Significance
Peptide ligases are important precision tools in biotechnology to modify and label proteins in a specific way. Asparaginyl ligases are a promising family for these applications. However, asparaginyl endopeptidases and ligases share a conserved structure, and the precise molecular basis for their opposite activities remains to be determined. Here we define the structural correlates of asparaginyl ligase activity using X-ray crystallography and functional comparisons between homologous plant enzymes extracted from Violaceae coupled to site-directed mutagenesis of their activity determinants.
Keywords: peptide ligase, data mining, ligase-activity determinant
Abstract
Asparaginyl endopeptidases (AEPs) are cysteine proteases which break Asx (Asn/Asp)–Xaa bonds in acidic conditions. Despite sharing a conserved overall structure with AEPs, certain plant enzymes such as butelase 1 act as a peptide asparaginyl ligase (PAL) and catalyze Asx–Xaa bond formation in near-neutral conditions. PALs also serve as macrocyclases in the biosynthesis of cyclic peptides. Here, we address the question of how a PAL can function as a ligase rather than a protease. Based on sequence homology of butelase 1, we identified AEPs and PALs from the cyclic peptide-producing plants Viola yedoensis (Vy) and Viola canadensis (Vc) of the Violaceae family. Using a crystal structure of a PAL obtained at 2.4-Å resolution coupled to mutagenesis studies, we discovered ligase-activity determinants flanking the S1 site, namely LAD1 and LAD2 located around the S2 and S1′ sites, respectively, which modulate ligase activity by controlling the accessibility of water or amine nucleophile to the S-ester intermediate. Recombinantly expressed VyPAL1–3, predicted to be PALs, were confirmed to be ligases by functional studies. In addition, mutagenesis studies on VyPAL1–3, VyAEP1, and VcAEP supported our prediction that LAD1 and LAD2 are important for ligase activity. In particular, mutagenesis targeting LAD2 selectively enhanced the ligase activity of VyPAL3 and converted the protease VcAEP into a ligase. The definition of structural determinants required for ligation activity of the asparaginyl ligases presented here will facilitate genomic identification of PALs and engineering of AEPs into PALs.
Ligases, enzymes which make peptide bonds, are useful biochemical and biotechnological tools. They enable linkage-specific and site-specific protein modifications and precision biomanufacturing of biotherapeutics such as antibody–drug conjugates. For these applications, three families of stand-alone and ATP-independent ligases have been identified. The first family is derived from the bacterial transpeptidases, which include sortase A (1–3). The second family contains modified subtilisins (4–7). However, a large and promising source of ligases was discovered within a third family: plant ligases producing ribosomally synthesized and posttranslationally modified peptides (RiPPs) (8). Many RiPPs contain a head-to-tail cyclic structure that requires either a cyclase or a ligase for posttranslational modification from the linear precursors (9–13). Cyclotide-processing enzymes such as asparaginyl endopeptidases (AEPs) (14) and cyclotide ligases, that are also asparaginyl-specific, were successfully isolated from the cyclotide-producing plant Clitoria ternatea and named butelase 1 and 2 (15). While butelase 2 is a protease, butelase 1 is an extremely efficient peptide asparaginyl ligase (PAL), with the highest reported catalytic efficiency, 1,340,000 M−1⋅s−1. Butelase 1 is a versatile protein-engineering tool for protein and peptide ligation, modification, cyclization, tagging, cyclooligomer formation, and live-cell labeling (16–22). Similarly, two PALs named OaAEP1b (23) and HeAEP3 (24), were later identified from other cyclotide-producing plants, Oldenlandia affinis and Hybanthus enneaspermus, respectively.
AEPs (or legumains) are cysteine proteases belonging to the subfamily C13 (EC 3.4.22.34) under clan CD (25, 26). They are expressed as inactive zymogens which contain a signal peptide, a prodomain, an active core domain, and a C-terminal cap domain. The cap domain is connected to the core domain by a flexible linker. The cap domain is also referred to as an activation peptide, supplemented by a legumain stabilization and activity modulation domain (27), which modulates or inhibits the core domain activity. Under acidic conditions, the zymogen undergoes autolytic activation to remove the proregion and cap domain on both termini of the core domain (28–31). Such autoactivation occurs in lysosomes or plant vacuoles at pH around 4.5–6 (32, 33). In vitro, AEP activations are usually performed at pH 4–5 (27, 34, 35). Notably, activation above pH 4.5 is reversible, as the reassociation of cap and core domains can occur when both domains remain intact and in close proximity after cleavage (34, 36).
Plant AEPs play important roles in protein degradation, maturation, programmed cell death, and host defense via their proteolytic activity triggered in the acidic environment of vacuoles (37–41). They are also known to exhibit ligase activity, although very rarely, such as in the maturation of Con A, by mediating circular permutation (42–44). CeAEP (jack bean; Canavalia ensiformis) (35), PxAEP3 (Petunia x hybrida) (24), and AtLEGγ (Arabidopsis thaliana) (45) catalyze both ligation and hydrolysis products from peptide substrates carrying AEP-recognition signals at near-neutral pH (6–7.5). Other AEPs, such as butelase 2, OaAEP2, and HaAEP1 (sunflower Helianthus annuus), display predominantly protease activity even at neutral pH, with a very low level of ligase activity (35, 45, 46). In contrast to these “bifunctional” or “predominant” AEPs, butelase 1 and OaAEP1b catalyze the formation of ligation products essentially devoid of any hydrolytic product at near-neutral pH, and their ligase activity is preponderant even under mild acidic conditions (pH < 6).
What are the molecular mechanisms differentiating AEPs and PALs? Despite the publication of several plant AEP crystal structures, including both proenzymes and active forms (34, 45–47), the structural determinants that underpin their nature as protease or ligase are still unresolved. Enzymes from both extremes share high structural similarities (e.g., OaAEP1b and HaAEP1). A recent mutagenesis study on the polymorphism of PxAEPs suggested that besides the gatekeeper residue, another motif named “marker of ligase activity” (MLA) played an important role (48). This was supported by the observation that the PxAEP3b ligase had a truncated MLA, while its protease isoform PxAEP3a had a longer and more hydrophilic MLA. However, the MLA region is located far away from the catalytic center, and is thus unlikely to solely influence activity. In the same report, the substitution of the MLA of OaAEP1b with the one from OaAEP2 only affected the reaction rate but not the cyclization/hydrolysis ratio. Hence, MLA is probably not the sole determinant that could explain the mechanism but rather appears as a correlate of ligase activity. Based on the observation that both AEPs and PALs share similar overall enzyme structure, we hypothesized that the enzymatic activity is controlled by subtle differences at key positions near the catalytic center. These local alterations are likely to control the access to the S-acyl enzyme intermediate of water molecules (leading to hydrolysis) or of incoming nucleophiles (leading to ligation).
Herein, we study a series of putative AEPs and PALs from two cyclotide-producing plants, Viola yedoensis (var. phillipica) and Viola canadensis (48), and use recombinant enzymes to investigate the molecular mechanisms responsible for ligase catalytic activity. We identified two putative ligase-activity determinants (LADs) and validated them by structural comparison, molecular dynamics (MD) simulation, and site-directed mutagenesis. Our results chiefly explain the molecular mechanism allowing the conversion of AEPs into PALs, and can be used for the discovery and engineering of new ligases.
Results
Mining AEPs in Violaceae Transcriptomes and Initial Classification Using the Gatekeeper Residue.
Violaceae is one of the major cyclotide-producing plant families, suggesting the presence of PALs in their genomes. With the hope of identifying PALs, we therefore performed data mining on two plants from this family, V. yedoensis (Vy) and V. canadensis (Vc).
