Abstract
This work describes the cloning and expression of the levocabastine-sensitive neurotensin (NT) receptor from mouse brain. The receptor protein comprises 417 amino acids and bears the characteristics of G-protein-coupled receptors. This new NT receptor (NTR) type is 39% homologous to, but pharmacologically distinct from, the only other NTR cloned to date from the rat brain and the human HT29 cell line. When the receptor is expressed in Xenopus laevis oocytes, the H1 antihistaminic drug levocabastine, like NT and neuromedin N, triggers an inward current. The pharmacological properties of this receptor correspond to those of the low-affinity, levocabastine-sensitive NT binding site described initially in membranes prepared from rat and mouse brain. It is expressed maximally in the cerebellum, hippocampus, piriform cortex, and neocortex of adult mouse brain.
Keywords: neurotensin, neuromedin N, receptor, levocabastine, cloning, low affinity, G-protein-coupled
The existence of multiple receptors for the neurotensin (NT)-related peptides was suggested initially by the description of two families of NT binding sites (KD1 = 0.17 nm;KD2 = 2 nm) in rat brain synaptic membranes (Mazella et al., 1983) and by the ability of the antihistaminic H1 drug levocabastine to totally inhibit NT binding to the low-affinity sites without affecting binding to the high-affinity sites (Schotte et al., 1986). The functional properties of NT receptors (NTRs) are numerous and include stimulations of intracellular cGMP production, turnover of inositol phosphates and Ca2+release, and inhibition of cAMP accumulation (for review, see Vincent, 1995). NT also produces several central and peripheral effects. For example, central administration of NT induces hypothermia and analgesia as well as an increase in dopamine turnover and release (for review, see Vincent, 1995).
Recently, an NTR cloned from rat brain (Tanaka et al., 1990) and from the human colon carcinoma HT29 cell line (Vita et al., 1993) was identified as being the high-affinity NT binding site insensitive to levocabastine (NTRH). This NTR was stably transfected in eukaryotic cells and was functionally coupled to phospholipase C and Ca2+ mobilization (Hermans et al., 1992, Watson et al., 1992, Chabry et al., 1994). More recently, the development of a nonpeptide NT antagonist, SR48692, exhibiting a high affinity for NTRH (Gully et al., 1993), demonstrated that this receptor type was involved in the dopamine-releasing effect of NT in guinea pig striatal slices and in the turning behavior induced by unilateral intrastriatal injection of NT in the mouse (Gully et al., 1993). SR48692, however, failed to antagonize NT-induced hypothermia and analgesia in the mouse and rat (Dubuc et al., 1994), suggesting the existence of other types of NT receptors. Since the cloning of the rat brain NTRH (Tanaka et al., 1990) and of its equivalent in the human HT29 cell line (Vita et al., 1993), no other NT receptor type has been identified.
We describe in this work the cloning and functional expression of the low-affinity levocabastine-sensitive NT receptor (NTRL). This receptor is poorly recognized by the nonpeptide NT antagonist SR48692. Its tissue distribution and ontogenic expression were analyzed and compared with those of the previously cloned NTRH.
MATERIALS AND METHODS
Materials. NT and neuromedin N (NN) were purchased from Peninsula Laboratories (Belmont, CA).dTrp11-NT, Trp11-NT, and xenin were from Bachem (Torrance, CA). SR48692 was from Sanofi. Levocabastine was a generous gift from Dr. Alain Schotte (Janssen Pharmaceutica, Beerse, Belgium). NT was iodinated and purified as described previously (Sadoul et al., 1984). The pcDNA3 expression vector was purchased from Invitrogen (San Diego, CA), DMEM and gentamycin from Life Technologies (Gaithersburg, MD), fetal calf serum from Boehringer Mannheim (Boehringer, Indianapolis, IN), and restriction endonucleases from Eurogentec.
cDNA cloning and expression of the mouse NTR. A cDNA library was constructed from mouse brain poly(A+) RNA into the UniZap XR vector (Stratagene, LaJolla, CA) according to the procedures described by the manufacturer. Clones (5 × 105) derived from the cDNA library were screened by hybridization with the total open reading frame of the rat NTR cDNA (1.27 kb) (Tanaka et al., 1990). Hybridization and filter washing were carried out at 60°C under previously described conditions (Sambrook et al., 1989). One hybridization-positive clone was isolated by repeated purification. Nucleotide sequences were determined in both strands by using the Abi-prime DNA sequencing kit (Applied Biosystems, Foster City, CA).
The 1.6 kb EcoRI-ApaI fragment of the NTRL was inserted into a eukaryotic expression vector (pcDNA3) containing the cytomegalovirus promoter and the resistance to G418 gene as a selective marker. Transient transfections were performed with 1 μg of recombinant pcDNA3 plasmid by the DEAE-dextran precipitation method (Cullen, 1987) onto semiconfluent COS-7 cells grown in 100 mm cell culture dishes. Binding and bioassays were performed ∼60 hr after transfection. Membranes from nontransfected COS-7 cells were totally devoid of specific 125I-NT binding.
