Abstract
Objective
The plasma-based methylated SEPTIN9 (mSEPT9) is a colorectal cancer (CRC) screening test for adults aged 50–75 years who are at average risk for CRC and have refused colonoscopy or faecal-based screening tests. The applicability of mSEPT9 for high-risk persons with Lynch syndrome (LS), the most common hereditary CRC condition, has not been assessed. This study sought preliminary evidence for the utility of mSEPT9 for CRC detection in LS.
Design
Firstly, SEPT9 methylation was measured in LS-associated CRC, advanced adenoma, and subject-matched normal colorectal mucosa tissues by pyrosequencing. Secondly, to detect mSEPT9 as circulating tumor DNA, the plasma-based mSEPT9 test was retrospectively evaluated in LS subjects using the Epi proColon 2.0 CE assay adapted for 1mL plasma using the “1/1 algorithm”. LS case groups included 20 peri-surgical cases with acolonoscopy-based diagnosis of CRC (stages I-IV), 13 post-surgical metastatic CRC, and 17 pre-diagnosis cases. The control group comprised 31 cancer-free LS subjects.
Results
Differential hypermethylation was found in 97.3% (36/37) of primary CRC and 90.0% (18/20) of advanced adenomas, showing LS-associated neoplasia frequently produce the mSEPT9 biomarker. Sensitivity of plasma mSEPT9 to detect CRC was 70.0% (95% CI, 48%-88%)in cases with a colonoscopy-based CRC diagnosis and 92.3% (95% CI, 64%-100%) inpost-surgical metastatic cases. In pre-diagnosis cases, plasma mSEPT9 was detected within two months prior to colonoscopy-based CRC diagnosis in 3/5 cases. Specificity in controls was 100% (95% CI 89%-100%).
Conclusion
These preliminary findings suggest mSEPT9 may demonstrate similar diagnostic performance characteristics in LS as in the average-risk population, warranting a well-powered prospective case–control study.
Keywords: colorectal cancer, lynch syndrome, SEPTIN9, plasma, circulating tumor dna
Summary box.
What is already known about this subject?
Lynch syndrome (LS) subjects are genetically predisposed to a high risk for colorectal cancer (CRC) and regular colonoscopy is currently the only CRC screening option recommended for these subjects, although compliance is suboptimal.
The plasma-based methylated SEPTIN9 (mSEPT9) CRC screening test available to adults in the general population aged ≥50 years who refuse colonoscopy or faecal-based screening has 68%–78% sensitivity and 80%–84% specificity to detect CRC. This test has not previously been assessed in LS subjects.
What are the new findings?
In this retrospective exploratory study we show two major new findings. First, we show the mSEPT9 biomarker is similarly frequent in Lynch-associated CRC (97%) as in sporadic CRC (96%), and is also found in 90% of advanced adenomas of various histologies. Second, although sample size was small, the plasma-based mSEPT9 test showed 70% sensitivity to detect CRC in patients with LS with a colonoscopy-based diagnosis of CRC.
How might it impact on clinical practice in the foreseeable future?
These encouraging preliminary findings suggest a well-powered prospective study of diagnostic performance is worthwhile to determine with high accuracy if the plasma-based mSEPT9 test would be applicable for CRC screening in subjects with LS. If so, this test could benefit subjects who are unaware they have LS, or LS subjects who are unable or unwilling to undergo frequent colonoscopy screening, or as an interval test between colonoscopies. This test may thus complement, but will not replace, ‘gold-standard’ colonoscopy screening in this high-risk group.
Introduction
The blood-based colorectal cancer (CRC) screening test Epi proColon 2.0 CE (Epigenomics, Berlin, Germany) provides an alternative screening test for the detection of asymptomatic CRC in average-risk adults aged 50–75 years who have refused colonoscopy and faecal-based screening tests.1 This test is based on the detection of methylated SEPTIN9 (mSEPT9) as circulating tumour (ct) DNA in plasma using sensitive ‘HeavyMethyl’ PCR technology.2 The mSEPT9 test was developed after the discovery of aberrant hypermethylation at an intragenic CpG island within the SEPT9 gene in ~97% of CRC, whereas methylation was low or absent in normal colorectal mucosa (NCM), peripheral blood lymphocytes (PBL), and other cancer types.3 4 A meta-analysis of pooled data from diagnostic performance studies of the mSEPT9 test in average-risk populations showed sensitivity to detect CRC (stages I–IV pooled) was 78% and specificity was 84%.5 In a prospective study, population sensitivity for CRC was 68.0% (95% CI 53% to 80%) and specificity was 80.0% (95% CI 78% to 82%).6 In a case–control study that compared the accuracy of the plasma mSEPT9 test with the faecal immunohistochemistry test (FIT) in paired samples from the same people, sensitivity to detect CRC was marginally superior for mSEPT9 at 72.2% (95% CI 63% to 80%) than for FIT at 68.0% (95% CI 58% to 77%). However, specificity at 80.8% (95% CI 75% to 86%) was inferior to FIT at 97.4% (95% CI 94% to 99%).7 The mSEPT9 test is currently contraindicated for use by those at high risk for CRC, including those with a hereditary cancer predisposition condition, since it has not been evaluated in this setting.
Lynch syndrome (LS) is the most common hereditary CRC syndrome, accounting for an estimated 2%–5% of all CRC.8 The population prevalence of LS is estimated to be 1 in 279 people in the USA,9 and higher in countries with founder populations, for example, 1 in 226 in Iceland.10 LS is caused by heterozygous germline mutations of one of the DNA mismatch repair (MMR) genes, MLH1, MSH2, MSH6 or PMS2.11 Mutation carriers are predisposed to the development of CRC (~10%–80% by age 70 years depending on gene mutated) as well as extracolonic cancers.12–14 They are advised to undergo colonoscopy at one to two yearly intervals starting by age 25 years.13 15 Regular colonoscopy screening has been shown to significantly reduce the incidence of CRC, tumour stage, and CRC-related mortality in LS subjects.16–18
Frequent lifelong screening by colonoscopy with the removal of precancerous lesions (PCL) is currently the only screening option presented to LS subjects for the prevention or early detection of CRC. However, it has been estimated that 95% of LS subjects are unaware that they carry an MMR gene mutation, hence are unaware of their cancer predisposition.19 Even among those who are aware that they carry an MMR mutation, a significant fraction are non-adherent with colonoscopy screening guidelines.18 20 21 One UK-based study found only 67% of mutation carriers underwent a colonoscopy within the recommended 2-year interval.18 LS subjects are younger, hence less tolerant of colonoscopy (more sedative is required), which impacts on future lifelong screening compliance.22 Furthermore, the occurrence of ‘interval’ CRC within the recommended screening period is frequent in LS, such that CRC arises even among patients who are compliant with regular colonoscopy screening recommendations.23–26 This has been attributed to various factors including a high miss rate for the detection of PCLs in patients with LS (these are often right sided and include flat and serrated histotypes that are more difficult to detect),27–33 and because progression from PCL to invasive CRC is highly accelerated in LS (<3 years) compared with sporadic CRC (6–10 years).29 34 The LS population could thus benefit from a minimally invasive CRC screening test to complement colonoscopy in those who are non-compliant with regular colonoscopy screening recommendations, unaware they have LS, and as an interval test between colonoscopies to identify subjects who should undergo colonoscopy sooner.
