Abstract
Autophagy and energy metabolism are known to follow a circadian pattern. However, it is unclear whether autophagy and the circadian clock are coordinated by common control mechanisms. Here, we show that the oscillation of autophagy genes is dependent on the nutrient‐sensitive activation of TFEB and TFE3, key regulators of autophagy, lysosomal biogenesis, and cell homeostasis. TFEB and TFE3 display a circadian activation over the 24‐h cycle and are responsible for the rhythmic induction of genes involved in autophagy during the light phase. Genetic ablation of TFEB and TFE3 in mice results in deregulated autophagy over the diurnal cycle and altered gene expression causing abnormal circadian wheel‐running behavior. In addition, TFEB and TFE3 directly regulate the expression of Rev‐erbα (Nr1d1), a transcriptional repressor component of the core clock machinery also involved in the regulation of whole‐body metabolism and autophagy. Comparative analysis of the cistromes of TFEB/TFE3 and REV‐ERBα showed an extensive overlap of their binding sites, particularly in genes involved in autophagy and metabolic functions. These data reveal a direct link between nutrient and clock‐dependent regulation of gene expression shedding a new light on the crosstalk between autophagy, metabolism, and circadian cycles.
Keywords: circadian rhythm, gene oscillation, MiT‐TFE, REV‐ERBα
Subject Categories: Autophagy & Cell Death, Metabolism, Transcription
Introduction
In mammals, most essential biological functions show a rhythmic pattern close to 24 h, including sleep‐wake and feeding‐non‐feeding cycles (Kalsbeek et al, 2010). These daily variations are controlled by the circadian clock located in the suprachiasmatic nucleus (SCN) and are mainly influenced by light. Secondary oscillators are present in many brain regions and peripheral organs (e.g., liver and muscle; Zvonic et al, 2006; Asher & Schibler, 2011). While in the SCN clock genes are synchronized by light, clock oscillations in peripheral tissues depend largely on feeding time (Damiola et al, 2000; Stokkan et al, 2001). The current model for the mammalian core clock involves transcriptional and translational feedback loops in which protein products of particular clock genes oscillate to induce or suppress transcription of other clock genes resulting in both positive and negative feedback loops (Harmer et al, 2001). Members of the bHLH‐PAS transcription factor family, CLOCK and BMAL1, form heterodimers and activate the expression of the Cryptochrome (Cry1 and Cry2) and Period (Per1, Per2, and Per3; Ueda et al, 2002) genes. PER:CRY heterodimers can translocate to the nucleus to inhibit E‐box‐mediated transcription by interacting with the CLOCK:BMAL1 complex (Ko & Takahashi, 2006). Moreover, BMAL1:CLOCK heterodimer activates the expression of orphan nuclear receptors REV‐ERBs (Rev‐erbα and Rev‐erbβ) and RORs (Rorα, Rorβ, and Rorγ). These receptors can in turn either repress or activate gene transcription by competing to the binding of RORE motifs in the promoter regions of their target genes, including Bmal1, thus contributing to the circadian oscillation of gene expression(Cho et al, 2012).
Autophagy is an evolutionarily conserved cellular pathway that delivers cytosolic components to lysosomes for degradation and recycling and is a key mechanism for cell survival in the absence of nutrients (Klionsky & Emr, 2000). Recent studies indicated that autophagy displays a robust circadian rhythmicity in mouse liver, skeletal muscle, heart, and kidney (Ma et al, 2011). Indeed, autophagy undergoes rhythmic variation following the circadian feeding pattern in adult mammals. However, the mechanisms underlying circadian regulation of autophagy are still under investigation.
The basic helix–loop–helix (bHLH) MiT‐TFE transcription factors (TFs) TFEB and TFE3 are master transcriptional controllers of lysosomal biogenesis and autophagy (Sardiello et al, 2009; Settembre et al, 2011; Martina et al, 2014). We and others previously reported that the mechanistic target of rapamycin (mTOR) negatively regulates the activity of both TFEB and TFE3 in a nutrient‐dependent manner, thus controlling the switch between anabolic and catabolic pathways in response to nutrient availability (Roczniak‐Ferguson et al, 2012; Settembre et al, 2012; Martina et al, 2014). Indeed, when nutrients are available, TFEB and TFE3 are phosphorylated by mTOR and retained in the cytoplasm. During starvation, inactivation of mTOR, as well as the concomitant activation of the phosphatase calcineurin, induces TFEB/TFE3 de‐phosphorylation and nuclear translocation (Settembre et al, 2013b; Medina et al, 2015). Interestingly, we recently reported a feedback loop by which MiT‐TFE TFs may in turn influence mTOR activity by directly regulating RragD expression (Di Malta et al, 2017).
The critical role of the MiT‐TFE genes in the regulation of autophagy (Settembre et al, 2011; Martina et al, 2014) and metabolism (Settembre et al, 2013a; Mansueto et al, 2017; Pastore et al, 2017) prompted us to investigate whether they also play a role in the circadian oscillation of the expression of genes involved in these processes, either directly or indirectly through the clock machinery. Here, we show that TFEB and TFE3 are rhythmically activated in peripheral tissues in a food‐dependent/clock‐independent manner, thus regulating the expression of genes involved in autophagy. By both in vitro and in vivo studies, we demonstrated that TFEB and TFE3 directly regulate Rev‐erbα expression, a transcriptional repressor component of the core clock machinery that controls the expression of genes involved in several biological processes including autophagy and lipid metabolism. We also found that TFEB/TFE3 and REV‐ERBα bind common promoter regions, suggesting that their intrinsic inductive and repressive roles, respectively, are responsible for the rhythmic expression of genes involved in autophagy and metabolism over the daily cycle. Thus, we identified a novel mechanism that links nutrient‐ and light‐induced control of circadian rhythm through the oscillation of gene expression.
Results
Nutrient‐dependent/clock‐independent rhythmic activation of MiT‐TFE transcription factors
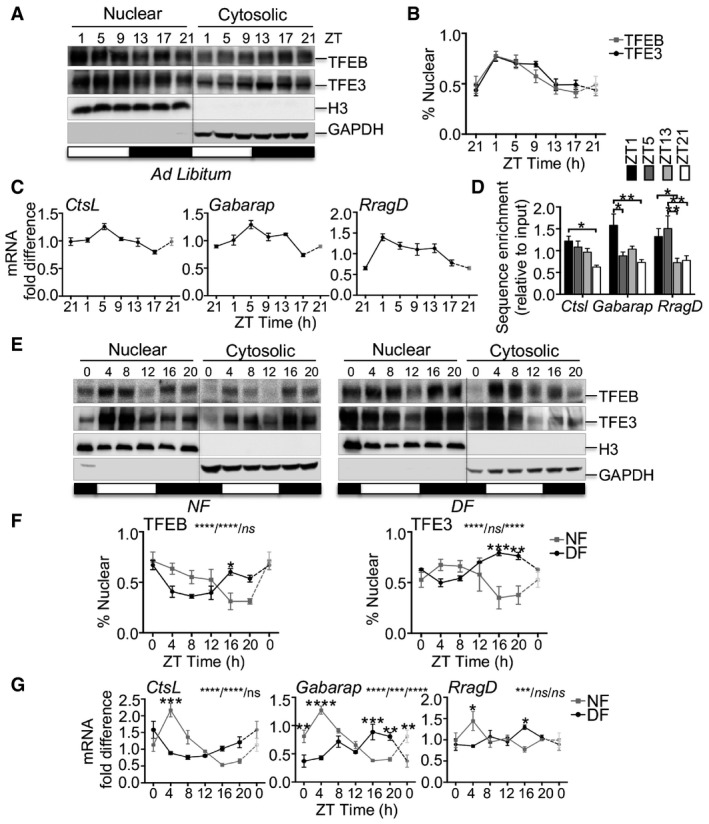
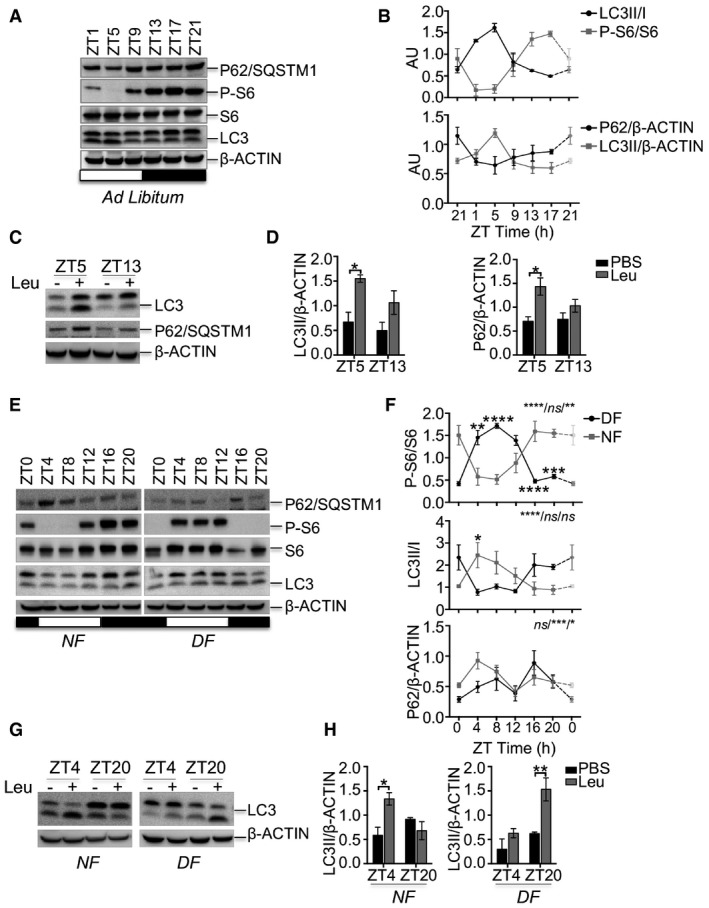
Mammals show a clear feeding rhythm: They eat during one part of the daily cycle and rest/sleep during the other. Consistent with the notion that the subcellular localization of TFEB and TFE3 TFs is controlled by the nutrient‐dependent activation of mTOR (Martina et al, 2012; Roczniak‐Ferguson et al, 2012; Settembre et al, 2012), we found that TFEB and TFE3 nuclear fraction in livers from wild‐type (WT) mice fed ad libitum peaked during the light phase (fasting period) and dropped during the night phase (feeding period; Fig 1A and B). Accordingly, the phosphorylation of mTORC1 downstream substrate S6 ribosomal protein peaks during the dark phase, when the mice are active, and drops during the light phase, when mice rest/sleep, as previously shown (Cornu et al, 2014; Fig EV1A and B). On the contrary, autophagy peaks during the light/fasting phase and undergoes diurnal rhythm over the 24‐h cycle, as demonstrated by LC3II and p62/SQSTM1 levels (Fig EV1A and B). To further analyze the autophagy flux, we treated mice with leupeptin, a lysosomal protease inhibitor, and harvested tissues 3 h following injection, as previously described (Ma et al, 2011). We analyzed liver samples at ZT5 and ZT13, when autophagy peaks and drops, respectively. We observed that the rate of LC3 conversion and the levels of p62 increase only at ZT5 after leupeptin injections, but it was modest in the dark phase, thus confirming that autophagy peaks during the light phase (Fig EV1C and D). Consistent with the timing of autophagy activation, the expression of TFEB/TFE3 direct targets (e.g., CtsL, Gabarap, and RragD) follows the same oscillatory pattern, thus peaking at ZT1‐9 (Fig 1C). Furthermore, chromatin immunoprecipitation (ChiP) experiments in mouse liver showed that the enrichment of TFE3 on CtsL, Gabarap, and RragD promoters follows its nuclear localization, thus peaking at ZT1‐5 and dropping at ZT13‐21 (Fig 1D), despite some variability among the analyzed genes. To further confirm the correlation of TFEB/TFE3 subcellular localization with feeding behavior, we performed time‐restricted/time‐switched feeding experiments. We subjected mice to night feeding (NF) for a period of 10 days and then switched half of them to day feeding (DF) for a period of 4 days. As expected, TFEB and TFE3 nuclear localization peaked during the starvation period (ZT4‐8 in NF and ZT16‐20 in DF; Fig 1E and F). Consistently, mTORC1 pathway activation, LC3II/I ratio, p62 levels, and the expression of the analyzed targets followed nutrient availability (Figs EV1E and F, and 1G). Interestingly, consistent with the direct transcriptional regulation of p62 gene by TFEB and TFE3 during starvation, p62 protein levels increase at the beginning of the fasting period (ZT4 in NF and ZT16 in DF) and rapidly drop when autophagy is fully activated (Fig EV1E and F).
