Abstract
Time-lapse fluorescence confocal microscopy was used to directly visualize the formation and dynamics of postsynaptic target structures (i.e., dendritic branches and spines) on pyramidal neurons within developing hippocampal tissue slices. Within a 2 week period of time, pyramidal neurons in cultured slices derived from early postnatal rat (postnatal days 2–7) developed complex dendritic arbors bearing numerous postsynaptic spines. At early stages (1–2 d in vitro), many fine filopodial protrusions on dendrite shafts rapidly extended (maximum rate ∼2.5 μm/min) and retracted (median filopodial lifetime, 10 min), but some filopodia transformed into growth cones and nascent dendrite branches. As dendritic arbors matured, the population of fleeting lateral filopodia was replaced by spine-like structures having a low rate of turnover. This developmental progression involved a transitional stage in which dendrites were dominated by persistent (up to 22 hr) but dynamic spiny protrusions (i.e., protospines) that showed substantial changes in length and shape on a timescale of minutes. These observations reveal a highly dynamic state of postsynaptic target structures that may actively contribute to the formation and plasticity of synaptic connections during CNS development.
Keywords: dendrite, dendritic spine, development, pyramidal cell, hippocampal slice, DiI, confocal imaging, time-lapse
Establishment of axodendritic synaptic connections within the mammalian CNS necessarily involves growth and contact of afferent axons and target cell dendrites. Growing axons often must traverse long distances and complex pathways to arrive at appropriate target regions, where individual axons subsequently engage a large number of potential target cells. With the advent of vital fluorescence staining and time-lapse imaging methodologies, the dynamics of axon growth and guidance in situ has been investigated intensively (Harris et al., 1987; O’Rourke and Fraser, 1990; Kaethner and Stuermer, 1992; Mason and Godement, 1992; Chien et al., 1993;Sretavan and Reichardt, 1993; Dailey et al., 1994; Halloran and Kalil, 1994; O’Rourke et al., 1994). Such studies have greatly elucidated the dynamic behavior of axons, and especially their growing terminals (i.e., growth cones), as they extend toward and grow into their target regions. A common theme is that growing axons explore their pathways and target regions by elaborating many dynamic, often transient protrusive filopodia and branchlets. Such highly protrusive behavior appears to be essential to the axon’s ability to pathfind and locate appropriate target cells (for review, see Bentley and O’Connor, 1994;Kater and Rehder, 1995).
Much less is known about the dynamics of postsynaptic target structures during developmental periods of targeting and synaptogenesis. Based on analysis of static images of fixed tissue taken from various regions in the nervous system, it is clear that target dendrites undergo a rather stereotypical sequence of morphological maturation concomitant with axon ingrowth and synaptogenesis (Morest, 1969a,b; Minkwitz and Holz, 1975; Minkwitz, 1976; Berry and Bradley, 1976a,b; Lund et al., 1977;Miller and Peters, 1981; Pokorny and Yamamoto, 1981; Phelps et al., 1983; Ramoa et al., 1987; Ulfhake and Cullheim, 1988). This dendritic maturation typically involves appearance of growth cones and fine lateral filopodia on dendrite shafts, coinciding with the elaboration of new dendritic branches. Subsequently, spine-like protrusions appear at virtually all sites of excitatory synaptic contact on dendrites. As development proceeds, stage-dependent differences in the morphology of dendritic spine-like protrusions are often observed (Purpura, 1975;Harris et al., 1989; Papa et al., 1995). Although these studies indicate that the structure of target dendrites evolves during developmental periods of synaptogenesis, the precise structural changes underlying the formation and plasticity of individual postsynaptic elements, especially the specialized postsynaptic spines, have remained poorly defined.
To elucidate in more detail the dynamics of dendrite growth and differentiation as they occur within a CNS tissue environment, we developed means for direct time-lapse visualization of growing neuronal dendrites in live, developing brain slice preparations. We examined hippocampal tissue derived from early postnatal rat at a stage when dendritic arbors of pyramidal cells are increasing in complexity and are studded with lateral filopodia and growth cones, but still lack classical dendritic spines (Minkwitz and Holz, 1975; Minkwitz, 1976). This stage is also characterized by a high rate of synaptogenesis (Bliss et al., 1974; Amaral and Dent, 1981; Buchs et al., 1993). We followed the progression of changes in dendrite structure and dynamics over a 2 week period of time as synaptic connections were forming with dendrites in developing brain slices in vitro. The development of dendritic structure was found to involve dynamic activities that varied with the state of differentiation. At early developmental stages, many filopodia and protospines actively protruded from dendrite shafts into the surrounding tissue. Postsynaptic spine-like structures that appeared subsequently were more stable, but there was nevertheless a low level of spine turnover. It seems likely that the active protrusion of dendritic filopodia and protospines during early synaptogenic periods may help initiate synaptic contacts, and that turnover of relatively stable dendritic spines at later stages may contribute to changes in the number or function of synaptic connections.
MATERIALS AND METHODS
Preparation and labeling of tissue slices
Hippocampal tissue slices were prepared from postnatal day 2 (P2) to P7 rat pups and cultured briefly (1–2 d), or slices derived from P5–P7 animals were cultured for up to 2 weeks, as described previously (Gähwiler, 1984; Dailey et al., 1994). We confined our observations to developing tissue slices prepared from neonates (P7 and younger animals) because (1) hippocampal slices derived from older animals do not remain viable for long periods of time under suchin vitro conditions (Gähwiler, 1988), and (2) at later developmental stages, the tissue isolation procedure is likely to significantly perturb target cells by deafferenting (i.e., removing axonal inputs to) dendrites and thereby altering the dynamics of postsynaptic structures. It is well established that axons of intrinsic neurons continue to develop and form synaptic contacts with pyramidal cell dendrites when hippocampal slices are derived from neonates (LaVail and Wolf, 1973; Gähwiler, 1984; Buchs et al., 1993;Muller et al., 1993; Dailey et al., 1994).
To prepare tissue slices, neonatal rats were killed according to institutional guidelines, and hippocampi were removed and transversely sliced (400 μm thick) using a manual tissue chopper (Stoelting). Slices were attached to alcohol-cleaned glass coverslips (11 × 22 mm) with a mixture of chicken plasma (10 μl; Cocalico) and bovine thrombin (10 μl; Sigma, St. Louis, MO). The slices were adherent within ∼10 min, at which point the coverslips were placed in a test tube with 1 ml of 25 mm HEPES-buffered culture media containing 25% serum. The tubes were kept in a warm room (37°C) and rotated at 12 revolutions/hr in a roller drum tilted at 5° with respect to horizontal (Gähwiler, 1984).
To label dendritic arbors of neurons in live tissue slices, cells were stained with a long-carbon-chain fluorescent membrane dye, DiIC18(3) [DiI] (Honig and Hume, 1986), or the red-shifted DiIC18(5) [DiD] (Agmon et al., 1995) (both dyes from Molecular Probes, Eugene, OR). Use of the longer wavelength dye, DiD, provided superior images of cells and dendrites located deep within the tissue, presumably because of reduced tissue scatter of red light (see Pawley, 1995). For acutely prepared slices, a 0.5% solution of dye in N,N-dimethylformamide (Sigma) was pressure-injected into the stratum oriens in area CA3 or CA1. Dye solution was delivered using a Picospritzer (General Valve, Fairfield, NJ) to present a series of brief pressure pulses (1 msec duration, 80 psi) to the back of a glass micropipette (2–3 μm tip diameter). After dye injection, slices were returned to the roller drum for a brief period to permit cellular uptake and diffusion of dye. Slices were labeled and examined within 2–24 hr after isolation from the brain, or they were cultured for 1–2 weeks to permit in vitro tissue development (LaVail and Wolf, 1973; Gähwiler, 1984; Dailey et al., 1994) before labeling and imaging. The cultured slices become somewhat thinner over time (∼100 μm thick at 2 weeks), and such slices were labeled by inserting and breaking off the tip of a glass micropipette coated with dye crystals. In some cases, slices were chemically fixed (2% formaldehyde in PBS) after 2 weeks in culture, then labeled with dye. Such fixed slices were maintained at 4°C for 4–8 weeks to permit adequate labeling and diffusion of the dye.
Confocal fluorescence imaging
Confocal fluorescence microscopy permitted high-resolution, three-dimensional imaging of neuronal structure up to 120 μm deep within the tissue slices. For time-lapse microscopic observation (Smith et al., 1990; Dailey, 1996), tissue was mounted in a perfusion chamber with culture media or saline and placed on the stage of a confocal microscope heated (35°C) with forced air. Tissue was continuously superfused (10–20 ml/hr) with media or saline. Imaging was performed using one of two custom-built, laser-scanning confocal microscopes designed by S.J.S. The 514 nm line of a 25 mW argon ion laser (Ion Laser Technology) was used for illumination when imaging the conventional form of DiI. To image neurons labeled with the longer wavelength dye, DiD, a 15 mW red helium/neon (HeNe) laser (HWK Laser) was used. The detector was a gallium/arsenide photomultiplier tube with a circular aperture placed in front to reject out-of-focus light. Microscope objectives used were a dry 20× numerical aperture (NA) 0.75 (Nikon), an oil–water–glycerin 25× NA 0.8 (Zeiss, Thornwood, NY), and an oil-immersion 40× NA 1.3 (Olympus). To clearly resolve small dendritic filopodia and spines, image data typically were collected at an additional electronic zoom factor of 2–3. The cells we examined were located 15–120 μm below the surface of the tissue slice. Because dendritic processes extended in three dimensions within the thick tissue slices, we usually collected multiple (5–7) optical sections spanning 10–30 μm in the Z-dimension to accommodate focus drift and to ensure that small structures spanning multiple confocal planes were fully captured. In most cases, stacks of images were collected at 5 min intervals. In a few experiments, individual confocal images were collected at 30 sec or 1 min intervals. All images were stored digitally using a network file server, and image stacks were later recombined using a maximum-brightness operation to produce extended focus images.
