Abstract
We describe the isolation of a molluscan (Lymnaea stagnalis) full-length complementary DNA that encodes a mature polypeptide (which we have named Lym-eGluR2) with a predicted molecular weight of 105 kDa that exhibits 44–48% identity to the mammalian kainate-selective glutamate receptor GluR5, GluR6, and GluR7 subunits. Injection of in vitro-transcribed RNA from this clone intoXenopus laevis oocytes results in the robust expression of homo-oligomeric cation channels that can be gated byl-glutamate (EC50 = 1.2 ± 0.3 μm) and several other glutamate receptor agonists; rank order of potency: glutamate ≫ kainate > ibotenate > AMPA. These currents can be blocked by the mammalian non-NMDA receptor antagonists 6,7-dinitroquinoxaline-2,3-dione, 6-cyano-7-nitroquinoxaline-2,3-dione, and 1-(4-chlorobenzoyl)piperazine-2,3-dicarboxylic acid. Ionic-replacement experiments have shown that the agonist-induced current is carried entirely by sodium and potassium ions. In situ hybridization has revealed that the Lym-eGluR2 transcript is present in all 11 ganglia of the Lymnaea CNS, including the 4-cluster motorneurons within the paired buccal ganglia. The pharmacological properties and deduced location of Lym-eGluR2 are entirely consistent with it being (a component of) the receptor, which has been identified previously on buccal motorneurons, that mediates the excitatory effects of glutamate released from neurons within the feeding central pattern generator.
Keywords: buccal ganglion, complementary DNA cloning, feeding behavior, in situ hybridization, ion channel, ionotropic glutamate receptor, kainate receptor, mollusc ( Lymnaea stagnalis ), Xenopus oocyte expression, 4-cluster motorneurons
Glutamate is an excitatory neurotransmitter that is present in both vertebrate and invertebrate nervous systems. In the mammalian CNS, this molecule mediates its depolarizing effects by binding to ionotropic glutamate receptors (GluRs) and promoting the influx of cations into the postsynaptic cell. In recent years, molecular biological studies have revealed a plethora of GluR subunits within the mammalian brain, and these have been distinguished pharmacologically based on their agonist selectivity. To date, four AMPA-selective subunits (GluR1–GluR4) (Hollmann et al., 1989;Keinänen et al., 1990; Nakanishi et al., 1990; Sakimura et al., 1990), five kainate-selective subunits (GluR5–GluR7 and KA-1 and KA-2) (Bettler et al., 1990, 1992; Egebjerg et al., 1991; Werner et al., 1991; Herb et al., 1992; Lomeli et al., 1992; Morita et al., 1992;Sakimura et al., 1992), five NMDA-selective subunits (NR1 and NR2A–NR2D) (Moriyoshi et al., 1991; Ikeda et al., 1992; Kutsuwada et al., 1992; Meguro et al., 1992; Monyer et al., 1992), and three GluR-like polypeptides of unknown function (δ1, δ2, and χ-1, which is also called NMDAR-L) (Yamazaki et al., 1992; Lomeli et al., 1993; Ciabarra et al., 1995; Sucher et al., 1995) have been identified. This pharmacological classification reflects the extent of sequence similarity seen between polypeptides, i.e., any two subunits having the same agonist preference will display greater sequence identity than any two subunits having different agonist selectivity.
In contrast to the situation for vertebrate GluRs, very little has been published on the structures and functions of the corresponding invertebrate channels. Three sequences have been described for GluR-like polypeptides of Drosophila melanogaster; one of these (DGluR-I) (Ultsch et al., 1992) most closely resembles mammalian AMPA-selective subunits; the second (DGluR-II) (Schuster et al., 1991) is only weakly related to vertebrate ionotropic GluR sequences; and the third (DNMDAR-I) (Ultsch et al., 1993) exhibits greatest identity to the mammalian NR1 subunit. In addition, an AMPA receptor-like sequence has been reported for the fresh-water mollusc Lymnaea stagnalis (Hutton et al., 1991). However, when expressed inXenopus laevis oocytes, these polypeptides either do not form functional ion channels (Hutton et al., 1991; Schuster et al., 1991; Ultsch et al., 1992, 1993) or do so only very inefficiently (Ultsch et al., 1992).
We have performed a detailed molecular analysis of molluscan glutamate-gated ion channels to investigate the relationship(s) between specific receptors and well characterized patterns of behavior in which glutamate is known to play a role (Quinlan and Murphy, 1991; Dale and Kandel, 1993; Quinlan et al., 1995). Here we report on the functional properties and location of an ionotropic GluR polypeptide fromLymnaea that displays greatest sequence similarity to the mammalian GluR5, GluR6, and GluR7 subunits. Taken together, our data strongly implicate the molluscan polypeptide in the motorneuron control of buccal muscle movement during feeding.
MATERIALS AND METHODS
Isolation of a full-length Lym-eGluR2 complementary DNA.A partial complementary DNA (cDNA) was obtained using the PCR and the following degenerate oligonucleotide primers: 5′-GCG GAATTCGT(C/G)(C/G)(C/G)GGA(C/T)GGXAA(A/G)TA(C/T)GG-3′ (where X = G, A, T, and C), which is based on the DNA sequences that encode a region amino-terminal of the first putative membrane-spanning domain, TM1, (I/L)(A/E/S/T)IV(G/P/S)DGKYG (single-letter amino-acid code), and 5′-GCC GGTACC(A/G/T)ATXA(A/G)XGT(A/G)AA(A/G)AACCACCA-3′, which is based on the DNA sequences that encode part of the second putative membrane-spanning domain (TM3) (Keinänen et al., 1990; Hollmann et al., 1994), WWFFTLIIIS, of the mammalian GluR1–GluR4 subunits. Lymnaea first-strand cDNA, synthesized from adult nervous system total RNA using both random hexamers and an oligo-dT17 primer, was used as template. Amplification was for 40 cycles of 94°C for 1 min (denaturation), 55°C for 1 min (annealing), and 72°C for 1 min (extension). Products were cloned into M13mp18 (Boehringer Mannheim, Mannheim, Germany), taking advantage of restriction endonuclease recognition sites (underlined) incorporated into the PCR primers, and sequenced.
Four different types of cDNA, encoding previously unidentified GluR-like polypeptides, were obtained. One of these (provisionally named clone #21) (Darlison et al., 1994) was used to screen 5 × 105 bacteriophage of an adult LymnaeaCNS cDNA library (a gift from Dr. Erno Vreugdenhil, Vrije Universiteit, Amsterdam), constructed in λZAPII, under conditions of high stringency. Hybridization was in 6× SSC (1× SSC = 0.15 m NaCl, 0.015 m sodium citrate, pH 7), 5× Denhardt’s solution, 0.5% (w/v) SDS, 10% (w/v) dextran sulfate, and 100 μg/ml yeast tRNA, at 65°C overnight. Library filters were washed in 0.1× SSC, 0.1% (w/v) SDS at 65°C for 2 × 15 min. Two positive plaques were identified and purified. These were found to contain identical inserts of ∼4.5 kb but, nevertheless, lacked sequences encoding the amino-terminal 230 amino acids.
The missing 5′ sequence was obtained using the rapid amplification of cDNA ends (RACE) (Frohman and Martin, 1989), essentially as described previously (Harvey et al., 1991), using random-nonamer primedLymnaea CNS first-strand cDNA as template. The first-stage PCR used the transcript-specific primer 21.2: 5′-CGTAATTGTGACGAAAATCTTCCATGTCTA-3′ in combination with RoRidT17and Ro (Harvey et al., 1991). Amplification was for 3 cycles of 94°C for 1 min, 42°C for 1 min, and 72°C for 1 min, followed by 38 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min. The second-stage PCR, which contained the transcript-specific primer 21.1: 5′-GAT TCTAGATCTAAATGAGTAACGATGTAA-3′ and Ri (Harvey et al., 1991), was for 38 cycles of 94°C for 1 min, 57°C for 1 min, and 72°C for 1 min. This yielded a product of ∼450 bp. The remaining 5′ sequence was obtained by a second RACE step in which primer 21.1 was used in the first-stage PCR with RoRidT17and Ro, and primer 21.4: 5′-TAG GGTACCAGGTCGGTGTAGGCCCGGCCC-3′ was used with Ri in the second-stage PCR. For the first-stage PCR, amplification was for 3 cycles of 94°C for 1 min, 42°C for 1 min, and 72°C for 3 min, followed by 37 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min. For the second-stage PCR, amplification was for 35 cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 2 min. Products were cloned into either M13mp18 or pBluescript II SK+ (Stratagene, Heidelberg, Germany), taking advantage of restriction endonuclease recognition sites (underlined) incorporated into the PCR primers, and sequenced.
