmembrane proteins. It is possible that this role of AP-1 has been previously overlooked, because the number of apical proteins studied in the context of AP-1 function so far has been relatively limited [i.e., p75, FcL receptor (9), TLR-2 (40), and gp135 (15)] and because apical sorting signals are less well defined than basolateral signals (3). Further characterization of apical signals is crucial to elucidating the trafficking mechanism mediated by AP-1. However, this is likely a complex issue, as this mechanism may not necessarily involve direct cargo–AP-1 interactions as described for basolateral proteins (13–16). For example, GPI-anchored proteins rely on AP-1 for apical localization, despite lacking a cytoplasmic tail that could be recognized by AP-1 (20).
The considerations discussed above highlight the need to search for additional partners of AP-1 that may provide directionality to the apical-basolateral trafficking mechanism mediated by AP-1. Candidate partners are vesicle fusion proteins, like syntaxin 3 and 4, which are apical and basolateral, respectively, in epithelia (41), and require AP-1 for their localization (20, 42). The search for additional partners of AP-1 may be facilitated by the identification of suitable cargoes, which should be empowered by the large-scale proteomics approach that we present here. For example, we noticed that some plasma membrane proteins displayed increased or decreased total surface expression levels on AP-1 silencing, whereas others did not (Fig. 4). Our identification via proteomics of such proteins provides an opportunity to study the underlying mechanism, which may entail changes in recycling affecting protein retrieval from the plasma membrane or intracellular compartments or alterations in protein turnover due to retromer activity. Additional research is required to clarify this point.
In summary, using a combined quantitative surface biotinylation/mass spectrometry approach, we determined the surface proteome of the prototypical epithelial cell line MDCK. Using this approach in conjunction with silencing of AP-1A and AP-1B, we uncovered global aspects of the trafficking role of these adaptors, in particular, a previously unrecognized role in maintaining polarity of transmembrane apical plasma membrane proteins. We identified selected subsets of apical and basolateral proteins that require AP-1A, AP-1B, or both to maintain their apical-basolateral distribution, indicating that these adaptors may control different sets of proteins. Hence, the assay that we have introduced should constitute a useful tool to screen for additional members of the epithelial trafficking machinery and to investigate unknown roles of existing members of this machinery.
Methods
Cell Culture and SILAC Labeling.
MDCK cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Corning,) supplemented with 5% fetal bovine serum (FBS) (Thermo Fisher Scientific). To obtain a polarized monolayer, cells were seeded at a density of 3 × 105 cells per 1 cm2 in polycarbonate permeable support in Transwell inserts (Corning) and grown for 4 d. For SILAC labeling, unless indicated otherwise, all media and reagents were from Cambridge Isotope Laboratories. Cells were cultured for at least six cell divisions in DMEM free of Lysine and Arginine supplemented with 5% dialyzed FBS and 150 μM Lysine and Arginine isotopes as follows: light (Lys0 + Arg0): l-Lysine + l-Arginine (Millipore-Sigma); medium (Lys4 + Arg6): D4-l-Lysine:2HCl + 13C6-l-Arginine:HCl; heavy (Lys8 + Arg10): 13C6 15N2-l-Lysine 2HCl + 13C6 15N4-l-Arginine:HCl. After labeling, cells were either plated for polarization or transfected with siRNAs as needed while kept in the corresponding SILAC medium until the surface biotinylation procedure was performed.
Blotting Conditions and Antibodies.
