Abstract
Recombinant brain, skeletal muscle, and heart voltage-gated Na+ channel α subunits differ in their functional responses to an accessory β1 subunit when coexpressed inXenopus oocytes. We exploited the distinct β1 subunit responses observed for the human heart (hH1) and human skeletal muscle (hSkM1) isoforms to identify determinants of this response. Chimeric α subunits were constructed by exchanging the S5–S6 interhelical loops of each domain between hH1 and hSkM1 and then examined for effects on inactivation induced by coexpressed β1 subunit in oocytes. Substitution of single S5–S6 loops in either domain 1 (D1/S5–S6) or domain 4 (D4/S5–S6) of hSkM1 by the corresponding segments of hH1 produced channels that exhibited an attenuated response to coexpressed β1 subunit. Substitutions of both D1/S5–S6 and D4/S5–S6 in hSkM1 by the corresponding loops from hH1 completely abolished the effects of the β1 subunit on inactivation. The reciprocal chimera in which both D1/S5–S6 and D4/S5–S6 from hSkM1 were transplanted into hH1 exhibited significant β1 responsiveness (accelerated inactivation). The region within D4/S5–S6 that conferred β1 responsiveness was determined to reside primarily within an extracellular loop between the putative pore-forming segment SS2 and D4/S6. Gating modulation was also demonstrated using a chimeric β subunit consisting of the extracellular domains of β1and the transmembrane and C-terminal domains of the rat brain β2 subunit. These results suggest that the D1/S5–S6 and D4/S5–S6 loops in the α subunit and the extracellular domain of the β1 subunit are important determinants of the β1 subunit-induced gating modulation in Na+channels.
Keywords: Na+ channel, ion channel gating, subunit interaction, electrophysiology, human ion channels, heart, skeletal muscle, hH1, hSkM1, SCN4A, SCN5A
Voltage-gated Na+ channels are heteromultimeric complexes of a large, heavily glycosylated α subunit and one or two smaller β subunits (Catterall, 1992). For heterologous expression of recombinant Na+ channels inXenopus oocytes, an α subunit alone is usually sufficient to form functional channels (Goldin et al., 1986; Trimmer et al., 1989;Cribbs et al., 1990; Klugbauer et al., 1995), whereas one or more β subunits may be required for normal gating (Messner et al., 1986; Isom et al., 1992, 1995a). Brain Na+ channel α subunits associate noncovalently with a β1 subunit and are disulfide-linked to a β2 subunit in vivo(Messner and Catterall, 1985; Gordon et al., 1988), whereas skeletal muscle Na+ channels are heterodimeric, consisting of only α and β1 subunits (Roberts and Barchi, 1987). The subunit composition of native cardiac Na+ channels is less clear (Lombet and Luzdunski, 1984; Cohen and Levitt, 1993).
Heterologous expression of recombinant rat or human β1subunits in Xenopus oocytes greatly accelerates the inactivation of brain (Isom et al., 1992; Patton et al., 1994) and skeletal muscle (Bennett et al., 1993; Cannon et al., 1993; Makita et al., 1994; Patton et al., 1994) Na+ channels. In some studies, the β1 subunit has been shown to increase the expression of functional brain Na+ channels on the cell surface as inferred by an increase in peak current amplitude or an increase in specific [3H]saxitoxin binding (Isom et al., 1995b). Additional effects of the β1 subunit on brain and skeletal muscle channels include acceleration of activation, speeding of recovery from inactivation, and changes in the steady-state voltage dependence of inactivation. In contrast to its dramatic effects on brain and skeletal muscle Na+ channel function, the β1 subunit has little or no effect on the gating of cloned cardiac Na+ channels (Kyle et al., 1993; Makita et al., 1994; Nuss et al., 1995; Qu et al., 1995), although peak current amplitude is increased.
The precise structural basis for α–β1 subunit interactions in voltage-gated Na+ channels is unknown. Because β1 modulates inactivation, one plausible hypothesis is that it interacts with α subunit structures that constitute the inactivation gate. Fast inactivation of Na+channels is postulated to occur via a mechanism in which the cytoplasmic interdomain 3–4 (ID3-4) region occludes the pore from the cytoplasmic side (Stühmer et al., 1989; West et al., 1992). In a previous study, we investigated the structural basis for the distinct β1 subunit effects on heart and skeletal muscle Na+ channels by using chimeras formed between the two α subunits. We demonstrated that none of the three predicted cytoplasmic interdomain regions nor the C terminus of the Na+ channel α subunit are responsible for isoform differences in the response to the β1 subunit, and that this functional difference was not confined to any single domain of the α subunit (Makita et al., 1996). It appears likely that Na+ channels require multiple regions of the α subunit for modulation of channel function by the β1 subunit.
In this study, human skeletal muscle isoform (hSkM1)/human cardiac isoform (hH1) chimeras were used to demonstrate that the β1 subunit response requires the combination of two short interhelical loops located between segments S5 and S6 (S5–S6) in α subunit domains 1 and 4. We also demonstrated by using a β1/β2 subunit chimera that the extracellular domain and the outermost residues in the transmembrane segment of the β1 subunit are sufficient to modulate the gating of hSkM1. These studies identify critical molecular determinants of the α–β1 functional interaction that localize to the extracellular surface of the channel.
MATERIALS AND METHODS
Construction of Na+ channel α subunit chimeras. Single S5–S6 loop chimeras were constructed using recombinant PCR (Higuchi, 1989), starting with modified expression constructs in which silent HindIII sites were inserted within D1/S6 of hSkM1 [nucleotide (nt) 1302] and hH1 (nt 1200) and changes made in the terminal 3′ untranslated regions of both cDNAs (XbaI site added to pSP64T–hSkM1 and a 0.9 kbSpeI fragment removed from hH1). Amino acid substitutions made in hSkM1 were as follows: hSkM1–P1, amino acids 278–422 of hSkM1 were replaced by hH1 residues 278–388; hSkM1–P2, residues 725–784 were replaced by hH1 804–921; hSkM1–P3, amino acids 1187–1273 were replaced by hH1 1361–1448; and hSkM1–P4, residues 1512–1573 were replaced by hH1 1678–1747. Amino acid substitutions in hH1 chimeras were as follows: hH1–P1, residues 278–388 were replaced by hSkM1 278–422 and hH1–P4, residues 1678–1747 were replaced by hSkM1 1512–1573.
Double hSkM1 S5–S6 loop chimeras were assembled by ligating pairs of the following cDNA fragments derived from the single S5–S6 loop chimeras; pSP64T–hSkM1–P1: 2.2 kb NotI–SphI (5′ cloning site to nt 2153); pSP64T–hSkM1–P2: 0.6 kbSphI–XhoI (nt 2153–2719); pSP64T–hSkM1–P3: 1.7 kb XhoI–SacII (nt 2719–4386); pSP64T–hSkM1–P4: 1.8 kb SacII/XbaI (nt 4386–3′ cloning site) (nt numbers correspond to the hSkM1 cDNA sequence). Chimera hH1–P14 was assembled by combining a 3.2 kbKpnI/XbaI pSP64T–hH1–P4 fragment with 7.5 kbKpnI/XbaI pSP64T–hH1–P1 fragment
Partial reversions of hH1–P14 were done by recombinant PCR and facilitated by the presence of a conserved EagI restriction site in the D4/SS2 region in both hSkM1 and hH1. Chimera hH1–P14N contains the hSkM1 D1/S5–S6 region in addition to hSkM1 residues 1521–1537 (N-terminal portion of D4/S5–S6). Chimera hH1–P14C contains the hSkM1 D1/S5–S6 region in addition to hSkM1 residues 1537–1573 (C-terminal portion of D4/S5–S6).
Correct assembly of each chimera was verified by restriction analysis and dideoxynucleotide sequencing of the junctional regions. Chimeric regions generated by PCR were sequenced completely to identify clones without polymerase errors. Functional expression studies were performed on multiple independent recombinants of each construct.