Figure 1. TFEB and TFE3 nuclear translocation correlates with nutrients availability.
-
A, BRhythmic activation of TFEB and TFE3 throughout the day determined by Western blot analysis of nuclear and cytosolic enriched liver fractions (A) and relative quantification (B) (n = 6 replicates/time point). The gray bars represent the dark cycle. Zeitgeber time ZT0: lights on; ZT14: light off. ZT21 is double‐plotted for visualization. Data are represented as mean ± SEM.
-
CmRNA expression levels of TFEB and TFE3 target genes in WT livers determined by qPCR analysis (n = 3 replicates/time point). ZT21 is double‐plotted for visualization. Data are represented as mean ± SEM.
-
DChIP analysis of liver from WT mice for the indicated promoters (n = 4 per group). Bar graph represents the amount of immunoprecipitated DNA as detected by qPCR assay. Values were normalized to the input. Data are presented as mean ± SEM. (two‐way ANOVA test followed by the Bonferroni post hoc test: *P ≤ 0.05; **P < 0.01).
-
E, FWestern blot analysis of nuclear and cytosolic liver fractions from mice fed exclusively during the night (NF) or during the day (DF) with relative quantification (n = 3 replicates/time point). The gray bars represent the dark cycle. Zeitgeber time ZT0: lights on; ZT12: light off. ZT0 is double‐plotted for visualization. Data are presented as mean ± SEM. (two‐way ANOVA test (interaction/time/group) followed by the Bonferroni post hoc test: ns non‐significant; *P ≤ 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
-
GGene expression analysis of TFEB and TFE3 target genes in WT livers fed during the night or during the day (n = 3 replicates/time point). ZT0 is double‐plotted for visualization. Data are presented as mean ± SEM. (two‐way ANOVA test (interaction/time/group) followed by the Bonferroni post hoc test: ns non‐significant; *P ≤ 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Source data are available online for this figure.
Figure EV1. Activation of autophagy and mTORC1 pathways depends on feeding time.
-
A, BAutophagy markers (LC3 and P62/SQSTM1) and mTORC1 activity as indicated by the phosphorylation of S6 ribosomal protein were measured during the 24‐h cycle in the liver from mice fed ad libitum. The plots represent average values of n = 3 for each time point expressed as ratio of LC3II/LC3I or LC3II/actin, p62/actin, and phosphorylated S6 versus pan‐S6. ZT21 is double‐plotted for visualization. Data are represented as mean ± SEM.
-
C, DAutophagy flux analysis in liver from WT mice fed ad libitum injected with leupeptin or PBS (C) and relative quantification (D) (n = 3 per group). Data are presented as mean ± SEM (two‐way ANOVA test followed by the Bonferroni post hoc test: *P < 0.05).
-
E, FAutophagy markers and mTORC1 activity as indicated by the phosphorylation of S6 ribosomal protein measured during the 24‐h cycle in the liver from mice fed during the night (NF) or during the day (DF). The plots represent average values of n = 3 for each time point. The gray bars represent the dark cycle. Zeitgeber time ZT0: lights on; ZT12: light off. ZT0 is double‐plotted for visualization. Data are presented as mean ± SEM (two‐way ANOVA test (interaction/time/group) followed by the Bonferroni post hoc test: (ns non‐significant; *P ≤ 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
-
G, HAutophagy flux analysis in liver from WT mice fed during the night (NF) or during the day (DF) injected with leupeptin or PBS (G) and relative quantification (H) (n = 3 per group). Data are presented as mean ± SEM. (two‐way ANOVA test followed by the Bonferroni post hoc test: *P < 0.05; **P < 0.01).
Source data are available online for this figure.
Autophagy flux experiments confirmed that autophagy activation follows nutrient availability and TFEB/TFE3 nuclear translocation. Indeed, LC3 conversion rate peaks at ZT4 and at ZT20 during the night feeding and the day feeding, respectively (Fig EV1G and H).
To test whether TFEB and TFE3 subcellular localization is under the control of the biological clock, we analyzed Bmal1 KO livers (Bmal1LiKO). Bmal1LiKO mice fed ad libitum showed the same diurnal nuclear localization of TFEB and TFE3 observed in Bmal1f/f littermates (Appendix Fig S1A–D), indicating the clock machinery has no effect on the subcellular localization of these transcription factors.
These data confirm earlier observations of the nutrient‐dependent activation of TFEB and TFE3 and demonstrate that the oscillation of their cytoplasmic/nuclear localization during the daily cycle is clock‐independent.
Impaired autophagy oscillation in TFEB/TFE3‐depleted mice
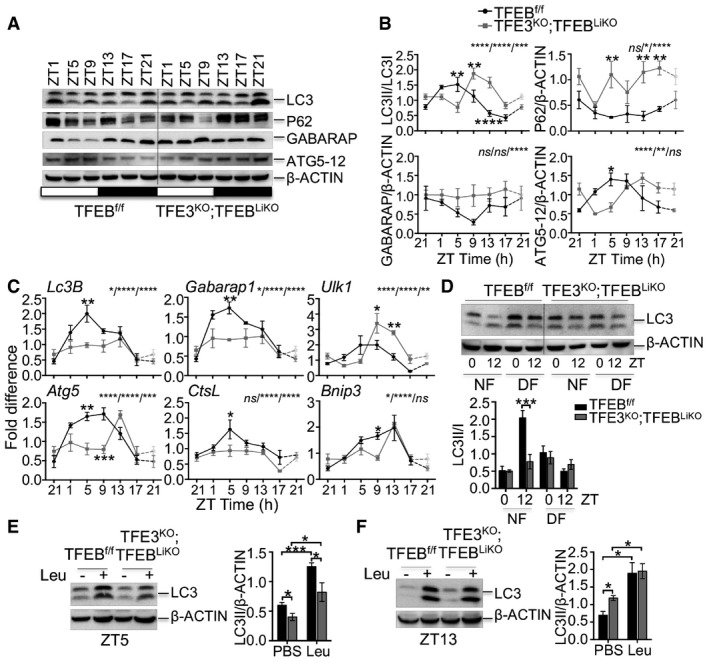
To further explore the role of TFEB/TFE3 in the circadian autophagy, we investigated the consequences of TFEB/TFE3 depletion on autophagy oscillation in vivo. Since Tcfeb KO mice show embryonic lethality (Steingrimsson et al, 1998), we analyzed TFE3KO;TFEBLiKO (Tcfeb liver‐specific KO) double KO mice. We found that depletion of both TFEB and TFE3 in the liver resulted in abnormal circadian pattern of LC3 lipidation, accumulation of p62 and GABARAP, and altered levels of ATG5 over the 24‐h cycle (Fig 2A and B). Consistently, TFE3KO;TFEBLiKO mice showed impaired oscillation of the expression of autophagic genes (e.g., Lc3B, Gabarap1, Ulk1, Atg5, CtsL, and Bnip3; Fig 2C). In particular, TFE3KO;TFEBLiKO mice show blunted induction of the analyzed genes compared with control mice during the light phase (ZT5), when autophagy is induced by TFEB and TFE3, and a shift in the oscillation pattern of Ulk1, Atg5, and Bnip3 (Fig 2C).
Figure 2. Rhythmic induction of autophagy in the liver is impaired in TFEB/TFE3‐depleted mice.
