Significance
There is a critical need to identify accessible stem cells that can form spontaneously beating cardiomyocytes (CMs) and enable regeneration. Here, we establish that intravenous delivery of placental Cdx2 cells resulted in directed homing, sustained engraftment, and differentiation into CMs and vascular cells in damaged hearts, significantly improving cardiac function. This study unveils a distinctive functional significance of Cdx2 beyond its established role in embryonic patterning. Therapeutic use of Cdx2 cells may represent a vital advance, as these cells are multipotent and immunologically naive, with a unique proteome, compared with embryonic stem cells. Moreover, they exhibit the ability to selectively home to sites of injury. These characteristics pave the way for novel allogeneic stem cell therapy for cardiac disease.
Keywords: Cdx2, placenta, cardiac regeneration, stem cells, cardiomyocytes
Abstract
The extremely limited regenerative potential of adult mammalian hearts has prompted the need for novel cell-based therapies that can restore contractile function in heart disease. We have previously shown the regenerative potential of mixed fetal cells that were naturally found migrating to the injured maternal heart. Exploiting this intrinsic mechanism led to the current hypothesis that Caudal-type homeobox-2 (Cdx2) cells in placenta may represent a novel cell type for cardiac regeneration. Using a lineage-tracing strategy, we specifically labeled fetal-derived Cdx2 cells with enhanced green fluorescent protein (eGFP). Cdx2-eGFP cells from end-gestation placenta were assayed for cardiac differentiation in vitro and in vivo using a mouse model of myocardial infarction. We observed that these cells differentiated into spontaneously beating cardiomyocytes (CMs) and vascular cells in vitro, indicating multipotentiality. When administered via tail vein to infarcted wild-type male mice, they selectively and robustly homed to the heart and differentiated to CMs and blood vessels, resulting in significant improvement in contractility as noted by MRI. Proteomics and immune transcriptomics studies of Cdx2-eGFP cells compared with embryonic stem (ES) cells reveal that they appear to retain “stem”-related functions of ES cells but exhibit unique signatures supporting roles in homing and survival, with an ability to evade immune surveillance, which is critical for cell-based therapy. Cdx2-eGFP cells may potentially represent a therapeutic advance in allogeneic cell therapy for cardiac repair.
The regenerative capacity of the adult mammalian heart is very limited, and this contributes to the extensive morbidity and mortality associated with cardiovascular disease (1, 2). It has also become increasingly apparent that adult mammalian hearts do not harbor endogenous stem cells of any physiological relevance that can regenerate injured myocardium. Despite exhaustive investigations with multiple cell types over 15 y, cell therapy results for cardiac repair have thus far been marginal at best (3–5). Coaxing the division of preexisting myocytes and discovering resident- or tissue-specific stem cells that can form cardiac cells are two of the major approaches that have been under investigation (6–11). In pursuit of the appropriate cell type for cardiac repair, we had previously reported that cells can “naturally” and “selectively” home from the placenta to injured maternal hearts and form functional cardiomyocytes (CMs) (12). The placenta acts as a reservoir of stem/progenitor cells that possess numerous advantages over adult stem cells in terms of stemness and plasticity (13, 14). In contrast to embryonic stem (ES) cells, teratoma initiation by placenta-derived stem cells has not been demonstrated in transplant settings (15), rendering them an attractive source for regenerative medicine, including cardiac cell therapy. Fetal microchimerism, the trafficking and persistence of fetal cells in the maternal circulation, is a common phenomenon during pregnancy involving cells that possess multilineage potential (16). Fetal trophoblasts are the initial cells that invade the maternal endometrium during placentation; however, the circulating fetal cells in the maternal body can migrate to various tissues and persist within them for decades (17). The exact function of fetal cells in the maternal body has not been well defined.
We had surmised that the fetal-derived cells homing to the injured maternal heart in the pregnancy model were stem or progenitor cells, and thereby analyzed numerous stem cell markers within these cells. Surprisingly, almost 40% of fetal placenta cells in the maternal heart expressed a homeodomain protein, Caudal-type homeobox-2 (Cdx2). Cdx2 is the master homeodomain protein that defines trophectoderm during development and is further involved in self-renewal of trophoblast stem cells that ultimately form placenta (18). During embryonic development, Cdx2 regulates cell polarity and lineage choices (19), where its postnatal function is reportedly limited to intestinal epithelial development (20). Our observation of the unique presence of Cdx2 within the fetal cells led us to hypothesize that these cells may represent a novel source of cell therapy for cardiac regeneration. To explore this hypothesis, we employed a lineage-tracing strategy to label Cdx2-expressing cells and their progeny using enhanced green fluorescent protein (eGFP), and then isolated and studied these cells from end-gestation mouse placentas and examined their function in vitro and in vivo in a mouse model of acute myocardial infarction (MI). Additionally, we analyzed the immune transcriptome of these cells to determine whether they express markers needed to elicit an immune response. Furthermore, to distinguish the properties of Cdx2 cells from ES cells, we compared the two proteomes and found that Cdx2 cells appear to retain the “stemness” of ES cells but exhibit unique proteins that facilitate homing and immune modulation.
Results
Cdx2 Expression and Isolation from End-Gestation Placenta.
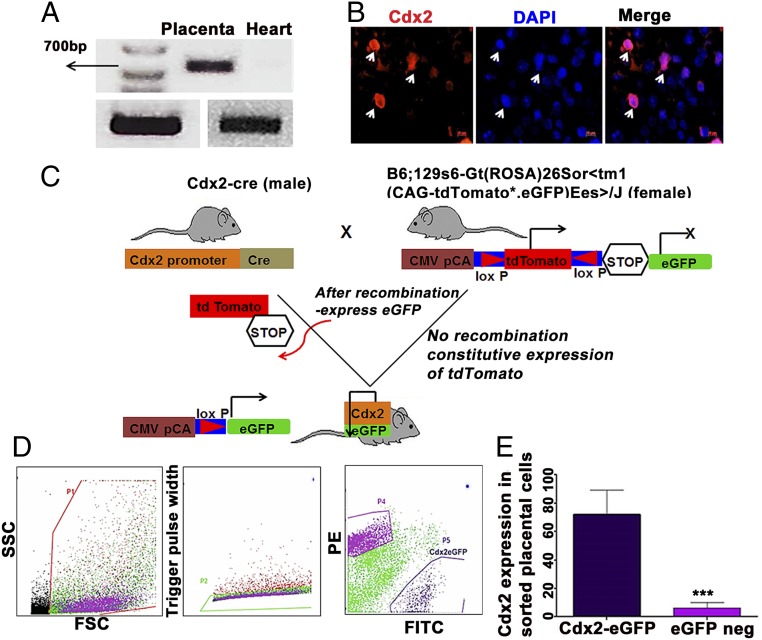
Since fetal-maternal trafficking is known to peak toward the end of gestation and immediately after delivery, embryonic day 18 (e18) was selected to isolate placentas as murine placentas are resorbed postdelivery. Placentas isolated from wild-type (WT) pregnant females were subjected to enzymatic digestion and processed to yield single cells. The presence of Cdx2 messenger RNA was confirmed by PCR (Fig. 1A), and isolated placenta cells were immunostained to identify the presence of cells expressing nuclear Cdx2 (Fig. 1B). Owing to the lack of known surface antigenic markers that can assist in isolation of placental Cdx2 cells, we utilized a lineage-tracing strategy that specifically selects Cdx2 cells and their progeny from end-gestation mouse placentas. To accomplish this, a Cre-Lox method was employed wherein fetal-derived Cdx2 cells were labeled with eGFP when female virgin B6;129S6-gt(ROSA)26Sor < tm1(CAG-tdTomato*-EGFP*) Ees>/J43 mice were crossed with male B6.Cg-Tg(CDX2-cre)101Erf/J mice (Fig. 1C). Cdx2-eGFP cells were subsequently purified using flow cytometry sorting as shown in Fig. 1D. The frequency of eGFP+ cells across placentas averaged 3.18% ± 0.265 as assessed by flow cytometry (SI Appendix, Table S1). The extent of Cdx2 enrichment within the isolated lineage cells (eGFP+ and eGFP−) was subsequently assessed (Cdx2-eGFP+: 72.23% ± 8.505 and eGFP−: 6.4% ± 2.088) using monoclonal Cdx2 antibody (21) followed by flow cytometry as shown in Fig. 1E. Additionally, to understand whether Cdx2-eGFP cells spontaneously populate fetal organs, we examined whole embryos and multiple fetal organs isolated from these embryos at two different time points (e13 and e18) during gestation. Consistent with the known developmental dynamics of Cdx2 expression, we could observe eGFP fluorescence along the caudal/hindgut regions of the embryos (SI Appendix, Fig. S1). This was further confirmed from the isolated embryonic tissue sections, where nuclear eGFP fluorescence was evident in the intestinal tissue sections from e13 and 18 embryos (SI Appendix, Fig. S2 A and B). Subsequently, nuclear eGFP fluorescence was not detected in other tissue sections, including fetal brain, heart, liver, lungs, spleen, and kidney. These experiments revealed that at these time points in gestation, Cdx2-eGFP cells were present in the fetal placenta and in the hind gut/caudal region of the embryo, with very limited or no expression in other sites/organs.