Construction of a β1/β2chimera. Rat brain β2 subunit (rβ2) cDNA was isolated by a reverse-transcription PCR (RT-PCR) strategy using published sequence data (Isom et al., 1995a). Total RNA was extracted from rat brain, primed with random hexamers, and reverse-transcribed using Superscript II reverse transcriptase (BRL, Bethesda, MD). A 714 bp cDNA containing the complete rβ2coding region was amplified using oligonucleotide primers rβ2-F (5′-GCCTAACATAGTCTCTGAA-3′) and rβ2-R (5′-GAGGAGACAGGACACAGGAA-3′). A chimeric β subunit (β1–2) was constructed by recombinant PCR. A 185 bp cDNA was amplified using hβ1 as a template with primers 5′-TCACCAATGTCACCTACAACCACTC-3′ and 5′-CACCACAGCTAGCACATACATCAT-3′, digested with AvaI/NheI, and gel purified. Similarly, a 200 bp cDNA fragment was amplified from rβ2using primers 5′-GGTTTGCTAGCTGTGGTCATCTTG-3′ and 5′-GCCGGAATTCGAGGAGACAGGACACAGGAA-3′, digested withNheI/EcoRI, and gel purified. These two fragments were subcloned back into the AvaI/EcoRI sites of plasmid pSP64T–hβ1. The resulting chimera β1–2 has the complete N terminus of hβ1extending five amino acids into the transmembrane domain (Met1 through Val164) joined to the transmembrane domain and C terminus of rβ2(Leu161 through stop216). Both rβ2 and β1–2 were directionally subcloned into the pSP64T vector (Krieg and Melton, 1987) for in vitrotranscription. All sequences were verified by dideoxynucleotide sequencing in the final constructs.
Expression in Xenopus oocytes and electrophysiology.All the cDNAs encoding wild-type and chimeric α and β subunits were transcribed in vitro from pSP64T constructs using SP6 RNA polymerase, and the resultant cRNAs were microinjected intoXenopus oocytes. Whole-cell currents were recorded from oocytes using the two-microelectrode voltage-clamp as described previously (Bennett et al., 1993; Makita et al., 1994, 1996).
To analyze the effect of the β1 subunit on Na+ inactivation, we fit the time course of inactivation (t = 0–30 msec, test voltage of −20 mV) with a two-exponential function:I(t)/Imax = A∞ + A1 · exp(−t/τf) + A2 · exp(−t/τs), where A∞is a constant value, A1 andA2 are fractions of fast- and slow-inactivating components, and τf and τs are the time constants of fast- and slow-inactivating components, respectively. Recovery from inactivation was assessed by a double-pulse protocol consisting of a 500 msec prepulse to +20 mV designed to fully inactivate all channels, followed by a variable duration interpulse interval (Δt) at −120 mV and a test pulse to −20 mV. The time interval between protocols was 30 sec. Recovery from inactivation was analyzed by fitting data using a nonlinear least-squares minimization method with a two-exponential equation:I(t)/Imax = A1 · exp(−t/τf) + A2 · exp(−t/τs) + A3, where t is the recovery time interval, and τf and τs are time constants of fast- and slow-recovering components, respectively.
Results were presented as means ± SE, and the statistical comparisons were made using the unpaired Student’s t test to evaluate the significance of the difference between means. Statistical significance was assumed for p < 0.05. Coefficients of variance were calculated by dividing the SD by the mean and multiplying the quotient by 100.
RESULTS
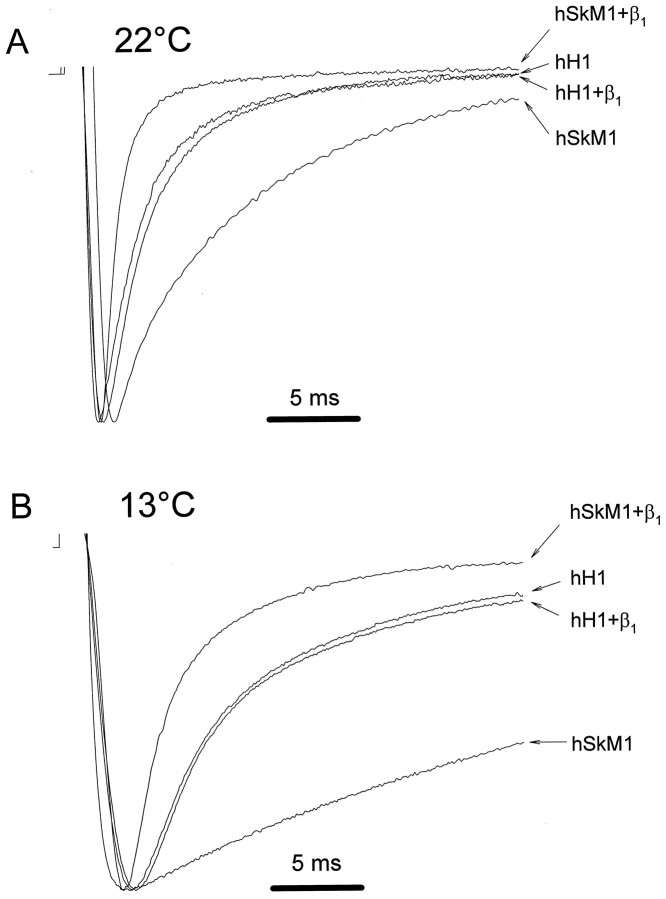
The marked difference in β1 subunit modulation of skeletal muscle compared with cardiac Na+ channels is shown in Figure 1A. This experiment illustrates the effect of a recombinant human β1 subunit (hβ1) on currents expressed in oocytes microinjected with RNA encoding either the hSkM1 or hH1 Na+ channel α subunits. Macroscopic current measurements at room temperature (Fig.1A) demonstrate a significant decrease in the time constants for inactivation (τf and τs) and a significant increase in the fraction of the fast-inactivating component when β1 is coexpressed with hSkM1 but not with hH1 (Table 1). Additional experiments at 13°C revealed similar results. Cooling was used to slow inactivation and alleviate concern that an effect of hβ1 on hH1 was missed because of the slow response time of the two-electrode voltage-clamp in oocytes (Fig. 1B). Coexpression of hβ1 also accelerates the time course of recovery from inactivation in oocytes expressing hSkM1 but not hH1 (Makita et al., 1994). These data suggest that intrinsic structural differences between the two α subunit isoforms are responsible for the distinct gating modulatory effects of the β1 subunit.
Fig. 1.
Effect of coexpressed hβ1 on hSkM1 and hH1 gating. A, Sodium currents were recorded inXenopus oocytes expressing either wild-type hSkM1 or hH1 in the presence or absence of hβ1. Representative current traces obtained during 50 msec test depolarizations to −20 mV from a holding potential of −120 mV at room temperature (22°C) are shown. Current amplitudes are scaled to unity in all traces. Horizontal bar indicates 5 msec. B, Same experiment except that the temperature was 13°C.
Table 1.
