ABSTRACT
After entering a cell, intracellular pathogens must evade destruction and generate a niche for intracellular replication. A strategy shared by multiple intracellular pathogens is the deployment of type III secretion system (T3SS)- and type IV secretion system (T4SS)-injected proteins (effectors) that subvert cellular functions. A subset of these effectors targets activities of the host cell’s endoplasmic reticulum (ER). Effectors are now appreciated to interfere with the ER in multiple ways, including capture of secretory vesicles, tethering of pathogen vacuoles to the ER, and manipulation of ER-based autophagy initiation and the unfolded-protein response. These strategies enable pathogens to generate a niche with access to cellular nutrients and to evade the host cell’s defenses.
INTRODUCTION
Multiple intracellular pathogens utilize the type III secretion system (T3SS) and type IV secretion system (T4SS) to target functions of the host cell’s endoplasmic reticulum (ER). While pathogens such as Legionella pneumophila and Brucella abortus have long been known to replicate in association with the ER (1, 2), the connection of vacuoles containing other intracellular pathogens, such as Coxiella burnetii (3, 4), Anaplasma spp. (5, 6), and Chlamydia trachomatis and its relatives (7, 8), with the ER has been recognized relatively recently. However, manipulation of ER function is not limited to pathogens that replicate within a vacuole, as cytosolic pathogens such as Orientia tsutsugamushi (9, 10) and Rickettsia rickettsii (11) also target ER-based functions via secreted effectors to promote their intracellular growth.
Recent progress in large-scale analyses of secreted proteins and in genetic analysis of previously intractable intracellular bacteria such as C. trachomatis, C. burnetii, and Ricksttsia spp. has led to an explosion in identification of new T3SS and T4SS effectors, and for some of these effectors, exciting recent advances have revealed how their interactions with host components contribute to the intracellular replication cycle of these organisms. This review focuses on recent progress in understanding how interactions with the ER mediated by secreted effectors (primarily of T4SS and T3SS) promote infection by intracellular bacteria.
THE ER: A BIOSYNTHESIS AND SIGNALING HUB OF THE CELL
The ER performs multiple functions that are critical to cellular homeostasis. Approximately one-third of the mammalian cell’s proteome is targeted to the ER, and accordingly, its best-characterized role is that of the “factory” for correct folding of proteins that ultimately function in the plasma membrane, the extracellular space, or secretory compartments such as the ER itself, the Golgi, secretory vesicles, and lysosomes. Within the ER lumen, protein folding is assisted by ER-resident chaperones, such as the Hsp70 chaperone BiP, which binds hydrophobic protein regions, thereby preventing their aggregation (reviewed in reference 12). The majority of secretory proteins are further modified by addition of glycans to asparagine residues, referred to as N-linked glycosylation. This modification increases the solubility and stability of hydrophobic proteins and promotes their cellular targeting and function (reviewed in reference 13). As protein folding proceeds, resident ER proteins and chaperones also perform quality control to ensure that misfolded or aggregated proteins do not accumulate, as they can disrupt ER function. If a protein is terminally misfolded and cannot be refolded to a functional conformation, it is targeted to the ER-associated degradation (ERAD) pathway, wherein the misfolded protein is extracted from the ER membrane to the cytosol while being tagged with polyubiquitin chains, resulting in proteosomal degradation (reviewed in reference 14).
In addition to its role in protein folding, the ER is site of lipid biosynthesis and central regulator of lipid levels throughout the cell (reviewed in reference 15). The ER produces the main phospholipids composing cellular membranes, as well as less abundant membrane components. Enzymes that synthesize cholesterol are also located in the ER. After their synthesis, these lipids are distributed from the ER to their sites of function in the cell via the secretory pathway or via membrane contact sites with other organelles (see below). Further, under conditions of excess nutrition, ER-localized enzymes synthesize triacylglycerides for energy storage within lipid droplets in the cell. Together, these ER-based functions are critical for maintaining cellular lipid homeostasis. As vacuolar pathogens replicate, their vacuole needs to expand, and thus an association with the ER could provide membrane lipids needed to enlarge the intracellular niche. Lipids produced by the ER might also provide biosynthetic material to intracellular pathogens for generation of membrane lipids or for energy (16).
Within the structure of the ER, specialized membrane domains are organized to carry out specific functions. Specific subdomains of the ER give rise to peroxisomes, organelles that sequester enzymes for β-oxidation of very-long-chain fatty acids as well as for metabolism of cholesterol, bile acids, and polyamines (17, 18). Another set of specialized ER domains are the membrane contact sites (MCS) that form between ER and other organelles in the cell, including mitochondria, the Golgi apparatus, the plasma membrane, endosomes, and peroxisomes (reviewed in reference 19). These are sites where organelles are tethered to each other via interactions between proteins in apposing membranes. The MCS between ER and mitochondria, for example, are extensive and play essential roles in mitochondrial division (20) and calcium signaling between the ER and mitochondria (21, 22). The ER proteins VAPA and VAPB tether multiple organelles in the cell to the ER via MCS, including the Golgi, endosomes, and the plasma membrane (19). Of particular interest for thinking about how pathogens could associate with the ER after uptake, it is now appreciated that endosomes associate with the ER, and these contacts become more extensive as endosomes mature; in fact, endosomes remain tightly associated with the ER throughout their trafficking (23), suggesting a potential point of contact between pathogen-containing endosomes and the ER that might be exploited by pathogens.
In response to a stimulus such as amino acid starvation, yet another specialized ER domain known as the omegasome forms, providing one of the pathways to initiate autophagy, a process in which cellular components are recycled to provide nutrition to the cell (reviewed in reference 24). The omegasome contains the protein DFCP1 and is enriched in phosphatidylinositol-3-phosphate, which is thought to increase the membrane curvature to initiate formation of the phagophore, the double membrane that is characteristic of autophagosomes (25).
BACTERIAL STRATEGIES FOR CO-OPTING ER FUNCTION
Recent work has identified how the ER functions outlined above can be subverted by intracellular bacterial pathogens to generate a replicative niche, gain nutrients for growth, or spread from cell to cell. While Brucella abortus was recently shown to replicate with the ER lumen (26), other pathogens, including Legionella pneumophila, Chamydia spp., Simkania negevensis, Anaplasma spp., and C. burnetii, reside in a vacuole that during some part of their replicative cycle is tethered to the ER via membrane contact sites between the pathogen-containing vacuole and the ER (2, 4, 6–8). Yet another group of pathogens, exemplified by R. rickettsii and Orientia tsutsugamushi, reside in the host cell’s cytosol and secrete effectors that target ER functions (11, 27). The following section reviews recent advances in our understanding of how pathogen effectors interact with the ER.