For Western blot, eluted surface proteins were run in gradient NuPAGE 4–12% Bis-Tris Protein gels (Thermo Fisher Scientific) and transferred to nitrocellulose membranes using the iBlot system according to the manufacturer’s instructions (Thermo Fisher Scientific). Membranes were blocked with Odyssey Blocking Buffer (Li-Cor) for 1 h at room temperature in rocking platform. Next, membranes were incubated for 1 h at room temperature with the corresponding primary antibody, washed three times with Tris-buffered saline (TBS)-0.1% Tween-20 buffer, then incubated 1 h with the corresponding infrared dye-tagged secondary antibody IRDye-680 or IRDye-800 (Li-Cor), and washed again. Membranes were scanned and bands were quantified with Odyssey imager system (Li-Cor). The primary antibodies sources and dilutions were the following: ADAM17 [chicken (1:2,000; R&D Systems)]; AE2 [rabbit (1:1,000; GeneTex)]; CD44 [rabbit (1:500; Developmental Studies Hybridoma Bank Product 5D2-27); deposited by J. T. August]; CDH1 (E-cadherin) [mouse (1:20,000; BD Transduction Laboratories)]; ENPEP [goat (1:1,000; R&D Systems)]; EphB4 [mouse (1:500; Developmental Studies Hybridoma Bank Product CPTC-EPHB4-1); deposited by Clinical Proteomics Technologies for Cancer]; CEACAM1 (gp114) [rabbit (1:1,000; rabbit polyclonal generated previously) (43)]; PODXL (gp135) [mouse (1:500; hybridoma from G. K. Ojakian and R. Schwimmer (State University of New York Downstate Medical Center, Brooklyn, New York) (44)]; GPRC5A [mouse (1:1,000; R&D Systems)]; HEPH [rabbit (1:500; GeneTex)]; ICAM1 [mouse (1:500; Developmental Studies Hybridoma Bank Product P2A4); deposited by E. A. Wayner and G. Vercellotti]; LSR [rabbit (1:500; Cell Signaling)]; MET [rabbit (1:500; Cell Signaling)]; ATP1B1 (Na/K ATPase) [mouse (1:1,000; Millipore-Sigma)]; SLITRK4 [mouse (1:1,000; R&D Systems)]; and STRA6 [rabbit (1:1,000; ProSci)].
Gene Silencing.
Silencing was performed as described previously using highly potent siRNAs against μ1A (GTGCTCATCTGCCGGAATT) (15) and μ1B (AACAAGCTGGTGACTGGCAAA) (12). Three rounds of silencing spaced by 3 d were performed in MDCK cells. Cells were suspended by trypsinization, and 4 × 106 cells were electroporated with 160 pmol of siRNA in Amaxa Nucleofector kit V (program T-23) following the manufacturer’s instructions (Lonza). All siRNA oligos, including a control against luciferase, were custom synthesized by Dharmacon. After the final round of silencing, cells were seeded at a density of 3 × 105 cells per 1 cm2 in Transwell inserts and cultured 4 d to achieve full polarization. Efficiency of silencing was corroborated by qRT-PCR as done routinely.
Surface Biotinylation.
Polarized MDCK cells were washed three times with biotinylation buffer [10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 130 mM NaCl, 2 mM MgSO4, 1 mM CaCl2, 5.5 mM glucose, pH 7.8–8.0] at 4 °C and kept on ice for the rest of the biotinylation procedure to stop protein trafficking. Surface protein was biotinylated at the apical or basolateral membrane with cell-impermeant EZ-Link Sulfo-NHS-SS-Biotin (Thermo Fisher Scientific) at 1 mg/mL by two consecutive incubations of 15 min each. Biotin was washed twice and quenched with 100 mM glycine to remove nonreacted Sulfo-NHS-SS-Biotin. Cells were lysed in buffer containing 150 mm NaCl, 50 mm HEPES, pH 7.5, 5 mm ethylenediaminetetraacetic acid (EDTA), 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), and Protease Inhibitor Mixture Set III (Thermo Fisher Scientific). Total protein was quantified by Bradford method, and 1 mg of protein (estimated to be contained in ∼1.2 × 107 cells at confluency) was incubated with high-capacity NeutrAvidin-coated agarose beads (Thermo Fisher Scientific) overnight at 4 °C to separate biotinylated proteins. Beads were washed twice in high-salt (1 M NaCl, 50 mm HEPES, 0.1% Triton X-100, pH 7.4) buffer and twice in no-salt (50 mm HEPES, pH 7.4) buffer. Proteins were eluted by 20-min incubations at room temperature in Laemmli buffer containing 5% SDS, 100 mM NaCl, 100 mM dithiothreitol (DTT), and 5% β-Mercapto-ethanol. Eluted surface proteins were loaded in polyacrylamide gels and run by electrophoresis for either Western blot or proteomics.