Quantitative kinetic data for onset of inactivation in Na+ channel chimeras
| Channel | +/− hβ1 | τf(msec) | p1_a | τs(msec) | p | Fast fraction | p | (n) |
|---|---|---|---|---|---|---|---|---|
| hSkM1 | − | 2.2 ± 0.3 | 13.2 ± 1.4 | 0.30 ± 0.03 | (20) | |||
| hSkM1 | + | 1.1 ± 0.1 | 1_150 | 8.4 ± 0.8 | 1_150 | 0.84 ± 0.01 | 1_150 | (28) |
| hSkM1–P14 | − | 1.0 ± 0.3 | 8.1 ± 0.8 | 0.20 ± 0.04 | (9) | |||
| hSkM1–P14 | + | 1.1 ± 0.1 | NS | 9.0 ± 0.8 | NS | 0.28 ± 0.04 | NS | (10) |
| hH1 | − | 1.2 ± 0.1 | 5.8 ± 0.6 | 0.78 ± 0.03 | (11) | |||
| hH1 | + | 1.0 ± 0.1 | NS | 5.8 ± 0.6 | NS | 0.75 ± 0.02 | NS | (11) |
| hH1–P1 | − | 1.3 ± 0.2 | 5.5 ± 0.1 | 0.43 ± 0.02 | (10) | |||
| hH1–P1 | + | 1.1 ± 0.1 | NS | 5.7 ± 0.1 | NS | 0.59 ± 0.04 | 1_150 | (10) |
| hH1–P4 | − | 1.3 ± 0.1 | 5.5 ± 0.1 | 0.74 ± 0.01 | (4) | |||
| hH1–P4 | + | 1.3 ± 0.1 | NS | 7.0 ± 0.7 | NS | 0.90 ± 0.02 | 1_150 | (4) |
| hH1–P14 | − | 1.4 ± 0.1 | 6.4 ± 0.3 | 0.57 ± 0.02 | (9) | |||
| hH1–P14 | + | 0.9 ± 0.1 | 1_150 | 4.4 ± 0.6 | 1_150 | 0.80 ± 0.04 | 1_150 | (12) |
| hH1–P14N | − | 1.6 ± 0.1 | 8.3 ± 0.4 | 0.49 ± 0.03 | (16) | |||
| hH1–P14N | + | 1.3 ± 0.1 | 1_150 | 7.3 ± 0.4 | NS | 0.71 ± 0.01 | 1_150 | (8) |
| hH1–P14C | − | 1.2 ± 0.1 | 7.3 ± 0.9 | 0.54 ± 0.03 | (10) | |||
| hH1–P14C | + | 0.9 ± 0.1 | 1_150 | 4.9 ± 0.3 | 1_150 | 0.81 ± 0.03 | 1_150 | (14) |
Significance level for comparisons between expression with (+) and without (−) hβ1. NS, Not significant;
F1_150: p < 0.05; τf, time constant for fast component of Na+ channel onset of inactivation; τs, time constant for slow component of Na+channel onset of inactivation.
Although coexpression of the β1 subunit does not modulate hH1 gating, it does exert significant effects on peak current amplitude for both hSkM1 and hH1. Coexpression of hβ1 significantly increased the absolute peak current amplitude of both hSkM1 and hH1 (hSkM1 alone: 1.6 ± 0.3 μA; hSkM1 + β1, 3.7 ± 0.4 μA, p < 0.001, n = 20; hH1 alone: 2.1 ± 0.3 μA; hH1 + β1: 4.6 ± 0.3 μA, p < 0.001, n = 20) and increased the peak current normalized for cell capacitance (hSkM1 alone: 3.7 ± 0.4 nA/nF; hSkM1 + β1: 14.8 ± 1.7 nA/nF; hH1 alone: 8.4 ± 0.9 nA/nF; hH1 + β1: 17.3 ± 1.1 nA/nF). However, the absolute values of peak current amplitude measured at a test potential of −20 mV varied widely from oocyte to oocyte regardless of the presence or absence of coexpressed hβ1(coefficient of variance, 50.7–69.5%). This variation is significantly larger than that of the microinjection volume calibrated by injecting [35S]methionine into control oocytes (53.0 ± 0.6 nl, n = 20, coefficient of variance, 5.6%). These observations indicate that a substantial degree of the peak current amplitude variation is attributable to intrinsic oocyte differences. Therefore, increased peak current amplitude in oocytes is not a reliable index of the β1 subunit effect, and we primarily used changes in gating as a measure of channel modulation.
Determinants of β1 subunit-induced gating modulation
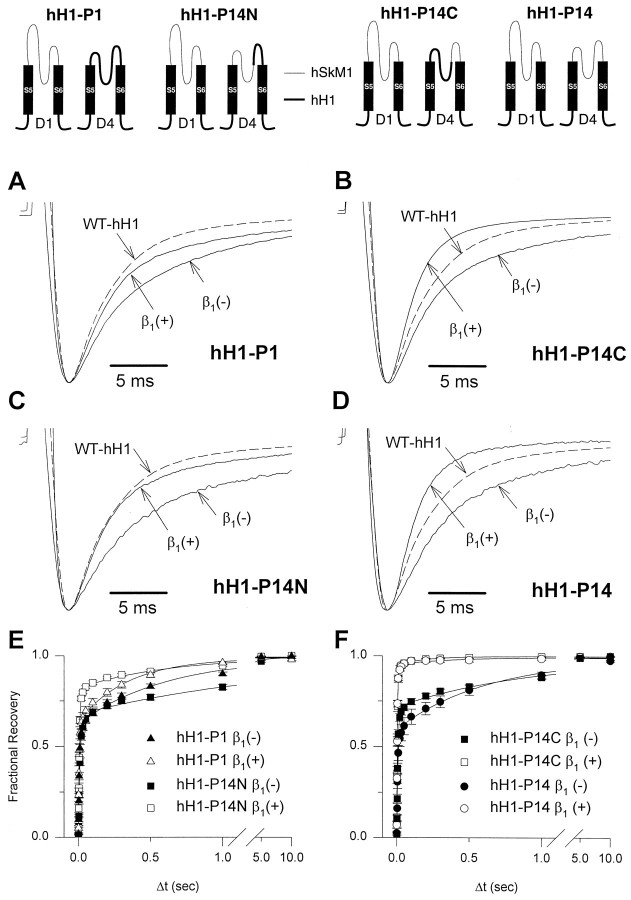
As part of a systematic analysis of α subunit domains that might contribute to isoform-specific differences in β1subunit-induced gating modulation, we examined the role of the S5–S6 interhelical regions. In current models of Na+ channels (Guy and Conti, 1990), these segments consist of two extracellular loops flanking an intramembranous structure (SS1/SS2 region) that contains residues critical for ion permeation and selectivity (Noda et al., 1989; Pusch et al., 1991; Heinemann et al., 1992). We first constructed chimeras in which single S5–S6 loops from hH1 were substituted for the corresponding regions in hSkM1 (Fig.2) and then tested whether the β1 response was abolished or diminished. We used the following criteria to indicate a β1 response: (1) a significant decrease in the time constants for onset of inactivation and recovery from inactivation (τf and τs for fast and slow components, respectively), and (2) a significant increase in the fraction of current exhibiting rapid onset of inactivation and recovery from inactivation. All single S5–S6 loop chimeras were functional when expressed in oocytes and all exhibited significant β1responses (Fig. 2). However, the β1 response of hSkM1–P4, which contains the D4/S5–S6 loop of hH1, was attenuated relative to wild-type hSkM1 and the other single S5–S6 loop chimeras.
Fig. 2.
Responses of wild-type and chimeric Na+ channels to coexpressed β1 subunit. Representative current tracings were recorded in the presence (asterisks) or absence (open circles) of coexpressed hβ1 during a voltage step to −20 mV from a holding potential of −120 mV. Current amplitudes are scaled and superimposed on the same time axis to illustrate the response of each channel to coexpressed β1 subunit. The composition of the α subunit chimeras constructed from hSkM1 and hH1 is illustrated next to each tracing. Filled boxes and thick lines represent structures from hH1, and open boxes and thin lines indicate hSkM1 segments. In the hH1 chimeras, vertical arrows point to the transferred hSkM1 sequences. Current decays of all the hSkM1 background chimeras except for hSkM1–P14 were significantly accelerated by coexpressed hβ1, subunit, and the hβ1subunit accelerated the inactivation kinetics of chimera hH1–P14.
