Abstract
The cellular glycocalyx controls many of the crucial signaling pathways involved in cellular development. Synthetic materials that can mimic the multivalency and three-dimensional architecture of native glycans serve as important tools for deciphering and exploiting the roles of these glycans. Here we describe a chemical approach for the engineering of growth-factor interactions at the surfaces of stem cells using synthetic glycomimetic materials, with an eye towards promoting their commitment towards specific cell lineages with therapeutic potential.
Keywords: Glycan microarrays, Glycopolymers, Glycosaminoglycans, Proteoglycans, Stem cells, Stem cell differentiation
1. Introduction
The interactions of growth factors with their cellular receptors are mediated by heparan sulfate proteoglycans (HSPGs) [1]. The glycosaminoglycan (GAG) appendages on proteoglycans recruit various growth factors to the cell surface and help organize their interactions with membrane receptors to activate intracellular signaling and gene expression. In mouse embryonic stem cells (mESCs), GAGs composed of alternating units of variously sul-fated glucosamine and uronic acid residues orchestrate the formation of complexes between fibroblast growth factors (FGFs) and their receptors (FGFRs) [2]. The sulfation patterns of GAGs are believed to be responsible for binding to various growth factors [3]. Subsequent phosphorylation of the Extracellular Signal-Regulated Kinases 1 and 2 (Erk1/2) and their associated downstream signaling events result in the differentiation of mESCs into neural precursor cells [4]. mESCs lacking exostosin 1 (Extl), an enzyme responsible for the biosynthesis of HS, fail to form functional ternary FGF-FGFR-HSPG complexes, and are arrested in their embryonic state [5].
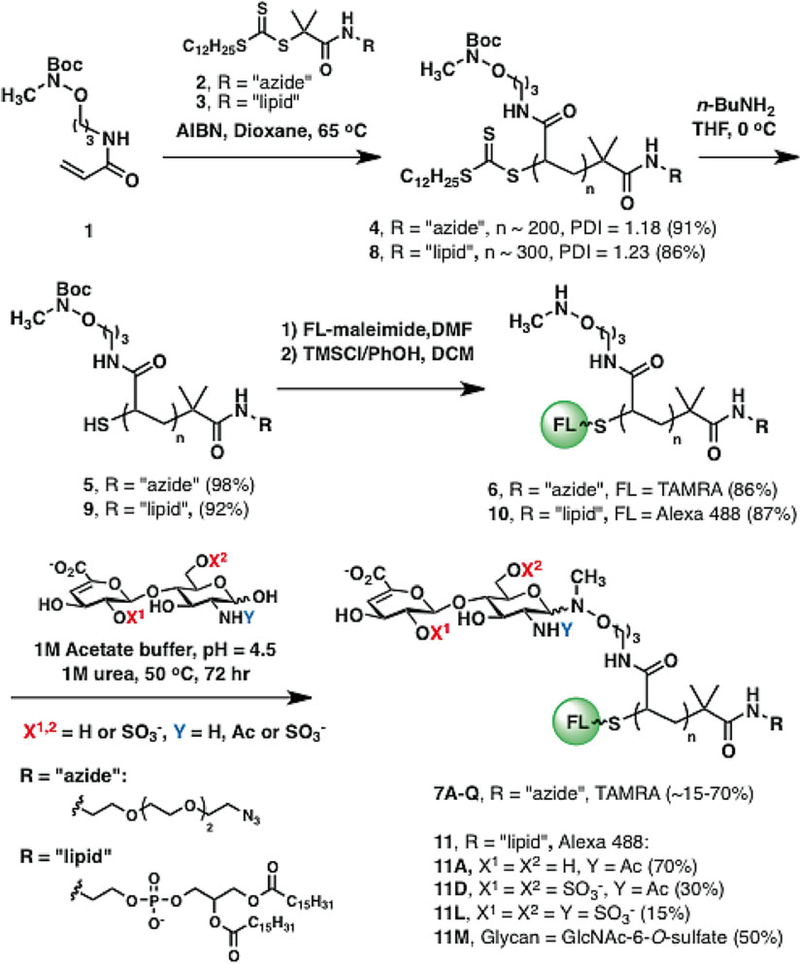
Herein, we present a strategy for the identification of synthetic GAG-mimetic polymers that bind FGF2 and a cell-surface engineering strategy to influence stem cell specification. Overall, the procedure begins with the synthesis of the glycopolymers from relevant starting materials using RAFT polymerization (Fig. 1) [6]. Polymer synthesis is modular, and the polymers can be designed to bear azido or lipid terminal functional groups. The azido-terminated polymers are useful for the “copper-free click” conjugation onto cyclooctyne-functionalized surfaces [7]. Libraries of such polymers are microarrayed onto glass slides and evaluated for their ability to bind desired proteins or growth factors. On the other hand, the lipid-functionalized polymers are useful towards cell surface re-engineering. The lipid tail allows for passive insertion onto cell surfaces [8]. Following identification of appropriate binders in the microarray, glycopolymers of the identified glycans are made into lipid-terminated polymers and tested in tissue culture for growth factor binding and stem cell differentiation.
Fig. 1.
Synthesis of fluorescently labeled neoproteoglycans (neoPGs). Polymer backbones can be prepared via RAFT polymerization to bear either azido or lipid end groups. Following trithiocarbonate deprotection, fluorophore conjugation, and Boc sidechain deprotection, the reactive aminooxy groups can be ligated to any glycan with a reducing end. Reprinted with permission from Huang, M.L., Smith, R.A.A., Trieger, G.W., Godula, K. Glycocalyx remodeling with proteoglycan mimetics promotes neural specification in embryonic stem cells. (2014) Journal of the American Chemical Society 136, 10565–10568. Copyright (2014) American Chemical Society
2. Materials
2.1. Chemicals
Azadibenzocyclooctyne amine.