Subversion of Vesicular Trafficking Between the ER and Golgi Apparatus
Brucella abortus, a zoonotic pathogen causing abortion in ruminants and febrile infections in humans, utilizes its T4SS to replicate intracellularly in multiple cell types, with the macrophage being the best studied (reviewed in reference 28). After uptake by macrophages, B. abortus is able to avoid degradation in lysosomes (reviewed in reference 29) and replicates within the ER (26). To establish this replicative niche, B. abortus utilizes its T4SS to interact with ER exit sites, where ER-to-Golgi transport is initiated, in a manner that is dependent on the small GTPase Sar1 (30), though the effectors mediating the association with ER exit sites have not yet been identified (Fig. 1). B. abortus also requires Golgi-to-ER transport for the maintenance of its replicative niche, as the small GTPase Rab2 contributes to intracellular replication of B. abortus (31). To date, approximately 15 T4SS effectors have been shown to be translocated into infected host cells (28). Recently, the T4SS effector BspB (Table 1) was found to alter secretory trafficking from both ER to Golgi and from the Golgi to the ERGIC (ER-to-Golgi intermediate compartment) to the ER by interacting with the conserved oligomeric Golgi (COG) complex (24). This interaction between BspB and the COG complex diverts Golgi-derived vesicles to Brucella’s replicative compartment, thereby promoting its intracellular replication—possibly by providing membrane for expansion of the bacterial niche (Fig. 1). In addition to BspB, several B. abortus effectors, including BspA, BspD, BspK, and VceC, accumulate in the ER after ectopic expression (32–35), suggesting that they may perturb the early secretory pathway, but how these effectors function remains to be determined.
FIGURE 1.
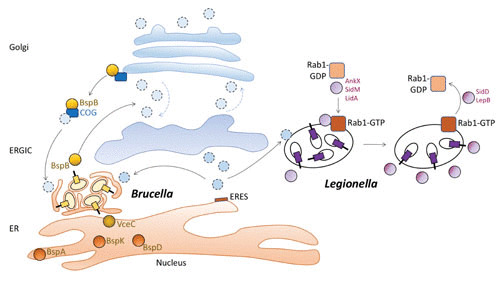
Hijacking of vesicular traffic between ER and Golgi by T4SS effectors of B. abortus (left) and L. pneumophila (right). It has been proposed that like B. abortus, L. pneumophila also intercepts Golgi-ER traffic (90). Abbreviations: ERGIC, ER-to-Golgi intermediate compartment; ERES, ER exit site; COG, conserved oligomeric Golgi complex.
TABLE 1.
Secreted pathogen effectors that localize to the ER or modulate its function
| Pathogen(secretion system) | Effector | Activity | Reference(s) |
|---|---|---|---|
| Brucella abortus(VirB T4ASS) | BspB | Impairs ER-to-Golgi secretory trafficking; interacts with the COG complex in the Golgi and redirects membrane vesicles from the Golgi to Brucella vacuole | 91 |
| VceC | Localizes to ER (ectopic expression); induces ER stress; interacts with BiP/GRP70 | 32–34 | |
| BtpA (TcpB/Btp1) | Induces ER stress; binds microtubules; inhibits TLR signaling | 75–78 | |
| BspA | Unknown; localizes to ER on ectopic expression | 35 | |
| BspD | Unknown; localizes to ER on ectopic expression | 35 | |
| BspK | Unknown; localizes to ER on ectopic expression | 35 | |
| Legionella pneumophila(Dot/Icm T4BSS) | Lgt1 | Inhibits the IRE1 pathway of the UPR; inhibits translation elongation by glucosylation of eukaryotic elongation factor 1A | 82–84 |
| Lgt2 | Inhibits the IRE1 pathway of the UPR; inhibits translation elongation by glucosylation of eukaryotic elongation factor 1A | 82, 84, 92 | |
| SidE | Localizes to the cytoplasmic face of the LCV; regulates ER tubule rearrangement and recruitment of ER markers to the LCV via modulating ubiquitination | 64, 93–95 | |
| SidC | Ubiquitin ligase and PI4P binding activity; promotes the association of LCVs with the ER by recruiting ER vesicles | 66, 68, 96 | |
| SdeA, -B, -C | Promote ER reorganization by progressive ADP-ribosylation of ubiquitin and transfer of phosphoribosyl moiety to Rtn4 | 64, 94, 95, 97 | |
| Ceg9 | Tethers the LCV to the ER via association with Rtn4 | 61 | |
| SidM/DrrA | Recruits Rab1 to LCV; acts as a GEF to recruit vacuoles to the LCV and as a GDF for Rab1; AMPylates Rab1 | 39, 41, 42, 47, 50, 98–100 | |
| LidA | Interacts with GTP-Rab1 to maintain it in the active conformation | 43, 45, 101 | |
| SidD | Catalyzes AMP release from Rab1 | 47 | |
| AnkX | Transfers phosphocholine to Rab1 | 50 | |
| RalF | Acts as a GEF to activate ARF | 37 | |
| Lem3 | Reverses activity of AnkX by removing phosphocholine from Rab1 | 52 | |
| SetA | Glycosylates Rab1 | 102, 103 | |
| LepB | Inactivates Rab1 via RabGAP activity; manipulates phosphoinositide composition of the Legionella-containing vacuole via phosphatidylinositide 4-kinase activity | 48, 104, 105 | |
| Lpg1137 | Cleaves syntaxin 17 at the mitochondrion-associated ER membrane and blocks autophagy | 106 | |
| RavZ | Delipidates Atg8 (LC3-II) at the phagophore to inhibit autophagosome formation | 70 | |
| LpSpl | Sphingosine-1-phosphate lyase disrupts host sphingolipid biosynthesis and inhibits autophagy during infection | 71 | |
| Chlamydia trachomatis(T3SS) | CT229 | C. pneumoniae homolog Cpn0585 recruits Rab1 from the ER to the inclusion membrane (effector that recruits Rab1 to C. trachomatis inclusion has yet to be identified) | 107 |
| IncD | Mediates contact with the ER at MCS via binding to ceramide transfer protein CERT | 54, 108 | |
| IncV | Tethers the C. trachomatis inclusion to the ER via interactions with VAPs | 55 | |
| MrcA | Interacts with the Ca2+ channel inositol-1,4,5-trisphosphate receptor, type 3, to promote release of bacteria from infected cells | 109 | |
| Coxiella burnetii(Dot/Icm T4BSS) | ElpA | Localizes to the ER on ectopic expression and blocks secretory traffic | 59 |
| Anaplasma spp.(VirB T4ASS) | Ats-1 | Nucleates autophagosomes by interacting with Beclin and recruitment of DFCP1 and ATG14L to the pathogen-containing vacuole | 5, 72 |
| Orientia tsutsugamushi(T1SS) | Ank4 | Interacts with eukaryotic chaperone Bat3 to transiently impede ER-associated protein degradation | 9 |
| Ank9 | Destabilizes the ER and Golgi by binding COPB2 and induces ATF4-dependent UPR | 27 | |
| Rickettsia rickettsii(Rvh T4ASS) | RARP-2 | Forms membranous structures in association with the ER; contributes to lysis of infected host cells. | 11 |
