Abstract
Viral infections in the immunocompetent host can cause both acute and chronic kidney disease either in direct damage to the infected kidney cells or as a consequence of systemic immune responses that impact kidney function. Viruses have evolved mechanisms to hijack signaling pathways of the infected cell including the mTOR pathway to support viral replication, and to evade the antiviral immune responses such as those mediated by miR-155 via microRNA mimetics expressed by the virus. At both the cellular and the systemic level, the host has also evolved mechanism to counter the viral subversion strategies in the evolutionary battle for mutual survival. In the era of genomic medicine, understanding individual genetic variations that leads to differences in susceptibilities to infection and variabilities in immune responses may open new avenues for treatment, such as the recently described functions of APOL1 risk alleles in HIV-associated nephropathy. In addition, state-of-the-art high throughput sequencing methods have discovered new viruses as the cause for chronic diseases not previously attributed to an infection. The potential application of these methods to idiopathic kidney diseases may reveal similar occult infections by unknown viruses. Precision medicine objectives to optimize host-directed and pathogen-directed therapies for kidney diseases associated with infectious causes will only develop through detailed understanding of genetic susceptibility associated with immune responses and viral tropism.
Keywords: Viruses, innate immunity, chronic kidney disease, gene-environment interactions, next-generation sequencing
Clinical summary
1. The evolutionary arms race between infectious pathogens and the human immune system has created elaborate viral mechanisms to escape immune clearing and equally complex host immune responses to counter the viral evasion strategies.
2. Novel host-directed and pathogen-directed therapies for chronic kidney diseases treatment are becoming possible with a better understanding of gene-environment interactions between host susceptibility to infection and viral tropism.
3. New high throughput nucleic acid sequencing methods can be used to investigate possible infectious etiologies for idiopathic kidney diseases.
Introduction
Viruses are obligate parasites and cannot complete their lifecycle outside a host organism, as they do not have the ability to synthesize new proteins or generate energy for enzymatic reactions on their own. With this dependency on the host, viruses do not intentionally cause disease and death, an event that would be counterproductive to their propagation. Much of the disease associated with viral infections is initiated by the response of the host’s immune system. Thus, many viruses have evolved immune evasion strategies to support viral replication but limit the damaging host response, achieving a mutually acceptable balance between the host and virus, allowing survival and proliferation of both. This careful balance can be lost in immunocompromised hosts, such as in transplant immunosuppression, and the impact of viral infections in the setting of immunodeficiency is the focus of other articles in this series.
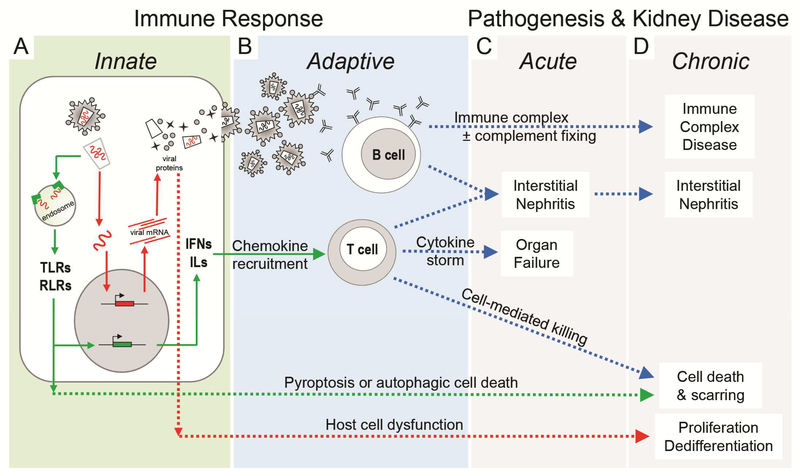
Viral-induced kidney injury can occur acutely or chronically as a consequence of both a direct impact on the infected cell and also from subsequent systemic and local responses of the innate and adaptive immune system.1 An overview of the interactions between the virus and the infected cell, and the cascade of the host’s systemic immune responses is shown in Figure 1. After entering the cells, viral nucleic acids are initially recognized by the intracellular pattern recognition receptors (Toll-like [TLR] and RIG-I-like receptors) of the innate immune system (Figure 1A) which may target the virus and viral genome to endocytic-lysosomal degradation or may engage cell survival pathways that induce apoptosis to eliminate the infected cell. The pattern recognition receptors also initiate post-translational signaling events that activate transcription of secreted interferons and interleukins that recruit and activate T and B cells of the adaptive immune system. The adaptive immune response (Figure 1B) may trigger massive immune cell recruitment, causing both a cytokine storm and severe interstitial inflammation that results in acute renal failure (Figure 1C). Chronic infections may follow from these same immune responses resulting in long term inflammatory and fibrotic changes resulting in chronic kidney disease (Figure 1D). A robust humoral response may also generate unmanageable antibody-antigen aggregates (with or without complement activation) resulting in immune complex deposition in the microvasculature. Additional adaptive immune responses may include recruitment of killer T cells or natural killer (NK) cells that exacerbates tissue damage through cell-mediated killing of infected kidney cells. Epithelial cell loss by either apoptosis or cell-mediated killing initiates typical injury repair processes, with deposition of provisional matrix and epithelial cell proliferation, recruitment of fibroblasts and matrix remodeling. Without resolution of the infection, this injury process will perpetuate as chronic inflammation resulting in progressive tissue scarring and subsequent loss of function. Within the kidney cell, viral proteins may hijack the cell machinery to repurpose cellular functions for viral replication, causing a profound dysregulation of cell homeostasis resulting in changes in cell proliferation and dedifferentiation.
