Significance
None of the single-cell RNA sequencing (scRNA-seq) studies published so far convincingly identified human γδ T lymphocytes despite their anticancer functions. To address this, we here report scRNA-seq of γδ T lymphocytes purified from human blood, and a signature identifying γδ T cells. The single-cell transcriptomes of TCRVδ1 and TCRVδ2 lymphocytes are intermediates resembing those of NK cells and T CD8, respectively, they reflect their respective maturation stage and their pseudo-time maturation trajectory unveils terminally differentiated cells with mitotic capacity. In human cancers, the γδ TIL, mostly expressing TCRVδ1, appear fewer in tumors than in blood and not correlated to αβ TIL abundance. This report is a landmark resource for future studies of γδ T lymphocytes at the single-cell level.
Keywords: γδ T lymphocyte, transcriptome, single-cell RNA-sequencing, human immunology, cancer
Abstract
γδ T lymphocytes represent ∼1% of human peripheral blood mononuclear cells and even more cells in most tissues of vertebrates. Although they have important anticancer functions, most current single-cell RNA sequencing (scRNA-seq) studies do not identify γδ T lymphocytes because their transcriptomes at the single-cell level are unknown. Here we show that high-resolution clustering of large scRNA-seq datasets and a combination of gene signatures allow the specific detection of human γδ T lymphocytes and identification of their T cell receptor (TCR)Vδ1 and TCRVδ2 subsets in large datasets from complex cell mixtures. In t-distributed stochastic neighbor embedding plots from blood and tumor samples, the few γδ T lymphocytes appear collectively embedded between cytotoxic CD8 T and NK cells. Their TCRVδ1 and TCRVδ2 subsets form close yet distinct subclusters, respectively neighboring NK and CD8 T cells because of expression of shared and distinct cytotoxic maturation genes. Similar pseudotime maturation trajectories of TCRVδ1 and TCRVδ2 γδ T lymphocytes were discovered, unveiling in both subsets an unattended pool of terminally differentiated effector memory cells with preserved proliferative capacity, a finding confirmed by in vitro proliferation assays. Overall, the single-cell transcriptomes of thousands of individual γδ T lymphocytes from different CMV+ and CMV− donors reflect cytotoxic maturation stages driven by the immunological history of donors. This landmark study establishes the rationale for identification, subtyping, and deep characterization of human γδ T lymphocytes in further scRNA-seq studies of complex tissues in physiological and disease conditions.
Single-cell level mRNA-sequencing (scRNA-seq) of heterogeneous cell populations has become the reference tool for establishing cellular lineages and composition of tissues from the human body (1). In addition, the development of novel and open-source computational tools for processing scRNA-seq datasets enables delineation of both broad and subtle differences in rare cell subsets present in complex mixtures, such as peripheral blood mononuclear cells (PBMC) (2, 3). Such developments are expected to build more knowledge about the cellular composition and states composing tumors. Recent scRNA-seq analyses in melanoma and colorectal cancer evidenced the various tumor microenvironments and exhaustion patterns of the tumor-infiltrating lymphocytes (4). Most of such studies currently focus on the cytotoxic CD8 T lymphocytes, which represent the main target of immune checkpoint blockade therapies, and are readily detected by their scRNA-seq profile. Other subsets of cytolytic T cells are also critical in this therapeutic perspective; however, currently they have never been characterized by scRNA-seq and are therefore missing from all current tumor microenvironments mapped by these technologies.
Human γδ T lymphocytes represent a peculiar lymphoid cell subset displaying hallmarks of both innate and adaptive immunity, and reacting to microbial pathogens and malignancies (5). These CD4− CD8− T cells express a somatically rearranged T cell receptor (TCR), as well as cytotoxicity receptors more typically expressed by NK cells. The diversity of γ- and δ-encoded TCR relies on VJ rearrangements of the TRG locus at 7p14 and VDJ rearrangements of the TRD locus at 14q11.2, respectively. In the human genome, the TRD locus contains three variable genes (TRDV1–3), three diversity genes (TRDD1–3), four joining genes (TRDJ1–4) and one constant TRDC gene, which rearrange to encode a TCRδ chain. The TRG locus that rearrange to encode a TCRγ chain contains 14 variable genes, of which only 6 are functional (TRGV2–5, TRGV8, and TRGV9), five joining genes (TRGJ1, J2, JP, JP1, JP2), and two constant genes (TRGC1, TRGC2). Despite this low number of TCR-encoding gene segments, the human TCR repertoire of γδ T cells is almost as diversified as that of αβ T lymphocytes. In addition, the repertoire of γδ TCR expressed at the cell surface of γδ T cells is biased according to their tissue localization. TRGC1 encodes for the γ-constant region of the cell surface TCRVγ9Vδ2 expressed by the most abundant γδ T lymphocytes in human adult blood, a subset of γδ T cells detecting microbial and tumoral metabolites called phosphoantigens (PAgs) (6) associated to the non-HLA butyrophilin-3 molecule (7). In contrast, TRGC2 encodes for the γ-constant region shared by all of the cell surface TCRVγ(non-9)Vδ2-, TCRVδ1-, and TCRVδ3-expressing γδ T cells, which are generally less frequent than Vγ9Vδ2 cells in adult blood but predominate in other tissues, and recognize different antigens. The TCRVδ1+ lymphocytes represent the prominent non-Vγ9Vδ2 γδ T cell subset and are mainly located in adult skin, lung, intestine, and colon epithelia, where they recognize antigens from virally infected and cancer cells (8).
Similarly, non-(TCRVδ2) γδ T cells are induced by environmental cytomegalovirus (CMV) (9), are associated with a reduced risk of cancer in immunosuppressed patients (10), and some of these lymphocytes recognize the endothelial protein C receptor overexpressed by carcinoma cells (11). TCRVδ3 cells represent a rarer γδ T cell subset in blood, and some TCRVγ8Vδ3 T lymphocytes recognize Annexin A2 from stressed and cancer cells (12). Hence, all of the TCR-based subsets of γδ T cells might participate to antitumor immunity, although by coreceptors and functions depending on the stage of maturation reached by these T lymphocytes. Upon antigenic stimulation, the γδ T lymphocytes successively mature from naïve (CD27+, CD62L+ CCR7+, CD45RA+) cells to central memory cells (CD27+, CD62L+ CCR7+, CD45RA−) with strong proliferative and low effector function. Upon further Ag stimulation, they may further mature into effector memory cells (CD27−, CD45RA− lymphocytes producing either IFN-γ or granzyme/perforin), and finally drive to terminally differentiated CD45RA-expressing terminally differentiated effector memory (TEMRA) cells (CD45RA+ CD16+) essentially mediating the ADCC-type of cytotoxic function. This maturation pathway, spanning from naïve to TEMRA cells, was identified in TCRVδ2+ γδ T lymphocytes, whose TCR activation precedes and progressively drives expression of cytotoxicity receptors shared with NK cells (13–16). Other γδ T cells, such as the CMV-reactive γδ T lymphocytes, also predominantly display a TEMRA and CD16+ phenotype with adaptive-like response to CMV (17–20). Most TCRVδ1+ γδ T cells may expand in a CDR3-independent (21), but AKT/γc cytokine-driven fashion (22), and progressively express cytolysis-inhibiting as well as natural cytotoxicity receptors. Hence whatever the TCR subset, this blend of innate and adaptive skills makes all γδ T lymphocytes with NK-like functions attractive candidates for controlling viral infections (23) and cancer (5, 24). Given the recent developments in adoptive γδ T cell therapies of cancer (25), it is important to know whether all subsets of γδ T cells mature similarly, but this remains unclear so far.