We sought evidence for the applicability of the mSEPT9 biomarker for CRC detection in LS subjects. First, we determined the frequency of SEPTIN9 methylation in archival specimens of CRC, advanced adenomas, and paired NCM from patients with LS. Second, we applied a modified version of the Epi proColon 2.0 CE mSEPT9 test to plasma samples from LS subjects with and without a diagnosis of CRC to determine if mSEPT9 ctDNA was detectable in the plasma of those with CRC.
Materials and methods
The Cancer Genome Atlas methylation data processing
Preprocessed The Cancer Genome Atlas (TCGA) methylation data generated using the Infinium Human Methylation 450K BeadChip array (Illumina), and clinical data, were downloaded from the colon and rectum adenocarcinoma (COADREAD) data set using the Firehose pipeline.35 Patients with more than 20% missing values for DNA methylation across the array were omitted. All remaining missing values were estimated using K-Nearest Neighbor (KNN) impute.36 Batch correction was performed using Combat.37 MethylMix38 was applied to classify abnormally methylated CpG sites across SEPT9, and to classify tumours binomially as having either a normal or hypermethylated state.
Patients
All patients included in this study had confirmed LS, as defined by carriage of a pathogenic germline mutation within one of the MMR genes.
Invasive CRC (adenocarcinoma of histological stages I–IV), advanced PCLs demonstrating one or more features associated with malignant potential (a villous/tubulovillous, serrated, or adenomatous cell type, a diameter of ≥1 cm, high-grade dysplasia), and macroscopically NCM were derived from archived formalin-fixed paraffin-embedded (FFPE) tissue blocks from patients who underwent either surgical resection for a diagnosis of CRC, or a polypectomy during colonoscopy (PCL), between December 1982 and December 2009.
Plasma samples were derived from peripheral blood drawn by venipuncture into K2EDTA tubes from LS subjects between March 2006 and February 2019. Plasma was separated from cellular components by centrifugation within 18 hours of blood collection, aliquoted into 1 mL volumes, stored at −80°C, and thawed immediately prior to mSEPT9 testing. Plasma samples from CRC cases were divided into three groups, depending on the timing of their blood drawn with respect to their CRC diagnosis and/or treatment: (1) cases with a colonoscopy-based diagnosis of CRC whose blood was drawn preoperatively or perioperatively; (2) cases whose blood was drawn up to 1 year prior to the colonoscopy procedure in which a CRC was subsequently diagnosed; and (3) cases diagnosed with metastatic CRC during postoperative surveillance for relapse. Plasmas from LS subjects without a prior diagnosis of cancer or preneoplastic lesions, and who did not go on to develop cancer within 2 years after the blood draw were included as controls.
Pathology and sampling of FFPE tissues
A 4 µM section of a representative FFPE tissue block for each patient was stained with H&E. American Joint Committee on Cancer stage was obtained from pathology reports. Immunohistochemistry (IHC) of all four MMR proteins was performed on serial 4 µM sections, as described either below or previously.10 All IHC staining protocols were fully automated with antigen retrieval. Mouse monoclonal antibodies included α-MLH1 and α-PMS2 (G168-15 and A16-4, respectively, from BD Pharmingen, San Diego, CA), α-MSH2 (G219-1129, Cell Marque, Rocklin, CA), and α-MSH6 (clone 44, Ventana, Tucson, AZ). IHC detection was performed on the Ventana benchmark Ultra instrument with Ultraview DAB detection for MLH1, MSH2 and MSH6 and the Leica Bond III instrument with Leica refine DAB detection for PMS2. All slides were subsequently counterstained with Mayer’s haematoxylin. IHC interpretation, histology, and demarcation of areas of maximum tumour cellularity representative of adenocarcinoma, areas with advanced histological features for PCL, and areas of NCM on each H&E slide were performed by a gastrointestinal pathologist.
With reference to the H&E slide, a 0.6/1 mm diameter core of each tissue type was excised from the respective demarcated areas on each block using a tissue microarrayer. Each FFPE core was deparaffinised in 1 mL Deparaffinization Solution (Qiagen), then DNA extracted using the QIAamp FFPE DNA Kit (Qiagen) according to the manufacturer’s instructions. DNA purity and quantification were determined by spectrophotometry.
Measurement of SEPT9 methylation levels in FFPE tissues
SEPT9 methylation was measured in bisulfite-converted DNA by CpG pyrosequencing. DNA (0.5–1 µg) from each FFPE core was bisulfite converted using the EZ-Methylation-Gold Kit (Zymo Research, Irvine, USA), and 100–200 ng was PCR amplified using 0.4 µM each of primers 5′-TAGTTGAGTTAGGGGGTTTAGGGGTTT-3′ and 5′-biotin-AAATCCRACATAATAACTAATAAACAAC-3′ with Platinum Taq Polymerase in a 50 µL volume containing 0.2 mM each dNTP and 3 mM MgCl2 for 40 cycles with annealing at 58°C. Products were purified on the PyroMark Vacuum Manifold (Qiagen), and pyrosequenced using internal primer GGTTAGTTTTGTATTGTAGG-3′ and nucleotide dispensation GATGTCAGTCAGTCAGTCAGTCGCTTA on the PyroMark ID96 instrument (Qiagen).
Methylation values were calculated as the mean percentage methylation across the five CpG sites interrogated. DNA from the RKO CRC cell line and PBL from a healthy control served as hypermethylated and unmethylated controls, respectively, and were included with each batch of patient samples.
Plasma-based mSEPT9 testing according to the ‘1/1 algorithm’
The standard operating procedure for the Epi proColon 2.0 CE clinical test requires 3.5 mL fresh plasma from each patient for the extraction and bisulfite conversion of cell-free (cf) DNA, which is then input into three duplex real-time PCR reactions of mSEPT9 and ACTB (cfDNA input control). By extrapolation, we calculated that bisulfite-converted cfDNA template derived from 0.875 mL plasma is input into each PCR reaction. Results are scored using the ‘1/3 algorithm’, whereby a positive test result is given if the mSEPT9 signal reaches a designated threshold in one or more of the three (ACTB-validated) PCR reactions.
For our retrospective study, just 1 mL plasma per patient was available for mSEPT9 testing. We modified the cfDNA extraction and bisulfite conversion steps of the Epi proColon 2.0 CE protocol by proportionally reducing reagent volumes to process 1 mL plasma, such that cfDNA template derived from 0.875 mL plasma was input into a single real-time PCR reaction. Results were scored using the ‘1/1 algorithm’, whereby a positive test result was designated if the mSEPT9 signal reached the specified threshold in the single (ACTB-validated) reaction, and negative if it did not. In validation tests on Epi proColon positive and negative control samples, we demonstrated equivalent results for each PCR reaction from 1 mL starting volume as for a 3.5 mL starting volume of sample divided into three reactions before applying the modified protocol to patient samples.