Figure 1.
Summary of cellular and systemic antiviral responses that impact kidney function. A. Immune response to viral infection (viral lifecycle in red with red arrows) begins with the nonspecific but immediate innate immune system (green box and green arrows). B. The innate immune response activates the adaptive immune system (blue box and blue arrows) resulting in the development of highly specific humoral and cell-mediated antiviral responses. C,D. Acute and chronic injury (dashed arrows) and pathogenesis may result as a consequence of cell damage or death from direct viral infection or secondarily from the robust antiviral immune response. TLRs, Toll-like receptors; RLRs, RIG-I-like receptors; IFNs, interferons; ILs, interleukins.
Subversion Mechanism
All viruses have a simple structure consisting of a genome, either single- or doubled-stranded RNA or DNA, surrounded by the viral capsid, a protein coat to protect the genome. Some viruses have a second coat consisting of proteins and lipids (enveloped viruses) that encase the capsid. Almost all viruses also encode additional “accessory” proteins that are not required for replication or for structural coat proteins, but are solely to subvert the host to support the viral lifecycle. The various subversion mechanisms, including mechanism to evade immune responses or hijacking cellular housekeeping functions, are mediated by these viral accessory proteins.
Immune evasion
One common immune evasion strategy is molecular mimicry, a process where the virus produces proteins that mimic host proteins.2 The viral genes for the mimetics were evolutionarily acquired directly from the host genome through recombination events during replication, and those that favored survival became fixed in the viral genome. These viral mimetics can have various functions, such as suppressing aspects of either the innate and adaptive immune systems or altering cellular homeostatic functions to support the viral life cycle. For example, the genome of the human β-herpesvirus cytomegalovirus (CMV) encodes up to 200 proteins but only ~60 are necessary for viral replication.3,4 The remaining viral proteins function solely to modify the immune response through mimicking cytokines (virokines), cytokine receptors (viroreceptors), and soluble cytokine-binding proteins that function as cytokine scavengers. In addition, viruses like CMV, with large DNA genomes and a nuclear replication cycle, are also known to use microRNAs (miRNAs) in mimicry strategies.5 The use of miRNAs allows the virus to modify host functions without producing an immunogenic protein, and like protein mimicry, viral miRNAs resemble host miRNAs and were also co-opted from the host genome. The human miR-155, an important regulator of inflammation, has been exploited by several herpesviruses. The Kaposi’s sarcoma-associated herpesvirus expresses a miR-155 mimetic that suppresses innate immune function through the TLR signaling mediators IRAK1 and MyD88.6,7 CMV encodes twelve host mimetic miRNAs that are expressed during its lytic phase and suppresses proteins involved in antigen presentation reducing NK and cytotoxic T cell recruitment.8 Some herpesviruses, such as Epstein-Barr virus (EBV), do not encode a miR-155 mimetic, but produce accessory proteins that induce the expression of the host miR-155. This induction causes cellular transformation, activating additional host pathways that also induces the expression of the host miR-155, resulting in a feed forward loop, elevating miR-155 expression by 200 fold.9,10
In addition to targeting the host transcriptome, viral miRNAs are also known to target their own transcriptome. Latency is another immune evasion strategy to maintain viral persistence in the host, which involves silencing viral replication to become dormant. Recent studies have shown many viruses use miRNAs against their own genes to control their switch from latency to active (lytic) replication. The EBV miRNA miR-BART2 downregulates the expression of its own viral DNA polymerase, inhibiting the key viral enzyme needed for lytic replication, contributing to the maintenance of latency.11,12 Exploiting this natural mechanism for maintaining viral latency would have important clinical applications, especially for the problematic viral reactivations that occur in the setting of transplant immunosuppression. Therapeutics are currently being developed to target the function of both viral-encoded and host-encoded miRNAs involved in immune system control as a treatment for several infectious diseases.13
Cellular Hijacking
Another common viral subversion strategy is hijacking cellular functions to support viral replication. This strategy does not kill the infected cell but dysregulates cellular homeostasis to the point of causing pathology. The most common cellular housekeeping functions intersected by the virus are post translational signaling pathways that include transcription factor activation, and cell cycle control functions that suppress apoptosis and enhance cell division.