In view of our previous findings that the difference in β1 subunit response between hSkM1 and hH1 were not attributable to a single domain (Makita et al., 1996), we next assembled chimeras in which pair-wise substitutions of hH1 S5–S6 loops were made in single hSkM1 constructs. Five of the six double S5–S6 loop chimeras exhibited a significant β1 response (Fig.2, Table 1), but hSkM1–P14, which contains both D1/S5–S6 and D4/S5–S6 of hH1, did not. The β1 response was absent completely in chimera hSkM1–P14 with respect to its effects on the time constants for both onset of inactivation and recovery from inactivation (Fig. 3A,C) and on the change in the fraction of current exhibiting rapid recovery from inactivation (Table 2). These data indicate that the β1 response phenotype of hH1 can be transferred to hSkM1 by the combination of D1/S5–S6 and D4/S5–S6.
Fig. 3.
Effect of coexpressed hβ1 on hSkM1–P14 or hH1–P14 Na+ channel chimeras.A, Representative current tracings illustrating failure of hβ1 to modulate inactivation in chimera hSkM1–P14. The response of hSkM1 to hβ1 in the same experiment is also shown. B, Representative current tracings illustrating accelerated time course of inactivation caused by coexpression of hβ1 with chimera hH1–P14.C, Plot of fractional recovery versus interpulse interval duration for hSkM1 (filled circles), hSkM1 + hβ1 (open circles), hSkM1–P14 (filled diamonds), and hSkM1–P14+hβ1 (open diamonds).D, Plot of fractional recovery versus interpulse interval duration (Δt) for hH1 (filled triangles), hH1–P14 (filled squares), and hH1–P14 + hβ1 (open squares). Recovery from inactivation in hH1+hβ1 is not shown but is identical to hH1 alone. (Results are represented for at least four cells; error bars are smaller than some data symbols.) Peak current amplitudes inA and B are scaled to unity. Recovery data in C and D are normalized to the current amplitude recorded after a recovery interval of 10 sec.
Table 2.
Quantitative kinetic data for recovery from inactivation in expressed Na+ channel chimeras
| Channel | +/− hβ1 | τf (msec) | τs(msec) | Fast fraction | (n) | pa |
|---|---|---|---|---|---|---|
| hSkM1 Chimeras | ||||||
| hSkM1 | − | 5.0 ± 2.2 | 1800 ± 727 | 0.37 ± 0.07 | (6) | |
| hSkM1 | + | 4.5 ± 0.8 | 162 ± 22 | 0.77 ± 0.02 | (7) | 2_150 |
| hSkM1–P1 | − | 3.9 ± 0.5 | 380 ± 26 | 0.42 ± 0.05 | (8) | |
| hSkM1–P1 | + | 2.3 ± 0.3 | 150 ± 20 | 0.83 ± 0.03 | (3) | 2_150 |
| hSkM1–P2 | − | 3.2 ± 0.3 | 1300 ± 240 | 0.14 ± 0.07 | (7) | |
| hSkM1–P2 | + | 1.4 ± 0.1 | 250 ± 140 | 0.96 ± 0.02 | (4) | 2_150 |
| hSkM1–P3 | − | 3.4 ± 0.8 | 730 ± 160 | 0.26 ± 0.09 | (3) | |
| hSkM1–P3 | + | 2.3 ± 0.8 | 130 ± 40 | 0.90 ± 0.02 | (4) | 2_150 |
| hSkM1–P4 | − | 205 ± 61 | 1500 ± 170 | 0.26 ± 0.05 | (12) | |
| hSkM1–P4 | + | 3.5 ± 0.7 | 720 ± 80 | 0.65 ± 0.06 | (5) | 2_150 |
| hSkM1–P12 | − | 2.4 ± 0.5 | 430 ± 44 | 0.48 ± 0.04 | (8) | |
| hSkM1–P12 | + | 2.5 ± 0.1 | 110 ± 31 | 0.82 ± 0.01 | (7) | 2_150 |
| hSkM1–P13 | − | 3.9 ± 0.3 | 740 ± 120 | 0.51 ± 0.04 | (6) | |
| hSkM1–P13 | + | 3.0 ± 0.7 | 120 ± 46 | 0.77 ± 0.03 | (4) | 2_150 |
| hSkM1–P23 | − | 3.7 ± 0.6 | 1100 ± 79 | 0.15 ± 0.02 | (9) | |
| hSkM1–P23 | + | 2.8 ± 0.1 | 270 ± 50 | 0.81 ± 0.03 | (7) | 2_150 |
| hSkM1–P24 | − | 6.8 ± 1.1 | 1300 ± 200 | 0.18 ± 0.02 | (8) | |
| hSkM1–P24 | + | 1.5 ± 0.2 | 580 ± 57 | 0.75 ± 0.03 | (4) | 2_150 |
| hSkM1–P34 | − | 5.3 ± 0.8 | 1700 ± 130 | 0.13 ± 0.02 | (7) | |
| hSkM1–P34 | + | 2.6 ± 0.2 | 600 ± 37 | 0.65 ± 0.03 | (11) | 2_150 |
| hSkM1–P14 | − | 3.9 ± 0.5 | 300 ± 42 | 0.37 ± 0.05 | (6) | |
| hSkM1–P14 | + | 3.3 ± 0.2 | 290 ± 33 | 0.29 ± 0.04 | (10) | NS |
| hH1 chimeras | ||||||
| hH1 | − | 6.5 ± 0.6 | 160 ± 45 | 0.81 ± 0.02 | (5) | |
| hH1 | + | 3.7 ± 0.4 | 110 ± 15 | 0.82 ± 0.02 | (8) | NS |
| hH1–P1 | − | 5.6 ± 0.4 | 550 ± 43 | 0.65 ± 0.02 | (6) | |
| hH1–P1 | + | 2.8 ± 0.4 | 310 ± 26 | 0.68 ± 0.03 | (9) | NS |
| hH1–P4 | − | 6.4 ± 0.3 | 140 ± 8 | 0.74 ± 0.01 | (5) | |
| hH1–P4 | + | 3.5 ± 0.6 | 48 ± 10 | 0.80 ± 0.01 | (4) | 2_150 |
| hH1–P14 | − | 5.7 ± 0.6 | 600 ± 78 | 0.66 ± 0.03 | (8) | |
| hH1–P14 | + | 3.1 ± 0.3 | 62 ± 7 | 0.92 ± 0.01 | (12) | 2_150 |
| hH1–P14C | − | 7.4 ± 0.5 | 950 ± 103 | 0.74 ± 0.02 | (10) | |
| hH1–P14C | + | 3.4 ± 0.3 | 139 ± 28 | 0.95 ± 0.01 | (14) | 2_150 |
| hH1–P14N | − | 10.2 ± 0.7 | 1419 ± 149 | 0.69 ± 0.02 | (16) | |
| hH1–P14N | + | 7.1 ± 0.8 | 652 ± 84 | 0.83 ± 0.01 | (8) | 2_150 |
Significance level for comparisons between fast fraction in the presence (+) or absence (−) of hβ1. NS, Not significant;
F2_150: p < 0.05; τf, time constant for fast component of Na+channel recovery; τs, time constant for slow component of Na+ channel recovery.
Conferring β1 subunit responsiveness to hH1
To confirm the observation that S5–S6 loops in both D1 and D4 are needed for the β1 response, we constructed reciprocal chimeras in which the hSkM1 D1/S5–S6, D4/S5–S6, or both, was inserted into hH1 (Fig. 2) and tested for the occurrence of a gain of β1 response. All hH1 chimeras expressed well in oocytes, and all exhibited inactivation kinetics that were more similar to wild-type hH1 than to hSkM1 (Tables 1, 2). Coexpression of hβ1 with either hH1–P1 or hH1–P4 caused a small but significant decrease in the time constants of both inactivation and recovery from inactivation, although changes in the fractions of rapid-inactivating and rapid-recovering current was significant in hH1–P4 (Fig. 2, Table 1). In the double S5–S6 loop chimera hH1–P14, a substantial β1 response was observed (Fig.3B,D). In this chimera, hβ1caused a significant change in τf, τs, and the fraction of the fast-inactivating component to values indistinguishable from that observed with the coexpression of wild-type hSkM1 with hβ1 (Table 1). Chimera hH1–P14 also exhibited significant decreases in the time constants for recovery from inactivation and a significant increase in the fraction of fast-recovering channels in response to the β1 subunit (Table 2). These data demonstrate that two S5–S6 interhelical loops of D1 and D4, which are separated by ∼750 amino acid residues in the primary sequence of the α subunit, are responsible for the β1 response differences between hSkM1 and hH1.