Briefly, to obtain the transcriptome of V. yedoensis, total RNA was extracted from fresh fruits and sequenced followed by assembly of the database (NCBI SRA accession no. PRJNA494974). Precursor sequences of butelase 1 and OaAEP1b were used to search for sequences homologous to AEPs. A total of 11 AEP precursors were found from the V. yedoensis transcriptome, including six complete sequences, three partial sequences containing an intact core domain, and two truncated sequences having an incomplete core domain that were discarded (SI Appendix, Fig. S1). The transcriptome of Vc is readily available in the 1KP database, and an AEP homolog (NJLF-2006002) named VcAEP was obtained by BLASTp using the butelase 1 sequence. To cluster the nine Vy sequences and VcAEP, we chose to use the nature of the gatekeeper residue as a criterion: It was previously observed that mutation of a Cys residue (Cys247) near the active site of OaAEP1b (PDB ID code 5H0I) to larger amino acids (Thr, Met, Val, Leu, Ile) reduced ligation catalytic efficiency, while mutations to smaller residues such as Ala resulted in over a hundred-fold improved ligation efficiency (figure 3D of ref. 47). Moreover, mutation of this “gatekeeper” residue into Gly results in an increased amount of hydrolysis product, suggesting that this site, located in the S2 substrate-binding pocket, plays an important role—although still elusive—in modulating enzyme function. Using the butelase 1 amino acid sequence to search for homologs in the NCBI databank returns more than 500 hits that share over 60% sequence identity, with 90% sequence coverage. Among them, more than 95% of sequences carry Gly at the gatekeeper site, including both proteases and dual-functional ligases, which agreed with the fact that PALs are rare in plant AEPs.
Using this criterion, four V. yedoensis sequences were classified as putative VyAEPs due to the presence of Gly as gatekeeper and designated VyAEP1–4. The other five, designated VyPAL1–5 as well as VcAEP from V. canadensis, were classified as putative VyPALs, as they contain Val (like butelase 1) or Ile as gatekeeper residues (Table 1 and SI Appendix, Fig. S2).
Table 1.
Sequence homology chart and nature of the gatekeeper of VyAEPs and VyPALs
| Enzyme | Gatekeeper | Core-domain identity, % | ||||||||||
| VyAEP | VyPAL | VcAEP | Butelase 1 | |||||||||
| 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | 5 | ||||
| VyAEP1 | Gly | 100.0 | ||||||||||
| VyAEP2 | Gly | 98.9* | 100.0 | |||||||||
| VyAEP3 | Gly | 66.1 | 65.7 | 100.0 | ||||||||
| VyAEP4 | Gly | 66.1 | 65.7 | 96.2* | 100.0 | |||||||
| VyPAL1 | Ile | 57.2 | 57.6 | 69.5 | 68.1 | 100.0 | ||||||
| VyPAL2 | Ile | 57.2 | 57.6 | 70.1 | 69.0 | 95.4 | 100.0 | |||||
| VyPAL3 | Val | 57.6 | 57.6 | 70.2 | 69.9 | 69.9 | 69.8 | 100.0 | ||||
| VyPAL4 | Ile | 56.8 | 57.2 | 69.1 | 67.7 | 99.7* | 95.0 | 69.9 | 100.0 | |||
| VyPAL5 | Ile | 56.8 | 57.2 | 69.8 | 68.7 | 95.0 | 99.6* | 69.8 | 95.4 | 100.0 | ||
| VcAEP | Val | 60.5 | 60.5 | 74.4 | 74.0 | 73.3 | 72.8 | 94.1 | 73.3 | 72.8 | 100.0 | |
| Butelase 1 | Val | 59.4 | 59.7 | 71.8 | 72.1 | 68.4 | 70.8 | 67.7 | 68.1 | 70.5 | 70.0 | 100.0 |
The pairs of isoforms are defined for a sequence identity greater than 96%.
Production of Active Recombinant VyAEPs and VyPALs.
Based on sequence identity, these putative AEPs and PALs could be partitioned into four groups: VyAEP1 and 2 (98.9%), VyAEP3 and 4 (96.2%), VyPAL1, 2, 4, and 5, (>95%), and VyPAL3. VyPAL3 only shares <70% core sequence identity with other putative VyPALs but is 94% identical to VcAEP (Table 1). We expressed VyAEP1, VyPAL1–3, and VcAEP for further studies. Recombinant expression was performed using both bacterial and insect cell systems, and the genes encoding complete amino acid sequences were cloned into the expression vectors, with the signal peptide substituted by a His tag for affinity purification. Following metal-affinity, ion-exchange, and size-exclusion chromatography (Materials and Methods), bacterial and insect cell systems yielded ∼0.5 and 10–20 mg/L purified proenzymes, respectively.
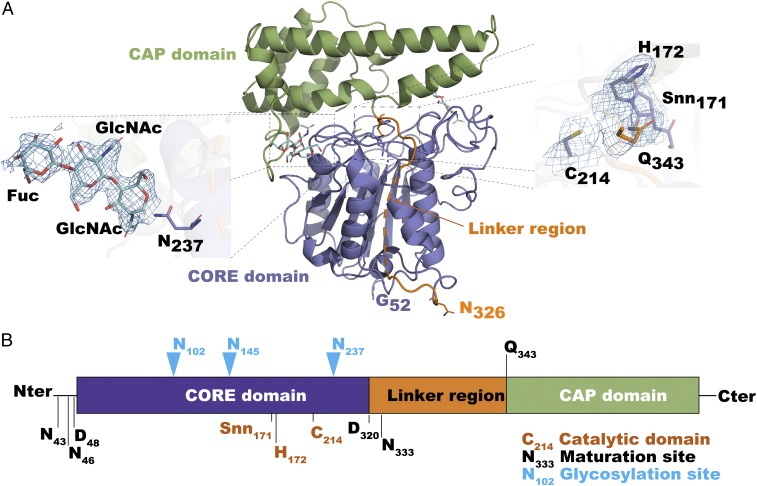
Following purification, proenzymes were subjected to 12- to 16-h activation at 4 °C, pH 4.5 in the presence of 0.5 mM N-lauroylsarcosine, 5 mM β-mercaptoethanol, and 1 mM EDTA (SI Appendix, Fig. S3A). Such mild but prolonged treatment allows cleavage and degradation of the cap domain, preventing cap-domain religation. Activated enzymes were further purified using size-exclusion chromatography (SI Appendix, Fig. S3B). The autoactivation sites of purified active VyPAL2 were determined by LC-MS/MS sequencing of the tryptic-digested active forms. The Asn/Asp cleavages sites at both ends of the core domain were found to be N43/N46/D48 in the N-terminal prodomain region and D320/N333 in the linker region. This confirmed the complete removal of the inhibitory cap domain and the production of a mixture containing active forms through protein processing at multiple sites (SI Appendix, Fig. S3C).
Ligase vs. Protease Activity of VyAEP1 and VyPAL1–3.