Legionella pneumophila, which naturally infects amoebae but causes opportunistic respiratory infections in humans, also uses its T4SS (called Dot/Icm) to target trafficking between the Golgi and the ER (reviewed in reference 36). Of the over 300 Dot/Icm effectors identified to date, a subset targets the function of the early secretory pathway. RalF, the first L. pneumophila T4SS effector to be identified, acts as a guanine nucleotide exchange factor (GEF) for ARF1, a small GTPase that regulates secretory membrane transport, primarily between the Golgi and ER (37). Several effectors target Rab1, the small, membrane-associated GTPase that regulates ER-to-Golgi vesicular transport. Rab1 cycles between GDP-bound (inactive) and GTP-bound (active) forms, with the assistance of multiple cellular factors (Fig. 1 and Table 1). GEFs activate Rab1 by converting GDP-Rab1 into GTP-Rab1. GTP-Rab1 then interacts with its target proteins in the membrane transport pathway to promote tethering and fusion of membrane vesicles (reviewed in reference 38). To inactivate GTP-Rab1, GTPase-activating proteins (GAPs) stimulate the GTPase activity of Rab1 to convert it to inactive GDP-Rab1. The interaction of GDP-Rab1 with membranes is regulated by GDP dissociation inhibitor (Rab-GDI), which extracts it from membranes, and by a GDI displacement factor (GDF), which targets Rab1-GDP to membranes to restart the Rab cycle. Rab1 is recruited to the Legionella-containing vacuole (LCV) via the activity of the Dot/Icm effector SidM (also known as DrrA [39]). Biochemical analysis of SidM has revealed multiple activities for modulating Rab1 activity (reviewed in references 36 and 40). A C-terminal phosphatidylinositol-4-phosphate (PI4P) binding domain of SidM mediates its association with the LCV after its secretion by the T4SS (41). The central domain of SidM has GDF/GEF activity for Rab1, which displaces Rab-GDI from Rab1-GDP and mediates GDP exchange for GTP, leading to its association with the membrane of the LCV. After recruiting Rab1 to the LCV, SidM uses its N-terminal domain, which contains adenylyltransferase activity, to covalently modify Rab1 by AMPylation (adenylylation) of a tyrosine residue. This modification of Rab1 prevents its interaction with GAPs, and as a result, Rab1 remains in its GTP-bound form and becomes constitutively active (42). Modification and recruitment of Rab1 by SidM are assisted by a second Dot/Icm effector, LidA (39, 43), which interacts with GTP-bound Rab1 (and with other Rab-GTP complexes as well [44, 45]). This manipulation of Rab1 activity enables L. pneumophila to recruit ER-derived vesicles, thereby remodeling its phagosome into a compartment supporting its replication (46).
Over the time of cellular infection by L. pneumophila, additional effectors are translocated that act in an antagonistic manner to SidM and LidA on Rab1 activity. SidD acts to de-AMPylate Rab1 (47), which restores its GTPase activity. Subsequently, LepB promotes hydrolysis of GTP by Rab1 (48), thereby enabling its extraction from the LCV by host Rab-GDI proteins. The time course of effector secretion and Rab1 modulation by L. pneumophila suggests that early during infection, recruitment and activation of Rab1 are beneficial for replication. However, prolonged activation of Rab1 may elicit cellular responses that are detrimental to intracellular infection, since ectopic expression of SidM/DrrA is cytotoxic to cells (42).
An additional covalent modification of Rab1, phosphocholination, is mediated by AnkX (49). Interestingly, while AMPylation targets a conserved tyrosine residue in Rab1, AnkX targets the adjacent serine residue. AnkX contains both ankyrin repeat domains and a FIC (filamentation induced by cyclic AMP) domain, which utilizes CDP-choline as a substrate for phosphocholination of Rab1 (50). It appears that Rab1 can only be either AMPylated or phosphocholinated at once, as only one or the other modification was identified per Rab1 molecule (50). Phosphocholination of Rab1 appears to promote its activity in a manner similar to AMPylation. Similarly to AMPylation, AnkX-mediated phosphocholination can be reversed by a second effector, Lem3 (51, 52). It was recently found that an endogenous host protein, transforming growth factor β-activated kinase (TAK1), regulates Rab1 by phosphorylation at the same site as modified by AnkX and SidM, suggesting that these T4SS effectors mimic the host’s own regulatory mechanism to co-opt Rab1 function (53).
One puzzling observation is that despite the multiple effectors that modulate Rab1 activity during Legionella infection, Rab1 itself appears to be dispensable for intracellular replication. A possible explanation for this finding is that a subset of effectors, such as SidM, AnkX, and LidA, appear to target multiple GTPases (44, 45, 50) and that these additional activities may act in parallel with perturbation of Rab1 function to promote the intracellular life cycle of L. pneumophila.
Tethering of Pathogen-Containing Vacuoles to the ER
Several intracellular pathogens, including L. pneumophila, C. burnetii (3, 4), Anaplasma spp. (5, 6), and Chlamydia spp., replicate in vacuoles that are closely associated to the ER but do not appear to fuse with it. This lifestyle is shared by Simkania negevensis, an organism related phylogenetically to Chlamydia and that naturally infects amoebae and, similar to L. pneumophila, causes opportunistic respiratory tract infection (7, 8). While the effectors mediating association with C. burnetii, Anaplasma spp., and S. negevensis with the ER remain to be identified, recent work has identified T3SS and T4SS effectors that promote association of vacuoles containing C. trachomatis and L. pneumophila with the ER (Fig. 2).
FIGURE 2.
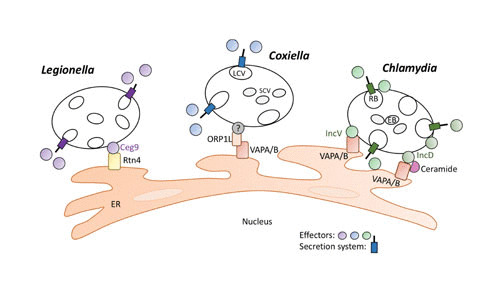
Role of pathogen effectors in tethering of pathogen-containing vacuoles to the ER. Abbreviations: Rtn4, reticulon 4; LCV, Coxiella large-cell variant; SCV, Coxiella small-cell variant; RB, Chlamydia reticulate body; EB, Chlamydia elementary body.
Chlamydia spp. are obligately intracellular pathogens that cause genital tract and ocular (C. trachomatis) or respiratory tract (C. pneumoniae and C. psittaci) infections. Both pathogens replicate within a vacuole termed an inclusion that has membrane contact sites with the ER. In C. trachomatis, the T3SS substrate IncD localizes to the inclusion membrane and mediates contact with the ER at membrane contact sites that also contain VAPA/B, the lipid transfer protein CERT, and the ER calcium sensor STIM1 (7, 54). A second T3SS effector, IncV, interacts with VAPA/B at the membrane contact sites between the inclusion and the ER (55). A C. trachomatis incV mutant exhibited decreased association of its inclusion with the ER but no overall intracellular growth defect, suggesting both the importance of IncV in ER tethering and the involvement of additional effectors in this process. Association of the chlamydial inclusion with the ER, especially with CERT, may promote acquisition of lipids to promote replication of Chlamydia either for nutrition or for expansion of the inclusion membrane during bacterial replication.