Azide chain transfer agent is synthesized as described in ref. [9].
α,α′-Azoisobutyronitrile (AIBN).
Chloroform.
Deuterium oxide (D2O).
Dichloromethane (DCM).
Diethyl ether.
Dimethylformamide (DMF).
Dioxane (anhydrous).
Lipid chain transfer agent is synthesized as described in [10].
Methanol.
N-butylamine.
Phenol.
Tetrahydrofuran (THF).
tert-butyl (3-acrylamidopropoxy)methyl carbamate monomer is synthesized as described in [10] (see Note 1).
Trimethylsilyl chloride (TMS-Cl).
2.2. Other Commercial Materials and Reagents
Amicon Ultra Centrifugal Filters (10 K MWCO, 0.5 mL capacity, Millipore).
Heparin sodium (Acros).
PD-10 columns (GE Healthcare).
ProLong Gold antifade reagent with DAPI (Cell Signaling Technology).
Recombinant Human Fibroblast Growth Factor-Basic (Gibco).
Superchip epoxy slides (Thermo Fisher).
2.3. Buffers and Coating Solutions
Reaction buffer: 1 M sodium acetate, 1 M urea, pH 4.5.
Deuterated PBS buffer: 100 mM sodium phosphate, 150 mM sodium chloride, in D2O, pD 7.4.
PBS buffer: 100 mM sodium phosphate, 2.7 mM potassium chloride, 137 mM NaCl, pH 7.4.
Passivating solution: 0.1 % BSA in PBS.
Blocking solution: 0.1 % (v/v) Tween in PBS.
Slide washing solution: 0.1 % (v/v) Triton X-100 in PBS, pH 7.4.
Assay buffer: 1 % BSA, 0.1 % Tween 20, PBS, pH 7.4.
ICC buffer: 2 % (v/v) goat serum, 3 % (w/v) BSA, 0.1 % (v/v) Triton X-100 in PBS.
2.4. Antibodies
Mouse anti-FGF2 monoclonal antibody, clone bFM-1 (Millipore).
Goat anti-mouse IgG, Cy5 (GE Healthcare).
Anti-nestin (Millipore cat. #MAB353).
Anti-Oct3/4 (Santa Cruz Biotechnology cat. #SC-25401).
Anti-mouse Alexa Fluor 555 (Cell Signaling Technology).
Anti-rabbit Alexa 488 (Cell Signaling Technology).
Anti-phospho-Erk1/2 (Cell Signaling Technology cat. #4370, used at 1:4000 for Western blot).
Anti-total Erk1/2 (Cell Signaling Technology cat. #9102 used at 1:6000 for Western blot).
Anti-alpha-tubulin (Abcam cat. # ab7750, used at 1:1000 for Western blot).
Anti-rabbit HRP (Cell Signaling Technology).
Anti-mouse HRP (Cell Signaling Technology).
2.5. Stem Cell Culture and Differentiation
Porcine gelatin powder (Sigma Aldrich).
Autoclavable glass media bottles.
Doubly distilled water.
Tissue culture plates (Corning).
Mouse embryonic stem cells wild-type E14TG2a and Ext1−/− cell lines [11].
Non-enzymatic cell dissociation buffer (Sigma).
Trypsin.
Maintenance medium: Knockout Dulbecco’s Modified Eagle’s Medium (KO-DMEM, Invitrogen) supplemented with 10 % (v/v) fetal bovine serum (Gibco), 20 mM L-glutamine (Gibco), 1 % (v/v) nonessential amino acids (Gibco), 55 μM 2-mercaptoethanol (Gibco), and 1000 U/mL leukemia inhibitor factor (LIF, Millipore ESGRO) (see Note 2).
Gelatin stock solution: 1 % (w/v) porcine gelatin in sterile water. Dissolve by autoclaving in glass media bottles (see Note 3).
Neural differentiation medium: 1:1 mixture of neurobasal medium and DMEM/F12 supplemented with 1 % (v/v) B27 (Gibco), 0.5 % (v/v) N2 (Gibco), 55 μM 2-mercaptoethanol, 500 μM L-glutamine, 50 μg/mL BSA, 100 U/mL penicillin, and 100 μg/mL streptomycin [12].
2.6. Western Blot
Lysis buffer: 1× cell lysis buffer (Cell Signaling Technology), 1× protease inhibitor cocktail (Cell Signaling Technology), and 1 mM PMSF (Cell Signaling Technology).
Laemmli buffer (1×): 2 % (v/v) SDS, 10 % (v/v) glycerol, 5 % (v/v) 2-mercaptoethanol, 0.002 % (w/v) bromophenol blue, 0. 0625 M Tris-HCl.
Protein ladder (New England Biolabs).
Pre-cast gels or self-casted (30 % (19:1) acrylamide-bis-acrylamide, TEMED, APS, Tris, SDS).
BCA Protein Quantification Kit (Pierce).
5× running buffer: 72 g glycine, 15 g Tris base in 1 L water.
10× TBS or Tris base solution: 80 g NaCl, 24.2 g of Tris base, 1 L water, pH 7.6.
1× running buffer: 200 mL 5× running buffer, 10 mL 10 % w/v SDS, 790 mL water.
1× transfer buffer: 200 mL 5× Running buffer, 200 mL methanol, 10 mL 10 % SDS, 590 mL water.