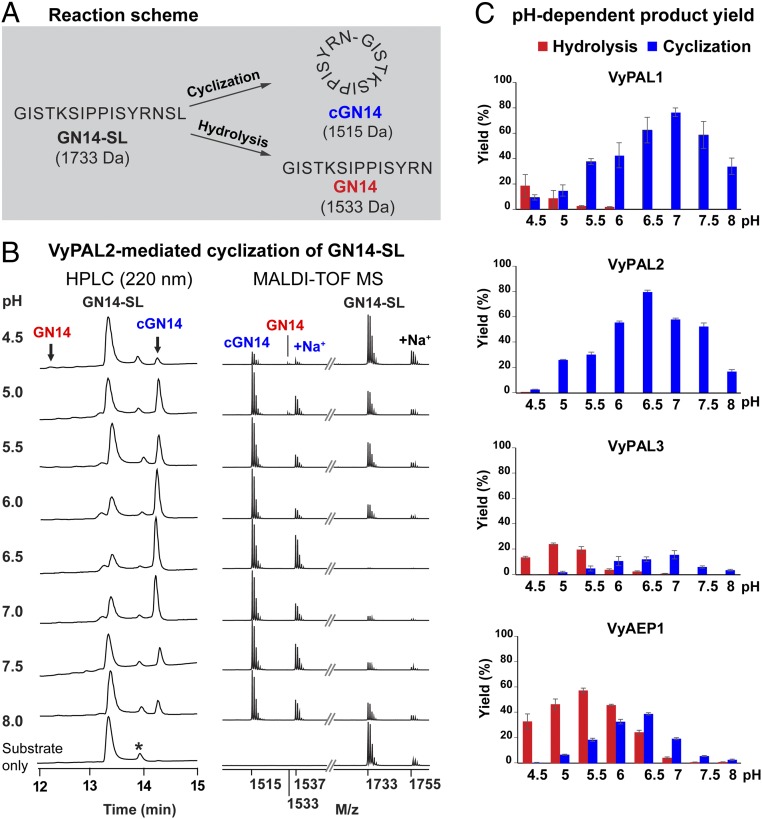
To determine the activity of VyAEP/PALs, we prepared a model peptide substrate termed “GN14-SL,” GISTKSIPPISYRNSL, with a molecular mass of 1,733 Da. GN14-SL contains the tripeptide-recognition motif NSL at its C terminus derived from the precursors of Vy cyclotides and analog of SFTI-1 (Fig. 1A). A fixed enzyme:substrate molar ratio (1:500) was used in all cyclization reactions, which were performed at 37 °C for 10 min at pH values ranging from 4.5 to 8.0 (at 0.5 intervals). The cyclization of GN14-SL was monitored using MALDI-TOF mass spectrometry. The yields of cyclic product cGN14 (molecular mass 1,515 Da) and linear product GN14 (molecular mass 1,533 Da) were quantified using RP-HPLC (Fig. 1B).
Fig. 1.
Enzymatic activity of recombinant VyPAL1–3 and VyAEP1. (A) Reaction scheme of ligase-mediated cyclization of GN14-SL. (B) Analytical HPLC and MALDI-TOF mass spectrometry data of VyPAL2-mediated cyclization under different reaction pH values. The asterisk indicates racemized synthetic GN14-SL. Note that MALDI-TOF MS was more sensitive against cyclic cGN14 than the linear species. (C) Quantitative summary of product ratio and reaction yield of each enzyme analyzed using RP-HPLC. For each reaction, a molar ratio of purified active enzyme:GN14-SL of 1:500 was mixed and reacted at 37 °C for 10 min. Average yield and SDs were calculated from experiments performed in triplicate.
Among the four PAL enzymes tested, VyPAL2 exhibited the best ligase activity, and did not produce any hydrolytic product at pH 5.5–8.0. At the optimal pH of 6.5, over 80% cyclization yield was observed (Fig. 1C). VyPAL1 also resulted in pure cyclization at pH 6–8 and, at the optimal pH of 7.0, about 80% cyclization yield was obtained. VyPAL3 displayed dominant hydrolysis activity at pH 4.5–5.5 and dominant ligase activity at pH 6.0–7.0. Its catalytic efficiency was the lowest among the three putative VyPALs, as only 20% of substrate was converted into cyclized product at the optimal pH, 7.0, in 10 min. As anticipated, the putative protease VyAEP1 displayed hydrolysis activity in the tested pH range of 4.5–8, although at the near-neutral and basic pH of 6.5–8 cyclization became noticeable. All four enzymes displayed varying degrees (2–40%; Fig. 1C) of protease activity at pH less than 5.0, reflecting the intrinsic proteolytic activity needed for acid-induced autoactivation.
Next, substrate specificity of VyPAL2 was studied, using three sets of peptide libraries (SI Appendix, Fig. S4). Efficient cyclization required a minimum of three residues as the C-terminal recognition signal Asn–P1′–P2′ [using Schechter and Berger nomenclature (49)]. At P1′, small amino acids, especially Gly and Ser, are favored, but not Pro. The P2′ position favors the presence of hydrophobic or aromatic residues, such as Leu/Ile/Phe. The catalytic efficiency of VyPAL2 was examined using the substrate GN14-SLAN (GISTKSIPPISYRNSLAN) that gave 274,325 M−1⋅s−1 when performing at pH 6.5 at 37 °C, which was 3.5-fold less than butelase 1 (971,936 M−1⋅s−1) (SI Appendix, Fig. S5).
Crystal Structure of VyPAL2.
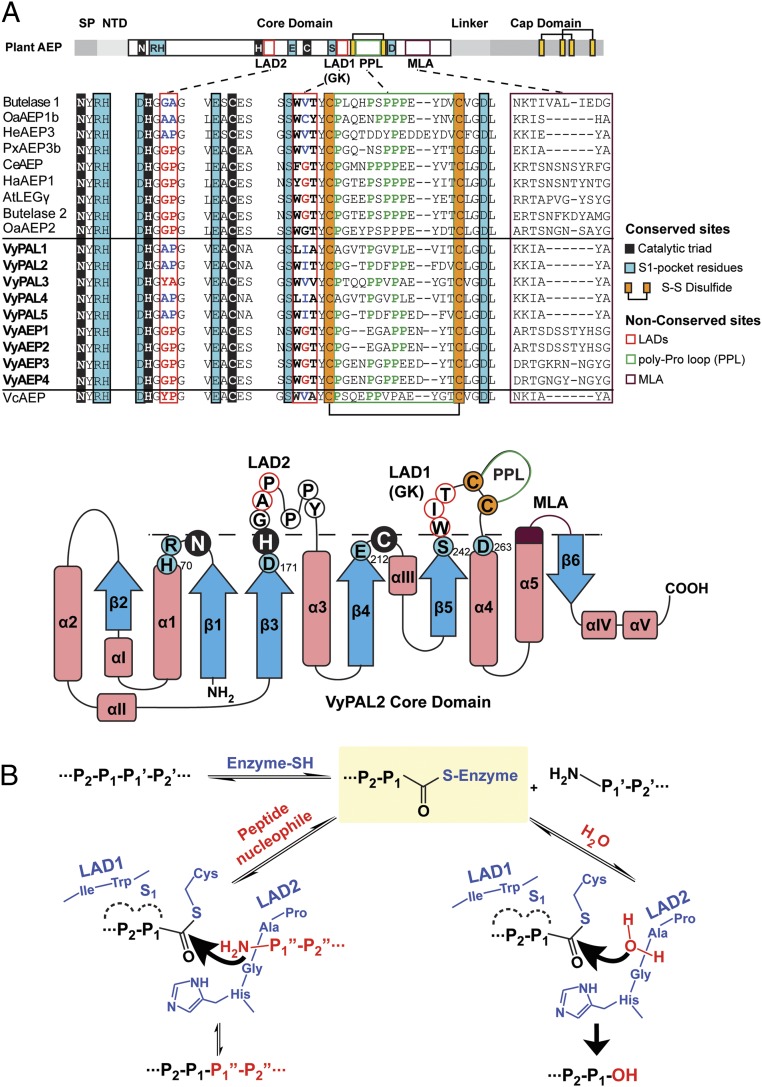
To understand the molecular mechanisms responsible for the differences in nature and efficiency between PALs and AEPs identified here, we obtained the crystal structure of the VyPAL2 proenzyme at a resolution of 2.4 Å (Fig. 2A and SI Appendix, Table S1). As expected, the structure displays the prolegumain fold with the active domain on the N terminus (residues 51–320) and the cap domain on the C terminus (residues 344–483) (Fig. 2A). These two domains are connected by a flexible linker (residues 321–343). The asymmetric unit contains two monomers of VyPAL2, forming a homodimer. In solution, this oligomeric form of VyPAL is present only at high protein concentrations (>5 mg/mL), as inferred from gel-filtration results. As the protein was expressed in insect cells, several asparagine residues on the surface of the protein are glycosylated (Fig. 2A) with one to three N-linked sugars [one N-acetylglucosamine (GlcNAc), two GlcNAcs, or two GlcNAcs and one fucose] on Asn102, Asn145, and Asn237, respectively. Members of the C13 subfamily share a conserved α–β–α sandwich structure and a His172–Cys214 catalytic dyad located in a well-defined oxyanion hole. Peptide-bond cleavage is catalyzed by the Cys thiol, which mediates an N- to S-acyl transfer to give the Asn–(S-)Cys thioester intermediate. The imidazole ring of His acts as a general base to accept a proton from the catalytic Cys (Fig. 2A).