Binding experiments were carried out on freshly prepared cell membrane homogenates as described previously (Chabry et al., 1994). Cell membranes (25 μg) were incubated in 250 μl of 50 mmTris-HCl, pH 7.5, containing 0.1% bovine serum albumin and 0.8 mm 1–10 phenanthroline (binding buffer) with increasing concentrations of 125I-Tyr3-NT alone (from 25 to 800 pm) or isotopically diluted by unlabeled NT (from 0.2 to 20 nm) for saturation experiments. Nonspecific binding was determined in parallel incubations containing 1 μm unlabeled NT. After 20 min at 25°C, incubation media were filtered through cellulose acetate filters (Sartorius, Bohemia, NY). Filters were rinsed twice with 2 ml of ice-cold binding buffer and counted in a Packard γ counter. Binding parameters [dissociation constant (KD) and maximal binding capacity (Bmax)] were derived from Scatchard analysis of the data. Competition experiments were performed by incubating membranes with 0.4 nm125I-Tyr3-NT and increasing concentrations of various synthetic or natural analogs of NT (from 10−11 to 10−6m) or with levocabastine or the nonpeptide NT antagonist SR48692. Incubations were terminated as for saturation experiments. IC50 values were determined from inhibition curves as the unlabeled ligand concentration inhibiting 50% of 125I-Tyr3-NT specific binding.
RNA blot hybridization analysis. RNAs were isolated from different tissues of adult male mice or from whole brain of 7-, 15-, and 35-d-old mice and rats by the guanidinium thiocyanate-phenol-chloroform method (Chomczynski and Sacchi, 1987). Northern analysis was carried out using 10 μg of oligo-dT purified poly(A+) RNAs. The rat cDNA probe used was the 1.27 kb fragment corresponding to the total reading frame of the NTRH; the mouse cDNA probe used was the 800 bp BstXI fragment of the NTRL inserted into the pcDNA3 vector.
Electrophysiological measurements. The pcDNA3 vector containing the cDNA of the mouse NTRL served as template to prepare cRNAs using the in vitro transcription kit from Stratagene. cRNAs (∼10 ng) were injected into Xenopus laevis oocytes. The oocytes were then incubated at 18°C for 2–4 d. Electrophysiological measurements were performed at 20°C according to the procedure described previously (Masu et al., 1987). Drugs were applied rapidly into the experimental chamber by a puffer pipette (200 μl). Responses to NT, NN, xenin, and levocabastine were recorded under voltage clamp at −60 mV.
In situ hybridization in mouse brain. A 600 bp cDNA fragment corresponding to nucleotides 660-1260 was inserted into the TA cloning vector (Invitrogen) by PCR and standard cloning techniques and used as a template to produce sense and antisense33P-labeled RNA probes. Paraformaldehyde-perfused mouse brain sagittal sections were incubated in 120 mm phosphate buffer, pH 7.2, containing 50% formamide, 4× SSC, 1× Denhardt’s solution, 10% dextran sulfate, and 0.6% sarcosyl with antisense and sense 33P-labeled RNA probes (3 × 105cpm/slice). After 15 hr at 50°C, slices were washed twice with 4× SSC and 1× SSC at room temperature, treated with RNase A (5 μg/ml) for 5 min at 37°C, and finally washed with 0.1× SSC and dehydrated in graded ethanol before radioautography for 5 d on Amersham Hyperfilm βmax.
RESULTS
Molecular cloning and nucleotide and amino acid sequence of the mouse NTR
A mouse brain cDNA library consisting of 5 × 105 clones was screened by hybridization with a cDNA probe corresponding to the open reading frame of the rat NTRH cDNA clone under low-stringency conditions. The only hybridization-positive clone was then purified. Sequence analysis revealed nucleic acid stretches that shared up to 70% homology with the rat NTRH cDNA.
Figure 1 shows the 1554 nucleotide sequence determined for the cloned cDNA. The amino acid sequence of the mouse NTRL was assigned from the longest open reading frame of the cDNA. The nucleotide sequence surrounding the putative initiation codon agrees with the consensus sequence for the eukaryotic translation sites (Kozak, 1987). The deduced NTRL sequence consists of 417 amino acid residues with a molecular weight of 46,509.
Fig. 1.

The cDNA sequence of the mouse NTRL and its deduced amino acid sequence. The amino acid sequence deduced for the NTRL is shown above the nucleotide sequence. Positions of the putative transmembrane domains I–VII of the NTRL are indicated above the amino acid sequence on the basis of hydropathicity profile (Kyte and Doolittle, 1982). These segments are marked as stretches of 18 consecutive residues, taking into account the possibility of adding four more residues on each side of the segments (Baldwin et al., 1993).Asterisks and black diamonds indicate potential phosphorylation sites by protein kinase C and casein kinase II, respectively.
The structure of NTRL indicates that it belongs to the large family of G-protein-coupled receptors (Probst et al., 1992), with some peculiarities. The hydropathicity profile analysis (Kyte and Doolittle, 1982) of NTRL is compatible with the possible existence of seven hydrophobic amino acid domains (data not shown). The third intracellular loop and the C-terminal region of NTRL bear many serine (Ser) and threonine (Thr) residues that could serve as possible phosphorylation sites. Moreover, these regions contain several protein kinase C and casein kinase II consensus sequences (Fig. 1); however, the N-terminal region of NTRL is devoid of potential N-glycosylation sites. This singularity is also observed in the case of the human α2–C2 adrenergic receptor (Lomasney et al., 1990). Except for this last point, amino acids that are conserved in most members of the G-protein-coupled receptors are present at the corresponding position in NTRL (Baldwin, 1993).
NTRL shows a high degree of sequence homology with the rat and human NTRH, as illustrated in Figure 2. The global amino acid homology is 39% and 36% with the rat and human NTRH, respectively. At the level of the transmembrane domains, however, this homology can rise up to 67% in TM III and 76% in TM VII with the rat NTRH. The first and third extracellular loops also show high degrees of sequence homology with the rat NTRH (68 and 59%, respectively). By contrast, the third intracellular loop is 19 amino acids longer than and weakly homologous to that of NTRH. It is noteworthy that NTRL does not show a high degree of sequence similarity with other G-protein-coupled receptors (Probst et al., 1992). The degree of amino acid sequence homology between the mouse NTRL and the NTRHs cloned previously indicates that NTRL belongs to the NTR family but corresponds to a new type of receptor.