Viral hijacking events can be particularly disruptive to the kidney due to the highly specialized, terminally differentiated epithelial cells such as the glomerular podocyte, and the unique structure-function relationship of the nephron. In HIV-associated nephropathy (HIVAN), the viral accessory protein Nef mediates most of the kidney pathology in animal models.14–17 Using in vitro studies with HIV-expressing and HIV-infected podocytes, Nef expression increases proliferation and dedifferentiation that is characteristic of HIVAN glomerular pathology.18–21 These changes largely involve the hijacking the STAT3 pathways via activation of Src family kinases, which form direct protein-protein interactions with Nef.18,22 In a mouse model of HIVAN, pharmacological suppression of the STAT3 pathway ameliorated renal disease and provides proof-of-concept that this type of host-directed therapy (e.g. targeting the host response to the viral protein and not the viral protein itself) has potential application for CKD treatment.22
Other common viral hijacking strategies redirect host metabolic resources for new protein and nucleic acid synthesis to support new virion production. The cellular mammalian target of rapamycin (mTOR) pathway is an important intracellular monitor of energy and nutrient status, and controls cellular metabolism to accommodate stress responses. Recent studies have shown retroviruses, polyomaviruses, and herpesviruses all encode proteins to manipulate mTOR signaling to facilitate viral protein production and to suppress autophagy and apoptosis to promote viral persistence.23 During viral infection, the typical cell stress responses is to repress mTORC1 kinase activity to conserve energy and resources, a process which is counter to the new DNA and protein synthetic demands of the viral lifecycle. To oppose this response, CMV produces the viral UL38 protein which bind and inactivates a key upstream signaling repressor (TSC proteins) and also propagates a downstream signaling event, the subcellular localization of the mTORC1 complex with the key activator Rheb-GTP at the perinuclear complex.24–26 Both of these hijacking events ensure the preservation of mTORC1 function during the stress response. Interestingly, this activation of the mTOR pathway by CMV is insensitive to Rapamycin, thus second generation mTOR inhibitors with different modes of action may have future uses in CMV suppressive strategies that complement Rapamycin effects.27,28
Gene-Environment Interactions
Individual genetic variation to both susceptibilities to infection and differences in the anti-viral immune response underlies person-to-person differences in disease occurrence and severity.29 These differences can be further modified by the unique environmental exposures from a wide range of factors such as diet, weather, occupation, and other infectious agents. The goal of precision medicine is to understand the interaction between the individual’s unique genetic attributes alongside the unique history of exposure to environment modifiers to develop an optimal therapy. Familial or genetic susceptibilities to kidney disease are well known, and genome-wide association studies have uncovered genetic variabilities that can alter either susceptibilities to infection or susceptibilities to the breadth and amplitude of immune responses that lead to pathology and disease.