Time-lapse imaging of live, fluorescently labeled cells can produce phototoxic effects in the imaged cells. Indeed, when well stained cells were imaged with very high incident illumination intensity, or imaged too frequently, we invariably saw changes in the structure and dynamics of dendritic branches and spines: dendrite shafts developed irregular varicosities, branch points became triangulated, and both branches and spiny structures consistently retracted (shortened or disappeared) within a period of 1–2 hr. However, when the incident illumination was sufficiently attenuated (typically <600 μJ/section over a 225 μm × 168 μm specimen field), labeled specimens could be imaged (1–5 min intervals) for many hours without signs of photodynamic damage. Several lines of evidence indicate that the labeling and imaging procedures we used did not have detrimental consequences on cell structure and dynamics: (1) the structure of dendritic branches and spines on live, vitally stained cells was essentially identical to that of cells in age-matched slices fixed before labeling and imaging (compare Figs. 1 and 5); (2) the structure of spiny protrusions on live cells was very similar at the beginning and end of time-lapse imaging sessions (see Fig. 6); and (3) the dynamic behavior of dendrites remained relatively constant over the period of time-lapse imaging: spine formation and resorption were evident throughout the time-lapse sequences, and some spines became more stable while others became more active along the same dendrite segment.
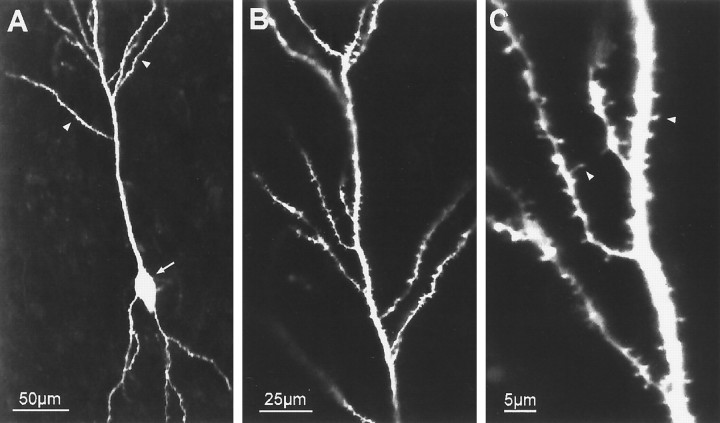
Fig. 1.
Dendrites of pyramidal cells grow and differentiate in in vitro tissue slices. The structure of a differentiated hippocampal CA1 pyramidal neuron is revealed by postfixation labeling with DiI after 2 weeks in slice culture.A, Low-magnification confocal image showing characteristic polarized pyramidal shape. Emerging from the cell soma (arrow) are several basal dendrites and a thick apical dendrite with several oblique secondary branches (arrowheads). B, Higher-magnification view of distal apical dendrites within the stratum radiatum showing secondary branches. Although their full extent is not evident in this single confocal image plane, these branches course throughout the thickness of the tissue slice. C, Dendritic branches are studded with numerous spine-like projections (arrowheads), 1–3 μm in length, presumed to be sites of synaptic contact. These features are characteristic of pyramidal neurons in vivo.
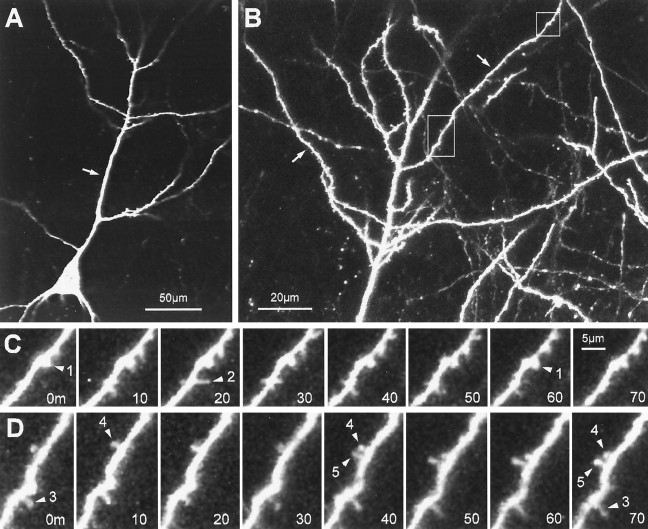
Fig. 5.
Dynamics of spine-like protrusions on well developed pyramidal cell dendrites (12 d in vitro).A, Single-focal-plane view of a CA1 neuron showing the characteristic pyramidal shape with a primary apical dendrite (arrow). The tissue was prepared from a P6 animal and cultured for 12 d. B, Higher-magnification, extended-focus view (combining 6 optical sections spanning 30 μm) of the distal portion of the apical dendrites. Note the complexity of the dendritic arbor and spiny appearance of the oblique dendrite branches (arrows). C, Time-lapse sequence of a region (top box) in B showing both stable, spine-like structures and transient filopodial extensions. A short, knobby spine-like protrusion (arrowhead 1) persists with little change in length or shape. Directly adjacent to this spine, a filopodia-like protrusion (arrowhead 2) extends transiently from the dendrite shaft. Time is shown in minutes. D, Time-lapse sequence of a region (lower box) inB showing rapid formation of a knobby, spine-like protrusion (arrowhead 5). The new spine forms and persists adjacent to an existing spine (arrowhead 4). Note also another spine (arrowhead 3) that persists for the entire observation period but does not change shape or length significantly. Scale bar inC also applies to D. The plots of length versus time for spiny protrusions 1, 2, and 5 correspond to traces 12, 9, and 11, respectively, in Figure 7.
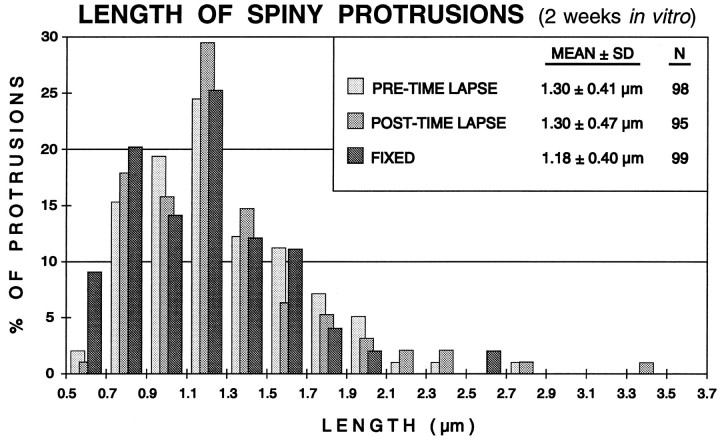
Fig. 6.
Distribution of spine lengths in fixed or live (time-lapse imaged) slices (2 weeks in vitro). The lengths of all spiny protrusions on dendrites in a live slice were determined at the beginning (PRE-TIME LAPSE) and end (POST-TIME LAPSE) of a standard 3 hr time-lapse imaging sequence (experiment shown in Fig. 5). There was no significant difference in the mean length or length distribution of spines before or after the imaging session, indicating that the time-lapse imaging did not induce changes in spine structure. Also, the mean length of spines on live cells was very similar to the distribution of spine lengths on dendrites in comparable slices fixed before labeling and imaging (FIXED). See Results for more details.
Quantitation of the lengths and lifetimes of spiny protrusions
Lengths. All clearly evident dendritic protrusions greater than 0.5 μm and less than 10 μm in length were classified as “spiny protrusions” and were included in these analyses. (On live cells, discrimination of protrusions shorter than ∼0.7 μm was not reliable.) The lengths (base to tip) of spiny protrusions were estimated from two-dimensional projections of three-dimensional image stacks (i.e., from extended-focus images) to ensure that the full lengths were captured. For quantitative analysis, the original digitized images (single pixel dimension = ∼0.3 μm in the specimen plane) were electronically zoomed an additional twofold. Measurements were made using custom-designed software using a mouse-controlled cursor.
Lifetimes. Lifetimes of spiny protrusions were estimated from time-lapse movies by determining the number of time points in which a given structure was evident, then multiplying that number by the time interval. For structures present at the beginning and/or end of the sequence, only a lower limit of lifetime could be determined. At later developmental stages, most spiny protrusions were evident for the entire observation period (3–22 hr). However, our quantitative analyses were limited by the length of time we could continuously observe any one dendrite segment, which varied from experiment to experiment. Consequently, to compare systematically the lifetime of spines between imaging sequences of different durations, it was necessary to truncate spine lifetimes that were longer than the shortest time-lapse sequence (e.g., 100 min for Fig. 8A and 180 min for Fig. 8B).