Fig. 2.
Crystal structure of VyPAL2. (A) The VyPAL2 monomer: Its cap region is colored in green, core in purple, and linker in orange. N-glycosylation sites are displayed as cyan sticks. (A, Insets) Active-site residues (Right) and one representative glycosylation site (Left). The electronic density map (with Fourier coefficients 2Fo − Fc and phases from the model) is overlaid and contoured at 1-sigma level. (B) Diagram of VyPAL2 protein domains. Blue arrowheads indicate glycosylation sites. Cleavage sites as determined by LC-MS/MS are indicated. Active-site residues (His172 and Cys214) as well as succinimide 171 are labeled.
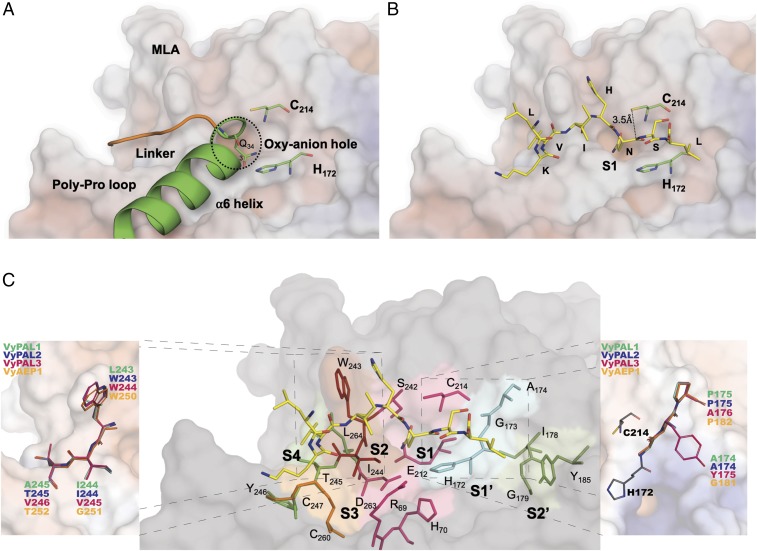
The structure is similar to other PALs and AEPs such as OaAEP1b (PDB ID code 5H0I), AtLEGγ (5NIJ, 5OBT), HaAEP1 (6AZT), or butelase 1 (6DHI), with an average root-mean-square deviation (rmsd) of atomic positions of 1.0 Å (SI Appendix, Table S2). Moreover, comparing the active domain alone returns rmsd values closer to an average of 0.7 Å, showing that the core-domain structure is strongly conserved. This further indicates that enzyme specificity is due to subtle variations in the substrate-binding pockets that influence the stability of the S-acyl intermediate and accessibility of the catalytic water molecule. In the present proenzyme form, helix α6 (the first helix in the cap domain) makes an angle of about 90° with the linker peptide (Fig. 3A). At the junction between the linker region and the α6 helix, Gln343 is anchored inside the oxyanion hole (or S1 pocket). In recent structures of active forms of HaAEP1 and AtLEGγ, the bound substrate or inhibitor is shifted by a distance of about 2.5 Å compared with the linker region and covalently linked to the catalytic cysteine via a thioester bond (see figure 2 of ref. 50).
Fig. 3.
Modeling of substrate LKVIHNSL binding in the core domain of VyPAL2. The electrostatic surface of the VyPAL2 core domain is represented with positive charges in blue and negative charges in red. Protein regions important for activity are labeled. Active-site residues His172 and Cys214 are represented as green sticks. (A) The linker region (orange), α6-helix (green), and substrate-binding pocket S1 are indicated. (B) Substrate residues (LKVIHNSL, yellow) are shown as sticks. The distance between the sulfur atom of Cys214 and the carbon of the carbonyl group of substrate–Asn is 3.5 Å. (C) Residues lining the substrate-binding pockets are depicted as sticks. The VyPAL2 core domain is represented as a gray surface. The S1 pocket (magenta) comprises residues R69, H70, Snn171, H172, G173, E212, A213, C214, S242, and D263. The S2 pocket (red) is shaped by W243 and I244. The S3 pocket (yellow) is lined by C247 and C260 that form a disulfide bond. The S4 pocket (lime) is shaped by T245, Y246, and L264. On the other side of the active site, the S1′ pocket is lined (cyan) by H172, G173, and A174, and the S2′ pocket (green) is open and comprises I178, G179, M180, and Y185. VyPAL polymorphism sites around the S1 pocket are highlighted (Insets). VyPAL1 residues are colored and labeled in green, VyPAL2 in blue, VyPAL3 in magenta, and VyAEP1 in orange. The catalytic residues Cys214 and His172 of VyPAL2 are shown as red and cyan sticks, respectively.
Modeling Substrate–Enzyme Interactions Using Energy Minimization.
Structures of ligand-bound active forms of both HaAEP1 (PDB ID code 5OBT) and AtLEGγ (6AZT) indicated that only small conformational changes occur after activation of the protein and cap release. We therefore modeled the active form of the VyPAL2 ligase using the present crystal structure of VyPAL2 and included residues Gly52–Asn326, which are clearly visible in the electron density. This is also in line with the boundaries of the VyPAL2 active form determined using LC-MS (SI Appendix, Fig. S3). To obtain an initial model of a peptide substrate bound to the active form of VyPAL2, we used the structure of the complex between AtLEGδ and a peptide inhibitor (50) having the sequence NH2–LKVIH–NSL–COOH. The N-terminal sequence of this peptide corresponds to the original linker sequence, and the C-terminal dipeptide is based on substrate-specificity studies presented in SI Appendix, Fig. S4. Energy minimization of the resulting complex with the peptide was then performed, constraining only the Cα-atoms of the active protein. The α-carbon atom of the P1 Asn residue was fixed at the position found in AtLEGγ (46) and used as an anchor to maintain the substrate in the S1 pocket. Upon MD equilibration of the system for 20 ns, the N-terminal portion of the substrate LKVIHN was shifted due to repulsion between I244 from VyPAL2 and the substrate. As a result, the α-carbon atom of the Ile at the substrate P3 position is displaced by 3 Å. The C-terminal SL dipeptide, on the other hand, becomes more extended, leading to a better fit of the peptide into the substrate-binding pockets (Fig. 3B and Movie S1). This more stable and energetically favorable position for the modeled substrate was used to map the S1′ and S2′ pockets that define the recognition motifs both for protease and ligase activities. By analyzing the interface with the model substrate, we could define residues of the active form of VyPAL2 that are lining the S4– to S2′ pockets (Fig. 3C). The composition of S4 agreed with earlier work on AtLEGγ, and involved residues from both the disulfide-clamped poly-Pro loop (PPL) equivalent to the c341 loop in caspase-1 and the MLA region (equivalent to the c381 loop in caspase-1) (Fig. 3A). On the other side of the S1 pocket, the S1′ pocket is shaped by the amide groups of H172, G173, and A174 that accommodate the backbone atoms of the P1′ and P2′ residues of the peptide. The S2′ pocket is lined by Y185 and backbone atoms of G179 and M180, which favors binding of hydrophobic residues at the P2′ position. MD simulation (Movie S1) shows that the interaction between the hydrophobic Leu side chain of the peptide and the phenol ring of Y185 is favored, which is in agreement with the preference for Ile/Val/Phe at P2′ observed in the specificity study (SI Appendix, Fig. S4C).