Because D1/S5–S6 and D4/S5–S6 consist of intramembranous and extracellular subsections, we attempted to localize the β1 response element further by examining additional chimeras in which these segments were subdivided. To accomplish this goal, we reverted small sections of D1/S5–S6 and D4/S5–S6 in hH1–P14 back to the wild-type hH1 sequence and then evaluated the β1 response. Chimeras in which subregions of the D1/S5–S6 loop were reverted back to hH1 were not functional, but two hH1 chimeras (hH1–P14N, hH1–P14C) having partial D4/S5–S6 reversions expressed well in oocytes (Fig. 4). The hH1–P14B chimera, which contains the N-terminal portion of the hSkM1 D4/S5–S6 loop, exhibits a β1 subunit response similar to that of hH1–P1 except for a small decrease in the fast time constant of inactivation and a significant increase in the fast fraction of recovery from inactivation (Tables 1, 2, Fig.4A,C). By contrast, hH1–P14C, which has the C-terminal portion of the hSkM1 D4/S5–S6 segment, exhibits a significant response to β1 that is indistinguishable from the response of the double S5–S6 loop chimera hH1–P14 (hH1–P14C alone: τf = 1.2 ± 0.1 msec, τs = 7.3 ± 0.9 msec, fast fraction = 0.54 ± 0.03, n = 10; hH1–P14C + h β1: τf = 0.9 ± 0.1 msec, τs = 4.9 ± 0.3 msec, fast fraction = 0.81 ± 0.03, n = 14) (Fig. 4B,D). The effect of the β1 subunit on recovery from inactivation of hH1–P14C closely resembles that observed for hH1–P14 (Table 2, Fig.4E,F). These results suggest that the major portion of the domain 4 β1 response element resides in a D4/S5–S6 extracellular loop. Differences in amino acid sequence between hSkM1 and hH1 within this region are restricted to a 31 residue stretch located between the intramembranous portion of the S5–S6 loop (SS1/SS2 region) and the D4/S6 segment (Fig.5). In this region, there is greater conservation of amino acid sequence between hSkM1 and the β1 responsive rat brain II Na+ channel (77% identity, 87% similarity) than between hSkM1 and hH1 (58% identity, 68% similarity).
Fig. 4.
Sublocalization of the β1 subunit response element in D4/S5–S6. Chimeras used to sublocalize the β1 response structure in D4/S5–S6 (hH1–P14N, hH1–P14C) are illustrated by the drawings at the top of the figure. Drawings were simplified by showing only the S5–S6 region of D1 and D4 for each chimera. Structures from hSkM1 are indicated bythin lines, and segments from hH1 are indicated bythick lines or filled rectangles.A–D, Representative current recordings from oocytes expressing hH1–P1 (A), hH1–P14N (B), hH1–P14C (C), or hH1–P14 (D) in the presence (+) or absence (−) of hβ1. Current sweeps of wild-type hH1 (WT–hH1, dashed lines) are provided as a reference. There were no statistical differences between time constants for hH1–P1 + β1 versus hH1–P14N + β1 and between hH1–P14C + β1 versus hH1–P14 + β1. Currents were recorded using the pulse protocol described in the legend for Figure 1, and their peak currents were scaled to unity. A representative tracing from oocytes expressing wild-type hH1 in the absence of hβ1 is shown in each figure part for comparison. E, F, Recovery from inactivation was determined as described in the legend for Figure 3 and plotted against a loga- rithmic time scale.E, Recovery curves for hH1–P14N in the presence (open squares) or absence (closed squares) of hβ1 are shown with data obtained from the hH1–P1 chimera in the presence (open triangles) or absence (closed triangles) of hβ1.F, Recovery curves for hH1–P14C in the presence (open squares) or absence (closed squares) of hβ1 are shown with data obtained from the hH1–P14 chimera in the presence (open circles) and absence (closed circles) of hβ1.
Fig. 5.
Amino acid sequence alignment of the D4/S5–S6 region. Alignment of amino acid sequences within the D4/S5–S6 region from hSkM1 (George et al., 1992), rat skeletal muscle Na+ channel μI (Trimmer et al., 1989), rat brain II Na+ channel (Auld et al., 1988), and hH1 (Gellens et al., 1992). Numbers indicate amino acid position at the beginning of each line. Residues identical to hSkM1 are shown asdashes in the aligned sequences, and the location of SS2 and D4/S6 segments is indicated by the thin lines. The β1 subunit response element is indicated by athick line. Residues within the hH1 sequence that differ from those in a consensus sequence of hSkM1, μI, and rat brain II Na+ channels are indicated byasterisks.
Functional domains of the β1 subunit
We considered next which regions of the β1 subunit are required for Na+ channel-gating modulation. The β1 subunit is predicted to have a single hydrophobic, membrane-spanning segment flanked by a predicted extracellular N terminus and cytoplasmic C-terminal domain. Chen and Cannon (1995) have demonstrated that deletion of the complete β1 subunit C terminus does not disturb its ability to accelerate the inactivation rate of the rat skeletal muscle Na+ channel. By contrast, small deletions in the extracellular domain result in β1subunits that have no effects on channel gating. However, it is not possible to rule out that protein misfolding or post-translational processing arrest is responsible for the observed lack of function in these β1 subunit deletion mutants and, thus, no solid conclusions can be made regarding the functional importance of this domain.
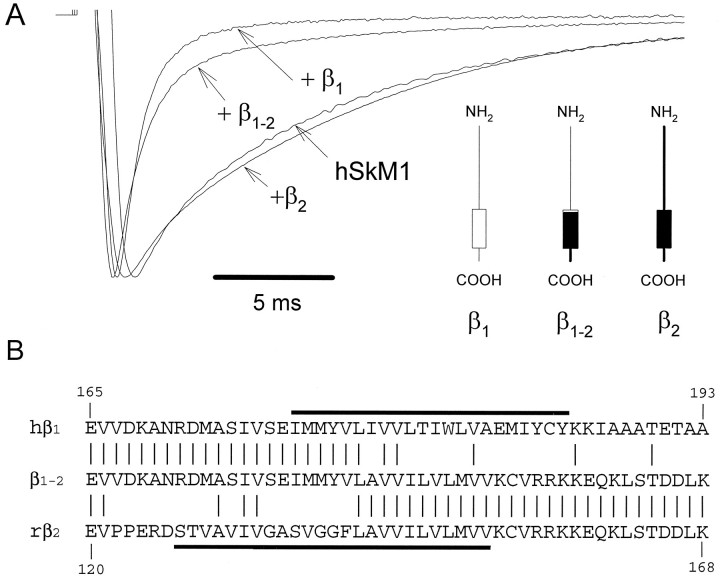
We used an alternative approach to evaluate the functional role of noncytoplasmic β1 subunit domains using a β subunit chimera formed by transplanting the extracellular portion of hβ1 onto the transmembrane domain and C terminus of the rβ2 subunit. The β2 subunit was selected to provide a surrogate transmembrane domain and C terminus because of its structural homology with the β1 subunit (Isom et al., 1995a) but lack of a functional effect on the gating of hSkM1 (see below). In these experiments, we tested the hypothesis that the extracellular domain of β1 is sufficient to modulate hSkM1 gating. If this is correct, then this functional property should be transferable to the β2 subunit. The rβ2subunit cDNA was isolated using RT-PCR based on the published sequence (Isom et al., 1995a), and its function was demonstrated by coexpression with the rat brain IIA Na+ channel (RBIIA) α subunit in oocytes. This recombinant rβ2 subunit significantly increased the peak current amplitude and accelerated the inactivation kinetics of RBIIA, as described by Isom et al. (1995a) (data not shown). When the rβ2 subunit was coexpressed with hSkM1, a significant increase in the peak current amplitude of both Na+ channel isoforms was observed. However, rβ2 had no significant effect on the inactivation kinetics of hSkM1 (hSkM1 alone, τf = 2.2 ± 0.3 msec, τs =13.2 ± 1.4 msec, n = 20; hSkM1 + rβ2, τf = 1.8 ± 0.7 msec, τs = 10.0 ± 0.6 msec, n = 6, not significant) (Fig. 6A, Table 1).
