Significance
One potential limitation of the understanding of high-altitude adaptation is that most previous research focused on individual species, without taking into account the phylogenetic context in which similar genetic adaptations may have evolved. We compared 3 pairs of closely related high- and low-altitude passerine birds. We showed that adaptive convergence in all 3 high-altitude species seldom occurred at the amino acid substitution level but was more common on positively selected genes. We also revealed an altitude-related expression pattern in differentially expressed genes between all 3 pairs and altitude-associated genes, which differed from the tissue-specific expression pattern across all genes.
Keywords: transcriptome, positive selection, gene expression pattern, high altitude, convergence
Abstract
High-altitude environments present strong stresses for living organisms, which have driven striking phenotypic and genetic adaptations. While previous studies have revealed multiple genetic adaptations in high-altitude species, how evolutionary history (i.e., phylogenetic background) contributes to similarity in genetic adaptations to high-altitude environments is largely unknown, in particular in a group of birds. We explored this in 3 high-altitude passerine birds from the Qinghai-Tibet Plateau and their low-altitude relatives in lowland eastern China. We generated transcriptomic data for 5 tissues across these species and compared sequence changes and expression shifts between high- and low-altitude pairs. Sequence comparison revealed that similarity in all 3 high-altitude species was high for genes under positive selection (218 genes) but low in amino acid substitutions (only 4 genes sharing identical amino acid substitutions). Expression profiles for all genes identified a tissue-specific expression pattern (i.e., all species clustered by tissue). By contrast, an altitude-related pattern was observed in genes differentially expressed between all 3 species pairs and genes associated with altitude, suggesting that the high-altitude environment may drive similar expression shifts in the 3 high-altitude species. Gene expression level, gene connectivity, and the interactions of these 2 factors with altitude were correlated with evolutionary rates. Our results provide evidence for how gene sequence changes and expression shifts work in a concerted way in a group of high-altitude birds, leading to similar evolution routes in response to high-altitude environmental stresses.
Species inhabiting high-altitude environments are subjected to severe selective pressure due to adverse environmental conditions, including hypoxia, low temperatures, and strong ultraviolet radiation (1). The Qinghai-Tibet Plateau (QTP) is the highest and largest plateau in the world, with an average elevation of 4,500 m above sea level (2). This harsh high-altitude environment has resulted in the evolution of similar phenotypic adaptations in humans, mammals, and birds, for example, hypoxic resistance, cold tolerance, enhanced metabolic capacity, and increased body masses (3–5). However, despite the substantial similarity of phenotypic adaptations identified in these species, genomic studies have shown that different organisms adapt to high altitudes via multiple genetic routes (1, 6–9). Considering that the similarity in genetic adaptations to high altitudes is largely subject to phylogenetic context (10–12), comparisons of phylogenetically distinct species in previous studies may have underestimated the actual degree of similarity in genomic evolution. Thus, studies on high-altitude adaptations of closely related species in a robust phylogenetic context will largely expand our current understanding of adaptive evolution to highly selective extreme environments.
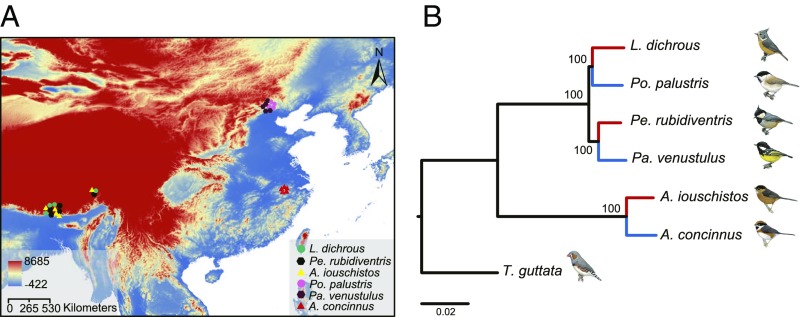
A group of passerine birds, tits, provide a unique opportunity to investigate high-altitude adaptations within a comparative framework (13). Three high-altitude tits, the gray-crested tit (Lophophanes dichrous), the rufous-vented tit (Periparus rubidiventris), and the black-throated tit (Aegithalos iouschistos), are residents of the high-altitude mountains on the QTP (over 2,300 m) (13–15). Their respective low-altitude relatives, the marsh tit (Poecile palustris), the yellow-bellied tit (Pardaliparus venustulus), and the rufous-fronted tit (Aegithalos concinnus), are residents of the lowland from sea level to low-altitude mountains (13–15). Previous phylogeny revealed that they are closely related species with similar divergence times between high- and low-altitude pairs (suggested by the similar genetic distance of mitochondrial nicotinamide adenine dinucleotide dehydrogenase subunit 2 gene, ∼0.11–0.14 between 3 pairs) (16, 17).