C. burnetii is a zoonotic pathogen that causes Q fever, which can manifest with both acute and chronic pathologies (56). It utilizes a T4SS related to the Legionella Dot/Icm apparatus to promote its intracellular replication (57, 58). Over 100 C. burnetii Dot/Icm substrates have been identified to date, but only a few have been characterized functionally (56). The vacuole containing C. burnetii, termed the parasitophorous vacuole, is decorated with calnexin and is tethered to the ER via membrane contacts that contain the host sterol-binding protein ORP1L, a protein that interacts with VAPA/B at ER MCS (4). While the effectors mediating this tethering remain to be identified, multiple T4SS effectors, including Cbu0372, Cbu1576, and ElpA, localize on ectopic expression to the ER (59, 60), and Cbu0635 interferes with the secretory pathway (58), suggesting that these and/or other T4SS effectors may play a role in interactions with the ER.
Recent evidence suggests tethering of vacuoles containing Legionella to the ER during the early stage of infection (Fig. 2). The Dot/Icm effector Ceg9 interacts with the ER protein reticulon 4 (Rtn4) shortly after uptake of bacteria, suggesting that recruitment of the ER helps to develop the replicative niche for L. pneumophila (61). In the host cell, Rtn4 helps generate the tubular morphology that is characteristic of the peripheral ER (62) and participates in the formation of plasma membrane-ER MCS that function in cellular Ca2+ homeostasis (63). Like Rab1, Rtn4 is targeted by multiple Dot/Icm effectors, including the SdeA to -C proteins, which modulate Rtn4 function via ubiquitination (64). Intriguingly, the Sde proteins perform a sequential set of reactions to transfer ubiquitin to Rtn4:ADP-ribosyl transfer to ubiquitin, followed by a nucleotidase/phosphohydrolase reaction that removes AMP and transfers phosphoribosylated ubiquitin to Rtn4 (64). The early targeting of Rtn4 after L. pneumophila entry to the cell and the localization of Rtn4 to plasma membrane-ER MCS raise the possibility that L. pneumophila could co-opt these MCS early during cellular infection to associate with the ER. Another Dot/Icm substrate involved in tethering the LCV to the ER is SidC, which is anchored to the cytosolic face of the LCV via binding of PI4P (65–67). Recruitment of ER proteins to the LCV by SidC requires a ubiquitin ligase activity in its N terminus (68). Taken together, these findings suggest that pathogens use multiple strategies to tether their replicative vacuoles to the ER.
Subversion of Autophagy Initiation at the ER
The L. pneumophila T4SS effector protein RavZ is secreted from the LCV and targets to omegasomes via its ability to interact with the lipid phosphatidylinositol-3-phosphate, which is enriched at these sites (69). There, the cysteine protease activity of RavZ irreversibly deconjugates lipids from ATG8 proteins (LC3-II) in the early-stage autophagosomal structures. As a result, the biogenesis of autophagosomes at the ER is inhibited (70). Since a ravZ mutant does not have a replication defect (70), it is unknown whether this activity promotes intracellular replication of L. pneumophila; however, RavZ may act in concert with other effectors, such as the inhibitor of sphingolipid biosynthesis LpSpl (71), to modulate autophagy. In contrast to L. pneumophila, Anaplasma phagocytophilum, which replicates within neutrophils, activates autophagy via a T4SS effector, Ats-1, to promote its replication (5). Ats-1 has an N-terminal domain that nucleates autophagosomes by interacting with Beclin 1, a protein crucial to initiation of autophagy, to recruit the ER-localized autophagy initiation proteins ATG14L and DFCP1 to the A. phagocytophilum inclusion (5). This subversion of autophagy initiation recruits autophagosomes to the inclusion, effectively delivering nutrients for intracellular replication of A. phagocytophilum (72).
Effector Modulation of the ER UPR
The unfolded-protein response (UPR) is a response to perturbation of ER function (broadly termed ER stress) that is initially cytoprotective and promotes return to homeostasis but can lead to apoptosis in the case of unresolved stress. The cellular response to ER stress is transmitted via three membrane sensors, IRE1α (inositol-requiring enzyme 1), ATF6 (activating transcription factor 6), and PERK (protein kinase RNA-like ER kinase). This response is linked to innate immunity via signaling through cytosolic pathways (reviewed in references 73 and 74). Secreted effectors have been identified that both induce and inhibit the UPR during intracellular infection. The B. abortus T4SS effector VceC localizes on ectopic expression to the ER, and during infection it activates IRE1α, initiating a proinflammatory arm of the UPR (32, 34) that activates NF-κB. This response could be beneficial to B. abortus in the bovine placenta, as placental inflammation in this context triggers abortion, driving transmission of the pathogen in its natural reservoir (32, 34). A second T4SS effector, BtpA (also called TcpB or Btp1) (75), triggers all three branches of the UPR during Brucella melitensis infection (76), but rather than localizing to the ER, BtpA binds microtubules (77, 78). It is not known how microtubule stabilization by BtpA links to UPR induction, but potential mechanisms could include altering interaction of integral ER membrane proteins with microtubules or effects on microtubule-dependent vesicular transport in the secretory pathway (79).
Like B. abortus, O. tsutsugamushi, the obligate intracellular agent of scrub typhus, activates the UPR (9, 27). Two T1SS-secreted effectors have been implicated (Table 1): Ank4, an ankyrin repeat protein, interacts at the cytosolic face of the ER with Bat3, a host cytosolic chaperone involved in ERAD (80), to inhibit UPR-induced ERAD during the early (nonreplicative) phase of O. tsutsugamushi infection. Later, Ank4 expression is downregulated, which releases repression of ERAD, making amino acids available for intracellular replication of O. tsutsugamushi, which provides an important source of nutrition, since this bacterium is auxotrophic for several amino acids (81). Ectopically expressed Ank9 binds the Golgi protein COPB2, involved in Golgi-to-ER vesicular trafficking, and Ank9 also traffics from Golgi to the ER, where it disrupts organelle morphology and induces the UPR. Ectopic Ank9 expression phenocopies disruption of the Golgi and ER, as well as inhibition of protein secretion, observed in cells infected with O. tsutsugamushi (27).