-
A, BWestern blot analysis of autophagy protein throughout the day in liver samples collected every 4 h for 24 h from control and TFE3KO;TFEBLiKO mice (A) and relative quantification (B) (n = 3 per group/time point). ZT21 is double‐plotted for visualization. Data are presented as mean ± SEM. (two‐way ANOVA test (interaction/time/group) followed by the Bonferroni post hoc test: ns non‐significant; *P ≤ 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
-
CDiurnal mRNA expression of genes involved in autophagy in WT and TFE3KO;TFEBLiKO livers determined by qPCR analysis (n = 3 per group/time point). ZT21 is double‐plotted for visualization. Data are presented as mean ± SEM. (two‐way ANOVA test (interaction/time/group) followed by the Bonferroni post hoc test: ns non‐significant; *P ≤ 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
-
DImmunoblotting of liver lysates from mice undergoing restricted feeding and relative quantification (n = 3 per group/time point). Data are presented as mean ± SEM. (two‐way ANOVA test followed by the Bonferroni post hoc test: ***P < 0.001).
-
E, FLC3 protein levels in livers from mice kept under constant darkness and injected with PBS or leupeptin at ZT5 (E) and ZT13 (F) and relative quantification (n = 3 per group/time point). Data are represented as mean ± SEM (*P ≤ 0.05; ***P < 0.001 Student's t‐test).
Source data are available online for this figure.
Restricted feeding experiments confirmed that impaired autophagy activation during the light phase was not due to a different habit in food intake between the two genotypes. Indeed, TFE3KO;TFEBLiKO mice showed reduced LC3II/I ratio at ZT12 when fed only during the night, confirming impaired autophagy activation during the fasting period (Fig 2D).
To assess the autophagy flux, we injected mice with leupeptin and collected livers at ZT5 and ZT13. Livers depleted for TFEB and TFE3 showed reduced LC3II accumulation after leupeptin injection at ZT5, a time point during which autophagy activation peaks (Fig 2E), while no differences were observed at ZT13 (Fig 2F), thus confirming a reduced autophagy flux mainly in the fasting phase.
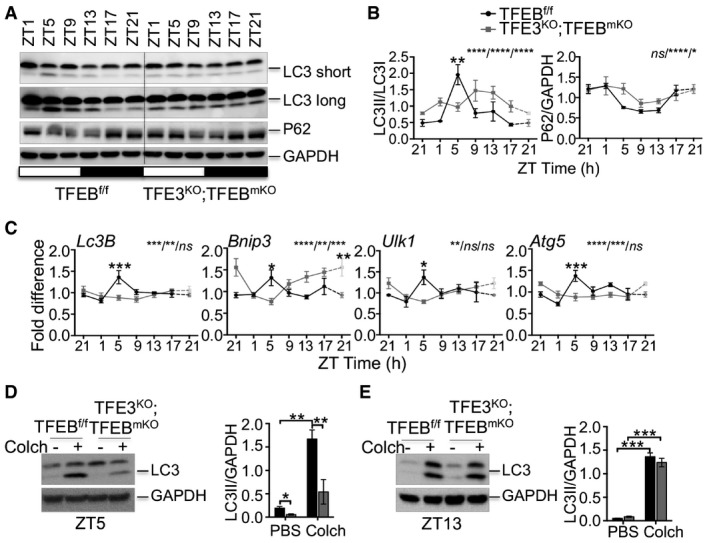
We also tested the effect of the depletion of both TFEB and TFE3 on autophagy activation in muscle. As in the liver, muscles from TFE3KO;TFEBmKO (Tcfeb muscle‐specific KO) double KO mice showed reduced induction of autophagy at ZT5, as indicated by reduced LC3II/I ratio and increased p62 protein levels (Fig EV2A and B). Moreover, they showed lower expression of autophagy genes (Lc3B, Bnip3, Ulk1, and Atg5; Fig EV2C). To analyze autophagy flux in muscle, we treated mice with the microtubule‐depolarizing agent colchicine. Two days of intraperitoneal injection of colchicine blocked autophagosome maturation to autolysosomes and increased LC3II protein levels in WT muscle, but not in TFE3KO;TFEBmKO muscle at ZT5 (Fig EV2D), with no main differences observed at ZT13 (Fig EV2E).
Figure EV2. Autophagy activation during the light phase is impaired in skeletal muscle from TFEB/TFE3‐deficient mice.
-
A, BWestern blot analysis of autophagy protein throughout the day in muscle samples collected every 4 h for 24 h from control and TFE3KO;TFEBmKO mice (A) and relative quantification (B) (n = 3 per group/time point). ZT21 is double‐plotted for visualization. Data are presented as mean ± SEM. (two‐way ANOVA test (interaction/time/group) followed by the Bonferroni post hoc test: ns non‐significant; *P ≤ 0.05; **P < 0.01; ****P < 0.0001).
-
CmRNA expression analysis at different time points of genes involved in autophagy in WT and TFE3KO;TFEBmKO muscle determined by qPCR. (n = 3 per group/time point). ZT21 is double‐plotted for visualization. Data are presented as mean ± SEM (two‐way ANOVA test (interaction/time/group) followed by the Bonferroni post hoc test: ns non‐significant; *P ≤ 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
-
D, ELC3 protein levels in muscle from mice kept under constant darkness and injected with PBS or colchicine at ZT5 (D) and ZT13 (E) (n = 3 per group). Data are presented as mean ± SEM. (two‐way ANOVA test followed by the Bonferroni post hoc test: *P ≤ 0.05; **P < 0.01; ***P < 0.001).
Source data are available online for this figure.
Altogether, these results strongly confirmed the involvement of TFEB and TFE3 in the circadian regulation of the autophagy rhythm.
TFEB and TFE3 regulate Rev‐erbα expression independently from the Bmal1 and Clock genes
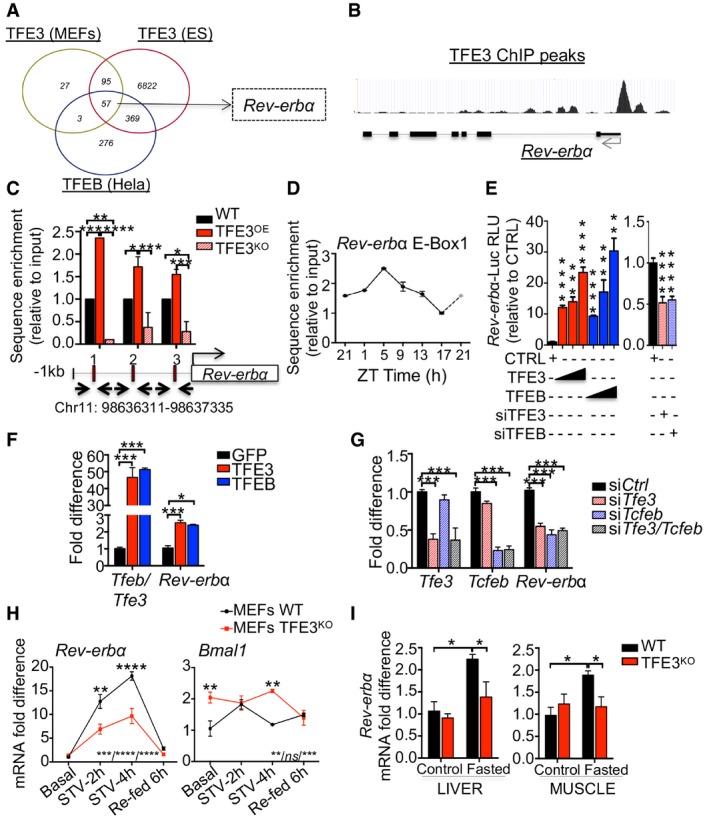
The rhythmic activation of TFEB and TFE3 prompted us to investigate whether their role in the circadian regulation of gene expression is also mediated by the direct regulation of clock‐controlled genes (CCGs). To test this hypothesis, we analyzed previously reported TFEB and TFE3 ChIP‐seq data (Feng et al, 2011; Palmieri et al, 2011; Betschinger et al, 2013). The Venn diagram reported in Fig 3A shows several common targets. Among them, Rev‐erbα, a component of the core clock machinery, showed several ChIP‐seq peaks for TFEB and TFE3 in its promoter (Fig 3B), suggesting a direct regulation of Rev‐erbα by TFEB/TFE3. This was confirmed by ChIP analysis, which showed a significant enrichment of TFE3 on Rev‐erbα promoter in TFE3‐overexpressing livers, which was lost in TFE3KO livers (Fig 3C). Moreover, TFE3 ChIP analysis in mouse liver at different time points during the day showed that TFE3 occupies Rev‐erbα promoter mainly during the day time (Fig 3D), consistent with TFE3 nuclear translocation pattern. Luciferase assay confirmed the direct activation of Rev‐erbα promoter by TFEB and TFE3 (Fig 3E).
Figure 3. TFEB and TFE3 directly regulate Rev‐erbα expression during the light phase.
-
AVenn diagrams showing cross‐comparison of TFEB‐ and TFE3‐bound genes based on published ChIP‐seq dataset.
-
BTFE3 ChIP‐seq peaks in the promoter of Rev‐erbα in ES cells (Betschinger et al, 2013).
-
CChIP analysis of livers from mice of the indicated genotypes (n = 2–4 per group). E‐Boxes in the promoter region of Rev‐erbα are indicated by squares. Bar graph represents the amount of immunoprecipitated DNA as detected by qPCR assay. Values were normalized to the input and plotted as relative enrichment over the WT control. Data are presented as mean ± SEM. (two‐way ANOVA test followed by the Bonferroni post hoc test: *P ≤ 0.05; **P < 0.01; ***P < 0.001).
-
DChIP analysis for Rev‐erbα in WT livers at different time points (n = 3 per group/time point). Values were normalized to the input and plotted as relative enrichment over the ZT1 time point. ZT21 is double‐plotted for visualization. Data are represented as mean ± SEM.
-
ELuciferase activity for Rev‐erbα promoter measured after transfection of increasing amount of TFE3‐GFP or TFEB‐GFP or with siRNA against TFE3 and TFEB (n = 3 per group). Data are represented as mean ± SEM (**P < 0.01; ***P ≤ 0.001; ****P ≤ 0.0001 Student's t‐test).
-
F, GQuantification of mRNA levels of Rev‐erbα in Hepa1‐6 overexpressing (F) or knock‐down (G) for TFEB and TFE3 (n = 3 per group). Data are represented as mean ± SEM (*P ≤ 0.05; ***P ≤ 0.001 by Student's t‐test).
