Abstract
Purpose: It has been reported that miRNA-124 inhibits hepatocellular carcinoma (HCC) progression, while lncRNA-UCA1 promotes HCC. The aim of this study is to find whether miRNA-124, as a tumor suppressor in HCC can inhibit lncRNA-UCA1 in HCC cell.
Methods: Tumor tissues and adjacent healthy tissues were obtained from 66 patients diagnosed with HCC in Binzhou Medical University Hospital from January 2011 to January 2013. Cell proliferation assay, in vitro cell migration and invasion assay were applied for the research.
Results: In the present study we found that miRNA-124 was downregulated, while lncRNA-UCA1 was upregulated in tumor tissues comparing to adjacent healthy tissues of HCC patients. Expression of miRNA-124 and lncRNA-UCA1 in tumor tissues were not affected by HBV or HCV infection. Analysis of followed-up data revealed that low miRNA-124 level and high lncRNA-UCA1 level were closely correlated with poor survival. Overexpression of miRNA-124 led to inhibited lncRNA-UCA1 expression in cells of HCC cell lines, while overexpression of lncRNA-UCA1 failed to significantly affect miRNA-124 expression. Expression levels of miRNA-124 and lncRNA-UCA1 were inversely and significantly correlated in tumor tissues but not in adjacent healthy tissues. Overexpression of miRNA-124 led to inhibited, while overexpression of lncRNA-UCA1 led to increased proliferation, migration and invasion rates of HCC cell lines. In addition, lncRNA-UCA1 overexpression attenuated the inhibitory effects of miRNA-124 overexpression on cancer cell proliferation, migration and invasion.
Conclusion: Therefore, miRNA-124 may inhibit the proliferation, migration and invasion of cancer cell in hepatocellular carcinoma by downregulating lncRNA-UCA1.
Keywords: hepatocellular carcinoma, lncRNA-UCA1, miRNA-124, survival
Introduction
Liver cancer is a deadly and highly prevalent malignancy in the world.1 Liver cancer affects about 800,000 new cases every year and the incidence is increasing trend.2 As the most common type of liver cancer, hepatocellular carcinoma accounts for about 80% cases of liver cancer.3 In spite of the efforts made on the treatment of HCC, recurrence rate is still high,4 leading to poor survival. At present, the overall 5-year survival rate of HCC is still below 20%.5
Non-coding RNAs (ncRNAs) are non-protein coding RNA transcripts with critical functions in both physiological and pathological processes.6,7 ncRNAs are divided into different subgroups according to their sizes and functions.6,7 Long non-coding RNAs, or lncRNAs, are non-coding RNA transcripts longer than 200 nucleotides.8 LncRNAs are key players in human cancers including HCC.9 It is known that lncRNAs may interact with microRNAs (miRNAs), another subgroup of ncRNAs, to participate in cancer biology.10 It is known that miRNA-124 inhibits hepatocellular carcinoma (HCC) progression,11,12 while lncRNA-UCA1 promotes HCC.13 Our data showed that miRNA-124 inhibited the proliferation, migration and invasion of cancer cell in hepatocellular carcinoma possibly by downregulating lncRNA-UCA1.
Materials and methods
Human specimens
Tumor tissues and adjacent healthy tissues were obtained from 66 patients diagnosed with HCC in Binzhou Medical University Hospital from January 2011 to January 2013. Inclusion criteria: 1) newly diagnosed HCC cases through pathological biopsies; 2) willing to and completed a 5-year follow-up after admission; 3) patients provided informed consent. Exclusion criteria: 1) patients who were not willing to receive liver biopsy; 2) patients who were diagnosed with multiple diseases; 3) death occurred by not by HCC. These patients included 36 males and 30 females (33–72 years, 49.4±4.8 years). HBV or HCV infections were detected by sensitive PCR. There were 30 cases of HBV-positive, 19 cases of HCV-positive and 17 cases of negative for both HBV and HCV. Ethics Committee of Binzhou Medical University Hospital approved this study.
Follow-up
A 5-year follow-up was performed after admission. Overall survival of patients was recorded to be used to plot survival curves.