Identification of Ligase-Activity Determinants in the S2 and S1′ Pockets.
Although classified and confirmed as PALs, VyPAL1–3 displayed various levels of ligase activity in terms of both cyclization/hydrolysis ratio and catalytic efficiency. Thus, we modeled the structures of VyPAL1 and VyPAL3 using the experimental crystal structure of VyPAL2 as template (SI Appendix, Fig. S6). The resulting models are likely to be accurate given the sequence identity between these three proteins. Mapping the polymorphic residues on the VyPAL1–3 structures indicates variations in the substrate-interacting surface located in the S2 and S1′ pockets. One variation lies in the first residue of S2, Leu243 in VyPAL1, in lieu of the aromatic and bulky Trp present in both VyPAL2 and VyPAL3. In the same region, position 244 of VyPAL2 is either Ile or Val, introducing little variation in local hydrophobicity. Finally, the side chain of the residue at position 245 is facing a direction opposite from the S1 pocket (and the backbone atoms of VyPAL1–3 completely overlap), suggesting that this residue has little impact on catalysis (Fig. 3C, Left Inset and SI Appendix, Fig. S8). However, on the other side of the S1 pocket, a more drastic difference is observed in the vicinity of S1′ and S2′: Ala174–Pro175 in both VyPAL1 and 2 is replaced by Tyr175–Ala176 in VyPAL3 (Fig. 3C, Right Inset).
Selectively Improving the Ligase Activity of VyPAL3 and VcAEP.
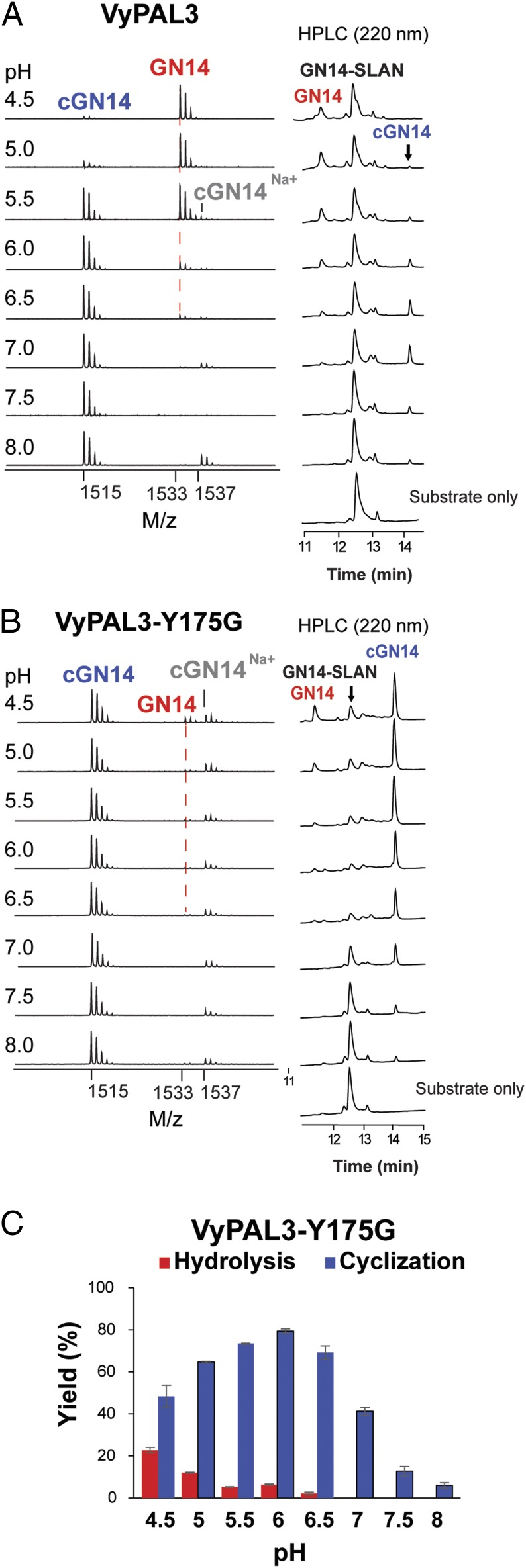
To validate experimentally these structural observations, we first targeted VyPAL3: We mutated the YA dipeptide in the S1′ region into GA, as found in the butelase 1 sequence. As anticipated, this Y175G point mutation resulted in a strong and selective increase of ligation activity observed at lower pH (4.5–6), compared with the wild-type VyPAL3 (Fig. 4). In addition, the catalytic efficiency was also significantly improved, with the maximum cyclization yield increasing from 20 to 80% (compare Figs. 1C and 4C).
Fig. 4.
Retroengineering in the S1′ pocket of VyPAL3. All reactions were performed in the pH range 4.5–8.0. MS peaks of the hydrolysis product GN14 are marked with a red dashed line. (A, Left) MS analysis of reactions catalyzed by the VyPAL3 wild type. (B, Left) MS spectra of reactions catalyzed by VyPAL3-Y175G. (A and B, Right) HPLC profiles of reactions catalyzed by VyPAL3 and VyPAL3-Y175G. (C) Quantitative summary of product ratio and reaction yield analyzed using RP-HPLC for VyPAL3-Y175G. Average yield and SDs were calculated from experiments performed in triplicate.
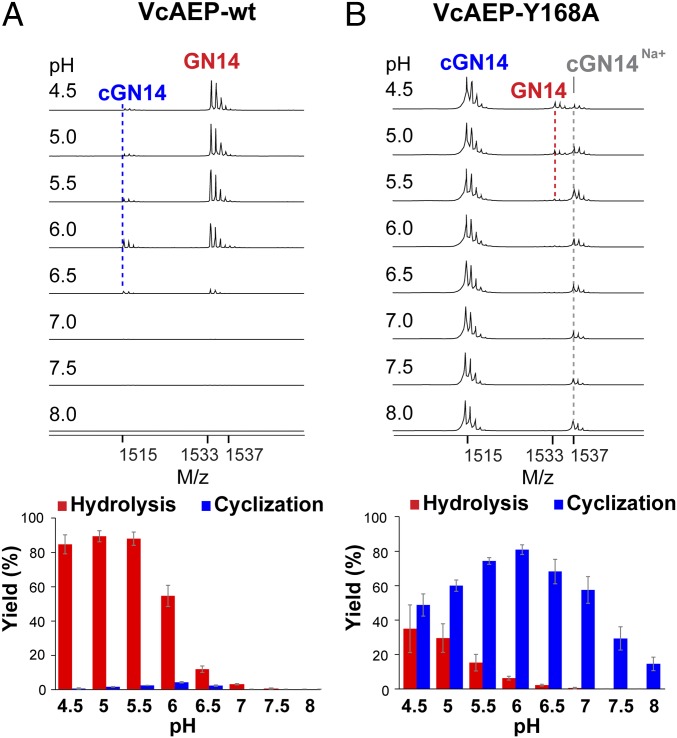
To further validate our hypothesis about the crucial role of the S1′ region in determining ligase activity, we targeted VcAEP with predominantly protease activity and virtually absent ligase activity (Fig. 5A). We performed the mutation Y168P169→A168P169 in its S1′ region (equivalent to Y175A176 in VyPAL3). The Y168A mutation drastically affected both the type of enzymatic activity and the catalytic efficiency (Fig. 5B) toward the GN14-SLDI substrate. The reaction with the wild-type VcAEP was performed using an enzyme:GN14-SLDI molar ratio of 1:200 for 5 h. In contrast, for VcAEP-Y168A the ratio was 1:2,000, and the reaction was quenched after 2-min incubation at 37 °C. At near-neutral pH, VcAEP-Y168A was able to convert over 60% substrate into its cyclic form, with less than 5% hydrolysis product formed (Fig. 5B).