Statistical analyses
The two-sample Wilcoxon rank-sum test was used to compare the median β values between COADREAD CRC and NCM tissues at each CpG site located within the SEPT9 gene. Probes with a Q-value (false discovery rate (FDR)-corrected p value) below 0.05 were considered significant. Analysis of variance was used to compare age at diagnosis between patient groups. For analyses of FFPE tissue-based methylation data, a receiver operating characteristic (ROC) curve and the area under the ROC curve (AUC) were estimated with a mixed-effects logistic regression model, taking into account subject-matched NCM samples to CRC or PCL samples and repeated measurements on the same NCM sample, and the corresponding 95% CI was estimated using a bootstrap method with 1000 replicates. To determine the optimal threshold value for methylation-positive test results to discriminate CRC or PCL samples from NCM samples, we examined the sensitivity and specificity for each point of the ROC curve, and the optimal threshold was returned by maximising the sum of sensitivity and specificity, which is comparable with Youden Index.39
The diagnostic performance of plasma-based mSEPT9 test was evaluated by examining the sensitivity and specificity of the test using the ‘1/1’ algorithm with the corresponding 95% exact binomial CI.40 Analyses were performed using the R package V.3.5.0 with two-sided tests and a significance level of 0.05.
Results
Development of a quantitative pyrosequencing assay to measure SEPT9 methylation levels in archival tissues
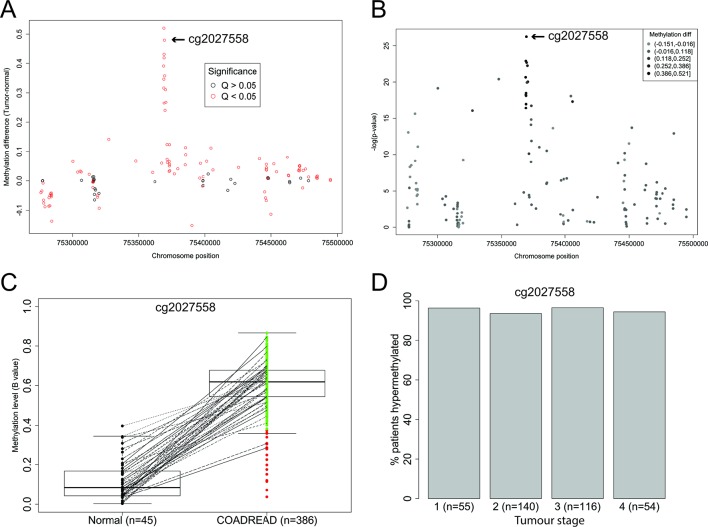
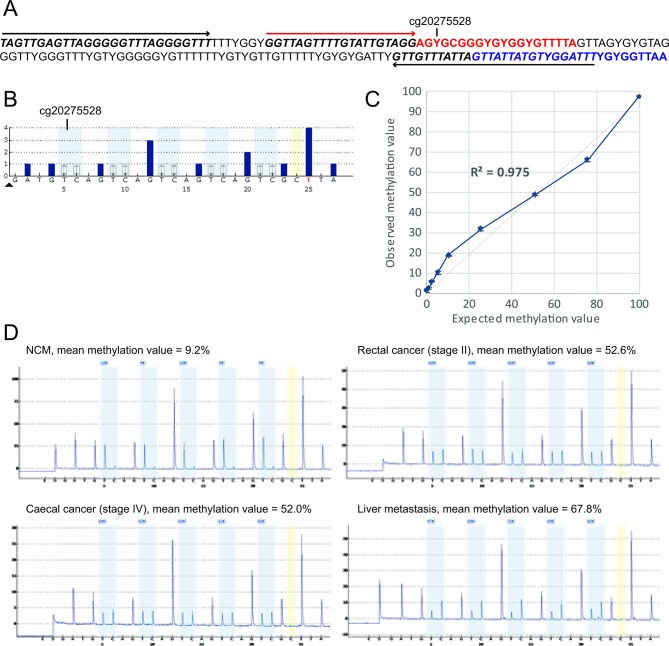
To compare the frequency of SEPT9 methylation between Lynch-associated CRC and ‘sporadic’ CRC, we developed a pyrosequencing assay to measure methylation levels in Lynch-associated colorectal neoplasia at a CpG site(s) that overlapped with data available for ‘sporadic’ CRC. We first undertook an analysis of the distribution and levels of methylation across the entire SEPT9 gene within CRC and NCM tissues from COADREAD data generated on the Infinium Human Methylation 450K Array by TCGA.41 Consistent with previous reports,3 4 the ‘V2 region’ targeted by the plasma-based Epi proColon 2.0 CE test had the most frequent and the highest levels of differential methylation between CRC and NCM tissues (figure 1A). The CpG site corresponding to Infinium probe cg20275528 had the most significant differential methylation between CRC and NCM (Wilcoxon rank-sum p=5.63e-27, figure 1B), which remained significant after FDR correction (Wilcoxon rank-sum Q=7.55e-25). Cg20275528 is the closest of the Infinium probes to the region assayed by the ‘MethylHeavy’ real-time PCR in the plasma-based mSEPT9 test.2 Cg20275528 was hypermethylated in 96% of CRC within the COADREAD data set (figure 1C), with similar frequency of methylation by tumour stage (figure 1D). Therefore, we developed a quantitative pyrosequencing assay that specifically measured methylation at five consecutive CpG sites including cg2027558 (figure 2A,B). In validation experiments of hypermethylated RKO diluted into unmethylated PBL DNA, the assay had a limit of detection down to 2.5% (coefficient of variance of 0.07), and strong linearity with methylation levels (r2>0.975), hence was unbiased with respect to methylation content (figure 2C). The assay proved robust for application to samples derived from FFPE tissues (figure 2D).
Figure 1.
Levels and prevalence of SEPT9 methylation in colon and rectal adenocarcinoma (COADREAD) compared with adjacent normal tissue in Infinium Human Methylation 450K Array data from The Cancer Genome Atlas (TCGA). (A) Dot plot of differential methylation levels (tumour-normal; y-axis) by genomic location (x-axis) according to the GRCh37/hg19 human genome assembly at each of the SEPT9 probes contained within the 450K array data from the TCGA COADREAD data set. The closest probe to the plasma-based Epi proColon 2.0 CE assay, cg2027558, located at Chr17: 75369484 is indicated. (B) Manhattan plot of the −log10 p values (Wilcoxon rank-sum test; y-axis) for a difference in methylation between tumour and normal tissues by genomic location (x-axis). Probe cg2027558, indicated, showed the most significant difference in tumour versus normal tissue. (C) Spaghetti plot of normalised levels of methylation (β values) at probe cg2027558 in normal tissue and COADREAD tumour tissue. Subject-matched sample pairs are linked by lines. Median, IQRs and 95% CI are indicated. Green=tumours showing hypermethylation, red=tumours with methylation levels in the normal range. (D) Histogram of the frequency of SEPT9 hypermethylation at cg2027558 in colorectal cancer (CRC) from COADREAD stratified by tumour stage.