Furthermore, for cancer therapy, determining the rate of tumor-infiltrating γδ lymphocytes (γδ TIL) from any tumor biopsy is critical. CIBERSORT is a recent algorithm deconvoluting the composition of TILs from microarrays of cancer biopsies (26), and its use to analyze 19,000 tumors concluded that rate of γδ TILs positively correlates with good outcome (27). Although encouraging, such results suffered of poor learning from too few (only two) γδ T cell transcriptomes, however, as CIBERSORT identifies erroneously most of CD8 T, NK cells, and γδ T lymphocytes (28). This problem reflects the massive gene multicollinearity of transcriptomes from these three closely related cell types (29), suggesting that deeper learning from many more γδ T cell transcriptomes is necessary. In addition to unfaithfully identifying γδ T cells as a whole, determining their subsets defined by cell surface TCR and stage of maturation is currently out of reach for the same reasons. Thus, a decisive milestone would be the straightforward identification of γδ T lymphocytes from scRNA-seq data. Such an achievement could allow us to determine their presence, their TCR, maturation stage, and activation/exhaustion status in the tumor microenvironment of a large panel of human cancers. Nevertheless, in this aim it remains necessary to identify γδ T lymphocytes from nonmalignant reference tissue samples, such as PBMC from healthy individuals. Because these lymphocytes are unfrequent (1–5% of PBMC) and their transcriptomes share cytotoxic hallmarks of both CD8 T and NK cells (29), they are not identified as such in current t-distributed stochastic neighbor embedding (t-SNE) plots of scRNA-seq datasets, which posit each cell within its “alikes” to construct clusters. Hence, in most t-SNE published so far, γδ T lymphocytes are generally embedded within larger clusters of CD8 T and NK cells without being correctly identified, despite their biological importance.
To overcome these problems, herein we produced scRNA-seq transcriptomes of blood γδ T lymphocytes of TCRVδ1+ and TCRVδ2+ subsets purified from three healthy donors, and depicted their respective gene-expression profiles at this high-definition level. These data represent a valuable resource for further studies delineating the landscape of γδ T lymphocytes in human physiology and disease.
Results
High Clustering Levels and High-Resolution t-SNE Plots of Large Datasets Are Required to Spot γδ T Lymphocytes from scRNA-Seq of Complex Tissues.
A CMV+ healthy donor #1 comprising 2.67% of γδ T lymphocytes in PBMC was selected for this study; ∼1,000 PBMC isolated from this donor were analyzed by scRNA-seq by using the 10x Genomics single-cell 3′ V2 chemistry, followed by sequencing of 96 million reads per library. After alignment on the GRCh38 transcriptome, a total of 15,309 genes were detected with an average of 120,000 reads for 1,314 unique genes per cell. Because γδ T cells represent only ∼1–5% of PBMC and cell resolution of scRNA-seq analyses increases with larger datasets, we also downloaded from 10x Genomics two additional scRNA-seq datasets produced by the same technology of 4k and 8k PBMC derived from one healthy donor (https://support.10xgenomics.com/single-cell-gene-expression/datasets). These latter datasets represent 4,340 PBMC with an average of 87,000 reads for 1,235 genes per cell (4k dataset), and 8,381 PBMC with an average of 95,000 reads for 1,297 genes per cell (8k dataset). These three datasets, totaling ∼13,000 PBMC, were integrated, aligned, log-normalized using the “Seurat alignment” workflow (2), and quality-controlled (QC) for number of genes per cell and mitochondrial gene content. Twelve thousand single cells passed the QC filter, with an average of 1,320 unique genes detected per cell. Unsupervised analysis of this 12k PBMC dataset identified 11 statistically significant principal components used by the Seurat package’s functions FindClusters and RunTSNE to partition the dataset into several clusters of cells. The nonlinear dimensionality reduction of this dataset by t-SNE (30) using a low (i.e., 0.6) resolution of this graph-based clustering produced eight clusters of cells visualized on a t-SNE map (Fig. 1A). The mRNA biomarkers identified these clusters as those of classic and intermediate monocytes (e.g., LYZ, S100A9, CD14), dendritic cells (DC) (LYZ, S100A9, FCER1A), intermediate monocytes (CD14−, LYZ, S100A9, FCGR3A), B cells (CD79A, CD79B, CD19), naïve and central memory CD4 and CD8 T cells (CD3E, CD4 or CD8A, SELL, CD27), effector memory CD4 and CD8 T cells (CD3E, CD4 or CD8A, CD27, PRF1, GNLY), and NK cells (CD3E-negativeNKG7 cells) (Fig. 1B). This classification was consistent with hematopoietic differentiation and the previously published t-SNE plots of PBMC scRNA-seq. (31–33).
Fig. 1.
Visualization of γδ T lymphocytes from PBMC scRNA-seq requires high resolution and clustering depth t-SNE plots of large datasets. (A) Unsupervised hierarchical clustering and correspondingly colored t-SNE plot at low resolution (granularity 0.6) of 12k PBMC partitions eight clusters lacking a γδ T lymphocyte cluster. (B) Expression levels of the specified biomarker genes (purple) on the same t-SNE plot. (C) Unsupervised hierarchical clustering and correspondingly colored t-SNE plot at high resolution (granularity 1.2) of the same dataset partition 17 clusters, of which one (boxed area) comprises cytotoxic CD8 T and γδ T cells. (D) Zoom-in of the boxed region (Top Right) with expression levels (purple) of the specified genes.
However, despite showing that transcriptomic signatures of blood T lymphocytes primarily reflect maturation stage rather than merely CD4 or CD8 lineages, this t-SNE plot did not evidence a γδ T cell cluster. To delineate such a cluster, expected to encompass infrequent CD3+CD4−CD8− cells expressing TRDC that encodes for the unique constant region of the TCRδ chain, the same dataset was analyzed as above and then visualized by t-SNE plots with a higher (i.e., 1.2) resolution. This partitioned 17 clusters that did not subdivide the previous NK cell cluster, but created six clusters of monocytes and DCs, two B cell clusters, and eight T cell clusters discriminating CD4 from CD8 T cells and their maturation stages (e.g., SELL, CD27, ITGAL, IL7R) (Fig. 1C). In these settings, the most mature cytotoxic T cell cluster (defined by FCGR3A and GNLY) included both CD3E+CD8A+ T cells and CD3E+CD4−CD8A− cells that expressed the TCRδ constant region-encoding segment TRDC, presumably corresponding to γδ T lymphocytes (Fig. 1D). Thus, high-resolution clustering of large scRNA-seq datasets and selected sets of genes may allow for the spotting of γδ T lymphocytes in complex cell mixtures from heterogeneous tissues, such as a bulk PBMC.
A Gene Signature to Detect Human γδ T Lymphocytes in Complex Cell Mixtures.