A complete cDNA, for expression purposes, was constructed by ligating part of the cDNA that was obtained from the library to a cDNA fragment that contained the 5′ end, which was generated using the PCR. The 5′ portion was amplified using primers 21.6: 5′-CGT GGATCCCTTGGACATACGATGACCAGC-3′, which represents the first three codons of the signal peptide and upstream 5′-untranslated sequence; and 21.7: 5′-AAG GAATTCAAGCTTGGACCCGATGCCCAC-3′, which recognizes a sequence that includes a naturally occurringHindIII restriction site (in italics) present within the library-derived cDNA fragment. Amplification was for 40 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 2.5 min. The resultant product (∼1.1 kb in size) was digested withBamHI and EcoRI at unique restriction endonuclease recognition sites (underlined) that were incorporated into primers 21.6 and 21.7, respectively, cloned into the corresponding sites in pBluescript II SK+, and sequenced. This plasmid was linearized with BamHI, dephosphorylated, and then digested withHindIII, which cleaves at the site within primer 21.7. The ∼4.5 kb library-derived 3′-cDNA fragment was partially digested withXbaI, and an ∼2.5 kb fragment was isolated and ligated into the XbaI site of pGEM-3Zf(+) (Serva Feinbiochemica, Heidelberg, Germany). This plasmid, which contains all of the 3′ coding sequence, was subsequently digested with BamHI (which recognizes a unique site within the vector polylinker), dephosphorylated, and finally digested with HindIII. The 5′ and 3′ fragments were then ligated together at the commonHindIII site, and the complete cDNA cloned as aBamHI fragment into the BamHI site of pGEMHE (Liman et al., 1992) to generate plasmid pleGluR2.
In vitro transcription, injection of oocytes, and electrophysiology. Capped RNA was transcribed in vitroafter linearization of plasmid pleGluR2 with NheI using T7 RNA polymerase. This was injected (at a concentration of 250 ng/μl; ∼50 nl per oocyte) into manually defolliculated stage V and VIXenopus oocytes; control oocytes were injected with ∼50 nl of sterile deionized water. Oocytes were maintained at 19°C in Barth’s medium [(in mm): 88 NaCl, 1 KCl, 0.41 CaCl2, 0.33 Ca(NO3)2, 0.82 MgSO4, 2.4 NaHCO3, and 10 HEPES, pH 7.6, supplemented with 100 μg/ml each of gentamycin, penicillin, and streptomycin] for 2–7 d.
Oocytes were voltage-clamped at room temperature (RT) using a standard two-microelectrode amplifier (Bio-logic, Claix, France), and recordings were made in standard frog Ringer’s solution (in mm): 115 NaCl, 2.5 KCl, 1.85 CaCl2, 5 HEPES, pH 7.2. Electrodes were filled with 3 m KCl and had resistances of between 2 and 5 MΩ. The bath was connected to a ground Ag/AgCl electrode through a 1 m KCl/agar bridge. Data were either recorded on a chart recorder or stored on an IBM PC-AT compatible computer using a Labmaster A-D converter (Scientific Solutions, Solon, OH). Oocytes were continuously perfused with frog Ringer’s at ∼5 ml/min and drugs, which were dissolved in either Ringer’s, dimethylsulfoxide, or ethanol, were bath-applied. At the dilutions used, the organic solvents had no effect on either the resting membrane potential or the membrane conductance. Concanavalin A type IV (Sigma, Deisenhofen, Germany) was dissolved in frog Ringer’s to yield a final concentration (for the tetramer) of either 1 or 10 μm, assuming a molecular weight for the monomer of 26.5 kDa.
All experiments were performed at a clamp potential of −60 mV unless otherwise stated. Current–voltage relationships and null potentials were determined using a 3 sec voltage ramp in the range of −120 mV to +40 mV; individual steps were separated by 3 sec intervals. The current in agonist-free solution was subtracted from that recorded in the presence of 1 μml-glutamate to obtain the agonist-sensitive current. To determine the ionic specificity of the channel, modified Ringer’s solutions containing decreasing concentrations of sodium ions were prepared by substitutingN-methyl-d-glucamine for NaCl, and adjusting the pH to 7.2 with HCl. A potassium-enriched solution (10 mm) was prepared by adding K2SO4 to the normal Ringer’s solution. A sodium- and potassium-free solution was made by iso-osmotically substitutingN-methyl-d-glucamine for NaCl and KCl, and adjusting the pH to 7.2 with HCl. To study permeation of calcium ions, the NaCl concentration of the Ringer’s was maintained at 115 mm, and the CaCl2concentration was either lowered to 0.185 mm (low calcium solution) or raised to 10 mm (high calcium solution).
Data were analyzed using pClamp 5.1 (Axon Instruments, Foster City, CA) and SigmaPlot 4.1 (Jandel Scientific, Erkrath, Germany) software. Concentration–response curves were fitted to the nonlinear Hill equation: I = 1/(1 + (EC50/x)nH, where I is the measured amplitude of the evoked current,x is the effector concentration, EC50is the effector concentration causing the half-maximal response, andnH is the Hill number.
Acetylcholine chloride, GABA, l-aspartic acid, l-cysteic acid, flufenamic acid,l-glutamic acid, l-glutamic acid diethyl ester (GDEE), glycine, l-homocysteic acid, kainic acid,N-methyl-dl-aspartic acid, niflumic acid, picrotoxin, and taurine were purchased from Sigma. (RS)-α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid, (d)-2-amino-3-phosphonopropionic acid (d-AP3), (d)-2-amino-4-phosphonobutyric acid (d-AP4), (d)-2-amino-5-phosphonopentanoic acid (d-AP5), 1-(4-chlorobenzoyl)piperazine-2,3-dicarboxylic acid (CBPD), 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX), 6,7-dinitroquinoxaline-2,3-dione (DNQX), domoic acid, ibotenic acid, philanthotoxin 343 (PhTX-343), and l-quisqualic acid were from Tocris Neuramin (Langford, Bristol, England).
In situ hybridization. Nervous systems from adultLymnaea (shell length ∼30 mm) were prepared for in situ hybridization as follows. Dissected nervous tissue was fixed in 1% (w/v) paraformaldehyde/1% (v/v) acetic acid overnight, then dehydrated through an ascending ethanol series. It was then incubated in ethanol/amylacetate (1:1) for 30 min, amylacetate for 2 × 30 min, and amylacetate/paraffin (1:1) for 30 min, and finally embedded in paraffin. Either 7 μm sections of whole nervous systems or 5 μm sections of dissected buccal ganglia were cut using a microtome, mounted onto 0.5% (w/v) KCr(SO4)2/0.5% (w/v) gelatin-coated slides using Eiweiss-Glycerin (Merck, Darmstadt, Germany), and dried at 37°C overnight. Before in situhybridization, sections were incubated at 60°C for 1 hr, immersed in xylene for 2 × 15 min, dehydrated through an ascending ethanol series, and air-dried for 30 min. The tissue was permeabilized by incubation in 0.02% (w/v) pepsin (Boehringer Mannheim) in 0.2 m HCl at 37°C for 7 min, post-fixed in 2% (w/v) paraformaldehyde in PBS (130 mm NaCl, 7 mmNa2HPO4, 3 mmNaH2PO4) at RT for 4 min, and then treated with 1% (w/v) hydroxylammoniumchloride (Merck) in PBS at RT for 15 min. Sections were subsequently washed in PBS at RT for 5 min, dehydrated through an ascending ethanol series, and stored in 96% (v/v) ethanol at 4°C until use.
A 45-base antisense oligonucleotide (5′-ACGCGGCTCAGACCCTGTGCCGTGCCATAGATGACCAGCATCTCC-3′; complementary to the nucleotide sequence encoding residues 139–154 of the mature Lym-eGluR2 polypeptide; Fig. 1) was labeled to high specific activity (≥109 cpm/μg) using [α35S]dATP (Du Pont de Nemours, Bad Homburg, Germany) and terminal deoxynucleotidyl transferase (Life Technologies, Eggenstein, Germany), as described previously (Wisden et al., 1991). Hybridization was in 50% (v/v) formamide, 4× SSC, 25 mm sodium phosphate buffer, pH 7, 1 mm sodium pyrophosphate, 20 mm dithiothreitol, 5× Denhardt’s solution, 100 μg/ml polyadenylic acid, 120 μg/ml heparin, 10% (w/v) dextran sulfate, and 200 μg/ml acid/alkali-denatured salmon sperm DNA, containing 1250 cpm probe/μl, at 42°C overnight. Slides were washed in 1× SSC at 55°C for 2 × 30 min, then dipped in Kodak NTB-3 photographic emulsion (Integra Biosciences, Fernwald, Germany). Sections were developed, as described previously (Harvey et al., 1994a), after between 50 and 140 d. In negative control experiments, the hybridization mixture contained, in addition, a 200-fold excess of the same unlabeled oligonucleotide.
Fig. 1.

Alignment of the Lymnaea Lym-eGluR2 sequence with those of the rat GluR5, GluR6, and GluR7 subunits. The amino acid sequences (shown in single-letter code) were aligned using the computer program PILEUP (Wisconsin Package, Version 8, September 1994, Genetic Computer Group, 575 Science Drive, Madison, WI);dots denote gaps that have been introduced to maximize the alignment. Positions at which all four of the sequences are identical are boxed. Amino acids are numbered from the proposed mature amino-terminal residues; the signal peptides (von Heijne, 1986) are indicated by negative numbering. The three putative membrane-spanning domains (TM1, TM3, and TM4) and the presumed membrane-associated segment (TM2) (Keinänen et al., 1990;Hollmann et al., 1994) are marked below the sequences bysolid and broken lines, respectively. For theLymnaea polypeptide, potential N-linked glycosylation sites (in the large amino-terminal presumed extracellular domain) and potential phosphorylation sites for protein kinase C and multifunctional calmodulin-dependent protein kinase type II (within presumed intracellular regions) are denoted by asterisks,open circles, and filled circles, respectively. Note that serine residue 593, which is a target for phosphorylation by multifunctional calmodulin-dependent protein kinase type II, is also part of a recognition sequence for protein kinase C. The mature sequences of the rat GluR5, GluR6, and GluR7 subunits have been taken from Bettler et al. (1992). The sequence of the cDNA, from which the sequence of the Lym-eGluR2 polypeptide was deduced, has been given the EMBL accession number X87404.