Fig. 1.
Cdx2 expression and lineage tracing. (A) Cdx2 messenger RNA (mRNA) expression in an e18 WT mouse placenta relative to the endogenous control gapdh (n = 3 mice). Heart mRNA at e18 served as the negative control. Please refer to additional data in SI Appendix for the original gel image file. (B) WT e18 placental cells positive for Cdx2. A representative immunofluorescence image depicting Cdx2 antibody staining on WT placental cells is shown. Cdx2 (red) and nuclei were counterstained by DAPI (blue) (n = 3 mice). (Scale bar: 10 μm.) (C) Illustration of Cre-Lox transgenic mouse strategy to trace Cdx2-derived cells. (D) Flow cytometry gating strategy and isolation of e18 eGFP–tagged placental Cdx2 cells. FITC, fluorescien isothiocyanate; FSC, forward scatter; PE, phycoerythrin; SSC, side scatter. (E) Cdx2-eGFP and eGFP− cells were immunostained using monoclonal Cdx2 antibody and analyzed by flow cytometry. Data are represented as mean ± SEM. ***P = 0.0013 from three different samples (n = 3).
Placental Cdx2-eGFP Cells Exhibit Clonal Differentiation.
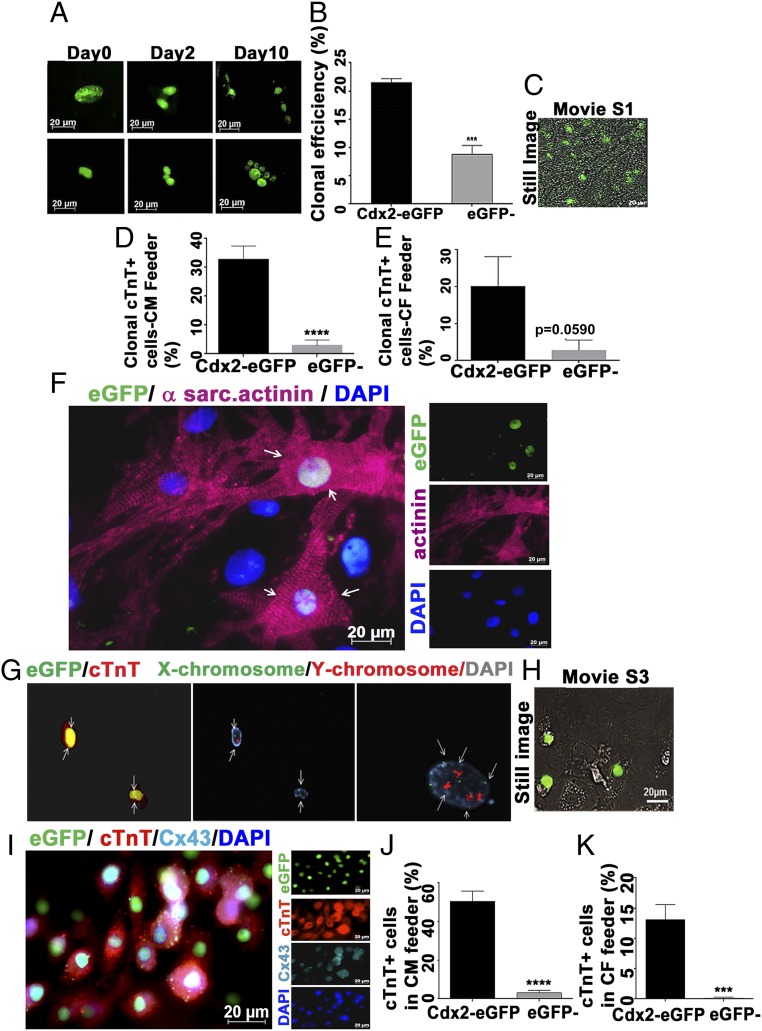
To understand the clonal nature of Cdx2-derived cells from placenta, single eGFP+ cells were sorted onto 96-well plates containing mitotically inactivated cardiac fibroblast (CF) feeders in standard culture conditions (Iscove’s Dulbecco’s modified Eagle’s medium + 10% fetal bovine serum). The Cdx2-eGFP cells were monitored and imaged sequentially to detect proliferation in vitro. We observed that the cells started to divide on day 2 and that the nuclear eGFP signal was equally distributed within each daughter cell. Subsequently, we observed higher numbers of eGFP+ cells within 10 d, out of the single cell plated on day 0, suggesting that Cdx2-eGFP cells from placenta can clonally proliferate in vitro (Fig. 2A). The viability of plated cells was 77.74% ± 2.6 from four different samples analyzed, whereas clonal efficiency was 21.49% ± 0.7338. Even though the viability was not different (77.77% ± 5.203), the clonal efficiency of the eGFP− population from the placenta was significantly lower compared with the Cdx2-eGFP population (8.80% ± 1.571; Fig. 2B and Table 1).
Fig. 2.
Clonal proliferation and cardiac differentiation of Cdx2-eGFP cells in vitro. (A) Single-cell sorting of eGFP cells was utilized to assess the clonal proliferation through live cell imaging for 10 d in a CF feeder. (B) Clonal efficiency of Cdx2-eGFP cells and eGFP− cells was calculated from the proliferative wells in the CF feeder (n ≥ 3 mice) (Table 1). Data are represented as mean ± SEM. ***P = 0.0005. (C) Still image from the Movie S1 depicting spontaneous beating of clonal cultures of Cdx2-eGFP cells after 4 wk in a CM feeder. (D and E) Cdx2-eGFP cells generated significantly more CMs in both CM (****P < 0.0001) and CF feeder systems. Data are represented as mean ± SEM (n = 3). (F) Representative field showing a differentiated Cdx2-eGFP cell expressing a sarcomeric structure (α-sarcomeric actinin, pink) when cultured on neonatal CM feeders for 5 wk. White arrows indicate the differentiated Cdx2-eGFP cell with sarcomere formation. (G) XY chromosome analysis (FISH) demonstrates a single set of sex chromosomes (Center; white arrows point to cells with either XY-red/green or XX-green chromosomes) in cTnT+ (red) eGFP cells (Left; white arrows), thus excluding cell fusion. (Right) Non-eGFP feeder CM with a tetraploid nucleus (white arrows). (H) Still image from Movie S3 showing spontaneous beating of Cdx2-eGFP CMs. (I) Connexin 43 (Cx43) expression is detectable in early-differentiating cTnT+ Cdx2-eGFP cells as early as 3 wk in culture [GFP, green; cTnT, red; Cx43, cyan; nuclei, blue (DAPI)]. Quantification of cardiac differentiation of Cdx2-eGFP and eGFP− cells on a CM feeder (J) and a CF feeder (K) is shown. Data are represented as mean ± SEM (n = 3). ****P < 0.0001, ***P = 0.0005.
Table 1.
Quantification of clonal efficiency of Cdx2-eGFP and eGFP− cell populations from the placenta
| Samples | Plated wells (single cell) | Proliferating wells | Dead cells/no fluorescence | Viability (%) | Clonality (%) |
| Cdx2-eGFP | |||||
| Mouse 1 | 24 | 4 | 7 | 70.8 | 23.5 |
| Mouse 2 | 18 | 3 | 3 | 83.3 | 20 |
| Mouse 3 | 18 | 3 | 4 | 77.7 | 21.42 |
| Mouse 4 | 24 | 4 | 5 | 79.16. | 21.05 |
| Mean ± SEM | 77.74 ± 2.6 | 21.49 ± 0.73 | |||
| eGFP− | |||||
| Mouse 1 | 24 | 2 | 3 | 87.5 | 9.52 |
| Mouse 2 | 24 | 2 | 6 | 75 | 11.1 |
| Mouse 3 | 24 | 1 | 7 | 70.8 | 5.88 |
| Mean ± SEM | 77.77% ± 5.2 | 8.80% ± 1.571 |
Clonally Expanding Cdx2 Cells Differentiate into Spontaneously Beating CMs in Vitro.