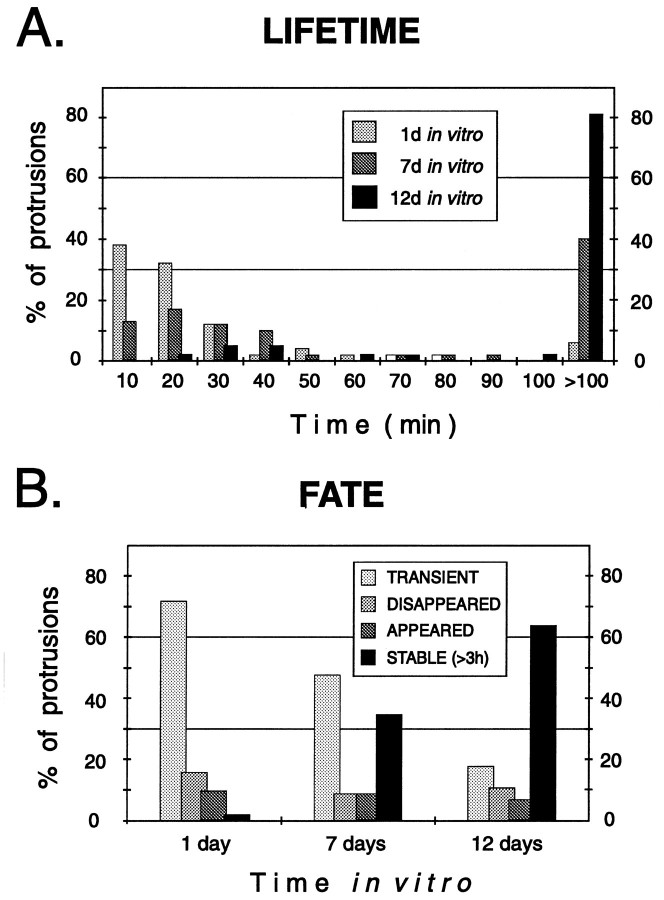
Fig. 8.
Stage-dependent differences in the lifetime and fate of spiny protrusions. A, The lifetimes of spiny protrusions increased progressively with time in vitro. Distribution of lifetimes of 146 spiny protrusions from apical dendrites of eight pyramidal cells in six tissue slices. B, Fate of spiny protrusions in tissue slices. All spiny structures observed during the first 3 hr of imaging were classified asTRANSIENT (came and went during the 3 hr observation period), DISAPPEARED (evident at the beginning but were resorbed during the 3 hr observation), APPEARED (formedde novo and persisted), or STABLE (evident throughout the 3 hr observation period). Note that a greater fraction of spines were stable on more mature (i.e., 12 d in vitro) dendrites, but there remained a low level of spine turnover.
RESULTS
Observations in fixed tissue slices
To obtain normative data free from possible biases of vital imaging techniques, the structure of pyramidal cell dendrites in tissue slices cultured for up to 14 d before fixation was visualized by post-fixation labeling with the fluorescent membrane dye DiI (Honig and Hume, 1986). Through-focus analysis with a confocal microscope indicated that, by 2 weeks in vitro, pyramidal cells had developed a complex, three-dimensional morphology with many oblique dendritic branches extending from the primary apical and basal processes (Fig. 1). At higher magnification, it was evident that dendritic branches were studded with numerous spine-like protrusions of varying shape and size (0.5–3 μm long). The morphological features of these spiny structures are essentially identical to those previously identified as sites of synaptic contact on hippocampal pyramidal cells both in vivo (Westrum and Blackstad, 1962; Harris et al., 1989) and in in vitro tissue slices (Deitch et al., 1991;Stoppini et al., 1991; Buchs et al., 1993; Pozzo Miller et al., 1993). Thus, within a 2 week period of time, dendrites of pyramidal cells inin vitro tissue slices acquire many structural features characteristic of their age-matched counterparts in vivo, including complex dendritic arbors and numerous postsynaptic spines.
Time-lapse imaging in live tissue slices
To examine directly the dynamics of dendrite structure during neuronal differentiation and synaptogenesis, neurons in tissue slices were labeled in the living state with a fluorescent membrane dye (DiI or DiD), then imaged over time with the confocal microscope. To study the development of single cells and subcellular structures, individual tissue slices were maintained on the microscope stage and imaged at intervals of 1–10 min for several hours. To test for stage-dependent variations in dendrite dynamics spanning a longer developmental period, we examined different slices cultured for progressively longer times (up to 2 weeks). The present time-lapse observations are based on imaging of 60 cells in 33 slices.
First days in culture
At early time points (1–2 d) in culture, dendrite branches were often tipped with growth cone-like structures (Fig.2A), suggesting active dendrite growth. Indeed, direct time-lapse imaging revealed that the dendritic arbor was highly dynamic. Branches of both apical and basal dendrites elongated within the tissue by advance of the leading growth cone, which involved cyclical protrusion and retraction of filopodia and lamellae. Growing dendritic branches extended within the tissue at a rate of 18.4 ± 10.5 μm/hr (mean ± SD, n = 12), which is comparable with the rate of axonal extension observed in these tissue slices (Dailey et al., 1994). Surprisingly, even as some dendritic branches elongated, other branches retracted (16.2 ± 6.6 μm/hr, n = 5) and were sometimes fully resorbed into the parent dendrite trunk.
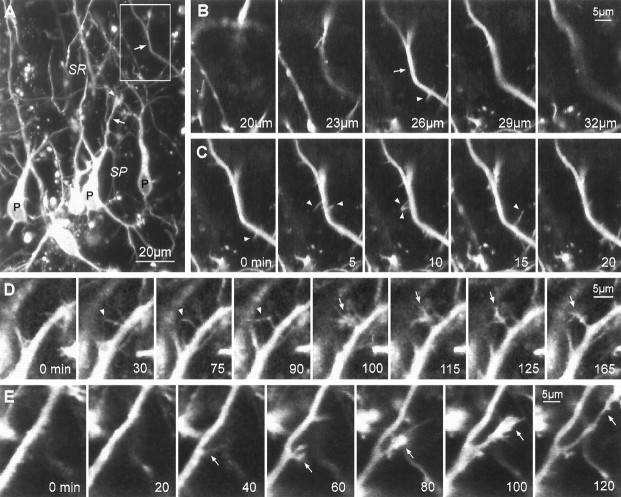
Fig. 2.
Sprouting of lateral filopodia, growth cones, and collateral branches from pyramidal cell dendrites during early stages of differentiation (1 d in vitro). A, Low-magnification image of DiI-labeled CA1 pyramidal neuron somata (P) and dendrites (arrows) in a live tissue slice taken from a P2 animal. This is an extended-focus image made by combining a stack of five optical sections spanning 15 μm in theZ-dimension. At early developmental stages, dendrites are often tipped by complex growth cones (arrowhead).SR, Stratum radiatum; SP, stratum pyramidale.B, Through-focus sequence of boxed regionin A showing a segment of an apical dendrite shaft (arrow) having fine, lateral filopodial protrusions (arrowhead). For time-lapse imaging, multiple optical sections were collected to ensure that small dendritic protrusions were captured in their entirety. C, Time-lapse sequence of the same field as in B showing that numerous lateral filopodia (arrowheads) extend from, and retract back to, the dendrite shaft. Most filopodia are resorbed within a few minutes after first appearing. These images are composites of two or three optical sections from the middle of a five-image stack (shown in B).D, Persisting lateral filopodia sometimes evolve into growth cones. Time-lapse sequence shows a lateral filopodium (arrowheads) sprouting from an apical dendrite shaft (0–30 min), persisting for more than an hour (30–90 min), then rapidly developing into a complex, growth cone-like structure (arrow; 90–100 min). E,De novo sprouting of a growth cone leading to formation of a collateral dendrite branch. A faint, filopodial structure is seen first (40 min), then a growth cone develops (60–80 min) and spins out a new branch that persisted and grew to a length of at least 35 μm (120 min). For C–E, elapsed time is shown in minutes.
Motile protrusions were not confined to the tips of growing branches: lateral filopodia and growth cone-like sprouts were commonly seen along developing dendrite shafts. Most of the filopodial protrusions (up to 10 μm long) extended (maximal rate: ∼2.5 μm/min) from dendrite shafts, then retracted back to the shaft within 30 min or less (median lifetime, 10 min) (Fig. 2B,C). Filopodia emerged from many different sites along the longitudinal extent of the apical and basal dendrites, although “hot spots” of filopodial protrusion were occasionally seen where filopodia repeatedly emerged and retracted from a localized site. Sometimes these hot spots of protrusive activity moved rapidly (114 ± 44 μm/hr, n = 4) toward the growing tip of the dendrite branch, giving the appearance of waves of activity that covered distances of 100 μm or more. After reaching the tip, the waves appeared to fuel rapid growth of the branch for several minutes (see Ruthel and Banker, 1992). The frequency of filopodial protrusions from the dendrite surface was highly variable from cell to cell and from branch to branch: we saw as many as 36 protrusions along a 50 μm dendrite segment within a 3 hr period of time. However, because the filopodial lifetimes were short and our sampling rate was relatively low (once every 5 min), the actual number of protrusive events is likely to be substantially higher.