Fig. 5.
Activity of VcAEP and the VcAEP-Y168A mutant (LAD2). MS analysis and HPLC-based quantitative summary of the reaction catalyzed by (A) VcAEP wild type and (B) VcAEP-Y168A mutant targeting the LAD2 region. All reactions were performed at pH values ranging from 4.5 to 8.0. MS peaks of the hydrolysis product GN14, cyclization product cGN14, and sodium ion adduct of cGN14 are marked with red, blue, or gray dashed lines. A dramatic improvement of ligase activity for the VcAEP-Y168A mutant is clearly visible. Average yield and SDs were calculated from experiments performed in triplicate.
Discussion
Discovery of PALs and AEPS from Violaceae.
Here we successfully identified ligases from plants of the Violaceae using homology with enzymes capable of ligase activity such as butelase 1. We named these enzymes “peptide asparagine ligases,” to highlight their specific transpeptidase activity and to differentiate them from AEPs. We purified and tested the activity of the corresponding recombinant enzymes and found that solely VyPAL2 has ligase activity at near-neutral pH and displays minimal hydrolase activity even at low pH, making it a recombinant PAL valuable for biotechnological applications, only 3.5 times less efficient than butelase 1. VyPAL1, despite being a good ligase, showed promiscuous activity, with some hydrolysis at acidic pH. VyPAL3 is characterized by an overall low catalytic efficiency together with a dominant hydrolysis activity at low pH. Moreover, the VyAEP1 protein, predicted to be a protease based on sequence homology, was indeed found to be a protease at low pH (Fig. 1). To reveal the molecular bases for the differences in activities between these enzymes, we obtained the crystal structure of VyPAL2 and used it as a template to model the structure of VyPAL protein isoforms. These comparisons pointed to two areas surrounding the S1 active-site pocket: the S2 and the S1′ pockets, that show striking variations between an AEP and a PAL.
Definition of Ligase-Activity Determinants.
One residue of OaAEP1b located in the S2 pocket was named the gatekeeper, as it was found to play an important role in controlling enzyme efficiency and protease versus ligase activity (46). Here, we found that using the nature of the gatekeeper residue as the only criterion is insufficient to explain the range of activities observed in VyPAL1–3 isoforms: VyPAL2, a very efficient PAL, and VyPAL3, a very poor enzyme, both have similar gatekeeper residues: Ile and Val, respectively (Fig. 6A). Moreover, VcAEP that has Val (like butelase 1) as a gatekeeper residue is a protease (Fig. 5A). Hence, we sought additional residues that could act as ligase-activity determinants. Accordingly, we identified sequence variations in two regions of VyPAL1–3: (i) the S2 pocket (LAD1) comprising residues W243, I244 (the gatekeeper), and T245 in VyPAL2, and (ii) the S2′ pocket (LAD2) including residues A174 and P175 (Fig. 6). The LAD2 regions of VyPAL1 and VyPAL2 are identical, while their gatekeeper region (LAD1) bears two variations (Fig. 6): The T245A substitution is likely to have little effect, since the side chain of residue 245 is oriented opposite the substrate-binding area. This suggests that the difference in activity observed between VyPAL1 and 2 is due to the other W243L substitution, making the enzyme “leakier” for hydrolysis and explaining the slight shift of VyPAL1 toward hydrolase at lower pH.
Fig. 6.
Ligase-activity determinants of PALs and the proposed catalytic mechanism. (A) Sequence alignment of PALs and AEPs studied in this work. The catalytic triad Asn–His–Cys is shaded in black. Residues belonging to the S1 pocket are shaded in blue. Proposed LAD residues are boxed in red. Residues of LAD1 and LAD2, that favor ligase activity, are highlighted in blue, and those favoring protease activity are highlighted in red. The conserved disulfide bond near LAD1 is highlighted in orange. The PPL is in a green box and the MLA loop is in purple. The nomenclature of secondary structures was adapted from ref. 48 with alterations according to the crystal structure of VyPAL2 (this work). Residues and motifs crucial for activity are labeled with the same color codes as used in the sequence alignment. Residues below the dashed line correspond to the oxyanion hole, and those above the dashed line correspond to the proposed activity determinants. (B) Schemes proposed for ligation and hydrolysis by VyPAL2 and the role of LAD1 and LAD2. The first step of the mechanism is identical for hydrolysis and ligation, leading to formation of the S-acyl enzyme intermediate, and is the rate-limiting step. Its main determinant is LAD1. LAD2 controls the nature of activity to either favor the nucleophilic attack by a peptide (ligation) or a water molecule (hydrolysis).
The Case of VyPAL3.
Compared with VyPAL1 and two ligases, VyPAL3 has variations at both LAD1 and LAD2. However, the conservative substitutions at LAD1, V245 instead of an Ile gatekeeper residue and V246 instead of Thr (VyPAL2), are unlikely to account for the drastic change of activity that we observe (Fig. 3C and SI Appendix, Fig. S8). Rather, on the other side of the active site, in LAD2, the AP dipeptide present in VyPAL1 and 2 is replaced by the bulkier YA dipeptide. This variation most likely contributes to the lower ligase activity observed in VyPAL3 compared with VyPAL1 and VyPAL2, as the bulky Tyr residue at this position could hamper the access of a peptidic nucleophile to the acyl-enzyme intermediate. Thus, inserting a smaller hydrophobic side chain such as Gly (or Ala) at the first position of LAD2 should suffice to significantly increase ligase efficiency, as seen in the corresponding VyPAL3 single Y175G mutant (Fig. 4). Importantly, we were able to confirm the involvement of the LAD2 region in controlling the ligase activity by introducing an equivalent mutation on another AEP from V. canadensis (compare Fig. 5 A and B). The volume occupied by Tyr at this position is likely to cause adverse effects such as accelerating the dissociation of the leaving group and slowing down the binding of an incoming peptide, that are essential steps to displace the catalytic water molecule and thus favor ligation over hydrolysis. This is in line with the importance of the interactions at the prime side that was proposed by Brandstetter and colleagues to favor cyclization by preventing premature thioester hydrolysis (50). On the other hand, the side chain of Tyr175 should not disturb the putative catalytic water molecule. This water molecule is presumably located right above Gly174 of VyPal3—a strictly conserved residue immediately following the catalytic His—as observed in the cases of AtLEGγ and other legumains (46, 50).
The Role of the Gatekeeper Residue.
The mechanism of AEPs and PALs can be decomposed into two steps: (i) acyl-enzyme thioester intermediate formation, which is likely the rate-limiting step, and (ii) nucleophilic attack by a water molecule (hydrolysis) or nucleophilic peptide (ligation) on the acyl-enzyme intermediate. Together with the known information on gatekeeper mutagenesis performed on OaAEP1b (46), our results suggest that hydrophobic residues such as Val/Ile/Cys/Ala at this central position favor ligation, while the presence of Gly favors proteolysis (Figs. 1 and 6). The gatekeeper could affect substrate positioning with an impact on enzyme activity, possibly by inducing some specific conformational strain in the substrate. Alternatively, the gatekeeper could also affect the dynamics of the enzyme. The side chain not only influences the ligase activity but greatly affects catalytic efficiency. Since changes at the gatekeeper mainly affect substrate binding and positioning, they will have a direct impact on intermediate formation and thus on the overall reaction rate. Conversely, changes in LAD2 would affect the nature and accessibility of the nucleophile, as proposed recently (50) and, as a consequence, be decisive on the nature of the overall reaction catalyzed.