-
HmRNA levels of Rev‐erbα in WT and TFE3KO MEFs in response to nutrient starvation/stimulation at the indicated time points (n = 3 per group). Data are presented as mean ± SEM. (two‐way ANOVA test (interaction/time/group) followed by the Bonferroni post hoc test: ns non‐significant; **P < 0.01; ***P < 0.001; ****P < 0.0001).
-
ITranscript levels of Rev‐erbα in liver and muscle tissues isolated from mice with the indicated genotypes fasted for 24 h (n = 4–5 per group). Values were normalized to ribosomal protein S16 (Rps16) gene and expressed as fold change relative to control fed mice. Bars represent means ± SEM (two‐way ANOVA test followed by the Bonferroni post hoc test: *P ≤ 0.05).
Consistent with these findings, overexpression of TFEB or TFE3 in Hepa1‐6 cells resulted in induction of Rev‐erbα mRNA levels (Fig 3F). Conversely, TFEB/TFE3 knock‐down resulted in significant reduction in Rev‐erbα (Fig 3G). Furthermore, viral‐mediated TFEB and TFE3 overexpression in liver and muscle of WT mice increased REV‐ERBα protein levels (Fig EV3A and C), while TFEB/TFE3‐depleted livers and muscles displayed reduced REV‐ERBα levels at ZT5 (Fig EV3B and D).
Figure EV3. TFEB and TFE3 regulate Rev‐erbα expression.
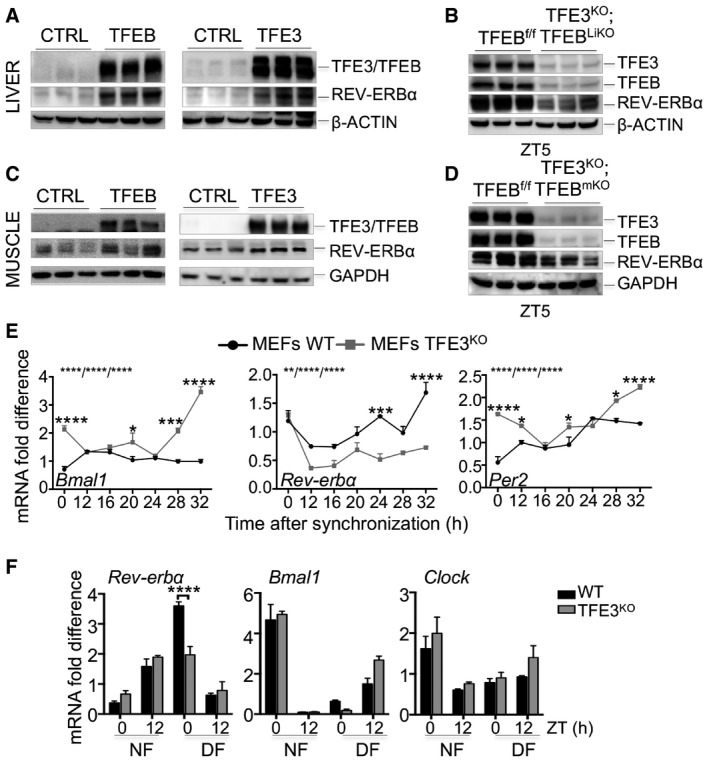
-
A–DTFEB and TFE3 overexpression results in increased levels of REV‐ERBα protein in liver (A) and muscle (C), while their depletion reduced REV‐ERBα protein amount (B, D).
-
ETime course expression of clock genes in WT and TFE3KO MEFs following dexamethasone synchronization (n = 3 per group/time point). Data are presented as mean ± SEM (two‐way ANOVA test (interaction/time/group) followed by the Bonferroni post hoc test: (*P ≤ 0.05; ***P < 0.001 ****P < 0.0001).
-
FqPCR analysis of total liver RNA isolated from control and TFE3KO mice subjected to night (NF) or day (DF) feeding as indicated in the method section (n = 3 per group/time point). Data are presented as mean ± SEM. (two‐way ANOVA test followed by the Bonferroni post hoc test: (***P < 0.001).
Source data are available online for this figure.
TFEB and TFE3 are activated by starvation (Settembre et al, 2011; Martina et al, 2014). Consistently, we observed an increase in Rev‐erbα mRNA levels during starvation in WT MEFs, which was blunted in TFE3KO MEFs (Fig 3H). Importantly, the levels of Bmal1 were increased after a 4‐h starvation in TFE3‐depleted MEFs compared with WT, coherent with the reduced levels of its repressor Rev‐erbα (Fig 3H). Similar data were obtained in vivo in starved mice that showed fasting‐induced increase in Rev‐erbα expression in liver and muscle, which was reduced in TFE3KO mice (Fig 3I).
In addition, TFE3KO MEFs exposed to dexamethasone (Dex) shock, which is able to induce circadian gene expression and oscillation in cultured fibroblasts (Balsalobre et al, 2000), exhibited abnormalities in the oscillation of clock gene expression with reduced induction of Rev‐erbα expression and increased levels of Bmal1 and Per2 (Fig EV3F).
Previous studies demonstrated that time‐restricted feeding can reset peripheral oscillators by uncoupling them from the master pacemaker in the suprachiasmatic nucleus (Damiola et al, 2000). We thus analyzed the expression of clock genes in WT and TFE3KO mice subjected to a night‐feeding/day‐feeding experiment. As expected, feeding switch leads to phase resetting of the expression of the core clock components Rev‐erbα, Bmal1, and Clock in control mice (Fig EV3F). Consistent with our findings, feeding‐induced resetting of Rev‐erbα expression was impaired in the day feeding in TFE3KO mice, with no significant differences in Bmal1 and Clock expression (Fig EV3F).
BMAL1 and CLOCK are HLH transcription factors, core components of the clock machinery that regulate the expression of several downstream genes, including Rev‐erbα, by binding the E‐boxes in its promoter region. Thus, we considered the possibility that TFEB/TFE3 may bind Rev‐erbα promoter by interacting with the BMAL1/CLOCK complex. However, immunoprecipitation assay from liver nuclear extracts and whole lysates at ZT5 failed to reveal an interaction between TFEB or TFE3 and BMAL1 (Fig EV4A and B). To test whether the effect of TFEB and TFE3 on clock genes was independent on the core clock components, we generated CRISPR/Cas9 BMAL1/CLOCK KO (B/C KO) Hepa1‐6 cells (Fig EV4C and D). TFEB and TFE3 overexpression in cells lacking the core clock components still resulted in increased Rev‐erbα expression (Fig EV4E), suggesting that these transcription factors regulate Rev‐erbα expression independently from the BMAL1/CLOCK pathway. Furthermore, ChIP analysis of Bmal1 KO livers confirmed that TFE3 binds the Rev‐erbα promoter in a clock‐independent manner at ZT5 (Fig EV4F).
Figure EV4. TFEB and TFE3 act in parallel and not in combination with BMAL1/CLOCK .
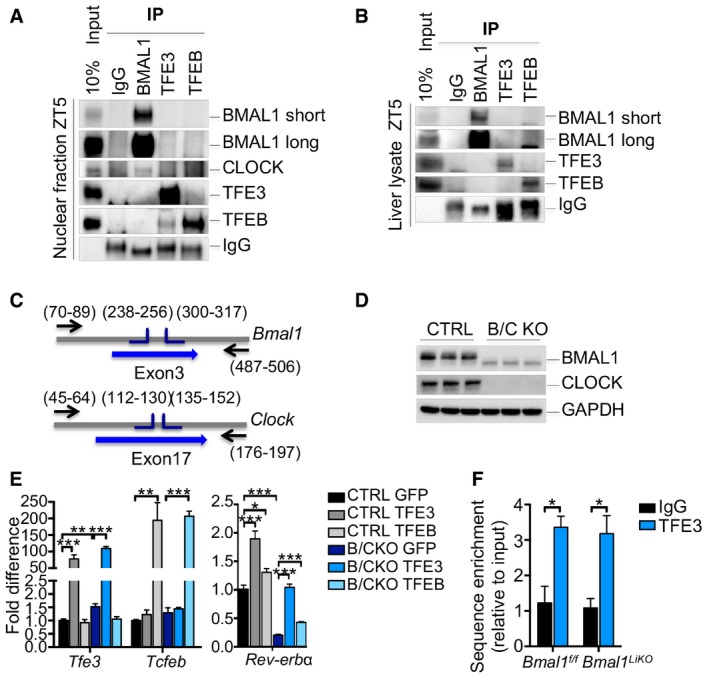
-
A, BCo‐immunoprecipitation analysis using the indicated antibodies in nuclear fraction (A) or whole lysate (B) from liver of WT mice fed ad libitum at ZT5.
-
CRepresentation of the strategy used for the generation of the BMAL1/CLOCK KO (B/C KO) Hepa1‐6 cells using CRISPR/Cas9 technology.
-
DValidation of the B/C KO cells by Western blot analysis.
-
EqPCR analysis of WT and B/C KO cells overexpressing TFEB and TFE3 (n = 3 per group). Data are represented as mean ± SEM (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 by Student's t‐test).
-
FChIP analysis for Rev‐erbα in liver from mice of the indicated genotypes (n = 3 per group). Bar graph represents the amount of immunoprecipitated DNA as detected by qPCR assay. Values were normalized to the input and plotted as relative enrichment over the IgG control. Data are represented as mean ± SEM (*P ≤ 0.05 by Student's t‐test).
Source data are available online for this figure.
Taken together, these results suggest that TFEB and TFE3 have an important role in the regulation of Rev‐erbα expression and act in parallel to the core clock components.