Mass Spectrometry.
Purified surface proteins were briefly run into a polyacrylamide gel by electrophoresis for 10 min at 90 V and stained with SimplyBlue SafeStain (Thermo Fisher Scientific) for 1 h at room temperature. Gel bands were cut into small pieces and destained in 25 mM NH4HCO3 and 50% acetonitrile. Proteins were reduced with DTT, then alkylated with iodoacetamide, and digested with 12.5 ng/μL trypsin in 25 mM NH4HCO3 overnight at 37 °C. Peptides were extracted twice with 5% formic acid and 50% acetonitrile followed by a final extraction with acetonitrile. Peptides were concentrated by vacuum centrifugation, desalted using C18 StageTips, and concentrated again. The liquid chromatography-mass spectrometry was performed in a Thermo Scientific EASY-nLC 1200 coupled to a Fusion Lumos Orbitrap mass spectrometer (Thermo Fisher Scientific). A self-packed 75-μm × 25-cm reversed-phase column (Reprosil C18, 3 μm) was used for peptide separation. Peptides were eluted by a gradient of 3–30% acetonitrile in 0.1% formic acid over 120 min at a flow rate of 300 nL/min at 45 °C. The Lumos was operated in data-dependent mode with a cycle time of 1.5 s. Survey scans were acquired in the Orbitrap at a resolution of 120,000 at m/z 200. Ions with two to six charges from the survey scan were selected with an isolation window of 1.6 Thomsons and fragmented by higher-energy collisional dissociation with normalized collision energies of 35. Fragment ions were acquired in the ion trap. The maximum ion injection times for the survey scan and the tandem mass spectrometry scans were 100 and 60 ms, respectively. The ion target values were set to 1,000,000 and 10,000, respectively.
Protein Identification and Quantitation.
The raw files were processed using the MaxQuant computational proteomics platform (version 1.5.5.1) for peptide identification and quantitation. The fragmentation spectra were searched against the UniProt canine database containing 25,539 protein sequences, allowing up to two missed tryptic cleavages. Carbamidomethylation of cysteine was set as a fixed modification, and oxidation of methionine, protein N-terminal acetylation, D4-Lysine, 13C6-Arginine, 13C6 15N2-Lysine, and 13C6 15N4-Arginine were used as variable modifications for database searching. The precursor and fragment mass tolerances were set to 5 ppm and 0.5 Da, respectively. Both peptide and protein identifications were filtered at 1% false discovery rate.
Bioinformatics Analysis.
Proteomics data were analyzed in R via the limma package. Proteins were cross-referenced with the UniProt knowledgebase to assign annotated subcellular location, presence of transmembrane domains, and Gene Ontology annotation. Proteins were filtered out before analysis if they had less than three replicates or were not plasma membrane associated as defined by the UniProt knowledgebase. SILAC apical/basolateral ratios were log2 transformed and median normalized before statistical analysis. Proteins were fit to linear models, and proteinwise sample variances were shrunk toward mean sample variance of all proteins in the sample. Pairwise moderated t tests were conducted between experimental conditions to determine differential protein expression. Pathway analysis was performed with Ingenuity Pathway Analysis software (Qiagen Bioinformatics).
Statistics.
Student’s t test for independent variables was used for comparisons between two means, and one-way ANOVA with Dunnett’s correction was used for multiple comparisons against control when required. Fisher’s exact test for small samples was utilized to compare the independent effects of μ1A and μ1B silencing in apical and basolateral shifts (2 × 2 contingency table). Density plots were obtained by kernel smoothing of the percentage frequency of the datasets over a continuous interval spanning the basolateral-apical distribution at the membrane. Linear regressions were performed with the least squares approach, setting the y and x intercepts at zero. Statistical analysis was performed with GraphPad Prism 7 Software. A significance level of 0.05 was used.