To further understand the clonal lineage commitment, single cells were sorted into 96-well plates containing CF feeders and additionally on neonatal CM feeders for an extended period of 5 wk. The CM feeders may mimic the cardiac microenvironment, and thus may provide a favorable milieu for specific differentiation. Clonal efficiency was found to be higher in CM feeders than in CF feeders, and clonally derived Cdx2-eGFP cells started to beat spontaneously within 4 wk in CM feeders (Fig. 2C, still image and Movie S1). The eGFP− cells (tdTomato+ fraction, note nuclear red fluorescence) exhibited limited clonal proliferation and did not show spontaneous beating with neighboring CM feeder cells (Movie S2). Clonal differentiation into CMs was also higher in CM feeders than in the CF feeder system. Clonally derived Cdx2-eGFP CMs [cardiac troponin T (cTnT+) cells] ranged from 32.63% ± 4.720 in CM feeders versus 20.13% ± 7.966 in CF feeders. We observed a negligible frequency of eGFP− cells that stained positive for cTnT (2.857% ± 1.941 and 2.77% ± 2.77 in CM and CF feeders, respectively) (Fig. 2 D and E), and this “cardiomyocyte-like” fraction may be attributed to the existing low level of maternal Cdx2 (placental decidua) within these cells (Fig. 1E) as opposed to the enriched fetal-Cdx2 eGFP cells from the placenta.
Cdx2 Cells Differentiate into CMs That Appear Rod-Shaped and Express Cardiac-Specific Markers in Vitro.
Cdx2-eGFP cells were also plated at a higher quantity (5,000 per well) after the single-cell experiments described above on neonatal CM feeders. These cells showed stable nuclear localization of eGFP that was sustained throughout the culture period, enabling the efficient tracing of Cdx2-eGFP cells from day 1 through the end of culture duration (SI Appendix, Fig. S3A). Cdx2-eGFP cells started to differentiate into CMs and expressed the cardiac structural protein troponin T as early as 2–3 wk in culture as shown in SI Appendix, Fig. S3B. Fig. 2F (also SI Appendix, Fig. S3 C and D) depicts an eGFP-derived CM displaying sarcomeric actinin (pink) after 5 wk in culture. At this point in time, rod-shaped morphology could be seen resembling that of the neighboring feeder myocytes. White arrowheads in the merged image (Fig. 2F) depict eGFP cells with sarcomere formation similar to adjacent feeder myocytes. The possibility of cell fusion with feeder CMs was excluded by in situ hybridization using mouse fluorescent XY probes [fluorescence in situ hybridization (FISH)] on cTnT+ eGFP cells. Fig. 2G, Center shows nuclei with DAPI (gray) and XY probes at different wavelengths (X: green, 520 nm; Y: red, 603 nm) whereas Fig. 2G, Left shows anti-eGFP signal in the same field at 488 nm and anti-cTnT Texas Red signal at 568 nm. Fig. 2G, Right shows detection of a tetraploid nucleus in feeder CMs using the same probe, indicating the ease with which tetraploid and diploid nuclei can be distinguished. FISH analysis clearly revealed the presence of only one set of sex chromosomes in eGFP cell nuclei, emphasizing that Cdx2-eGFP–derived CMs did not exhibit cell fusion.
Furthermore, live cell imaging revealed spontaneous beating of Cdx2-eGFP (note nuclear eGFP) cells in the syncytium with neighboring feeder cells (Fig. 2H, still image and Movies S3 and S4). As described in the clonal experiments, we did not observe any events of spontaneous beating in eGFP− cells in bulk culture even after the 5-wk duration (Movies S5 and S6), with few cells expressing cTnT (SI Appendix, Fig. S3E). Additionally, as early as 3 wk, differentiating cTnT+ CMs from CDx2-eGFP cells also expressed connexin 43, suggesting the presence of functional gap junctions in these cells (Fig. 2I and SI Appendix, Fig. S3F). The effect of a CM-independent microenvironment in cardiac differentiation of Cdx2-eGFP cells was also studied using mitotically inactivated CF feeders as shown in Fig. 2A. The generation of distinct eGFP+ CMs that expressed cTnT was noted, highlighting a cardiac differentiation pathway for Cdx2 cells even in the presence of CFs in place of CMs (SI Appendix, Fig. S3G). However, as noted before, in CM feeders, Cdx2-eGFP cells differentiated into CMs with a 10-fold higher frequency (50.4% ± 5.376) compared with the eGFP− cell population (5.015% ± 2.049; Fig. 2J). Overall, the frequency of CMs in CF feeders was significantly lower compared with that in CM feeders. In CF feeders, Cdx2-eGFP derived CM-like cells comprised ∼13.1% ± 2.505, whereas cTnT+ eGFP− cells were rarely present (0.1429% ± 0.1429; Fig. 2K).
Cdx2-eGFP Cells also Differentiate into Vascular Lineages in Vitro.
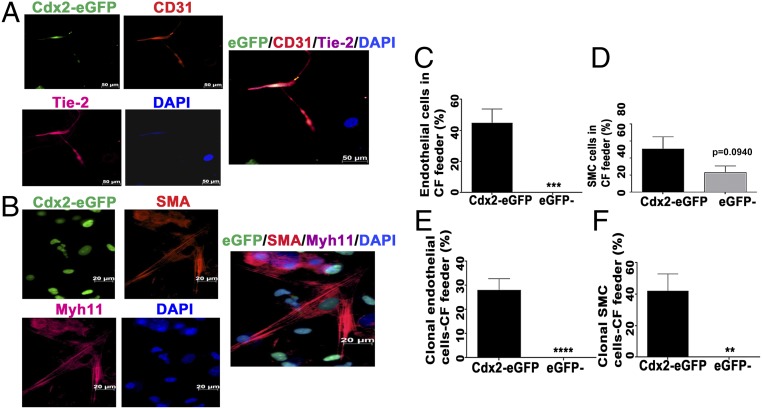
Vascular differentiation of cultured Cdx2-eGFP and eGFP− cells was studied using mitotically inactivated CF feeders for 3 wk. We observed the emergence of Cdx2-eGFP–derived endothelial-like cells expressing CD31 and Tie-2 (Fig. 3A) and smooth muscle cells expressing αSMA and myosin heavy chain 11 (Myh11) (Fig. 3B) in the CF feeder system (also SI Appendix, Fig. S4 A and B), corroborating the vascular identity of these cells originating from Cdx2-eGFP. We did not observe any discernible endothelial differentiation in eGFP− cells even though they did form some cells expressing smooth muscle actin (Fig. 3D). The results from the single-cell and the bulk culture methods of proliferation and differentiation suggested that fetal Cdx2-eGFP cells may represent a novel multipotent cell type capable of both cardiomyogenic and vasculogenic potential.
Fig. 3.
Vascular differentiation of Cdx2-eGFP cells in vitro. Cdx2-eGFP cells differentiated into endothelial (CD31/Tie-2) cells (A) and smooth muscle (αSMA/myh11) cells (B) in vitro (CD31 and SMA, red; Tie-2 and Myh11, pink; DAPI, blue). (Scale bar: 20 μm.) Quantification of Cdx2-eGFP cells derived from endothelial cells (C) and smooth muscle (D) cells in vitro in CF feeders. (E and F) Quantification of clonal differentiation of Cdx2-eGFP cells into vascular lineages. However, eGFP− cells did not display clonal vascular commitment compared with the Cdx2-eGFP cell population. Data are represented as mean ± SEM (n = 3 independent experiments). **P = 0.0065, ***P = 0.005, ****P < 0.0001.
Cdx2 Cells’ Transcriptome Supports the Ability to Evade Host Immune Surveillance.