TFEB/TFE3‐mediated activation of Rev‐erbα contributes to the circadian oscillation of gene expression
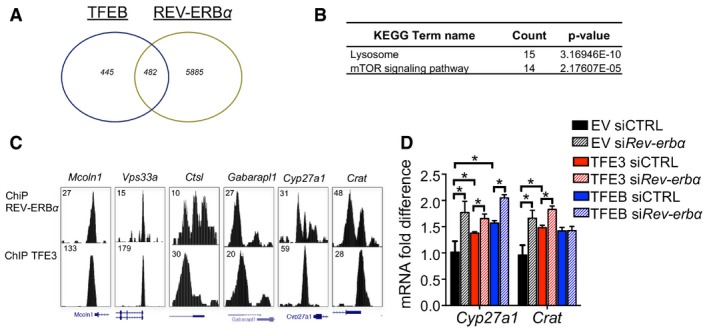
The nuclear receptor REV‐ERBα is a transcriptional repressor expressed in a circadian manner and mainly involved in the regulation of lipid metabolism (Zhang et al, 2015). Previous studies revealed that Rev‐erbα can also regulate the expression of several autophagy genes involved in vesicle nucleation and expansion, autophagosome formation, and lysosomal enzymatic activities resulting in decreased H3K27 and H3K9 acetylation, consistent with its repressive role on gene transcription (Ma et al, 2011; Woldt et al, 2013; Huang et al, 2016; Sulli et al, 2018). Thus, we decided to evaluate whether Rev‐erbα is a downstream mediator of the effect of TFEB and TFE3 on the regulation of the circadian autophagy. A comparison of the published REV‐ERBα, TFE3, and TFEB binding sites as determined by ChIP‐seq analysis (Feng et al, 2011; Palmieri et al, 2011; Betschinger et al, 2013) revealed that 52% of TFE3 and TFEB peaks were shared with REV‐ERBα (Figs 4A and EV5A). Pathway analysis of the shared peaks (Appendix Table S1) revealed an enrichment in lysosome and autophagy pathways as well as in mTOR signaling pathway (Figs 4B and EV5B). As expected, binding of both TFE3 and REV‐ERBα was enriched in promoter regions, mostly within the first 500 bp upstream of the transcriptional start sites (Fig 4C). As expected, enrichment of REV‐ERBα binding sites was observed in the promoter regions of gene involved in the circadian rhythm as well as in autophagy and lysosome biogenesis, such as Lamp1, Mcoln1, Vps33a, Atg3, Atg5, Ctsl, and Gabarapl1 (Figs 4D and EV5C). The presence of both E‐boxes/CLEAR sites and REV‐ERBα‐responsive elements (ROREs) in the autophagy gene promoters (Fig 4E) strongly suggests that they cooperate to coordinately regulate cell homeostasis. To test this hypothesis, we overexpressed TFEB and TFE3 in Hepa1‐6 cells with the concomitant knock‐down of the endogenous Rev‐erbα and analyzed the expression of genes bound by both TFEB/TFE3 and REV‐ERBα. As expected, TFEB and TFE3 induced the expression of the analyzed genes involved in the autophagy pathway (Atg3, Atg5, Atp6v0c, Bnip3, Lc3B, and Gabarap) and silencing of Rev‐erbα further enhanced their expression (Fig 4F). Consistent with these results, autophagy flux was further increased in TFEB/TFE3‐overexpressing cells in the context of Rev‐erbα knock‐down (Fig 4G). We also analyzed genes involved in lipid metabolism such as Cyp27a1 and Crat, which are known TFE3 and REV‐ERBα targets, confirming the same results (Fig EV5D).
Figure 4. TFEB/TFE3‐mediated regulation of Rev‐erbα contributes to the oscillation of gene expression.
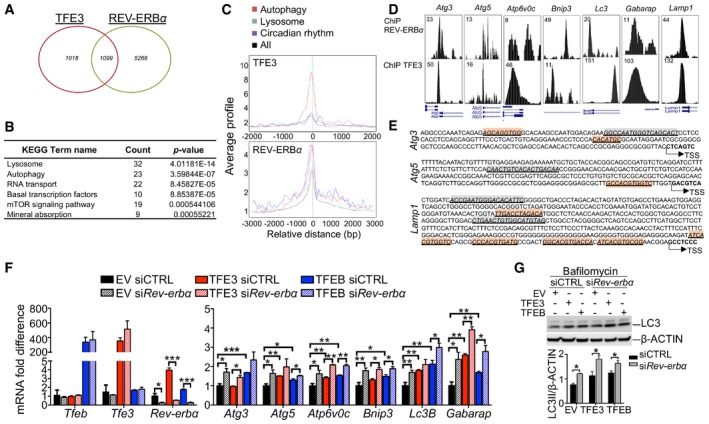
- Venn diagram depicting the unique and shared TFE3 and REV‐ERBα binding peaks identified as reported in the Materials and Methods section. Fisher's exact test P‐value < 2.2e‐16.
- KEGG analysis showing that shared TFE3 and REV‐ERBα peaks are enriched for genes involved in lysosome and metabolic pathway.
- Average TFE3 and REV‐ERBα signals from −1 kb to +1 kb to the transcriptional start site (TSS).
- TFE3 cistromes significantly overlap with REV‐ERBα in the promoter region of genes involved in autophagy and lysosome. Tag counts are shown in the corner.
- E‐boxes/CLEAR (red) sites and RORE (green) location in the promoter of their target genes. Arrows indicate the number from transcriptional start site.
- Expression analysis of autophagy‐related genes in Hepa1‐6 overexpressing TFEB or TFE3 and depleted for Rev‐erbα (n = 3 per group). mRNA levels were normalized using Rps16 and expressed as relative to cells transfected with scramble siRNA and empty vector. Data are represented as mean ± SEM (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 by Student's t‐test).
- Western blot analysis of Hepa1‐6 overexpressing TFEB or TFE3 and depleted for Rev‐erbα (n = 3 per group). Cells were treated with bafilomycin for 4 h before collection to analyze the autophagy flux. Data are presented as mean ± SEM. (two‐way ANOVA test followed by the Bonferroni post hoc test: *P ≤ 0.05).
Source data are available online for this figure.
Figure EV5. REV‐ERBα binds the promoter regions of several TFEB/TFE3 target genes.
- Venn diagram showing the unique and common TFEB and REV‐ERBα bindings. Fisher's exact test P‐value < 2.2e‐16.
- KEGG analysis of the shared TFEB and REV‐ERBα peaks.
- TFE3 and REV‐ERBα cistromes in the promoter region of genes involved in autophagy and lysosome. Tag counts are shown in the corner.
- Expression analysis of circadian‐ and metabolism‐related genes in Hepa1‐6 overexpressing TFEB or TFE3 and depleted for Rev‐erbα (n = 3 per group). mRNA levels were normalized using Rps16 and expressed as relative to cells transfected with scramble siRNA and empty vector. Data are represented as mean ± SEM (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 by Student's t‐test).
These findings confirm that TFEB/TFE3 and REV‐ERBα coordinate the regulation of their target genes establishing their rhythmic pattern of expression.
Altered circadian physiology in TFEB‐ and TFE3‐depleted mice
Our data indicate that TFEB and TFE3 activation oscillates over the 24‐h cycle and are directly involved in the oscillation of gene expression. Coherent with our findings, qPCR analysis in liver and muscle from TFE3KO;TFEBcKO mice showed reduced expression of Rev‐erbα at ZT5 and a deregulated expression of the core components of the circadian clock, as evident from altered levels of Npas2, Per2, Bmal1, and Dbp (Fig 5A). Thus, we postulated that TFEB/TFE3 deficiency could result in circadian misalignment of energy utilization and consequently may affect general physiology and behavior. To test this hypothesis, we monitored the circadian wheel‐running activity in TFE3KO, TFEBcKO, TFE3KO;TFEBcKO, and control mice individually in circadian activity‐monitoring chambers. Mice were initially maintained in a 14 h of light: 10 h of dark (LD) cycle for 2 weeks. In the LD conditions, we could not detect any differences in the free‐running period (tau) between the analyzed genotypes (Appendix Fig S2A and B). After releasing constant darkness (DD) for 2 weeks, control animals showed a free‐running period of approximately 23.7 h, whereas TFE3KO and TFE3KO;TFEBcKO mice showed a 24.0‐h free‐running period, indicating that TFEB and TFE3 influence the period length of the circadian system (Appendix Fig S2A and B). Moreover, overall activity was lower in TFE3KO mice during the dark period compared with control mice and was distributed throughout the night and day time (Fig 5B–D). TFEB depletion in the liver did not alter the tau or the wheel‐running activity in mice, while TFE3KO;TFEBLiKO mice showed the same behavior as the TFE3KO mice (Fig 5B and C, and Appendix Fig S2A), suggesting that the depletion of TFEB only in the liver has no main effect on the adaptation to the circadian cycle. TFEBmKO mice showed increased, despite not significant, tau compared with controls (Appendix Fig S2B), and reduced activity during the dark phase (Fig 5B and D). Interestingly, the TFE3KO;TFEBmKO phenotype was more severe than the single KO mice, suggesting an additive effect (Fig 5B and D).
Figure 5. Disruption of circadian rhythm in TFEB/TFE3‐depleted mice.
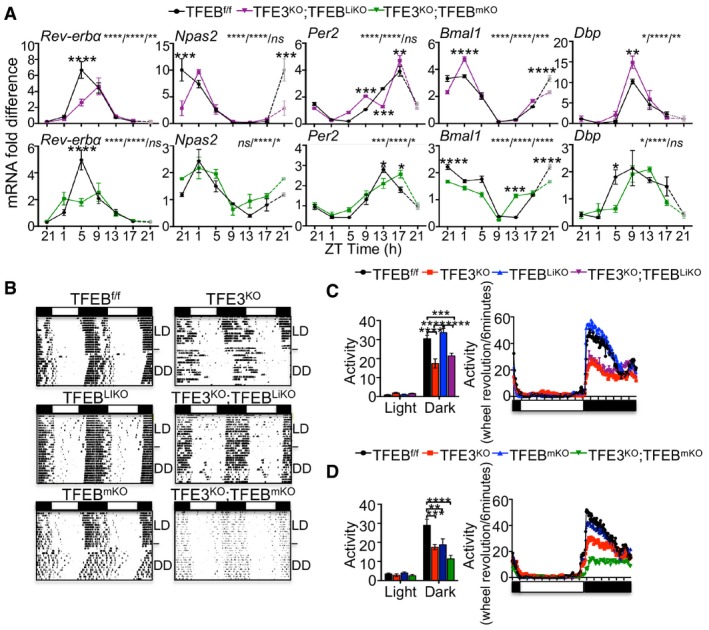
-
AqPCR analysis of gene expression in liver (A) and muscle (B) from TFE3KO; TFEBcDKO and control mice over the 24‐h cycle (n = 3 per group/time point). ZT21 is double‐plotted for visualization. Data are presented as mean ± SEM (two‐way ANOVA test (interaction/time/group) followed by the Bonferroni post hoc test: ns non‐significant; *P ≤ 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
-
BRepresentative activity records (actograms) of mice of the indicated genotypes under normal light:dark cycles 14:10 (LD) or constant darkness (DD). Data were plotted in duplicate columns in each panel.