Growth cone-like sprouts were also observed to emerge de novo from proximal regions of dendrite shafts. Formation of growth cone sprouts typically involved transformation from a single lateral filopodium (Fig. 2D). Although many of these sprouts were resorbed within a few minutes, others continued extending and generated new collateral branches (Fig. 2E). Thus, a common sequence of events leading to collateral branch formation included: first, emergence of a single filopodium; then, conversion of that filopodium into a collateral growth cone; and finally, advance of the growth cone and consolidation of the new outgrowth to form a cylindrical dendrite branch.
These observations indicate that the developing dendritic arbor is highly dynamic, and that some of the lateral dendritic filopodia and growth cones are precursors to new dendrite branches. However, most lateral dendritic filopodia extend transiently and are fully resorbed within a few minutes. We found little change in the dynamic properties of dendrites in slices prepared from animals during the first postnatal week: over the first few days in culture, slices from P2–P7 animals were all dominated by highly dynamic, transient filopodia.
One week in culture
When slices of developing hippocampus (derived from P5–P7 animals) were cultured for several more days (i.e., 6–8 d total) to allow synaptogenesis and dendritic differentiation to proceed in vitro (Gähwiler, 1984; Buchs et al., 1989; Muller et al., 1993), dendritic arbors were found to have increased in complexity (Fig. 3A). Many more oblique branches had extended from primary dendrite trunks. In addition, dendrite branches bore numerous spiny protrusions (1.8 ± 0.9 μm) having simple filiform or complex shapes (Fig. 3B). Some of these lateral “spine-like” protrusions had short (∼1 μm) stalks and knobby heads, features that are characteristic of postsynaptic spines on mature dendrites (Harris et al., 1992; Papa et al., 1995;Trommald et al., 1995).
Fig. 3.
Dynamics of spiny protrusions on differentiating pyramidal cell dendrites (7 d in vitro). Tissue was taken from a P5 rat and cultured for 1 week. A, Low-magnification image of a live, DiI-labeled CA1 pyramidal cell (arrow) showing extensive apical dendrite arbors that have developed in vitro. This image is a Z-axis composite of 15 separate confocal images spanning a tissue depth of 45 μm. Because of the greater volume of tissue sampled, dendritic arbors appear more complex in Z-axis composite images than in single focal plane images. The complexity of dendritic arbors at 7 d in vitro was intermediate to that of neurons at 1 d (compare Fig.2A) and 12 d (compare Figs. 1A, 5A)in vitro. B, Higher-magnification image of boxedregion in A showing many simple and branched (arrows) filopodia as well as spine-like protrusions with bulbous tips (arrowheads). Schaffer axons from CA3 pyramidal cells, which are retained in the tissue slices, normally synaptically terminate on spines in this region of the dendrite. Scale bar inB applies to B–D. C, Selected images of a time-lapse sequence of a region in B (rightbox) showing extension and partial retraction of a filopodium (arrow). By contrast, the length of a spine-like protrusion with a bulbous tip (arrowhead) is stable over the same time period. D, Time-lapse sequence of a region inB (left box) showing dynamic changes in structure of a spine-like protrusion. The protrusion starts with a bulbous head (arrow) that appears to bifurcate (arrowheads). Images in C and D, shown at 1 min intervals, are single-focal-plane scans selected from an original data set taken at 30 sec intervals.
Time-lapse imaging revealed that both filopodia- and spine-like protrusions on dendrites were structurally very dynamic at 1 weekin vitro. Filopodial protrusions (without knobby heads) often exhibited rounds of rapid extension and retraction (Fig.3C). The spine-like protrusions (with heads) generally persisted for longer periods of time and were more stable in length than filopodia (Fig. 3C), although some spines elongated and retracted, and spine heads were observed to change shape from minute to minute (Fig. 3D). A few spines showed no detectable change in length or shape over a several hour period of observation. Thus, with regard to both morphology and structural dynamics, there was a heterogeneous population of spiny structures on developing dendrites at this stage.
To assess better the long-term stability and dynamics of the various spiny protrusions, we collected confocal images of single dendrite branches for lengthy periods of time (up to 22 hr). In contrast to earlier time points in vitro, many of the spiny protrusions at this stage were not transient but instead persisted for long periods of time. The time-lapse observations also revealed a variety of dynamic behaviors on individual dendrite branches (Fig.4A,B), including: (1) de novo formation of spiny structures (Fig.4B3,C3); (2) resorption of some spiny structures; (3) persistence of some spiny structures in a dynamic state (Fig. 4A2,C2); and (4) conversion of some dynamic spiny structures to a more stable state (Fig. 4C1,C3). Analysis of the length of individual spiny protrusions over time revealed that many of the persisting structures had a dynamic distal segment that protruded rapidly (up to 35 μm/hr) and cyclically from a relatively stable, proximal stump (0.5–4 μm) (Fig. 4C). The apparent length of the short persisting stump also changed somewhat over time, but at a much slower rate (generally <1 μm/hr). These observations are in contrast to earlier developmental time points at which transient filopodia appeared to protrude directly from, and to be fully resorbed back to, the surface of the dendrite shaft.
Fig. 4.
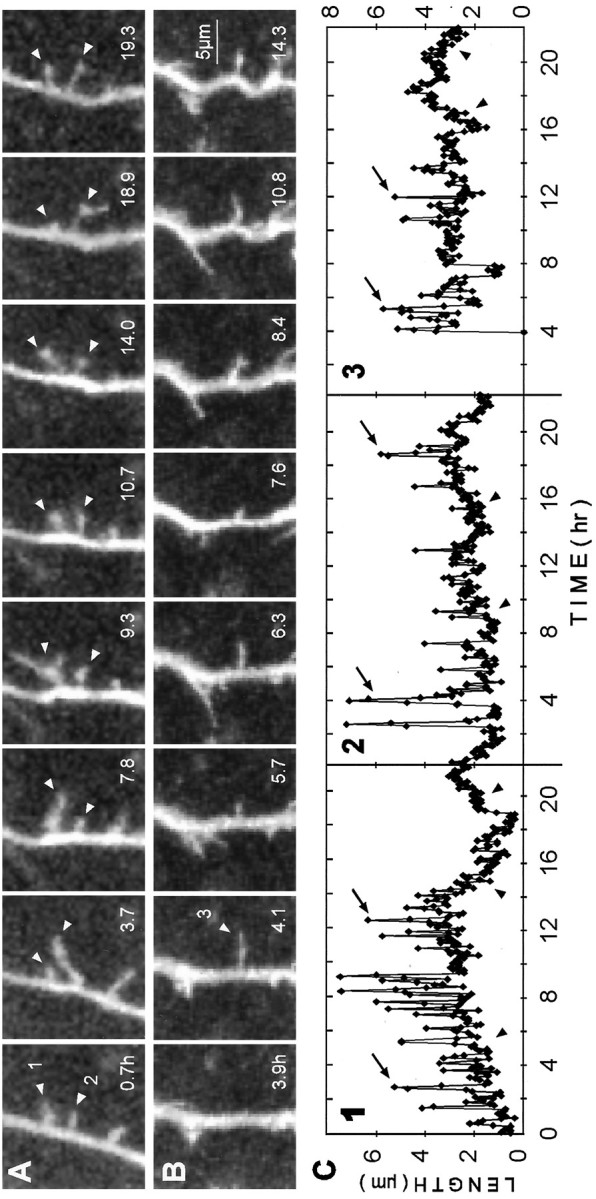
Dynamics of long-lived, spiny protrusions on differentiating dendrites (7 d in vitro). These extended time-lapse sequences show changes in length and shape of spiny protrusions (arrowheads) over a period of up to 18.6 hr. All images are composites of two adjacent focal planes selected from a five-image stack to ensure that the full lengths of the structures were captured. Images were selected from an original data set collected at 5 min intervals. Elapsed time is shown in hours. A, Two of the spiny protrusions (1 and 2) were highly dynamic but persisted for the entire observation period. Length plots for these protrusions are shown in C (traces 1 and2). B, De novo appearance of a spiny protrusion (arrowhead) that persisted after being formed. Length plot for this protrusion is shown in C (trace3). C, Plots of length versus time for spiny protrusions shown above. Measurements of protrusion length were made at each time point (5 min intervals). Traces 1 and2 correspond to the protrusions indicated in A. Note that transient length excursions (arrows), which can effectively double or triple the length of the protrusion, are made from relatively stable bases (arrowheads). Reduction in the transient length excursions of protrusions 1 and3 near the end of the sequences suggests a further stabilization of these protrusions. In contrast, the transient length excursions persist in trace 2.