To explore high-altitude adaptations within this phylogenetic context, we compared transcriptomic data of high-altitude tits with those of their low-altitude relatives by integrating protein-coding sequence comparison and gene expression analysis across multiple tissues. Unlike most comparative studies on high-altitude adaptation that have only focused on mammals (6, 9, 18–22), fishes (23–26), and plants (27), this work investigated transcriptomic changes in a group of bird species native to the high-altitude environment by collecting a large number of tissue samples (e.g., 128 samples from 5 tissues of multiple individuals of multiple species). The large-scale analysis presented in this study has only been conducted for model species like humans, rats, mice, and monkeys (9, 28–32). Our sequence comparison revealed that the 3 high-altitude tits were highly similar for positively selected genes but were different in amino acid substitutions of convergent genes. The expression profiles in general revealed a tissue-clustered pattern, but altitude-associated genes and differentially expressed genes between all 3 pairs have shifted their expression patterns in an altitude-clustered pattern. Evolutionary rates were indirectly correlated to altitude via the interaction between altitude and gene expression and via the interaction between altitude and gene connectivity. This suggested that sequence changes and gene expression shifts may work together in the 3 high-altitude species. Our results provide insightful understanding of how species respond to high-altitude environments by combining protein-coding sequence changes and gene expression shifts.
Results
De Novo Transcriptome Assembly and Quality Assessment.
We sequenced 128 samples from 5 tissues (cardiac muscle, flight muscle, liver, lung, and kidney) across 6 tit species (Fig. 1A and SI Appendix, Tables S1 and S2). These tissues are known to be critical to metabolic performance and oxygen utilization (33, 34). After quality control, ∼1,433 gigabase pairs were kept for subsequent analyses. Transcriptional reads were assembled into transcripts for each species using Trinity (35) and CD-HIT (36). The contig ExN50 values across the 6 species had similar distributions, with peak lengths ranging from 2,223 to 3,790 base pairs and corresponding numbers of transcripts ranging from 27,581 to 36,505 (SI Appendix, Fig. S1 and Table S3). Samples were aligned with their own transcriptomes, with mapping rates ranging from 78.75 to 90.81% across 128 samples: the mean alignment rates were 85.53, 84.61, 85.23, 83.92, 85.28, and 83.54% for L. dichrous, P. palustris, P. rubidiventris, P. venustulus, A. iouschistos, and A. concinnus, respectively (SI Appendix, Table S2). Based on 4,915 conserved avian orthologs, our Benchmarking Universal Single-Copy Orthologs (BUSCO) assessment (37) of transcriptome completeness identified 4,467 (90.9%) complete and fragmented BUSCOs in L. dichrous, 4,427 (90.1%) in P. palustris, 4,404 (89.6%) in P. rubidiventris, 4,344 (88.4%) in P. venustulus, 4,389 (89.3%) in A. iouschistos, and 4,393 (89.4%) in A. concinnus (SI Appendix, Table S4). Thus, our results indicated that the 6 transcriptomes were well assembled and relatively complete. To further filter out erroneous and nonexpressed transcripts, we used RSEM (38) to quantify the expression of each transcript in each sample. For each species, we kept only transcripts with fragments per kilobase of exon per million fragments mapped >1 in at least half of all samples from any one tissue.
Fig. 1.
Localities and phylogenetic relationships of the 6 tit species. (A) Sampling locations. The high-altitude species (Lophophanes dichrous, Periparus rubidiventris, and Aegithalos iouschistos) were sampled from altitudes of 2730–3900 m above sea level on the Qinghai-Tibet Plateau. The low-altitude species (Poecile palustris, Pardaliparus venustulus, and A. concinnus) were sampled from areas below 500 m above sea level in eastern China. (B) Maximum likelihood phylogeny of the 6 tit species with the zebra finch (Taeniopygia guttata) as an out-group based on 4-fold-degenerate sites of one-to-one orthologous coding sequences. Maximum likelihood bootstrap support values for all high- and low-altitude pairs were 100%. The scale bar indicates the number of substitutions per site. Red branches indicate the high-altitude species; blue branches indicate the low-altitude species. Bird illustrations reprinted with permission from ref. 15.
Ortholog Identification, Annotation, and Phylogenetic Analysis.
We used the reciprocal best-hit method to identify orthologous genes between the 6 tits and the out-group (zebra finch, Taeniopygia guttata). We identified 7,915 one-to-one putatively orthologous genes across 7 species based on the respective best Basic Local Alignment Search Tool hits between the 6 tits and the zebra finch. Of them, we identified 7,914 (99.99%) matches in the National Center for Biotechnology Information nonredundant protein database (NR), 7,875 (99.49%) in Swiss-Prot, 7,901 (99.82%) in Interpro, 6,574 (83.06%) in Gene Ontology (GO), and 5,953 (75.21%) in the Kyoto Encyclopedia of Genes and Genomes (KEGG) (SI Appendix, Fig. S2). After sequence alignment and filtration, the 7,048 high-confidence alignments were retained for downstream analyses. We constructed a maximum likelihood (ML) phylogenetic tree of the 6 tits and the zebra finch based on 4-fold-degenerate (4D) sites of these gene alignments. The 6 species were clustered into 3 well-supported high- and low-altitude pairs (ML bootstrap values = 100% for all nodes): L. dichrous (high) and P. palustris (low), P. rubidiventris (high) and P. venustulus (low), and A. iouschistos (high) and A. concinnus (low) (Fig. 1). The topology of our tree was congruent with previous phylogenies based on fewer nuclear and mitochondrial markers (13, 16, 17), confirming that high- and low-altitude pairs are closely related.
Coding Sequence Change Involved in High-Altitude Adaptation.