1× TBST: 100 mL 10× TBS, 900 mL water, 1 mL Tween 20.
Luminata Forte HRP detection reagent (Millipore).
ECL hyperfilm (GE).
Immobilon-FL 0.45 μM PVDF membrane (Millipore).
Restore PLUS Western blot stripping buffer (Thermo Scientific).
3. Methods
3.1. Preparation of Polymers
3.1.1. RAFT Polymerization
Prepare apparatus consisting of a flame-dried 10 mL Schlenk flask equipped with a magnetic stirrer, inlet tube provided with a stream of nitrogen, and connected to a vacuum pump.
Charge the flame-dried Schlenk flask with the desired (azide or lipid) chain transfer agent (6.2 mg, 0.50 mol% relative to monomer), a solution of AIBN (0.2 g, 0.05 mol% relative to monomer), monomer (570.2 mg), and anhydrous dioxane (443.3 mg) (see Note 4).
Equip the flask with a rubber septum and fill with a stream of nitrogen.
Degas the yellow solution with three freeze-pump-thaw cycles.
Allow the flask to warm to room temperature, then immerse in an oil bath preheated to 65 °C. Let the solution stir for 12 h (see Note 5).
Dilute the reaction with ether and precipitate with excess hexanes by vigorous stirring. Repeat three times after isolation of the solid residue (by simple decantation) at each step.
Remove residual hexanes by successive dissolution and evaporation in chloroform for three times. Dry under high vacuum overnight at room temperature (see Note 6).
3.1.2. Analysis by GPC
GPC is performed on a Hitachi Chromaster system equipped with an RI detector and a 5 μm, mixed bed, 7.8 mm I.D. × 30 cm TSKgel column (Tosoh Bioscience).
Polymer is analyzed using an isocratic method with a flow rate of 0.7 mL/min in DMF (0.2 % LiBr, 70 °C) (see Note 7).
Based on the polymer elution profiles, molecular weight distribution differential curves are analyzed using standard approaches to enable the calculation of the polymer molecular weight characteristics (Mn and DI = Mw/Mn).
3.1.3. Fluorophore Conjugation to Polymer Backbones
Charge a Schlenk flask equipped with a stir bar with the polymer backbone. Add a 100 mg/mL equivalent of a degassed solution of n-butylamine in THF (20 mM).
Submerge the Schlenk flask in an ice bath and allow the solution to react for 2 h.
Dilute the reaction mixture in ether and precipitate into excess hexanes with vigorous stirring. Perform this step three times, isolating the solid residue at each step.
Concentrate the end-deprotected polymer from chloroform three times to remove excess residual hexanes. Dry under vacuum overnight (see Note 8).
Dissolve the polymer (6.35 mg) in a solution of the desired fluorophore-maleimide conjugate in DMF (e.g., TAMRA-maleimide, 2 mM, 1.3 eq, 81.2 μL).
Degas the solution with three freeze-pump thaw cycles, and let the reaction stir overnight at room temperature in the dark.
Dilute the reaction mixture in ether and precipitate in excess hexanes. Repeat three times, concentrate from chloroform as before, and dry under high vacuum.
Store the polymer residue in the freezer until ready for the next step (see Note 9).
3.1.4. Preparation of Synthetic Glycopolymers
To the labeled polymer backbone, add 0.5 mL of freshly prepared solution of TMS-Cl (1 M) and phenol (3 M) in anhydrous DCM. Stir 2 h, in the dark, at room temperature.
Add ether (20 mL) to precipitate the Boc-deprotected polymer, and wash three times with fresh ether by centrifugation at 1000 ×g for 3 min and decantation (see Note 10).
After removing the last ether wash, briefly dry the residue at room temperature with a stream of air or nitrogen.
Redissolve the residue in water and pass through a PD-10 column to isolate the polymer from other products.
Collect, freeze, and lyophilize the polymer (monitored as the colored band along the column or visible via a UV lamp) (see Note 11).
Redissolve the dried polymer in reaction buffer to form a 200 mM (by side chain) solution. Vortex vigorously and let stand in the dark for a few hours to overnight to dissolve (see Note 12).
Weigh desired amount of glycan (e.g., 1 mg) into a PCR tube. Add the appropriate amount of polymer solution to the PCR tube, such that there is 1.1 eq of glycan per 1 eq side chain. Vortex and quick-spin the mixture to dissolve (see Note 13).
Let the mixture react in a PCR thermocycler for 72 h at 50 °C.
Transfer the reaction mixture to an Amicon Ultra Centrifugal Filter, and spin dialyze four times at 6000 × g, for 10 min using deuterated phosphate buffer. Discard the flow through and refill the inner tube to 0.5 mL during each wash (see Note 14).
Collect the final solution from the inner filter by inverting into a new conical tube and centrifugation at low speed.
Resuspend the sample to a final volume of 0.5–0.6 mL and characterize by 1H NMR and UV analysis to determine ligation efficiencies and polymer concentrations, respectively (see Note 15).
3.2. Microarray Fabrication
In a Coplin jar, rinse the epoxy glass slides with doubly distilled water and spin-dry in a centrifuge (500 rpm, 5 min).
Prepare a solution of dibenzyclooctyne-amine (1 mM) and DIPEA (10 mM) in anhydrous DMF. Prepare enough to completely submerge the glass slide in a Coplin jar or petri dish.
Let the slides incubate in this solution overnight at room temperature, with a gentle shaking or rocking motion.
Remove slides, sonicate in methanol twice for 15 min. Rinse with water and spin-dry. Store at 4 °C until next step.
Passivate slides in passivation buffer for 30 min at RT prior to microarray printing.