Genetic susceptibility to infection
Few kidney diseases are known to be caused by direct viral infection of renal parenchymal cells. Some kidney-tropic viruses, including the polyomaviruses BK and JC and the herpesvirus CMV, are common in the general population world-wide. These infections typically achieve a harmless, lifelong latency and only become disease-causing if they reactivate in the setting of immunodeficiency. In immunocompetent hosts, CKD caused by direct viral infection has been best described for HIV-1 infection and HIVAN, a rapidly progressive collapsing glomerulopathy that is associated with infection of podocytes and tubular epithelial cells.30–32 Although HIV infection also is associated with acquired immunodeficiency syndrome (AIDS), HIVAN development is not entirely dependent on immunodeficiency.32–34
An interesting observation regarding the incidence of HIVAN is that it primarily occurs in individuals of African ancestry. African Americans are more susceptible to CKD in general, and recent genome-wide association studies have identified a causal genetic link with African ancestry and HIVAN, idiopathic focal segmental glomerulosclerosis, and hypertension-attributed CKD.35,36 This genetic association was attributed to two polymorphisms (referred to as risk alleles G1 and G2) in the gene for the innate immune protein apolipoprotein L1 (APOL1) that are present only in African ancestral populations.37,38 Although the association of CKD risk and carriage of two risk alleles is strong, the majority of individuals with risk alleles do not develop CKD. Thus, APOL1 risk alleles are believed to be susceptible genes that are only disease causing under an inducing environmental condition or stressor. Thus, HIV-1 infection appears to be a strong environmental stressor that induces CKD in individuals with the APOL1 genetic susceptibility. It has been estimated HIV-1 infected individuals with the at-risk APOL1 genotype (and not receiving antiviral treatment) have a 50% lifetime risk for developing HIVAN.39
Although APOL1 is believed to have evolved as part of our innate immune system to primarily combat parasitic infections,40 several in vitro studies provide evidence APOL1 also may be a host restriction factor for HIV-1.41,42 Host restriction factors are intracellular proteins with functions that disable some aspect of the viral lifecycle and prevents the virus from replicating and establishing a chronic infection. They are part of the innate immune system, but have separate functions from the intracellular pathogen recognition receptors such as the TLRs. Recent studies have found APOL1 expressed in T cells and podocytes was able to partially block HIV-1 replication.41,43 However, expression of APOL1 risk alleles failed to suppress HIV replication. Since HIVAN is associated with direct infection of renal cells, it is possible the APOL1 risk alleles cause CKD by failing to block viral replication in the infected kidney cells.
Other reports have similarly observed individuals with APOL1 risk alleles may be more susceptible to kidney cell infections of other viruses resulting in CKD. In a recent case report, an immunocompetent pregnant women homozygous for APOL1 risk alleles developed collapsing glomerulopathy as a result of an acute B19 parvovirus infection.44 In this individual, there was evidence of parvovirus proteins in tubular epithelial cells, suggesting a direct kidney cell infection. Similarly, two case reports of transplant patients without APOL1 risk alleles receiving kidneys from donors homozygous for APOL1 risk alleles rapidly developed a collapsing glomerulopathy. These patients were found to have either CMV or a polyomavirus infection in their kidneys.45,46 Although more mechanistic studies of APOL1 function and viral replication control are needed, there appears to be a connection between the expression of APOL1 and the ability of the host to control a renal cell infection, which is lost with the APOL1 risk alleles.
Genetic susceptibility to aberrant immune responses
The complex interactions between host genetics, immune responses, and infectious agents are gaining acceptance as critical pathogenic events that underlie many chronic conditions, including those not etiologically associated with an infection but are more typically classified as autoimmune.47 For example, Crohn’s disease, a chronic inflammatory bowel disease (IBD), has a known genetic susceptibility gene (ATG16L1, an autophagy protein) where individuals with a polymorphism in ATG16L1 have an elevated risk for IBD. Using mouse models, IBD can be triggered by the infection of Paneth cells by an enteric virus.48 Mice engineered with the human ATG16L1 susceptibility polymorphism have an altered response to the virus, resulting in damage to the epithelial barrier and release of cytokines that subsequently alters the commensal bacterial population of the gut. Thus, this loss in symbiotic homeostasis between the commensal bacteria and the host immune system as a combined consequence of the enteric viral infection and the ATG16L1 susceptibility show the complexity of host-virus interactions in autoimmune diseases.