We identified positively selected genes using the branch-site likelihood ratio test in PAML (39) and obtained 379 genes across all 3 high-altitude species: 307 in L. dichrous, 294 in P. rubidiventris, and 317 in A. iouschistos (SI Appendix, Table S5). Out of them, 321 genes (84.7%) were shared between any 2 of the 3 high-altitude species, and 218 genes (57.5%) were shared among the 3 high-altitude species (SI Appendix, Fig. S3). We further explored genes with common amino acid substitutions in the 3 high-altitude species using Jones, Taylor, and Thorton (JTT)-fgene amino acid substitution models (40). We identified 280 convergent genes with common amino acid substitutions in any 2 of the 3 high-altitude species (SI Appendix, Table S6). Of these genes, we found 99 adaptively convergent genes that not only have been subject to positive selection but also have undergone nonrandom convergent changes for any 2 of the 3 high-altitude species (SI Appendix, Table S7). Of them, only 4 genes, CYP2R1, L2HGDH, HN1L, and IFNAR2, had common amino acid substitutions in all 3 high-altitude species (SI Appendix, Table S8). CYP2R1 is involved in cardiomyocyte proliferation and hypertrophy (41), L2HGDH contributes to electron transport and glycolysis (42), HN1L affects the systolic function of the left ventricle (43), and IFNAR2 is related to angiogenesis (44). Together, by examining coding sequence changes of the 3 high-altitude species, we demonstrated that the convergence more likely occurred on positively selected genes but less likely in common amino acid substitutions.
Gene Expression Profiles Across Tissues and Species.
De novo transcriptome assembly may lead to differences in the transcript lengths and numbers among different species, which would cause bias in cross-species gene expression analysis. We thus trimmed the 7,048 orthologs across the 6 species to the same lengths and calculated the expression levels using RSEM (38). We normalized gene expression data using trimmed mean of M-value normalization in edgeR (45) to consider variations among biological replicates and using scaling factors based on comparisons of the median expression levels of the top 1,000 most conserved genes to consider variations among tissues and species (see SI Appendix, Materials and Methods for details). To check if gene expression data after normalization performed better than nonnormalized data, we compared the coefficient of variance (CV) of gene expression levels among all samples based on all genes, conserved genes, and nonconserved genes and found that the normalized data had a significantly lower CV than the nonnormalized data (SI Appendix, Fig. S4 and Table S9). Furthermore, Spearman’s correlation coefficients between all pairs of samples for each tissue were significantly higher in normalized data for the trimmed orthologs than for the raw orthologs (P = 4.21e-4, Mann–Whitney U test) (SI Appendix, Fig. S5). These results suggested that the normalized expression data have significantly reduced bias due to different assemblies, biological replicates, tissues, and species, and we subsequently performed gene expression analyses based on the normalized data.
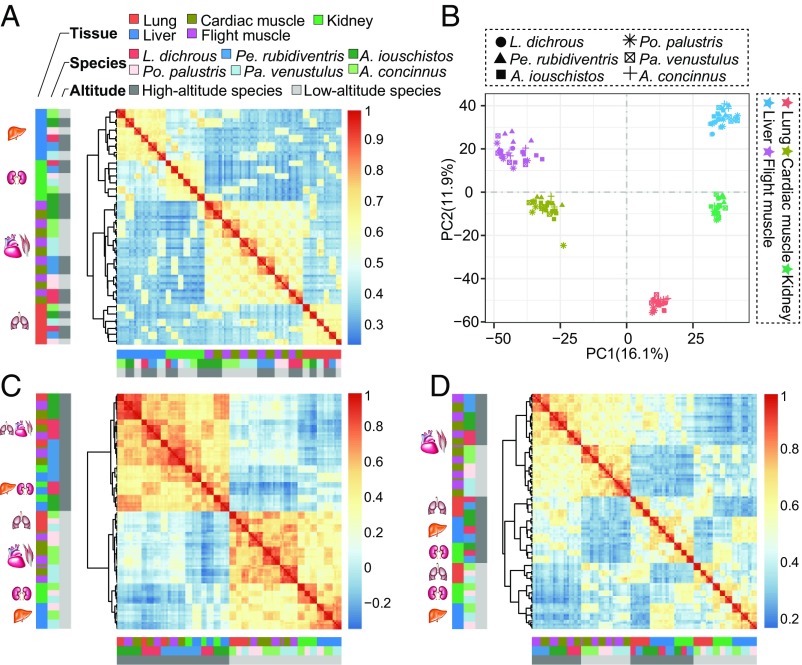
In general, 2 patterns, tissue-specific expression and species-specific expression, have been observed in multiple tissues across different species. The tissue-specific expression pattern suggests that gene expression should be more similar in the same tissue among different species than in different tissues within the same species (30–32, 46, 47). The species-specific expression pattern suggests that gene expression should be more similar in different tissues within the same species than in the same tissue among species (9, 48). We investigated patterns of gene expression across all tissues with a hierarchal-clustering expression profile analysis based on Spearman’s correlation coefficients of gene expression between pairs of samples. Samples of the same tissue from different species clustered together, suggesting a general tissue-specific expression pattern (Fig. 2A). The principal component analysis (PCA) also revealed the tissue-specific expression pattern: samples across all 6 species clustered by tissue (Fig. 2B). This pattern was further supported by ANOVA (SI Appendix, Table S10). Our results indicated that differences among tissues were more significant than those among species, suggesting that tissue differentiation might precede species differentiation (32). One exception to this was that samples of flight muscle and cardiac muscle were clustered together in each species (species-specific expression), suggesting that muscular tissues (e.g., cardiac and flight muscles) may have similar expression profiles (Fig. 2A).