L. pneumophila activates the UPR at the transcriptional level (82) but suppresses the downstream translation of UPR target transcripts by translocating five T4SS effectors (Lgt1 to -3, SidI, and SidL) that inhibit translation elongation. The L. pneumophila effectors function by glycosylation of a conserved serine residue in host elongation factor 1A (83). Translation inhibition effectively reduces the basal load of protein entering the ER for protein folding, which is a physiologic activator of IRE1α (82, 84); thus, the outcome of this interaction is to inhibit the IRE1α pathway. The ability to block IRE1α signaling is shared by S. negevensis, which encodes both the T3SS and T4SS in its genome; however, the effectors that mediate this activity remain to be discovered (8). Blockade of the IRE1α pathway may be beneficial in the context of bacterial infection either to reduce innate immune signaling downstream of this pathway or to block induction of apoptosis in response to uncontrolled ER stress. Interestingly, blockade of the UPR is a strategy shared by viral pathogens, which, via their subversion of the ER for production of virions, trigger ER stress (85).
CONCLUSIONS AND PERSPECTIVE
The biosynthetic capacity of the ER and its extensive network of contacts with other cellular organelles make it a logical target for exploitation by T3SSs and T4SSs of different intracellular pathogens. Pathogens such as L. pneumophila and C. burnetii have dedicated a substantial part of their genome coding capacities to secreted effectors that modulate their host cells, which highlights the importance of these interactions to their biology (56, 86, 87). Recent progress in understanding the biology of T3SS and T4SS effectors has revealed novel mechanisms utilized by intracellular bacteria to co-opt multiple functions of the ER, including protein secretion, lipid biosynthesis, membrane tethering, and autophagy initiation, to promote their replication. While our understanding of how individual effectors modulate ER function is growing, for the majority of effectors, the molecular mechanisms of action remain unknown.
One of the challenges to identifying effector functions and understanding their roles in the context of infection has been the redundancy of effector function. However, elegant approaches have been employed in C. trachomatis and Mycobacterium tuberculosis to generate interaction networks (88, 89) for secreted effectors that, together with newly developed methodologies for genetic manipulation of the obligate intracellular pathogens, will facilitate functional and mechanistic studies of effector proteins and uncover new strategies by which they manipulate ER biology.
REFERENCES
- 1.Anderson TD, Cheville NF. 1986. Ultrastructural morphometric analysis of Brucella abortus-infected trophoblasts in experimental placentitis. Bacterial replication occurs in rough endoplasmic reticulum. Am J Pathol 124:226–237. [PMC free article] [PubMed] [Google Scholar]
- 2.Swanson MS, Isberg RR. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun 63:3609–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campoy EM, Zoppino FC, Colombo MI. 2011. The early secretory pathway contributes to the growth of the Coxiella-replicative niche. Infect Immun 79:402–413. 10.1128/IAI.00688-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Justis AV, Hansen B, Beare PA, King KB, Heinzen RA, Gilk SD. 2017. Interactions between the Coxiella burnetii parasitophorous vacuole and the endoplasmic reticulum involve the host protein ORP1L. Cell Microbiol 19:e12637. 10.1111/cmi.12637. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niu H, Xiong Q, Yamamoto A, Hayashi-Nishino M, Rikihisa Y. 2012. Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proc Natl Acad Sci U S A 109:20800–20807. 10.1073/pnas.1218674109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Truchan HK, Cockburn CL, Hebert KS, Magunda F, Noh SM, Carlyon JA. 2016. The pathogen-occupied vacuoles of Anaplasma phagocytophilum and Anaplasma marginale interact with the endoplasmic reticulum. Front Cell Infect Microbiol 6:22. 10.3389/fcimb.2016.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derré I. 2015. Chlamydiae interaction with the endoplasmic reticulum: contact, function and consequences. Cell Microbiol 17:959–966. 10.1111/cmi.12455. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehlitz A, Karunakaran K, Herweg JA, Krohne G, van de Linde S, Rieck E, Sauer M, Rudel T. 2014. The chlamydial organism Simkania negevensis forms ER vacuole contact sites and inhibits ER-stress. Cell Microbiol 16:1224–1243. 10.1111/cmi.12278. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Rodino KG, VieBrock L, Evans SM, Ge H, Richards AL, Carlyon JA. 2017. Orientia tsutsugamushi modulates endoplasmic reticulum-associated degradation to benefit its growth. Infect Immun 86:e00596-17. 10.1128/IAI.00596-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.VieBrock L, Evans SM, Beyer AR, Larson CL, Beare PA, Ge H, Singh S, Rodino KG, Heinzen RA, Richards AL, Carlyon JA. 2015. Orientia tsutsugamushi ankyrin repeat-containing protein family members are type 1 secretion system substrates that traffic to the host cell endoplasmic reticulum. Front Cell Infect Microbiol 4:186. 10.3389/fcimb.2014.00186. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehman SS, Noriea NF, Aistleitner K, Clark TR, Dooley CA, Nair V, Kaur SJ, Rahman MS, Gillespie JJ, Azad AF, Hackstadt T. 2018. The rickettsial ankyrin repeat protein 2 is a type IV secreted effector that associates with the endoplasmic reticulum. mBio 9:e00975-18. 10.1128/mBio.00975-18. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCaffrey K, Braakman I. 2016. Protein quality control at the endoplasmic reticulum. Essays Biochem 60:227–235. 10.1042/EBC20160003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Helenius A, Aebi M. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem 73:1019–1049. 10.1146/annurev.biochem.73.011303.073752. [PubMed] [DOI] [PubMed] [Google Scholar]
- 14.Preston GM, Brodsky JL. 2017. The evolving role of ubiquitin modification in endoplasmic reticulum-associated degradation. Biochem J 474:445–469. 10.1042/BCJ20160582. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacquemyn J, Cascalho A, Goodchild RE. 2017. The ins and outs of endoplasmic reticulum-controlled lipid biosynthesis. EMBO Rep 18:1905–1921. 10.15252/embr.201643426. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toledo A, Benach JL. 2015. Hijacking and use of host lipids by intracellular pathogens. Microbiol Spectr 3:VMBF-0001-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. 2006. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol 173:521–532. 10.1083/jcb.200601036. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joshi AS, Zhang H, Prinz WA. 2017. Organelle biogenesis in the endoplasmic reticulum. Nat Cell Biol 19:876–882. 10.1038/ncb3579. [PubMed] [DOI] [PubMed] [Google Scholar]