RESULTS
Structure of Lym-eGluR2
Degenerate PCR, using primers designed on the basis of conserved amino acid sequences amino-terminal of the first (TM1) and within the second (TM3) (Keinänen et al., 1990; Hollmann et al., 1994) presumed membrane-spanning domain of mammalian GluR1–GluR4 subunits, resulted in the amplification of four different types ofLymnaea cDNA. All of these encode GluR-like polypeptides (provisionally named clone #2, #13, #19, and #21) (Darlison et al., 1994), and these are distinct from the previously describedLymnaea GluR sequence (Hutton et al., 1991). A full-length clone encoding one of these (clone #21) has now been obtained using a combination of conventional cDNA library screening and 5′ RACE. This cDNA contains an open reading frame that specifies a protein of 953 amino acids (Fig. 1), the first 24 of which have the properties of a signal peptide (von Heijne, 1986). The mature polypeptide is predicted to have a molecular weight of 105 kDa, and exhibits all of the features of mammalian glutamate-gated cation-channel subunits, including a long amino-terminal presumed extracellular domain, three putative membrane-spanning domains (TM1, TM3, and TM4), an assumed membrane-associated segment (TM2) (Hollmann et al., 1994), and a carboxyl terminus that is thought to be intracellular. In addition, it displays strong sequence identity to mammalian non-NMDA GluR subunits: 38–39% to GluR1–GluR4, 44–48% to GluR5–GluR7, and 37% to KA-1 and KA-2. The molluscan polypeptide is less related in sequence to the δ1 and δ2 subunits (30 and 28% identity, respectively), to χ-1 (23% identity), to NR1 (25% identity), and to NR2A–NR2D (22–24% identity). It also exhibits 37, 35, 32, and 27% identity, respectively, to the original Lymnaea GluR polypeptide and the Drosophila DGluR-I, DGluR-II, and DNMDAR-I subunits. Surprisingly, the new Lymnaea sequence displays 37–40% identity to the avian, amphibian, and piscine kainate-binding proteins (Gregor et al., 1989; Wada et al., 1989; Kimura et al., 1993; Wo and Oswald, 1994), which do not form functional ion channels and which are considerably smaller in size (i.e., ∼50 kDa) than ionotropic GluRs.
We have named the Lymnaea polypeptide, which we describe here, Lym-eGluR2; the letter “e” stands for excitatory. Inhibitory ionotropic GluRs, also known to exist in molluscs (Yarowsky and Carpenter, 1976; Kehoe, 1978; Ikemoto and Akaike, 1988), when identified at the molecular level will be given the prefix “i.” The originally described Lymnaea GluR polypeptide (Hutton et al., 1991), which has been named LymGluR by others (Hollmann and Heinemann, 1994), will henceforth be known as Lym-eGluR1.
As with mammalian ionotropic GluR subunits, the Lym-eGluR2 polypeptide is predicted to be glycosylated. Six consensus sites for N-linked sugar attachment are located in the amino-terminal presumed extracellular domain. Three of these (N242, N380, and N395; Fig. 1) are found at the equivalent positions in GluR5, GluR6, and GluR7; one site (N395) is conserved in all non-NMDA GluR sequences. The Lymnaeasequence also has five consensus motifs for phosphorylation by protein kinase C and one for multifunctional calmodulin-dependent protein kinase type II (Woodgett et al., 1986; Kemp and Pearson, 1990) (Fig. 1) in regions of the polypeptide that are predicted to be intracellular (i.e., either between TM1 and TM2 or TM2 and TM3, or in the carboxyl-terminal segment after TM4).
Ionic properties of homo-oligomeric Lym-eGluR2 channels
Injection of Xenopus oocytes with in vitro-transcribed RNA for the Lymnaea polypeptide resulted in the appearance of functional glutamate-gated ion channels in the cell membrane. Large inward currents (at a holding potential of −60 mV) could be reliably and readily detected; for example, 1 μml-glutamate elicited responses, with an average amplitude of ∼200 nA (n ∼ 100 oocytes) 2 d postinjection, that reached ∼1.5 μA after 5–7 d. The glutamate-evoked currents (IGlu) were dose-dependent (Fig. 2A) and had a fast time course of both onset and offset. There was no evidence under our experimental conditions of desensitization, i.e., no significant reduction in current was observed upon agonist application for a period of up to 3 min (data not shown). No responses were evoked by the nonglutamatergic compounds acetylcholine, GABA, glycine, and taurine in oocytes that did respond to glutamate. Control oocytes, which were injected instead with deionized water, did not respond to glutamate (tested at up to 1 mm).
Fig. 2.
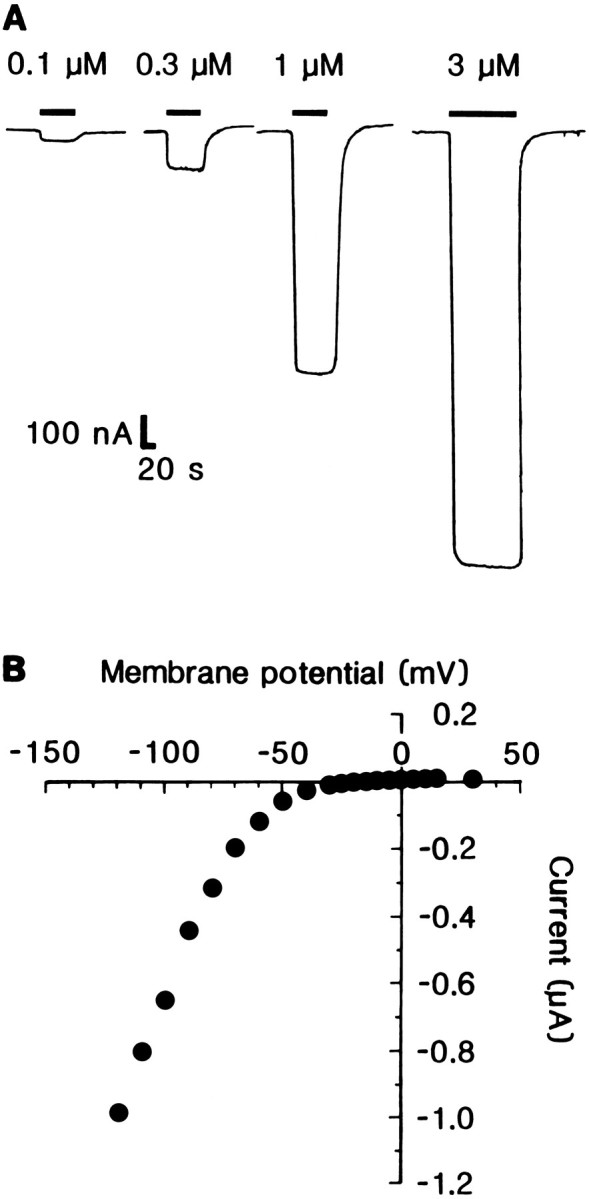
Dose-dependence and current–voltage relationship of glutamate-evoked responses at Lym-eGluR2 channels expressed in Xenopus oocytes. A, Effect of four different concentrations of l-glutamate on a single oocyte 5 d postinjection. Downward deflections denote inward currents, and horizontal bars indicate the duration of agonist application. B, Current–voltage relationship of the response elicited by 1 μml-glutamate from an oocyte 5 d postinjection.
The current–voltage (I/V) relationship of the expressed channels for glutamate-induced responses showed a pronounced voltage dependence (Fig. 2B). Between −120 mV and approximately −60 mV, the I/V curve was linear; however, at more positive voltages, it exhibited strong inward rectification. No outward currents were observed at positive membrane potentials. Because it is impossible to determine the reversal potential from this type of curve, we determined the null potential for IGlu, i.e., the potential at which zero current was observed for the first time; this was −11.0 ± 4.5 mV.
To elucidate the nature of the ions responsible for the current flow through the glutamate-gated channels, we examined the effect of altering the ionic gradient for specific ions on the I/Vrelationship. When sodium ions in the Ringer’s were replaced byN-methyl-d-glucamine, the null potential for IGlu shifted in the hyperpolarizing direction (Fig. 3A) and the amplitude of the current was dramatically reduced (Fig. 3B), showing that sodium ions participate in the agonist-induced current. At concentrations between 10 and 115 mm, the null potential was linearly related to the log of the extracellular sodium ion concentration (Fig. 3A); the slope of this relationship was 34 mV per 10-fold change in sodium ion concentration. This value is smaller than that expected from the Nernst equation, which predicts 58 mV per 10-fold change if only sodium ions are permeable. Thus, some other ion(s) must also penetrate the channel. This notion is supported by the observation that the null potential deviates from the linear relationship at sodium ion concentrations close to 1 mm (Fig. 3A), where the contribution of other ions such as potassium would be more prominent. When the extracellular potassium ion concentration was increased from 2.5 to 10 mm, in the presence of sodium ions, the null potential shifted in the depolarizing direction by ∼10 mV (data not shown). Replacement of both sodium and potassium ions byN-methyl-d-glucamine completely eliminated the response to glutamate (Fig. 3B).
Fig. 3.