Immunoglobulin A (IgA) nephropathy, an example of an immune complex disease, is the most common form of glomerulonephritis worldwide, with enhanced susceptibilities in Asian populations but rare occurrences in African populations.49 Similar to the etiology of Crohn’s disease described above, IgA nephropathy also is a complex disease trait that requires interaction of genetic and environmental factors. As such, the variability in presentation of IgA nephropathy likely results from the convergence of the individual’s unique genetic variabilities and exposure to environmental triggers. Production of IgA is an normal humoral response to infections in mucosal membranes, and disease risk is associated with defects in IgA O-glycosylation resulting in autoantibodies generated to the incorrectly glycosylated IgA. This pathogenic cascade, however, begins with a viral or bacterial infections that initiates the production of IgA. Although well-defined environmental triggers may not be clear in all cases of IgA nephropathy, studies have documented viral antigens in immune complexes including HIV-1, EBV, CMV, and hepatitis B, indicating viral infections are contributing events for immune complex formation.50 Common treatments for IgA nephropathy typically use general immunosuppressants, but with a better understanding of host susceptibilities, novel therapies addressing either the genetic variants or the initiating mucosal immune event have potential for future clinical use.51
The Future Outlook: Viral Etiologies for Idiopathic CKDs
The list of noncommunicable chronic diseases or diseases of unknown etiology that can now be attributed to an infectious agent is expanding.47 A high profile case in point is the Nobel-prize winning observation that stomach ulcers, widely accepted by the medical communtiy to be caused by diet and stress, are in fact cause by a treatable bacterial (Helicobacter pylori) infection. Idiopathic forms of chronic kidney disease (CKD), especially those with ineffective treatment options, may also benefit from an unconventional perspective regarding their etiological mechanism. The combination of genetic disease susceptibilities driven by gene-environment interactions and the new pathogen discovery methods using genome-wide interrogation techniques makes pathogen discovery studies both justifiable and feasible. Importantly, idenfication of potential infectious contributors to idiopathic CKDs can open alternative strategies for disease prevention and treatment. Pathogen-directed therapies such as vaccines and anti-viral medications may be effective new tools to treat and prevent CKD. For example, the development of suppressive antiretroviral drugs for HIV-1 infection has been effective at both reducing the incidence of HIVAN and restoring renal function in those with HIVAN.52
Next-generation sequencing and the metagenome
DNA extracted from a tissue, such as a renal biopsy, includes DNA representing the host genome and also the metagenome, the genomes of environmental organisms present in the tissue. High throughput DNA and RNA sequencing technologies, collectively known as next-generation sequencing (NGS), produce a comprehensive catalog of DNAs and/or RNAs in clinical specimens with the sensitivity to detect rare, low abundance nucleic acids such as those originating from an infectious agent. In addition to pathogen discovery, some of the newer NGS technologies are inexpensive and provide real time analysis of clinical samples that can be used in any setting, making them the future for infectious disease diagnostics.
For pathogen discovery, use of NGS methods recently identified an infectious cause for a poorly understood spontaneous CKD that occurs in some immunocompetent mouse strains.53 This progressive CKD causes death at 200–300 days of age and is characterized clinically by elevated serum creatinine, blood urea nitrogen and anemia, and pathologically by shrunken kidneys, tubulointerstitial fibrosis, and tubular degeneration. The insidious nature of this murine CKD and predilection to a few inbred strains is not unlike forms of human CKD that are more common in some racial or ethnic groups and characterized by progressive tubular atrophy and fibrosis.54 For decades, the sporadic occurrence of the murine CKD had eluded explanation. Like all modern research animal facilities, rigorous screening for infectious and parasitic agents was routine and consistently negative. NGS of diseased mouse kidney tissue identified a viral sequence of a never-before identified parvovirus, of the same family as the better known B19 parvovirus, but highly divergent from any known member of the Parvoviridea family. Using the sequence data for the new virus, screening reagents were developed and revealed affected mouse kidneys were highly positive for the virus in tubular epithelium, in addition to high viral titers in serum and virus being shed into urine. Examination of other mouse colonies with similarly affected mice, from Australia to the United States, identified the same virus. This study illustrates several important concepts in new pathogen discovery with NGS. First, it is possibility to find novel infectious agents that directly cause CKD in immunocompetent hosts. Second, these novel viruses can be easily missed using standard clinical screening tests. And third, these novels viruses, once reagents are developed to screen for them, may in fact be common and wide spread.