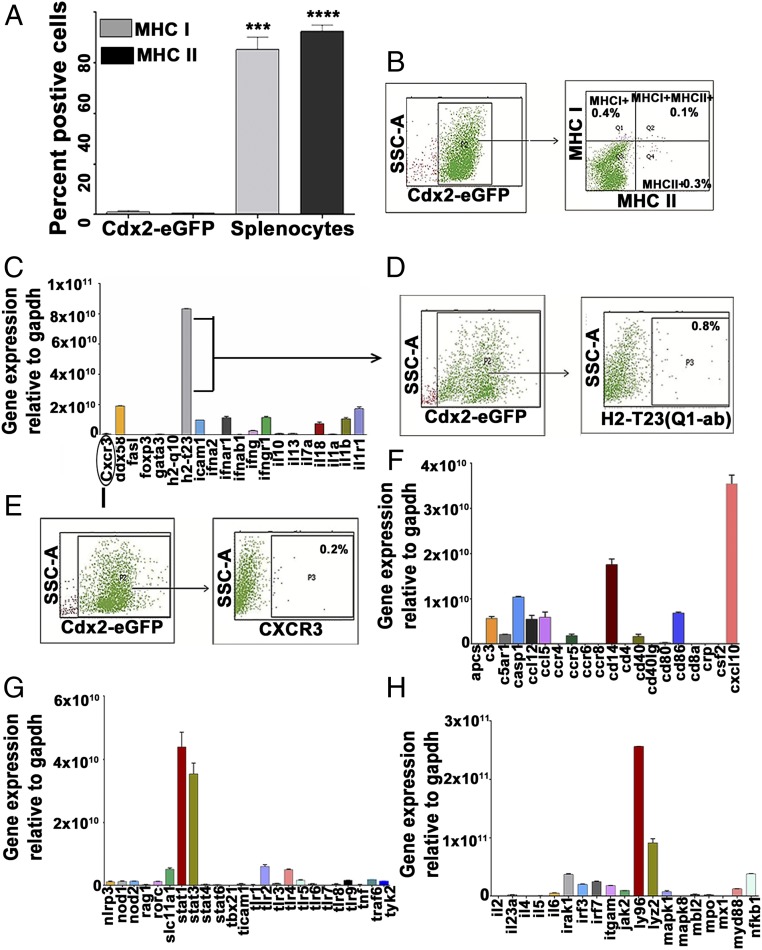
Use of placental stem/progenitor cells for regenerative therapy would necessitate that they exhibit favorable immunomodulatory characteristics (22). Immunologically relevant markers expressed by Cdx2-eGFP cells were examined to assess their potential for use in allogeneic cell therapy. Major histocompatibility complex (MHC) molecules, MHC class I and MHC class II, are the genes that encode cell surface proteins which control adaptive immune responses that involve T cell interactions (23). Surface expression of MHC class I and class II proteins was extremely low in isolated Cdx2-eGFP cells (Fig. 4 A and B), indicating that these cells may harbor an immunologically naive phenotype. Furthermore, we performed an RNA array that detected the major 84 genes involved in adaptive and innate immune responses. We noticed that the majority of these genes were minimally expressed even after preamplification of cDNA using panel-specific primers. Another MHC class I member, H2-Q10, was not detectable at the RNA level, but we noticed a transcriptional abundance for MHC class Ib subtype H2-T23 (Qa-1 or HLA-E in humans). Flow cytometry analysis, however, revealed a lack of surface expression for this marker on Cdx2 cells (Fig. 4 C and D). Cdx2 cells lacked the expression of cognate ligand Cxcr3 (Fig. 4 C and E) despite a transcriptional abundance of the receptor Cxcl10 (Fig. 4F), suggesting it may be functionally insignificant. Additional immune signaling markers displayed by Cdx2 cells were Ly96, Stat1, and Stat3, which are important mediators of Toll-like receptor (TLR) signaling (Fig. 4 G and H). Interestingly, the Cdx2-eGFP transcriptome did not show the expression of TLRs (1–9) (Fig. 4G), suggesting that a functionally relevant TLR-signaling cascade may be inconsequential in these circumstances. The immune marker transcriptome thus implied that isolated Cdx2 cells may be able to evade host immune surveillance, enabling a role in allogeneic cell therapy.
Fig. 4.
Immune marker expression in placental Cdx2-eGFP cells. (A and B) Flow cytometry analysis of e18 placental Cdx2-eGFP cells demonstrated very low expression of MHC class I (1.1% ± 0.51; n = 3) and class II (0.466% ± 0.16; n = 3) molecules compared with the mouse splenocytes (MHC class I: 65.3 ± 4.8, MHC class II: 72.5 ± 2.5) as a positive control. Data are represented as mean ± SEM (n = 3). SSC-A, side scatter area. ***P = 0.0004, ****P < 0.0001. (C–H) Sorted Cdx2-eGFP cells were analyzed for the expression of 84 genes involved in innate and adaptive immunity. Low expression of CXCR3 [cognate ligand of cxcl10 panel (F) and surface H2-T23 (C–E)]. G and H show an increase in stat3/4 messenger RNA with negligible expression of TLRs and ly96 on sorted Cdx2 cells. Data are represented as mean ± SEM (n = 3).
Cdx2 Cells Exhibit a Unique Proteome Reflecting Enriched Functions Regarding Cell Migration and Survival.
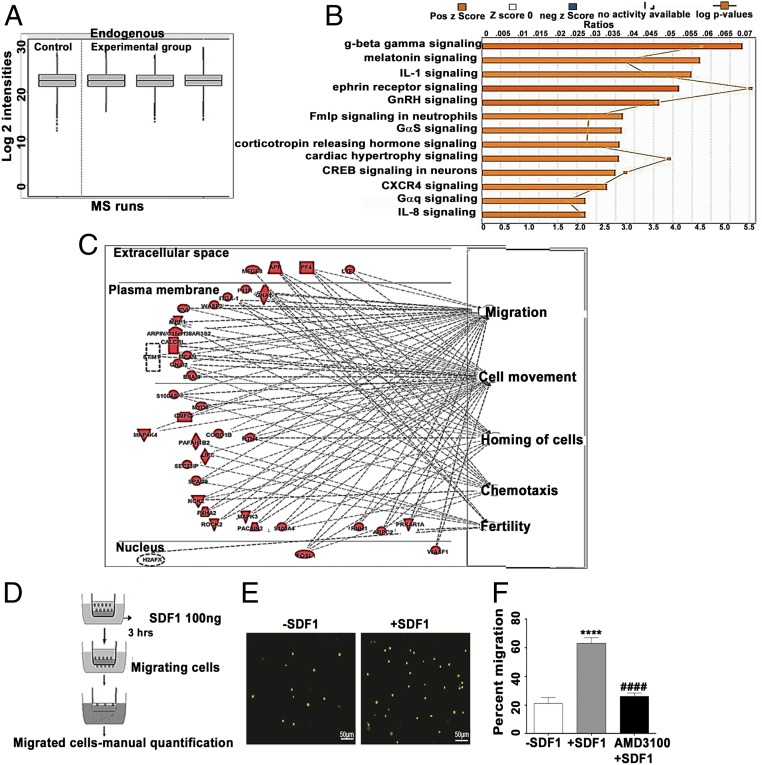
To further analyze cell functions and processes reflected by protein expression patterns, the whole-cell proteome of Cdx2-eGFP cells was studied. Since the eGFP− fraction showed limited “stemness” in the studies detailed above, we decided to compare the proteome of Cdx2-eGFP cells with undifferentiated murine ES cells, obtained from the inner cell mass (ICM) of blastocysts, as they definitively lack Cdx2 expression (24). Mass spectrometric analysis demonstrated 6,646 unique peptides (8,085 precursor scans, 24,135 transitions), with 1,504 proteins (0.95 confidence) differentially expressed in Cdx2-eGFP cells versus ES cells. Approximately 145 proteins were uniquely identified in the Cdx2 samples (SI Appendix). Interestingly, no proteins were observed to be unique to ES cells. These data imply that the protein network governing stem cell-related functions appears to be conserved in Cdx2 cells. Remarkably, ∼19 proteins in the Cdx2-eGFP cells were found to positively regulate cardiac actin dynamics and related functions (Dataset S1). The remaining ∼1,359 proteins were found to have quantitative values in both groups. The mass spectrometry data displayed similar intensity profiles across runs, suggesting equivalent protein loading and allowing valid comparisons (Fig. 5A). Average fold change values were inferred from the quantified protein level values as the quotient of the experimental and control groups. These expression differences were subject to ingenuity pathway analysis (IPA). This analysis found that Cdx2-eGFP protein expression patterns reflect an increased activation in many cell functions (Fig. 5B), the top five of which pertain to cell movement, migration of cells, fertility, homing of cells, and chemotaxis (Fig. 5C). The processes or diseases that were predicted to reflect a decreased activation by this dataset were seizures, growth failure, and organismal death, thus indicating that these cells may be primed for survival. Trafficking and survival of progenitor cells in the host microenvironment is an important parameter for successful regenerative therapies. Probing further into the homing function suggested by proteomics, we checked whether placental Cdx2 cells were “primed” for such a response in the steady state. Injury and subsequent inflammation release SDF1, which attracts stem cells toward the site of injury (25). We examined the effect of chemokine SDF1α (CXCL-12) on Cdx2-eGFP cell migration in vitro using a transwell assay (Fig. 5D). Cdx2-eGFP cells spontaneously migrated (in the absence of SDF1) at a frequency of 20.98% ± 4.36, whereas 63.15% ± 3.99 of cells migrated in response to SDF1 stimulation, demonstrating functionally active SDF1 signaling in these cells (Fig. 5 E and F). To examine the specificity of the SDF1-mediated homing response in Cdx2-eGFP cells, we used AMD3100 (Plerixafor 8HCl), an antagonist of CXCR4 (SDF1 receptor), to precondition isolated CDX2-eGFP cells before chemotaxis. The blocking of CXCR4 significantly reduced the ability of Cdx2-eGFP cells to migrate toward the chemokine SDF1α compared with the untreated Cdx2-eGFP cells (26.25% ± 2.156; Fig. 5F), suggesting that homing/chemotaxis of Cdx2-eGFP cells may involve a functional SDF1-CXCR4 axis.