Rinse slides with water and spin-dry (centrifugation at low speed, e.g., 300 rpm, 5 min).
Print the microarrayed azido-terminated glycopolymers (10 μM in 0.005 % Tween/PBS, pH 7.4) in replicate spots at 80–85 % relative humidity using a robotic spotter. Scan slide at the appropriate wavelength to inspect spot morphology (see Note 16).
Let slides react overnight at 4 °C in the dark.
Wash excess polymers off the slide by vigorously plunging in Slide Washing Solution for 2 min, followed by rocking for 15 min, at room temperature.
Wash the slide twice in PBS for 10 min with rocking, rinse with water, and spin-dry.
Scan the slide to determine polymer grafting efficiencies (see Note 17).
Fluorescence intensity at 535 nm was measured via a Molecular Devices GenePix 4000B microarray scanner.
3.3. Microarray Evaluation
Draw hydrophobic boundaries around each sub-array using the PAP pen, being careful not to draw over the printed spots.
To eliminate nonspecific binding, briefly block the wells with assay buffer for 30 min at room temperature.
Add a solution of growth factor FGF2 (0.1–1.0 μM) in assay buffer to the desired wells, and incubate at room temperature for 90 min.
Wash the well carefully with assay buffer using the edge of a pipet tip and a corner of the well. Repeat four times.
Conduct subsequent incubations with the desired primary (mouse anti-FGF2, 1:1000) and secondary antibodies (Cy5-goat-anti-mouse, 1:1000) in assay buffer (see Note 18).
Following all incubations, wash the slide twice for 15 min in PBS, rinse with water, spin-dry, and scan (see Note 19) (Fig. 2).
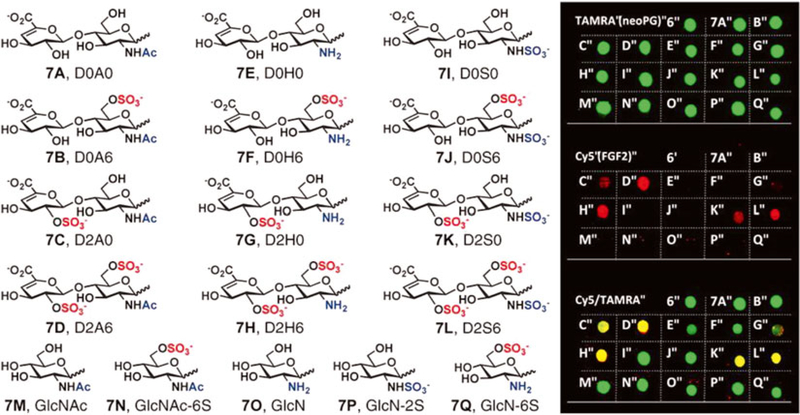
Fig. 2.
Microarray analysis of a library of TAMRA-labeled neoproteoglycans (neoPGs) representing most naturally occurring heparan sulfate sulfation motifs identified neoPGs with specificity for FGF2. Reprinted with permission from Huang, M.L., Smith, R.A.A., Trieger, G.W., Godula, K. Glycocalyx remodeling with proteoglycan mimetics promotes neural specification in embryonic stem cells. Journal of the American Chemical Society 136, 10565–10568. Copyright (2014) American Chemical Society
3.4. Tissue Culture
Prepare gelatin-coated tissue culture plates by incubating plates or wells with 0.1 % gelatin in PBS solution (diluted 1:10 in sterile PBS or deionized water from gelatin stock solution) for at least 10 min at room temperature. Short-term storage under sterile conditions is also acceptable. Remove gelatin solution prior to use.
mESCs are thawed and propagated using maintenance medium in gelatinized sterile tissue-culture-treated well plates.
Under normal passaging conditions, cells are split 1:3–1:8 ratios onto new gelatinized well plates, and passaged every 2 days or at 70–80 % confluency.
For expansion, cells are split using trypsin or cell dissociation buffer into T-75 or T-175 vented flasks.
3.5. Analysis of neoPG Incorporation
3.5.1. Analysis of neoPG Incorporation by Flow Cytometry
A BD FACSCalibur flow cytometer equipped with BD FACSStation is used. Data analysis is conducted using FlowJo (TreeStar). Typical analyses are conducted with 10,000 events in a single run.
Detach a (60–80 %) confluent flask of Ext1−/− cells with trypsin and resuspend to 1 × 106 cells/mL in maintenance medium. Seed each well of a 12-well gelatin-coated plate with 1 mL of the cell suspension overnight at 5 % CO2, 37 °C.
Wash each well with PBS (1 mL) and incubate with the desired solution of lipid-terminated neoPG (0.5 mL) resuspended in serum-free KO-DMEM for desired periods of time (e.g., 1 h. see Note 20).
Wash cells 2× with PBS and detach the cells from each well using cell dissociation buffer (1 mL).
Harvest the cells from each well into sterile microtubes, and wash the cells 2× with PBS by centrifugation at 300 ×g for 3–5 min.
Fix the cells by resuspending the cell pellet in 300 μL of 4 % paraformaldehyde in PBS for 30–60 min on ice. Wash the cells 2× with PBS.
Resuspend the cells in 0.1 % BSA/PBS for flow cytometry analysis.