- 19.Wu H, Carvalho P, Voeltz GK. 2018. Here, there, and everywhere: the importance of ER membrane contact sites. Science 361:eaan5835. 10.1126/science.aan5835. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. 2011. ER tubules mark sites of mitochondrial division. Science 334:358–362. 10.1126/science.1207385. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cárdenas C, Miller RA, Smith I, Bui T, Molgó J, Müller M, Vais H, Cheung KH, Yang J, Parker I, Thompson CB, Birnbaum MJ, Hallows KR, Foskett JK. 2010. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell 142:270–283. 10.1016/j.cell.2010.06.007. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280:1763–1766. 10.1126/science.280.5370.1763. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Friedman JR, Dibenedetto JR, West M, Rowland AA, Voeltz GK. 2013. Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol Biol Cell 24:1030–1040. 10.1091/mbc.e12-10-0733. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller C, Celli J. 2016. Avoidance and subversion of eukaryotic homeostatic autophagy mechanisms by bacterial pathogens. J Mol Biol 428:3387–3398. 10.1016/j.jmb.2016.07.007. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182:685–701. 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sedzicki J, Tschon T, Low SH, Willemart K, Goldie KN, Letesson JJ, Stahlberg H, Dehio C. 2018. 3D correlative electron microscopy reveals continuity of Brucella-containing vacuoles with the endoplasmic reticulum. J Cell Sci 131:jcs210799. 10.1242/jcs.210799. [PubMed] [DOI] [PubMed] [Google Scholar]
- 27.Beyer AR, Rodino KG, VieBrock L, Green RS, Tegels BK, Oliver LD, Jr, Marconi RT, Carlyon JA. 2017. Orientia tsutsugamushi Ank9 is a multifunctional effector that utilizes a novel GRIP-like Golgi localization domain for Golgi-to-endoplasmic reticulum trafficking and interacts with host COPB2. Cell Microbiol 19:e12727. 10.1111/cmi.12727. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dehio C, Tsolis RM. 2017. Type IV effector secretion and subversion of host functions by Bartonella and Brucella species. Curr Top Microbiol Immunol 413:269–295. 10.1007/978-3-319-75241-9_11. [PubMed] [DOI] [PubMed] [Google Scholar]
- 29.Celli J. 2015. The changing nature of the Brucella-containing vacuole. Cell Microbiol 17:951–958. 10.1111/cmi.12452. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Celli J, Salcedo SP, Gorvel JP. 2005. Brucella coopts the small GTPase Sar1 for intracellular replication. Proc Natl Acad Sci U S A 102:1673–1678. 10.1073/pnas.0406873102. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fugier E, Salcedo SP, de Chastellier C, Pophillat M, Muller A, Arce-Gorvel V, Fourquet P, Gorvel JP. 2009. The glyceraldehyde-3-phosphate dehydrogenase and the small GTPase Rab 2 are crucial for Brucella replication. PLoS Pathog 5:e1000487. 10.1371/journal.ppat.1000487. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Jong MF, Starr T, Winter MG, den Hartigh AB, Child R, Knodler LA, van Dijl JM, Celli J, Tsolis RM. 2013. Sensing of bacterial type IV secretion via the unfolded protein response. mBio 4:e00418-12. 10.1128/mBio.00418-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Jong MF, Sun YH, den Hartigh AB, van Dijl JM, Tsolis RM. 2008. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol Microbiol 70:1378–1396. 10.1111/j.1365-2958.2008.06487.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keestra-Gounder AM, Byndloss MX, Seyffert N, Young BM, Chávez-Arroyo A, Tsai AY, Cevallos SA, Winter MG, Pham OH, Tiffany CR, de Jong MF, Kerrinnes T, Ravindran R, Luciw PA, McSorley SJ, Bäumler AJ, Tsolis RM. 2016. NOD1 and NOD2 signalling links ER stress with inflammation. Nature 532:394–397. 10.1038/nature17631. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myeni S, Child R, Ng TW, Kupko JJ III, Wehrly TD, Porcella SF, Knodler LA, Celli J. 2013. Brucella modulates secretory trafficking via multiple type IV secretion effector proteins. PLoS Pathog 9:e1003556. 10.1371/journal.ppat.1003556. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherwood RK, Roy CR. 2016. Autophagy evasion and endoplasmic reticulum subversion: the yin and yang of Legionella intracellular infection. Annu Rev Microbiol 70:413–433. 10.1146/annurev-micro-102215-095557. [PubMed] [DOI] [PubMed] [Google Scholar]
- 37.Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679–682. 10.1126/science.1067025. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10:513–525. 10.1038/nrm2728. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Machner MP, Isberg RR. 2006. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell 11:47–56. 10.1016/j.devcel.2006.05.013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 40.Neunuebel MR, Machner MP. 2012. The taming of a Rab GTPase by Legionella pneumophila. Small GTPases 3:28–33. 10.4161/sgtp.18704. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brombacher E, Urwyler S, Ragaz C, Weber SS, Kami K, Overduin M, Hilbi H. 2009. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem 284:4846–4856. 10.1074/jbc.M807505200. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller MP, Peters H, Blümer J, Blankenfeldt W, Goody RS, Itzen A. 2010. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329:946–949. 10.1126/science.1192276. [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Derré I, Isberg RR. 2005. LidA, a translocated substrate of the Legionella pneumophila type IV secretion system, interferes with the early secretory pathway. Infect Immun 73:4370–4380. 10.1128/IAI.73.7.4370-4380.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.So EC, Schroeder GN, Carson D, Mattheis C, Mousnier A, Broncel M, Tate EW, Frankel G. 2016. The Rab-binding profiles of bacterial virulence factors during infection. J Biol Chem 291:5832–5843. 10.1074/jbc.M115.700930. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schoebel S, Cichy AL, Goody RS, Itzen A. 2011. Protein LidA from Legionella is a Rab GTPase supereffector. Proc Natl Acad Sci U S A 108:17945–17950. 10.1073/pnas.1113133108. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arasaki K, Toomre DK, Roy CR. 2012. The Legionella pneumophila effector DrrA is sufficient to stimulate SNARE-dependent membrane fusion. Cell Host Microbe 11:46–57. 10.1016/j.chom.2011.11.009. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neunuebel MR, Chen Y, Gaspar AH, Backlund PS Jr, Yergey A, Machner MP. 2011. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333:453–456. 10.1126/science.1207193. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ingmundson A, Delprato A, Lambright DG, Roy CR. 2007. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450:365–369. 10.1038/nature06336. [PubMed] [DOI] [PubMed] [Google Scholar]