In peripheral blood of adults, γδ T lymphocytes usually represent 1–4% of PBMC and present a [CD3+TCRγδ+CD4−CD8−] cell surface phenotype. Nevertheless, here the corresponding [CD3E+TRDC+CD4−CD8A−] gene-expression criterion identified n = 862 cells (7.2%) of the 12k PBMC dataset (Fig. 2A). This probably overestimated γδ T lymphocytes, as n = 487 of these cells were likely false-positives dispersed in the myeloid, B cells, NK cells, CD4, and CD8 T lymphocytes clusters. Consistent with the transcriptome hallmarks of bulk γδ T lymphocytes (29), however, n = 375 γδ T cell candidates were mapped between the cytotoxic CD8 T and NK cell clusters. Nevertheless, the single-cell transcriptomes of cytotoxic CD8 T and NK cells perplexed the delineation of γδ T lymphocytes. On the one hand, frequent single cells of the NK cluster from this PBMC dataset (SI Appendix, Fig. S1A) and from other datasets of purified NK cells (34) (SI Appendix, Table S1) strongly express the γδ TCR-encoding gene segments TRDC, and more weakly the TRGC1 and TRGC2 segments. On the other hand, cytotoxic CD8 T lymphocytes frequently and strongly express TRGC2, but weakly TRDC and TRGC1 (SI Appendix, Fig. S1B).
Fig. 2.
Gene signatures identifying γδ T lymphocytes. (A) In the 12k PBMC dataset, n = 862 (7%) PBMC express the CD3E and TRDC but not CD4 and not CD8A genes (red), including n = 375 cells mapped between the cytotoxic CD8 T and NK cell clusters as expected for γδ T lymphocytes. (B) In the same dataset, n = 515 (4%) PBMC are detected by the combined signature score (SI Appendix, Material and Methods), including n = 418 cells mapped between the cytotoxic CD8 T and NK cell clusters. (C) In the dataset integrating both 12k PBMC and 1,333 purified γδ T lymphocytes from CMV+ donor #1, n = 1,683 cells are identified as γδ T lymphocytes. These include the above 418 PBMC-derived γδ T embedded in the n = 1,265 purified γδ T lymphocytes. (D) ROC curves for γδ T cell detection in the 13k dataset, using the genes depicted in A, or the signature combination used for B.
Furthermore, blood γδ T lymphocytes comprise variable rates of cells with TCRVδ2 encoded by the TRGC1 and TRDC gene segments, and of cells with TCRVδ1 encoded by the TRGC2 and TRDC gene segments, although both subsets express some level of each other’s TRGC1 and TRGC2 (SI Appendix, Fig. S1B). Therefore, to identify γδ T lymphocytes exhaustively and without NK and T CD8 false-positives, we designed a more performant γδ signature combining two gene sets (SI Appendix, Material and Methods) that was scored for each single cell and visualized in the t-SNE by Single-Cell Signature Explorer (35). In the 12k PBMC dataset, this γδ signature score was >0.35 for n = 515 cells, including 418 (3.4% of PBMC) cells mapped between the T CD8 and NK cell clusters (Fig. 2B). To validate this γδ signature, highly pure (>99%) (SI Appendix, Material and Methods) γδ T lymphocytes were positively sorted by anti-TCRVδ1 and anti-TCRVδ2 mAbs from the same CMV+ blood donor #1 as for the above PBMC. After scRNA-seq, preprocessing, and QC filtering as above, the pooled transcriptomes of n = 1,333 of 1,529 purified TCRγδ+ T lymphocytes were obtained, with a mean of 263k reads for 12,412 unique genes per cell. After integration, alignment, and processing with the above 12k PBMC dataset, the resulting 13k dataset was visualized by high-resolution t-SNE plot. Most (n = 1,265 of 1,333, 95%) purified γδ T lymphocytes colocalized with the 418 PBMC between T CD8 and NK, and (n = 1,020 of 1,265, 80%) were positive for the γδ signature (Fig. 2C). When the performances of the [CD3E+TRDC+CD4−CD8A−] gene set and the γδ signature were compared for detecting γδ T lymphocytes within the 13k dataset, their ROC curves showed that the γδ signature was superior (Fig. 2D). These and additional results (see below) validated the identification of γδ T lymphocytes by the combination signature.
Using the same processing and combination signature, the rate of γδ T cells was determined in additional scRNA-seq datasets of PBMC from healthy donors and tumor biopsies from cancer patients. This approach identified 3.2% and 5% of γδ T lymphocytes in PBMC from two other healthy donors (Fig. 3 A and B). Although TILs are less abundant in tumors, some γδ TILs could nevertheless be detected in such samples. Among n = 2,933 total blood cells from a relapsed chronic lymphocytic leukemia (CLL) patient, n = 23 TILs, including n = 3 γδ T lymphocytes, were detected (Fig. 3C). In the tumor biopsy from a freshly diagnosed follicular lymphoma (FL) (36), n = 13 γδ T lymphocytes of 472 TILs (3.3%) were detected (Fig. 3D and SI Appendix, Fig. S2), consistent with the 5% γδ TILs rate reported in this malignancy (27, 28, 37). In a nonsmall cell lung cancer (NSCLC) tumor sample comprising 252 TILs, only n = 3 γδ T lymphocytes (1.2% of TIL) were detected, while n = 10 γδ T of 620 TILs (1.6% of TIL) were detected in a lung adenocarcinoma (LUAD) biopsy (Fig. 3 E and F and SI Appendix, Table S2).
Fig. 3.
Identifying γδ T lymphocytes in t-SNE plots of other PBMC and tumor samples. (A and B) Same visualization as above for γδ T lymphocytes (orange-red dots) from other examples of PBMC from two healthy donors, (C) blood cells from a relapsed CLL patient (present study), (D) tumor biopsy from a diagnosed FL patient (36), (E) and tumor biopsies from a patient with NSCLC, and (F) from a patient with LUAD (66). Arrows: γδ T cells. The cell clusters of PBMC are specified.
Distinct Clusters for TCRVδ1 and TCRVδ2 γδ T Lymphocytes.
The distinct partitions of TRGC1 and TRGC2 gene expression in the 12k PBMC t-SNE plot (SI Appendix, Fig. S1A) suggested that blood γδ T lymphocytes expressing TCRVγ9Vδ2 (encoded by TRGC1 and TRDC) and those expressing TCRVγ(non-9)Vδ1 (encoded by TRGC2 and TRDC) have different transcriptomes. To validate this observation, the above 1,333 purified γδ T lymphocytes were analyzed separately according to their cell surface TCRVδ1 or TCRVδ2. These encompassed n = 570 of 641 purified TCRVδ1 γδ T lymphocytes with a mean of 155k reads for 1,374 unique genes per cell, and n = 763 of 888 purified TCRVδ2 γδ T lymphocytes, with a mean of 108k reads for 1,323 unique genes per cell. In the high-resolution t-SNE plot of the integrated 13k dataset, these cells segregated in neighboring but distinct regions: the TCRVδ2 T lymphocytes mapped next to the mature CD8 T cells, whereas the TCRVδ1 T lymphocytes were closer to the NK cells. Unsupervised hierarchical clustering of the 13k dataset confirmed this partition (SI Appendix, Fig. S3). Of note, the expression levels of TRGC1 and TRGC2 was inversed between the two subsets of purified γδ T cells [TRGC1 mean (log2 unique molecular identifier [UMI] = 0.9 in TCRVδ1+ cells versus 1.7 in TCRVδ2+ cells, TRGC2 mean = 2.3 in TCRVδ1+ cells versus 0.8 in TCRVδ2+ cells)]. Thus, their significantly different (TRGC1-TRGC2) expression values (Student P = 10−235) characterized the TCRVδ1 and TCRVδ2 subtypes of γδ T cells identified in the above-depicted datasets (SI Appendix, Table S2). Hence, the 515 γδ T cells from the 12k PBMC comprised n = 195 TCRVδ1, n = 244 TCRVδ2, and n = 76 TCR-unassigned γδ T lymphocytes. From this and three additional datasets totaling 28,339 PBMC (SI Appendix, Table S2), there were a total of n = 263 TCRVδ1 cells (on average 0.9% of PBMC), n = 460 TCRVδ2 (on average 1.6% of PBMC), and n = 197 TCR-unassigned γδ T lymphocytes (∼0.6% of PBMC), totaling n = 920 γδ T lymphocytes or 3.2% of PBMC as expected in normal blood. However, in 26 cancer samples totaling 25,658 cells, the γδ TIL were consistently more scarce, n = 115 γδ T lymphocytes (0.4% of total cells), and were mostly of TCRVδ1+ subtype (n = 52 lymphocytes, 0.9% of total cells; n = 8 TCRVδ2 cells; n = 55 TCR-unassigned γδ T). In addition, although γδ TIL counts and subsets varied between patients, the γδ T lymphocytes detected in lung cancer samples often came from their normal adjacent tissue. Finally, across these cancer samples, the rates of γδ TIL and αβ TILs were not correlated (Pearson r = 0.12) (SI Appendix, Table S2), as reported previously (28).