Fig. 6.
Effect of a β subunit chimera on hSkM1 gating.A, Scaled current recordings obtained from oocytes expressing hSkM1 alone or in the presence of wild-type hβ1 (+β1), wild-type rβ2(+β2), or a chimeric β1/β2subunit (+β1–2). Recording conditions were the same as in Figure 1A. The figure in theinset depicts the proposed membrane topology of wild-type and chimeric β subunits. Thin lines andopen boxes indicate structures from hβ1, whereas thick black lines and filled boxes represent structures from rβ2.B, Alignment of partial amino acid sequences of hβ1, rβ2, and chimeric β1–2. The location of putative transmembrane domains of hβ1 and rβ2 is depicted by thick lines.
We constructed a chimeric β1/β2 subunit (β1–2) in which the transmembrane and C-terminal domains of hβ1 were replaced by the corresponding regions of rβ2 to test the functional importance of the extracellular domain in gating (Fig. 6B). The β1–2 chimera significantly decreased both hSkM1 inactivation time constants (τf: 2.2 ± 0.3–0.6 to 0.1 msec, p < 0.05; τs: 13.2 ± 0.3 to 7.7 ± 0.2 msec, p < 0.05, n = 16). The time constant of the fast-inactivating component observed with hSkM1 + β1–2 was comparable to that seen with hSkM1 + hβ1, although the fraction of the fast-inactivating component was smaller for hSkM1 + β1–2 (0.66 ± 0.03) than that of hSkM1 + hβ1 (0.84 ± 0.01). These data demonstrate that the extracellular domain and adjacent residues within the transmembrane segment of the β1 subunit are sufficient to induce a near-complete modulation of hSkM1 gating. These findings are quite consistent with the results obtained from our α subunit chimeric studies indicating that structures located largely on the extracellular side of the membrane are responsible for the effect of the β1 subunit.
DISCUSSION
Defining the structural elements required for specific ion channel functions can be facilitated by studying chimeras constructed between similar channel isoforms that exhibit distinct properties. Such an approach depends on the existence of a clear quantitative or qualitative functional difference between two parent channels and the ability to transfer this functional difference by transplanting simple structures from one channel to another.
Using such an approach, we have defined the regions within the Na+ channel α subunit required for gating modulation by the β1 subunit. These results indicate that the β1 subunit effects on the kinetics of inactivation and recovery from inactivation requires two widely spaced regions (S5–S6 loops of D1 and D4) in the primary sequence of the α subunit molecule, along with the extracellular domain and a portion of the membrane spanning segment of the β1 subunit. There are two possible interpretations of these data: (1) extracellular domains of the β1 subunit interact with a discontinuous epitope formed by D1/S5–S6 and D4/S5–S6 of the α subunit and (2) the β1 subunit interacts elsewhere on the α subunit, but the D1/S5–S6 and D4/S5–S6 loops are required for changes in gating behavior. At this point, without additional biochemical mapping of the α–β1 subunit contact regions, we can only speculate about the mechanism of subunit-induced gating modulation. However, knowledge of the structural requirements for the α–β1functional effects does contribute greatly to localizing candidate interaction domains.
A direct interaction between β1 and the D1/S5–S6 and D4/S5–S6 loops, rather than an allosteric interaction, has been suggested by previous biochemical studies of brain Na+channels. Photoreactive derivatives of α-scorpion toxin V fromLeiurus quinquestriatus have been shown to covalently label both α and β1 Na+ channel polypeptides in rat brain synaptosomal membranes (Beneski and Catterall, 1980), suggesting that the toxin binds near the subunit contact region (Catterall, 1988). The binding site of α-scorpion toxin has been further localized by demonstrating that its effects on brain Na+ channels can be blocked by site-specific antibodies directed against epitopes within D1/S5–S6 and D4/S5–S6 (Tejedor and Catterall, 1988; Thomsen and Catterall, 1989). These two observations support the hypothesis that the β1 subunit associates with the α subunit in a region that either overlaps or is adjacent to the α-scorpion toxin binding site in D1/S5–S6 and D4/S5–S6. In view of the fact that α-scorpion toxin and other site 3 neurotoxins can affect Na+ channel inactivation from the extracellular side of the membrane (Catterall, 1988), it is conceivable that their mechanism of action involves disturbing this α–β1subunit interaction.
Topology of α–β1 subunit interaction
On which side of the membrane does the Na+channel α–β1 subunit interaction occur? In studies of other voltage-gated ion channels, accessory subunits have been found to modulate gating kinetics through cytoplasmic interactions. In voltage-gated potassium and calcium channels, small hydrophilic β subunits interact with the principal channel subunit from the cytoplasmic side of the membrane and accelerate inactivation (Pragnell et al., 1994; Rettig et al., 1994). We have determined previously that cytoplasmic structures in hSkM1 and hH1 do not participate in β1 subunit-induced gating modulation (Makita et al., 1996). Similarly, the predicted cytoplasmic domain of hβ1is not necessary for its function in modulating the inactivation kinetics or current amplitude increase of hSkM1 (Chen and Cannon, 1995)
We now present positive evidence of a functional role for the extracellular β1 subunit domain in modulating Na+ channel gating using β1/β2subunit chimeras. These results indicate an essential role of the extracellular domain and the outermost residues making up the transmembrane segment of the β1 subunit in gating modulation. Other α subunit domains may contribute to the apparent enhancement in cell surface expression induced by the β1subunit. This idea is supported by the observed dissociation of β1 subunit effects on gating modulation and expression in hH1. It is not clear at this time whether enhanced expression and accelerated inactivation are the result of a single α–β1 interaction or whether multiple contact regions are responsible for these effects.
Implications for Na+ channel tertiary structure
Several lines of evidence point toward a three-dimensional model of the Na+ channel in which D1 and D4 are adjacent. Support for this model comes from observations made on the interaction of α-scorpion toxin and brevetoxin with rat brain Na+channels. As discussed above, binding of α-scorpion toxin can be blocked by antibodies directed against regions of the D1/S5–S6 and D4/S5–S6 (Tejedor and Catterall, 1988; Thomsen and Catterall, 1989). Specifically, these antibodies recognize epitopes located in the predicted extracellular loops immediately flanking the intramembranous portion of D1/S5–S6 and the S5–SS1 region of D4/S5–S6. These results were taken as evidence that D1/S6 and D1/S5 are adjacent in the tertiary structure of the channel. Similar conclusions were derived from studies using photoreactive brevetoxin, which was found to bind at the intramembranous interface between D1/S6 and D4/S5 (Trainer et al., 1994). This pattern of α subunit folding is also suggested by apparent interactions between the N terminus and C terminus of the rat skeletal muscle Na+ channel (Sun et al., 1995). Taking these data together with our findings, it is possible to propose a tertiary structural model for α–β1 subunit interaction in which β1 resides in close proximity to D1/S6 and D4/S5, which are adjacent to one another in the folded structure of the Na+ channel. In this position, both extracellular and intramembranous portions of β1 can interact with both D1/S5–S6 and D4/S5–S6. The interaction of β1 with the short extracellular loop adjacent to the S6 segment in D4 suggests a possible mechanism of action of the β1 subunit. Interaction of the β1 subunit with this region may have a direct influence on the conformation or position of the D4/S6 segment, which may form part of the inactivation gate receptor (McPhee et al., 1994). A similar mechanism may be operating in D1 and affect movement or conformation of D1/S6, which may also contain residues important for Na+ channel inactivation (Zhang et al., 1995). Our data defining the dependence of Na+ channel subunit interaction on a composite structure formed by regions that are widely spaced in the primary sequence of the α subunit molecule are strong evidence that Na+ channel α–β1 subunit interaction depends on a three-dimensional conformation of the channel. These results also support the notion that D1 and D4 are contiguous in the folded structure of the Na+ channel.