Ionic permeability of Lym-eGluR2 channels expressed in Xenopus oocytes. A, Relationship of the null potential for glutamate-evoked currents to the external concentration of sodium ions. The data derive from two oocytes 5 d postinjection. B, Current–voltage relationships of the response elicited by 1 μml-glutamate from an oocyte 6 d postinjection in normal Ringer’s (open circles), in a Ringer’s that contained only 20% of the normal sodium ion concentration (filled squares), or in a sodium- and potassium-free Ringer’s (inverted filled triangles). C, Current–voltage relationships of the response evoked by 1 μml-glutamate from an oocyte 6 d postinjection in either normal frog Ringer’s (open circles) or a low calcium (0.185 mm) Ringer’s (inverted filled triangles). D, Current–voltage relationships of the responses elicited by 1 μml-glutamate from an oocyte 6 d postinjection in either normal frog Ringer’s (open circles) or a high calcium (10 mm) Ringer’s (filled triangles). In B–D, responses have been normalized to that evoked by 1 μm glutamate in normal Ringer’s at a holding potential of −120 mV.
To confirm that IGlu was carried by only sodium and potassium ions and that there was no significant contribution by calcium ions, which might also activate endogenous oocyte calcium-dependent chloride channels (Dascal, 1987), we examined the effect of niflumic acid and flufenamic acid (Vernino et al., 1992). Incubation of oocytes with these two chloride-channel blockers (both at 400 μm) for 15 min had no effect on either the size or shape of glutamate-induced currents (data not shown). In addition, either reducing (to 0.185 mm) or increasing (to 10 mm) the external calcium ion concentration had no effect on either the amplitude or voltage-dependence of IGlu or the null potential (Fig. 3C,D).
Agonist selectivity of homo-oligomeric Lym-eGluR2 channels
The EC50 for channel activation byl-glutamate was 1.2 ± 0.3 μm (n = 10 oocytes; Fig.4A), and the Hill number was 2.1 ± 0.4, indicating that more than one agonist molecule is required for channel opening. Several other glutamatergic ligands were tested for their ability to evoke currents, and kainate, ibotenate, and AMPA were found to be powerful agonists (Fig. 4A,B). Note that control oocytes did not respond to any of these compounds (each tested at up to 1 mm). The rank order of potency of the operant agonists was: glutamate ≫ kainate > ibotenate > AMPA. The EC50 values for kainate, ibotenate, and AMPA were 22.3 ± 9.0 μm (n = 10 oocytes), 37.4 ± 10.0 μm (n = 8), and 67.2 ± 10.3 μm (n = 10), respectively. As with glutamate, no significant reduction in current was observed upon the application of any of these agonists for a period of up to 3 min (data not shown), although the response evoked by kainate appeared to exhibit a slightly slower onset than those elicited by the other agonists (Fig. 4B).
Fig. 4.
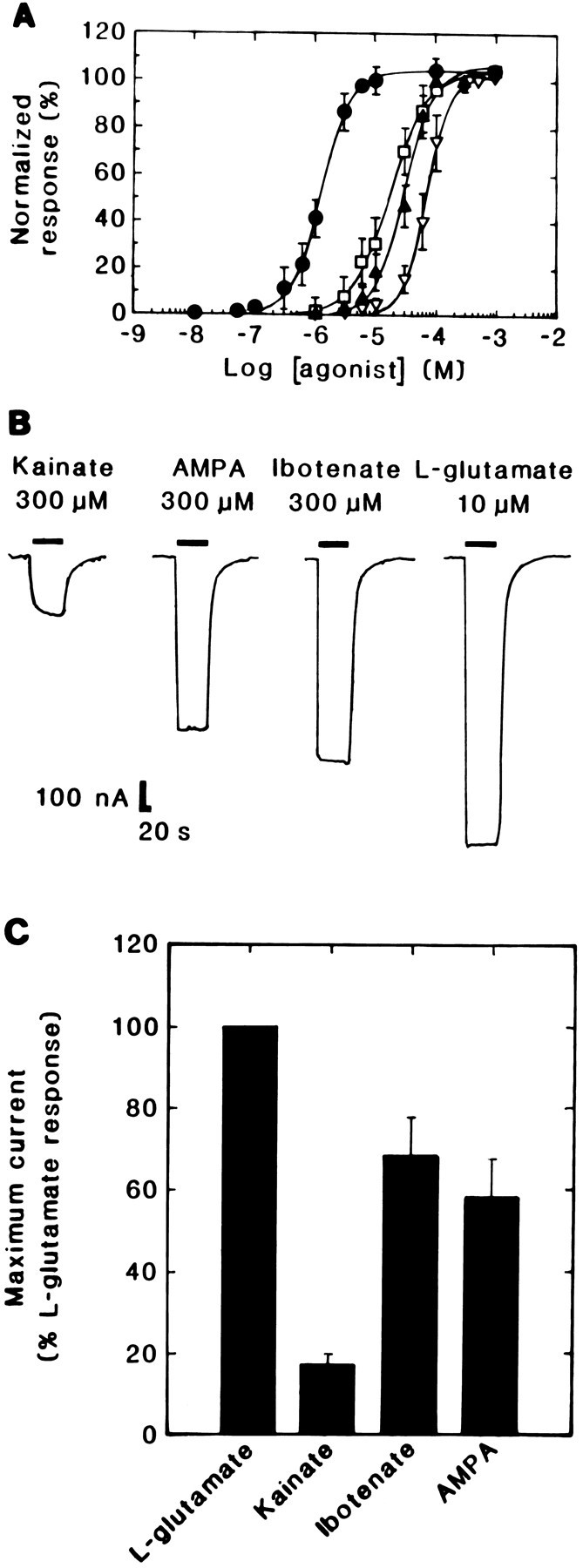
Agonist pharmacology of Lym-eGluR2 channels expressed in Xenopus oocytes. A, Dose–response curves for l-glutamate (filled circles), kainate (open squares), ibotenate (filled triangles), and AMPA (inverted open triangles). Each curve was determined on several oocytes (for glutamate, n = 12; for kainate, n = 12; for ibotenate, n = 10; and for AMPA, n = 12). Each point represents the mean ± SD. Note that the response to each concentration of a given agonist was normalized to the maximal response (denoted as 100%) to that ligand. B, Selected traces of responses evoked by maximally effective concentrations of various glutamatergic agonists (Kainate, 300 μm; AMPA, 300 μm; Ibotenate, 300 μm; l-glutamate, 10 μm) on a single oocyte 6 d postinjection.Downward deflections denote inward currents andhorizontal bars indicate the duration of agonist application. C, A bar graph showing the efficacies of various glutamatergic agonists. All agonists were tested at their maximally effective concentrations on the same oocyte 5 d postinjection. In this case, all responses were normalized to the average peak current elicited by 10 μml-glutamate; the bars represent the mean ± SD of between 4 and 10 experiments. The concentrations of agonists used were: l-glutamate, 10 μm; Kainate, 300 μm; Ibotenate, 300 μm; AMPA, 300 μm; these were applied with a 10 min interval between applications.
GDEE, a drug that has been reported to inhibit native vertebrate quisqualate receptors (Foster and Fagg, 1984) and to act as a weak agonist at the rodent homo-oligomeric GluR1 receptor (Hollmann et al., 1989; Sakimura et al., 1990), appeared to act as an agonist at the molluscan receptor (data not shown) having an EC50 of 64.0 ± 7.4 μm(n = 4 oocytes). However, because the main breakdown product of GDEE is glutamate, the activation seen may be a consequence of low-level (1–2%) contamination by the latter. Other glutamatergic agonists (each tested at up to 100 μm), such asl-aspartate, l-cysteine, domoate, l-homocysteate, NMDA (either alone or in the presence of between 10 and 300 μm glycine), and quisqualate, were unable to gate the Lym-eGluR2 channels.
When dose–response data for the various ligands were normalized to the maximal glutamate response rather than to the maximal current evoked by each individual agonist (Fig. 4A), it became evident that kainate, ibotenate, and AMPA elicited smaller responses thanl-glutamate (data not shown). This was particularly striking for kainate. We therefore examined this effect further by comparing the responses, induced by maximally effective concentrations of each compound, on the same oocyte. As shown in Figure4C, kainate, ibotenate, and AMPA were observed to be less efficacious than glutamate as agonists; kainate, for example, elicited only ∼20% of the current evoked byl-glutamate. To determine whether the lower efficacies of kainate, ibotenate, and AMPA are the result of differences in agonist-induced desensitization, we repeated these experiments in the presence of the plant lectin Concanavalin A, which suppresses the desensitization of both vertebrate (Mayer and Vyklicky, 1989) and invertebrate (Mathers and Usherwood, 1976; Kehoe, 1978) GluRs. Under our conditions, this lectin had no significant effect on either the magnitude or the shape of the responses induced by any of the agonists (data not shown).
Antagonism of homo-oligomeric Lym-eGluR2 channels
We also examined the effects of various mammalian ionotropic GluR antagonists on IGlu. Currents induced by 1 μml-glutamate were reversibly inhibited by the following non-NMDA receptor competitive antagonists (Fig.5A,B): DNQX (IC50 = 1.4 ± 0.4 μm; n = 4 oocytes), CNQX (IC50 = 2.2 ± 0.7 μm;n = 5), and CBPD (IC50 = 2.5 ± 1.4 μm; n = 5). Agonist responses were fully restored after a 10 min perfusion with frog Ringer’s. The classical NMDA receptor competitive antagonistd-AP5 also inhibitedIGlu, albeit very weakly (Fig.5A,B). Other phosphonic acid derivatives such asd-AP3 and d-AP4 had no effect (data not shown). PhTX-343, a synthetic derivative of the polyamine amide wasp toxin philanthotoxin, was a comparatively poor blocker of the molluscan receptor (IC50 = 45.0 ± 3.3 μm; n = 3 oocytes; Fig.5B).