Real-time quantitative PCR (RT-qPCR)
To detect miRNA-124, mirVana miRNA isolation Kit (Thermo Fisher Scientific) was used to extract miRNAs, and reverse transcriptions were performed using TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific). PCR mixtures were made using Agilent miRNA QRT-PCR Detection Kit (Agilent). To detect the expression of lncRNA-UCA1, MPure™ Total RNA Extraction Kit (117022160, MP Biomedicals) was used to extract total RNA and MMLV Reverse Transcriptase 1st-Strand cDNA Synthesis Kit (Lucigen) was used to perform reverse transcription. PCR mixtures were made using SYBR® Green Quantitative RT-qPCR Kit (Sigma-Aldrich). Primers of miRNA-124, lncRNA-UCA1 as well as endogenous control β-actin and U6 were designed and synthesized by GenePharma (Shanghai, China). Sequences of primers were: 5ʹ-TCGGGTAACTCTTACGGT-3ʹ (forward) and 5ʹ-GGTCCATTGAGGCTGTAG-3ʹ (reverse) for UCA1; 5ʹ-GACCTCTATGCCAACACAGT-3ʹ (forward) and 5ʹAGTACTTGCGCTCAGGAGGA-3ʹ (reverse) for β-actin; 5ʹ-CTAGCCTGCAGGCGTGCTG-3ʹ (forward) for miRNA-124. miRNA-124 reverse primer and U6 primers were included in the qPCR kit. Using 2−ΔΔCT method, lncRNA-UCA1 was normalized to endogenous control β-actin and miRNA-124 expression was normalized to endogenous control U6.
HCC cell lines, vectors and cell transfection
Cells of SNU-398 and SNU-449 HCC cell lines were from ATCC (USA). RPMI-1640 Medium (Catalog No. 30-2001) containing 10% heat-inactivated fetal bovine serum (FBS) was used as cell culture medium. Vectors expressing lncRNA-UCA1 and ROCK1 were purchased from Sangon (Shanghai, China). miR-124 mimic and Scrambled negative control miRNA were bought for Sigma-Aldrich. Lipofectamine 3000 reagent (Thermo Fisher Scientific) was used to achieve transient transfections. All operations were performed according to the manufacturer’s instructions. Doses of miRNA and vectors were 40 and 10 nM, respectively. Cells with no transfections were control cells (control, C). Empty vector- or Scrambled negative control miRNA-transfected cells were negative control cells (negative control, NC).
Cell proliferation assay
Expression of miRNA-124 and lncRNA-UCA1 was detected by RT-qPCR. Cell proliferation was detected only in cases of overexpression rates of miRNA-124 and lncRNA-UCA1 were higher than 200%. Single cell suspensions (3×104 cells/mL) were prepared using RPMI-1640 Medium (10% FBS). Cell suspensions were transferred to each well of a 96-well plate with 0.1 mL for each well and cells were cultivated under normal conditions, followed by addition of CCK-8 solution (10 µL) every 24 hrs for 4 times. After that, cells were cultivated for further 4 hrs and cell proliferation was represented by OD values at 450 nm.
In vitro cell migration and invasion assay
Expression of miRNA-124 and lncRNA-UCA1 was detected by RT-qPCR. Cell migration and invasion were detected only in cases of overexpression rates of miRNA-124 and lncRNA-UCA1 were higher than 200%. Single cell suspensions (3×104 cells/mL) were prepared using RPMI-1640 Medium (1% FBS). Cell suspensions were transferred to the upper chamber with 0.1 mL for each well, and RPMI-1640 Medium containing 20% FBS was used to fill the lower chamber. Cell culture was performed under normal conditions for 24 hrs and 0.5% crystal violet (Sigma-Aldrich, USA) was performed at room temperature for 10 mins. The same protocol was used for both migration and invasion assay, but the upper chamber was coated with Matrigel (356,234, Millipore, USA) prior to invaison assay. Cell migration and invasion were normalized to cell proliferation. Control group was set to 100%, and all other groups were normalized to this group.
Statistical analysis
All statistical analyses were performed by GraphPad Prism 6 software. Correlations were analyzed by Pearson’s correlation coefficient. Based on follow-up data, Kaplan–Meier method and log-rank test were used to plot and compare survival curves. Comparisons of expression levels of miRNA-124 and lncRNA-UCA1 were compared with paired t-test. Comparisons between 2 groups were compared by unpaired t-test. One-way ANOVA and Tukey test was used for comparisons among 3 groups. p<0.05 was statistically significant.