Fig. 2.
Gene expression patterns across the 6 tit species: Lophophanes dichrous, Periparus rubidiventris, Aegithalos iouschistos, Poecile palustris, Pardaliparus venustulus, and A. concinnus. (A) Symmetrical heat map of Spearman’s correlation coefficients between all pairs of samples across all genes. (B) PCA of the log-transformed normalized expression levels of all orthologs across all species and tissues. Species are represented by point shape; tissues are represented by point color. (C) Symmetrical heat map of Spearman’s correlation coefficients between all pairs of samples across genes differentially expressed between all 3 high- and low-altitude pairs (see SI Appendix, Materials and Methods for details). Key is as in A. (D) Symmetrical heat map of Spearman’s correlation coefficients between all pairs of samples across genes associated with altitude (see SI Appendix, Materials and Methods for details). Key is as in A. Data are hierarchically clustered (complete method, correlation distance) in A, C, and D.
We explored the gene expression pattern for each tissue because previous studies showed that high-altitude environments can drive expressional changes in these tissues (21, 22). Our neighbor-joining trees based on pairwise expression distance matrices revealed that these tissues generally clustered according to phylogenetic relationship (SI Appendix, Fig. S6). The results of PCA in these tissues did not detect any clear clustering patterns, except that high- and low-altitude tits were separated in the lung and kidney along the second principal component (SI Appendix, Fig. S7).
Gene Expression Shift Accompanying High-Altitude Environment.
To detect expression shifts in response to the high-altitude environment, we identified differentially expressed genes between each of the 3 high- and low-altitude pairs using a combination of DESeq2 (49), edgeR (45), reproducibility-optimized test statistic (ROTS) (50), and limma (51). We identified 69 differentially expressed genes in the lung (34 up-regulated and 35 down-regulated in the high-altitude species), 53 in the cardiac muscle (15 up-regulated and 38 down-regulated in the high-altitude species), 89 in the kidney (35 up-regulated and 54 down-regulated in the high-altitude species), 51 in the liver (16 up-regulated and 35 down-regulated in the high-altitude species), and 56 in the flight muscle (27 up-regulated and 29 down-regulated in the high-altitude species) (SI Appendix, Figs. S8–S10 and Dataset S1). The expression profiles of a combination of all differentially expressed genes showed a different pattern from those of all genes (e.g., tissue-specific expression) by separating high-altitude tits from low-altitude tits (Fig. 2C), suggesting that the 3 high-altitude tits have similarly shifted their expression profiles.
To classify groups of genes interacting at the network level, we used weighted gene coexpression network analysis (WGCNA) (52) to determine modules. Module expression was summarized using the first principal component of gene expression for each module and regressed against the altitude of the sample. We identified 28 modules in the lung, 26 in the cardiac muscle, 23 in the kidney, 24 in the liver, and 25 in the flight muscle. Out of these modules, 5 in the lung, 3 in the cardiac muscle, 5 in the kidney, 6 in the liver, and 5 in the flight muscle were significantly correlated with altitude (SI Appendix, Figs. S11–S14). We considered genes within the altitude-related modules to be altitude-associated genes. The expression profiles of these altitude-associated genes revealed that muscular tissues deviated from other tissues, with samples of cardiac and flight muscles being one clade and those of the remaining tissues (liver, lung, and kidney) being the other clade (Fig. 2D). Within each clade, high-altitude tits were separated from low-altitude tits (Fig. 2D). These results suggested that muscular tissues and other tissues may have different regulatory shifts accompanying the high-altitude environment.
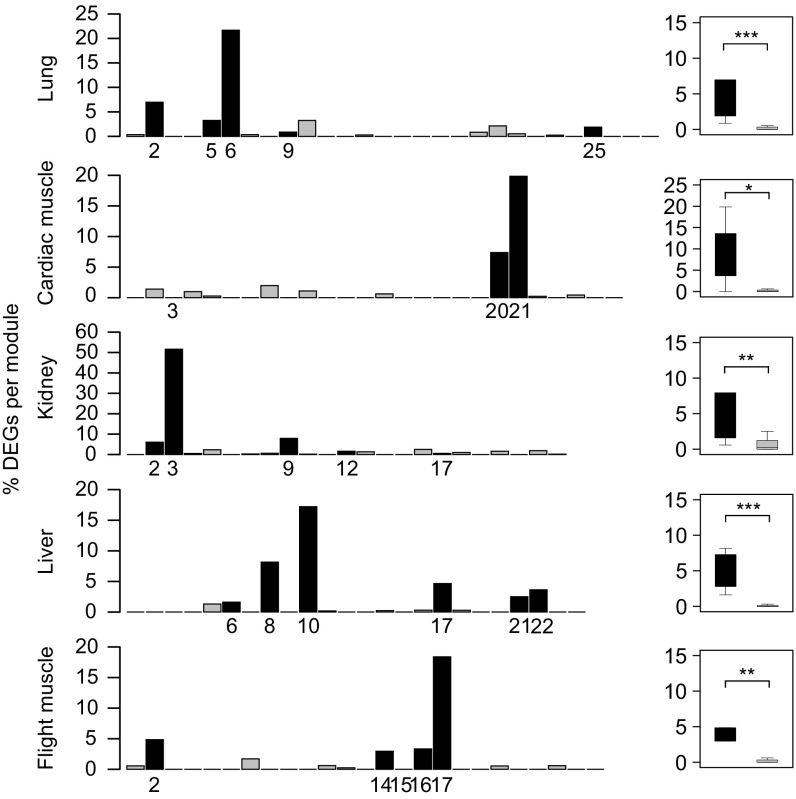
Interestingly, we found a significantly higher proportion of differentially expressed genes in the altitude-related modules than in the other modules (P < 0.05, Mann–Whitney U test; Fig. 3), suggesting that the high-altitude environment has promoted expressional changes. Subsequently, we tested correlations between module expression and phenotypic traits. Most or all of the altitude-related modules were significantly correlated with body length and tarsus length (SI Appendix, Table S11), suggesting that genes in these modules may be involved in these phenotypic changes. A previous study on functional traits revealed similar phenotypic changes (53), indicating that high-altitude species generally have a larger body size to increase cold resistance and a longer tarsus to facilitate locomotion and foraging in open habitats at high altitudes (54).