Fig. 5.
Comparison of whole-cell proteome of Cdx2-eGFP cells vs. ES cell control (ESC). (A) Quality control plot of mass spectrometry (MS) data. Quartiles and intensities were aligned between files to ensure that samples were similarly loaded. Run 1, ESC; Run 2, Cdx2-eGFP cells mouse 1; Run 3, Cdx2-eGFP cells mouse 2; Run 4, Cdx2-eGFP cells mouse 3. Six micrograms of protein was loaded on the column for each run. (B) IPA z-scores reflecting functions up-regulated in Cdx2-eGFP cells. (C) Top five cell functions predicted increased activation in Cdx2-eGFP cells relative to ESC. (Right) Cell function. (Left) Up-regulated proteins, graphed according to subcellular localization. (D) Schema depicting the transwell chemotaxis experiment. (E) Representative images of migrating Cdx2-eGFP cells in the absence (−SDF1) and presence (+SDF1) of SDF1. (Scale bars: 50 μm; Magnification: 20×). (F) Quantification of migration of Cdx2-eGFP cells to SDF1α in the presence and absence of the CXCR4 inhibitor AMD3100, showing the possible role of SDF1-CXCR4 signaling in the migration of Cdx2-eGFP cells. Data are represented as mean ± SEM (n = 4) −SDF1 vs. +SDF1: ****P ≤ 0.0001, +SDF1 vs. AMD3100, ####P ≤ 0.0001.
Intravenous Delivery of Cdx2-eGFP Cells Regenerates Infarcted Myocardium.
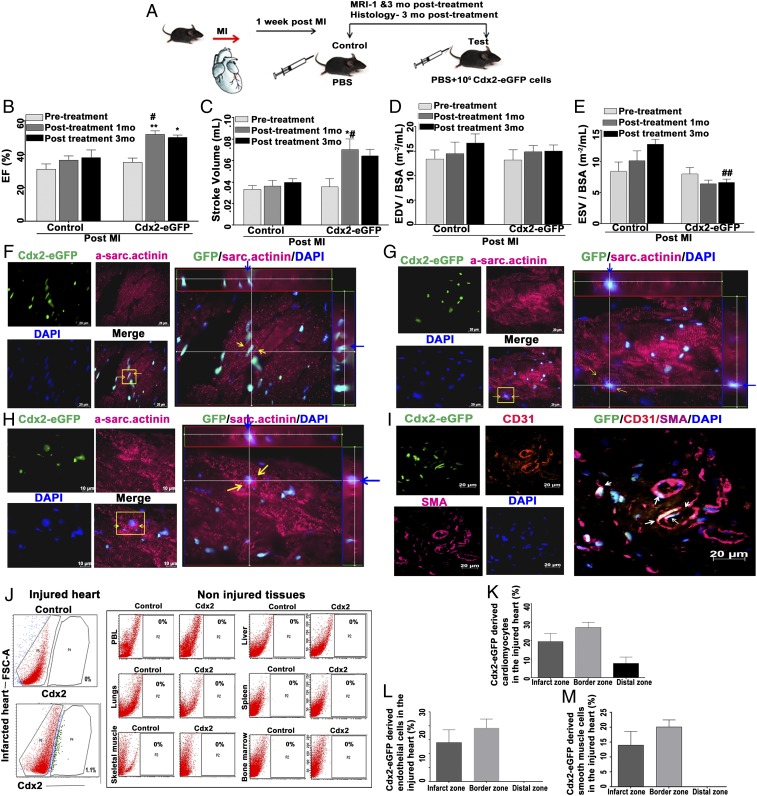
We examined the regenerative potential of Cdx2 cells utilizing an intravenous (i.v.) route to deliver these cells in vivo in a mouse model of MI. WT male mice were subjected to experimental MI via permanent ligation of the left anterior descending artery (LAD) as we described previously (12, 26). The experimental design schema is illustrated in Fig. 6A. Mice were blindly randomized, and the experimental groups of infarcted mice were injected with 1 million Cdx2-eGFP cells in phosphate-buffered saline (PBS) through tail veins, while the control mice received an equal volume of PBS. Cardiac MRI was carried out before (pretreatment post-MI) and after (1 mo and 3 mo post–MI-PBS, Cdx2-eGFP cells) treatment in both sets to confirm the presence of MI and then to analyze cardiac function at these time points. MRI analyses revealed a significant increase in contractility as measured by ejection fraction (EF) in the cell-treated group versus controls [Fig. 6B; test EF: 51.89 ± 2.296 vs. controls 36.44 ± 2.618 in 1 mo and test EF: 50.03 ± 1.582 vs. controls 38.12 ± 4.507 in 3 mo (*P = 0.0263)] (SI Appendix, Table S2 and Movies S7–S11). Quantification of left ventricular functional parameters by MRI suggested that delivery of Cdx2-eGFP cells may reduce the adverse remodeling after MI. We observed a significant increase in stroke volume (SV) within the cell-treated group before and after treatment [Fig. 6C; test SV: 0.0355 ± 0.0078 mL before 1 mo vs. 0.0701 ± 0.01009 mL after 1 mo (*P = 0.0121) and 0.064 ± 0.00627 mL after 3 mo]. It was noteworthy that Cdx2 cell-treated mice exhibited a significantly improved SV at 1 mo in comparison to control mice at the same time point [test SV: 0.0701 ± 0.01009 mL after 1 mo vs. control SV: 0.036 ± 0.03413 mL after 1 mo (*P < 0.05)]. End-diastolic volume measurements indexed to the body surface area did not show much difference between the control and cell-treated groups (Fig. 6D). However, Cdx2 cell delivery decreased the end-SV (ESV) significantly at 3 mo post-MI compared with the control mice [Fig. 6E; test ESV: 6.638 ± 0.5402 m−2/mL after 3 mo vs. control ESV: 12.8 ± 0.7836 m−2/mL after 3 mo (**P = 0.0045)]. These observations suggested that Cdx2 cell delivery reduced the progressive systolic dysfunction after infarction and improved contractility. To understand the contribution of differentiation toward improved cardiac function, we examined the heart tissues at 3 mo after Cdx2-eGFP cell treatment and confirmed the engraftment (SI Appendix, Fig. S5A) and differentiation of Cdx2-eGFP cells into CMs of mature appearance (Fig. 6 F–H and SI Appendix, Fig. S5 B and C). Cdx2-eGFP cells also contributed to endothelial cell differentiation and coexpressed smooth muscle actin, lining the blood vessels (Fig. 6I). Since eGFP fluorescence is localized to the nucleus, we further confirmed that the eGFP nuclei were embedded within the α-sarcomeric actinin+ CM using Z-stack analyses (Movies S12–S14). The hearts from infarcted mice were then examined to quantify the percentage of Cdx2-eGFP cells after 3 mo of cell delivery. Flow cytometry analysis of isolated total cardiac cells revealed that eGFP cells were exclusively present in the injured myocardium (with 1.1% of total heart cells composed of eGFP+ cells at 3 mo; Fig. 6J) with no trafficking observed to any noninjured organs (Fig. 6K), demonstrating the specificity of these cells in “homing” and retention thereafter. We also corroborated the lack of homing of Cdx2-eGFP cells in sections of noninjured (sham) heart tissues (SI Appendix, Fig. S5D). To understand the identity of Cdx2-eGFP–derived cells in vivo, we quantified the spatial distribution of the percentage of eGFP-derived CMs and vascular cells in the injury zone, border zone, and distal zone. Cdx2-eGFP cells contributed to 19.5% ± 4.484, 27.2% ± 2.837, and 7.77% ± 3.409 of cTnT+ actinin+ cells in the infarct zone, border zone, and distal zone, respectively, in post-MI heart after 3 mo of cell delivery (Fig. 6K). This suggests that the majority of eGFP cells were localized to the injury and border zone, and thus could be able to presumably augment cardiac function as noted on MRI. In the border (periinfarct zone), eGFP cells displayed more mature CM morphology with the presence of a developed sarcomeric structure (Movies S12–S14 correspond to Fig. 6 F–H, which displays the Z–stack three-dimensional view with sarcomeres). In vivo CM differentiation from Cdx2-eGFP cells is illustrated in SI Appendix, Fig. S5 B and C. Cdx2-derived endothelial cells also coexpressed Ang-1 receptor Tie-2 (SI Appendix, Fig. S5E) and smooth