Shared and Distinct Cytotoxic Genes of TCRVδ1 and TCRVδ2 γδ T Lymphocytes.
Together, the n = 1,265 purified TCRVδ1 and TCRVδ2 γδ T cells mapped in the γδ cell area of the t-SNE had 470 differentially expressed genes compared with the (n = 7,071) αβ T lymphocytes (both CD4 and CD8 T cell clusters from the 13k integrated dataset), with significantly higher expression of the cell cytotoxicity genes. They also had 570 differentially expressed genes relative to the (n = 493) NK cells from the NK cluster, with significantly stronger expression of the T cell-specifying genes (SI Appendix, Tables S3 and S4), consistent with earlier microarray studies (29).
At the single-cell level, few genes were selectively expressed by most γδ T cells relative to all of the rest of the PBMC. These included FGFBP2 encoding for the cytotoxic serum protein fibroblast growth factor-binding protein 2, MT2A encoding for metallothionein 2A, CST7 encoding for the cysteine protease inhibitor Cystatin F, HOPX encoding for the homeobox transcription repressor X, GZMM encoding for granzyme M, and KLRD1 encoding for the cell cytotoxicity coreceptor CD94, although these genes were also expressed by some cells from other subsets of PBMC (SI Appendix, Fig. S4). Typically, 99% of the purified γδ T lymphocytes from donor #1, but also 99% of NK cells and 93% of mature cytotoxic (FCGR3A+) CD8 T lymphocytes from the 13k dataset expressed NKG7 encoding for the NK cell granule protein 7 (Fig. 4A). Similar results were observed in additional PBMC datasets (SI Appendix, Table S2). Most purified γδ T lymphocytes also expressed mRNA encoding for several others typical cytotoxic but not γδ T cell-specific mediators, such as GZMM (87% of cells), the granulysin-encoding GNLY (78% of cells), and the perforin-encoding PRF1 (66% of cells), to quote a few. Similar expression rates (GZMM: 80%, GNLY: 91%, PRF1: 86%), were found for the bulk γδ T lymphocytes from the 12k PBMC scRNA-seq dataset. Consistent with the TCRVδ2 T lymphocyte maturation pathway depicted previously (13, 14), both FACS- purified TCRVδ1 and TCRVδ2 cells expressed variable levels of the maturation marker genes ITGAL (CD11b) and SELL (CD62L) characterizing naïve T cells, as well as CD27, CD28, and CD44 shared by central and effector memory T cells, and FCGR3A (encoding for CD16) typifying the TEMRA T lymphocytes (Fig. 4B and SI Appendix, Figs. S4 and S5).
Fig. 4.
Distinctive gene-expression patterns of TCRVδ1 and TCRVδ2 γδ T lymphocytes from CMV+ donor #1. (A) Zoom-in of the same cytotoxic cell area of t-SNE plot as in Fig. 1, shown with purple color-encoded expression level of NKG7 gene across the NK cells, CD8 T lymphocytes, TCRVδ1, and TCRVδ2 γδ T lymphocytes. (B) Same as in A for expression of the specified genes. (C) Representative genes differentially expressed between TCRVδ1 and TCRVδ2 γδ T lymphocytes. (D) Functional enrichment of the specified signatures in each single cell from the integrated PBMC and purified γδ T cells dataset is shown by featuring the single-cell enrichment score (i.e., the proportion of UMI for genes from the signature and expressed by each cell divided by the total UMI of the cell) (SI Appendix, Material and Methods), and contour plots of the region with differentially enriched signature over the t-SNE plot (region of cells having signature score >2× mean score of t-SNE). They comprise a negative control signature (12 randomly selected genes, RGS407_12 from this study), B cell activation [132 genes, gene ontology (GO)], dendritic cell differentiation (33 genes, GO), NK cell activation (55 genes, GO), regulation of fever generation (11 genes, GO), regulation of macrophage chemotaxis (16 genes, GO), positive regulation of T cell cytotoxicity (16 genes, GO), cytosolic DNA sensing: (56 genes, Kyoto Encyclopedia of Genes and Genomes). The corresponding genes are listed in SI Appendix, Table S6.
Overall, 38 genes were differentially expressed between the (n = 570) purified TCRVδ1 and (n = 763) purified TCRVδ2 γδ T lymphocytes from CMV+ donor #1. These comprised 25 genes, such as KLRC2 (NKG2A), KLRF1 (NKp80), FCGR3A (CD16), GZMB, and GZMH, which were up-regulated in TCRVδ1 cells, and 13 genes, including KLRC1 (NKG2C), GZMK, LTB, and IL7R, which were up-regulated in TCRVδ2 cells (Fig. 4 B and C and SI Appendix, Table S5). These gene-expression profiles were consistent with the immunostaining and flow cytometry phenotypes of the γδ T lymphocytes from the same CMV+ PBMC donor (SI Appendix, Fig. S5). Nevertheless, the clear-cut segregation of KLRC2/KLRC1 expression by the TCRVδ subsets from this CMV+ PBMC donor #1 could not be generalized, as it disappeared when examining the single γδ T cell transcriptomes from additional scRNA-seq datasets (SI Appendix, Table S2) and when phenotyping the γδ PBMC from 15 additional CMV+ kidney transplant recipients (SI Appendix, Fig. S6).
The gene-expression programs of all cells from the 13k PBMC dataset were finally visualized by scoring each single cell for expression of functionally defined pathways, and comparing these single-cell scores over the t-SNE. In control tests, no subset of PBMC showed up-regulation of random gene signatures, while the B cell activation signature was found in B cell clusters, and the DC differentiation signature was found in myeloid cell clusters, as expected. This analysis revealed that among the most salient signatures of the γδ T lymphocytes, and to some extent of the surrounding NK and CD8 T cell clusters, were “NK cell cytotoxicity,” “positive regulation of T cell cytotoxicity,” “regulation of fever generation,” “regulation of macrophage chemotaxis,” and “cytosolic DNA sensing” signatures (Fig. 4D and SI Appendix, Table S6). In contrast, no curated signature was found specific for either of the purified TCRVδ1 or the purified TCRVδ2 γδ T cell subsets in these analyses. Thus, these two subsets of γδ T lymphocytes share a cytotoxic signature comprising both common and distinct genes.