Fig. 3.
Relative numbers of differentially expressed genes (DEGs) identified in each module for each tissue. Black bars represent altitude-associated modules, and gray bars represent all other modules. Altitude-associated modules are labeled. The box plot shows the relative number of DEGs in altitude-associated modules compared with the other modules. The P value above each pair indicates the significance of the difference in relative number of DEGs between altitude-associated and non-altitude-associated modules (Mann–Whitney U test). Displayed significant scores are ***P < 0.001, **P < 0.01**, and *P < 0.05.
Evolutionary Rate Was Indirectly Correlated with Altitude.
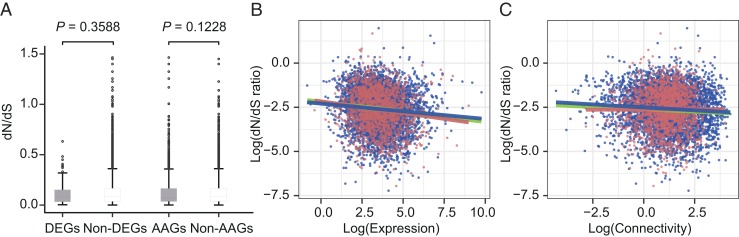
The ratio of nonsynonymous substitutions per nonsynonymous site to synonymous substitutions per synonymous site (dN/dS) represents the evolutionary rate of a protein-coding gene (55). We found that dN/dS ratios did not differ between differentially expressed genes and nondifferentially expressed genes (P = 0.3588, Mann–Whitney U test) or between altitude-associated genes and non-altitude-associated genes (P = 0.1228, Mann–Whitney U test) (Fig. 4A). Likewise, we also did not find significant correlation between dN/dS ratio and altitude (P = 0.211, linear model) (SI Appendix, Table S12), but we identified significant negative correlations between gene expression and dN/dS ratio (P < 2.2e-16, linear model), as well as between gene connectivity and dN/dS ratio (P = 7.75e-6, linear model) (Fig. 4 B and C; SI Appendix, Table S12). We found significant correlations between expression–altitude (the interaction between expression and altitude) and dN/dS ratio (P = 0.0474, linear model), as well as between connectivity–altitude (the interaction between connectivity and altitude) and dN/dS ratio (P = 9.31e-3, linear model) (Fig. 4 B and C and SI Appendix, Table S12). These results indicated that rates of genetic evolution were correlated with gene expression and connectivity and were indirectly correlated with altitude via the interactions of these factors with altitude, thus suggesting that protein-coding sequence changes and gene expression shifts may work together in the 3 high-altitude tits.
Fig. 4.
The correlation between evolutionary rates (dN/dS ratios) and altitude, gene expression, and connectivity. (A) Box plots showing the dN/dS ratios across all pairs of complementary gene sets: DEGs and non-DEGs and altitude-associated genes (AAGs) and non-AAGs. The P value above each pair indicates the significance of the difference in dN/dS ratio between the 2 complementary gene sets (Mann–Whitney U test). (B and C) dN/dS as a function of (B) gene expression and (C) connectivity. All lines represent the results of smoothing functions for different sets of genes. The green line represents all orthologous genes, the pink line represents altitude-associated genes, and the blue line represents non-altitude-associated genes. Ortholog expression and dN/dS ratio are negatively correlated, as are connectivity and dN/dS ratio.
Discussion
By systematically investigating 3 high-altitude passerine birds and their low-altitude relatives within a phylogenetic context, our comparative transcriptomics expanded our current understanding of how organisms respond to a highly selective environment. The 3 high-altitude birds were genetically convergent on positively selected genes (218 genes), but their amino acid substitutions mostly occurred at divergent sites, with only 4 genes sharing common amino acid changes. In gene expression profiles, we found that the high-altitude environment may have promoted substantial expression shifts in the 3 high-altitude birds since expression profiles of differentially expressed genes and altitude-associated genes (altitude-clustered pattern) largely differed from those of all genes (tissue-clustered pattern). Few genes under positive selection for all 3 high-altitude species (2.3%, 5 out of 218) overlapped with differentially expressed genes, but the interaction between gene expression and altitude and the interaction between gene connectivity and altitude were correlated with evolutionary rates. These findings suggested that the 3 high-altitude birds may evolve in a concerted way through both sequence changes and expression shifts.