muscle markers in the injury zone. However, we identified smooth muscle-specific myh11 and smooth muscle actin cells additionally (SI Appendix, Fig. S5F) in the border zone of the post-MI heart. Overall, these newly formed vascular cells were predominantly limited to the injury zone and border zone from the site of infarct [EC + SMC: 14.02 ± 4.601 in the infarct zone versus EC: 19.23% ± 3.26 and SMC: 20.14 ± 4.601 in the border zone; Fig. 6 L and M]. Furthermore, eGFP− cell delivery did not result in homing to infarcted hearts, and eGFP− cells did not augment cardiac function post-MI. Cardiac MRI was utilized in a similar manner, and we did not observe any improvement in left ventricular EF in mice receiving eGFP− cells (EF pretreatment: 27.57% ± 7.203 vs. after 1 mo: 18.22 ± 4.385; SI Appendix, Fig. S5G). Immunostaining of heart tissues was also carried out to examine the presence/differentiation of cTnT+ actinin+ eGFP− cells, which was not observed (SI Appendix, Fig. S5H). These results demonstrate that i.v. delivery of fetal placental Cdx2-eGFP cells resulted in specific “homing” directly to the injury and border zones to facilitate cardiovascular regeneration, and this was associated with significant enhancement in contractile function and reduced adverse cardiac remodeling.
Fig. 6.
Cdx2-eGFP cells contributed to cardiac repair in vivo. (A) Schema illustrating MI induction and Cdx2 cell delivery in vivo. (B) Cardiac MRI analyses showing a significant increase in EF in Cdx2 cell-injected mice versus the control animals at 1-mo and 3-mo time points compared with the pretreatment timeline (Cdx2 group, n = 8; control) group, n = 5). Data are represented as mean ± SEM. #P = 0.0263 (control vs. test at 1 mo), **P = 0.0095 (test before treatment vs. test at 1 mo post-MI and *P = 0.0269 test 3 mo post-MI). (C–E) Left ventricular functional parameters, including SV, LVEDV, and LVESV, as measured in control and cell-treated groups showed a reduction in adverse remodeling. (C) SV is significantly higher in the test group at 1 mo versus the control group at 1 mo post-MI (#P < 0.05). SV within the test cohort before treatment and after Cdx2-eGFP cell injection: before treatment test vs. test at 1 mo post-MI (*P = 0.0121). (D) LVEDV indexed to bovine serum albumin (BSA) did not show significant change in control and test mice at any time point. (E) Cdx2-eGFP cell delivery significantly reduced ESV compared with the control mice at 3 mo post-MI (##P = 0.00045). (F–H) Cdx2-eGFP cells “homed” to injured heart and differentiated into CMs in vivo. CMs expressing α-sarcomeric actinin derived from Cdx2-eGFP cells in the border zone are shown. The panels show striated CMs with nuclear eGFP [Alexa 647, pink; nuclei, blue (DAPI)]. (Scale bar: 10 μm.) (Right) Yellow arrows indicate Cdx2-derived CMs highlighted in the Z-stack view. Blue arrows indicate the same highlighted cell within the Z-stacks, demonstrating nuclear eGFP signal within an α-sarcomeric actinin+ cell (Movies S12–S14). (I) Cdx2-eGFP cells contributed to blood vessel formation in vivo. (Scale bar: 20 μm.) (Right) Border zone shows CD31+ SMA+ cells with nuclear eGFP incorporated into blood vessels highlighted with white arrows. (J) Flow cytometry analysis revealed that the homing and retention of Cdx2-eGFP cells are specific to the injured myocardium and not to any noninjured organs. Quantification of the percentage of Cdx2-eGFP–derived CMs (K) and vascular cells (L and M) is shown in heart sections in vivo in the infarct zone, border zone, and distal zone, demonstrating differentiation into cardiovascular lineages. Data are presented as mean ± SEM.
Discussion
Our data establish the existence of multipotent cells capable of cardiac and vascular differentiation that can be isolated from mouse placenta. Originating from the Cdx2 lineage, these cells can be identified as a distinct subtype within the placenta. Of note, an array of stem cell types, including resident cardiac progenitor cells, mesenchymal stem cells (MSCs), and bone marrow stem cells, have been studied as possible sources for cardiomyogenic differentiation (27–29). The concept of resident cKit+ cells being true cardiac progenitor cells has recently been disproven. Comprehensive lineage-tracing studies by independent groups contradict earlier results and suggest that these cells may have a very limited ability (0.05%) to replace damaged myocytes (30, 31). On the other hand, adult hematopoietic progenitor-derived “CMs” apparently lacked contractile function in vitro and failed to form mature CMs in vivo (32, 33). Studies of MSCs in cardiomyogenesis have resulted in conflicting reports suggesting a lack of functional coupling following transplantation and subsequent failure in forming mature CMs (33, 34). ES cells do form beating clusters in vitro; however, feasibility, ethical concerns, and the propensity of teratoma initiation by these cells in vivo have dampened enthusiasm for clinical applications (35, 36). Induced pluripotent stem cells (iPSCs) have been studied extensively for their role as an autologous patient-specific cell source; however, the risk of arrhythmias (37) and teratoma formation (38, 39) is of significant concern. More recently, it was demonstrated that despite the improvement in ventricular function, transplantation of ESC/iPSC-derived CMs displays limited engraftment, suggesting a paracrine mode of action similar to MSCs and other adult stem cells (40). However, in placenta-derived cells, chorionic plate cells from human placenta have been shown to form CMs with action potentials in vitro, suggesting that placenta may contain cells with cardiomyogenic capability (41). After we had noted a prevalence of Cdx2 cells within mixed populations of migrating fetal cells to maternal hearts, Li et al. (42) reported that trophoblasts isolated from murine blastocysts were superior to MSCs for cardiac repair in a mouse model of MI.