Supplementary Material
Acknowledgments
We thank the Proteomics and Bioinformatics Core facilities from Weill Cornell Medical College (New York, NY) for their support with the proteomics experiments and technical expertise. We thank the Developmental Studies Hybridoma Bank (Iowa City, IA) for making available the antibodies against CD44, ICAM1, and EphB4. This research was supported by an NIH supplement for minority scientists (to P.S.C.) and NIH Grant R01-GM34107 (to E.R.-B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821076116/-/DCSupplemental.
References
- 1.Mellman I., Nelson W. J., Coordinated protein sorting, targeting and distribution in polarized cells. Nat. Rev. Mol. Cell Biol. 9, 833–845 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez-Boulan E., Kreitzer G., Müsch A., Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell Biol. 6, 233–247 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Weisz O. A., Rodriguez-Boulan E., Apical trafficking in epithelial cells: Signals, clusters and motors. J. Cell Sci. 122, 4253–4266 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deborde S., et al. , Clathrin is a key regulator of basolateral polarity. Nature 452, 719–723 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohno H., et al. , Mu1B, a novel adaptor medium chain expressed in polarized epithelial cells. FEBS Lett. 449, 215–220 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Diaz F., et al. , Clathrin adaptor AP1B controls adenovirus infectivity of epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 106, 11143–11148 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schreiner R., et al. , The absence of a clathrin adapter confers unique polarity essential to proximal tubule function. Kidney Int. 78, 382–388 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen I. B., Mogensen E. N., Damkier H. H., Praetorius J., Choroid plexus epithelial cells express the adhesion protein P-cadherin at cell-cell contacts and syntaxin-4 in the luminal membrane domain. Am. J. Physiol. Cell Physiol. 314, C519–C533 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fölsch H., Ohno H., Bonifacino J. S., Mellman I., A novel clathrin adaptor complex mediates basolateral targeting in polarized epithelial cells. Cell 99, 189–198 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Fölsch H., Pypaert M., Maday S., Pelletier L., Mellman I., The AP-1A and AP-1B clathrin adaptor complexes define biochemically and functionally distinct membrane domains. J. Cell Biol. 163, 351–362 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan Y., McGraw T. E., Rodriguez-Boulan E., The epithelial-specific adaptor AP1B mediates post-endocytic recycling to the basolateral membrane. Nat. Cell Biol. 4, 605–609 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Gravotta D., et al. , AP1B sorts basolateral proteins in recycling and biosynthetic routes of MDCK cells. Proc. Natl. Acad. Sci. U.S.A. 104, 1564–1569 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugimoto H., et al. , Differential recognition of tyrosine-based basolateral signals by AP-1B subunit mu1B in polarized epithelial cells. Mol. Biol. Cell 13, 2374–2382 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvajal-Gonzalez J. M., et al. , Basolateral sorting of the coxsackie and adenovirus receptor through interaction of a canonical YXXPhi motif with the clathrin adaptors AP-1A and AP-1B. Proc. Natl. Acad. Sci. U.S.A. 109, 3820–3825 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gravotta D., et al. , The clathrin adaptor AP-1A mediates basolateral polarity. Dev. Cell 22, 811–823 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo X., et al. , The adaptor protein-1 μ1B subunit expands the repertoire of basolateral sorting signal recognition in epithelial cells. Dev. Cell 27, 353–366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillard G., et al. , Control of E-cadherin apical localisation and morphogenesis by a SOAP-1/AP-1/clathrin pathway in C. elegans epidermal cells. Development 142, 1684–1694 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Zhang H., et al. , Clathrin and AP-1 regulate apical polarity and lumen formation during C. elegans tubulogenesis. Development 139, 2071–2083 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hase K., et al. , AP-1B-mediated protein sorting regulates polarity and proliferation of intestinal epithelial cells in mice. Gastroenterology 145, 625–635 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Castillon G. A., Burriat-Couleru P., Abegg D., Criado Santos N., Watanabe R., Clathrin and AP1 are required for apical sorting of glycosyl phosphatidyl inositol-anchored proteins in biosynthetic and recycling routes in Madin-Darby canine kidney cells. Traffic 19, 215–228 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Mathias R. A., et al. , Tandem application of cationic colloidal silica and Triton X-114 for plasma membrane protein isolation and purification: Towards developing an MDCK protein database. Proteomics 11, 1238–1253 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Sargiacomo M., Lisanti M., Graeve L., Le Bivic A., Rodriguez-Boulan E., Integral and peripheral protein composition of the apical and basolateral membrane domains in MDCK cells. J. Membr. Biol. 107, 277–286 (1989). [DOI] [PubMed] [Google Scholar]