Results
miRNA-124 was downregulated and lncRNA-UCA1 was upregulated in HCC
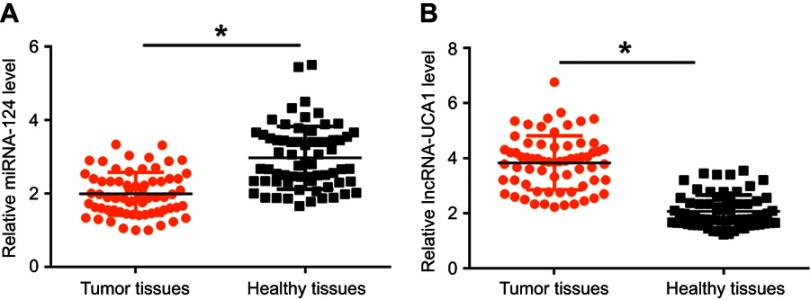
miRNA-124 and lncRNA-UCA1 in 66 patients with HCC were detected by RT-qPCR. It was observed that miRNA-124 was significantly downregulated (Figure 1A) and lncRNA-UCA1 was significantly upregulated (Figure 1B) in tumor tissues of HCC patients compared to healthy tissues (p<0.05).
Figure 1.
MiRNA-124 was downregulated and lncRNA-UCA1 was upregulated in tumor tissues of HCC patients. RT-qPCR results showed that miRNA-124 was significantly downregulated (A), and lncRNA-UCA1 was significantly upregulated (B) in tumor tissues than in adjacent healthy tissues of HCC patients (*p<0.05).
Expression of miRNA-124 and lncRNA-UCA1 in tumor tissues was not affected by HBV or HCV infection
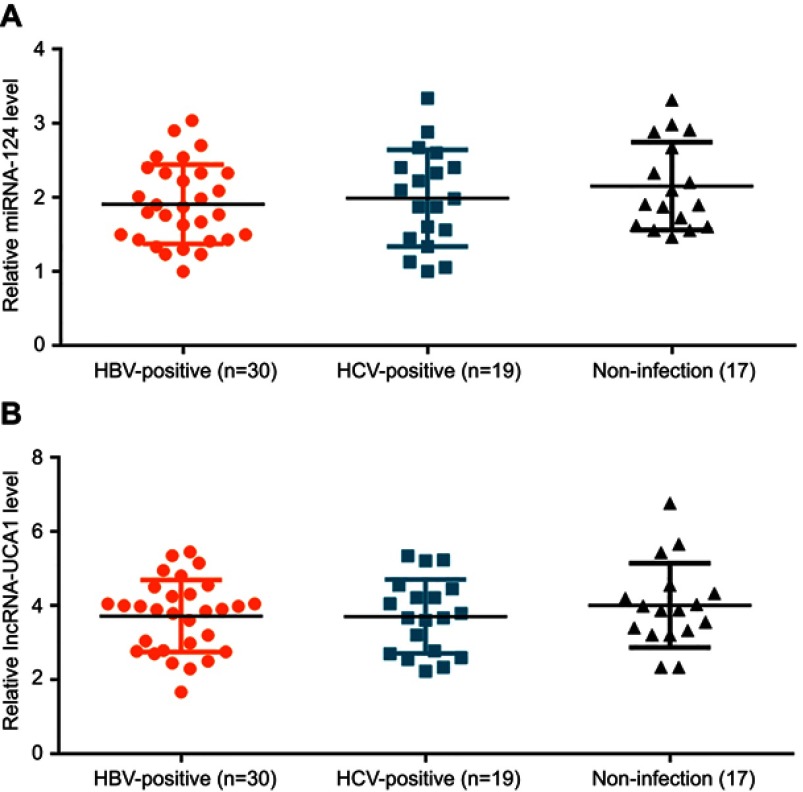
Among the 66 patients with HCC, there were 30 cases of HBV-positive, 19 cases of HCV-positive and 17 cases of negative for both HBV and HCV. As shown in Figure 2, no significant differences in expression levels of miRNA-124 (Figure 2A) and lncRNA-UCA1 (Figure 2B) were found among HBV-positive, HCV-negative and non-infection groups.