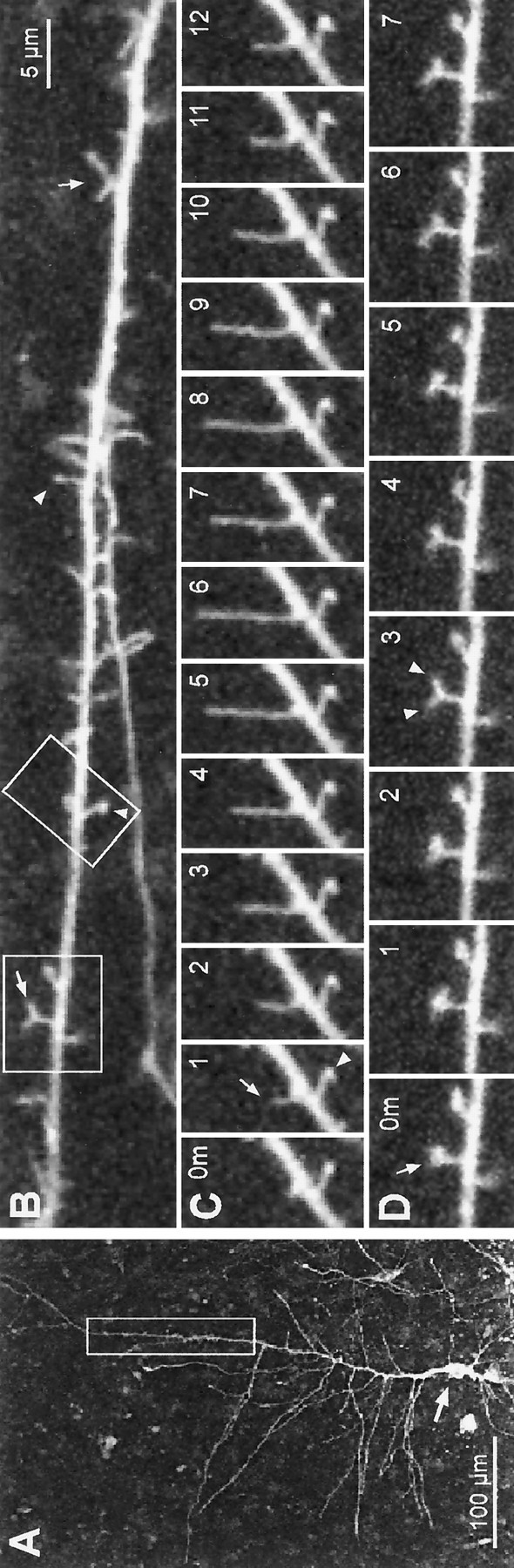
Two weeks in culture
When tissue slices were cultured for ∼2 weeks (12–14 d) before observation, dendrites had very elaborate arbors that were more uniformly covered with short (0.5–2 μm), spiny protrusions (Fig.5A,B). Time-lapse observations revealed that the structure of these well differentiated dendrites was more stable than at earlier times in vitro. We observed neither substantial elongation or retraction of existing branches, nor formation of new branches. Moreover, many of the spine-like protrusions persisted for the entire observation period and did not exhibit detectable changes in shape or length (Fig. 5D). Nevertheless, we occasionally observed filopodia-like protrusions transiently extending to a length of 2–3 μm before retracting (Fig.5C), and some spines rapidly emerged de novo(Fig. 5D) while others appeared to be entirely resorbed. These observations indicate that some stable spines can form on more mature dendrites in the absence of dynamic filopodial and protospine precursors, and that even at the latest stage examined (2 weeksin vitro) there is turnover of some spine-like protrusions.
Quantitative analysis of spine structure and dynamics
Spines on live versus fixed dendrites
The structure of spiny protrusions on dendrites in the live, 2-week-old tissue slices looked very similar to those previously observed in fixed (nonimaged), age-matched slice cultures (compare Figs. 1 and 5). To assess and compare spine structure in the living and fixed states quantitatively, we measured the length of all spiny protrusions on dendrites in fixed tissue and at two time points in live tissue slices (both before and after time-lapse imaging). Although the distribution of spine lengths for live and fixed dendrites was quite similar (Fig. 6), statistical analysis revealed a significant difference (p = 0.017; Mann–Whitney rank sum test) between these groups when the minimum threshold length for spine detection was set at 0.5 μm. The mean length of spines on live dendrites (before imaging) was 1.30 ± 0.41 μm (mean ± SD;n = 98), and for spines on fixed cells, 1.18 ± 0.40 μm (n = 99). This difference was attributable to the greater number of spines scored in the 0.5–0.7 μm range on fixed cells compared with live cells. The apparent increase in the frequency of the shortest spines (0.5–0.7 μm) on fixed dendrites could be attributable to tissue shrinkage associated with fixation (Deutsch and Hillman, 1977), or to the different imaging conditions. Our ability to reliably detect the shortest spines was dependent in part on the intensity of the illuminating laser light (i.e., higher laser intensity increases the signal-to-noise ratio and improves spatial resolution). Because we used a lower intensity on the live cells to minimize phototoxic effects, we were not able to resolve clearly the shortest spines on live cells as well as we could on fixed cells. It is likely, then, that we could not detect reliably some of the shortest spines (<0.7 μm) on the live dendrites under the imaging conditions we used. Indeed, when the minimum spine length threshold was set to a more conservative value (0.7 μm), there was no statistical difference between spine lengths on the live and fixed cells (p = 0.091).
Before and after imaging
We also compared spine lengths on individual live dendrites before and after time-lapse imaging sessions (see Fig. 6). The lengths of spines after a standard 3 hr imaging session (mean spine length, 1.30 ± 0.47 μm; n = 95) were found to be not statistically different (p = 0.598) from that at the beginning of the imaging session. This supports the idea that the labeling and time-lapse imaging procedures we used did not have detrimental effects on dendrite structure in the live brain slices.
Developmental differences in spine dynamics
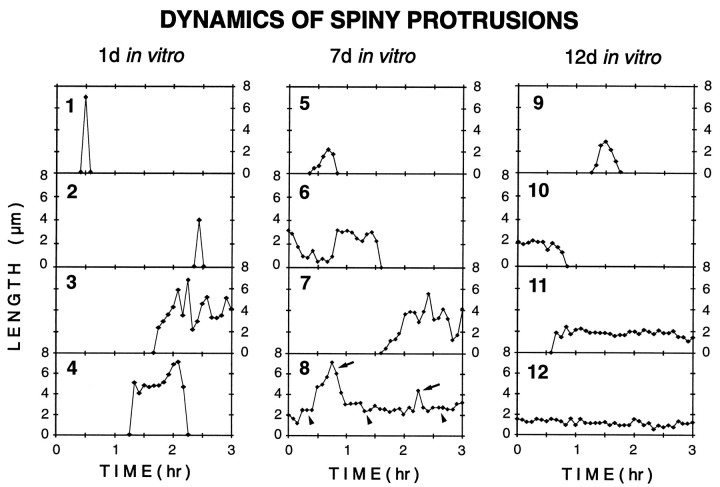
Our time-lapse observations on dendrites at different developmental times suggested stage-dependent differences in the dynamic properties of spiny structures. To more systematically assess and compare the dynamics of spiny protrusions, the lengths of individual spiny structures were determined and plotted for every time point during the first 3 hr of observation on cells cultured for 1, 7, and 12 d. These plots clearly demonstrated the various motile behaviors of spiny structures, including transient protrusion, resorption,de novo formation, and stable persistence (Fig.7). They also illustrated that transientspiny protrusions were found at all developmental stages examined, and that persisting spiny structures were significantly more dynamic at 7 versus 12 d in vitro. In particular, at 7 din vitro many spiny structures made transient length changes (compare Fig. 4), whereas at 12 d in vitro persisting spines generally lacked such length changes. These observations thus reveal a transient developmental period in which individual spiny structures embody features characteristic of both fleeting filopodia and stable spines.
Fig. 7.
Stage-dependent differences in the dynamics of spiny protrusions. Representative plots of length versus time for spiny dendritic protrusions at 1 d (traces 1–4), 7 d (traces 5–8), and 12 d (traces 9–12) in vitro. The traces illustrate a range of behaviors, including transient protrusion (1, 2, 4, 5, 9), resorption of preexisting structures (6, 10), de novo formation and persistence (3, 7, 11), and persistence of preexisting structures (8, 12). Transient filopodia-like protrusions (1, 2, 5, 9) were observed at all times in vitrobut were most prominent at early stages (i.e., 1 d in vitro). At 7 d in vitro, persisting spiny protrusions often showed transient length excursions (arrows) from short, stable bases (arrowheads) (compare trace 8 and Fig. 4C). Most persisting spines at 12 d in vitro showed little change in length (e.g., trace 12), although there was some turnover of spines (e.g., traces 10, 11). (Note different time base of Figs. 4C and7.)
Although a variety of protrusive structures was evident at the various developmental stages we examined, the persisting (>30 min) spiny structures appeared to predominate at later stages. Indeed, analysis of the lifetimes of spiny protrusions revealed that the fraction of persisting protrusions progressively increased over timein vitro (Fig. 8). On the first day in vitro, most (94%) of the spiny protrusions had a lifetime of <100 min. However, by 12 d in vitro, 81% of the spiny protrusions persisted for longer than 100 min, and over half (64%) persisted for 3 hr or more. Thus, as dendritic arbors matured and synaptic connections were established, spiny dendritic protrusions became progressively more stable.