- 23.Ong S. E., et al. , Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Steinberg F., et al. , A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell Biol. 15, 461–471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNally K. E., et al. , Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat. Cell Biol. 19, 1214–1225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kammers K., Cole R. N., Tiengwe C., Ruczinski I., Detecting significant changes in protein abundance. EuPA Open Proteom. 7, 11–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phipson B., Lee S., Majewski I. J., Alexander W. S., Smyth G. K., Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 10, 946–963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savas J. N., Stein B. D., Wu C. C., Yates J. R. 3rd, Mass spectrometry accelerates membrane protein analysis. Trends Biochem. Sci. 36, 388–396 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vuckovic D., Dagley L. F., Purcell A. W., Emili A., Membrane proteomics by high performance liquid chromatography-tandem mass spectrometry: Analytical approaches and challenges. Proteomics 13, 404–423 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Weekes M. P., et al. , Comparative analysis of techniques to purify plasma membrane proteins. J. Biomol. Tech. 21, 108–115 (2010). [PMC free article] [PubMed] [Google Scholar]
- 31.Wollscheid B., et al. , Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 27, 378–386 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nunomura K., et al. , Cell surface labeling and mass spectrometry reveal diversity of cell surface markers and signaling molecules expressed in undifferentiated mouse embryonic stem cells. Mol. Cell. Proteomics 4, 1968–1976 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Hofmann A., et al. , Proteomic cell surface phenotyping of differentiating acute myeloid leukemia cells. Blood 116, e26–e34 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Bausch-Fluck D., et al. , A mass spectrometric-derived cell surface protein atlas. PLoS One 10, e0121314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.da Cunha J. P. C., et al. , Bioinformatics construction of the human cell surfaceome. Proc. Natl. Acad. Sci. U.S.A. 106, 16752–16757 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathias R. A., et al. , Secretome-based proteomic profiling of Ras-transformed MDCK cells reveals extracellular modulators of epithelial-mesenchymal transition. J. Proteome Res. 8, 2827–2837 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Cortes L. K., et al. , Proteomic identification of mammalian cell surface derived glycosylphosphatidylinositol-anchored proteins through selective glycan enrichment. Proteomics 14, 2471–2484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doyon J. B., et al. , Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat. Cell Biol. 13, 331–337 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibson T. J., Seiler M., Veitia R. A., The transience of transient overexpression. Nat. Methods 10, 715–721 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Takahashi D., et al. , The epithelia-specific membrane trafficking factor AP-1B controls gut immune homeostasis in mice. Gastroenterology 141, 621–632 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Low S. H., et al. , Differential localization of syntaxin isoforms in polarized Madin-Darby canine kidney cells. Mol. Biol. Cell 7, 2007–2018 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reales E., Sharma N., Low S. H., Fölsch H., Weimbs T., Basolateral sorting of syntaxin 4 is dependent on its N-terminal domain and the AP1B clathrin adaptor, and required for the epithelial cell polarity. PLoS One 6, e21181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Bivic A., Sambuy Y., Mostov K., Rodriguez-Boulan E., Vectorial targeting of an endogenous apical membrane sialoglycoprotein and uvomorulin in MDCK cells. J. Cell Biol. 110, 1533–1539 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ojakian G. K., Schwimmer R., The polarized distribution of an apical cell surface glycoprotein is maintained by interactions with the cytoskeleton of Madin-Darby canine kidney cells. J. Cell Biol. 107, 2377–2387 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.