Fig. 2.

Alignment of the amino acid sequences between mouse, human, and rat NTR. The mouse NTRL (mNTRL) sequence is compared with the rat (rNTRH) (Tanaka et al., 1990) and the human (hNTRH) (Vita et al., 1993) NTRH. The boxed amino acid residues represent residues that are identical. The global sequence homology is 39 and 36% with the rat and the human NTRH, respectively. Positions of the putative transmembrane segments I–VII of the NTRs are indicated.
Biochemical properties of the mouse NTRL
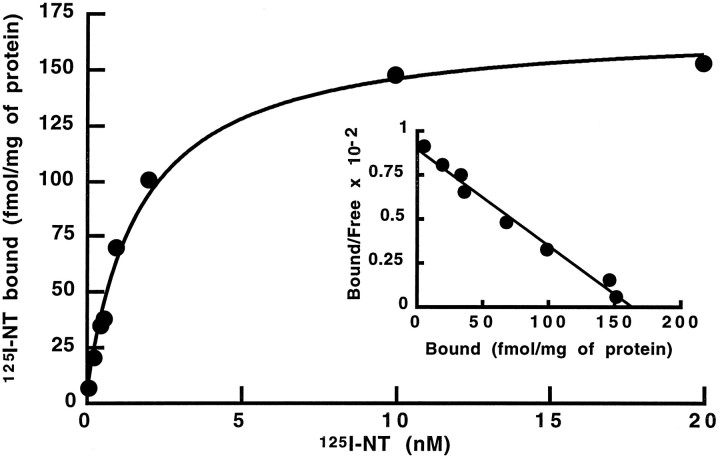
The binding properties of the mouse NTRL were characterized by expression after transient transfection of the cloned cDNA in eukaryotic COS-7 cells. 125I-Tyr3-NT bound to membranes prepared from cells transfected with the NTRL cDNA in a specific and saturable manner (Fig. 3). No binding was detected with membranes prepared from untransfected cells or cells transfected with the vector alone (data not shown). Scatchard plot analysis of 125I-Tyr3-NT binding (Fig. 3,inset) showed a single class of sites with aKd value of 2.45 ± 1.04 nm(n = 6). This value is very close to that reported for the low-affinity NT binding site in the brain of mammals (Vincent, 1995).
Fig. 3.
Saturation of specific125I-Tyr3-NT binding to membranes prepared from NTRL cDNA-transfected COS-7 cells. Experimental details are described in Materials and Methods. Results are shown from one of three independent experiments. Inset, Scatchard analysis of the data. In this experiment, Kd andBmax values were 2.2 nm and 160 fmol/mg protein, respectively.
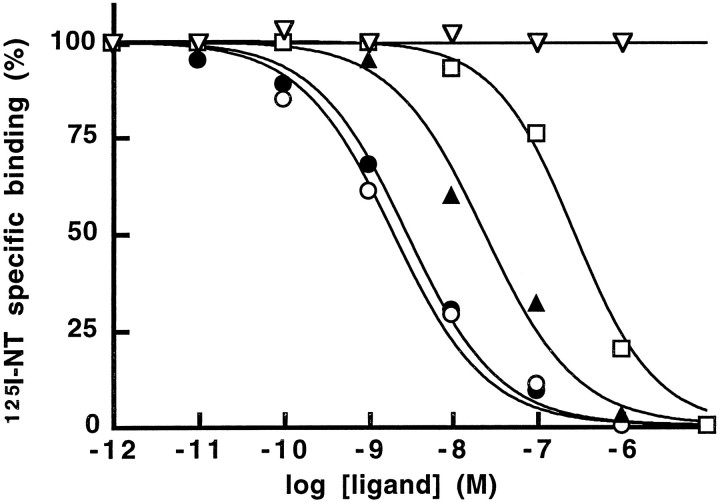
The ability of various unlabeled peptide and nonpeptide compounds to inhibit the binding of 125I-Tyr3-NT to membranes from COS-7 cells transfected with NTRL is shown in Figure4 and Table 1. With an IC50of 1–2 nm, the antihistaminic drug levocabastine was as potent as unlabeled NT in inhibiting the binding of labeled NT (Fig.4). By contrast, histamine lacked the ability to compete for binding to NTRL (Table 1). NN, Trp11-NT, and the mammalian xenopsin analog xenin (Feurle et al., 1992) had potencies comparable to that of NT, whereas d-Trp11-NT exhibited an IC50 value of 25 nm. NT (1–10) was totally devoid of NT binding activity, and dynorphin (1–13), which binds to the levocabastine-sensitive NT binding site in rat brain (Pettibone et al., 1988), inhibited 125I-Tyr3-NT binding to NTRL with an IC50 of 200–300 nm. The nonpeptide antagonist SR48692 had an IC50 value of 300 nm. We also measured the effect of cations on125I-Tyr3-NT binding and observed that Na+ and K+ inhibited the binding with IC50 values of 250 and 300 mm, respectively, whereas Mg2+ at concentrations up to 100 mm had no effect on NT binding (Table 1). Altogether, these data strongly support the assumption that the newly cloned NT receptor corresponds to the low-affinity, levocabastine-sensitive binding site described previously in rat and mouse brain.
Fig. 4.
Displacement of125I-Tyr3-NT bound to membranes prepared from NTRL cDNA-transfected COS-7 cells by NT, levocabastine,d-Trp11-NT, SR48692, and NT (1–10). Experimental details are described in Materials and Methods. The binding of 125I-Tyr3-NT was measured in the presence of increasing concentrations of NT (closed circles), levocabastine (open circles),d-Trp11-NT (closed triangles), SR48692, (open squares), and NT (1–10) (open triangles). Each point represents the mean of two separate experiments.