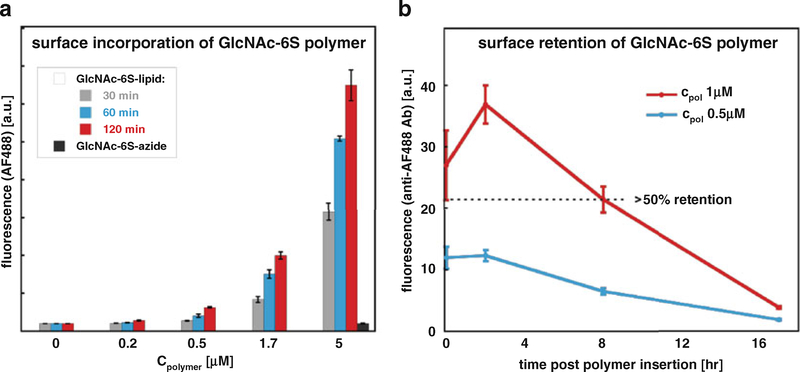
For each condition, conduct at least two replicates, and repeat the assay at least twice (Fig. 3a).
Fig. 3.
Flow cytometry analysis of neoPG (a) cell surface incorporation and (b) cell surface retention. A dose-dependent increase in fluorescence is observed with increasing concentrations of a lipid-terminated polymer (GlcNAc6S-lipid), whereas azide-terminated polymers (GlcNAc-6S-azide) displayed minimal fluorescence. Greater than 50% of fluorescence is retained after 8 h following glycocalyx remodeling. Reprinted with permission from Huang, M.L., Smith, R.A.A., Trieger, G.W., Godula, K. Glycocalyx remodeling with proteoglycan mimetics promotes neural specification in embryonic stem cells. Journal of the American Chemical Society 136, 10565–10568. Copyright (2014) American Chemical Society
3.5.2. Analysis of neoPG Cell Surface Retention
Seed cells and incubate with neoPG and controls as before in a twelve-well plate (see Subheading 3.5, steps 2 and 3).
Wash cells 2× with PBS, then incubate with fresh complete maintenance medium for desired periods of time (e.g., 0, 2, 8, and 17 h) at 37 °C, 5 % CO2.
Following this incubation, repeat steps for harvesting and fixing cells (see Subheading 3.5.1, steps 4–8) (Fig. 3b).
3.6. Analysis of FGF2 Binding Via Immunocytochemistry
Seed 10,000 cells/cm2 of a gelatin-coated 24-well plate. and incubate with neoPG and controls as before in a 24-well plate (see Subheading 3.5, steps 2 and 3).
Incubate with lipid-terminated neoPG solution in KO-DMEM for desired period of time.
Remove media and wash cells 2× with PBS. Fix with 4 % paraformaldehyde in PBS solution for 1 h at 4 °C.
Wash cells 2× PBS, block with 2% BSA/PBS for 1 h at 4 °C.
Incubate cells with FGF2 (450 nM in 2% BSA/PBS) for 1 h at 4 °C.
Wash cells 2× with PBS. Immunostain with rabbit anti-FGF2 (1:1000) and corresponding secondary antibody (1:1000, Alexa Fluor 555 anti-rabbit antibody) for 1 h (each incubation) at 4 °C, in the dark.
After final wash with PBS, wash once with water, and add a drop of anti-fade mounting medium with DAPI. Incubate for 3 h at room temperature or store in the dark at 4 °C until ready for analysis.
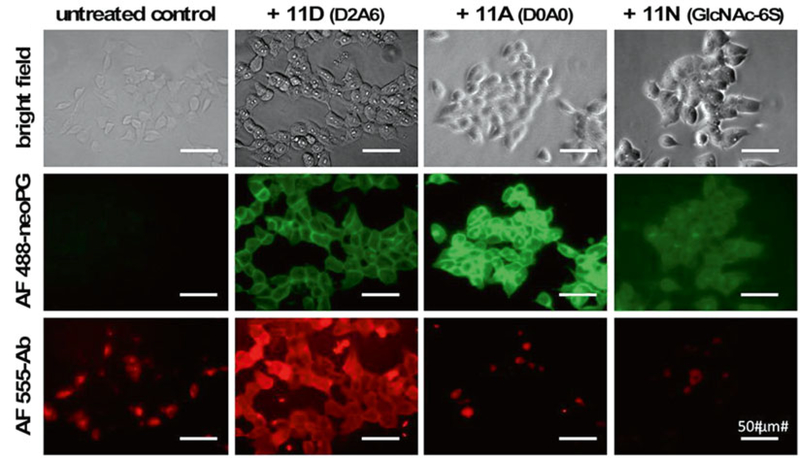
Image under a fluorescence microscope with relevant filter sets (Fig. 4).
Fig. 4.
Fluorescence microscopy analysis of FGF2 binding to cells remodeled with various neoPGs. Cells treated with AF488-labeled neoPGs display green fluorescence. Cells remodeled with FGF2-binding neoPGs (11D) show significant fluorescence compared to non-binding neoPGs (11A and 11N) upon treatment with exogenous FGF2 and immunostaining with anti-FGF2 and AF555-labeled secondary antibody. Adapted with permission from Huang, M.L., Smith, R.A.A., Trieger, G.W., Godula, K. Glycocalyx remodeling with proteoglycan mimetics promotes neural specification in embryonic stem cells. Journal of the American Chemical Society (2014), 136, 10565–10568. Copyright (2014) American Chemical Society
3.7. Neural Differentiation of mESCs
Harvest cells at 70–80 % confluence.
Count and seed cells overnight at 8000 cells/cm in mESC medium onto gelatin-coated tissue culture-treated well plates (for microscopic evaluation, use 24-well plates).
Remove medium the following day and replace with neural differentiation medium with or without neoPGs, washing the monolayer twice with PBS prior to media replacement (day 0). Incubate with neoPGs for desired periods of time (e.g., 1 h) at 37 °C, 5 % CO2, and wash cells three times with PBS. Replace PBS with neural differentiation medium (see Note 21).