Figure 2.
Expression of miRNA-124 and lncRNA-UCA1 in tumor tissues were not affected by HBV or HCV infection. No significant differences in expression levels of miRNA-124 (A) and lncRNA-UCA1 (B) were found among HBV-postive, HCV-negative and non-infection groups.
Low miRNA-124 level and high lncRNA-UCA1 level were closely correlated with poor survival of HCC patients
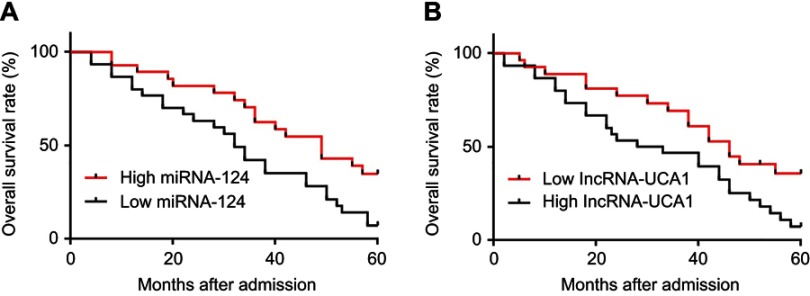
Patients were divided into low (n=35) and high (n=31) lncRNA-UCA1 (cutoff value =4.12) as well as low (n=32) and high (n=34) miR-124 (2.04) groups. Survival curve analyses were performed. As shown in Figure 3, overall survival rate was significantly lower in low miRNA-124 level (Figure 3A) and high lncRNA-UCA1 level group (Figure 3B).
Figure 3.
Low miRNA-124 level and high lncRNA-UCA1 level were closely correlated with poor survival of HCC patients. ROC curve analysis showed that low miRNA-124 level (A) and high lncRNA-UCA1 level (B) were closely correlated with poor survival of HCC patients.
Expression levels of miRNA-124 and lncRNA-UCA1 were inversely and significantly correlated in HCC
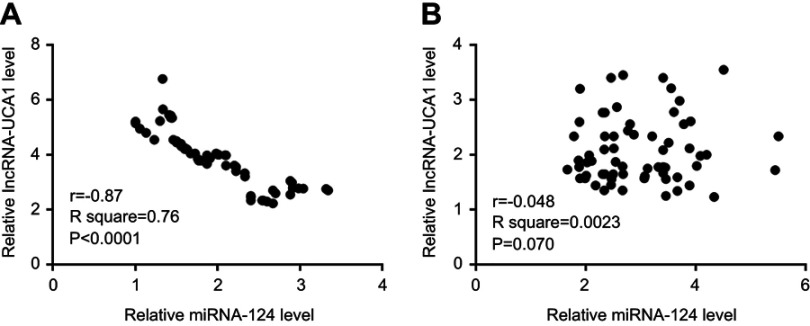
Correlations between miRNA-124 and lncRNA-UCA1 were analyzed by Pearson’s correlation coefficient. As shown in Figure 4, expression levels of miRNA-124 and lncRNA-UCA1 were inversely and significantly correlated in tumor tissues (Figure 4A) but not in adjacent healthy tissues (Figure 4B).
Figure 4.
Expression levels of miRNA-124 and lncRNA-UCA1 were inversely and significantly correlated in tumor tissues. Pearson’s correlation coefficient analysis showed that expression levels of miRNA-124 and lncRNA-UCA1 were inversely and significantly correlated in tumor tissues (Figure 4A) but not in adjacent healthy tissues (Figure 4B).