DISCUSSION
The present observations revealed a high level of structural dynamics of dendrites during development, including both active growth and dynamic turnover (resorption) of dendritic branches and spiny protrusions. A striking feature of developing dendrites was the high level of protrusive activity along proximal dendrite shafts. Many lateral dendritic growth cones and filopodia were transitory, whereas others were precursors to more stable dendritic branches and spine-like projections. This suggests that much of dendritic structure (i.e., branches and spines) in developing CNS tissue is generated by the selective stabilization of highly motile protrusive structures.
Dynamics of spine formation
Our time-lapse observations revealed several features of spine formation and dynamics that could not be determined from static images of dendrite structure, including: (1) the highly dynamic nature (i.e., rapid appearance and disappearance) of filopodial protrusions on developing dendrites in CNS tissue; (2) a transitional developmental phase involving conversion of dynamic filopodia to more stable, spine-like protrusions; and (3) turnover (addition and resorption) of some spine-like protrusions on more fully differentiated dendrite branches.
Time-lapse analysis revealed a heterogeneous population of spiny dendritic protrusions with regard to structural dynamics. Based on their dynamic properties, we propose a threefold classification of spiny dendritic protrusions: (1) transient filopodia that show rapid changes in length and shape but are short-lived (generally <0.5 hr); (2) metastable protospines that persist for longer periods of time (0.5 to >22 hr) but remain structurally dynamic; and (3) stable spines that have a low rate of turnover and show relatively little change in structure. Fine spiny protrusions from each of the different classes can coexist on single dendrite branches, but there is a clear developmental progression. At early stages of differentiation, dendrites predominately bear the filopodia, whereas the mature dendrites are dominated by spines. The protospines appear at an intermediate stage and are transient developmental structures that share properties of both the filopodia and spines: they persist like the spines but also show rapid changes in length like the filopodia.
The three spiny populations may represent separate structures that are ontogenetically unrelated. Alternatively, dynamic structures may evolve into progressively more stable structures. This latter idea is consistent with the transient appearance of protospines, which share features of both the filopodia and spines, at an intermediate developmental stage preceded by a filopodia-dominated stage and followed by a spine-dominated stage. Moreover, our time-lapse observations revealed that individual spiny structures can make transitions from a relatively dynamic state to a more stable configuration (compare Fig. 4C). However, in our older cultures we also observed rapid appearance of stable spines without dynamic filopodia or protospines immediately preceding the spines (e.g., Fig. 5D). Thus, stable spines may emerge on developing dendrites via two different mechanisms: (1) conversion from more dynamic spiny structures, and (2) rapid and de novoextension of a stable spine directly from dendrite shafts.
The stability of spiny protrusions on well differentiated dendrites in our older slice cultures is consistent with previous time-lapse observations of more mature dendrites in acutely isolated hippocampal tissue (Hosokawa et al., 1992, 1994). In slices prepared from 3-week-old animals, spiny structures likely to be postsynaptic elements often persisted for >5 hr, although a small proportion of spines appeared or disappeared over the course of the 5 hr observations (Hosokawa et al., 1992). The present observations extend the lower limit of spine lifetime to at least 22 hr, and additionally indicate that long-lived spines can rapidly (<5 min) appear and disappear, and reversibly change shape, at least during developmental periods.
The morphological characteristics of the stable spiny protrusions we observed (i.e., the protospines and spines) are very similar to spine-like protrusions observed on mature pyramidal cell dendrites from fixed hippocampal tissue (Harris et al., 1989, 1992; Trommald et al., 1995). Previous ultrastructural studies have shown that such spine-like protrusions, both in vivo and in in vitrotissue slices, are virtually always postsynaptic elements (Westrum and Blackstad, 1962; Gottlieb and Cowan, 1972; Harris et al., 1989,1992; Deitch et al., 1991; Stoppini et al., 1991; Buchs et al., 1993;Pozzo Miller et al., 1993). Indeed, the time course of emergence of stable spines in our slices is consistent with the known course of synapse formation and accumulation on dendritic spines in comparable slice cultures (LeVail and Wolf, 1973; Gähwiler, 1984; Dailey et al., 1994; Buchs et al., 1993; Muller et al., 1993). In stratum radiatum of area CA1, there is a 15-fold increase in the density of synapses over the first 2 weeks in culture, and virtually all of the new synapses that appear are located on synaptic spines (Buchs et al., 1993). Taken together, these observations strongly suggest that the stable spiny protrusions we observed are sites of synaptic contact. However, in the future it will be important to develop methods to determine definitively which of the dynamic or stable dendritic protrusions described here correspond to, or evolve into, actual postsynaptic spines bearing synaptic contacts.
Possible relation of dendritic growth and protrusion to synapse formation
If, as we suspect, many or all of the stable spiny structures bear synaptic contacts, our observations would be consistent with either of two scenarios of synaptogenesis: (1) synapses are first formed on dendrite shafts, then emerge with spines as they extend from dendrite shafts; or (2) synapses are first formed on protruding filopodia that then retract back to the dendrite or transform into stable, synapse-bearing spines. If synaptogenic contacts are in fact made by protrusive dendritic filopodia, the active resorption of some of these protrusions during early stages of differentiation may serve to draw nascent synaptic contacts onto dendritic shafts (Povlishock, 1974;Ulfhake and Cullheim, 1988; Saito et al., 1992). Alternatively, those filopodia that do not synaptogenically engage axons may be resorbed, whereas those that synaptogenically interact with axons may be stabilized and remain extended.
The different classes of spiny protrusions thus may represent progressive stages in synaptogenesis. We propose the following model of synapse formation: (1) protrusive filopodia make the initial synaptogenic interactions with axons; (2) filopodia with nascent synaptic contacts may be drawn back to the dendrite shaft, or may be converted to metastable protospines that could hold developing synapses off the dendrite shafts in a structurally and functionally plastic configuration; and (3) relatively stable spine synapses appear as shaft synapses reemerge with stable spines, or by conversion (further stabilization) of protospines bearing physiologically validated synapses. This model does not exclude the possibility that stable spines (with synapses) may emerge from shafts by mechanisms unrelated to dynamic filopodia and protospines (Juraska and Fifkova, 1979; Miller and Peters, 1981; Steward et al., 1988; Harris et al., 1989), especially at later developmental stages (Papa et al., 1995). However, during peak periods of axonal ingrowth and synaptogenesis, dendritic filopodia may serve to enhance the ability of target cells to attract and compete for axonal inputs (Cooper and Smith, 1992).
The idea that dendritic filopodia may be specialized sites of synaptogenic interaction with axons has a long history (see Vaughn, 1989). Dendritic filopodia are known to be abundant during periods of synaptogenesis (Morest, 1969a,b; Minkwitz and Holz, 1975; Berry and Bradley, 1976a,b; Minkwitz, 1976; Miller and Peters, 1981; Phelps et al., 1983; Ramoa et al., 1987; Ulfhake and Cullheim, 1988), and they are often found by electron microscopy to be postsynaptic elements (Hinds and Hinds, 1972; Hayes and Roberts, 1973; Skoff and Hamburger, 1974; Vaughn et al., 1974; Pappas et al., 1975; Saito et al., 1992). Moreover, time-lapse studies indicate that filopodial interactions precede formation of stable contacts between neuronal processes (O’Connor et al., 1990; Baird et al., 1992; Cooper and Smith, 1992). Together, these observations suggest that synaptic contacts are initiated at actively growing regions on target cells. This idea is consistent with a synaptogenic filopodial theory (Vaughn et al., 1974;Berry and Bradley, 1976a,b; Vaughn, 1989), which posits that synaptic contacts between neurons in the CNS are initiated predominantly between filopodia on the growing distal tips of axonal and dendritic branches. However, in many CNS regions including the hippocampus, the spatiotemporal pattern of afferent axon ingrowth and dendrogenesis has seemed incompatible with such an interaction between distaltips (Andersen, 1979): later generated afferents often contact progressively more proximal dendrite segments on target cells (Bayer, 1980; Bayer and Altman, 1987). Our observations may resolve this apparent difficulty, because they indicate that even proximal dendritic segments can exhibit abundant outgrowth of lateral growth cones and filopodia during synaptogenic periods.
The abundance of dendritic protrusive motions observed in our studies supports the hypothesis (Saito et al., 1992) that dendrites are active partners in the initiation of synaptogenic cell–cell contacts, not merely passive targets for axonal exploration. The relatively short lifetimes (∼10 min) of most of the filopodia at early stages in development also suggest that any single image (e.g., from a fixed tissue specimen) would drastically under-represent the total number of filopodial eruptions that would occur during a typical developmental synaptogenesis period extending over several days. This consideration might reconcile the relatively small number of filopodia classically observed on a given, fixed dendrite (e.g., Morest, 1969a,b) with a much larger number of synapses eventually formed per dendrite. This reconciliation, in turn, permits a hypothesis that dendritic filopodia might actually be involved in establishing all axo-dendritic synaptic contacts. In the future, it will be important to learn much more about how the motility of the developing dendrite is generated and regulated, and how protrusive motility relates to the array of dendritic surface molecules necessary to permit the recognition of appropriate axonal partners and the initiation of synaptogenesis.