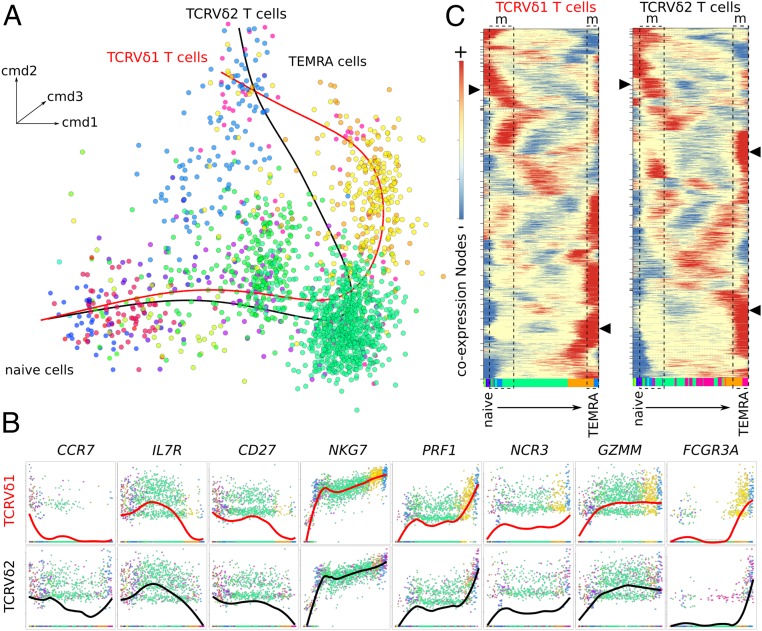
Similar Pseudotemporal Maturation Trajectories of TCRVδ1 and TCRVδ2 γδ T Lymphocytes Unveil Mature γδ T Cells with Proliferative Capacity.
The fate of peripheral γδ T lymphocytes relies on a maturation process driven by antigen and cytokine stimulation. This maturation starts from naïve resting cells that evolve to central memory cells able to proliferate, and may further mature into effector memory cells, which produce either IFN-γ or Perforin, and finally terminally differentiated, CD45RA+CD16+ cytotoxic cells with short telomeres, named TEMRA (13–15). Whether this seemingly linear maturation trajectory is validated by an unsupervised algorithmic derivation of high-resolution differentiation trajectories deserved clarification. Furthermore, whether Vδ1 and Vδ2 γδ T cells follow the same scheme is unclear (22, 38). To address these issues, the purified γδ T cells of the 13k PBMC dataset and the bulk cells colocalized within the purified γδ T lymphocyte clusters from t-SNE plot (n = 1,683 lymphocytes) were selected and their TCRVδ lineage was inferred on the basis of TRGC1 and TRGC2 expressions. A pseudotemporal trajectory of maturation was then reconstructed using the FateID package (39), which orders topologically the successive expression profiles, and draws a differentiation trajectory fitting to all ordered cells, visualized by a self-organizing pseudotemporal map. This revealed that TCRVδ1 and TCRVδ2 γδ T lymphocytes display quite parallel maturation trajectories (Fig. 5A). Both subsets expressed the maturation-defining genes CCR7, IL7R, SELL, ITGAL, and CD27 at the earliest stages only, while the cytotoxicity genes NKG7, PRF1, NCR3, GZMM, and FGFBP2 increased at later time points. Both subsets also encompassed FCGR3A-expressing mature cells, although more frequently among the TCRVδ1 than the TCRVδ2 γδ T lymphocytes from CMV+ donor #1, suggesting their more advanced cytotoxic maturation (Fig. 5B).
Fig. 5.
Pseudotemporal reconstruction of the γδ T cell maturation trajectory. (A) Tridimensional t-SNE representation of γδ T cell data after classic multidimensional scaling and dimensionality reduction. Data shown represent n = 1,683 cells, and principal curves fitted to either the TCRVδ1 γδ T lymphocytes (red line) or the TCRVδ2 γδ T lymphocytes (black line). Cells are colored according to trajectory subclusters. (B) Gene-to-gene scatterplots of marker genes expression in the TCRVδ1 γδ T lymphocytes and TCRVδ2 γδ T lymphocytes from CMV+ donor #1 across their respective maturation trajectories. The average level is shown in bold line. (C) Self-organizing map of z-score–transformed pseudotemporal coexpression nodes profiled along the respective maturation trajectories of TCRVδ1 γδ T lymphocytes and TCRVδ2 γδ T lymphocytes. The areas (dotted rectangle) labeled “m” show maturation stages with coexpression nodes (arrows) significantly enriched for mitosis/cell cycle signatures. The bottom color bar shows the appearance order of each maturation subcluster of cells, using the same color keys as in A.
Chronological ordering of the z-score–transformed pseudotemporal expression profiles of the TCRVδ1 and TCRVδ2 γδ T lymphocytes identified several nodes of gene coexpression (Fig. 5C). For each cell subset, the gene signatures significantly enriched at all these nodes were identified as above. This analysis revealed two mitotic windows at early, presumably naïve/central memory stages, and at late mature/TEMRA stage (Fig. 5C). Consistent with this finding, flow cytometry analysis of far red cell tracker (CFTR) dilution by PBMC stimulated in vitro with either PHA/IL-2 as TCRVδ1-stimulus (22) or BrHPP/IL-2 as TCRVδ2-stimulus (40) confirmed that some TEMRA γδ T lymphocytes had proliferated after 8 d of in vitro culture. Although in each subset the divided cells were mostly naïve and central memory lymphocytes, as expected from earlier literature (13–15), both PHA/IL-2–stimulated TCRVδ1 and BrHPP/IL-2–stimulated TCRVδ2 γδ T lymphocytes from healthy donors comprised some terminally differentiated γδ lymphocytes with mitotic capacity, as evidenced by the diluted intracellular CFTR of such cells with intermediate CD45RA phenotype after 14 d of culture (SI Appendix, Fig. S7). To confirm that TEMRA cells are able to proliferate, CMV-infected kidney graft recipient’s PBMC with TCRVδ1 γδ T cells composed of more than 90% of TEMRA cells were cultured with IL-2 and IL-15. After 7 d of culture, the number of TCRVδ1 T cells was increased by 10-fold (SI Appendix, Fig. S8A) and these were still predominantly TEMRA cells (SI Appendix, Fig. S8B). CFSE [5-(and 6)-Carboxyfluorescein diacetate succinimidyl ester] staining of PBMC and gating of TEMRA cells only at day 7 confirmed that TEMRA TCRVδ1 cells were able to proliferate in the presence of IL-2 and IL-15 but did not proliferate in the absence of activation (SI Appendix, Fig. S8C).
Hence, the pseudotemporal maturation trajectories correctly identified the terminally differentiated TCRVδ1 and TCRVδ2 γδ T lymphocytes, which maintained a proliferative capacity.
Maturation Stages of TCRVδ1 and TCRVδ2 γδ T Lymphocytes from CMV+ and CMV− Donors.