Fig. 5.
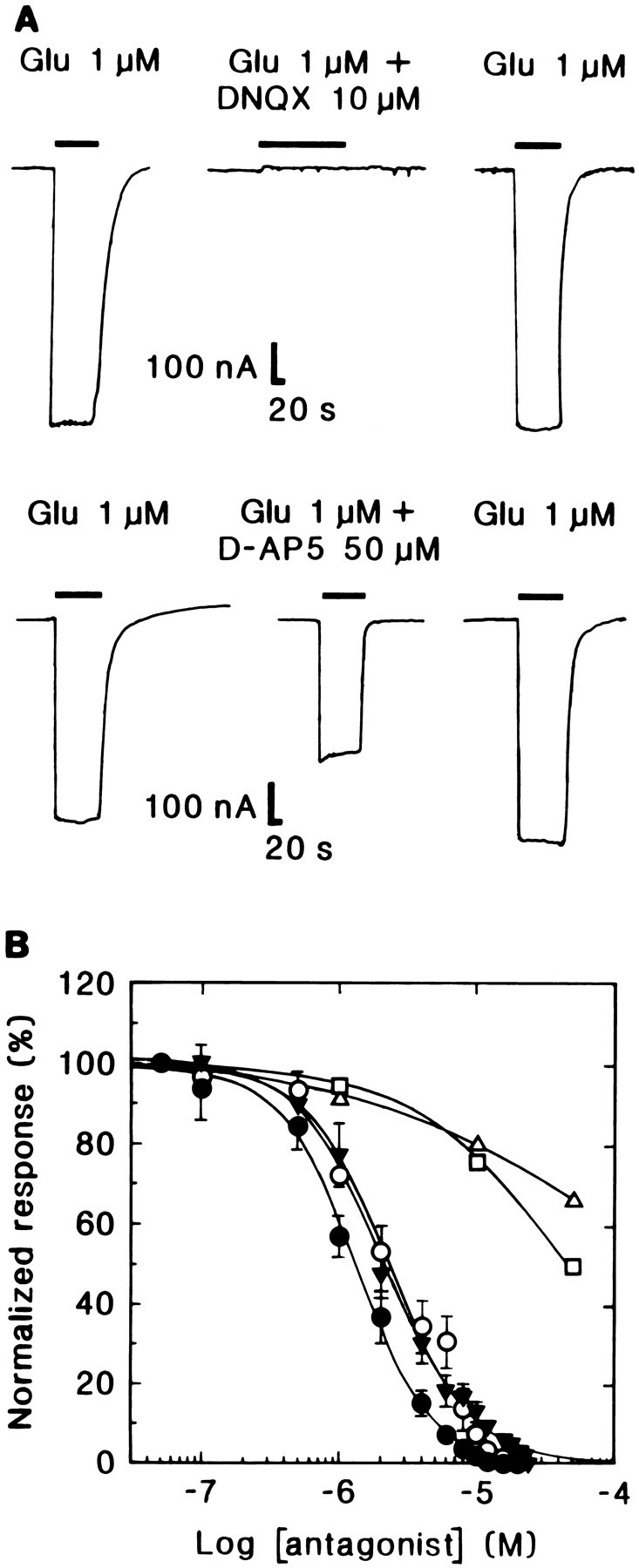
Antagonist pharmacology of Lym-eGluR2 channels expressed in Xenopus oocytes. A, Selected traces illustrating the inhibitory effect of 10 μmDNQX and 50 μmd-AP5 on responses evoked by 1 μml-glutamate on a single oocyte 4 d postinjection.Downward deflections denote inward currents, andhorizontal bars indicate the duration of agonist (and antagonist) application. Note that in both cases, the glutamate response returned to normal after a washout period of 10 min.B, Inhibition curves for various mammalian GluR antagonists. Each point represents the mean ± SD of the percentage inhibition of the current evoked by 1 μml-glutamate; the current elicited in the absence of antagonist is denoted as 100%. Filled circles, DNQX;open circles, CNQX; inverted filled triangles, CBPD; open squares, PhTX-343; open triangles,d-AP5. Each curve was determined on several oocytes (for DNQX, n = 4; for CNQX, n = 5; for CBPD, n = 5; for PhTX-343, n = 3; and ford-AP5, n = 3).
Localization of Lym-eGluR2 transcripts in the Lymnaeanervous system
In situ hybridization, using a specific 45-base oligonucleotide probe, was used to reveal Lym-eGluR2 transcripts within the CNS of adult Lymnaea. Hybridization-positive neurons were detected in all of the nine central ganglia (i.e., right and left cerebral ganglia, right and left parietal ganglia, right and left pedal ganglia, right and left pleural ganglia, and the visceral ganglion) and in the paired buccal ganglia. Figure 6, A andB, shows the labeling pattern in a representative section through seven of these ganglia. Higher-power magnification of the visceral ganglion (Fig. 6C) and the left parietal ganglion (Fig. 6D) demonstrates the specificity and cytoplasmic location of the hybridization signal; note also that control hybridizations (see Materials and Methods) did not yield any signal. Detailed analysis of hybridized sections reveals that a total of 350–400 neurons per nervous system express the Lym-eGluR2 gene. The corresponding mRNA is found in heterogeneous populations of neurons, the cell bodies of which range in size between 15 and 70 μm in diameter; none of these are identifiable peptidergic cells. Within each buccal ganglion, the Lym-eGluR2 transcript is detected in ∼50 cells. These include a cluster of ∼10 cells, most of which are large (40–70 μm in diameter), and are located dorsoposteriorly (Fig.7). Based on their size and position, these cells are most likely the 4-cluster motorneurons (Goldschmeding et al., 1977;Rose and Benjamin, 1979; Kemenes et al., 1991).
Fig. 6.
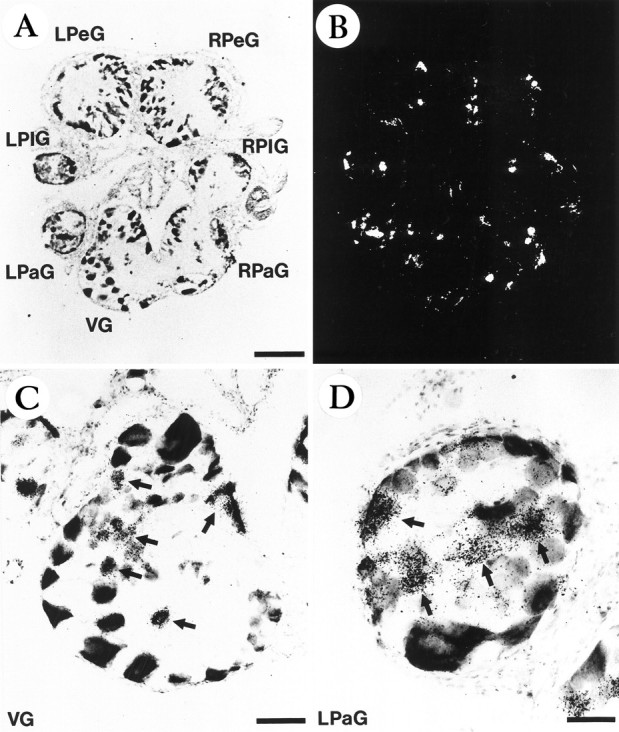
In situ hybridization localization of the Lym-eGluR2 transcript in the adult Lymnaea nervous system. Light-field (A) and corresponding dark-field (B) photomicrographs are shown of a representative section. Note that only 7 of the 11 ganglia are present in this section; the right and left cerebral ganglia and the paired buccal ganglia do not appear. Also shown are higher-magnification light-field photomicrographs of the visceral ganglion (C) and the left parietal ganglion (D). LPaG, Left parietal ganglion; LPeG, left pedal ganglion; LPlG, left pleural ganglion; RPaG, right parietal ganglion;RPeG, right pedal ganglion; RPlG, right pleural ganglion; VG, visceral ganglion. The arrows inC and D point to strongly labeled cells or clusters of cells. Scale bars: 260 μm (A andB), 96 μm (C), and 49 μm (D).
Fig. 7.