Figure 2.
Measurement of SEPTIN9 methylation levels by quantitative CpG pyrosequencing in archival Lynch syndrome (LS)-associated colorectal cancer (CRC), advanced adenoma, and paired normal colorectal mucosa (NCM) tissues. (A) Assay design. Sequence coordinates GRCh37/hg19 assembly Chr17: 75369414–75369587 are shown as bisulfite converted. YG indicates CpG sites where the cytosine may be methylated (CG) or unmethylated (TG). PCR primer-binding sites are italicised and indicated by black arrows. Pyrosequencing primer-binding site is italicised and indicated by a red arrow. The sequence analysed (red text) contains five CpG sites, of which the first corresponds to Infinium probe cg20275528, located 92 bp upstream of the fluorescent probe-binding sequence within the plasma-based Epi proColon 2.0 CE assay (blue text). (B) Simulated pyrosequencing assay. Grey shading shows the five CpG sites interrogated. Yellow shading shows the single cytosine control for bisulfite conversion efficiency. (C) Analytical sensitivity and linearity of the quantitative CpG pyrosequencing assay to measure methylation levels at SEPTIN9. The titration curve shows the observed (±1 SD) versus expected methylation values for the hypermethylated RKO CRC cell line DNA (expected value 100% methylated) diluted into the peripheral blood DNA from a healthy control (0% expected methylation value) in specific proportions. (D) Illustrative pyrograms obtained from formalin-fixed paraffin-embedded (FFPE) samples of primary CRC, a liver metastasis, and paired NCM from a Lynch syndrome patient with an MSH2 mutation.
SEPT9 hypermethylation is frequent in Lynch-associated CRC and PCL and distinguishable from NCM
Key clinicopathological features of LS cases whose neoplasia was tested for SEPTIN9 methylation are summarised in table 1. Thirty-six cases developed 37 CRCs, of which 30 CRCs from 29 patients showed clear evidence that the tumour was Lynch associated through absence of IHC staining of the MMR protein(s) predicted by the causative germline mutation or the finding of a somatic ‘second hit’ in the tumour via mutation profiling, undertaken herein or as previously published10 (online supplementary table 1). Seven CRCs had no IHC data and insufficient FFPE tissue available for testing. Fourteen patients developed 20 PCLs (11 tubular adenomas, 3 tubulovillous adenomas, 2 serrated adenomas, 2 sessile serrated adenomas, 2 tubular adenomas with high-grade dysplasia), all of which were classified histologically as ‘advanced adenomas’ (online supplementary table 1). Four patients developed synchronous CRC and advanced adenoma and were represented in both groups. Pathology review revealed no NCM tissue was present within the FFPE blocks for nine CRC and four adenoma cases.
Table 1.
Lynch syndrome cases of CRC and advanced adenomas tested for SEPTIN9 methylation in tissues
| CRC group (%) | Adenoma group (%) | |
| All unique cases | 36 | 14 |
| Both CRC and adenoma | 4 | 4 |
| Mean age at diagnosis, years (±SD) | 55.28 (±15.63) | 46.7 (±11.0) |
| Sex | ||
| Males | 26 (72.2%) | 7 (50%) |
| Females | 10 (27.8%) | 7 (50%) |
| Gene mutated | ||
| MLH1 | 10 (27.8%)3 | 9 (64.3%)3 |
| MSH2 | 5 (13.9%) | 4 (28.6%) |
| MSH6 | 10 (27.8%)1 | 1 (7.1%)1 |
| PMS2 | 11 (30.6%) | 0 (0.0%) |
| Neoplasia, n* | 37 | 20 |
| Location | ||
| Left sided | 13 (35.1%) | 15 (75%) |
| Right sided | 24 (64.9%) | 5 (25%) |
| Stage | NA | |
| I | 8 (21.6%) | |
| II | 17 (46.0%) | |
| III | 5 (13.5%) | |
| IV | 4 (10.8%) | |
| Unknown | 3 (8.1%) | |
| Immunohistochemistry | ||
| MLH1 | 3 IHC loss, 1 LOH, 6 NT | NT |
| MSH2 | 4 IHC loss, 1 NT | NT |
| MSH6 | 7 IHC loss, 3 IHC weak+somatic mutation† | NT |
| PMS2 | 11 IHC loss† | NT |
| Matched NCM | 29 (28)‡ | 13 (10)‡ |
Numbers in superscript indicate the number of cases who developed synchronous CRC and adenoma.
*Includes one CRC case developing two synchronous CRCs, and five adenoma cases developing two or more synchronous adenomas.
†Immunohistochemistry (IHC) or identification of a somatic second hit in tumour, as previously published.10
‡Number of matched pairs of tumour-NCM with unique NCM samples (some cases developed more than one synchronous neoplasm).
CRC, colorectal cancer; LOH, loss of heterozygosity of normal allele; NA, not applicable; NCM, normal colorectal mucosa; NT, not tested.
bmjgast-2019-000299supp001.docx (33.8KB, docx)
SEPT9 methylation values were obtained for all 37 primary CRC samples (median 54.0%, range 1.6%–86.8%), all 20 advanced adenoma samples (median 63.6%, range 6.6%–88.2%), and 33 paired NCM samples (median 4.6%, range 0.0%–16.6%) (figure 3A). In one MSH2 germline mutation carrier who developed two synchronous primary CRCs, SEPTIN9 methylation status was concordant between the two primary CRCs and a liver metastasis (figure 2C).
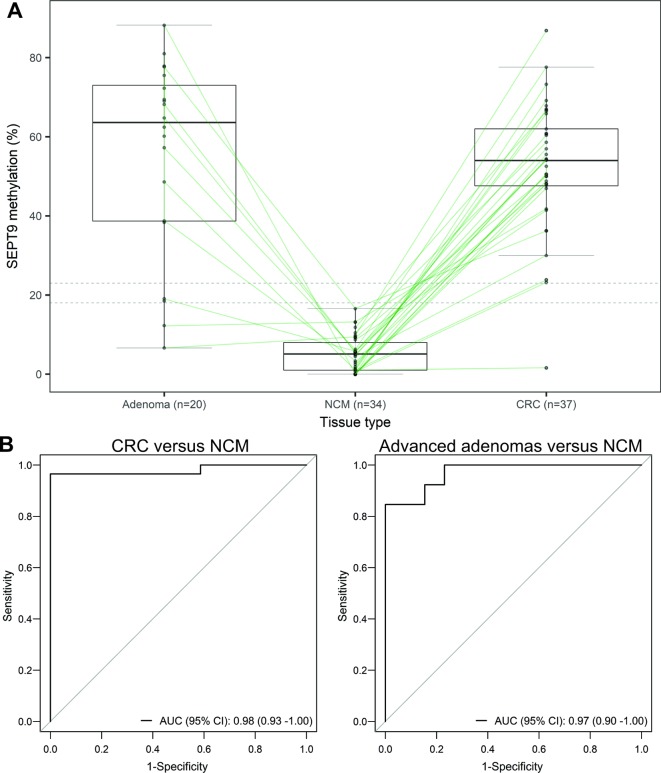
Figure 3.