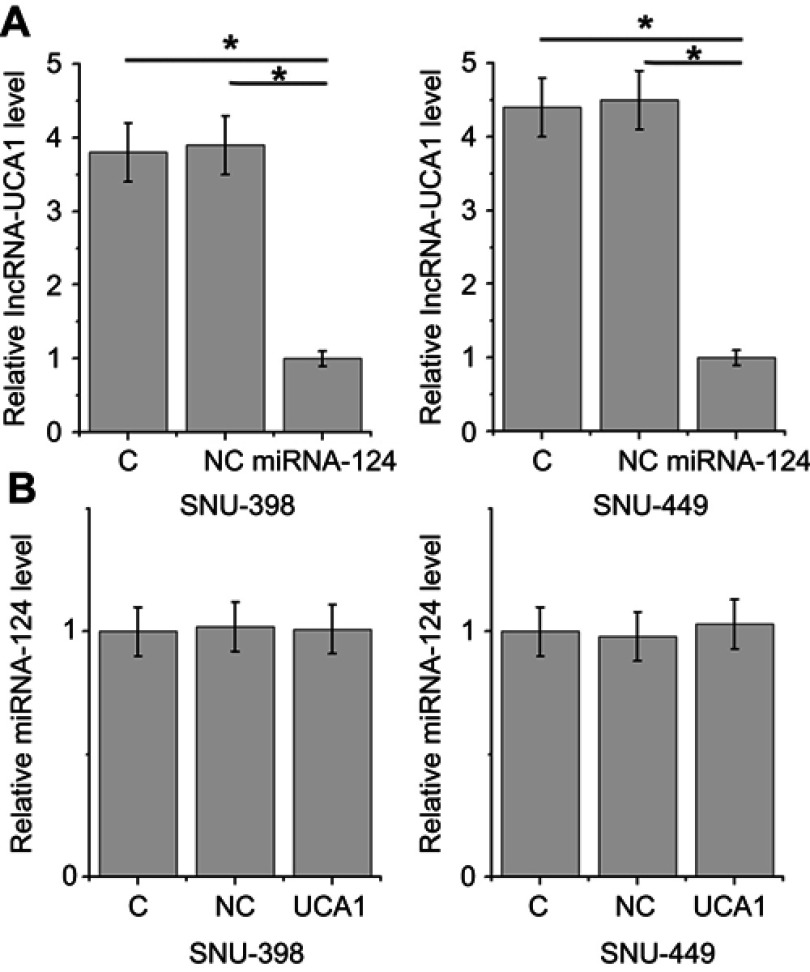
Overexpression of miRNA-124 led to inhibited lncRNA-UCA1 expression
To further confirm the correlation between miRNA-124 and lncRNA-UCA1, miRNA-124 mimic and lncRNA-UCA1 expression vectors were transfected into cells of both SNU-398 and SNU-449 HCC cell lines. Overexpression rates of miRNA-124 (255–375%) and lncRNA-UCA1 (230–340%) were higher than 200% (data not shown). Comparing to C and NC groups, overexpression of miRNA-124 led to inhibited lncRNA-UCA1 expression in cells of both HCC cell lines (Figure 5A, p<0.05), while overexpression of lncRNA-UCA1 failed to significantly affect miRNA-124 expression (Figure 5B).
Figure 5.
Overexpression of miRNA-124 led to inhibited lncRNA-UCA1 expression. Overexpression of miRNA-124 led to inhibited lncRNA-UCA1 expression in cells of both HCC cell lines (A) (*p<0.05), while overexpression of lncRNA-UCA1 failed to significantly affect miRNA-124 expression (B).