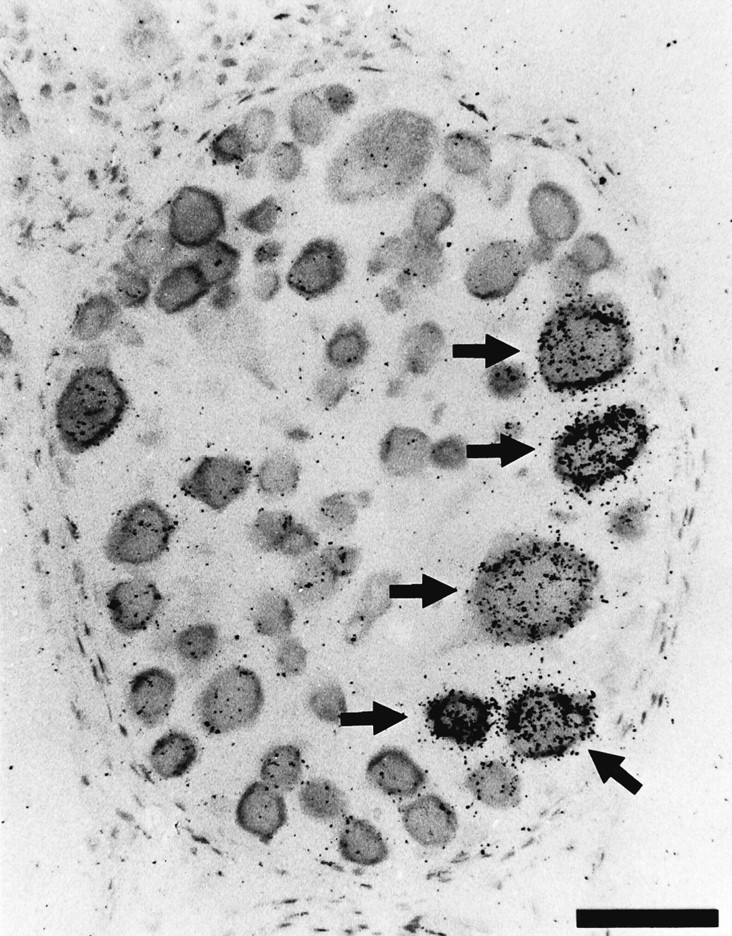
In situ hybridization localization of the Lym-eGluR2 transcript in adult Lymnaea buccal ganglia. A light-field photomicrograph is shown of a section through a single buccal ganglion. The arrows indicate five of the hybridization-positive 4-cluster motorneurons that appear in this section. Scale bar, 67 μm.
DISCUSSION
Lym-eGluR2 is structurally related to the mammalian GluR5, GluR6, and GluR7 subunits, but has a distinct pharmacology
The work described here represents the most detailed electrophysiological and pharmacological characterization of any cloned invertebrate glutamate-gated cation channel. The polypeptide that we have identified in the fresh-water snail Lymnaea clearly resembles in sequence (44–48% identity) the mammalian kainate-selective GluR5–GluR7 family of subunits. Injection ofin vitro-transcribed RNA for the molluscan polypeptide intoXenopus oocytes results in the appearance in the cell membrane of homo-oligomeric channels that can be gated by low micromolar concentrations of l-glutamate, kainate, ibotenate, and AMPA.
Although Lym-eGluR2 is very similar in sequence to the mammalian GluR5–GluR7 subunits, the molluscan channel exhibits some functional differences compared with homo-oligomeric GluR5 and GluR6 channels. For example, although all three receptors respond to kainate, the rank order of potency for Lym-eGluR2 is glutamate ≫ kainate > ibotenate > AMPA; that for GluR5 is domoate ≫ kainate ≫ glutamate > AMPA (Sommer et al., 1992); and that for GluR6 is domoate > kainate ≫ quisqualate > glutamate (Bettler et al., 1992; Hollmann and Heinemann, 1994). Note that ibotenate was not tested at GluR5 and GluR6 receptors. Although the GluR7 subunit does not form functional homo-oligomeric channels, it does bind various GluR ligands; the rank order of potency for displacing [3H]kainate has been reported to be: domoate ≫ glutamate > quisqualate (Bettler et al., 1992). Thus, there are two major pharmacological distinctions between Lym-eGluR2 and the GluR5–GluR7 polypeptides: Lym-eGluR2 is insensitive to domoate and quisqualate, and the EC50 for channel activation of Lym-eGluR2 receptors is ∼20-fold greater for kainate (22.3 μm) than for glutamate (1.2 μm). The lack of response of Lym-eGluR2 channels to quisqualate is consistent with electrophysiological studies on molluscs that demonstrate that this compound activates inhibitory GluRs (Bolshakov et al., 1991; Quinlan and Murphy, 1991). Furthermore, although ibotenate has been shown to activate GluRs on molluscan neurons that mediate hyperpolarization (Roberts et al., 1982; Katz and Levitan, 1993), this molecule also induces excitatory responses when applied to leech (Hirudo medicinalis) (Roberts et al., 1982;Mat Jais et al., 1984), horseshoe crab (Limulus polyphemus), and molluscan (Helix aspersa) neurons (Roberts et al., 1982).
The Lym-eGluR2 channel is permeable to sodium and potassium ions but not to calcium ions, a feature that is characteristic of invertebrate excitatory GluRs (Jan and Jan, 1976; Kehoe, 1978; Mat Jais et al., 1984). This is in contrast to the situation for the mammalian homo-oligomeric GluR6 receptor. When expressed in either human embryonic kidney 293 cells (Köhler et al., 1993) orXenopus oocytes (Egebjerg and Heinemann, 1993), the GluR6 subunit yields channels that are permeable to calcium. This polypeptide exists in different forms, which arise by enzymatic modification of the corresponding mRNA (RNA editing) (Sommer et al., 1991; Köhler et al., 1993). This results in subunits having amino-acid differences at three positions (two in TM1 and one in TM2), which affect the extent of calcium permeability. Although the Lym-eGluR2 polypeptide has a valine (V528; Fig. 1) and a tyrosine (Y532) in TM1 and a glutamine (Q582) in TM2 (residues that are found in a hemi-edited GluR6 subunit), we have no evidence whatsoever for editing of the molluscan RNA at any position.
Relationship of Lym-eGluR2 to other cloned invertebrate glutamate-gated cation channels
The sequences of four invertebrate GluR-like polypeptides have been described previously; three of these (DGluR-I, DGluR-II, and DNMDAR-I) derive from Drosophila and one (Lym-eGluR1, formerly called LymGluR) emanates from Lymnaea. However, we consider none of these to be capable of forming bona fide glutamate-gated cation channels. The first publishedDrosophila sequence, DGluR-II (Schuster et al., 1991), which displays no >33% identity to any mammalian GluR polypeptide, was reported to form homo-oligomeric channels when the corresponding cDNA was expressed in Xenopus oocytes. However, these could only be gated by l-glutamate andl-aspartate, and then only at very high concentrations (the EC50 values for channel activation were ∼35 and ∼50 mm, respectively). Subsequently, the same group (Ultsch et al., 1992) noted that artefactual (i.e., non-receptor) responses were observed when concentrations of glutamate >10 mm were applied to oocytes. Another Drosophila polypeptide, DGluR-I, which is most closely related in sequence (41–44% identity) to the mammalian AMPA-selective GluR1–GluR4 subunits, was reported (Ultsch et al., 1992) to form homo-oligomeric channels (albeit inefficiently; maximum currents were of the order of 10 nA) that could be blocked by CNQX and PhTX-343. However, surprisingly, these could be gated only by kainate; the presumptive natural ligand, glutamate, was ineffective. The two other invertebrate GluR-like polypeptides that have been described are DNMDAR-I (Ultsch et al., 1993), which exhibits greatest similarity (46% identity) to the mammalian NR1 subunit, and Lym-eGluR1 (Hutton et al., 1991), which most closely resembles (43–46% identity) the mammalian AMPA-selective GluR1–GluR4 subunits. Neither DNMDAR-I nor Lym-eGluR1 form functional homo-oligomeric glutamate-gated channels when the corresponding in vitro-transcribed RNA is injected into Xenopus oocytes.
The polypeptide described here, Lym-eGluR2, exhibits 35, 32, and 27% identity to DGluR-I, DGluR-II, and DNMDAR-I, respectively; thus, it does not appear to be the species homolog of any of theDrosophila GluR polypeptides. Furthermore, it is the only invertebrate GluR protein that forms robust homo-oligomeric channels that can be gated by micromolar concentrations of a variety of glutamatergic agonists and blocked by several mammalian non-NMDA receptor antagonists. It therefore represents an excellent tool with which to dissect those polypeptide domains that are responsible for the functional differences between this receptor and vertebrate GluRs. It is unclear why Lym-eGluR2 readily forms glutamate-gated channels and Lym-eGluR1, DGluR-I, DGluR-II, and DNMDAR-I do not. However, it is well known that Drosophila ligand-gated ion-channel polypeptides rarely form functional homo-oligomers (Bertrand et al., 1994; Harvey et al., 1994b). In contrast, of seven full-length ligand-gated ion-channel cDNAs that this laboratory has isolated from Lymnaea, four have, to date, been shown to form functional GABA or glutamate receptors (Harvey et al., 1991; Darlison et al., 1994; Darlison et al., unpublished observations; this study).
Relationship of Lym-eGluR2 to a native receptor on buccal motorneurons
In situ hybridization has revealed that the Lym-eGluR2 transcript is distributed throughout the Lymnaea nervous system, being present in many cells within each of the nine central ganglia and the paired buccal ganglia; these data strongly suggest that Lym-eGluR2 has an important function in excitatory neurotransmission in this animal. Furthermore, in the buccal ganglia, the Lym-eGluR2 gene is expressed in an identifiable group of cells, namely the 4-cluster motorneurons, which are known to play a role in the feeding rhythm (Goldschmeding et al., 1977; Rose and Benjamin, 1979). Although it is currently unclear whether Lym-eGluR2 exists as a homo-oligomer in vivo, it is evident that the corresponding gene is not frequently coexpressed with any of the other four Lymnaea GluR-like genes that we have identified to date (i.e., those encoding Lym-eGluR1, clone #2, clone #13, and clone #19; data not shown).