Frequent hypermethylation of SEPT9 in Lynch syndrome (LS)-associated colorectal cancer (CRC) and advanced adenomas. (A) Spaghetti plot showing actual values of SEPT9 methylation measured by pyrosequencing at five CpG sites, including cg20275528, in CRC, advanced adenomas, and normal colorectal mucosa (NCM) from patients with LS (each black dot represents a single tissue sample and subject-matched samples are linked by lines), overlaid with box and whisker plot indicating the median value (horizontal black line) and IQRs (boxes). Dotted horizontal lines indicate the threshold values of 23% and 18% for methylation-positive test results to discriminate CRC and adenoma samples from NCM samples, respectively. Accompanying clinicopathological and SEPTIN9 methylation data are provided in online supplementary tables 1 and 2. (B) Receiver operating characteristic (ROC) curve for SEPT9 methylation values in CRC versus paired NCM (left), and advanced adenomas versus paired NCM (right). The diagonal lines represent the non-informative prediction model with an area under the ROC curve (AUC) of 0.5. ROC analyses were performed for the subset of neoplasms with a subject-matched NCM sample, including 29 CRC samples with 28 unique paired NCM samples, and 13 adenoma samples with 10 unique paired NCM samples.
ROC analysis performed on the SEPTIN9 methylation values for the primary CRC and paired NCM samples showed SEPTIN9 methylation was capable of discriminating CRC from NCM, with an AUC of 0.98 (95% CI 0.93 to 1.00) (figure 3B). With an optimal threshold level of 23% considered ‘methylation-positive’, the frequency of SEPTIN9 hypermethylation in CRC was 97.3% (36/37), as detailed in online supplementary table 1. One CRC sample, a caecal stage IV, mucinous adenocarcinoma, from an MLH1 mutation carrier aged 32 years was negative for SEPTIN9 methylation. No IHC or microsatellite instability data were available for this tumour. ROC analysis on the 13 advanced adenomas with paired NCM showed SEPTIN9 methylation was capable of discriminating advanced adenomas from NCM, with an AUC of 0.97 (95% CI 0.90 to 1.00) (figure 3B). With an optimal threshold level of 18% considered ‘methylation-positive’, the frequency of SEPTIN9 hypermethylation in advanced adenomas was 90.0% (18/20) across various histotypes (tubular, tubulovillous, serrated, sessile serrated, and high-grade dysplasia), as detailed in online supplementary table 2. A tubulovillous adenoma and a flat serrated adenoma were methylation negative.
Exploratory analyses of sensitivity and specificity to detect CRC in LS using the plasma-based Epi proColon 2.0 CE test with a ‘1/1 algorithm’
A pilot study of the performance characteristics of the plasma-based mSEPT9 ctDNA test to detect CRC in LS was undertaken in a retrospective study of cryopreserved plasma samples in CRC cases and controls. For exploratory estimates of sensitivity to detect CRC, plasma samples from LS ‘CRC cases’ were separated into three groups, based on the timing of the blood draw with respect to their CRC diagnosis and treatment, as follows: (Case group 1) Patients with a colonoscopy-based diagnosis of CRC with blood drawn prior to, or circa, surgical resection of the primary tumour were termed ‘pre-surgical CRC cases’. This was our primary case group of interest, which aimed to assess whether mSEPT9 was detectable in plasma samples taken around the time of usual diagnosis by colonoscopy. Sample selection for inclusion in case group 1 most closely resembled the timing of sample collection in previously published case–control studies of the plasma mSEPT9 test in average-risk groups.5 This group included 20 patients whose blood draw ranged from 138 days to 1 day prior to surgery, with the exception of one patient whose blood was drawn 4 days after surgery. The delay between the blood draw and surgery was accounted for by several patients with rectal cancer who underwent presurgical chemo/radiotherapy treatment. (Case group 2) Patients whose blood was drawn within 1 year prior to a colonoscopy-based diagnosis of CRC were included as ‘pre-diagnosis CRC cases’. This group was of interest to determine if mSEPT9 was detectable before usual diagnosis, and thus potentially serve as an interval test between colonoscopies. This group included 18 patients whose blood draw ranged from 328 days to 20 days prior to the colonoscopy screen in which their CRC was detected. (Case group 3) Patients with stage IV CRC whose blood was drawn any time after surgical resection of the primary tumour were included as ‘post-surgical metastatic CRC cases’. This group was of interest to determine if plasma-based mSEPT9 testing might have utility for postoperative monitoring of cases diagnosed with stage II/III disease for progression to metastatic disease. Finally, for exploratory analyses of specificity, a fourth group of 34 LS subjects (germline mutation carriers) with no personal history of cancer and who did not go on to develop a colorectal neoplasia within 2 years of the blood draw (the maximum recommended colonoscopy screening interval) were included as cancer-free ‘controls’ in our case–control study.
Results of mSEPT9 plasma-based testing are summarised in table 2. All 20 plasma samples from the ‘pre-surgical CRC case’ group 1 yielded valid test results and 14 were positive for mSEPT9 (online supplementary table 3). Sensitivity of the plasma-based mSEPT9 test to detect CRC (stages I–IV) around the time of usual diagnosis was thus 70.0% (95% CI 48% to 88%). Of the 18 plasma samples from the ‘pre-diagnosis CRC case’ group 2, a valid test result was obtained for 17, and three patients were positive for mSEPT9 (online supplementary table 4). The three mSEPT9-positive patients were among five patients with CRC stages I–III whose blood had been drawn up to 63 days (~2 months) prior to their colonoscopy-based diagnosis of CRC, with sensitivity of 60% (95% CI 15% to 95%) to detect CRC ~2 months prior to diagnosis. All 13 plasma samples from the ‘post-surgical metastatic CRC case’ group 3 yielded a valid test result and 12 were positive for mSEPT9 (online supplementary table 5). Sensitivity of the plasma-based mSEPT9 test to detect metastatic CRC was 92.3% (95% CI 64% to 100%). Of the 34 plasma samples from the cancer-free ‘control’ group, a valid test result was obtained for 31 and all were negative for mSEPT9 (online supplementary table 6). Hence, specificity of mSEPT9 in cancer-free LS subjects was 100% (95% CI 89% to 100%).
Table 2.