The Role of LAD2.
LAD2 seems to be a crucial determinant for the nature of the activity catalyzed: A bulky residue on this side of the active site, such as Tyr at the first position of the YA dipeptide in VyPAL3 and YP in VcAEP, would facilitate the departure of the cleaved peptide group, which thus results in recruitment of the catalytic water and exposing the acyl-enzyme thioester to nucleophilic water molecules. This mechanism is in line with the study by Brandstetter and colleagues (50), who showed that a cleaved peptide group remaining in the S1′ and S2′ pockets displaced the nucleophilic water molecule and thus favors ligation over hydrolysis. Moreover, a bulky residue oriented in the direction where the incoming nucleophilic peptide would bind will hamper access to the acyl-enzyme intermediate, and thus severely reduce the rate of ligation. Conversely, small hydrophobic dipeptides like GA/AA/AP in LAD2 would retain the departing group (blocking access to the thioester bond), until another peptide acts as a nucleophile, leading to ligase activity as described previously. However, we note that mutations of both sites of VyPAL2 (SI Appendix, Fig. S7) to engineer an AEP did not result in an efficient and drastic conversion into a protease like OaAEP2 or butelase 2, suggesting the existence of other determinants for proteolysis, besides LAD1 and LAD2. One attractive possibility is that residues within LAD1 (the gatekeeper), LAD2 (this work), and MLA cooperate to determine protease vs. ligase activity. In this respect, we note that the presence of a truncated MLA alone (24) does not necessarily imply a ligase activity, because VcAEP, which possesses a truncated MLA (Fig. 6), displays mainly protease activity (Fig. 5). Further examples need to be studied through site-directed mutagenesis to more comprehensively delineate the relative importance of these determinants or markers of protease vs. ligase activity.
Conclusion
In summary (Fig. 6), we propose that the molecular determinants governing asparaginyl endopeptidase and ligase activity are primarily found in the amino acid composition of the substrate-binding grooves flanking the S1 pocket, in particular the gatekeeper (within the LAD1 region) and LAD2 that are centered around the S2 and S1′ pockets, respectively. Combining structural analysis and mutagenesis study, we uncovered that, for an efficient peptide asparaginyl ligase, the first position of LAD1 is preferably bulky and aromatic, such as Trp/Tyr, and the second position is hydrophobic, such as Val/Ile/Cys/Ala but not Gly. For LAD2, we found that GlyAla/AlaAla/AlaPro dipeptides are favored. A bulky residue such as Tyr is disadvantageous at the first position of LAD2, as it may destabilize the acyl-enzyme intermediate by affecting the binding affinity of substrates and controlling the accessibility of water molecules and by increasing the dissociation rate of the cleaved peptide tail after the Asn/Asp residue. Therefore, a small residue such as Gly or Ala at the first position of this dipeptide is a necessary, although not always sufficient, condition for ligase activity. As long as this condition is met, a natural AEP is amenable to becoming a PAL through mutations or changes at other locations such as LAD1 (gatekeeper) or more remote regions like the MLA.
Materials and Methods
Detailed materials and methods can be found in SI Appendix.
RNA Extraction, Construction of the Vy Transcriptome, and Search of AEP Analogs.
RNA extracts of fresh fruits were sequenced. The butelase 1 amino acid sequence was used for homology search. A search using the butelase 1 proenzyme sequence resulted in over 500 hits with >60% sequence identity and >90% sequence coverage.
Cloning and Recombinant Expression, Purification, and Autoactivation.
cDNA sequences without the predicted signal peptides were synthesized and cloned into expression vectors [pET28a(+) for bacterial expression, or pFB for sf9 insect cell expression]. Protein purification was performed in three steps with IMAC affinity purification followed by ion-exchange and size-exclusion chromatography. The protein was then concentrated and stored at 4 °C.
Activation was performed by acidification at pH 4.5 (50 mM sodium citrate buffer, 1 mM DTT, 1 mM EDTA, 0.1 M NaCl) at 4 °C for 12–16 h with 0.5 mM N-lauroylsarcosine. Subsequently, active enzymes were purified on a size-exclusion chromatography column (S100 16/60) (GE Life Sciences) preequilibrated at pH 4.0 in SEC buffer (20 mM sodium citrate buffer, 1 mM EDTA, 5 mM β-mercaptoethanol, 5% glycerol, 0.1 M NaCl).
Characterization of Enzyme Activity.
Reaction mixtures contained 40 nM active enzyme and 20 µM substrate in the reaction buffer and were incubated at 37 °C for 10 min before quenching the reaction. Reaction results were analyzed by MALDI-TOF and RP-HPLC. For kinetic studies, the cyclization reactions were conducted at pH 6.5 at 37 °C with a fixed concentration of active enzymes (10 nM) and various concentrations (2–20 µM) of the substrate. The yield of cyclization product cGN14 was quantified by RP-HPLC at every 20-s interval and, the initial rate V0 (μM/s) was plotted against substrate concentration [S] (μM) to obtain the Michaelis–Menten curve to analyze the kinetic parameters (kcat and KM) of each enzyme.
Crystallization, Data Collection, and Structure Determination.
VyPAL2 was concentrated to 10 mg/mL and screened against commercial screens. X-ray diffraction was collected and processed using XDS (51), and the structure was solved using 5H0I as a model in Phaser (CCP4) (52) and refined using BUSTER TNT (53) (GlobalPhasing Ltd.). Processing and refinement statistics are presented in SI Appendix, Table S1. The VyPAL2 structure was deposited in the Protein Data Bank under PDB ID code 6IDV.
Molecular Dynamics Simulation.
Using the core domain of theVyPAL2 crystal structure and the AtLEGγ enzyme inhibitor complex as a model, a substrate was modeled and subjected to molecular dynamics using NAMD 2.12 (54). The system was simulated for a total of 20 ns with the backbone atoms of the protein ligase, as well as the Cα-atom of N343 of the peptide, constrained. Such constraints allow the side chains of VyPAL2 and the rest of the peptide substrate to move freely. All simulations were performed under the NPT ensemble assuming the CHARMM36 force field (55) for the protein.
Supplementary Material
Acknowledgments
We thank the scientists and beamline staff at the Proxima 2A, SOLEIL (Proposal 20180290), and MX2 (Australian Synchrotron) for their expert assistance. We thank Professor Lloyd Ruddock (University of Oulu) for providing the disulfide-promoting plasmids. This research was supported by Academic Research Grant Tier 3 (MOE2016-T3-1-003) from the Singapore Ministry of Education (MOE) to the J.P.T., J.L., and C.-F.L. laboratories, and NMRC Grants CBRG/0028/2014 and NRF2016NRF-CRP001-063.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structure of VyPAL2 reported in this paper has been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6IDV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818568116/-/DCSupplemental.