Our data reveal an additional functional property of the Na+ channel S5–S6 loop. This region has been implicated in several diverse functional properties of mammalian Na+channels including ion selectivity (Heinemann et al., 1992), permeation (Pusch et al., 1991), block by divalent cations (Backx et al., 1992), and interaction with two distinct families of neurotoxins (Noda et al., 1989; Thomsen and Catterall, 1989; Terlau et al., 1991; Satin et al., 1992; Dudley et al., 1995). Localization of a β1 subunit interaction domain to D1/S5–S6 and D4/S5–S6 indicates specialization among the four S5–S6 loops and distinguishes Na+ channels from the perfect fourfold symmetry in homotetrameric potassium channels.
Footnotes
This work was supported by National Institutes of Health (NS32387), American Heart Association Tennessee Affiliate, and the Lucille P. Markey Charitable Trust. P.B.B. and A.L.G. are Established Investigators of the American Heart Association. A.L.G. is a Lucille P. Markey Scholar. We thank Brady Palmer for oocyte injections; Dr. Alan Goldin for providing the rat brain IIA plasmid; and Dick Horn, Lee Limbird, and Dan Roden for their critical reviews of this manuscript.
Correspondence should be addressed to Dr. Alfred L. George Jr., S-3223 MCN, Vanderbilt University Medical Center, 21st Avenue South at Garland, Nashville, TN 37232-2372.
Dr. Makita’s present address: Department of Cardiovascular Medicine, Hokkaido University School of Medicine, Kita-15, Nishi-7, Kita-Ku, Sapporo 060, Japan.
REFERENCES
- 1.Auld VJ, Goldin AL, Krafte DS, Marshall J, Dunn JM, Catterall WA, Lester HA, Davidson N, Dunn RJ. A rat brain Na+ channel α subunit with novel gating properties. Neuron. 1988;1:449–461. doi: 10.1016/0896-6273(88)90176-6. [DOI] [PubMed] [Google Scholar]
- 2.Backx PH, Yue DT, Lawrence JH, Marban E, Tomaselli GF. Molecular localization of an ion-binding site within the pore of mammalian sodium channels. Science. 1992;257:248–251. doi: 10.1126/science.1321496. [DOI] [PubMed] [Google Scholar]
- 3.Beneski DA, Catterall WA. Covalent labeling of protein components of the sodium channel with a photactivable derivative of scorpion toxin. Proc Natl Acad Sci USA. 1980;77:639–643. doi: 10.1073/pnas.77.1.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett PB, Makita N, George AL., Jr A molecular basis for gating mode transitions in human skeletal muscle sodium channels. FEBS Lett. 1993;326:21–24. doi: 10.1016/0014-5793(93)81752-l. [DOI] [PubMed] [Google Scholar]
- 5.Cannon SC, McClatchey AI, Gusella JF. Modification of the Na+ current conducted by the rat skeletal muscle α subunit by coexpression with a human brain β subunit. Pflügers Arch. 1993;423:155–157. doi: 10.1007/BF00374974. [DOI] [PubMed] [Google Scholar]
- 6.Catterall WA. Structure and function of voltage-sensitive ion channels. Science. 1988;242:50–61. doi: 10.1126/science.2459775. [DOI] [PubMed] [Google Scholar]
- 7.Catterall WA. Cellular and molecular biology of voltage-gated sodium channels. Physiol Rev. 1992;72:S15–S48. doi: 10.1152/physrev.1992.72.suppl_4.S15. [DOI] [PubMed] [Google Scholar]
- 8.Chen C, Cannon SC. Modulation of Na+ channel inactivation by the β1 subunit: a deletion analysis. Pflügers Arch. 1995;431:186–195. doi: 10.1007/BF00410190. [DOI] [PubMed] [Google Scholar]
- 9.Cohen SA, Levitt LK. Partial characterization of the rH1 sodium channel protein from rat heart using subtype-specific antibodies. Circ Res. 1993;73:735–742. doi: 10.1161/01.res.73.4.735. [DOI] [PubMed] [Google Scholar]
- 10.Cribbs LL, Satin J, Fozzard HA, Rogart RB. Functional expression of the rat heart I Na+ channel isoform. FEBS Lett. 1990;275:195–200. doi: 10.1016/0014-5793(90)81470-9. [DOI] [PubMed] [Google Scholar]
- 11.Dudley SC, Jr, Todt H, Lipkind G, Fozzard HA. A μ-conotoxin-insensitive Na+ channel mutant: possible localization of a binding site at the outer vestibule. Biophys J. 1995;69:1657–1665. doi: 10.1016/S0006-3495(95)80045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gellens ME, George AL, Chen L, Chahine M, Horn R, Barchi RL, Kallen RG. Primary structure and functional expression of the human cardiac tetrodotoxin-insensitive voltage-dependent sodium channel. Proc Natl Acad Sci USA. 1992;89:554–558. doi: 10.1073/pnas.89.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.George AL, Komisarof J, Kallen RG, Barchi RL. Primary structure of the adult human skeletal muscle voltage-dependent sodium channel. Ann Neurol. 1992;31:131–137. doi: 10.1002/ana.410310203. [DOI] [PubMed] [Google Scholar]
- 14.Goldin AL, Snutch T, Lubbert H, Dowsett A, Marshall J, Auld V, Downey W, Fritz LC, Lester HA, Dunn R, Catterall WA, Davidson N. Messenger RNA coding for only the α subunit of the rat brain Na channel is sufficient for expression of functional channels in Xenopus oocytes. Proc Natl Acad Sci USA. 1986;83:7503–7507. doi: 10.1073/pnas.83.19.7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon D, Merrick D, Wollner DA, Catterall WA. Biochemical properties of sodium channels in a wide range of excitable tissues studied with site-directed antibodies. Biochemistry. 1988;27:7032–7038. doi: 10.1021/bi00418a054. [DOI] [PubMed] [Google Scholar]
- 16.Guy HR, Conti F. Pursuing the structure and function of voltage-gated channels. Trends Neurosci. 1990;13:201–206. doi: 10.1016/0166-2236(90)90160-c. [DOI] [PubMed] [Google Scholar]
- 17.Heinemann SH, Terlau H, Stühmer W, Imoto K, Numa S. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 1992;356:441–443. doi: 10.1038/356441a0. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi R. Using PCR to engineer DNA. In: Erlich HA, editor. PCR technology. Stockton; New York: 1989. pp. 61–70. [Google Scholar]
- 19.Isom LL, De Jongh KS, Patton DE, Reber BFX, Offord J, Charbonneau H, Walsh K, Goldin AL, Catterall WA. Primary structure and functional expression of the β1 subunit of the rat brain sodium channel. Science. 1992;256:839–842. doi: 10.1126/science.1375395. [DOI] [PubMed] [Google Scholar]
- 20.Isom LL, Ragsdale DS, De Jongh KS, Westenbroek RE, Reber BFX, Scheuer T, Catterall WA. Structure and function of the β2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell. 1995a;83:433–442. doi: 10.1016/0092-8674(95)90121-3. [DOI] [PubMed] [Google Scholar]
- 21.Isom LL, Scheuer T, Brownstein AB, Ragsdale DS, Murphy BJ, Catterall WA. Functional co-expression of the β1 and type IIA α subunits of sodium channels in a mammalian cell line. J Biol Chem. 1995b;270:3306–3312. doi: 10.1074/jbc.270.7.3306. [DOI] [PubMed] [Google Scholar]