Recently, a GluR has been characterized on buccal motorneurons of the closely related gastropod Helisoma trivolvis (Quinlan and Murphy, 1991; Quinlan et al., 1995). These neurons are excited by S2 interneurons of the tripartite feeding central pattern generator, which control the second phase of the bite cycle, namely the scraping of food particles from the substrate and the retraction of the radula (the first phase is the protraction of the radula to contact the food substrate, and the third phase is radular tensing to extrude the food down the esophagus). This excitation of the follower motorneurons could be mimicked by the application of either glutamate or kainate and blocked by CNQX (Quinlan and Murphy, 1991); quisqualate could not activate the excitatory GluR but could imitate the hyperpolarizing effect of glutamate on the same and other S2 follower motorneurons. Interestingly, the excitatory receptor did not show evidence of desensitization. This property might be expected to be of great importance for neurons that receive tonic input and which must rhythmically fire, such as those involved in patterned behaviors. In conclusion, the deduced location and functional properties of Lym-eGluR2 strongly suggest that it is the species homolog of (a component of) the excitatory GluR that has been identified onHelisoma buccal motorneurons (Quinlan and Murphy, 1991).
Footnotes
This work was supported in part by a research grant (BIO2-CT93-0243 to M.G.D.) and a concerted action (BIO2-CT93-0169) from the European Commission under the Biotechnology Programme 1992–1994, a European Research Grant from the European Science Foundation (M.G.D. and J.v.M.), and an international postdoctoral fellowship from the Wellcome Trust, United Kingdom (M.A.). We thank Hye-Chin Kim for expert technical assistance, Dr. Erno Vreugdenhil for the LymnaeacDNA library, Dr. Emily Liman for pGEMHE, Günter Ellinghausen and Hans-Hinrich Hönck for the preparation and injection of oocytes, Sönke Harder for DNA sequencing, Marion Däumigen-Kullmann for oligonucleotide synthesis, and Dr. Michael Pusch and Professors Thomas Jentsch and Dietmar Richter for help and access to electrophysiological equipment. M.G.D. also thanks Professor Dietmar Richter for his continued support.
Correspondence should be addressed to Dr. Mark G. Darlison, Institut für Zellbiochemie und Klinische Neurobiologie, Universitäts-Krankenhaus Eppendorf, Universität Hamburg, Martinistrasse 52, 20246 Hamburg, Germany.
REFERENCES
- 1.Bertrand D, Ballivet M, Gomez M, Bertrand S, Phannavong B, Gundelfinger ED. Physiological properties of neuronal nicotinic receptors reconstituted from the vertebrate β2 subunit and Drosophila α subunits. Eur J Neurosci. 1994;6:869–875. doi: 10.1111/j.1460-9568.1994.tb00997.x. [DOI] [PubMed] [Google Scholar]
- 2.Bettler B, Boulter J, Hermans-Borgmeyer I, O’Shea-Greenfield A, Deneris ES, Moll C, Borgmeyer U, Hollmann M, Heinemann S. Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron. 1990;5:583–595. doi: 10.1016/0896-6273(90)90213-y. [DOI] [PubMed] [Google Scholar]
- 3.Bettler B, Egebjerg J, Sharma G, Pecht G, Hermans-Borgmeyer I, Moll C, Stevens CF, Heinemann S. Cloning of a putative glutamate receptor: a low affinity kainate-binding subunit. Neuron. 1992;8:257–265. doi: 10.1016/0896-6273(92)90292-l. [DOI] [PubMed] [Google Scholar]
- 4.Bolshakov VY, Gapon SA, Magazanik LG. Different types of glutamate receptors in isolated and identified neurones of the mollusc Planorbarius corneus . J Physiol (Lond) 1991;439:15–35. doi: 10.1113/jphysiol.1991.sp018654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciabarra AM, Sullivan JM, Gahn LG, Pecht G, Heinemann S, Sevarino KA. Cloning and characterization of χ-1: a developmentally regulated member of a novel class of the ionotropic glutamate receptor family. J Neurosci. 1995;15:6498–6508. doi: 10.1523/JNEUROSCI.15-10-06498.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dale N, Kandel ER. l-Glutamate may be the fast excitatory transmitter of Aplysia sensory neurons. Proc Natl Acad Sci USA. 1993;90:7163–7167. doi: 10.1073/pnas.90.15.7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darlison MG, Stühmer T, Harvey RJ, Dautzenberg FM, Kim H-C, Zimmermann C, Amar M, Bermudez I, van Minnen J. Molecular characterisation and functional expression of molluscan ion-channel receptors that can be activated by either γ-aminobutyric acid or l-glutamate. Neth J Zool. 1994;44:473–485. [Google Scholar]
- 8.Dascal N. The use of Xenopus oocytes for the study of ion channels. CRC Crit Rev Biochem. 1987;22:317–387. doi: 10.3109/10409238709086960. [DOI] [PubMed] [Google Scholar]
- 9.Egebjerg J, Heinemann SF. Ca2+permeability of unedited and edited versions of the kainate selective glutamate receptor GluR6. Proc Natl Acad Sci USA. 1993;90:755–759. doi: 10.1073/pnas.90.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egebjerg J, Bettler B, Hermans-Borgmeyer I, Heinemann S. Cloning of a cDNA for a glutamate receptor subunit activated by kainate but not AMPA. Nature. 1991;351:745–748. doi: 10.1038/351745a0. [DOI] [PubMed] [Google Scholar]
- 11.Foster AC, Fagg GE. Acidic amino acid binding sites in mammalian neuronal membranes: their characteristics and relationship to synaptic receptors. Brain Res Rev. 1984;7:103–164. doi: 10.1016/0165-0173(84)90020-1. [DOI] [PubMed] [Google Scholar]
- 12.Frohman MA, Martin GR. Rapid amplification of cDNA ends using nested primers. Technique. 1989;1:165–170. [Google Scholar]
- 13.Goldschmeding JT, Bruins HJ, Everts WM. Topography of buccal and cerebral neurones involved in feeding in the freshwater snail Lymnaea stagnalis . Proc K Ned Acad Wet. 1977;80:83–96. [Google Scholar]
- 14.Gregor P, Mano I, Maoz I, McKeown M, Teichberg VI. Molecular structure of the chick cerebellar kainate-binding subunit of a putative glutamate receptor. Nature. 1989;342:689–692. doi: 10.1038/342689a0. [DOI] [PubMed] [Google Scholar]
- 15.Harvey RJ, Vreugdenhil E, Zaman SH, Bhandal NS, Usherwood PNR, Barnard EA, Darlison MG. Sequence of a functional invertebrate GABAA receptor subunit which can form a chimeric receptor with a vertebrate α subunit. EMBO J. 1991;10:3239–3245. doi: 10.1002/j.1460-2075.1991.tb04887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvey RJ, Kim H-C, Darlison MG. Expression patterns of the GABAA receptor α1-, β2-, γ2- and γ4-subunit genes in chick retina. Neurosci Res Commun. 1994a;15:95–102. [Google Scholar]
- 17.Harvey RJ, Schmitt B, Hermans-Borgmeyer I, Gundelfinger ED, Betz H, Darlison MG. Sequence of a Drosophila ligand-gated ion-channel polypeptide with an unusual amino-terminal extracellular domain. J Neurochem. 1994b;62:2480–2483. doi: 10.1046/j.1471-4159.1994.62062480.x. [DOI] [PubMed] [Google Scholar]
- 18.Herb A, Burnashev N, Werner P, Sakmann B, Wisden W, Seeburg PH. The KA-2 subunit of excitatory amino acid receptors shows widespread expression in brain and forms ion channels with distantly related subunits. Neuron. 1992;8:775–785. doi: 10.1016/0896-6273(92)90098-x. [DOI] [PubMed] [Google Scholar]
- 19.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 20.Hollmann M, O’Shea-Greenfield A, Rogers SW, Heinemann S. Cloning by functional expression of a member of the glutamate receptor family. Nature. 1989;342:643–648. doi: 10.1038/342643a0. [DOI] [PubMed] [Google Scholar]
- 21.Hollmann M, Maron C, Heinemann S. N-glycosylation site tagging suggests a three transmembrane domain topology for the glutamate receptor GluR1. Neuron. 1994;13:1331–1343. doi: 10.1016/0896-6273(94)90419-7. [DOI] [PubMed] [Google Scholar]
- 22.Hutton ML, Harvey RJ, Barnard EA, Darlison MG. Cloning of a cDNA that encodes an invertebrate glutamate receptor subunit. FEBS Lett. 1991;292:111–114. doi: 10.1016/0014-5793(91)80846-u. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda K, Nagasawa M, Mori H, Araki K, Sakimura K, Watanabe M, Inoue Y, Mishina M. Cloning and expression of the ε4 subunit of the NMDA receptor channel. FEBS Lett. 1992;313:34–38. doi: 10.1016/0014-5793(92)81178-o. [DOI] [PubMed] [Google Scholar]
- 24.Ikemoto Y, Akaike N. The glutamate-induced chloride current in Aplysia neurones lacks pharmacological properties seen for excitatory responses to glutamate. Eur J Pharmacol. 1988;150:313–318. doi: 10.1016/0014-2999(88)90012-x. [DOI] [PubMed] [Google Scholar]