Sensitivity and specificity of the plasma-based mSEPT9 test to detect CRC in LS using the Epi proColon 2.0 CE assay with the ‘1/1 algorithm’
| Group | Presurgical CRC cases (case group 1) |
2 months’ prediagnosis CRC cases (case group 2) | Postsurgical metastatic CRC cases (case group 3) |
Cancer-free controls |
| All cases | 20 | 5 | 13 | 31 |
| Mean age (±SD) | 49.3 (±14.1) | 43.2 (±5.3) | 48.0 (±12.3) | 50.45 (±17.8) |
| Years* | P=0.80 | P=0.062 | P=0.65 | Reference |
| Gene mutated | ||||
| MLH1 | 3 (15.0%) | 1 | 5 (38.5%) | 8 (25.8%) |
| MSH2 | 6 (30.0%) | 3 | 6 (46.1%) | 7 (22.6%) |
| MSH6 | 7 (35.0%) | 1 | 1 (7.7%) | 6 (19.4%) |
| PMS2 | 4 (20.0%) | 0 | 1 (7.7%) | 10 (32.3%) |
| Positive test† | 13/19 | 3/5 | 12/13 | 0/31 |
| Stage I | 4/8 | 1/2 | NA | |
| Stage II | 3/3 | 0/0 | NA | |
| Stage III | 4/6 | 1/2 | NA | |
| Stage IV | 3/3 | 0/0 | 12/13 | |
| Unknown | 0/0 | 1/1 | NA | |
| Sensitivity (95% CI)‡ | 0.70 (0.48 to 0.88) | 0.60 (0.15 to 0.95) | 0.92 (0.64 to 1.00) | NA |
| Specificity (95% CI) | NA | NA | NA | 1.00 (0.89 to 1.00) |
The number of samples for which a valid test result was obtained for the 1 mL plasma sample is included.
*The mean age (±SD) is shown at the time of CRC diagnosis (case groups) or the blood draw (cancer-free control group). The control group showed no significant difference in age from any of the case groups.
†The proportion of positive test results to number of cases with valid test results is shown for each CRC case group, pooled and stratified by stage, and for the cancer-free controls.
‡Sensitivity is provided for all cases within each CRC case group combined by gene mutated and disease stage.
CRC, colorectal cancer; LS, Lynch syndrome; NA, not applicable.
Discussion
LS subjects are genetically predisposed to a high risk for CRC. Regular colonoscopy is currently the only CRC screening option recommended for LS subjects, and compliance is suboptimal. The plasma-based mSEPT9 is a screening test for CRC in adults aged ≥50 years at average risk for CRC, but has not been assessed in the LS population. We sought preliminary evidence for the utility of the mSEPT9 biomarker for CRC screening in LS.
First, we sought to determine if the mSEPT9 biomarker was produced by Lynch-associated colorectal neoplasia and found evidence in support of this. Elevated levels of SEPTIN9 methylation occurred with similarly high frequency in Lynch-associated CRC (97.3%) as in ‘sporadic’ CRC (96% in TCGA COADREAD data), and also at high frequency (90.0%) in advanced adenomas of various histologies, suggesting SEPTIN9 methylation is a common and early event in the neoplastic process of Lynch-associated CRC. Further testing of larger sample sizes of advanced PCL of different histologies is needed to determine if SEPTIN9 methylation is universal among the various histologies observed in LS, or more common among particular subtypes of PCL.
Second, we tested the plasma of LS subjects with and without CRC in an exploratory assessment of the sensitivity and specificity of the mSEPT9 ctDNA test to detect CRC in this high-risk group. Two key limitations of this aspect of the study are noted. First, the sample size available for study within each group of CRC cases and the cancer-free controls was too small for precise estimates of sensitivity and specificity, and plasma was not available from LS subjects with advanced PCL. A second limitation was that only 1 mL plasma was available for testing, which may have led to an underestimation of sensitivity, and an overestimation of specificity, of mSEPT9 to detect CRC. Prior studies in which the sensitivity and specificity of the ‘1/1 algorithm’ and the Food and Drug Administration-approved ‘1/3 algorithm’ were compared have shown the ‘1/3 algorithm’ had higher sensitivity and lower specificity than the ‘1/1 algorithm’ in the setting of CRC screening.42 43 This is presumably because testing of a larger volume of plasma which provides a greater likelihood for yielding a positive test result in mSEPT9 is present; the ‘1/3 algorithm’ tests 3.5 mL plasma in three PCR reactions,5 43 whereas just 1 mL plasma was available to us for a single PCR reaction. Nevertheless, using the ‘1/1 algorithm’, we found sensitivity of the plasma mSEPT9 test to detect CRC in our main case group of interest (case group 1), patients with LS with a prior colonoscopy-based diagnosis of CRC, was 70.0%. Given the use of the ‘1/1 test algorithm’, this may represent a conservative estimate of sensitivity. A meta-analysis of prior CRC case–control studies of average-risk populations using the mSEPT9 assay with the ‘1/3 test algorithm’ showed sensitivity was 78% and specificity was 84%.5 A large prospective study using the ‘1/3 algorithm’ showed sensitivity for CRC was 68% (95% CI 53% to 80%) and specificity was 80.0% (95% CI 78% to 82%).6 Our study suggests that sensitivity to detect CRC in patients with LS around the time of usual diagnosis could thus be similar to that of the average-risk population, having fallen within these two ranges. In patients with LS whose blood was drawn within 1 year prior to a colonoscopy-based diagnosis of CRC of stages I–III, mSEPT9 ctDNA was detected in 60% (3/5) patients within ~2 months prior to their diagnosis. Although this sample size is too small to form conclusions, this finding is nevertheless encouraging in suggesting the mSEPT9 ctDNA may be detectable in patients with LS prior to their usual colonoscopy-based diagnosis. A large, longitudinal, prospective study would be required to determine if the plasma mSEPT9 test would have utility for interval testing between colonoscopies. We found sensitivity to detect metastatic CRC in LS cases who had undergone surgical resection of the primary tumour was high at 92.0%, although the sample size was small. Others have found 75% or higher sensitivity to detect metastatic CRC in population-based patients,44 45 exceeding that of the serum protein-based carcinoembryonic antigen marker.5 Therefore, the plasma-based mSEPT9 test may also be worthy of formal exploration for application to postoperative monitoring of Lynch-associated CRC cases diagnosed with stage II/III disease for relapse during the recommended 5-year postoperative surveillance period. All 31 of the cancer-free control subjects with LS were negative for the plasma-based mSEPT9 test, showing specificity of 100%. This may represent an overestimate due to the small sample size and use of the ‘1/1 algorithm’. The median age of our case and control groups was only marginally younger than the age at which the mSEPT9 plasma test has received approval in the average-risk population (50 years and above), on account of the inclusion of cases and controls with MSH6 and PMS2 germline mutations, which are associated with an older age of CRC onset.46 Increased age (≥65 years) has been associated with increased non-specificity for the mSEPT9 test.43 Given the LS populations are advised to begin CRC screening from a younger age (typically 25 years), age is unlikely to adversely impact on test characteristics, if applied to this high-risk group. Nevertheless, we argue that sensitivity for any liquid biopsy-based test to detect CRC is more important than specificity in the context of high-risk groups.