- 22.Klugbauer N, Lacinova L, Flockerzi V, Hofmann F. Structure and functional expression of a new member of the tetrodotoxin-sensitive voltage-activated sodium channel family from human neuroendocrine cells. EMBO J. 1995;14:1084–1090. doi: 10.1002/j.1460-2075.1995.tb07091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krieg PA, Melton DA. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–415. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- 24.Kyle JW, Chang SY, Satin J, Fang JM, Fozzard HA, Rogart RB. Rat brain β1 Na channel subunit interactions with rat brain IIa, skeletal muscle (μ1), and rat heart (RH1) α-subunits. Biophys J. 1993;64:A88. [Google Scholar]
- 25.Lombet A, Luzdunski M. Characterization, solubilization, affinity labeling and purification of the cardiac Na+ channel using Tityus toxin gamma. Eur J Biochem. 1984;141:651–660. doi: 10.1111/j.1432-1033.1984.tb08241.x. [DOI] [PubMed] [Google Scholar]
- 26.Makita N, Bennett PB, Jr, George AL., Jr Voltage-gated Na+ channel β1 subunit mRNA expressed in adult human skeletal muscle, heart, and brain is encoded by a single gene. J Biol Chem. 1994;269:7571–7578. [PubMed] [Google Scholar]
- 27.Makita N, Bennett PB, George AL., Jr Multiple domains contribute to the distinct inactivation properties of human heart and skeletal muscle Na+ channels. Circ Res. 1996;78:244–252. doi: 10.1161/01.res.78.2.244. [DOI] [PubMed] [Google Scholar]
- 28.McPhee JC, Ragsdale DS, Scheuer T, Catterall WA. A mutation in segment IVS6 disrupts fast inactivation of sodium channels. Proc Natl Acad Sci USA. 1994;91:12346–12350. doi: 10.1073/pnas.91.25.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messner DJ, Catterall WA. The sodium channel from rat brain. Separation and characterization of subunits. J Biol Chem. 1985;260:10597–10604. [PubMed] [Google Scholar]
- 30.Messner DJ, Feller DJ, Scheuer T, Catterall WA. Functional properties of rat brain sodium channels lacking the β1 or β2 subunit. J Biol Chem. 1986;261:14882–14890. [PubMed] [Google Scholar]
- 31.Noda M, Suzuki H, Numa S, Stühmer W. A single point mutation confers tetrodotoxin and saxitoxin insensitivity on the sodium channel II. FEBS Lett. 1989;259:213–216. doi: 10.1016/0014-5793(89)81531-5. [DOI] [PubMed] [Google Scholar]
- 32.Nuss HB, Chiamvimonvat M, Perez-Garcia MT, Tomaselli GF, Marban E. Functional association of the β1 subunit with human cardiac (hH1) and rat skeletal muscle (μI) sodium channel α subunits expressed in Xenopus oocytes. J Gen Physiol. 1995;106:1171–1191. doi: 10.1085/jgp.106.6.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patton DE, Isom LL, Catterall WA, Goldin AL. The adult rat brain β1 subunit modifies activation and inactivation gating of multiple sodium channel α subunits. J Biol Chem. 1994;269:17649–17655. [PubMed] [Google Scholar]
- 34.Pragnell M, De Waard M, Mori Y, Tanabe T, Snutch TP, Campbell KP. Calcium channel β-subunit binds to a conserved motif in the I–II cytoplasmic linker of the α1-subunit. Nature. 1994;368:67–70. doi: 10.1038/368067a0. [DOI] [PubMed] [Google Scholar]
- 35.Pusch M, Noda M, Stühmer W, Numa S, Conti F. Single point mutations of the sodium channel drastically reduce the pore permeability without preventing its gating. Eur Biophys J. 1991;20:127–133. doi: 10.1007/BF01561134. [DOI] [PubMed] [Google Scholar]
- 36.Qu Y, Isom LL, Westenbroek RE, Rogers JC, Tanada TN, McCormick KA, Scheuer T, Catterall WA. Modulation of cardiac Na+ channel expression in Xenopus oocytes by β1 subunits. J Biol Chem. 1995;270:25696–25701. doi: 10.1074/jbc.270.43.25696. [DOI] [PubMed] [Google Scholar]
- 37.Rettig J, Heinemann SH, Wunder F, Lorra C, Parcej DN, Dolly JO, Pongs O. Inactivation properties of voltage-gated K+ channels altered by presence of β-subunit. Nature. 1994;369:289–294. doi: 10.1038/369289a0. [DOI] [PubMed] [Google Scholar]
- 38.Roberts RH, Barchi RL. The voltage-sensitive sodium channel from rabbit skeletal muscle. Chemical characterization of subunits. J Biol Chem. 1987;262:2298–2303. [PubMed] [Google Scholar]
- 39.Satin J, Kyle JW, Chen M, Bell P, Cribbs LL, Fozzard HA, Rogart RB. A mutant of TTX-resistant cardiac sodium channels with TTX-sensitive properties. Science. 1992;256:1202–1205. doi: 10.1126/science.256.5060.1202. [DOI] [PubMed] [Google Scholar]
- 40.Stühmer W, Conti F, Suzuki H, Wang X, Noda M, Yahagi N, Kubo H, Numa S. Structural parts involved in activation and inactivation of the sodium channel. Nature. 1989;339:597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]
- 41.Sun W, Barchi RL, Cohen SA. Probing sodium channel cytoplasmic domain structure. Evidence for the interaction of the rSkM1 amino and carboxyl termini. J Biol Chem. 1995;270:22271–22276. doi: 10.1074/jbc.270.38.22271. [DOI] [PubMed] [Google Scholar]
- 42.Tejedor FJ, Catterall WA. Site of covalent attachment of α-scorpion toxin derivatives in domain I of the sodium channel α subunit. J Biol Chem. 1988;85:8742–8746. doi: 10.1073/pnas.85.22.8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terlau H, Heinemann SH, Stühmer W, Pusch M, Conti F, Imoto K, Numa S. Mapping the site of block by tetrodotoxin and saxitoxin of sodium channel II. FEBS Lett. 1991;293:93–96. doi: 10.1016/0014-5793(91)81159-6. [DOI] [PubMed] [Google Scholar]
- 44.Thomsen WJ, Catterall WA. Localization of the receptor site for α-scorpion toxins by antibody mapping: implications for sodium channel topology. Proc Natl Acad Sci USA. 1989;86:10161–10165. doi: 10.1073/pnas.86.24.10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trainer VL, Baden DG, Catterall WA. Identification of peptide components of the brevetoxin receptor site of rat brain sodium channels. J Biol Chem. 1994;269:19904–19909. [PubMed] [Google Scholar]
- 46.Trimmer JS, Cooperman SS, Tomiko SA, Zhou J, Crean SM, Boyle MB, Kallen RG, Sheng Z, Barchi RL, Sigworth FJ, Goodman RH, Agnew WS, Mandel G. Primary structure and functional expression of a mammalian skeletal muscle sodium channel. Neuron. 1989;3:33–49. doi: 10.1016/0896-6273(89)90113-x. [DOI] [PubMed] [Google Scholar]
- 47.West JW, Patton DE, Scheuer T, Wang Y, Goldin AL, Catterall WA. A cluster of hydrophobic amino acid residues required for fast Na+-channel inactivation. Proc Natl Acad Sci USA. 1992;89:10910–10914. doi: 10.1073/pnas.89.22.10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang JF, Ellinor PT, Tsien RW, Aldrich RW. Molecular determinants of sodium channel inactivation include residues near segment IS6. Biophys J. 1995;68:A361. [Google Scholar]