The above results from a single γδ donor of CMV+ status questioned their generalization. To address this, the circulating TCRVδ1 and TCRVδ2 cells were purified from two additional donors of known CMV status, and their scRNA-seq were performed and analyzed as above. For the CMV+ donor #2, these encompassed n = 1,775 of 1,800 purified TCRVδ1 γδ T lymphocytes totalizing 13,241 unique genes and a median of 1,311 unique genes per cell, as well as n = 1,277 of 1,306 purified TCRVδ2 γδ T lymphocytes, totaling 12,787 unique genes and a median of 1,370 unique genes per cell. For the CMV− donor #3, these encompassed n = 1,803 of 1,825 purified TCRVδ1 γδ T lymphocytes totaling 13,335 unique genes and a median of 1,197 unique genes per cell, as well as n = 1,720 of 1,750 purified TCRVδ2 γδ T lymphocytes totaling 13,283 unique genes and a median of 1,419 unique genes per cell. Altogether, these (n = 6,575) additional purified γδ T lymphocyte datasets were integrated, aligned, and processed with the previous 13k dataset, yielding a 20k dataset that was visualized by high-resolution t-SNE plot, as above. Scoring and projecting the γδ signature across the 20k t-SNE, which included 7,150 γδ T cells, both 6,575 purified and the 575 γδ T already in the 12k PBMC (SI Appendix, Table S7) identified a total of n = 5,995 γδ T lymphocytes (84% sensitivity). As previously, these cells neighbored both CD8 T and NK cells and each donor yielded two distinct clusters of γδ T lymphocytes corresponding to each TCRV subset (SI Appendix, Fig. S9A).
In this t-SNE, the cytotoxic hallmark genes NKG7, GZMH, GZMK, KLRC1, and FCGR3A were expressed by the γδ T lymphocytes from all donors, except KLRC2, which was only expressed by the FCGR3A (TEMRA) TCRVδ1 cells from donor #1 (SI Appendix, Fig. S9B). For donors #2 and #3, the genes differentially expressed between TCRVδ1 and TCRVδ2 γδ T lymphocytes (SI Appendix, Tables S8 and S9) were reminiscent of those reported above for donor #1, and reflected their respective maturation stage in each donor. In t-SNE of purified γδ T cells alone, the embedding of TCRVδ1 and TCRVδ2 γδ T cells from the two CMV+ donors #1 and #2 indicated these subsets share transcriptional hallmarks that TCRVδ1 and TCRVδ2 γδ T cells from the CMV− donors #3 do not (SI Appendix, Fig. S9C). The respective cell counts of the naïve, central memory, effector memory, and effector memory RA cells in each TCRVδ subset from each donor accounted for the above results (SI Appendix, Table S10). These data unveiled the prominence of mature TCRVδ1 cells in CMV+ donor #1 and of mature TCRVδ2 cells in CMV− donor #3, while CMV+ donor #2 shared both hallmarks with each respective donor (SI Appendix, Fig. S9D).
Thus, in healthy individuals, the single-cell transcriptomes of circulating TCRVδ1 and TCRVδ2 γδ T lymphocytes merely reflect their immune and CMV-driven maturation.
Discussion
The identification of human γδ T lymphocytes is a major issue for cancer immunotherapy, as these pleiomorphic cells infiltrating most healthy organs are often misidentified when not unidentified at all. In this aim, detection of γδ T lymphocytes classically relies on a combination of cell surface markers labeled by immunostaining and visualized by flow cytometry or immunohistochemistry. These techniques require access to fresh tumor samples and, furthermore, immunohistochemical spotting of cells expressing the γδ TCR remains largely hampered by the paucity of anti-TCR reagents compatible with staining of formalin-fixed paraffin-embedded samples. However, the recent developments in scRNA sequencing are updating these methodologies, and they provide us with the finest level of resolution for the gene-expression patterns of large sets of cells, unveiling much finer aspects of cells and allowing their classifications from human tissues.
We report here scRNA-seq datasets of human γδ T lymphocytes as bulk cells among unsorted PBMC, and as highly purified lymphocytes. These transcriptomes were integrated to those of a pool of several bulk PBMC, because t-SNE plots from larger datasets provide a wider and more comprehensive view of all cell components from complex tissues. In addition, t-SNE hyperparameters, such as clusterization level, determine the visualization of the underlying data output by this nondeterminist algorithm. These two features raise the issue of spotting in large t-SNE maps those rare subsets of cells, like γδ T lymphocytes, which transcriptomes resemble those of different and more abundant cell types, and without any selectively identifying gene. This issue was exemplified here by the strikingly high TRDC gene expression by NK cells, a hitherto unreported (34) transcriptional hallmark supporting the innateness gradient of lymphoid cells (41). In the absence of techniques such as the recently developed CITE-Seq combining scRNA-seq and detection of cells displaying surface markers (33), which fails at detecting the γδ TCR, spotting the γδ T lymphocytes was elusive, and actually never done in published scRNA-seq studies. This pitfall is typically exemplified by recently improved scRNA-seq processing algorithms, which outperform the other softwares for execution time, clustering accuracy, and detectability of minor cell subtypes (SI Appendix, Material and Methods), but still fail at spotting any γδ T lymphocytes among ∼68,000 PBMC scRNA-seq (42). We show here that identification of γδ T lymphocytes requires not only large datasets and highly granular t-SNE plots, but also a γδ signature combining γδ T-scoring with NK and T CD8-discarding genes. This allowed us to detect the γδ T lymphocytes and identify their subsets in healthy individuals PBMC and in tumor biopsies from cancer patients. Our study represents a resource to identify γδ T lymphocytes from human tissues, as undertaken by the human cell atlas project (1), and to determine their fonctional status in cancer patients under immunotherapies.
The second conclusion of this study is the signature of γδ T from PBMC that still remains at the single-cell level a profound blend in of the cytotoxic signatures of mature CD8 T and NK cells. Collectively, this cytotoxic hallmark is today well documented through microarrays and phenotypic studies of bulk TCRVδ1 T lymphocytes and TCRVδ2 T lymphocytes, taken separately (29, 43–49). As a consequence, several clinical trials in cancer are currently aimed at assessing the therapeutic activity of allogeneic or autologous TCRVδ1 or TCRVδ2 γδ T cell transplants produced by various methodologies. Nevertheless, these earlier studies had demonstrated that TCRVδ1 T lymphocytes and TCRVδ2 T lymphocytes mediate distinct cytotoxic responses. In particular, cytotoxicity-controlling receptors, such as NKG2A, NKG2C, NKG2D, NKp30, and CD85, were reported as a differential hallmark of these two main subsets of γδ T cells (22, 24, 50). Indeed, although a segregation of KLRC1, 2, and GZMB, H expressions according to the TCRVδ subset can be occasionally seen as in CMV+ donor #1, this differential hallmark was not a general rule. Instead, the NK receptor pattern displayed by the various γδ T cell subsets could reflect distinct maturation stages driven by prior exposure to activating ligands (reviewed in ref. 24), as shown by the gene-expression profiles along maturation trajectories. Pioneering studies (48, 51) have demonstrated age-related changes in the γδ repertoire. TCRVδ2 γδ T cells are a minority of blood γδ T cells at birth, but expand later to constitute the majority of γδ T cells in adults blood while TCRVδ1 γδ T cells have no such expansion. Recent RNA-seq studies (19, 20) have shown that while the TCRVδ2 repertoire remains almost equivalent in cord and adult blood, the private and unfocused TCRVδ1 repertoire in cord blood strongly focuses in few high-frequency adult clonotypes. Such clonal expansions are accompanied by maturation from naive to effector phenotypes, consistent with the gene-expression profiles promoted by physiological immune processes (52), exemplified here by the different TCRVδ1 transcriptomes of the CMV+ and CMV− donors.