A conceptual hurdle in making a connection between an idiopathic kidney disease and an occult infection is that an immunocompetent patient may lack the typical clinical presentation of an infection such as a fever and high lymphocyte counts. As described above, viruses have evolved a resourceful collection of mechanisms to suppress host immune responses. Thus, viruses may target selective anatomic sites, such as the kidney, and “silent” infections may persist with little clinical evidence of an infection. HIV-1 infection is an example of chronic viral infections that can be asymptomatic for years without any obvious signs of illness55, for which there are also human polymorphisms in viral receptors that can either block or enhance infection.56,57 Thus, as the NGS methodologies are becoming more mainstream, their use to reevaluate idiopathic CKDs for an infectious cause is both realistic and practical. In the event of a new virus discovery, established pathogen-directed therapies such as vaccines and antiviral medication could become valuable new clinical tools to prevent and treat these CKDs. In addition, characterizing the biology of these newly identified viruses may reveal additional human genetic variations associated with susceptibilities to infection, potentially enhancing our understanding of racial predilections to CKD.
Conclusions
Infectious diseases remain a major cause of death worldwide, ranking highly with other chronic non-communicable diseases such as cancer and cardiovascular disease.58 Kidney injury caused by direct viral infection or from secondary damage due to immune response to systemic infections remain a significant cause of acute and chronic kidney disease in both resource-rich and resource-limited health care settings.1,59 Ongoing research to understand genetic susceptibilities for kidney diseases is revealing the complex co-evolution of pathogens and the human immune system, and intricate viral interactions with susceptibility genes to modulate disease occurrence and severity. Continuing research in the era of NGS and other “omics” technologies will allow for comprehensive system biology approaches toward the investigation of disease mechanisms. Integrating Individual genotype-phenotyping relationships and new virus discovery would be paradigm shifting to generate new tools for prevention, control, and treatment of CKD, especially for CKDs where pathogenesis is incompletely understood or where effective treatment options are limited.
Acknowledgments
LAB is supported by NIH grants DK097836, DK108329, and AI35434.
Footnotes
Conflict of interest/financial disclosure
The author declares no conflicts of interest, financial or otherwise, related to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kupin WL. Viral-Associated GN: Hepatitis B and Other Viral Infections. Clin J Am Soc Nephrol. 2017;12(9):1529–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beachboard DC, Horner SM. Innate immune evasion strategies of DNA and RNA viruses. Curr Op Microbiol. 2016;32:113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Picarda G, Benedict CA. Cytomegalovirus: Shape-Shifting the Immune System. J Immunol. 2018;200(12):3881–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mocarski ES, Jr., Immune escape and exploitation strategies of cytomegaloviruses: impact on and imitation of the major histocompatibility system. Cellular Microbiol. 2004;6(8):707–717. [DOI] [PubMed] [Google Scholar]
- 5.Cullen BR. MicroRNAs as mediators of viral evasion of the immune system. Nat Immunol. 2013;14(3):205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skalsky RL, Samols MA, Plaisance KB, et al. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81(23):12836–12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abend JR, Ramalingam D, Kieffer-Kwon P, Uldrick TS, Yarchoan R, Ziegelbauer JM. Kaposi’s sarcoma-associated herpesvirus microRNAs target IRAK1 and MYD88, two components of the toll-like receptor/interleukin-1R signaling cascade, to reduce inflammatory-cytokine expression. J Virol. 2012;86(21):11663–11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stark TJ, Arnold JD, Spector DH, Yeo GW. High-resolution profiling and analysis of viral and host small RNAs during human cytomegalovirus infection. J Virol. 2012;86(1):226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu F, Weidmer A, Liu CG, Volinia S, Croce CM, Lieberman PM. Epstein-Barr virus-induced miR-155 attenuates NF-kappaB signaling and stabilizes latent virus persistence. J Virol. 2008;82(21):10436–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin Q, McBride J, Fewell C, et al. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol. 2008;82(11):5295–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfeffer S, Zavolan M, Grasser FA, et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. [DOI] [PubMed] [Google Scholar]