Although species evolution relies on random processes like mutations, the probability of evolving similar adaptive strategies may also be influenced by evolutionary history. We documented a high similarity in positively selected genes shared among the 3 high-altitude tits. In broader comparisons with other high-altitude species, we found very few genes shared by the 3 high-altitude tits and other species (56, 57). This suggested that genetic background (i.e., genetic contingency) may constrain subsequent changes during adaptive evolution.
Previous efforts to study high-altitude adaptation have focused on genetic changes in the genome, whereas dynamics and mechanisms of transcriptomic changes have received much less attention (but see refs. 58 and 59). Our transcriptomic comparison across multiple tissues from 3 pairs of birds with similar phylogenetic divergence provides insights into how a high-altitude environment can drive transcriptomic changes in these organisms. Even though high-altitude adaptation can be an important factor driving these phenotypic changes, similar expression variations observed in the 3 high-altitude tits may also be partially contributed by habitat-specific responses or plastic reactions to environmental changes. Currently, evaluating how gene expression plasticity might have contributed to adaptation and acclimation to high-altitude environments would require challenging transplanting experiments and is only possible for species with broad altitudinal distributions (e.g., deer mice and rufous-collared sparrows) (60, 61). Future studies integrating whole-animal physiological measurements and transcriptomic comparisons based on transplanting experiments in natural populations could potentially provide us with a more unified picture of the regulatory circuits driving high-altitude adaptation.
In addition, expressional changes can also be driven by noncoding regulatory changes, which we could not explore using the transcriptomic data collected in this study. There is emerging evidence that noncoding regulatory regions, such as cis-regulatory elements, could be an important factor driving expressional divergence (62, 63). With weaker pleiotropic effects than coding genes, noncoding regions can have much higher evolutionary flexibility and have been found to be involved in several cases of adaptive convergent evolution (64, 65). Future studies with well-annotated genomes and resequencing data from multiple individuals could effectively explore adaptive evolution in noncoding regions more thoroughly. Despite several limitations discussed above, our study provides a glimpse into this interesting landscape of transcriptomic rewiring in high-altitude adaptation in avian species and provides an important basis for many promising studies in the coming years.
Materials and Methods
Details of the ethics statement, specimen collection, RNA extraction, transcriptome sequencing and assembly, quantification, ortholog determination, phylogenetic analysis, convergence identification at the sequence level, and gene expression pattern analysis can be found in the SI Appendix, Materials and Methods.
Specimen Collection and De Novo Transcriptome Assembly.
We captured and killed 28 male and female adult birds (SI Appendix, Table S1). We sequenced the transcriptomes of 128 samples, including 5 tissues (cardiac muscle, flight muscle, liver, lung, and kidney) for 28 birds. We used Trinity (35) to assemble de novo transcriptomes for the 6 species. Contig ExN50 length was calculated to estimate the quality of each assembly. After the removal of redundant sequences, we used Bowtie 2 (66) to map reads back to their respective transcriptomes. We then used BUSCO (37) to determine the transcriptome assembly completeness based on conserved avian orthologs. We quantified the expression of each transcript in each sample using RSEM (38) and retained only those transcripts that were expressed in at least half of all samples from any one tissue of each species.
Ortholog Identification and Phylogenetic Analysis.
Using the reciprocal best-hit method with an E value cutoff of 1e-10 and a minimum percentage identity of 30%, we identified genes that were orthologous across all 6 species and the out-group (Taeniopygia guttata). Genes were functionally annotated against NR, Swiss-Prot, InterPro, GO, and KEGG. Well-aligned orthologous coding sequences and protein sequences from the 7 species were obtained using Exonerate (67), Prank (68), and Gblocks (69). A phylogenetic tree based on 4D sites from all orthologs was reconstructed using RAxML (70).
Convergence at the Sequence Level.
Positively selected genes were identified using the branch-site likelihood ratio test, setting 1 to 3 high-altitude species as foreground branches in PAML (39). Convergent genes were obtained by using JTT-fgene amino acid substitution models, coupled with Poisson tests, to examine the convergence between any pair of the 3 high-altitude species (40). Genes identified as both a positively selected gene and a convergent gene for any 2 of the 3 high-altitude species were considered adaptively convergent genes.
Gene Expression Pattern Analysis.
Gene expression levels for the trimmed orthologs from the 5 tissues across all 6 species were calculated using RSEM (38) and normalized and then log transformed for principal component analysis. We identified differentially expressed genes within each high- and low-altitude pair using DESeq2 (49), edgeR (45), ROTS (50), and limma (51). Especially, we used DESeqDataSetFromMatrix and DESeq in DESeq2 (49) to conduct differential expression analysis. For edgeR (45), we used calcNormFactors to calculate normalization factors, estimateDisp to estimate common dispersion and tagwise dispersion, and exactTest to determine expression differentiation. For ROTS (50), we used calcNormFactors in edgeR (45) to calculate normalization factors, voom in limma (44) to transform data and ROTS to perform differential expression analysis. For limma (51), we used lmFit to fit a linear model and eBayes to compute moderated t statistics for differential expression. A weighted gene coexpression network analysis was performed using the R package WGCNA (52) to classify genes into modules. Genes in altitude-related modules were considered altitude-associated genes. We determined the correlation in gene expression levels between each pair of samples using Spearman’s correlation. This analysis was performed for all genes, differentially expressed genes, and altitude-associated genes. We performed the hierarchical clustering of Spearman’s correlation coefficients for each pair of samples for each gene set using the complete-linkage agglomerative method and the correlation distance metric. Symmetrical heat maps were generated. We determined the correlations between phenotypic traits (including body length, body weight, wing length, tail length, tarsus length, and culmen length) and the expression levels of the altitude-related modules.