Although no plasma-based screening test is ever likely to replace colonoscopy screening in high-risk setting such as LS, a minimally invasive screening test could complement colonoscopy in certain circumstances. In LS subjects who are non-compliant with colonoscopy screening recommendations, a positive test result from a minimally invasive screening test could serve as the impetus needed to undergo a follow-up colonoscopy. In the average-risk population, subjects who chose to undergo an FIT or mSEPT9 screening test which yields a positive result are recommended to undergo a follow-up diagnostic colonoscopy. If the mSEPT9 test were in future to be implemented in the LS population as a screening test, the same advice would be given. Furthermore, a positive plasma-based test result in subjects choosing to undergo additional interval screening between biennial colonoscopies would indicate the need to schedule an earlier colonoscopy. Whether or not the plasma-based mSEPT9 test could serve as a minimally invasive test for CRC detection in the LS population will require a well-powered prospective case–control study to determine the diagnostic performance characteristics of this test with high confidence. This would need to include sensitivity estimates of LS subjects with advanced adenoma as well, since early detection of colorectal neoplasia in this high-risk setting is key to CRC prevention. Despite the limitations of our study, outlined above, our findings of frequent SEPTIN9 hypermethylation in colorectal neoplasia and the detection of mSEPT9 ctDNA in a subset of plasma samples from CRC cases suggest a formal prospective case–control study of the diagnostic performance of the plasma mSEPT9 test in the LS population is warranted. This will determine with high accuracy if the blood-based mSEPT9 test would be applicable for CRC screening in subjects with LS. Such a study would be important for determining if this blood-based test could be used to complement colonoscopy-based screening for CRC detection in this high-risk group, in subjects unwilling or unable to undergo regular lifelong colonoscopy screening, as an interval screening test between colonoscopies, and to provide an alternative screening test to those unaware they even have LS.
Acknowledgments
We thank Jan Lockman of OSU for logistical support.
Footnotes
RWH and HTL contributed equally.
Contributors: MPH and RWH contributed to the concept and design, and supervised the study. MPH, IPV, SH, NZ, BM, RA, XY, MG, AEH, MFK, JDV, JMC, AdelaC, HH, RP, MC, CS, SJL, and HTL contributed to the acquisition of data. MPH, IPV, KB, BM, SJL, RWH, and HTL contributed to the analysis and interpretation of data. MPH, KB and SK contributed to statistical analyses. MPH, IPV, KB, SK, and RWH contributed to drafting of the manuscript. All authors provided critical review of the manuscript. MPH was responsible for critical revision of the manuscript.
Funding: This project was funded in part by the Chen-Yang Foundation, the Liz’s Legacy fund through Kicks for a Cure, the Samuel Oschin Comprehensive Cancer Center at Cedars-Sinai Medical Center, and by revenue from Nebraska's excise tax on cigarettes awarded to Creighton University through the Nebraska Department of Health & Human Services (DHHS). HTL is partially funded through the Charles F and Mary C Heider Chair in Cancer Research, which he holds at Creighton University. We acknowledge support from the Biobank & Translational Research Core at CSMC, which was funded in part through institutional support and in part through NIH grant G20 RR030860.
Disclaimer: The contents represent the views of the authors and do not necessarily represent the official views of the State of Nebraska or DHHS.
Competing interests: None declared.
Patient consent for publication: Obtained.
Ethics approval: The Hereditary Cancer Center, Creighton University (No 05-13877), The Icelandic Population-based Cancer Registry (Icelandic National Bioethics Committee No VSNb2013010033/03.15 and Data Protection Authority No 2013010109TS), The Ohio State University Comprehensive Cancer Center from the prior ‘Columbus-area HNPCC-study’ and ‘Ohio State Colorectal Cancer Prevention Initiative’ (No 2019C0064), The Cleveland Clinic, Ohio (No 2884), and Cedars-Sinai Medical Center (CSMC), California (No Pro00049624). Designated use of the retrospectively collected samples from these sources for SEPT9 methylation testing was subsequently approved by the Icelandic National Bioethics Committee (No 13-024 S1), Stanford University (No 120715 NOT-H3), and CSMC (No Pro00051525).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Data are available upon reasonable request.
References
- 1. Administration USFaD. Premarket approval (PMA) for EPI proColon, 2016. [Google Scholar]
- 2. deVos T, Tetzner R, Model F, et al. . Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem 2009;55:1337–46. 10.1373/clinchem.2008.115808 [DOI] [PubMed] [Google Scholar]
- 3. Lofton-Day C, Model F, Devos T, et al. . DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem 2008;54:414–23. 10.1373/clinchem.2007.095992 [DOI] [PubMed] [Google Scholar]
- 4. Wasserkort R, Kalmar A, Valcz G, et al. . Aberrant septin 9 DNA methylation in colorectal cancer is restricted to a single CpG island. BMC Cancer 2013;13 10.1186/1471-2407-13-398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Song L, Jia J, Peng X, et al. . The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: a meta-analysis. Sci Rep 2017;7 10.1038/s41598-017-03321-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Potter NT, Hurban P, White MN, et al. . Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem 2014;60:1183–91. 10.1373/clinchem.2013.221044 [DOI] [PubMed] [Google Scholar]
- 7. Johnson DA, Barclay RL, Mergener K, et al. . Plasma Septin9 versus fecal immunochemical testing for colorectal cancer screening: a prospective multicenter study. PLoS One 2014;9:e98238 10.1371/journal.pone.0098238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hampel H, Frankel WL, Martin E, et al. . Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 2005;352:1851–60. 10.1056/NEJMoa043146 [DOI] [PubMed] [Google Scholar]
- 9. Win AK, Jenkins MA, Dowty JG, et al. . Prevalence and penetrance of major genes and polygenes for colorectal cancer. Cancer Epidemiol Biomarkers Prev 2017;26:404–12. 10.1158/1055-9965.EPI-16-0693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haraldsdottir S, Rafnar T, Frankel WL, et al. . Comprehensive population-wide analysis of Lynch syndrome in Iceland reveals founder mutations in MSH6 and pms2. Nat Commun 2017;8 10.1038/ncomms14755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lynch HT, Snyder CL, Shaw TG, et al. . Milestones of Lynch syndrome: 1895-2015. Nat Rev Cancer 2015;15:181–94. 10.1038/nrc3878 [DOI] [PubMed] [Google Scholar]
- 12. Barrow E, Hill J, Evans DG. Cancer risk in Lynch syndrome. Fam Cancer 2013;12:229–40. 10.1007/s10689-013-9615-1 [DOI] [PubMed] [Google Scholar]