Monitor cells daily. Replace with fresh neural differentiation medium whenever necessary (e.g., when media turns yellow) by first washing cells once with PBS (see Note 22).
Harvest cells for microscopy analysis (generally at day 6 or onwards) by fixation with 4 % paraformaldehyde for 10 min at RT, then blocking/permeabilizing with ICC buffer for 1 h at room temperature.
Immunostain for embryonic and neural differentiation markers. Perform primary (anti-Oct3/4 at 1:100 dilution and antinestin at 1:250 dilution) and secondary antibody incubations in ICC buffer, for 1 h at room temperature (see Note 23).
Follow steps 7 and 8 of Subheading 3.6 (Fig. 5).
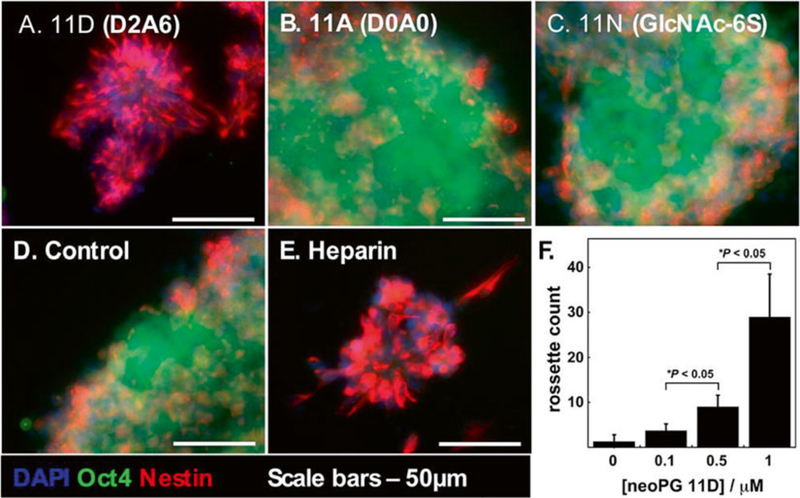
Fig. 5.
Neural differentiation of neoPG-remodeled Ext1−/− mouse embryonic stem cells remodeled with neoPGs. (a) NeoPG 11D with D2A6 sulfation pattern, with affinity for FGF2, promoted differentiation into neural rosettes, whereas cells treated with neoPGs 11A (b) and 11N (c), and untreated cells (d), retained their embryonic characteristics. (e) Soluble heparin was also found to promote neural differentiation into rosettes. (f) Increasing dosages of neoPG 11D increased neural rosette count. Reprinted with permission from Huang, M.L., Smith, R.A.A., Trieger, G.W., Godula, K. Glycocalyx remodeling with proteoglycan mimetics promotes neural specification in embryonic stem cells. Journal of the American Chemical Society (2014) 136, 10565–10568. Copyright (2014) American Chemical Society
3.8. FGF2 Stimulation Analysis Via Western Blotting
3.8.1. FGF2 Stimulation
Seed cells in gelatin-coated six-well plates at a density of1 × 105 cells/cm2 in maintenance medium (see Note 24).
Aspirate medium and replace with maintenance medium with out FBS to serum-starve the cells. Incubate overnight.
Aspirate medium and wash cells three times with PBS.
Add lipid-neoPGs in KO-DMEM (or KO-DMEM alone as negative control) at desired concentration (e.g., 1 μM). Incubate for 1 h at 37 °C, 5 % CO2.
Remove solutions, wash three times with PBS.
Perform stimulation by adding 25 ng/mL of FGF2 in KO-DMEM with or without heparin (5 μg/mL). Incubate for 15 min at 5 % CO2, 37 °C.
Place plate directly on ice. Aspirate medium and wash cells twice with ice-cold PBS. Aspirate PBS.
Add 50 μL of ice-cold lysis buffer to each well, and detach cells from plate with a cell scraper.
Collect the suspension into an eppendorf, and incubate on ice for 10 min.
Centrifuge the suspension at 4 °C, for 10 min at 20,000 ×g.
Transfer the supernatant to fresh eppendorf. Store at −80 °C if needed.
3.8.2. Western Blot
Quantify protein content using an aliquot of the previous lysate using a standard BCA assay protocol.
Add protein sample (5 μg total) to a new eppendorf tube and dilute to 10 μL total volume with doubly distilled water.
Add 10 μL of 2× Laemmli buffer to each sample. Briefly vortex at low speed and centrifuge for a few seconds.
Transfer eppendorf tubes to a heat block at 100 °C for 5 min to reduce proteins (see Note 25).
Return the boiled samples to ice and centrifuge at low speed upon cooling.
Load sample and protein ladder solution (10 μL) in a pre-cast gel (or self-casted 10 % SDS-PAGE) on a running apparatus. Run gel at 100 V in 1× running buffer for 90–120 min.
Transfer the proteins from the PAGE gel onto a nitrocellulose membrane using a transfer cassette. Run transfer at constant 350 mA/90 V for 1 h in transfer buffer (see Note 26).
Once complete, block the nitrocellulose membrane in 5 % BSA in 1× TBST for 1 h at room temperature.
Remove blocking solution and add primary antibody solution (anti-phospho-Erk1/2, anti-total Erk1/2, anti-alpha-tubulin) in 5 % BSA in 1× TBST. Incubate overnight at 4 °C, overnight (see Note 27).
Wash membranes three times with 1× TBST. Remove last wash.
Add HRP-conjugated secondary antibodies (anti-rabbit HRP, anti-mouse HRP) in 5 % BSA in 1× TBST for 1 h at room temperature.