-
C, DQuantification of the overall activity during the LD period of mice at the indicated genotypes (n = 6–9 per group). Data are presented as mean ± SEM. (two‐way ANOVA test followed by the Bonferroni post hoc test: **P < 0.01; ***P < 0.001; ****P < 0.0001).
The altered circadian physiology and expression of the components of the clock machinery in TFEB‐ and TFE3‐depleted mice further support the role of these TFs in the regulation of the circadian rhythm.
Discussion
Many biological processes in mammals exhibit diurnal rhythms, especially pathways involved in nutrient and energy metabolism (Bass & Takahashi, 2010). The rhythmicity of metabolism enables organisms to synchronize their behavior and physiology to feeding and activity cycles. A tight and synchronized regulation of metabolism and autophagy is necessary to optimize nutrient storage or consumption. Previous studies showed that the number of autophagic vacuoles and the levels of lysosomal enzymes activity exhibit rhythmicity during the day (Pfeifer & Scheller, 1975; Bhattacharya & von Mayersbach, 1976), while recently a temporal restricted activation of autophagy was described in liver, heart, and skeletal muscle (Ma et al, 2011). However, the molecular mechanism that regulates autophagy oscillation during the 24 h still requires further investigation. TFEB and TFE3, members of the MiT‐TFE family of transcription factors, are master regulators of autophagy and lysosomal biogenesis (Sardiello et al, 2009; Settembre et al, 2011; Martina et al, 2014) and participate in crucial metabolic pathways such as glucose and lipid regulation in liver and muscle (Settembre et al, 2013a; Mansueto et al, 2017; Pastore et al, 2017). The critical role of TFEB and TFE3 in linking environmental cues to metabolic adaptation prompted us to examine whether these genes are involved in the regulation of the circadian clockwork. The circadian clock is a light‐entrainable system based on several positive and negative feedback loops generating oscillation in sleep, locomotor activity, and metabolism (Harmer et al, 2001). While the light activates the clock machinery in the brain, food can regulate the circadian rhythm in the peripheral organs.
In this study, we show that MiT‐TFE genes have a critical role in the regulation of circadian autophagy, metabolism, and clock machinery in peripheral tissues. First, we observed that the nuclear/cytoplasmic localization of MiT‐TFE TFs has a cyclic pattern which is food‐dependent and clock‐independent. Moreover, we found that depletion of both TFEB and TFE3 resulted in altered locomotor activity and adaptation to time‐restricted feeding in mice, together with impaired autophagy activation during the light/fasting phase. By a combination of in vitro and in vivo studies, we demonstrated that overexpression of TFEB/TFE3 is sufficient to stimulate the expression of the core component of the clock machinery Rev‐erbα, whereas their depletion impaired Rev‐erbα expression and oscillation. Finally, we found that TFEB and TFE3 directly regulate Rev‐erbα expression by binding its promoter region, thus revealing a connection between the transcriptional regulation of gene expression and the circadian clockwork.
The nuclear receptor REV‐ERBα is expressed in tissues such as liver and adipose tissue where it is involved in lipid, bile acid and glucose metabolism (Yin et al, 2007; Duez et al, 2008; Bugge et al, 2012; Cho et al, 2012; Solt et al, 2012), adipogenesis (Fontaine et al, 2003; Wang & Lazar, 2008), and macrophage inflammatory response (Fontaine et al, 2008). REV‐ERBα interacts with NCOR1 and chromatin modifiers (e.g., histone deacetylase 3) forming a complex that represses target gene transcription (Feng et al, 2011). REV‐ERBα also plays a key role in regulating the oxidative capacity of the muscle and exercise endurance by increasing mitochondrial biogenesis. Its expression is higher in oxidative compared with more glycolytic muscles and increases upon exercise training (Woldt et al, 2013). Interestingly, REV‐ERBα binds the regulatory regions and inhibits the expression of several autophagy and autophagy‐related genes such as Ulk1, Ulk3, Bnip3, Park2, Atg5, Atg7, Bcln1, and Ctsl (Woldt et al, 2013; Huang et al, 2016; Sulli et al, 2018). Consistently, REV‐ERBα overexpression inhibits the autophagic flux. Interestingly, we observed the presence of both E‐boxes/CLEAR sites and ROREs in the promoters of autophagy genes, suggesting that TFEB and TFE3 regulate autophagy rhythm by the temporal regulation of autophagy genes, as well as Rev‐erbα, a transcriptional repressor of the same genes. Thus, the repressive and inductive activities of REV‐ERBα and TFEB/TFE3, respectively, during the 24‐h cycle, are able to establish the rhythmic expression of target genes involved in autophagy and cell homeostasis (Fig 6). Our data show that some genes are strongly responsive to TFEB and TFE3 (e.g., Gabarap and Lc3), and therefore, their expression is dampened in TFEB/TFE3 KO mice during the light phase compared with normal mice. Interestingly, Ulk1, Atg5, and Bnip3, which are known targets of REV‐ERBα, show a phase shifting. We postulate that the reduced expression of Rev‐erbα at ZT5 in TFE3;TFEBcKO mice results in increased expression of its preferential targets at later time points.
Figure 6. Model of TFEB/TFE3 regulation of the oscillation of gene expression.
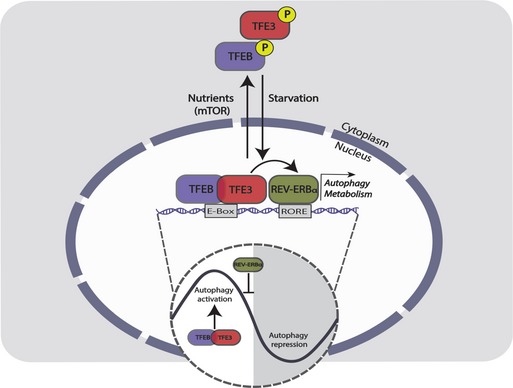
In the absence of nutrients (light phase), TFEB and TFE3 translocate to the nucleus and regulate the expression of genes involved in autophagy and metabolism by binding their promoter at E‐Boxes/CLEAR sites. On the other hand, TFEB and TFE3 regulate the expression of REV‐ERBα that binds ROREs in the promoter of its target genes, including TFEB/TFE3 target genes. Since REV‐ERBα displays a robust oscillation of its protein levels peaking at around ZT8, the repression of autophagy by REV‐ERBα results in a strong day–night transition. This temporal balance of autophagy stimulation (due to TFEB and TFE3 activation) and inhibition (due to the repressor REV‐ERBα) results in its circadian oscillation.
Therefore, our study suggests that a dynamic balance between TFEB/TFE3 and REV‐ERBα is responsible for the modulation and the oscillation of autophagy activation.
Our finding that TFEB and TFE3 are able to bind the E‐Boxes/CLEAR sites in the promoters of CCG is consistent with other studies showing that other HLH transcription factors, e.g., USF1 (Shimomura et al, 2013) and MYC (Altman et al, 2015), can also compete with BMAL1/CLOCK for the same regulatory elements.
A previous study showed that TFEB regulates Per3 expression by forming a complex with BMAL1 and CLOCK (Luo et al, 2016). In our hands, we could not detect any binding of TFEB and TFE3 to BMAL1 and CLOCK, which are therefore able to transactivate the expression of Rev‐erbα independently of the core clock machinery. However, we cannot completely rule out the formation of the complex due to the limitation of the co‐IP experiment, which relies on the efficiency of the antibodies used.
In summary, our study identified TFEB and TFE3 as modulators of the circadian oscillation of autophagy, thus revealing an interplay between TFEB/TFE3 and REV‐ERBα in the regulation of gene expression. Given that deregulation of the circadian machinery has been associated with a variety of clinical conditions (e.g., hypertension, obesity, diabetes, depression, and cancer), the understanding of novel mechanisms that regulate the clock machinery may provide new insights into the mechanisms underlying these disorders.
Materials and Methods
Mouse experiments
Tfe3 KO and Tcfeb flox/flox transgenic mouse line generation has been previously described (Steingrimsson et al, 2002; Settembre et al, 2012). Bmal1 flox/flox mice were obtained from Dr. Vijay K. Yechoor at Baylor College of Medicine. Wild‐type, Albumin‐CRE, and Mlc‐CRE mouse lines were obtained from the Jackson Laboratory (Bar, Harbor, ME). All mice used in experiments were males and were maintained on a C57BL/6 strain background. All experiments were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee (IACUC) and conformed to the legal mandates and federal guidelines for the care and maintenance of laboratory animals. Standard food and water were given ad libitum. For restricted feeding, mice were fed exclusively at night for 10 days and then switched to a day feeding for 4 days. For fasting experiments, mice were starved overnight and samples collected at ZT5.
Locomotor activity analysis
Assessment of circadian locomotor activity was performed in the Behavioral Core at the Neurological Research Institute, Houston, Texas. A ClockLab system with mouse running wheel cages was utilized. Mice were housed individually and maintained for 2 weeks in a 14‐h:10‐h light:dark (LD) cycle. The LD cycle was then changed to constant dark for 2 weeks. Animals were checked for health and food status daily.
RNA extraction and RT–PCR
Total RNA was extracted in TRIzol reagent (Life Technologies, Carlsbad, CA) using RNeasy Kit (Qiagen, Hilden, Germany). RNA was reverse‐transcribed using a first‐strand complementary deoxyribonucleic acid kit with random primers according to the manufacturer's protocol (Applied Biosystems). The RT–PCRs were performed using the CFX96 Real‐Time System (Bio‐Rad, Hercules, CA). The PCR was performed using iTaq SYBR Green Supermix (Bio‐Rad, Hercules, CA) with the following thermocycler conditions: pre‐heating, 5 min at 95°C; cycling, 40 cycles of 15 s at 95°C, 15 s at 60°C, and 25 s at 72°C. Results were expressed in terms of cycle threshold (C t). The C t values were averaged for each duplicate. The Hprt1, actin, and ribosomal protein S16 or Beta2‐microglobulin genes were used as endogenous controls (reference markers). Differences between the mean Ct values of the tested genes and those of the reference gene were calculated as ∆C tgene = C tgene − C treference. Relative fold increase in expression level was determined as 2−∆∆Ct. Primers used for RT–PCR are listed in Appendix Table S2.