Table 1.
IC50 values of NT and its related compounds in competition experiments with 125I-Tyr3-NT on membranes from COS-7 cells transiently transfected with the mNTR, and inhibitory effect of NA+, K+, and Mg2+
| Compound | Structure | IC50 values (nm) |
|---|---|---|
| NT | pELYENKPRRPYIL | 2–3 |
| NN | KIPYIL | 1.5–2.5 |
| Levocabastine | Nonpeptidic | 1–2 |
| Xenin | MLTKFETKSARVKGLSFHPKRPWIL | 1.5–2 |
| Trp11-NT | pELYENKPRRPWIL | 1–1.5 |
| d-Trp11-NT | pELYENKPRRP[d-Trp]IL | 24–30 |
| SR48692 | Nonpeptidic | 200–300 |
| Dynorphin (1–13) | YGGFLRRIRPKLK | 220–300 |
| NT (1–10) | pELYENKPRRP | >10,000 |
| Histamin | Bioamine | >10,000 |
| Ions | ||
| Na+ | 250–300 | |
| K+ | 300–320 | |
| Mg2+ | >100 |
The indicated values were obtained from two independent experiments performed in triplicate.
Functional properties of the mouse NTRL
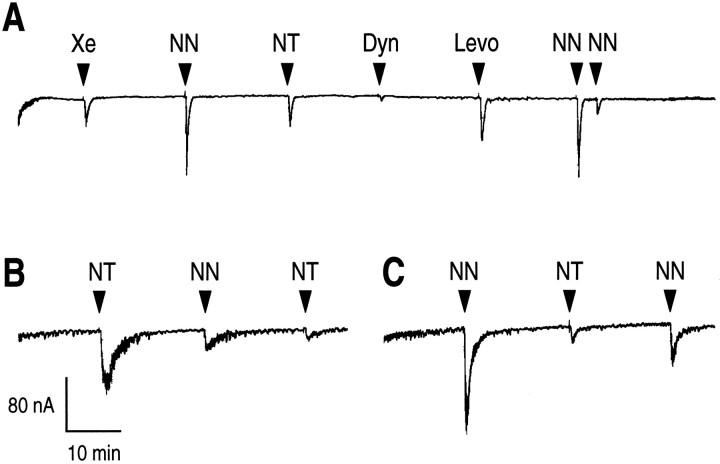
To investigate the functional properties of NTRL, we expressed this receptor type into Xenopus laevis oocytes. Figure5A shows examples of electrophysiological responses recorded after application of NT, NN, xenin, and levocabastine to oocytes injected with cRNAs encoding the receptor. The application of 10−7m xenin, NT, or NN induced a Ca2+-activated Cl− current recorded under voltage clamp at −60 mV. The response was greater for NN than for NT itself. The response to 10−7m dynorphin (1–13) was very weak, whereas levocabastine induced a current similar to that evoked by NT. The time necessary to recover the total response was rapid, because applications could be repeated every 20 min without loss of efficiency, as can be seen with the second application of NN. When NN was applied again 5 min later (third application), however, a markedly reduced response was recorded (19 ± 6% of the first response; n = 6; p = 0.007). We observed that the period of desensitization varied for each oocyte. In Figure 5B, an example is given in which a second application of NT or NN after washing for 20 min showed a reduced current amplitude, indicating a desensitization phenomenon. The same oocyte recorded after 5 hr of washing gave a better response to NN than to NT but was also desensitized. We determined that noninjected oocytes or oocytes injected with antisense mouse NTRL cRNAs were unable to respond to applications of the compounds tested above (data not shown). It is important to note that all oocytes injected with cRNAs encoding the NTRL displayed a recorded membrane potential of −23 ± 5 mV (n = 12), whereas noninjected oocytes or oocytes injected with cRNAs encoding the rat NTRH (Tanaka et al., 1990) presented a membrane potential of −55 ± 6 mV (n = 12). The Cl− equilibrium potential of −20 mV may indicate that NTRL elicits a basal Ca2+ release that activates the Ca2+-dependent Cl− current in the absence of agonist.
Fig. 5.
Current traces recorded fromXenopus oocytes injected with the in vitro synthesized NTRL mRNA. Experimental details are described in Materials and Methods. A, 10−7m xenin (Xe), neurotensin (NT), neuromedin N (NN), dynorphin (1–13) (Dyn), or levocabastine (Levo) was applied every 20 min. B, Applications of 10−7m NT, and then 10−7m NN, and again 10−7m NT at 20 min time intervals. C, The same oocyte as in B was washed for 5 hr, and then 10−7m NN, 10−7m NT, and 10−7m NN were applied every 20 min.Downward deflections indicate Ca2+-activated Cl− currents.
Tissue distribution and ontogeny in the brain of NTRL mRNA
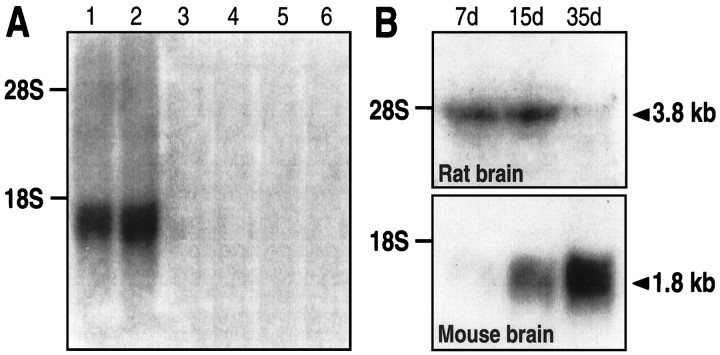
The level of expression of NTRL mRNA in brain and peripheral tissues was examined by blot hybridization analysis. As shown in Figure6A, mouse poly(A+) RNAs isolated from the brain and the cerebellum were labeled on a single band with an estimated mRNA size of ∼1.8 kilonucleotides. Note that NTRL expression was slightly higher in the cerebellum. Surprisingly, no labeling of poly(A+) RNAs isolated from large intestine, heart, testis, and liver was observed under the hybridization conditions used.