References
- 1.Mazmanian S. K., Liu G., Ton-That H., Schneewind O., Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285, 760–763 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Mao H., Hart S. A., Schink A., Pollok B. A., Sortase-mediated protein ligation: A new method for protein engineering. J. Am. Chem. Soc. 126, 2670–2671 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Piotukh K., et al. , Directed evolution of sortase A mutants with altered substrate selectivity profiles. J. Am. Chem. Soc. 133, 17536–17539 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Chang T. K., Jackson D. Y., Burnier J. P., Wells J. A., Subtiligase: A tool for semisynthesis of proteins. Proc. Natl. Acad. Sci. U.S.A. 91, 12544–12548 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toplak A., Nuijens T., Quaedflieg P. J. L. M., Wu B., Janssen D. B., Peptiligase, an enzyme for efficient chemoenzymatic peptide synthesis and cyclization in water. Adv. Synth. Catal. 358, 2140–2147 (2016). [Google Scholar]
- 6.Schmidt M., et al. , Omniligase-1: A powerful tool for peptide head-to-tail cyclization. Adv. Synth. Catal. 359, 2050–2055 (2017). [Google Scholar]
- 7.Weeks A. M., Wells J. A., Engineering peptide ligase specificity by proteomic identification of ligation sites. Nat. Chem. Biol. 14, 50–57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnison P. G., et al. , Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haase J., Lanka E., A specific protease encoded by the conjugative DNA transfer systems of IncP and Ti plasmids is essential for pilus synthesis. J. Bacteriol. 179, 5728–5735 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barber C. J. S., et al. , The two-step biosynthesis of cyclic peptides from linear precursors in a member of the plant family Caryophyllaceae involves cyclization by a serine protease-like enzyme. J. Biol. Chem. 288, 12500–12510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chekan J. R., Estrada P., Covello P. S., Nair S. K., Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants. Proc. Natl. Acad. Sci. U.S.A. 114, 6551–6556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mann G., et al. , The structure of the cyanobactin domain of unknown function from PatG in the patellamide gene cluster. Acta Crystallogr. F Struct. Biol. Commun. 70, 1597–1603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo H., et al. , Peptide macrocyclization catalyzed by a prolyl oligopeptidase involved in α-amanitin biosynthesis. Chem. Biol. 21, 1610–1617 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conlan B. F., Gillon A. D., Craik D. J., Anderson M. A., Circular proteins and mechanisms of cyclization. Biopolymers 94, 573–583 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Nguyen G. K. T., et al. , Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nat. Chem. Biol. 10, 732–738 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Nguyen G. K. T., et al. , Butelase 1: A versatile ligase for peptide and protein macrocyclization. J. Am. Chem. Soc. 137, 15398–15401 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Cao Y., Nguyen G. K. T., Tam J. P., Liu C.-F., Butelase-mediated synthesis of protein thioesters and its application for tandem chemoenzymatic ligation. Chem. Commun. (Camb.) 51, 17289–17292 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Nguyen G. K. T., et al. , Butelase-mediated cyclization and ligation of peptides and proteins. Nat. Protoc. 11, 1977–1988 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Hemu X., Qiu Y., Nguyen G. K. T., Tam J. P., Total synthesis of circular bacteriocins by butelase 1. J. Am. Chem. Soc. 138, 6968–6971 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Nguyen G. K. T., Hemu X., Quek J.-P., Tam J. P., Butelase-mediated macrocyclization of d-amino-acid-containing peptides. Angew. Chem. Int. Ed. Engl. 55, 12802–12806 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Bi X., et al. , Enzymatic engineering of live bacterial cell surfaces using butelase 1. Angew. Chem. Int. Ed. Engl. 56, 7822–7825 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Hemu X., Zhang X., Tam J. P., Ligase-controlled cyclo-oligomerization of peptides. Org. Lett. 21, 2029–2032 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Harris K. S., et al. , Efficient backbone cyclization of linear peptides by a recombinant asparaginyl endopeptidase. Nat. Commun. 6, 10199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson M. A., et al. , Molecular basis for the production of cyclic peptides by plant asparaginyl endopeptidases. Nat. Commun. 9, 2411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J. M., Rawlings N. D., Stevens R. A., Barrett A. J., Identification of the active site of legumain links it to caspases, clostripain and gingipains in a new clan of cysteine endopeptidases. FEBS Lett. 441, 361–365 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Barrett A. J., Rawlings N. D., Evolutionary lines of cysteine peptidases. Biol. Chem. 382, 727–733 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Dall E., Brandstetter H., Mechanistic and structural studies on legumain explain its zymogenicity, distinct activation pathways, and regulation. Proc. Natl. Acad. Sci. U.S.A. 110, 10940–10945 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halfon S., Patel S., Vega F., Zurawski S., Zurawski G., Autocatalytic activation of human legumain at aspartic acid residues. FEBS Lett 438, 114–118 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Hiraiwa N., Nishimura M., Hara-Nishimura I., Vacuolar processing enzyme is self-catalytically activated by sequential removal of the C-terminal and N-terminal propeptides. FEBS Lett 447, 213–216 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Chen J. M., Fortunato M., Barrett A. J., Activation of human prolegumain by cleavage at a C-terminal asparagine residue. Biochem. J. 352, 327–334 (2000). [PMC free article] [PubMed] [Google Scholar]
- 31.Kuroyanagi M., Nishimura M., Hara-Nishimura I., Activation of Arabidopsis vacuolar processing enzyme by self-catalytic removal of an auto-inhibitory domain of the C-terminal propeptide. Plant Cell Physiol. 43, 143–151 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Shen J., et al. , Organelle pH in the Arabidopsis endomembrane system. Mol. Plant 6, 1419–1437 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Mindell J. A., Lysosomal acidification mechanisms. Annu. Rev. Physiol. 74, 69–86 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Zhao L., et al. , Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage. Cell Res. 24, 344–358 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernath-Levin K., et al. , Peptide macrocyclization by a bifunctional endoprotease. Chem. Biol. 22, 571–582 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Dall E., Fegg J. C., Briza P., Brandstetter H., Structure and mechanism of an aspartimide-dependent peptide ligase in human legumain. Angew. Chem. Int. Ed. Engl. 54, 2917–2921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatsugai N., et al. , A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305, 855–858 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Hara-Nishimura I., Hatsugai N., Nakaune S., Kuroyanagi M., Nishimura M., Vacuolar processing enzyme: An executor of plant cell death. Curr. Opin. Plant Biol. 8, 404–408 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Hatsugai N., Yamada K., Goto-Yamada S., Hara-Nishimura I., Vacuolar processing enzyme in plant programmed cell death. Front. Plant Sci. 6, 234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Souza Cândido E., et al. , Plant storage proteins with antimicrobial activity: Novel insights into plant defense mechanisms. FASEB J 25, 3290–3305 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Müntz K., Shutov A. D., Legumains and their functions in plants. Trends Plant Sci. 7, 340–344 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Carrington D. M., Auffret A., Hanke D. E., Polypeptide ligation occurs during post-translational modification of concanavalin A. Nature 313, 64–67 (1985). [DOI] [PubMed] [Google Scholar]
- 43.Bowles D. J., et al. , Posttranslational processing of concanavalin A precursors in jackbean cotyledons. J. Cell Biol. 102, 1284–1297 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Min W., Jones D. H., In vitro splicing of concanavalin A is catalyzed by asparaginyl endopeptidase. Nat. Struct. Biol. 1, 502–504 (1994). [DOI] [PubMed] [Google Scholar]
- 45.Zauner F. B., et al. , Crystal structure of plant legumain reveals a unique two-chain state with pH-dependent activity regulation. Plant Cell 30, 686–699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haywood J., et al. , Structural basis of ribosomal peptide macrocyclization in plants. eLife 7, e32955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang R., et al. , Engineering a catalytically efficient recombinant protein ligase. J. Am. Chem. Soc. 139, 5351–5358 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Trabi M., et al. , Variations in cyclotide expression in Viola species. J. Nat. Prod. 67, 806–810 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Schechter I., Berger A., On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27, 157–162 (1967). [DOI] [PubMed] [Google Scholar]
- 50.Zauner F. B., Elsässer B., Dall E., Cabrele C., Brandstetter H., Structural analyses of Arabidopsis thaliana legumain γ reveal differential recognition and processing of proteolysis and ligation substrates. J. Biol. Chem. 293, 8934–8946 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabsch W., XDS. Acta Crystallogr. D Struct. Biol. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Struct. Biol. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bricogne G., et al. , BUSTER (Global Phasing Ltd., Cambridge, UK, 2016). [Google Scholar]
- 54.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Best R. B., et al. , Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.