Western blotting
Liver, muscle, and cells samples were homogenized in RIPA buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1% Triton X‐100, 1 mM EDTA pH 8.0, 0.1% SDS) containing complete protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Samples were incubated for 20 min at 4°C and centrifuged at 16,000 × g for 10 min. The pellet was discarded, and cell lysates were used for Western blot analysis. Ten to twenty micrograms of liver protein was run on a 4–20% SDS–PAGE. After transfer to a PVDF membrane, the blots were blocked in TBS‐Tween 20 containing 5% non‐fat milk for 1 h at RT, then exposed to primary antibody overnight at 4°C. Anti‐rabbit IgG or anti‐mouse IgG conjugated with horseradish peroxidase (GE Healthcare, Little Chalfont, UK) and ECL (Pierce, Thermo Fisher Scientific, Wilmington, DE) was used for detection. Antibodies used for immunoblots are listed in Appendix Table S3.
Cell culture, plasmids, and transfection reagents
Wild‐type and Tfe3 KO murine embryonic fibroblasts (MEFs; Pastore et al, 2017), and Hepa1‐6 and HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin in 5% CO2 at 37°C. For dexamethasone (Dex) shock, MEFs were treated with 100 nM Dex for 15 min and then switched to a complete medium and collected at the indicated time points. For starvation experiments, MEFs were treated with HBSS1x for the indicated time and then switched to a complete medium for refeeding. Hepa1‐6 cells were transfected with pCMV‐GFP, pCMV‐TFEB‐GFP, or pEGFP‐N1‐TFE3 (Addgene plasmid # 38120) using Lipofectamine LTX reagent (Invitrogen) according to the manufacturer's instructions. For silencing experiments, siRNA for Tfe3, Tcfeb, or Rev‐erbα (Dharmacon) was transfected at 100 nM and the cells were collected after 24–48 h for RNA analysis. For Western blot analysis, cells were treated with 200 nM bafilomycin for 4 h.
Generation of Bmal1/Clock KO Hepa1‐6 through CRISPR‐Cas9
Two sgRNAs targeting adjacent regions of Bmal1 exon 4 and Clock exon 17 were designed and synthetized according to Gundry et al (2016) with minimal modification. Briefly, forward primers containing the T7 promoter sequence, the proto‐spacer sequence, and the sgRNA scaffold overlap sequence were designed through CRISPRscan (Moreno‐Mateos et al, 2015). Full‐length sgRNA scaffold was obtained through an overlap PCR with the universal scaffold reverse primer.
Bmal1sgRNA1 forward: taatacgactcactataGGGTGGACTGCAATCGCAAGgttttagagctagaaatagc
Bmal1sgRNA2 forward: taatacgactcactataGGGAGACATGAAGTCGCTGAgttttagagctagaaatagc
Universal scaffold reverse: agcaccgactcggtgccactttttcaagttgataacggactagccttattttaacttgctatttctagctctaaaac
PCR products were used for in vitro transcription (IVT). IVT was performed using the HiScribe T7 High Yield RNA Synthesis Kit (NEB) following manufacturer instructions. Purified sgRNAs were pre‐complexed with spCas9 protein (IDT). Finally, sgRNA‐Cas9 ribonucleoproteins (RNPs) containing the two sgRNAs were simultaneously transfected into primary mouse hepatoblasts using the NEON Transfection System (Thermo Fisher) and the following electroporation conditions: Buffer R, 1,400 V, 10 ms, three pulses. Deletion efficiency was assessed through PCR using the following primers: Bmal1 forward: GCCTCATTCTTCCACGGGTA; Bmal1 reverse: GTGCTGCTGGCCATTTAAGA. Clock forward: GGCTGTGCAGTTCGCTAAAG; Clock reverse: TTCTGGTCTCTGTTGACTGTCT.
Cellular fractionation
Enriched nuclear and cytosolic cellular subfractions were isolated by differential centrifugation, as previously described (Pastore et al, 2017). Briefly, the liver was minced on ice and homogenized using a Teflon pestle and mortar and suspended in mitochondrial isolation buffer (MIB; 250 mM sucrose, 20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA) supplemented with protease and phosphatase inhibitor cocktails (Complete and PhosSTOP Roche; Roche Diagnostics, Basel, Switzerland). The homogenates were then centrifuged at 1,000 g for 10 min at 4°C to pellet the nuclei while mitochondrial and cytosolic fractions were contained within the supernatant. The supernatant fraction was re‐centrifuged twice at 16,000 g for 20 min at 4°C to pellet the mitochondria, and supernatant containing cytosolic subfraction was collected. Pellets containing nuclei were re‐suspended in nuclear lysis buffer (1.5 mM MgCl2, 0.2 mM EDTA, 20 mM HEPES, 0.5 M NaCl, 20% glycerol, 1% Triton X‐100), incubated on ice for 30 min, and then sonicated 3 Å ~ 10 s followed by a final centrifugation step of 15 min at 16,000 g. The supernatant was collected to obtain the enriched nuclear fraction. Fraction purity was determined by Western blot analysis.
Autophagy flux
For the autophagy flux analysis in the liver, mice were injected i.p. with PBS or leupeptin (40 mg/kg single injection) and collected 3 h after injection. For muscle flux analysis, PBS or colchicine (0.4 mg/kg) was injected every 24 h for 2 days and muscle samples were collected at 3 h after the last injection for Western blot analysis.
Co‐immunoprecipitation analysis
Liver nuclear fraction or whole lysate at ZT5 was subjected to co‐immunoprecipitation experiments. 50 μl of Protein A Sepharose beads (Sigma‐Aldrich) was added to 500 μg of lysate and incubated at 4°C for 30 min. The samples were centrifuged at 16,000 g for 5 min at 4°C and the supernatant incubated with the primary antibody at 4°C overnight. 50 μl of Protein A Sepharose beads was added to the samples and incubated at 4°C for 3 h. The beads were precipitated at 16,000 g for 30 s at 4°C, washed three times with lysis buffer (RIPA), and re‐suspended in sample buffer for immunoblot analysis.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed using livers of 2‐month‐old WT mice as previously described (Settembre et al, 2013a). Primers used for RT–PCR are listed in Appendix Table S4.
Luciferase assay
The Per2‐luciferase and Bmal1‐luciferase plasmids were kindly provided by Dr. Zhaoyang Zhao at UT Health Medical School. The Rev‐erbα promoter was amplified from mouse genomic DNA and cloned into the pGL4 plasmid (Promega, Madison, MI). The cells were transfected with the promoter‐reporter luciferase construct and pRL‐CMV (Promega, Madison, MI) along with TFEB, TFE3, or REV‐ERBα‐expressing plasmids. Cells were harvested 24 h after transfection and subjected to luminescence detection by dual‐luciferase assay using the Dual‐Luciferase Reporter Assay System (Promega, Madison, MI). A Turner Designs Luminometer (DLReady; Promega, Madison, MI) was used to measure luminescence normalized against Renilla luciferase activity.
Vector production and injections
HDAd‐PEPCK‐TFE3 and HDAd‐PEPCK‐TFEB viruses were generated as previously described (Settembre et al, 2013a; Pastore et al, 2017). HDAds were produced in 116 cells with the helper virus AdNG163 as described in detail elsewhere (Ng et al, 2002; Palmer & Ng, 2003). Hepatic transduction was achieved by intravenous administration (retro‐orbital) of approximately 200 μl at a dose of 5 × 1012 viral particles per kg. Livers were collected 1 month after injection. AAV vectors were produced and characterized by the Telethon Institute of Genetics and Medicine (TIGEM, Naples, Italy) AAV Vector Core. pAAV2/1‐CMV‐EGFP, pAAV2/1‐CMV‐TFE3 (Pastore et al, 2017), and pAAV2/1‐CMV‐mTFEB‐3xFlag (Mansueto et al, 2017) were triple‐transfected in subconfluent 293 cells along with pAd‐Helper and pack2/1 packaging plasmids as previously described (Xiao et al, 1999). Recombinant vectors were purified by two rounds of CsCl gradient centrifugation, as previously described (Xiao et al, 1999). Vector titers, expressed as genome copies per milliliter (GC/ml), were assessed by both PCR quantification and dot‐blot analysis. Mice were injected intramuscularly with 100 μl at a total dose of 1011 GC of AAV2.1 vector preparation. Muscles were collected 1 month after injection.
ChIP‐seq bioinformatic analyses
TFE3 and REV‐ERBα ChIP‐seq data were downloaded from GEO (Accession Nos. GSE39815 and GSE26345). Since REV‐ERBα data were originally aligned on a different mouse build, raw sequences from the corresponding Short Read Archive entry (SRA Accession Nos. SRR351393 and SRR351397) were downloaded and pre‐processed according to ENCODE pipeline (https://github.com/ENCODE-DCC/chip-seq-pipeline2): Reads were aligned on mouse mm9 reference genome using BWA (Li & Durbin, 2009), sorted with samtools (Li et al, 2009), and duplicated reads were removed with MarkDuplicates (http://broadinstitute.github.io/picard/). Peaks were called with MACS (Zhang et al, 2008; FDR < 0.01) and annotated using CEAS (Ji et al, 2006) after building custom references using each factor corresponding input as background and current RefSeq genes from UCSC genome browser (Haeussler et al, 2018). Genes showing a binding site within a distance of 2,000 bp upstream and 1,000 bp downstream the TSS were defined targets, respectively, in mm9 for TFE3 and REV‐ERBα and hg19 for TFEB, for which mouse homologs were determined using NCBI Homologene (Coordinators, 2016). Sequence‐binding signatures in promoters were searched with TRANSFAC (Wingender, 2008).
Statistical analyses
Data are expressed as averages ± standard error. Statistical significance was computed using Student's two‐tailed t‐test or ANOVA test followed by the Bonferroni post hoc test as indicated in the figure legends. A P‐value < 0.05 is considered statistically significant.