Comparison of Evolutionary Rates and Detection of Explanatory Variables.
We used Mann–Whitney U tests to compare evolutionary rates between each gene set and its complementary gene set, that is, differentially expressed genes and the remaining genes and altitude-associated genes and the remaining genes. We also used linear models to test the effects of altitude, gene expression, and gene connectivity, as well as altitude plus gene expression and altitude plus gene connectivity, on evolutionary rates.
Supplementary Material
Acknowledgments
We thank Yongbin Chang and Yongjie Wu for assistance with field work, Zhengting Zou and Qi Wu for assistance with data analysis, Weiwei Zhai for revising the manuscript, and Yong E. Zhang for discussion. We also thank the editor and 2 anonymous reviewers for valuable comments and suggestions to improve the manuscript. This study was funded by grants from the Strategic Priority Research Program, Chinese Academy of Sciences [XDB13020300 (to F.L.)], and the National Science Foundation of China [31672275 (to Y.Q.), 31630069 (to F.L.), and 31572249 (to F.L.)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All sequencing data reported in this paper have been deposited into the National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/ (BioProject ID PRJNA470787).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819657116/-/DCSupplemental.
References
- 1.Qu Y., et al. , Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat. Commun. 4, 2071 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Ruddiman W. F., Kutzbach J. E., Plateau uplift and climate change. Sci. Am. 264, 66–74 (1991).1711238 [Google Scholar]
- 3.Monge C., Leon-Velarde F., Physiological adaptation to high altitude: Oxygen transport in mammals and birds. Physiol. Rev. 71, 1135–1172 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Wu T., Kayser B., High altitude adaptation in Tibetans. High Alt. Med. Biol. 7, 193–208 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Beall C. M., Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc. Natl. Acad. Sci. U.S.A. 104 (suppl. 1), 8655–8660 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li M., et al. , Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 45, 1431–1438 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Qu Y., et al. , Genetic responses to seasonal variation in altitudinal stress: Whole-genome resequencing of great tit in eastern Himalayas. Sci. Rep. 5, 14256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z., et al. , Convergent evolution of rumen microbiomes in high-altitude mammals. Curr. Biol. 26, 1873–1879 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Yu L., et al. , Genomic analysis of snub-nosed monkeys (Rhinopithecus) identifies genes and processes related to high-altitude adaptation. Nat. Genet. 48, 947–952 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Scott G. R., Egginton S., Richards J. G., Milsom W. K., Evolution of muscle phenotype for extreme high altitude flight in the bar-headed goose. Proc. Biol. Sci. 276, 3645–3653 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns K. J., et al. , Phylogenetics and diversification of tanagers (Passeriformes: Thraupidae), the largest radiation of neotropical songbirds. Mol. Phylogenet. Evol. 75, 41–77 (2014). [DOI] [PubMed] [Google Scholar]
- 12.McGuire J. A., et al. , Molecular phylogenetics and the diversification of hummingbirds. Curr. Biol. 24, 910–916 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Zhu X., et al. , Divergent and parallel routes of biochemical adaptation in high-altitude passerine birds from the Qinghai-Tibet Plateau. Proc. Natl. Acad. Sci. U.S.A. 115, 1865–1870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G., Zheng B., Liu G., Fauna Sinica: Aves (Science Press, Beijing, 1982). [Google Scholar]
- 15.del Hoyo J., Elliott A., Sargatal J., Christie D., de Juana E., Handbook of the Birds of the World Alive (Lynx Edicions, Barcelona, 2018). [Google Scholar]
- 16.Päckert M., et al. , Horizontal and elevational phylogeographic patterns of Himalayan and Southeast Asian forest passerines (Aves: Passeriformes). J. Biogeogr. 39, 556–573 (2012). [Google Scholar]
- 17.Johansson U. S., Nylinder S., Ohlson J. I., Tietze D. T., Reconstruction of the late Miocene biogeographical history of tits and chickadees (Aves: Passeriformes: Paridae): A comparison between discrete area analyses and probabilistic diffusion approach. J. Biogeogr. 45, 14–25 (2018). [Google Scholar]
- 18.Qiu Q., et al. , The yak genome and adaptation to life at high altitude. Nat. Genet. 44, 946–949 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Ge R. L., et al. , Draft genome sequence of the Tibetan antelope. Nat. Commun. 4, 1858 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gou X., et al. , Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 24, 1308–1315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K., et al. , Different gene expressions between cattle and yak provide insights into high-altitude adaptation. Anim. Genet. 47, 28–35 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Qi X., et al. , The transcriptomic landscape of yaks reveals molecular pathways for high altitude adaptation. Genome Biol. Evol. 11, 72–85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., et al. , Evidence for adaptation to the Tibetan Plateau inferred from Tibetan loach transcriptomes. Genome Biol. Evol. 7, 2970–2982 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L., Wang Y., Zhang Z., He S., Comprehensive transcriptome analysis reveals accelerated genic evolution in a Tibet fish, Gymnodiptychus pachycheilus. Genome Biol. Evol. 7, 251–261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Yang L., Wu B., Song Z., He S., Transcriptome analysis of the plateau fish (Triplophysa dalaica): Implications for adaptation to hypoxia in fishes. Gene 565, 211–220 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Ma X., Dai W., Kang J., Yang L., He S., Comprehensive transcriptome analysis of six catfish species from an altitude gradient reveals adaptive evolution in Tibetan fishes. G3 (Bethesda) 6, 141–148.