- 12.Barth S, Pfuhl T, Mamiani A, et al. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36(2):666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drury RE, O’Connor D, Pollard AJ. The Clinical Application of MicroRNAs in Infectious Disease. Front Immunol. 2017;8:1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuo Y, Matsusaka T, Zhong J, et al. HIV-1 genes vpr and nef synergistically damage podocytes, leading to glomerulosclerosis. J Am Soc Nephrol. 2006;17(10):2832–2843. [DOI] [PubMed] [Google Scholar]
- 15.Hanna Z, Kay DG, Rebai N, Guimond A, Jothy S, Jolicoeur P. Nef harbors a major determinant of pathogenicity for an AIDS-like disease induced by HIV-1 in transgenic mice. Cell. 1998;95(2):163–175. [DOI] [PubMed] [Google Scholar]
- 16.Hanna Z, Priceputu E, Hu C, Vincent P, Jolicoeur P. HIV-1 Nef mutations abrogating downregulation of CD4 affect other Nef functions and show reduced pathogenicity in transgenic mice. Virology. 2006;346(1):40–52. [DOI] [PubMed] [Google Scholar]
- 17.Hanna Z, Weng X, Kay DG, Poudrier J, Lowell C, Jolicoeur P. The pathogenicity of human immunodeficiency virus (HIV) type 1 Nef in CD4C/HIV transgenic mice is abolished by mutation of its SH3-binding domain, and disease development is delayed in the absence of Hck. J Virol. 2001;75(19):9378–9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He JC, Husain M, Sunamoto M, et al. Nef stimulates proliferation of glomerular podocytes through activation of Src-dependent Stat3 and MAPK1,2 pathways. J Clin Invest. 2004;114(5):643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Husain M, D’Agati VD, He JC, Klotman ME, Klotman PE. HIV-1 Nef induces dedifferentiation of podocytes in vivo: a characteristic feature of HIVAN. AIDS. 2005;19(17):1975–1980. [DOI] [PubMed] [Google Scholar]
- 20.Sunamoto M, Husain M, He JC, Schwartz EJ, Klotman PE. Critical role for Nef in HIV-1-induced podocyte dedifferentiation. Kid Int. 2003;64(5):1695–1701. [DOI] [PubMed] [Google Scholar]
- 21.Barisoni L, Bruggeman LA, Mundel P, D’Agati VD, Klotman PE. HIV-1 induces renal epithelial dedifferentiation in a transgenic model of HIV-associated nephropathy. Kid Int. 2000;58(1):173–181. [DOI] [PubMed] [Google Scholar]
- 22.Feng X, Lu TC, Chuang PY, et al. Reduction of Stat3 activity attenuates HIV-induced kidney injury. J Am Soc Nephrol. 2009;20(10):2138–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Sage V, Cinti A, Amorim R, Mouland AJ. Adapting the Stress Response: Viral Subversion of the mTOR Signaling Pathway. Viruses. 2016;8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clippinger AJ, Maguire TG, Alwine JC. Human cytomegalovirus infection maintains mTOR activity and its perinuclear localization during amino acid deprivation. J Virol. 2011;85(18):9369–9376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10(8):935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe. 2008;3(4):253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kudchodkar SB, Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus infection alters the substrate specificities and rapamycin sensitivities of raptor- and rictor-containing complexes. Proc Natl Acad Sci (USA). 2006;103(38):14182–14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moorman NJ, Shenk T. Rapamycin-resistant mTORC1 kinase activity is required for herpesvirus replication. J Virol. 2010;84(10):5260–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet. 2005;6(4):287–298. [DOI] [PubMed] [Google Scholar]
- 30.Bruggeman LA, Ross MD, Tanji N, et al. Renal epithelium is a previously unrecognized site of HIV-1 infection. J Am Soc Nephrol. 2000;11(11):2079–2087. [DOI] [PubMed] [Google Scholar]
- 31.Marras D, Bruggeman LA, Gao F, et al. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat Med. 2002;8(5):522–526. [DOI] [PubMed] [Google Scholar]
- 32.Winston JA, Bruggeman LA, Ross MD, et al. Nephropathy and establishment of a renal reservoir of HIV type 1 during primary infection. N Engl Med. 2001;344(26):1979–1984. [DOI] [PubMed] [Google Scholar]
- 33.Bruggeman LA, Dikman S, Meng C, Quaggin SE, Coffman TM, Klotman PE. Nephropathy in human immunodeficiency virus-1 transgenic mice is due to renal transgene expression. J Clin invest. 1997;100(1):84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Laroche M, Desbuissons G, Rouvier P, et al. APOL1 variants may induce HIV-associated nephropathy during HIV primary infection. J Antimicrobial Chemother. 2017;72(5):1539–1541. [DOI] [PubMed] [Google Scholar]