Our current study highlights a regenerative role specifically for fetal-derived placental Cdx2 cells from end-gestation placentas, in that they are able to selectively and specifically “home” through the circulation to injured hearts and effect significant and sustained enhancement of left ventricular function in a mouse model of MI. Adverse remodeling after MI can progressively lead to heart failure. In this study, we noticed that Cdx2-eGFP cells reduced adverse remodeling (especially that of end-systolic volume), improved SV, and significantly enhanced the left ventricular EF. Cdx2-derived cells from placenta could thus be a unique, clinically viable, plentiful, and expandable cell source for cardiac repair with an inherent homing ability that favors a less invasive i.v. route of delivery. We observed that Cdx2-eGFP cells differentiated largely into CMs, followed by endothelial cells and smooth muscle cells, in the injured myocardium. In the border zone, Cdx2-eGFP–derived CMs displayed a rod-shaped appearance with striated sarcomeres, suggesting maturation similar to the adjacent endogenous myocytes. As regenerative mechanisms warrant a greater metabolic rate and blood supply in the injured areas, a proangiogenic mechanism additionally favors the functional recovery of injured myocardium. It is important that Cdx2-eGFP cells exhibited CM differentiation and vascular commitment in vivo, signifying a wider implication in cardiac functional recovery post-MI. The eGFP− cells from the placenta did not home to the heart, and they did not show cardiac repair ability in vivo; however, a few cells did show “CM-like” staining for cTnT in vitro owing to the possible lower retention of maternal Cdx2 (Fig. 1E) in them. However, our transgenic strategy relied on the isolation of placental fetal-derived Cdx2 lineage cells, as we have observed previously that fetal cells can home naturally to the injured heart.
It is interesting to note that the expression of Cdx2 has not so far been associated with a cardiac fate choice during early embryonic patterning but has been implicated as a master regulator of trophoblast stem cells (18). Our findings may therefore represent a paradigmatic shift in the way we approach early embryonic lineages. Our data underscore that Cdx2-derived cells can be isolated from placenta and differentiate into CMs in vitro and in vivo, leading to significant improvement of cardiac function in a mouse model of acute MI. Moreover, they appear to be multipotent in nature with the ability to form endothelial and smooth muscle cells, which are essential components of the myocardial structural matrix (1). It may be quite plausible to speculate that this particular characteristic feature may have wider implications in other organ injury settings as well.
Placental Cdx2 cells also exhibit the ability to clonally proliferate in vitro, supporting their “stem cell” functions. Stem/progenitor cell therapy is a very promising approach for cardiac repair provided the donor cells do not evoke a strong immune response in the host environment. Low or absent MHC expression on Cdx2 cells is a major benefit for any allogeneic cell therapy strategy. The placenta is crucial for fetomaternal tolerance; hence, the immunological properties of placenta-derived stem cells are of special interest. We observed that placental Cdx2 cells lack the surface expression of MHC class I, MHC class II, and nonclassical H2-T23 (Qa-1) molecules, indicating an immunologically naive phenotype. We have shown that the presence of costimulatory molecules, interleukins, and TLRs was very low in isolated Cdx2 cells. CXCl10, a proinflammatory cytokine implicated in tissue injury and immune rejection, was highly expressed. However, we found that CXCR3, the putative ligand required for functional CXCL10 interactions (43), was not present at either the transcriptional or surface protein level. Thus, with respect to innate and adaptive immunity markers, this suggests that Cdx2 cells may represent an ideal approach for allogeneic cell therapy. Additionally, the robust engraftment, proliferation, and differentiation of Cdx2-eGFP cells in the host myocardium noted even after 3 mo of cell therapy, mostly in the infarct and periinfarct zones of the heart, highlighted their safe retention, survival, and function in vivo.
Fetal stem cells derived from placentas may possess some characteristic features similar to ES cells. ES cells are derived from the ICM of the blastocyst, which is essentially of non-Cdx2 lineage, whereas the epiblast or outer layer is uniquely marked by Cdx2 expression (18, 24). Considering this developmental cue, we examined the characteristics of Cdx2-eGFP cells compared with ES cells. Mass spectrometry and subsequent IPA demonstrated that fetal Cdx2-derived cells possess unique proteins compared with ES cells. The enrichment of proteins that also have a role in the dynamics of cardiac sarcomere organization could imply a potential procardiac signature in Cdx2-eGFP cells. Noteworthy was the expression of β-parvin (affixin), an adaptor protein that is important for actin dynamics and is also reported to exhibit cardioprotective effects when bound to STAT3 (transcript abundance of stat3 noted in isolated Cdx2-eGFP cells) (44). Additionally, proteins like sorcin, involved in calcium handling and in rhythmic contractility (45, 46), were uniquely identified in the Cdx2-eGFP cell population. Based on these findings, one can rationally assume that the proteome signature of Cdx2-eGFP cells could have exerted a conducive and synergistic effect on regeneration and functional recovery in a post-MI setting. We further note that the up-regulated proteome of Cdx2 cells reflected roles in migration, chemotaxis, cell movement, and survival/antiapoptosis. Also noteworthy was the specific positive correlation of the proteins that modulate cardiac hypertrophic signaling. Moreover, Cdx2-eGFP cells responded efficiently to SDF1 stimulation in vitro with an approximate migration efficiency of 63%. Moreover, blocking the functional SDF1-CXCR4 signaling adversely affected the Cdx2-eGFP cell response to chemokine SDF1α, suggesting that homing responses are, in part, mediated by the SDF1-CXCR4 axis. This functional implication uncovered by our proteomics data was also substantiated by identifying Cdx2-eGFP cells entirely in the injured myocardium and adjacent areas, and not in any noninjured tissues in vivo.
As we move toward translation of these data and begin to characterize human CDX2 cells, a number of objectives will need to be addressed. Since we are able to isolate CDX2 cells from human term placentas, we must first identify the unique surface proteins of human CDX2 cells to develop a clinical strategy to isolate live cells for use in cell therapy. Furthermore, we must exclude the propensity of teratoma formation by CDX2 cells. These studies, along with the critical proteomics profiling of human CDX2 cells, are underway. Maternal fetal cell transfer is a phenomenon that has been reported for decades, and our laboratory has demonstrated a functional significance for the flux of fetal cells into the maternal circulation (12). Due to a dearth of clinically feasible cell types for actual therapeutic use in cardiac repair, we sought to explore whether this “naturally” occurring biological pathway could be exploited for cardiac regenerative purposes. In so doing, we have identified a cell type in placenta that can clonally proliferate and differentiate into spontaneously beating CM and vascular cells in vitro, can “naturally” and robustly home to injured myocardium in a selective manner, is able to evade host immune surveillance, and can generate CMs and vascular cells in vivo to result in significant and sustained enhancement of cardiac contractility. We can now envision translation of these findings to develop a human allogeneic cell therapy for cardiac repair that may be delivered via an i.v. route.
Methods
Mouse Models.
Adult WT C57/BL6J (JAX:000664) mice were purchased from The Jackson Laboratory. For lineage-tracing experiments, we crossed 8-w-old female virgin B6; 129S6-gt (ROSA) 26Sor tm1 (CAG-tdTomato*, EGFP*)Ees>/J43 (JAX stock no. 023035) with 10- to 12-wk-old male B6.Cg-Tg(CDX2-cre)101Erf/J(JAX stock no. 009350) to induce pregnancy (Fig. 1C and SI Appendix).
MI and Cdx2 Cell Infusion.
MI was induced in 10-wk-old male C57/BL6J mice by permanent ligation of the LAD (details are provided in SI Appendix). At 1 wk post-MI, MRI was carried out and the mice were blindly randomized to control and test groups. The test groups received 1 million Cdx2-eGFP cells in PBS through the tail vein, and the control groups received PBS. The animals were monitored, and MRI was carried out using a 7T Bruker Biospec 70/30 scanner (Bruker Corporation) 1 wk post-MI, and 1 mo and 3 mo posttreatment to analyze cardiac function. Cardiologists who were blinded to the treatment groups analyzed cardiac functional parameters from MRI. The left ventricular end-diastolic volume (LVEDV) and left ventricular end-systolic volume (LVESV) were measured, and the SV was calculated as the difference between the LVEDV and LVESV. The EF was measured from SV/LVEDV × 100. In parallel experiments, eGFP− cells were also delivered i.v., and cardiac MRI was carried out after 1 mo to analyze cardiac function.
Statistical Analysis.
One-way ANOVA with a post-Bonferroni test was used to compare datasets with more than two groups (Fig. 6 B–E, K, and L and SI Appendix, Fig. S4A). An unpaired t test was used to analyze the rest of the datasets. Data are presented as mean ± SEM unless otherwise indicated. The number of biological and technical replicates is indicated in figure legends wherever appropriate. A two-tailed alpha level of 0.05 was used, and a P value of ≤0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Drs. Mone Zaidi, Debra Wolgemuth, and Judith Swain for critical review of the data and manuscript. We thank the Flow Cytometry Core Facility, Imaging Core Facility, Pathology Core Facility, and Center for Comparative Medicine and Surgery at the Icahn School of Medicine at Mount Sinai for technical assistance. This study was supported by New York Stem Cell Board funds (IIRP Contract C029565 to H.W.C.).