Author contribution
NP, AV, NJH, and TH performed the experiments. LB helped with the generation of CRISPR/Cas9 clones. TJK performed ChIP experiments. MM performed the bioinformatic analysis. PA and NB‐P provided HDAd viruses. KK and PS‐C contributed to the interpretation of the results. AB and NP designed the overall study, interpreted the results, supervised the work, and wrote the article.
Conflict of interest
A. Ballabio is a co‐founder of CASMA Therapeutics, Boston, MA, USA.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 4
Acknowledgements
We thank Gennaro Napolitano and Antonella De Matteis for critical reading of the article. This work was supported by a grant from the US National Institutes of Health (R01‐NS078072), AIRC (Italian Association for Cancer Research; IG 2015‐17639), Fondation Louis‐Jeantet, and Telethon Foundation to AB. K.K. was supported by a postdoctoral fellowship from the Japan Society for the Promotion of Science (JSPS). The project was supported in part by Baylor College of Medicine IDDRC Grant (U54HD083092‐02) from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health and Human Development or the National Institutes of Health.
The EMBO Journal (2019) 38: e101347
See also: RC Brooks & CV Dang (June 2019)
Contributor Information
Nunzia Pastore, Email: nunzia.pastore@bcm.edu.
Andrea Ballabio, Email: ballabio@tigem.it.
References
- Altman BJ, Hsieh AL, Sengupta A, Krishnanaiah SY, Stine ZE, Walton ZE, Gouw AM, Venkataraman A, Li B, Goraksha‐Hicks P et al (2015) MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab 22: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Schibler U (2011) Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 13: 125–137 [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U (2000) Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289: 2344–2347 [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS (2010) Circadian integration of metabolism and energetics. Science 330: 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A (2013) Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153: 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya R, von Mayersbach H (1976) Histochemistry of circadian changes of some lysosomal enzymes in rat liver. Acta Histochem Suppl 16: 109–115 [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA (2012) Rev‐erbalpha and Rev‐erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev 26: 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK et al (2012) Regulation of circadian behaviour and metabolism by REV‐ERB‐alpha and REV‐ERB‐beta. Nature 485: 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coordinators NR (2016) Database resources of the national center for biotechnology information. Nucleic Acids Res 44: D7–D19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu M, Oppliger W, Albert V, Robitaille AM, Trapani F, Quagliata L, Fuhrer T, Sauer U, Terracciano L, Hall MN (2014) Hepatic mTORC1 controls locomotor activity, body temperature, and lipid metabolism through FGF21. Proc Natl Acad Sci USA 111: 11592–11599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury‐Olela F, Schibler U (2000) Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14: 2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Malta C, Siciliano D, Calcagni A, Monfregola J, Punzi S, Pastore N, Eastes AN, Davis O, De Cegli R, Zampelli A et al (2017) Transcriptional activation of RagD GTPase controls mTORC1 and promotes cancer growth. Science 356: 1188–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duez H, van der Veen JN, Duhem C, Pourcet B, Touvier T, Fontaine C, Derudas B, Bauge E, Havinga R, Bloks VW et al (2008) Regulation of bile acid synthesis by the nuclear receptor Rev‐erbalpha. Gastroenterology 135: 689–698 [DOI] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA (2011) A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331: 1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine C, Dubois G, Duguay Y, Helledie T, Vu‐Dac N, Gervois P, Soncin F, Mandrup S, Fruchart JC, Fruchart‐Najib J et al (2003) The orphan nuclear receptor Rev‐Erbalpha is a peroxisome proliferator‐activated receptor (PPAR) gamma target gene and promotes PPARgamma‐induced adipocyte differentiation. J Biol Chem 278: 37672–37680 [DOI] [PubMed] [Google Scholar]
- Fontaine C, Rigamonti E, Pourcet B, Duez H, Duhem C, Fruchart JC, Chinetti‐Gbaguidi G, Staels B (2008) The nuclear receptor Rev‐erbalpha is a liver X receptor (LXR) target gene driving a negative feedback loop on select LXR‐induced pathways in human macrophages. Mol Endocrinol 22: 1797–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundry MC, Brunetti L, Lin A, Mayle AE, Kitano A, Wagner D, Hsu JI, Hoegenauer KA, Rooney CM, Goodell MA et al (2016) Highly efficient genome editing of murine and human hematopoietic progenitor cells by CRISPR/Cas9. Cell Rep 17: 1453–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Hinrichs AS, Gonzalez JN et al (2018) The UCSC Genome Browser database: 2019 update. Nucleic Acids Res 47: D853–D858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Panda S, Kay SA (2001) Molecular bases of circadian rhythms. Annu Rev Cell Dev Biol 17: 215–253 [DOI] [PubMed] [Google Scholar]
- Huang G, Zhang F, Ye Q, Wang H (2016) The circadian clock regulates autophagy directly through the nuclear hormone receptor Nr1d1/Rev‐erbalpha and indirectly via Cebpb/(C/ebpbeta) in zebrafish. Autophagy 12: 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Li W, Song J, Wei L, Liu XS (2006) CEAS: cis‐regulatory element annotation system. Nucleic Acids Res 34: W551–W554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsbeek A, Yi CX, La Fleur SE, Fliers E (2010) The hypothalamic clock and its control of glucose homeostasis. Trends Endocrinol Metab 21: 402–410 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD (2000) Autophagy as a regulated pathway of cellular degradation. Science 290: 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS (2006) Molecular components of the mammalian circadian clock. Hum Mol Genet 15(Spec No 2): R271–R277 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Ma S, Yang Y, Wang C, Zhang D, Zhang Q, Liu Y, Liu Z (2016) TFEB regulates PER3 expression via glucose‐dependent effects on CLOCK/BMAL1. Int J Biochem Cell Biol 78: 31–42 [DOI] [PubMed] [Google Scholar]
- Ma D, Panda S, Lin JD (2011) Temporal orchestration of circadian autophagy rhythm by C/EBPbeta. EMBO J 30: 4642–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansueto G, Armani A, Viscomi C, D'Orsi L, De Cegli R, Polishchuk EV, Lamperti C, Di Meo I, Romanello V, Marchet S et al (2017) Transcription factor EB controls metabolic flexibility during exercise. Cell Metab 25: 182–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, Puertollano R (2012) MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8: 903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N, Puertollano R (2014) The nutrient‐responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 7: ra9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto‐Rosato A, Prezioso C, Forrester A et al (2015) Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17: 288–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno‐Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ (2015) CRISPRscan: designing highly efficient sgRNAs for CRISPR‐Cas9 targeting in vivo . Nat Med 12: 982–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P, Parks RJ, Graham FL (2002) Preparation of helper‐dependent adenoviral vectors. Methods Mol Med 69: 371–388 [DOI] [PubMed] [Google Scholar]
- Palmer D, Ng P (2003) Improved system for helper‐dependent adenoviral vector production. Mol Ther 8: 846–852 [DOI] [PubMed] [Google Scholar]
- Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A (2011) Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 20: 3852–3866 [DOI] [PubMed] [Google Scholar]
- Pastore N, Vainshtein A, Klisch TJ, Armani A, Huynh T, Herz NJ, Polishchuk EV, Sandri M, Ballabio A (2017) TFE3 regulates whole‐body energy metabolism in cooperation with TFEB. EMBO Mol Med 9: 605–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer U, Scheller H (1975) A morphometric study of cellular autophagy including diurnal variations in kidney tubules of normal rats. J Cell Biol 64: 608–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak‐Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM (2012) The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 5: ra42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS et al (2009) A gene network regulating lysosomal biogenesis and function. Science 325: 473–477 [DOI] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P et al (2011) TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC et al (2012) A lysosome‐to‐nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ et al (2013a) TFEB controls cellular lipid metabolism through a starvation‐induced autoregulatory loop. Nat Cell Biol 15: 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, Ballabio A (2013b) Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura K, Kumar V, Koike N, Kim TK, Chong J, Buhr ED, Whiteley AR, Low SS, Omura C, Fenner D et al (2013) Usf1, a suppressor of the circadian Clock mutant, reveals the nature of the DNA‐binding of the CLOCK:BMAL1 complex in mice. Elife 2: e00426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R et al (2012) Regulation of circadian behaviour and metabolism by synthetic REV‐ERB agonists. Nature 485: 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Tessarollo L, Reid SW, Jenkins NA, Copeland NG (1998) The bHLH‐Zip transcription factor Tfeb is essential for placental vascularization. Development 125: 4607–4616 [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, Copeland NG, Jenkins NA (2002) Mitf and Tfe3, two members of the Mitf‐Tfe family of bHLH‐Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl Acad Sci USA 99: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M (2001) Entrainment of the circadian clock in the liver by feeding. Science 291: 490–493 [DOI] [PubMed] [Google Scholar]
- Sulli G, Rommel A, Wang X, Kolar MJ, Puca F, Saghatelian A, Plikus MV, Verma IM, Panda S (2018) Pharmacological activation of REV‐ERBs is lethal in cancer and oncogene‐induced senescence. Nature 553: 351–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S et al (2002) A transcription factor response element for gene expression during circadian night. Nature 418: 534–539 [DOI] [PubMed] [Google Scholar]
- Wang J, Lazar MA (2008) Bifunctional role of Rev‐erbalpha in adipocyte differentiation. Mol Cell Biol 28: 2213–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E (2008) The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform 9: 326–332 [DOI] [PubMed] [Google Scholar]
- Woldt E, Sebti Y, Solt LA, Duhem C, Lancel S, Eeckhoute J, Hesselink MK, Paquet C, Delhaye S, Shin Y et al (2013) Rev‐erb‐alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med 19: 1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Chirmule N, Berta SC, McCullough B, Gao G, Wilson JM (1999) Gene therapy vectors based on adeno‐associated virus type 1. J Virol 73: 3994–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB et al (2007) Rev‐erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W et al (2008) Model‐based analysis of ChIP‐Seq (MACS). Genome Biol 9: R137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE et al (2015) GENE REGULATION. Discrete functions of nuclear receptor Rev‐erbalpha couple metabolism to the clock. Science 348: 1488–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM (2006) Characterization of peripheral circadian clocks in adipose tissues. Diabetes 55: 962–970 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 4