- 49.Pan X, Lührmann A, Satoh A, Laskowski-Arce MA, Roy CR. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651–1654. 10.1126/science.1158160. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukherjee S, Liu X, Arasaki K, McDonough J, Galán JE, Roy CR. 2011. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477:103–106. 10.1038/nature10335. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goody PR, Heller K, Oesterlin LK, Müller MP, Itzen A, Goody RS. 2012. Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J 31:1774–1784. 10.1038/emboj.2012.16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan Y, Arnold RJ, Luo ZQ. 2011. Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc Natl Acad Sci U S A 108:21212–21217. 10.1073/pnas.1114023109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levin RS, Hertz NT, Burlingame AL, Shokat KM, Mukherjee S. 2016. Innate immunity kinase TAK1 phosphorylates Rab1 on a hotspot for posttranslational modifications by host and pathogen. Proc Natl Acad Sci U S A 113:E4776–E4783. 10.1073/pnas.1608355113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Derré I, Swiss R, Agaisse H. 2011. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog 7:e1002092. 10.1371/journal.ppat.1002092. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stanhope R, Flora E, Bayne C, Derré I. 2017. IncV, a FFAT motif-containing Chlamydia protein, tethers the endoplasmic reticulum to the pathogen-containing vacuole. Proc Natl Acad Sci U S A 114:12039–12044. 10.1073/pnas.1709060114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Schaik EJ, Chen C, Mertens K, Weber MM, Samuel JE. 2013. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat Rev Microbiol 11:561–573. 10.1038/nrmicro3049. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, Cockrell DC, Howe D, Voth DE, Heinzen RA. 2011. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2:e00175-11. 10.1128/mBio.00175-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carey KL, Newton HJ, Lührmann A, Roy CR. 2011. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog 7:e1002056. 10.1371/journal.ppat.1002056. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Graham JG, Winchell CG, Sharma UM, Voth DE. 2015. Identification of ElpA, a Coxiella burnetii pathotype-specific Dot/Icm type IV secretion system substrate. Infect Immun 83:1190–1198. 10.1128/IAI.02855-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weber MM, Chen C, Rowin K, Mertens K, Galvan G, Zhi H, Dealing CM, Roman VA, Banga S, Tan Y, Luo ZQ, Samuel JE. 2013. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol 195:3914–3924. 10.1128/JB.00071-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haenssler E, Ramabhadran V, Murphy CS, Heidtman MI, Isberg RR. 2015. Endoplasmic reticulum tubule protein reticulon 4 associates with the Legionella pneumophila vacuole and with translocated substrate Ceg9. Infect Immun 83:3479–3489. 10.1128/IAI.00507-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124:573–586. 10.1016/j.cell.2005.11.047. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Jozsef L, Tashiro K, Kuo A, Park EJ, Skoura A, Albinsson S, Rivera-Molina F, Harrison KD, Iwakiri Y, Toomre D, Sessa WC. 2014. Reticulon 4 is necessary for endoplasmic reticulum tubulation, STIM1-Orai1 coupling, and store-operated calcium entry. J Biol Chem 289:9380–9395. 10.1074/jbc.M114.548602. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kotewicz KM, Ramabhadran V, Sjoblom N, Vogel JP, Haenssler E, Zhang M, Behringer J, Scheck RA, Isberg RR. 2017. A single Legionella effector catalyzes a multistep ubiquitination pathway to rearrange tubular endoplasmic reticulum for replication. Cell Host Microbe 21:169–181. 10.1016/j.chom.2016.12.007. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luo ZQ, Isberg RR. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A 101:841–846. 10.1073/pnas.0304916101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ragaz C, Pietsch H, Urwyler S, Tiaden A, Weber SS, Hilbi H. 2008. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol 10:2416–2433. 10.1111/j.1462-5822.2008.01219.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 67.Weber SS, Ragaz C, Reus K, Nyfeler Y, Hilbi H. 2006. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog 2:e46. 10.1371/journal.ppat.0020046. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsu F, Luo X, Qiu J, Teng YB, Jin J, Smolka MB, Luo ZQ, Mao Y. 2014. The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling. Proc Natl Acad Sci U S A 111:10538–10543. 10.1073/pnas.1402605111. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horenkamp FA, Kauffman KJ, Kohler LJ, Sherwood RK, Krueger KP, Shteyn V, Roy CR, Melia TJ, Reinisch KM. 2015. The Legionella anti-autophagy effector RavZ targets the autophagosome via PI3P- and curvature-sensing motifs. Dev Cell 34:569–576. 10.1016/j.devcel.2015.08.010. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choy A, Dancourt J, Mugo B, O’Connor TJ, Isberg RR, Melia TJ, Roy CR. 2012. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338:1072–1076. 10.1126/science.1227026. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rolando M, Escoll P, Nora T, Botti J, Boitez V, Bedia C, Daniels C, Abraham G, Stogios PJ, Skarina T, Christophe C, Dervins-Ravault D, Cazalet C, Hilbi H, Rupasinghe TW, Tull D, McConville MJ, Ong SY, Hartland EL, Codogno P, Levade T, Naderer T, Savchenko A, Buchrieser C. 2016. Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy. Proc Natl Acad Sci U S A 113:1901–1906. 10.1073/pnas.1522067113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rikihisa Y. 2017. Role and function of the type IV secretion system in Anaplasma and Ehrlichia species. Curr Top Microbiol Immunol 413:297–321. 10.1007/978-3-319-75241-9_12. [PubMed] [DOI] [PubMed] [Google Scholar]
- 73.Byndloss MX, Keestra-Gounder AM, Bäumler AJ, Tsolis RM. 2016. NOD1 and NOD2: new functions linking endoplasmic reticulum stress and inflammation. DNA Cell Biol 35:311–313. 10.1089/dna.2016.3396. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Celli J, Tsolis RM. 2015. Bacteria, the endoplasmic reticulum and the unfolded protein response: friends or foes? Nat Rev Microbiol 13:71–82. 10.1038/nrmicro3393. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salcedo SP, Marchesini MI, Lelouard H, Fugier E, Jolly G, Balor S, Muller A, Lapaque N, Demaria O, Alexopoulou L, Comerci DJ, Ugalde RA, Pierre P, Gorvel JP. 2008. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog 4:e21. 10.1371/journal.ppat.0040021. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith JA, Khan M, Magnani DD, Harms JS, Durward M, Radhakrishnan GK, Liu YP, Splitter GA. 2013. Brucella induces an unfolded protein response via TcpB that supports intracellular replication in macrophages. PLoS Pathog 9:e1003785. 10.1371/journal.ppat.1003785. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Felix C, Kaplan Türköz B, Ranaldi S, Koelblen T, Terradot L, O’Callaghan D, Vergunst AC. 2014. The Brucella TIR domain containing proteins BtpA and BtpB have a structural WxxxE motif important for protection against microtubule depolymerisation. Cell Commun Signal 12:53. 10.1186/s12964-014-0053-y. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Radhakrishnan GK, Harms JS, Splitter GA. 2011. Modulation of microtubule dynamics by a TIR domain protein from the intracellular pathogen Brucella melitensis. Biochem J 439:79–83. 10.1042/BJ20110577. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gurel PS, Hatch AL, Higgs HN. 2014. Connecting the cytoskeleton to the endoplasmic reticulum and Golgi. Curr Biol 24:R660–R672. 10.1016/j.cub.2014.05.033. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Claessen JH, Ploegh HL. 2011. BAT3 guides misfolded glycoproteins out of the endoplasmic reticulum. PLoS One 6:e28542. 10.1371/journal.pone.0028542. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Min CK, Yang JS, Kim S, Choi MS, Kim IS, Cho NH. 2008. Genome-based construction of the metabolic pathways of Orientia tsutsugamushi and comparative analysis within the Rickettsiales order. Comp Funct Genomics 2008:623145. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Treacy-Abarca S, Mukherjee S. 2015. Legionella suppresses the host unfolded protein response via multiple mechanisms. Nat Commun 6:7887. 