Footnotes
Conflict of interest statement: H.W.C. is the founder and an equity holder in VentriNova, Inc. and is the inventor on a pending patent regarding Cdx2 cells and cardiac repair.
This article is a PNAS Direct Submission.
Data deposition: The mass spectrometry proteomics data have been deposited on the ProteomeXchange Consortium via the PRIDE partner repository (accession ID PXD011783).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811827116/-/DCSupplemental.
References
- 1.Xin M, Olson EN, Bassel-Duby R (2013) Mending broken hearts: Cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol 14:529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmann O, et al. (2009) Evidence for cardiomyocyte renewal in humans. Science 324:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stamm C, et al. (2003) Autologous bone-marrow stem-cell transplantation for myocardial regeneration. Lancet 361:45–46. [DOI] [PubMed] [Google Scholar]
- 4.White IA, Sanina C, Balkan W, Hare JM (2016) Mesenchymal stem cells in cardiology. Methods Mol Biol 1416:55–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida Y, Yamanaka S (2017) Induced pluripotent stem cells 10 years later: For cardiac applications. Circ Res 120:1958–1968. [DOI] [PubMed] [Google Scholar]
- 6.Shapiro SD, et al. (2014) Cyclin A2 induces cardiac regeneration after myocardial infarction through cytokinesis of adult cardiomyocytes. Sci Transl Med 6:224ra27. [DOI] [PubMed] [Google Scholar]
- 7.Bersell K, Arab S, Haring B, Kühn B (2009) Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138:257–270. [DOI] [PubMed] [Google Scholar]
- 8.Tokita Y, et al. (2016) Repeated administrations of cardiac progenitor cells are markedly more effective than a single administration: A new paradigm in cell therapy. Circ Res 119:635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang XL, et al. (2016) Long-term outcome of administration of c-kit(pos) cardiac progenitor cells after acute myocardial infarction: Transplanted cells do not become cardiomyocytes, but structural and functional improvement and proliferation of endogenous cells persist for at least one year. Circ Res 118:1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguirre A, et al. (2014) In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell 15:589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, et al. (2015) Microrna-34a plays a key role in cardiac repair and regeneration following myocardial infarction. Circ Res 117:450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kara RJ, et al. (2012) Fetal cells traffic to injured maternal myocardium and undergo cardiac differentiation. Circ Res 110:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Coppi P, et al. (2007) Isolation of amniotic stem cell lines with potential for therapy. Nat Biotechnol 25:100–106. [DOI] [PubMed] [Google Scholar]
- 14.Trounson A, McDonald C (2015) Stem cell therapies in clinical trials: Progress and challenges. Cell Stem Cell 17:11–22. [DOI] [PubMed] [Google Scholar]
- 15.Herberts CA, Kwa MS, Hermsen HP (2011) Risk factors in the development of stem cell therapy. J Transl Med 9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khosrotehrani K, Bianchi DW (2005) Multi-lineage potential of fetal cells in maternal tissue: A legacy in reverse. J Cell Sci 118:1559–1563. [DOI] [PubMed] [Google Scholar]
- 17.Bianchi DW, Zickwolf GK, Weil GJ, Sylvester S, DeMaria MA (1996) Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci USA 93:705–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ralston A, Rossant J (2008) Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol 313:614–629. [DOI] [PubMed] [Google Scholar]
- 19.Jedrusik A, et al. (2008) Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev 22:2692–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verzi MP, et al. (2010) TCF4 and CDX2, major transcription factors for intestinal function, converge on the same cis-regulatory regions. Proc Natl Acad Sci USA 107:15157–15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niwa H, et al. (2005) Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123:917–929. [DOI] [PubMed] [Google Scholar]
- 22.Gharibi T, Ahmadi M, Seyfizadeh N, Jadidi-Niaragh F, Yousefi M (2015) Immunomodulatory characteristics of mesenchymal stem cells and their role in the treatment of multiple sclerosis. Cell Immunol 293:113–121. [DOI] [PubMed] [Google Scholar]
- 23.Neefjes J, Jongsma ML, Paul P, Bakke O (2011) Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol 11:823–836. [DOI] [PubMed] [Google Scholar]
- 24.Sritanaudomchai H, et al. (2009) CDX2 in the formation of the trophectoderm lineage in primate embryos. Dev Biol 335:179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penn MS, Pastore J, Miller T, Aras R (2012) SDF-1 in myocardial repair. Gene Ther 19:583–587. [DOI] [PubMed] [Google Scholar]
- 26.Cheng RK, et al. (2007) Cyclin A2 induces cardiac regeneration after myocardial infarction and prevents heart failure. Circ Res 100:1741–1748. [DOI] [PubMed] [Google Scholar]
- 27.Bondue A, et al. (2008) Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell 3:69–84. [DOI] [PubMed] [Google Scholar]
- 28.Boyle AJ, McNiece IK, Hare JM (2010) Mesenchymal stem cell therapy for cardiac repair. Methods Mol Biol 660:65–84. [DOI] [PubMed] [Google Scholar]
- 29.Hatzistergos KE, et al. (2010) Bone marrow mesenchymal stem cells stimulate cardiac stem cell proliferation and differentiation. Circ Res 107:913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Berlo JH, Molkentin JD (2016) Most of the dust has settled: Ckit+ progenitor cells are an irrelevant source of cardiac myocytes in vivo. Circ Res 118:17–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sultana N, et al. (2015) Resident c-kit(+) cells in the heart are not cardiac stem cells. Nat Commun 6:8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murry CE, et al. (2004) Haematopoietic stem cells do not transdifferentiate into cardiac myocytes in myocardial infarcts. Nature 428:664–668. [DOI] [PubMed] [Google Scholar]
- 33.Siegel G, et al. (2012) Bone marrow-derived human mesenchymal stem cells express cardiomyogenic proteins but do not exhibit functional cardiomyogenic differentiation potential. Stem Cells Dev 21:2457–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karantalis V, Hare JM (2015) Use of mesenchymal stem cells for therapy of cardiac disease. Circ Res 116:1413–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nussbaum J, et al. (2007) Transplantation of undifferentiated murine embryonic stem cells in the heart: Teratoma formation and immune response. FASEB J 21:1345–1357. [DOI] [PubMed] [Google Scholar]
- 36.Chung J, et al. (2011) In vivo molecular MRI of cell survival and teratoma formation following embryonic stem cell transplantation into the injured murine myocardium. Magn Reson Med 66:1374–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiba Y, et al. (2016) Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 538:388–391. [DOI] [PubMed] [Google Scholar]
- 38.Choi HW, et al. (2015) In vivo reprogrammed pluripotent stem cells from teratomas share analogous properties with their in vitro counterparts. Sci Rep 5:13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z, et al. (2013) The tumourigenicity of iPS cells and their differentiated derivates. J Cell Mol Med 17:782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tachibana A, et al. (2017) Paracrine effects of the pluripotent stem cell-derived cardiac myocytes salvage the injured myocardium. Circ Res 121:e22–e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okamoto K, et al. (2007) ‘Working’ cardiomyocytes exhibiting plateau action potentials from human placenta-derived extraembryonic mesodermal cells. Exp Cell Res 313:2550–2562. [DOI] [PubMed] [Google Scholar]
- 42.Li G, et al. (2017) Cardiac repair in a mouse model of acute myocardial infarction with trophoblast stem cells. Sci Rep 7:44376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piper KP, et al. (2007) CXCL10-CXCR3 interactions play an important role in the pathogenesis of acute graft-versus-host disease in the skin following allogeneic stem-cell transplantation. Blood 110:3827–3832. [DOI] [PubMed] [Google Scholar]
- 44.Luedde M, et al. (2011) Affixin (β-parvin) promotes cardioprotective signaling via STAT3 activation. J Mol Cell Cardiol 50:919–923. [DOI] [PubMed] [Google Scholar]
- 45.Farrell EF, Antaramian A, Rueda A, Gómez AM, Valdivia HH (2003) Sorcin inhibits calcium release and modulates excitation-contraction coupling in the heart. J Biol Chem 278:34660–34666. [DOI] [PubMed] [Google Scholar]
- 46.Collis LP, et al. (2007) Expression of a sorcin missense mutation in the heart modulates excitation-contraction coupling. FASEB J 21:475–487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.