Footnotes
This work was supported by grants from the National Institutes of Health (NS28587, NS09027) and the National Institute of Mental Health Silvio Conte Center for Neuroscience Research (MH48108). We thank Drs. Nancy O’Rourke and Noam Ziv for discussion and critical comments on this manuscript.
Correspondence should be addressed to Dr. Michael E. Dailey, Department of Molecular and Cellular Physiology, Beckman Center, Stanford University Medical Center, Stanford, CA 94305-5426.
REFERENCES
- 1.Agmon A, Yang LT, Jones EG, O’Dowd DK. Topological precision in the thalamic projection to neonatal mouse barrel cortex. J Neurosci. 1995;15:549–561. doi: 10.1523/JNEUROSCI.15-01-00549.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaral D, Dent JA. Development of the mossy fibers of the dentate gyrus. I. A light and electron microscopic study of the mossy fibers and their expansions. J Comp Neurol. 1981;195:51–86. doi: 10.1002/cne.901950106. [DOI] [PubMed] [Google Scholar]
- 3.Andersen P. Factors influencing functional connectivity during hippocampal development. Prog Brain Res. 1979;51:139–147. doi: 10.1016/S0079-6123(08)61302-3. [DOI] [PubMed] [Google Scholar]
- 4.Baird DH, Hatten ME, Mason CA. Cerebellar target neurons provide a stop signal for afferent neurite extension in vitro. J Neurosci. 1992;12:619–634. doi: 10.1523/JNEUROSCI.12-02-00619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayer SA. The development of the hippocampal region in the rat. I. Neurogenesis examined with [3H]thymidine autoradiography. J Comp Neurol. 1980;190:87–114. doi: 10.1002/cne.901900107. [DOI] [PubMed] [Google Scholar]
- 6.Bayer S, Altman J. Directions in neurogenetic gradients and patterns of anatomical connections in the telencephalon. Prog Neurobiol. 1987;29:57–106. doi: 10.1016/0301-0082(87)90015-3. [DOI] [PubMed] [Google Scholar]
- 7.Bentley D, O’Connor TP. Cytoskeletal events in growth cone steering. Curr Opin Neurobiol. 1994;4:43–48. doi: 10.1016/0959-4388(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 8.Berry M, Bradley PM. The application of network analysis to the study of branching patterns of large dendritic fields. Brain Res. 1976a;109:111–132. doi: 10.1016/0006-8993(76)90383-8. [DOI] [PubMed] [Google Scholar]
- 9.Berry M, Bradley P. The growth of the dendritic trees of Purkinje cells in the cerebellum of the rat. Brain Res. 1976b;112:1–35. doi: 10.1016/0006-8993(76)90331-0. [DOI] [PubMed] [Google Scholar]
- 10.Bliss TVP, Chung SH, Stirling RV (1974) Structural and functional development of the mossy fibre system in the hippocampus of the post-natal rat. J Physiol (Lond) 239:92P–94P. [PubMed]
- 11.Buchs P-A, Stoppini L, Muller D. Structural modifications associated with synaptic development in area CA1 of rat hippocampal organotypic cultures. Dev Brain Res. 1993;71:81–91. doi: 10.1016/0165-3806(93)90108-m. [DOI] [PubMed] [Google Scholar]
- 12.Chien C-B, Rosenthal DE, Harris WA, Holt CE. Navigational errors made by growth cones without filopodia in the embryonic Xenopus brain. Neuron. 1993;11:237–251. doi: 10.1016/0896-6273(93)90181-p. [DOI] [PubMed] [Google Scholar]
- 13.Cooper MW, Smith SJ. A real-time analysis of growth cone-target cell interactions during the formation of stable contacts between hippocampal neurons in culture. J Neurobiol. 1992;23:814–828. doi: 10.1002/neu.480230704. [DOI] [PubMed] [Google Scholar]
- 14.Dailey ME. Dynamic optical imaging of neuronal structure and physiology: confocal fluorescence microscopy in living brain slices. In: Toga A, Mazziotta J, editors. Brain mapping: the Methods. Academic; San Diego: 1996. pp. 29–54. [Google Scholar]
- 15.Dailey ME, Buchanan J, Bergles DE, Smith SJ. Mossy fiber growth and synaptogenesis in rat hippocampal slices in vitro . J Neurosci. 1994;14:1060–1078. doi: 10.1523/JNEUROSCI.14-03-01060.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deitch JS, Smith KL, Swann JW, Turner JN. Ultrastructural investigation of neurons identified and localized using the confocal scanning laser microscope. J Electron Microsc Tech. 1991;18:82–90. doi: 10.1002/jemt.1060180112. [DOI] [PubMed] [Google Scholar]
- 17.Deutsch K, Hillman H. The effect of six fixatives on the areas of rabbit neurons and rabbit and rat cerebral slices. J Microsc. 1977;109:303–309. doi: 10.1111/j.1365-2818.1977.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 18.Gähwiler BH. Development of the hippocampus in vitro : cell types, synapses, and receptors. Neuroscience. 1984;11:751–760. doi: 10.1016/0306-4522(84)90192-1. [DOI] [PubMed] [Google Scholar]
- 19.Gähwiler BH. Organotypic cultures of neural tissue. Trends Neurosci. 1988;11:484–489. doi: 10.1016/0166-2236(88)90007-0. [DOI] [PubMed] [Google Scholar]
- 20.Godement P, Wang LC, Mason CA. Retinal axon divergence in the optic chiasm: dynamics of growth cone behavior at the midline. J Neurosci. 1994;14:7024–7039. doi: 10.1523/JNEUROSCI.14-11-07024.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb DI, Cowan WM. On the distribution of axonal terminals containing spheroidal and flattened synaptic vesicles in the hippocampus and dentate gyrus of the rat and cat. Z Zellforsch. 1972;129:413–429. doi: 10.1007/BF00307297. [DOI] [PubMed] [Google Scholar]
- 22.Halloran MC, Kalil K. Dynamic behaviors of growth cones extending in the corpus callosum of living cortical brain slices observed with video microscopy. J Neurosci. 1994;14:2161–2177. doi: 10.1523/JNEUROSCI.14-04-02161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris KM, Jensen FE, Tsao BH. Ultrastructure, development, and plasticity of dendritic spine synapses in area CA1 of the rat hippocampus: extending our vision with serial electron microscopy and three-dimensional analyses. In: Chan-Palay V, Köhler C, editors. The hippocampus—new vistas. Liss; New York: 1989. pp. 33–52. [Google Scholar]
- 24.Harris KM, Jensen FE, Tsao BH. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic plasticity and long-term potentiation. J Neurosci. 1992;12:2685–2705. doi: 10.1523/JNEUROSCI.12-07-02685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris WA, Holt CE, Bonhoeffer F. Retinal axons with and without their somata, growing to and arborizing in the tectum of Xenopus embryos: a time-lapse video study of single fibres in vivo . Development. 1987;101:123–133. doi: 10.1242/dev.101.1.123. [DOI] [PubMed] [Google Scholar]
- 26.Hayes BP, Roberts A (1973) Synaptic junction development in the spinal cord of an amphibian embryo: an electron microscope study. Z Zellforsch. 137:251–269. [DOI] [PubMed]
- 27.Hinds JW, Hinds PL. Reconstruction of dendritic growth cones in neonatal mouse olfactory bulb. J Neurocytol. 1972;1:169–187. doi: 10.1007/BF01099183. [DOI] [PubMed] [Google Scholar]
- 28.Honig MG, Hume RI. Fluorescent carbocyanine dyes allow living neurons of identified origin to be studied in long-term culture. J Cell Biol. 1986;103:171–186. doi: 10.1083/jcb.103.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosokawa T, Bliss TVP, Fine A. Persistence of individual dendritic spines in living brain slices. NeuroReport. 1992;3:477–480. doi: 10.1097/00001756-199206000-00005. [DOI] [PubMed] [Google Scholar]
- 30.Hosokawa T, Bliss TVP, Fine A. Quantitative three-dimensional confocal microscopy of synaptic structures in living brain tissue. Microsc Res Tech. 1994;29:290–296. doi: 10.1002/jemt.1070290405. [DOI] [PubMed] [Google Scholar]
- 31.Juraska JM, Fifkova E. A Golgi study of the early postnatal development of the visual cortex of the hooded rat. J Comp Neurol. 1979;183:247–256. doi: 10.1002/cne.901830203. [DOI] [PubMed] [Google Scholar]
- 32.Kaethner RJ, Stuermer CAO. Dynamics of terminal arbor formation and target approach of retinotectal axons in living zebrafish embryos: a time-lapse study of single axons. J Neurosci. 1992;12:3257–3271. doi: 10.1523/JNEUROSCI.12-08-03257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kater SB, Rehder V. The sensory-motor role of growth cone filopodia. Curr Opin Neurobiol. 1995;5:68–74. doi: 10.1016/0959-4388(95)80089-1. [DOI] [PubMed] [Google Scholar]