- 13. Vasen HFA, Tomlinson I, Castells A. Clinical management of hereditary colorectal cancer syndromes. Nat Rev Gastroenterol Hepatol 2015;12:88–97. 10.1038/nrgastro.2014.229 [DOI] [PubMed] [Google Scholar]
- 14. Baglietto L, Lindor NM, Dowty JG, et al. . Risks of Lynch syndrome cancers for Msh6 mutation carriers. J Natl Cancer Inst 2010;102:193–201. 10.1093/jnci/djp473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Network NCC Genetic/Familial high-risk assessment: colorectal. NCCN Clinical Practice Guidelines in Oncology, 2018. [Google Scholar]
- 16. Järvinen HJ, Aarnio M, Mustonen H, et al. . Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 2000;118:829–34. 10.1016/S0016-5085(00)70168-5 [DOI] [PubMed] [Google Scholar]
- 17. de Jong AE, Hendriks YMC, Kleibeuker JH, et al. . Decrease in mortality in Lynch syndrome families because of surveillance. Gastroenterology 2006;130:665–71. 10.1053/j.gastro.2005.11.032 [DOI] [PubMed] [Google Scholar]
- 18. Newton K, Green K, Lalloo F, et al. . Colonoscopy screening compliance and outcomes in patients with Lynch syndrome. Colorectal Dis 2015;17:38–46. 10.1111/codi.12778 [DOI] [PubMed] [Google Scholar]
- 19. Blue Ribbon Panel NCI Cancer Moonshot blue ribbon panel report, 2016. Available: https://www.cancer.gov/research/key-initiatives/moonshot-cancer-initiative/blue-ribbon-panel/blue-ribbon-panel-report-2016.pdf
- 20. Stoffel EM, Mercado RC, Kohlmann W, et al. . Prevalence and predictors of appropriate colorectal cancer surveillance in Lynch syndrome. Am J Gastroenterol 2010;105:1851–60. 10.1038/ajg.2010.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aktan-Collan K, Kääriäinen H, Järvinen H, et al. . Psychosocial consequences of predictive genetic testing for Lynch syndrome and associations to surveillance behaviour in a 7-year follow-up study. Fam Cancer 2013;12:639–46. 10.1007/s10689-013-9628-9 [DOI] [PubMed] [Google Scholar]
- 22. de Vos tot Nederveen Cappel WH, Järvinen HJ, Lynch PM, Engel C, et al. . Colorectal surveillance in Lynch syndrome families. Fam Cancer 2013;12:261–5. 10.1007/s10689-013-9631-1 [DOI] [PubMed] [Google Scholar]
- 23. Vasen HF, Nagengast FM, Khan PM. Interval cancers in hereditary non-polyposis colorectal cancer (Lynch syndrome). Lancet 1995;345:1183–4. 10.1016/S0140-6736(95)91016-6 [DOI] [PubMed] [Google Scholar]
- 24. Lanspa SJ, Jenkins JX, Cavalieri RJ, et al. . Surveillance in Lynch syndrome: how aggressive? Am J Gastroenterol 1994;89:1978–80. [PubMed] [Google Scholar]
- 25. Møller P, Seppälä T, Bernstein I, et al. . Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: first report from the prospective Lynch syndrome database. Gut 2017;66 10.1136/gutjnl-2015-309675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stuckless S, Green JS, Morgenstern M, et al. . Impact of colonoscopic screening in male and female Lynch syndrome carriers with an MSH2 mutation. Clin Genet 2012;82:439–45. 10.1111/j.1399-0004.2011.01802.x [DOI] [PubMed] [Google Scholar]
- 27. Jass JR, Smyrk TC, Stewart SM, et al. . Pathology of hereditary non-polyposis colorectal cancer. Anticancer Res 1994;14:1631–4. [PubMed] [Google Scholar]
- 28. Lindgren G, Liljegren A, Jaramillo E, et al. . Adenoma prevalence and cancer risk in familial non-polyposis colorectal cancer. Gut 2002;50:228–34. 10.1136/gut.50.2.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rijcken FEM, Hollema H, Kleibeuker JH. Proximal adenomas in hereditary non-polyposis colorectal cancer are prone to rapid malignant transformation. Gut 2002;50:382–6. 10.1136/gut.50.3.382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rondagh EJA, Gulikers S, Gómez-García EB, et al. . Nonpolypoid colorectal neoplasms: a challenge in endoscopic surveillance of patients with Lynch syndrome. Endoscopy 2013;45:257–64. 10.1055/s-0032-1326195 [DOI] [PubMed] [Google Scholar]
- 31. van Rijn JC, Reitsma JB, Stoker J, et al. . Polyp miss rate determined by tandem colonoscopy: a systematic review. Am J Gastroenterol 2006;101:343–50. 10.1111/j.1572-0241.2006.00390.x [DOI] [PubMed] [Google Scholar]
- 32. Stoffel EM, Turgeon DK, Stockwell DH, et al. . Missed adenomas during colonoscopic surveillance in individuals with Lynch syndrome (hereditary nonpolyposis colorectal cancer). Cancer Prev Res 2008;1:470–5. 10.1158/1940-6207.CAPR-08-0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Niv Y, Moeslein G, Vasen HFA, et al. . Quality of colonoscopy in Lynch syndrome. Endosc Int Open 2014;2:E252–E255. 10.1055/s-0034-1377920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Edelstein DL, Axilbund J, Baxter M, et al. . Rapid development of colorectal neoplasia in patients with Lynch syndrome. Clin Gastroenterol Hepatol 2011;9:340–3. 10.1016/j.cgh.2010.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Samur MK. RTCGAToolbox: a new tool for exporting TCGA Firehose data. PLoS One 2014;9:e106397 10.1371/journal.pone.0106397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Troyanskaya O, Cantor M, Sherlock G, et al. . Missing value estimation methods for DNA microarrays. Bioinformatics 2001;17:520–5. 10.1093/bioinformatics/17.6.520 [DOI] [PubMed] [Google Scholar]
- 37. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–27. 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 38. Gevaert O. MethylMix: an R package for identifying DNA methylation-driven genes. Bioinformatics 2015;31:1839–41. 10.1093/bioinformatics/btv020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Youden WJ. Index for rating diagnostic tests. Cancer 1950;3:32–5. [DOI] [PubMed] [Google Scholar]
- 40. Collett D, Data MB. Modelling binary data. Boca Raton Florida: Hall/CRC Ca, 1999. [Google Scholar]
- 41. Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Song L, Li Y, Jia J, et al. . Algorithm optimization in methylation detection with multiple RT-qPCR. PLoS One 2016;11:e0163333 10.1371/journal.pone.0163333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Song L, Jia J, Yu H, et al. . The performance of the mSEPT9 assay is influenced by algorithm, cancer stage and age, but not sex and cancer location. J Cancer Res Clin Oncol 2017;143:1093–101. 10.1007/s00432-017-2363-0 [DOI] [PubMed] [Google Scholar]
- 44. Fu B, Yan P, Zhang S, et al. . Cell-free circulating methylated SEPT9 for noninvasive diagnosis and monitoring of colorectal cancer. Dis Markers 2018;2018:1–11. 10.1155/2018/6437104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee HS, Hwang SM, Kim TS, et al. . Circulating methylated septin 9 nucleic acid in the plasma of patients with gastrointestinal cancer in the stomach and colon. Transl Oncol 2013;6:290–IN4. 10.1593/tlo.13118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Møller P, Seppälä TT, Bernstein I, et al. . Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: a report from the Prospective Lynch Syndrome Database. Gut 2018;67:1306–16. 10.1136/gutjnl-2017-314057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjgast-2019-000299supp001.docx (33.8KB, docx)