Wash membranes three times with 1× TBST. Remove last wash.
Develop membrane using HRP detection reagent and developing film (see Note 28).
Perform densitometry using an appropriate software such as Adobe Photoshop or ImageJ (see Note 29) (Fig. 6).
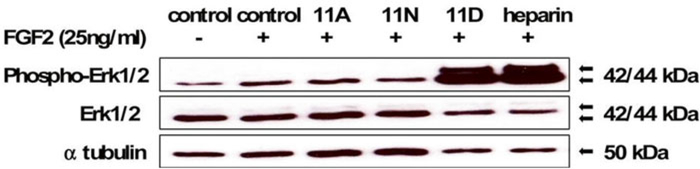
Fig. 6.
Glycocalyx remodeling of Ext1−/− mESCs rescued FGF2-mediated signaling. Similar to heparin, cells remodeled with neoPG 11D significantly promoted Erk phosphorylation compared to controls and neoPGs 11A and 11N. Adapted with permission from Huang M.L., Smith, R.A.A., Trieger, G.W., Godula, K. Glycocalyx remodeling with proteoglycan mimetics promotes neural specification in embryonic stem cells. Journal of the American Chemical Society (2014), 136, 10565–10568. Copyright (2014) American Chemical Society
4. Notes
Characterization data for carbamate monomer: 1H NMR (500 MHz, CDCl3) δ (ppm): 7.63 (br s, 1H), 6.28 (dd, 1H, J = 17 Hz, J = 2 Hz), 6.11 (dd, 1H, J = 17 Hz, J = 10 Hz), 5.56 (dd, 1H, J = 10 Hz, J = 2 Hz), 3.91 (t, J = 6 Hz, 2H), 3.50 (m, 2H), 3.09 (s, 3H), 1.78 (dd, 2H, J = 10 Hz, J = 2 Hz), 1.47 (s, 9h). 13C NMR (125 MHz, CDCl3) δ (ppm): 165.6, 156.9, 131.6, 125.5, 81.9, 72.4, 36.9, 28.3, 26.9. Calculated C12H22N2O4, 258.16, [M+Na]+: 281.15. HRMS m/z found: 281.1470.
To prepare maintenance medium, add all components together except 2-mercaptoethanol and LIF, and sterile filter through a 0.22 μm-membrane filtration device. Add the last two components after filtration.
It is critical to ensure that glass media bottles are washed extensively with doubly distilled water to remove detergent when used for preparing gelatin stock solutions. It is suggested to use a dedicated glass media bottle for this purpose that does not encounter soap.
It is crucial that all components of this reaction are well- dissolved in the solution. Avoid leaving solids on the side of the flask. The ratio of monomer to chain transfer agent can be tuned to achieve the desired degree of polymerization (DP). In our hands, a 300:1 ratio of monomer to lipid chain transfer agent generated polymers of DP~300.
The reacted monomer (to form the polymer) is expected to be significantly viscous in form relative to the starting mixture.
A pale yellow solid is often isolated in 90% yield. Characterization data for both azide and lipid-terminated polymer products: 1H NMR (CDCl3, 500 MHz) δ (ppm): 3.90–3.65 (bs, 2H), 3.35–2.80 (bm, 5H), 1.80–1.05 (bm, 16H).
For the azide-terminated polymer: Mw = 47.3 kDa, Mn = 40.1 kDa, DI = 1.18, DP ≈ 200. Lipid-terminated polymer: Mw = 78.5 kDa, Mn = 63.9 kDa, DI = 1.23, DP ≈ 300.
A white solid is isolated in ~95 % yield. Successful end-deprotection can be monitored as the reduction of UV absorbance at 310 nm.
Determine polymer labeling efficiencies by UV-Vis absorbance at the appropriate fluorophore wavelength. For TAMRA (λmax = 510 nm), we observed 98% labeling efficiencies.
At this stage, the polymer precipitates from ether as a colored fibrous-like residue. Centrifugation helps to clump the residue, such that simple decantation of the solvent is possible.
Characterization data for side-chain deprotected polymer. 1H NMR (D2O, 300 MHz) δ (ppm): 3.94 (bs, 2H), 3.13 (bs, 2H), 2.82 (bs, 3H), 1.75 (6H, bm). It is best to leave the polymer on the lyophilizer right before dissolution in reaction buffer for the next step.
A benchtop UV-lamp can be used to monitor the dissolution of clumped polymer residue in reaction buffer. Vortex or shake as needed to dissolve.
Glycans used to generate mimics of heparan sulfate were purchased from commercial vendors. These glycans are generated by enzymatic digestion of heparan sulfate. For a description of diGAG nomenclature, see reference [13]. We conduct these reactions in a PCR thermocycler, mainly because of the small scale of the reaction, and the convenience of weighing small amounts in PCR tubes. Other formats may be acceptable.
The centrifugal filters often come with a small amount of glycerol present, which can obscure subsequent NMR analysis. This can be minimized by washing the filter 3× with water (with centrifugation), and leaving the filter to soak overnight with water, and re-centrifugation.
Residual acetate peaks from the reaction buffer are often detectable in the NMR analysis of the final product. These are present as a singlet at 1.88 ppm. For diGAGs containing α,β-unsaturated uronic acids, ligation efficiency can be determined by integrating the glycan olefin signal (5.8 ppm) relative to the polymer backbone methyl protons (2.4–2.8 ppm). For monosaccharides, such as GlcNAc–6S, ligation efficiency can be determined by subtracting polymer backbone protons from the total integration in the region 2.5–4.5 ppm, and dividing this difference by the number of glycan protons. Ligation efficiencies from 15 to 70 % have been observed. Highly charged glycans are observed to cause lower ligation efficiencies, due to charge repulsion effects. Some cleavage of the fluorophore from the polymer backbone has been observed (depending on the fluorophore). It is best to re-characterize the polymer at this stage by UV–Vis to determine polymer labeling efficiencies using known weights of the polymer.