- 34.LaVail JH, Wolf MK. Postnatal development of the mouse dentate gyrus in organotypic cultures of the hippocampal formation. Am J Anat. 1973;137:47–66. doi: 10.1002/aja.1001370105. [DOI] [PubMed] [Google Scholar]
- 35.Lund JS, Boothe RG, Lund RD. Development of neurons in the visual cortex (area 17) of the monkey (Macaca nemestrina ): a Golgi study from fetal day 127 to postnatal maturity. J Comp Neurol. 1977;176:149–188. doi: 10.1002/cne.901760203. [DOI] [PubMed] [Google Scholar]
- 36.Mason CA, Godement P. Growth cone form reflects interactions in visual pathways and cerebellar targets. In: Letourneau PC, Kater SB, Macagno ER, editors. The nerve growth cone. Raven; New York: 1992. pp. 405–423. [Google Scholar]
- 37.Miller M, Peters A. Maturation of rat visual cortex. II. A combined Golgi-electron microscope study of pyramidal neurons. J Comp Neurol. 1981;203:555–573. doi: 10.1002/cne.902030402. [DOI] [PubMed] [Google Scholar]
- 38.Minkwitz H-G, Holz L. Die ontogenetische entwicklung von pyramidenneuronen aus dem hippocampus (CA1) der ratte. J Hirnforsch. 1975;16:37–54. [PubMed] [Google Scholar]
- 39.Minkwitz H-G. Zur entwicklung der neuronenstruktur des hippocampus wahrend der pra- und postnatalen ontogenese der albinoratte. I. Mitteilung: neurohistologische darstellung der entwicklung langaxoniger neurone aus den regionen CA3 und CA4. J Hirnforsch. 1976;17:213–231. [PubMed] [Google Scholar]
- 40.Morest DK. The differentiation of cerebral dendrites: a study of the post-migratory neuroblast in the medial nucleus of the trapezoid body. Z Anat Entwicklung-Gesch. 1969a;128:271–289. doi: 10.1007/BF00522528. [DOI] [PubMed] [Google Scholar]
- 41.Morest DK. The growth of dendrites in the mammalian brain. Z Anat Entwicklung-Gesch. 1969b;128:290–317. doi: 10.1007/BF00522529. [DOI] [PubMed] [Google Scholar]
- 42.Muller D, Buchs P-A, Stoppini L. Time course of synaptic development in hippocampal organotypic cultures. Dev Brain Res. 1993;71:93–100. doi: 10.1016/0165-3806(93)90109-n. [DOI] [PubMed] [Google Scholar]
- 43.O’Connor TP, Duerr JS, Bentley D. Pioneer growth cone steering decisions mediated by single filopodial contacts in situ. J Neurosci. 1990;10:3935–3946. doi: 10.1523/JNEUROSCI.10-12-03935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Rourke NA, Fraser SE. Dynamic changes in optic fiber terminal arbors lead to retinotypic map formation: an in vivo confocal microscopic study. Neuron. 1990;5:159–171. doi: 10.1016/0896-6273(90)90306-z. [DOI] [PubMed] [Google Scholar]
- 45.O’Rourke NA, Cline HT, Fraser SE. Rapid remodeling of retinal arbors in the tectum with and without blockade of synaptic transmission. Neuron. 1994;12:921–934. doi: 10.1016/0896-6273(94)90343-3. [DOI] [PubMed] [Google Scholar]
- 46.Papa M, Bundman MC, Greenberger V, Segal M. Morphological analysis of dendritic spine development in primary cultures of hippocampal neurons. J Neurosci. 1995;15:1–11. doi: 10.1523/JNEUROSCI.15-01-00001.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pappas GD, Fox GQ, Masurovsky EB, Peterson ER, Crain SM. Neuronal growth cone relationships and their role in synaptogenesis in the mammalian central nervous system. Adv Neurol. 1975;12:163–180. [PubMed] [Google Scholar]
- 48.Pawley J. Plenum; New York: 1995. Handbook of biological confocal microscopy, 2nd Ed. . [Google Scholar]
- 49.Phelps PE, Adinolfi AM, Levine MS. Development of the kitten substantia nigra: a rapid Golgi study of the early postnatal period. Brain Res. 1983;312:1–19. doi: 10.1016/0165-3806(83)90116-5. [DOI] [PubMed] [Google Scholar]
- 50.Pokorny J, Yamamoto T. Postnatal ontogenesis of hippocampal CA1 area in rats. I. Development of dendritic arborisation in pyramidal neurons. Brain Res Bull. 1981;7:113–120. doi: 10.1016/0361-9230(81)90075-7. [DOI] [PubMed] [Google Scholar]
- 51.Povlishock JT. The presence of perisomatic processes during maturation of the hypoglossal, vagal and red nuclei of the rat. Brain Res. 1974;1:272–278. doi: 10.1016/0006-8993(74)90605-2. [DOI] [PubMed] [Google Scholar]
- 52.Pozzo Miller LD, Petrozzino JJ, Mahanty NK, Connor JA. Optical imaging of cytosolic calcium, electrophysiology, and ultrastructure in pyramidal neurons of organotypic slice cultures from rat hippocampus. NeuroImage. 1993;1:109–120. doi: 10.1006/nimg.1993.1004. [DOI] [PubMed] [Google Scholar]
- 53.Purpura D. Dendritic differentiation in human cerebral cortex: normal and aberrant developmental patterns. Adv Neurol. 1975;12:91–116. [PubMed] [Google Scholar]
- 54.Ramoa AS, Campbell G, Shatz CJ. Transient morphological features of identified ganglion cells in living fetal and neonatal retina. Science. 1987;237:522–525. doi: 10.1126/science.3603038. [DOI] [PubMed] [Google Scholar]
- 55.Ruthel G, Banker G. “Waves”: growth cone-like structures that travel down the processes of cultured hippocampal neurons. Soc Neurosci Abstr. 1992;18:1286. [Google Scholar]
- 56.Saito Y, Murakami F, Song W-J, Okawa K, Shimono K, Katsumaru H. Developing corticorubral axons of the cat form synapses on filopodial dendritic protrusions. Neurosci Lett. 1992;147:81–84. doi: 10.1016/0304-3940(92)90779-7. [DOI] [PubMed] [Google Scholar]
- 57.Skoff RP, Hamburger V. Fine structure of dendritic and axonal growth cones in embryonic chick spinal cord. J Comp Neurol. 1974;153:107–148. doi: 10.1002/cne.901530202. [DOI] [PubMed] [Google Scholar]
- 58.Smith SJ, Cooper M, Waxman A. Schattauer Verlag; Stuttgart: 1990. Laser microscopy of subcellular structure in living neocortex: Can one see dendritic spines twitch? XXIII Symposia Medica Hoechst: The Biology of Memory (Squire LR, Lindenlaub E, eds), pp 49–71. . [Google Scholar]
- 59.Sretavan D, Reichardt LF. Time-lapse video analysis of retinal ganglion cell axon pathfinding at the mammalian optic chiasm: growth cone guidance using intrinsic chiasm cues. Neuron. 1993;10:761–777. doi: 10.1016/0896-6273(93)90176-r. [DOI] [PubMed] [Google Scholar]
- 60.Steward O, Davis L, Dotti C, Phillips LL, Rao A, Banker G. Protein synthesis and processing in cytoplasmic microdomains beneath postsynaptic sites on CNS neurons. A mechanism for establishing and maintaining a mosaic postsynaptic receptive surface. Mol Neurobiol. 1988;2:227–261. doi: 10.1007/BF02935634. [DOI] [PubMed] [Google Scholar]
- 61.Stoppini L, Buchs P-A, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 62.Terasaki M, Dailey ME. Confocal microscopy of living cells. In: Pawley JB, editor. Handbook of biological confocal microscopy, 2nd Ed. Plenum; New York: 1995. pp. 327–346. [Google Scholar]
- 63.Trommald M, Jensen V, Andersen P. Analysis of dendritic spines in rat CA1 pyramidal cells intracellularly filled with a fluorescent dye. J Comp Neurol. 1995;353:260–274. doi: 10.1002/cne.903530208. [DOI] [PubMed] [Google Scholar]
- 64.Ulfhake B, Cullheim S. Postnatal development of the cat hind limb motoneurons. II. In vivo morphology of dendritic growth cones and the maturation of dendrite morphology. J Comp Neurol. 1988;278:88–102. doi: 10.1002/cne.902780106. [DOI] [PubMed] [Google Scholar]
- 65.Vaughn JE. Fine structure of synaptogenesis in the vertebrate central nervous system. Synapse. 1989;3:255–285. doi: 10.1002/syn.890030312. [DOI] [PubMed] [Google Scholar]
- 66.Vaughn JE, Henrikson CK, Grieshaber JA. A quantitative study of synapses on motor neuron dendritic growth cones in developing mouse spinal cord. J Cell Biol. 1974;60:664–672. doi: 10.1083/jcb.60.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Westrum LE, Blackstad TW. An electron microscope study of the stratum radiatum of the rat hippocampus (regio superior, CA1) with particular emphasis on synaptology. J Comp Neurol. 1962;119:281–292. doi: 10.1002/cne.901190303. [DOI] [PubMed] [Google Scholar]