Fig. 6.
Blot hybridization analysis of poly(A+) RNAs isolated from various mouse tissues (A) and from rat and mouse brains of different ages (B). Experimental details are described in Materials and Methods. The poly(A+) RNAs used were isolated from the following tissues: mouse in A, lane 1, cerebral cortex;lane 2, cerebellum; lane 3, testis;lane 4, heart; lane 5, large intestine; and lane 6, liver; in B, whole brain of rat (top) or mouse (bottom) age 7 d (7d), 15 d (15d), and 35 d (35d). The hybridization was carried out with probes from NTRL in A and B (top) and from NTRH in B (bottom).
To identify definitely the cloned mouse NTRL as the low-affinity binding site characterized previously in rat and mouse brain, the cerebral expression of the NTRL mRNA at different ages was analyzed and compared with the cerebral expression of the rat NTRH at corresponding ages by blot hybridization. Figure 6B demonstrates that the mouse NTRL was poorly expressed in 7-d-old brain and that the expression increased at day 15 to reach a maximal level in 35-d-old brain. By contrast, the rat NTRH was expressed maximally in 7-d-old brain, and its expression decreased progressively until adulthood (35-d-old brain). These results are totally in accordance with results obtained in binding experiments with rat and mouse brain that described the transient high expression of the NTRH between 7 and 10 d after birth, whereas the NTRL appeared later and were expressed maximally in adult brain (Schotte and Laduron, 1987; Zsürger et al., 1992).
Distribution of NTRL in the mouse brain
The regional distribution of this NT receptor type was examined byin situ hybridization analysis. The labeling obtained with the antisense probe is illustrated in Figure7A–C. NTRL mRNAs were expressed in discrete regions of the mouse brain. The most important labeling was observed in the cerebellar cortex, particularly in the layer of Purkinje cells (Fig. 7A–C). The labeling was also important in the CA1–CA3 fields of the hippocampus, in the dentate gyrus, and at the level of the piriform cortex (Fig. 7B,C). A more diffuse but important labeling was also detected throughout the cerebral neocortex. Note the absence of labeling in the substantia nigra and the hypothalamic regions. A negative control obtained with the sense probe is shown in Figure 7D.
Fig. 7.
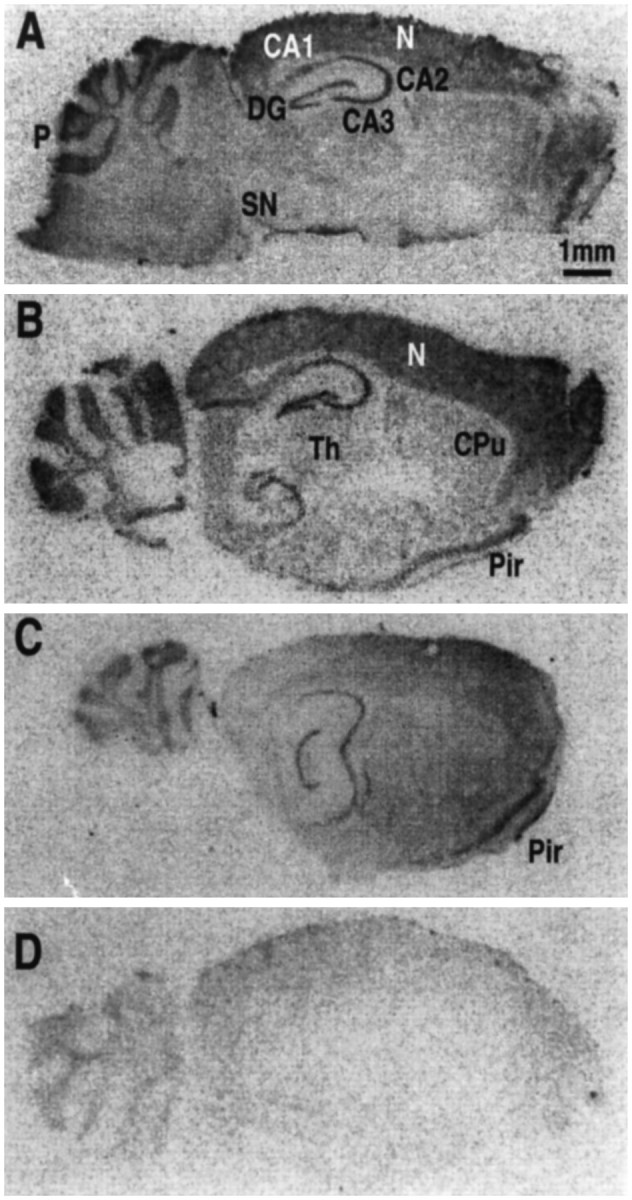
Localization of the NTRL in the mouse brain byin situ hybridization analysis. Experimental details are described in Materials and Methods. Sagittal brain slices were hybridized with the antisense (A, L1.45;B, L2.2; C, L2.45) or with the sense (D) mNTR probe. CA1, CA2, CA3, Fields of the hippocampus; CPu, caudate putamen;DG, dentate gyrus; P, Purkinje cell layer; N, neocortex; SN, substantia nigra; Th, thalamus; Pir, piriform cortex. Scale bar, 1 mm.