10.1038/ncomms8887. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Belyi Y, Niggeweg R, Opitz B, Vogelsgesang M, Hippenstiel S, Wilm M, Aktories K. 2006. Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A. Proc Natl Acad Sci U S A 103:16953–16958. 10.1073/pnas.0601562103. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hempstead AD, Isberg RR. 2015. Inhibition of host cell translation elongation by Legionella pneumophila blocks the host cell unfolded protein response. Proc Natl Acad Sci U S A 112:E6790–E6797. 10.1073/pnas.1508716112. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith JA. 2014. A new paradigm: innate immune sensing of viruses via the unfolded protein response. Front Microbiol 5:222. 10.3389/fmicb.2014.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gomez-Valero L, Rusniok C, Carson D, Mondino S, Pérez-Cobas AE, Rolando M, Pasricha S, Reuter S, Demirtas J, Crumbach J, Descorps-Declere S, Hartland EL, Jarraud S, Dougan G, Schroeder GN, Frankel G, Buchrieser C. 2019. More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells. Proc Natl Acad Sci U S A 116:2265–2273. 10.1073/pnas.1808016116. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ensminger AW. 2016. Legionella pneumophila, armed to the hilt: justifying the largest arsenal of effectors in the bacterial world. Curr Opin Microbiol 29:74–80. 10.1016/j.mib.2015.11.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 88.Mirrashidi KM, Elwell CA, Verschueren E, Johnson JR, Frando A, Von Dollen J, Rosenberg O, Gulbahce N, Jang G, Johnson T, Jäger S, Gopalakrishnan AM, Sherry J, Dunn JD, Olive A, Penn B, Shales M, Cox JS, Starnbach MN, Derre I, Valdivia R, Krogan NJ, Engel J. 2015. Global mapping of the Inc-human interactome reveals that retromer restricts Chlamydia infection. Cell Host Microbe 18:109–121. 10.1016/j.chom.2015.06.004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Penn BH, Netter Z, Johnson JR, Von Dollen J, Jang GM, Johnson T, Ohol YM, Maher C, Bell SL, Geiger K, Golovkine G, Du X, Choi A, Parry T, Mohapatra BC, Storck MD, Band H, Chen C, Jager S, Shales M, Portnoy DA, Hernandez R, Coscoy L, Cox JS, Krogan NJ. 2018. An Mtb-human protein-protein interaction map identifies a switch between host antiviral and antibacterial responses. Mol Cell 71:637–648.e635. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bärlocher K, Welin A, Hilbi H. 2017. Formation of the Legionella replicative compartment at the crossroads of retrograde trafficking. Front Cell Infect Microbiol 7:482. 10.3389/fcimb.2017.00482. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miller CN, Smith EP, Cundiff JA, Knodler LA, Bailey Blackburn J, Lupashin V, Celli J. 2017. A Brucella type IV effector targets the COG tethering complex to remodel host secretory traffic and promote intracellular replication. Cell Host Microbe 22:317–329.e317. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Belyi Y, Tabakova I, Stahl M, Aktories K. 2008. Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J Bacteriol 190:3026–3035. 10.1128/JB.01798-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jeong KC, Sexton JA, Vogel JP. 2015. Spatiotemporal regulation of a Legionella pneumophila T4SS substrate by the metaeffector SidJ. PLoS Pathog 11:e1004695. 10.1371/journal.ppat.1004695. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qiu J, Sheedlo MJ, Yu K, Tan Y, Nakayasu ES, Das C, Liu X, Luo ZQ. 2016. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533:120–124. 10.1038/nature17657. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sheedlo MJ, Qiu J, Tan Y, Paul LN, Luo ZQ, Das C. 2015. Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination. Proc Natl Acad Sci U S A 112:15090–15095. 10.1073/pnas.1514568112. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weber S, Wagner M, Hilbi H. 2014. Live-cell imaging of phosphoinositide dynamics and membrane architecture during Legionella infection. mBio 5:e00839-13. 10.1128/mBio.00839-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bardill JP, Miller JL, Vogel JP. 2005. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol 56:90–103. 10.1111/j.1365-2958.2005.04539.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 98.Hubber A, Arasaki K, Nakatsu F, Hardiman C, Lambright D, De Camilli P, Nagai H, Roy CR. 2014. The machinery at endoplasmic reticulum-plasma membrane contact sites contributes to spatial regulation of multiple Legionella effector proteins. PLoS Pathog 10:e1004222. 10.1371/journal.ppat.1004222. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Machner MP, Isberg RR. 2007. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318:974–977. 10.1126/science.1149121. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. 2006. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol 8:971–977. 10.1038/ncb1463. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Neunuebel MR, Mohammadi S, Jarnik M, Machner MP. 2012. Legionella pneumophila LidA affects nucleotide binding and activity of the host GTPase Rab1. J Bacteriol 194:1389–1400. 10.1128/JB.06306-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Z, McCloskey A, Cheng S, Wu M, Xue C, Yu Z, Fu J, Liu Y, Luo ZQ, Liu X. 2018. Regulation of the small GTPase Rab1 function by a bacterial glucosyltransferase. Cell Discov 4:53. 10.1038/s41421-018-0055-9. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Heidtman M, Chen EJ, Moy MY, Isberg RR. 2009. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol 11:230–248. 10.1111/j.1462-5822.2008.01249.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen J, Reyes M, Clarke M, Shuman HA. 2007. Host cell-dependent secretion and translocation of the LepA and LepB effectors of Legionella pneumophila. Cell Microbiol 9:1660–1671. 10.1111/j.1462-5822.2007.00899.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 105.Dong N, Niu M, Hu L, Yao Q, Zhou R, Shao F. 2016. Modulation of membrane phosphoinositide dynamics by the phosphatidylinositide 4-kinase activity of the Legionella LepB effector. Nat Microbiol 2:16236. 10.1038/nmicrobiol.2016.236. [PubMed] [DOI] [PubMed] [Google Scholar]
- 106.Arasaki K, Mikami Y, Shames SR, Inoue H, Wakana Y, Tagaya M. 2017. Legionella effector Lpg1137 shuts down ER-mitochondria communication through cleavage of syntaxin 17. Nat Commun 8:15406. 10.1038/ncomms15406. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cortes C, Rzomp KA, Tvinnereim A, Scidmore MA, Wizel B. 2007. Chlamydia pneumoniae inclusion membrane protein Cpn0585 interacts with multiple Rab GTPases. Infect Immun 75:5586–5596. 10.1128/IAI.01020-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Agaisse H, Derré I. 2014. Expression of the effector protein IncD in Chlamydia trachomatis mediates recruitment of the lipid transfer protein CERT and the endoplasmic reticulum-resident protein VAPB to the inclusion membrane. Infect Immun 82:2037–2047. 10.1128/IAI.01530-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nguyen PH, Lutter EI, Hackstadt T. 2018. Chlamydia trachomatis inclusion membrane protein MrcA interacts with the inositol 1,4,5-trisphosphate receptor type 3 (ITPR3) to regulate extrusion formation. PLoS Pathog 14:e1006911. 10.1371/journal.ppat.1006911. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