Beyond cytotoxic hallmarks, however, our study also detected enrichment of a gene signature dedicated to regulation of fever generation in the single cells of γδ T lymphocytes. This finding was unattended for human blood γδ T cells, but fully consistent with the recent discovery that mice lacking γδ T lymphocytes cannot regulate their core body temperature (53). Taken together, those observations raise the emerging possibility that, like their invariant NK T cell counterparts, the γδ T cells might fulfill physiological functions of immune-mediated regulation of body metabolism (54).
Despite the current model stating that terminally differentiated γδ T lymphocytes have progressively lost their proliferative capacity (13–15), the third conclusion of this study is that some terminally differentiated TEMRA cells from both TCRVδ1 and TCRVδ2 subsets of γδ T lymphocytes have kept a significant proliferative capacity. This proliferative capacity remains in a CD45RAintermediate subpopulation of TEMRA cells, however, fully consistent with a recent report depicting two populations of TEMRA TCRVδ2 cells in solid cancer patients (55). This unattended finding arose from the unbiased algorithmic spotting of mitotic gene signatures in cells positioned at the end of chronologically ordered maturation trajectories, and was validated by further flow cytometric analyses and in vitro proliferation assays. On the one hand, such results illustrate the resolutive power of the scRNA-seq technologies that were not available when the first γδ maturation model was elaborated, and on the other hand they prompt new maturation models. These findings also reconciles the previously unexplained—although largely used—capacity to grow in vitro large-scale and clinical grade batches of terminally differentiated TCRVδ2 γδ T lymphocytes for cell-therapy purposes in patients with metastatic renal cell carcinoma (56), of TCRVδ1 γδ T lymphocytes for cell therapy of chronic lymphocytic leukemia (25), or total γδ T cells for cell therapy in neuroblastoma (57) or in ovarian cancer (58). Beyond cancer, mature γδ T lymphocytes contribute to control infections such as HIV disease (59) and CMV reactivation (60, 61). Therefore, terminally differentiated TCRVδ1 and TCRVδ2 γδ T lymphocytes can improve the prevention of early relapse after allogeneic hematopoietic stem cell transplantation (62), and are developed for cell therapy by haploidentical mature γδ T cell infusions (63). Newer developments of expanding cell batches of terminally differentiated γδ T lymphocytes are also emerging for CAR-T cell therapy (64), and will benefit of the present in-depth characterization of these γδ T lymphocytes at the single cell level (65).
Materials and Methods
Informed consent and ethical approval was obtained for blood samples (Bordeaux University Hospital; ethical committee agreement 2013/57). The isolation and phenotyping of PBMC and purified γδ T lymphocytes, in vitro proliferation assays, scRNA sequencing and analyses, including single-cell signatures and scores, maturation trajectories, additional datasets, and statistical analyses are described in SI Appendix, Material and Methods.
Supplementary Material
Acknowledgments
We thank J.-J.F. team members and Cancer Research Center of Toulouse colleagues for stimulating discussions; Asaf Madi and Aviv Regev (Broad Institute, Boston) for sharing their R scripts; and Sagar (Max Planck Institute, Freiburg) for advice on trajectory computations. This work was supported in part by institutional grants from INSERM, Université Toulouse III: Paul Sabatier, CNRS, Laboratoire d’Excellence Toulouse Cancer (Contract ANR11-LABX), Programme Hospitalo-Universitaire en Cancérologie CAPTOR (Contract ANR11-PHUC0001), SIRIC Bordeaux Research in Oncology, Institut Carnot Lymphome (CALYM), Ligue Nationale Contre le Cancer (J.-D.M.), Fondation pour la Recherche Médicale (H.K.), University of Palermo, and Humanitas University (G.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.K.B. is a guest editor invited by the Editorial Board.
Data deposition: The scRNASeq data were deposited in the NCBI Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE128223).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818488116/-/DCSupplemental.
References
- 1.Regev A., et al. ; Human Cell Atlas Meeting Participants , The human cell atlas. eLife 6, e27041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papalexi E., Satija R., Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 18, 35–45 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Tirosh I., et al. , Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonneville M., O’Brien R. L., Born W. K., Gammadelta T cell effector functions: A blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 10, 467–478 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Fournié J. J., Bonneville M., Stimulation of gamma delta T cells by phosphoantigens. Res. Immunol. 147, 338–347 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Harly C., et al. , Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human γδ T-cell subset. Blood 120, 2269–2279 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halary F., et al. , Shared reactivity of Vdelta2(neg) gammadelta T cells against cytomegalovirus-infected cells and tumor intestinal epithelial cells. J. Exp. Med. 201, 1567–1578 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couzi L., Pitard V., Moreau J. F., Merville P., Déchanet-Merville J., Direct and indirect effects of cytomegalovirus-induced γδ T cells after kidney transplantation. Front. Immunol. 6, 3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couzi L., et al. , Cytomegalovirus-induced gammadelta T cells associate with reduced cancer risk after kidney transplantation. J. Am. Soc. Nephrol. 21, 181–188 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willcox C. R., et al. , Cytomegalovirus and tumor stress surveillance by binding of a human γδ T cell antigen receptor to endothelial protein C receptor. Nat. Immunol. 13, 872–879 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Marlin R., et al. , Sensing of cell stress by human γδ TCR-dependent recognition of annexin A2. Proc. Natl. Acad. Sci. U.S.A. 114, 3163–3168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dieli F., et al. , Differentiation of effector/memory Vdelta2 T cells and migratory routes in lymph nodes or inflammatory sites. J. Exp. Med. 198, 391–397 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Angelini D. F., et al. , FcgammaRIII discriminates between 2 subsets of Vgamma9Vdelta2 effector cells with different responses and activation pathways. Blood 104, 1801–1807 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Caccamo N., et al. , Differential requirements for antigen or homeostatic cytokines for proliferation and differentiation of human Vgamma9Vdelta2 naive, memory and effector T cell subsets. Eur. J. Immunol. 35, 1764–1772 (2005). [DOI] [PubMed] [Google Scholar]
- 16.von Lilienfeld-Toal M., et al. , Activated gammadelta T cells express the natural cytotoxicity receptor natural killer p 44 and show cytotoxic activity against myeloma cells. Clin. Exp. Immunol. 144, 528–533 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitard V., et al. , Long-term expansion of effector/memory Vdelta2-gammadelta T cells is a specific blood signature of CMV infection. Blood 112, 1317–1324 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couzi L., et al. , Antibody-dependent anti-cytomegalovirus activity of human γδ T cells expressing CD16 (FcγRIIIa). Blood 119, 1418–1427 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Davey M. S., et al. , Clonal selection in the human Vδ1 T cell repertoire indicates γδ TCR-dependent adaptive immune surveillance. Nat. Commun. 8, 14760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravens S., et al. , Human γδ T cells are quickly reconstituted after stem-cell transplantation and show adaptive clonal expansion in response to viral infection. Nat. Immunol. 18, 393–401 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Boullier S., Cochet M., Poccia F., Gougeon M. L., CDR3-independent gamma delta V delta 1+ T cell expansion in the peripheral blood of HIV-infected persons. J. Immunol. 154, 1418–1431 (1995). [PubMed] [Google Scholar]