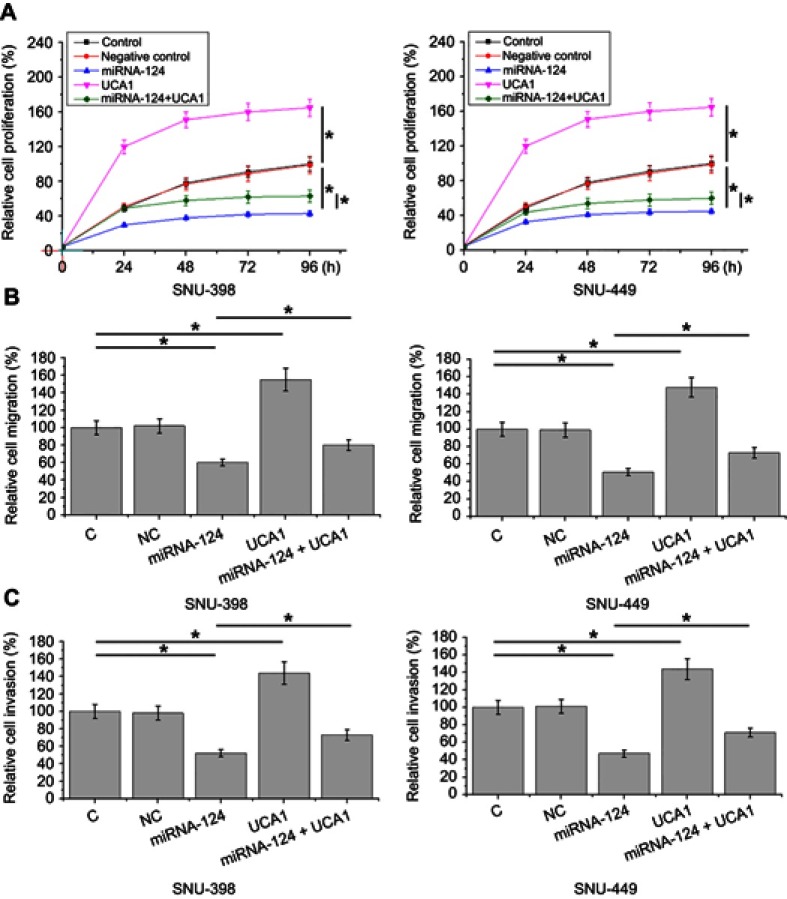
miRNA-124 led to inhibited proliferation, migration and invasion of HCC cells by inhibiting lncRNA-UCA1
Comparing to C and NC groups, overexpression of miRNA-124 led to inhibited, while overexpression of lncRNA-UCA1 led to increased proliferation (Figure 6A), migration (Figure 6B) and invasion (Figure 6C) rates of cells of HCC cell lines SNU-398 and SNU-449 (p<0.05). In addition, lncRNA-UCA1 overexpression attenuated the inhibitory effects of miRNA-124 overexpression (p<0.05).
Figure 6.
MiRNA-124 led to inhibited proliferation, migration and invasion of HCC cells by inhibiting lncRNA-UCA1. Overexpression of miRNA-124 led to inhibited, while overexpression of lncRNA-UCA1 led to accelerated proliferation (A), migration (B) and invasion (C) of cells of HCC cell lines SNU-398 and SNU-449. In addition, lncRNA-UCA1 overexpression attenuated the inhibitory effects of miRNA-124 overexpression on cancer cell proliferation, migration and invasion (*p<0.05).
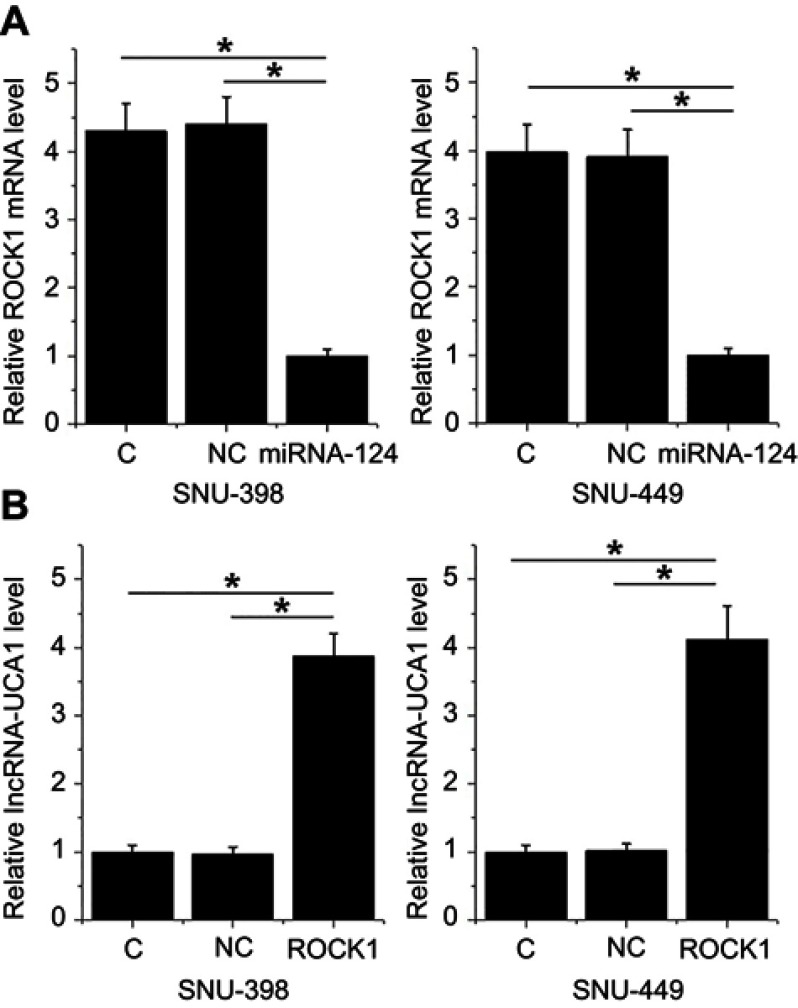
miRNA-124 downregulated ROCK1, which upregulated UCA1
Comparing to two controls, overexpression of ROCK1 resulted in the downregulation of ROCK1 mRNA in cells of HCC cell lines SNU-398 and SNU-449 (Figure 7A, p<0.05). In contrast, ROCK1 overexpression mediated the upregulation of UCA1 in cells of both cell lines (Figure 7B, p<0.05).