DISCUSSION
The present study identifies in the mouse brain a NT receptor that shares 39% and 36% homology with the previously cloned rat and human NTRH, respectively. Such a relatively low degree of homology makes it likely that the newly cloned receptor is not really the mouse counterpart of the NTRH, but rather represents a new NTR type. Actually, several lines of evidence indicate that the mouse brain receptor characterized here corresponds to the low-affinity, levocabastine-sensitive NT binding site described previously in the rat and mouse brain (Schotte et al., 1986). Indeed, the pharmacological properties described here for the mouse receptor are comparable to those reported previously for the low-affinity NT binding site. This includes a relatively low affinity (2–3 nm) of NT for the cloned NTRL, the recognition of this receptor type by levocabastine (IC50 = 1–2 nm), and the fact that the antagonist SR48692 possesses a very low affinity (IC50 = 300 nm). Furthermore, the sensitivity to cations of the NTRL as described here is similar to that reported for the low-affinity NT binding sites in rat brain (Kitabgi and Vincent, 1986).
The functional consequences of levocabastine binding to NTRL remained unknown until now. The cloning and expression of NTRL in Xenopus leavis oocytes made it possible to address this issue. Interestingly, it was found here that levocabastine behaves as an agonist of the NTRL in the oocyte expression system. It will now be necessary and of interest to investigate further the effects of levocabastine in mammalian systems that express the NTRL, whether normally or on transfection. Because no evidence of coupling to transduction mechanisms has been established for the low-affinity sites, they were considered for a long time as NT acceptor sites devoid of pharmacological function (Laduron, 1995). We now demonstrate that the NT/levocabastine receptor site is a member of the G-protein-coupled receptor family that is coupled functionally to phospholipase C in the oocyte expression system. Levocabastine could now possibly serve as a specific effector of the NTRL for investigating its central physiological roles. NN, which is produced from a common precursor with NT (Dobner et al., 1987), displays the same affinity as NT on the NTRL, whereas NN is five times less potent than NT on the NTRH (Tanaka et al., 1990). The relatively good affinity of NN for the low-affinity receptor, added to a greater ability to induce inward currents in the Xenopus laevis oocytes expressing the NTRL, could suggest that this receptor is somewhat specific for NN.
Several additional data identify this newly cloned NTRL as the low-affinity component of NT binding sites. First, the comparison of the cerebral ontogenic expression of the mouse NTRL with the cerebral expression of the NTRH is totally in accordance with the ontogenic expression of the low- and high-affinity NT binding components, respectively, determined in the rat and mouse brain using binding experiments (Schotte and Laduron, 1987; Zsürger et al., 1992). Second, the cerebral pattern of the NTRL expression, particularly in the neocortical region and the CA1 and CA2 fields of the hippocampal formation, has been evidenced previously by radioautography studies performed with 125I-Tyr3-NT in the absence or presence of levocabastine (Kitabgi et al., 1987). In the latter study, however, the detection of the levocabastine-sensitive NT binding site in CA3 field, dentate gyrus, and piriform cortex was not observed. This is probably attributable to the technique used, on the basis of the different labeling obtained with or without levocabastine. Interestingly, the presence of the low-affinity NT binding component in the cerebellum, particularly in the Purkinje cell layer, has already been observed in a radioautographic study on the ontogeny of rat brain NT receptors, but only 21 d after birth (Palacios et al., 1988). Overall, the pattern of NTRL expression is clearly distinct from that of NTRH, which is observed mainly in the suprachiasmatic nucleus, supramammillary area, substantia nigra, and ventral tegmental area (Elde et al., 1990), regions that are essentially devoid of NTRL.
Some peculiar structural characteristics of the cloned NTRL need to be emphasized. This receptor clearly belongs to the family of G-protein-coupled receptors, because it possesses seven hydrophobic domains and is coupled in the oocyte expression system; however, the N-terminal sequence of the protein is devoid of a putative N-glycosylation site. This rare property, observed only in the case of the α2–c2 adrenergic receptor (Lomasney et al., 1990), could explain the relatively low level of expression after transfection into COS-7 cells (100–150 fmol/mg). The other distinct feature corresponds to the absence in TM II of an aspartate (Asp) residue that has been implicated in the Na+ sensitivity of other G-protein-coupled receptors such as the somatostatin receptor 2a (Kong et al., 1993). The lack of Asp residue could be responsible for the low sensitivity of NTRL to Na+ ions (IC50 = 250 mm). Furthermore, this together with the extremely rich composition in Ser/Thr residues (23 and 6, respectively) of the third intracytoplasmic loop and the C terminus of the protein might explain its relatively low-affinity state for natural ligand(s). The strong desensitization of the receptor observed in Xenopus oocytes could be related to the presence of these residues rather than being a consequence of a temporary depletion of Ca2+ from intracellular stores. Indeed, the abundance of Ser/Thr residues might play a role in desensitizing the NTRL by allowing the numerous protein kinase C and casein kinase phosphorylation sites to maintain the receptor protein under a basal phosphorylation state that could desensitize the effector system. A similar mechanism has been proposed to explain the high rate of desensitization observed with the NTRH (Tanaka et al., 1990).