(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo X., et al. , The genomes of two Eutrema species provide insight into plant adaptation to high altitudes. DNA Res. 25, 307–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khaitovich P., et al. , Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309, 1850–1854 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Oldham M. C., Horvath S., Geschwind D. H., Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc. Natl. Acad. Sci. U.S.A. 103, 17973–17978 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brawand D., et al. , The evolution of gene expression levels in mammalian organs. Nature 478, 343–348 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Barbosa-Morais N. L., et al. , The evolutionary landscape of alternative splicing in vertebrate species. Science 338, 1587–1593 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Merkin J., Russell C., Chen P., Burge C. B., Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338, 1593–1599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hochachka P. W. Mechanism and evolution of hypoxia-tolerance in humans. J. Exp. Biol. 201, 1243–1254 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Hochachka P. W., Rupert J. L., Monge C., Adaptation and conservation of physiological systems in the evolution of human hypoxia tolerance. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 124, 1–17 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Haas B. J., et al. , De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu L., Niu B., Zhu Z., Wu S., Li W., CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Zou Z., Zhang J., Are convergent and parallel amino acid substitutions in protein evolution more prevalent than neutral expectations? Mol. Biol. Evol. 32, 2085–2096 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T. J., et al. , Common genetic determinants of vitamin D insufficiency: A genome-wide association study. Lancet 376, 180–188 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oldham W. M., Clish C. B., Yang Y., Loscalzo J., Hypoxia-mediated increases in L-2-hydroxyglutarate coordinate the metabolic response to reductive stress. Cell Metab. 22, 291–303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasan R. S., et al. , Genetic variants associated with cardiac structure and function: A meta-analysis and replication of genome-wide association data. JAMA 302, 168–178 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarty M. F., Bielenberg D., Donawho C., Bucana C. D., Fidler I. J., Evidence for the causal role of endogenous interferon-α/β in the regulation of angiogenesis, tumorigenicity, and metastasis of cutaneous neoplasms. Clin. Exp. Metastasis 19, 609–615 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng-Bradley X., Rung J., Parkinson H., Brazma A., Large scale comparison of global gene expression patterns in human and mouse. Genome Biol. 11, R124 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilad Y., Mizrahi-Man O., A reanalysis of mouse ENCODE comparative gene expression data. F1000 Res. 4, 121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sousa A. M. M., et al. , Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suomi T., Seyednasrollah F., Jaakkola M. K., Faux T., Elo L. L., ROTS: An R package for reproducibility-optimized statistical testing. PLOS Comput. Biol. 13, e1005562 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ritchie M. E., et al. , Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shao S., et al. , Evolution of body morphology and beak shape revealed by a morphometric analysis of 14 Paridae species. Front. Zool. 13, 30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kennedy J. D., et al. , Ecological limits on diversification of the Himalayan core Corvoidea. Evolution 66, 2599–2613 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Axelsson E., et al. , Natural selection in avian protein-coding genes expressed in brain. Mol. Ecol. 17, 3008–3017 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Zhang W., et al. , Hypoxia adaptations in the grey wolf (Canis lupus chanco) from Qinghai-Tibet Plateau. PLoS Genet. 10, e1004466 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Y. B., et al. , Species groups distributed across elevational gradients reveal convergent and continuous genetic adaptation to high elevations. Proc. Natl. Acad. Sci. U.S.A. 115, E10634–E10641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitehead A., Galvez F., Zhang S., Williams L. M., Oleksiak M. F., Functional genomics of physiological plasticity and local adaptation in killifish. J. Hered. 102, 499–511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kenkel C. D., Matz M. V., Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat. Ecol. Evol. 1, 14 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Cheviron Z. A., Bachman G. C., Connaty A. D., McClelland G. B., Storz J. F., Regulatory changes contribute to the adaptive enhancement of thermogenic capacity in high-altitude deer mice. Proc. Natl. Acad. Sci. U.S.A. 109, 8635–8640 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scott G. R., Elogio T. S., Lui M. A., Storz J. F., Cheviron Z. A., Adaptive modifications of muscle phenotype in high-altitude deer mice are associated with evolved changes in gene regulation. Mol. Biol. Evol. 32, 1962–1976 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Signor S. A., Liu Y., Rebeiz M., Kopp A., Genetic convergence in the evolution of male-specific color patterns in Drosophila. Curr. Biol. 26, 2423–2433 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Osada N., Miyagi R., Takahashi A., Cis- and trans-regulatory effects on gene expression in a natural population of Drosophila melanogaster. Genetics 206, 2139–2148 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.King M. C., Wilson A. C., Evolution at two levels in humans and chimpanzees. Science 188, 107–116 (1975). [DOI] [PubMed] [Google Scholar]
- 65.Sackton T. B., et al. , Convergent regulatory evolution and loss of flight in paleognathous birds. Science 364, 74–78 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Slater G. S., Birney E., Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6, 31 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Löytynoja A., Goldman N., Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320, 1632–1635 (2008). [DOI] [PubMed] [Google Scholar]
- 69.Castresana J., Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- 70.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.