- 35.Kao WH, Klag MJ, Meoni LA, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Gen. 2008;40(10):1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopp JB, Smith MW, Nelson GW, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40(10):1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genovese G, Friedman DJ, Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Genovese G, Tonna SJ, Knob AU, et al. A risk allele for focal segmental glomerulosclerosis in African Americans is located within a region containing APOL1 and MYH9. Kid Int. 2010;78(7):698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kopp JB, Nelson GW, Sampath K, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22(11):2129–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kruzel-Davila E, Wasser WG, Skorecki K. APOL1 Nephropathy: A Population Genetics and Evolutionary Medicine Detective Story. Sem Nephrol. 2017;37(6):490–507. [DOI] [PubMed] [Google Scholar]
- 41.Taylor HE, Khatua AK, Popik W. The innate immune factor apolipoprotein L1 restricts HIV-1 infection. J Virol. 2014;88(1):592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLaren PJ, Gawanbacht A, Pyndiah N, et al. Identification of potential HIV restriction factors by combining evolutionary genomic signatures with functional analyses. Retrovirology. 2015;12:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikulak J, Oriolo F, Portale F, et al. Impact of APOL1 polymorphism and IL-1beta priming in the entry and persistence of HIV-1 in human podocytes. Retrovirology. 2016;13(1):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Besse W, Mansour S, Jatwani K, Nast CC, Brewster UC. Collapsing glomerulopathy in a young woman with APOL1 risk alleles following acute parvovirus B19 infection: a case report investigation. BMC Nephrol. 2016;17(1):125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang JH, Husain SA, Santoriello D, et al. Donor’s APOL1 Risk Genotype and “Second Hits” Associated With De Novo Collapsing Glomerulopathy in Deceased Donor Kidney Transplant Recipients: A Report of 5 Cases. Am J Kid Diseases. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah PB, Cooper JE, Lucia MS, Boils C, Larsen CP, Wiseman AC. APOL1 Polymorphisms in a Deceased Donor and Early Presentation of Collapsing Glomerulopathy and Focal Segmental Glomerulosclerosis in Two Recipients. Am J Transplant. 2016;16(6):1923–1927. [DOI] [PubMed] [Google Scholar]
- 47.Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157(1):142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cadwell K, Patel KK, Maloney NS, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141(7):1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiryluk K, Julian BA, Wyatt RJ, et al. Genetic studies of IgA nephropathy: past, present, and future. Ped Nephrol. 2010;25(11):2257–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyatt RJ, Julian BA. IgA nephropathy. N Engl J Med. 2013;368(25):2402–2414. [DOI] [PubMed] [Google Scholar]
- 51.Barratt J, Tang SCW. Treatment of IgA Nephropathy: Evolution Over Half a Century. Sem Nephrol. 2018;38(5):531–540. [DOI] [PubMed] [Google Scholar]
- 52.Mallipattu SK, Salem F, Wyatt CM. The changing epidemiology of HIV-related chronic kidney disease in the era of antiretroviral therapy. Kid Int. 2014;86(2):259–265. [DOI] [PubMed] [Google Scholar]
- 53.Roediger B An atypical parvovirus drives chronic tubulointerstitial nephropathy and kidney fibrosis. Cell. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schelling JR. Tubular atrophy in the pathogenesis of chronic kidney disease progression. Ped Nephrol. 2016;31(5):693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabin CA, Lundgren JD. The natural history of HIV infection. Curr Op HIV AIDS. 2013;8(4):311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He W, Neil S, Kulkarni H, et al. Duffy antigen receptor for chemokines mediates transinfection of HIV-1 from red blood cells to target cells and affects HIV-AIDS susceptibility. Cell Host Microbe. 2008;4(1):52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang Y, Paxton WA, Wolinsky SM, et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2(11):1240–1243. [DOI] [PubMed] [Google Scholar]
- 58.Global, regional, and national age-sex-specific mortality and life expectancy, 1950–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1684–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.George C, Mogueo A, Okpechi I, Echouffo-Tcheugui JB, Kengne AP. Chronic kidney disease in low-income to middle-income countries: the case for increased screening. BMJ Global Health. 2017;2(2):e000256. [DOI] [PMC free article] [PubMed] [Google Scholar]