- 22.Correia D. V., et al. , Differentiation of human peripheral blood Vδ1+ T cells expressing the natural cytotoxicity receptor NKp30 for recognition of lymphoid leukemia cells. Blood 118, 992–1001 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Gougeon M. L., Boullier S., Colizzi V., Poccia F., NKR-mediated control of gammadelta T-cell immunity to viruses. Microbes Infect. 1, 219–226 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Correia D. V., Lopes A., Silva-Santos B., Tumor cell recognition by γδ T lymphocytes: T-cell receptor vs. NK-cell receptors. OncoImmunology 2, e22892 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almeida A. R., et al. , Delta one T cells for immunotherapy of chronic lymphocytic leukemia: Clinical-grade expansion/differentiation and preclinical proof of concept. Clin. Cancer Res. 22, 5795–5804 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Newman A. M., et al. , Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gentles A. J., et al. , The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tosolini M., et al. , Assessment of tumor-infiltrating TCRVγ9Vδ2 γδ lymphocyte abundance by deconvolution of human cancers microarrays. OncoImmunology 6, e1284723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pont F., et al. , The gene expression profile of phosphoantigen-specific human γδ T lymphocytes is a blend of αβ T-cell and NK-cell signatures. Eur. J. Immunol. 42, 228–240 (2012). [DOI] [PubMed] [Google Scholar]
- 30.van der Maaten L., Hinton G., Visualising data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
- 31.Fan X., et al. , Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16, 148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng C., et al. , Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169, 1342–1356.e16 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Stoeckius M., et al. , Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crinier A., et al. , High-dimensional single-cell analysis identifies organ-specific signatures and conserved NK cell subsets in humans and mice. Immunity 49, 971–986.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pont F., Tosolini M., Fournie J. J., Single-cell signature explorer for comprehensive visualization of single cell signatures across scRNA-seq data sets. bioRxiv:10.1101/621805 (29 April 2019). [DOI] [PMC free article] [PubMed]
- 36.Milpied P., et al. , Human germinal center transcriptional programs are de-synchronized in B cell lymphoma. Nat. Immunol. 19, 1013–1024 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Rossi C., et al. , Boosting γδ T cell-mediated antibody-dependent cellular cytotoxicity by PD-1 blockade in follicular lymphoma. OncoImmunology 8, 1554175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hudspeth K., et al. , Engagement of NKp30 on Vδ1 T cells induces the production of CCL3, CCL4, and CCL5 and suppresses HIV-1 replication. Blood 119, 4013–4016 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Herman J. S., Sagar D., Grün D., FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat. Methods 15, 379–386 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Espinosa E., et al. , Chemical synthesis and biological activity of bromohydrin pyrophosphate, a potent stimulator of human gamma delta T cells. J. Biol. Chem. 276, 18337–18344 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Gutierrez-Arcelus M., et al. , Lymphocyte innateness defined by transcriptional states reflects a balance between proliferation and effector functions. Nat. Commun. 10, 687 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinha D., Kumar A., Kumar H., Bandyopadhyay S., Sengupta D., dropClust: Efficient clustering of ultra-large scRNA-seq data. Nucleic Acids Res. 46, e36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fahrer A. M., et al. , Attributes of gammadelta intraepithelial lymphocytes as suggested by their transcriptional profile. Proc. Natl. Acad. Sci. U.S.A. 98, 10261–10266 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kress E., Hedges J. F., Jutila M. A., Distinct gene expression in human Vdelta1 and Vdelta2 gammadelta T cells following non-TCR agonist stimulation. Mol. Immunol. 43, 2002–2011 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Vermijlen D., et al. , Distinct cytokine-driven responses of activated blood gammadelta T cells: Insights into unconventional T cell pleiotropy. J. Immunol. 178, 4304–4314 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dieli F., et al. , Targeting human gammadelta T cells with zoledronate and interleukin-2 for immunotherapy of hormone-refractory prostate cancer. Cancer Res. 67, 7450–7457 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beetz S., et al. , Innate immune functions of human gammadelta T cells. Immunobiology 213, 173–182 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Vermijlen D., et al. , Human cytomegalovirus elicits fetal gammadelta T cell responses in utero. J. Exp. Med. 207, 807–821 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dimova T., et al. , Effector Vγ9Vδ2 T cells dominate the human fetal γδ T-cell repertoire. Proc. Natl. Acad. Sci. U.S.A. 112, E556–E565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lança T., et al. , The MHC class Ib protein ULBP1 is a nonredundant determinant of leukemia/lymphoma susceptibility to gammadelta T-cell cytotoxicity. Blood 115, 2407–2411 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Parker C. M., et al. , Evidence for extrathymic changes in the T cell receptor gamma/delta repertoire. J. Exp. Med. 171, 1597–1612 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huygens A., Dauby N., Vermijlen D., Marchant A., Immunity to cytomegalovirus in early life. Front. Immunol. 5, 552 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohlgruber A. C., et al. , γδ T cells producing interleukin-17A regulate adipose regulatory T cell homeostasis and thermogenesis. Nat. Immunol. 19, 464–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lynch L., et al. , iNKT cells induce FGF21 for thermogenesis and are required for maximal weight loss in GLP1 therapy. Cell Metab. 24, 510–519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Odaira K., et al. , CD27(-)CD45(+) γδ T cells can be divided into two populations, CD27(-)CD45(int) and CD27(-)CD45(hi) with little proliferation potential. Biochem. Biophys. Res. Commun. 478, 1298–1303 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Bennouna J., et al. , Phase-I study of Innacell gammadeltatrade mark, an autologous cell-therapy product highly enriched in gamma9delta2 T lymphocytes, in combination with IL-2, in patients with metastatic renal cell carcinoma. Cancer Immunol Immunother 57, 1599–1609 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fisher J. P., et al. , Neuroblastoma killing properties of Vδ2 and Vδ2-negative γδT cells following expansion by artificial antigen-presenting cells. Clin. Cancer Res. 20, 5720–5732 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deniger D. C., et al. , Activating and propagating polyclonal gamma delta T cells with broad specificity for malignancies. Clin. Cancer Res. 20, 5708–5719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pauza C. D., Poonia B., Li H., Cairo C., Chaudhry S., γδ T cells in HIV disease: Past, present, and future. Front. Immunol. 5, 687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaminski H., et al. , Surveillance of γδ T cells predicts cytomegalovirus infection resolution in kidney transplants. J. Am. Soc. Nephrol. 27, 637–645 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scheper W., et al. , γδT cells elicited by CMV reactivation after allo-SCT cross-recognize CMV and leukemia. Leukemia 27, 1328–1338 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Airoldi I., et al. , γδ T-cell reconstitution after HLA-haploidentical hematopoietic transplantation depleted of TCR-αβ+/CD19+ lymphocytes. Blood 125, 2349–2358 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Witte M. A., et al. , Early reconstitution of NK and γδ T cells and its implication for the design of post-transplant immunotherapy. Biol. Blood Marrow Transplant. 24, 1152–1162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du S. H., et al. , Co-expansion of cytokine-induced killer cells and Vγ9Vδ2 T cells for CAR T-cell therapy. PLoS One 11, e0161820 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fournié J. J., Single cell RNAseq of human TCRVdelta 1 and TCRVdelta 2 gammadelta T lymphocytes purified from healthy adults blood. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128223. Deposited 13 March 2019.
- 66.Lambrechts D., et al. , Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.