The differential tissue distribution of NTRL and NTRH suggests that these receptors might subserve distinct NT effects. Pharmacological data also support this hypothesis. In this context, it is worth noting that the recently developed nonpeptide antagonist SR48692 is much less potent on the new NT receptor (IC50 = 300 nm) than on the rat brain NTRH (IC50 = 5.6 nm) (Gully et al., 1993). The fact that SR48692 failed to antagonize the hypothermic and analgesic effects of intracerebroventricular injections of NT in the mouse and rat (Dubuc et al., 1994) could suggest that the newly cloned NTRL described in this work mediates these effects. Supporting this contention further is the finding that thed-Trp11-NT analog, which is one of the most potent effectors on hypothermia and analgesia, displays a relatively good affinity for the mouse NTRL (IC50 = 25 nm) by comparison with its poor affinity for the rat NTRH (IC50 = 320 nm) (Labbé-Jullié et al., 1994). Structure–activity studies with a number of peptide and pseudopeptide NT analogs have revealed clear, distinct pharmacological profiles for the analgesic and hypothermic responses on the one hand and binding to the NTRH on the other hand (Al-Rhodan et al., 1991;Labbé-Jullié et al., 1994). In particular, some analogs were shown to behave as potent agonists of the hypothermic and analgesic responses, whereas they had low affinity for the NTRH. It will be of great interest to assess the binding properties of these analogs on the NTRL. This could provide definitive evidence for a role of the NTRL in mediating the analgesic and hypothermic effects of NT and might open the way for designing potent analgesic compounds devoid of the NT effects exerted through the NTRH. It could also lead to the development of high-affinity ligands specific for the NTRL that could provide useful tools to characterize further the pharmacological properties and cerebral distribution of this new receptor.
In summary, this work demonstrates that at least two different types of functional NT receptors exist. The new NT receptor cDNA described here should help to define more accurately the different pathways leading to the expression of the various central and peripheral properties of NT.
These authors contributed equally to this work.
This work was supported by Centre National de la Recherche Scientifique. We thank Dr. Jean-Philippe Hugnot, Gisèle Jarretou, and Gilles Toumaniantz for fruitful discussion and technical comments. We also thank Franck Aguila for excellent artwork, Nicole Zsürger for her help in histology, and Dr. Patrick Kitabgi for carefully reading this manuscript.
GenBank accession number for the nucleotidic sequence of mouse NTRL:U51908.
Correspondence should be addressed to Jean Mazella, Institut de Pharmacologie Moléculaire et Cellulaire, Unité Propre de Recherche 411, Centre National de la Recherche Scientifique, 660 Route des Lucioles, Sophia Antipolis, 06560 Valbonne, France.
REFERENCES
- 1.Al-Rodhan NRF, Richelson E, Gilbert JA, McCormick DJ, Kanba KS, Pfenning MA, Nelson A, Larson EW, Yaksh TL. Structure-antinociceptive activity of neurotensin and some novel analogues in the periaqueductal gray region of the brainstem. Brain Res. 1991;557:227–235. doi: 10.1016/0006-8993(91)90139-m. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin JM. The probable arrangement of the helices in G protein-coupled receptors. EMBO J. 1993;12:1693–1703. doi: 10.1002/j.1460-2075.1993.tb05814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chabry J, Labbé-Jullié C, Gully D, Kitabgi P, Vincent JP, Mazella J. Stable expression of the cloned rat brain neurotensin receptor into fibroblasts: binding properties, photoaffinity labeling, transduction mechanisms, and internalization. J Neurochem. 1994;63:19–27. doi: 10.1046/j.1471-4159.1994.63010019.x. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.Cullen BR. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–695. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 6.Dubuc I, Costentin J, Terranova JP, Barnouin MC, Soubrié P, Le Fur G, Rostène W, Kitabgi P. The nonpeptide neurotensin antagonist, SR 48692, used as a tool to reveal putative neurotensin receptor subtypes. Br J Pharmacol. 1994;112:352–354. doi: 10.1111/j.1476-5381.1994.tb13077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobner PR, Barber DL, Villa-Komaroff L, McKierman C. Cloning and sequence analysis of cDNA from the canine neurotensin/neuromedin N precursor. Proc Natl Acad Sci USA. 1987;84:3516–3520. doi: 10.1073/pnas.84.10.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elde R, Schalling M, Ceccatelli S, Nakanishi S, Hökfelt T. Localization of neuropeptide mRNA in rat brain: initial observations using probes for neurotensin and substance P receptors. Neurosci Lett. 1990;120:134–138. doi: 10.1016/0304-3940(90)90187-e. [DOI] [PubMed] [Google Scholar]
- 9.Feurle GE, Hamscher G, Kusiek R, Meyer HE, Metzger JW. Identification of xenin, a xenopsin-related peptide, in the human gastric mucosa and its effects on exocrine pancreatic secretion. J Biol Chem. 1992;267:22305–22309. [PubMed] [Google Scholar]
- 10.Gully D, Canton M, Boigegrain R, Jeanjean F, Molimard JC, Poncelet M, Gueudet C, Heaulme M, Leyris R, Brouard A, Pelaprat D, Labbé-Jullié C, Mazella J, Soubrié P, Maffrand JP, Rostène W, Kitabgi P, Le Fur G. Biochemical and pharmacological profile of a potent and selective nonpeptide antagonist of neurotensin receptor. Proc Natl Acad Sci USA. 1993;90:65–69. doi: 10.1073/pnas.90.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermans E, Maloteaux JM, Octave JN. Phospholipase C activation by neurotensin and neuromedin N in chinese hamster ovary cells expressing the rat neurotensin receptor. Mol Brain Res. 1992;15:332–338. doi: 10.1016/0169-328x(92)90126-v. [DOI] [PubMed] [Google Scholar]