Figure 7.
MiRNA-124 downregulated ROCK1, which upregulated UCA1. Comparing to two controls, overexpression of ROCK1 resulted in the downregulation of ROCK1 mRNA in cells of HCC cell lines SNU-398 and SNU-449 (A). In contrast, ROCK1 overexpression mediated the upregulation of UCA1 in cells of both cell lines (B) (*p<0.05).
Discussion
It is well known that miRNAs and lncRNAs are essential players in cancer biology. The involvement of miRNA–lncRNA interactions in cancer has also been reported. We first showed that miRNA-124, as a tumor suppressor in HCC, may inhibit lncRNA-UCA1, an oncogene in HCC, to through ROCK1 to inhibit HCC.
MiRNA-124 is a well-characterized tumor suppression miRNA in many different types of human cancers, including HCC.11,12,14 Our study further confirmed that miRNA-124 was downregulated in HCC tissues than in adjacent tissues, and the low expression level of miRNA-124 is closely correlated with poor overall survival of HCC patients. In addition, overexpression of miRNA-124 led to inhibited behaviors of HCC cells. Our study further confirmed the role of miRNA-124 in HCC and suggested that miRNA may serve as a prognostic marker for HCC.
MiRNA-124 participates in cancer biology through the interactions with multiple signaling molecules.11,12,14 A recent study reported that miRNA-124 interacted with lncRNA MALAT1 to regulate the growth of tongue cancer.15 LncRNA-UCA1 exerts oncogenic effects on different types of cancers, and it has been reported that lncRNA-UCA1 may inhibit miR-216b,13 miR-193a-3p16 and miR-204-5p17 to promote cancer development. However, the inhibition of lncRNA-UCA1 by miRNAs has not been reported. In this study, we first reported that miRNA-124 may serve as an upstream inhibitor of lncRNA-UCA1, and the inhibition of lncRNA-UCA1 by miRNA-124 is involved in the regulation of behaviors of HCC cells. However, significantly inverse correlation between lncRNA-UCA1 and miRNA-124 was only observed in tumor tissues but not in adjacent cancer tissues. Therefore, the interaction between lncRNA-UCA1 and miRNA-124 may be disease-specific. In addition, our study also observed that expression of lncRNA-UCA1 and miRNA-124 in tumor tissues was not affected by HBV or HCV infections, which are common causes of HCC.18 Therefore, lncRNA-UCA1 and miRNA-124 may interact to participate in HCC through HBV- and HCV-independent pathways. It has been reported that TGF-β pathway can induce the expression of lncRNA-UCA1,19 while which TGF-β pathway branch mediates this action is unknown. ROCK1, a branch of TGF-β pathway, is a direct target of miR-124.20 In the present study, we showed that ROCK1 overexpression resulted in upregulated lncRNA-UCA1, while miR-124 overexpression caused downregulated ROCK1 in HCC cells. Therefore, we characterized a novel miR-124/ROCK1/lncRNA-UCA1 pathway in HCC.