A standard robotic printer can be used to array spots of the glycopolymers onto the glass slides. We used a non-contact ultrasonic spotter equipped with glass capillary dispensers (GIX Microplotter, Sonoplot, Middleton, WI, uSa).
Polymer grafting efficiencies can be calculated as the percentage of fluorescence signal retained versus printed onto the slide.
Make sure to include negative controls in the experiment to account for background binding of the primary and secondary antibodies. In addition, a control experiment wherein FGF2 is pre-incubated with 1 mg/mL heparin for 15 min can be included.
Growth factor binding to each spot/polymer can be calculated as the average ratio of fluorescence intensities at Cy5/TAMRA.
The neoPG solution in serum-free KO-DMEM can be sterilized by passage through a low retention volume 0.22 pm sterile syringe filter.
Experiments with neoPG are conducted in 1 μM polymer in a 1:1 mixture of neurobasal medium and DMEM/F12. Control experiments with heparin are prepared as a 5 μg/mL solution of heparin in neural differentiation medium.
It is recommended that media changes be conducted around the same time each day to minimize cell death. The first media change can be expected to be required around day 2, and daily after then. Cell death is commonly observed at days 3–4.
In our hands, the listed antibodies displayed high efficiency, based on the protocol. Other sources of antibodies may also be applicable.
We generally allow seeding for 8–10 h for this experiment prior to serum starvation.
The lids will tend to pop open during this step. Place a heavy object atop the eppendorf tubes to prevent them from doing so.
Gels and membranes must be handled with tweezers at all times. Pre-wet the nitrocellulose/PVDF membranes (appropriately cut to match the dimensions of the gel and cassette) prior to use. Prepare transfer cassette by encasing the nitrocellulose membrane and gel with four sheets of thick filter paper and fiber pads (outer). Add a frozen block and magnetic stirrer to transfer tank to prevent overheating.
For sequential antibody staining, strip membranes of primary or secondary antibodies using a Western blot stripping buffer for 15 min at room temperature. Wash membranes three twice with 1× TBST, and block with 5% BSA in TBST for 1 h at room temperature before subsequent antibody staining.
Membranes are incubated for 2 min with 1–2 mL of developing reagent. Excess reagent is drained and membranes transferred to a developing cassette. Membranes are developed using ECL Hyperfilm for a period of 10s to 1 min, dependent on signal strength. Films are finally developed using a photo-developer in a dark room.
Normalize both Phospho-Erk1/2 and total Erk1/2 levels to alpha-tubulin levels, then normalize phospho-Erk1/2 levels to total Erk1/2 levels.
References
- 1.Varki A, Freeze HH, Vacquier VD (2009) Essentials of glycobiology, 2nd edn. CSHL Press, New York, pp. 531–536. [Google Scholar]
- 2.Yayon A, Klagsbrun M, Esko JD et al. (1991) Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 64:841–848. [DOI] [PubMed] [Google Scholar]
- 3.Kreuger J, Spillman D, Li JP et al. (2006) Interactions between heparan sulfate and proteins: the concept of specificity. J Chem Biol 174:323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunath T, Saba-El-Leil MK, Almousailleakh M et al. (2007) FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134:2895–2902. [DOI] [PubMed] [Google Scholar]
- 5.Johnson CE, Ward CM, Wilson V et al. (2007) Essential alterations of heparan sulfate during the differentiation of embryonic stem cells to Sox1-enhanced green fluorescent proteinexpressing neural progenitor cells. Stem Cells 25:1913–1923. [DOI] [PubMed] [Google Scholar]
- 6.Chiefari J, Chong YK, Ercole F et al. (1998) Living free-radical polymerization by reversible addition-fragmentation chain transfer: the RAFT process. Macromolecules 31:5559–5562 [Google Scholar]
- 7.Kuzmin A, Poloukhtine A, Wolfert MA et al. (2010) Surface functionalization using catalystfree azide-alkyne cycloaddition. Bioconjugate Chem 21:2076–2085. [DOI] [PubMed] [Google Scholar]
- 8.Rabuka D, Forstner MB, Groves JT et al. (2008) Noncovalent cell surface engineering: incorporation of bioactive synthetic glycopolymers into cellular membranes. J Am Chem Soc 130:5947–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang ML, Smith RAA, Trieger GW et al. (2014) Glycocalyx remodeling with proteoglycan mimetics promotes neural specification in embryonic stem cells. J Am Chem Soc 136:10565–10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godula K, Umbel ML, Rabuka D et al. (2009) Control of the molecular orientation of membrane-anchored biomimetic glycopoly-mers. J Am Chem Soc 131:10263–10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xin L, Wei G, Shi Z et al. (2000) Disruption of gastrulation and heparan sulfate biosynthesis in Ext1-deficient mice. Dev Biol 224:299–311. [DOI] [PubMed] [Google Scholar]
- 12.Ying Q-L, Stavridis M, Griffiths D et al. (2003) Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21:183–186. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence R, Lu H, Rosenberg RD et al. (2008) Disaccharide structure code for the easy representation of constituent oligosaccharides from glycosaminoglycans. Nat Methods 5:291–292. [DOI] [PubMed] [Google Scholar]