- 12.Kitabgi P, Vincent JP. Effects of cations and nucleotides on neurotensin binding to rat brain synaptic membranes. In: Moody TW, editor. Neural and endocrine peptides and receptors. Plenum; New York: 1986. pp. 313–319. [Google Scholar]
- 13.Kitabgi P, Rostène W, Dussaillant M, Schotte A, Laduron PM, Vincent JP. Two populations of neurotensin binding sites in murine brain: discrimination by the antihistamine levocabastine reveals markedly different radioautographic distribution. Eur J Pharmacol. 1987;140:285–293. doi: 10.1016/0014-2999(87)90285-8. [DOI] [PubMed] [Google Scholar]
- 14.Kong H, Raynor K, Yasuda K, Bell GI, Reisine T. Mutation of an aspartate at residue 79 in the SRIF receptor subtype SSTR2 prevents Na+ regulation of agonist binding but does not affect apparent receptor/G protein association. Mol Pharmacol. 1993;44:380–384. [PubMed] [Google Scholar]
- 15.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 17.Labbé-Jullié C, Dubuc I, Brouard A, Doulut S, Bourdel E, Pelaprat D, Mazella J, Martinez J, Rostène W, Costentin J, Kitabgi P. In vivo and in vitro structure-activity studies with peptide and pseudopeptide neurotensin analogs suggest the existence of distinct central neurotensin receptor subtypes. J Pharmacol Exp Ther. 1994;268:328–336. [PubMed] [Google Scholar]
- 18.Laduron PM. Functional consequences of retrograde axonal transport of receptor-bound neurotensin. Trends Pharmacol Sci. 1995;16:338–343. doi: 10.1016/s0165-6147(00)89067-7. [DOI] [PubMed] [Google Scholar]
- 19.Lomasney JW, Lorenz W, Allen LF, King K, Regan JW, Yang FTL, Caron MG, Lefkowitz RJ. Expansion of the alpha 2-adrenergic receptor family: characterization of a human alpha 2-adrenergic receptor gene for which is located on chromosome 2. Proc Natl Acad Sci USA. 1990;87:5094–5098. doi: 10.1073/pnas.87.13.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masu Y, Nakayama K, Tamaki H, Harada Y, Kuno M, Nakanishi S. cDNA cloning of bovine substance-K receptor through oocyte expression system. Nature. 1987;329:836–838. doi: 10.1038/329836a0. [DOI] [PubMed] [Google Scholar]
- 21.Mazella J, Poustis C, Labbé C, Checler F, Kitabgi P, Granier C, Van Rietschoten J, Vincent JP. Monoiodo Trp11-neurotensin, a highly radioactive ligand of neurotensin receptors: preparation, biological activity, and binding properties to rat brain synaptic membranes. J Biol Chem. 1983;258:3476–3481. [PubMed] [Google Scholar]
- 22.Palacios JM, Pazos A, Dietl MM, Schlumpf M, Lichtensteiger W. The ontogeny of brain neurotensin receptors studied by autoradiography. Neuroscience. 1988;25:307–317. doi: 10.1016/0306-4522(88)90028-0. [DOI] [PubMed] [Google Scholar]
- 23.Pettibone DJ, Totaro JA, Harris E, Robinson FM. Heterogeneity of [3H] neurotensin binding: studies with dynorphin, L-156,903 and levocabastine. Brain Res. 1988;457:212–218. doi: 10.1016/0006-8993(88)90688-9. [DOI] [PubMed] [Google Scholar]
- 24.Probst WC, Snyder LA, Schuster DI, Brosius J, Sealfon SC. Sequence alignment of the G-protein coupled receptor family. DNA Cell Biol. 1992;11:1–20. doi: 10.1089/dna.1992.11.1. [DOI] [PubMed] [Google Scholar]
- 25.Sadoul JL, Mazella J, Amar S, Kitabgi P, Vincent JP. Preparation of neurotensin selectively iodinated on the tyrosine 3 residue: biological activity and binding properties on mammalian neurotensin receptors. Biochem Biophys Res Commun. 1984;120:812–819. doi: 10.1016/s0006-291x(84)80179-5. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch EF, Maniatis T. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. Molecular cloning: a laboratory manual. . [Google Scholar]
- 27.Schotte A, Laduron P. Different postnatal ontogeny of two [3H] neurotensin binding sites in rat brain. Brain Res. 1987;408:326–328. doi: 10.1016/0006-8993(87)90398-2. [DOI] [PubMed] [Google Scholar]
- 28.Schotte A, Leysen JE, Laduron PM. Evidence for a displaceable non-specific [3H]neurotensin binding site in rat brain. Arch Pharmacol. 1986;333:400–405. doi: 10.1007/BF00500016. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka K, Masu M, Nakanishi S. Structure and functional expression of the cloned rat neurotensin receptor. Neuron. 1990;4:847–854. doi: 10.1016/0896-6273(90)90137-5. [DOI] [PubMed] [Google Scholar]
- 30.Vincent JP. Neurotensin receptors: binding properties, transduction pathways, and structure. Cell Mol Neurobiol. 1995;15:501–512. doi: 10.1007/BF02071313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vita N, Laurent P, Lefort S, Chalon P, Dumont X, Kaghad M, Gully D, Le Fur G, Ferrara P, Caput D. Cloning and expression of a complementary DNA encoding a high affinity human neurotensin receptor. FEBS Lett. 1993;317:139–142. doi: 10.1016/0014-5793(93)81509-x. [DOI] [PubMed] [Google Scholar]
- 32.Watson MA, Yamada M, Yamada M, Cusak B, Veverka K, Bolden-Watson C, Richelson E. The rat neurotensin receptor expressed in chinese hamster ovary cells mediates the release of inositol phosphates. J Neurochem. 1992;59:1967–1970. doi: 10.1111/j.1471-4159.1992.tb11035.x. [DOI] [PubMed] [Google Scholar]
- 33.Zsürger N, Chabry J, Coquerel A, Vincent JP. Ontogenesis and binding properties of high-affinity neurotensin receptors in human brain. Brain Res. 1992;586:303–310. doi: 10.1016/0006-8993(92)91640-z. [DOI] [PubMed] [Google Scholar]