Conclusion
In conclusion, our study confirmed the roles of lncRNA-UCA1 and miRNA-124 in HCC and revealed that miRNA-124 may serve as an upstream inhibitor of lncRNA-UCA1 to inhibit the proliferation, migration and invasion of HCC cells.
Acknowledgments
We received the financial support from Natural Science Foundation of China (No. 81502069), Natural Science Fund Project of Shandong Province (No. BS2015YY025 and ZR2015HL084), and Scientific Research Foundation of Binzhou Medical University (BY2014KYQD35).
Ethical approval and informed consent
We followed ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Ryerson AB, Eheman CR, Altekruse SF, et al. Annual report to the nation on the status of cancer, 1975-2012, featuring the increasing incidence of liver cancer. Cancer. 2016;122(9):1312–1337. doi: 10.1002/cncr.29936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63(5):844–855. doi: 10.1136/gutjnl-2013-306627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torres HA, Vauthey JN, Economides MP, Mahale P, Kaseb A. Hepatocellular carcinoma recurrence after treatment with direct-acting antivirals: first, do no harm by withdrawing treatment. J Hepatol. 2016;65(4):862–864. doi: 10.1016/j.jhep.2016.05.034 [DOI] [PubMed] [Google Scholar]
- 5.Chapman BC, Paniccia A, Hosokawa PW, et al. Impact of facility type and surgical volume on 10-year survival in patients undergoing hepatic resection for hepatocellular carcinoma. J Am Coll Surg. 2017;224(3):362–372. doi: 10.1016/j.jamcollsurg.2016.11.011 [DOI] [PubMed] [Google Scholar]
- 6.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046 [DOI] [PubMed] [Google Scholar]
- 7.Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9(1):3–12. doi: 10.4161/epi.27473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 9.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi: 10.4161/rna.20481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing miRNA-lncRNA interactions In: Feng Y, Zhang L, editors. Long Non-Coding RNAs. New York, NY: Humana Press; 2016:271–286. [DOI] [PubMed] [Google Scholar]
- 11.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31(5):766–776. doi: 10.1093/carcin/bgp250 [DOI] [PubMed] [Google Scholar]
- 12.Lang Q, Ling C. MiR-124 suppresses cell proliferation in hepatocellular carcinoma by targeting PIK3CA. Biochem Biophys Res Commun. 2012;426(2):247–252. doi: 10.1016/j.bbrc.2012.08.075 [DOI] [PubMed] [Google Scholar]
- 13.Wang F, Ying HQ, He BS, et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6(10):7899–7917. doi: 10.18632/oncotarget.3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie C, Han Y, Liu Y, Han L, Liu J. miRNA-124 down-regulates SOX8 expression and suppresses cell proliferation in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7(10):6534–6542. [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang TH, Liang LZ, Liu XL, et al. Long non-coding RNA MALAT1 interacts with miR-124 and modulates tongue cancer growth by targeting JAG1. Oncol Rep. 2017;37(4):2087–2094. doi: 10.3892/or.2017.5445 [DOI] [PubMed] [Google Scholar]
- 16.Nie W, Ge HJ, Yang XQ, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99–106. doi: 10.1016/j.canlet.2015.11.024 [DOI] [PubMed] [Google Scholar]
- 17.Bian Z, Jin L, Zhang J, et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892. doi: 10.1038/srep23892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waziry R, Grebely J, Amin J, et al. Trends in hepatocellular carcinoma among people with HBV or HCV notification in Australia (2000-2014). J Hepatol. 2016;65(6):1086–1093. doi: 10.1016/j.jhep.2016.08.010 [DOI] [PubMed] [Google Scholar]
- 19.Zuo ZK, Gong Y, Chen XH, et al. TGFbeta1-induced lncRNA UCA1 upregulation promotes gastric cancer invasion and migration. DNA Cell Biol. 2017;36(2):159–167. doi: 10.1089/dna.2016.3553 [DOI] [PubMed] [Google Scholar]
- 20.Hu CB, Li QL, Hu JF, Zhang Q, Xie JP, Deng L. miR-124 inhibits growth and invasion of gastric cancer by targeting ROCK1. Asian Pac J Cancer Prev. 2014;15(16):6543–6546. [DOI] [PubMed] [Google Scholar]