- 25.Jan LY, Jan YN. l-Glutamate as an excitatory transmitter at the Drosophila larval neuromuscular junction. J Physiol (Lond) 1976;262:215–236. doi: 10.1113/jphysiol.1976.sp011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katz PS, Levitan IB. Quisqualate and ACPD are agonists for a glutamate-activated current in identified Aplysia neurons. J Neurophysiol. 1993;69:143–150. doi: 10.1152/jn.1993.69.1.143. [DOI] [PubMed] [Google Scholar]
- 27.Kehoe J. Transformation by concanavalin A of the response of molluscan neurones to l-glutamate. Nature. 1978;274:866–869. doi: 10.1038/274866a0. [DOI] [PubMed] [Google Scholar]
- 28.Keinänen K, Wisden W, Sommer B, Werner P, Herb A, Verdoorn TA, Sakmann B, Seeburg PH. A family of AMPA-selective glutamate receptors. Science. 1990;249:556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- 29.Kemenes G, Daykin K, Elliott CJH. Photoinactivation of neurones axonally filled with the fluorescent dye 5(6)-carboxyfluorescein in the pond snail, Lymnaea stagnalis . J Neurosci Methods. 1991;39:207–216. doi: 10.1016/0165-0270(91)90099-l. [DOI] [PubMed] [Google Scholar]
- 30.Kemp BE, Pearson RB. Protein kinase recognition sequence motifs. Trends Biochem Sci. 1990;15:342–346. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- 31.Kimura N, Kurosawa N, Kondo K, Tsukada Y. Molecular cloning of the kainate-binding protein and calmodulin genes which are induced by an imprinting stimulus in ducklings. Mol Brain Res. 1993;17:351–355. doi: 10.1016/0169-328x(93)90022-h. [DOI] [PubMed] [Google Scholar]
- 32.Köhler M, Burnashev N, Sakmann B, Seeburg PH. Determinants of Ca2+permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron. 1993;10:491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- 33.Kutsuwada T, Kashiwabuchi N, Mori H, Sakimura K, Kushiya E, Araki K, Meguro H, Masaki H, Kumanishi T, Arakawa M, Mishina M. Molecular diversity of the NMDA receptor channel. Nature. 1992;358:36–41. doi: 10.1038/358036a0. [DOI] [PubMed] [Google Scholar]
- 34.Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 35.Lomeli H, Wisden W, Köhler M, Keinänen K, Sommer B, Seeburg PH. High-affinity kainate and domoate receptors in rat brain. FEBS Lett. 1992;307:139–143. doi: 10.1016/0014-5793(92)80753-4. [DOI] [PubMed] [Google Scholar]
- 36.Lomeli H, Sprengel R, Laurie DJ, Köhr G, Herb A, Seeburg PH, Wisden W. The rat delta-1 and delta-2 subunits extend the excitatory amino acid receptor family. FEBS Lett. 1993;315:318–322. doi: 10.1016/0014-5793(93)81186-4. [DOI] [PubMed] [Google Scholar]
- 37.Mat Jais AM, Kerkut GA, Walker RJ. The ionic mechanisms associated with the excitatory response of kainate,l-glutamate, quisqualate, ibotenate, AMPA and methyltetrahydrofolate on leech Retzius cells. Comp Biochem Physiol [C] 1984;77:115–126. doi: 10.1016/0742-8413(84)90139-7. [DOI] [PubMed] [Google Scholar]
- 38.Mathers DA, Usherwood PNR. Concanavalin A blocks desensitisation of glutamate receptors on insect muscle fibres. Nature. 1976;259:409–411. doi: 10.1038/259409a0. [DOI] [PubMed] [Google Scholar]
- 39.Mayer ML, Vyklicky L. Concanavalin A selectively reduces desensitization of mammalian neuronal quisqualate receptors. Proc Natl Acad Sci USA. 1989;86:1411–1415. doi: 10.1073/pnas.86.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meguro H, Mori H, Araki K, Kushiya E, Kutsuwada T, Yamazaki M, Kumanishi T, Arakawa M, Sakimura K, Mishina M. Functional characterization of a heteromeric NMDA receptor channel expressed from cloned cDNAs. Nature. 1992;357:70–74. doi: 10.1038/357070a0. [DOI] [PubMed] [Google Scholar]
- 41.Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- 42.Morita T, Sakimura K, Kushiya E, Yamazaki M, Meguro H, Araki K, Abe T, Mori KJ, Mishina M. Cloning and functional expression of a cDNA encoding the mouse β2 subunit of the kainate-selective glutamate receptor channel. Mol Brain Res. 1992;14:143–146. doi: 10.1016/0169-328x(92)90023-5. [DOI] [PubMed] [Google Scholar]
- 43.Moriyoshi K, Masu M, Ishii T, Shigemoto R, Mizuno N, Nakanishi S. Molecular cloning and characterization of the rat NMDA receptor. Nature. 1991;354:31–37. doi: 10.1038/354031a0. [DOI] [PubMed] [Google Scholar]
- 44.Nakanishi N, Shneider NA, Axel R. A family of glutamate receptor genes: evidence for the formation of heteromultimeric receptors with distinct channel properties. Neuron. 1990;5:569–581. doi: 10.1016/0896-6273(90)90212-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan EM, Murphy AD. Glutamate as a putative neurotransmitter in the buccal central pattern generator of Helisoma trivolvis . J Neurophysiol. 1991;66:1264–1271. doi: 10.1152/jn.1991.66.4.1264. [DOI] [PubMed] [Google Scholar]
- 46.Quinlan EM, Gregory K, Murphy AD. An identified glutamatergic interneuron patterns feeding motor activity via both excitation and inhibition. J Neurophysiol. 1995;73:945–956. doi: 10.1152/jn.1995.73.3.945. [DOI] [PubMed] [Google Scholar]
- 47.Roberts CJ, Nielsen E, Krogsgaard-Larsen P, Walker RJ. The actions of ibotenate, homoibotenate analogues and AMPA on central neurons of Hirudo , Limulus and Helix . Comp Biochem Physiol [C] 1982;73:439–444. [Google Scholar]
- 48.Rose RM, Benjamin PR. The relationship of the central motor pattern to the feeding cycle of Lymnaea stagnalis . J Exp Biol. 1979;80:137–163. doi: 10.1242/jeb.80.1.137. [DOI] [PubMed] [Google Scholar]
- 49.Sakimura K, Bujo H, Kushiya E, Araki K, Yamazaki M, Yamazaki M, Meguro H, Warashina A, Numa S, Mishina M. Functional expression from cloned cDNAs of glutamate receptor species responsive to kainate and quisqualate. FEBS Lett. 1990;272:73–80. doi: 10.1016/0014-5793(90)80452-o. [DOI] [PubMed] [Google Scholar]
- 50.Sakimura K, Morita T, Kushiya E, Mishina M. Primary structure and expression of the γ2 subunit of the glutamate receptor channel selective for kainate. Neuron. 1992;8:267–274. doi: 10.1016/0896-6273(92)90293-m. [DOI] [PubMed] [Google Scholar]
- 51.Schuster CM, Ultsch A, Schloss P, Cox JA, Schmitt B, Betz H. Molecular cloning of an invertebrate glutamate receptor subunit expressed in Drosophila muscle. Science. 1991;254:112–114. doi: 10.1126/science.1681587. [DOI] [PubMed] [Google Scholar]
- 52.Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 53.Sommer B, Burnashev N, Verdoorn TA, Keinänen K, Sakmann B, Seeburg PH. A glutamate receptor channel with high affinity for domoate and kainate. EMBO J. 1992;11:1651–1656. doi: 10.1002/j.1460-2075.1992.tb05211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sucher NJ, Akbarian S, Chi CL, Leclerc CL, Awobuluyi M, Deitcher DL, Wu MK, Yuan JP, Jones EG, Lipton SA. Developmental and regional expression of a novel NMDA receptor-like subunit (NMDAR-L) in the rodent brain. J Neurosci. 1995;15:6509–6520. doi: 10.1523/JNEUROSCI.15-10-06509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ultsch A, Schuster CM, Laube B, Schloss P, Schmitt B, Betz H. Glutamate receptors of Drosophila melanogaster : Cloning of a kainate-selective subunit expressed in the central nervous system. Proc Natl Acad Sci USA. 1992;89:10484–10488. doi: 10.1073/pnas.89.21.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ultsch A, Schuster CM, Laube B, Betz H, Schmitt B. Glutamate receptors of Drosophila melanogaster : Primary structure of a putative NMDA receptor protein expressed in the head of the adult fly. FEBS Lett. 1993;324:171–177. doi: 10.1016/0014-5793(93)81387-f. [DOI] [PubMed] [Google Scholar]
- 57.Vernino S, Amador M, Luetje CW, Patrick J, Dani JA. Calcium modulation and high calcium permeability of neuronal nicotinic acetylcholine receptors. Neuron. 1992;8:127–134. doi: 10.1016/0896-6273(92)90114-s. [DOI] [PubMed] [Google Scholar]
- 58.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wada K, Dechesne CJ, Shimasaki S, King RG, Kusano K, Buonanno A, Hampson DR, Banner C, Wenthold RJ, Nakatani Y. Sequence and expression of a frog brain complementary DNA encoding a kainate-binding protein. Nature. 1989;342:684–689. doi: 10.1038/342684a0. [DOI] [PubMed] [Google Scholar]
- 60.Werner P, Voigt M, Keinänen K, Wisden W, Seeburg PH. Cloning of a putative high-affinity kainate receptor expressed predominantly in hippocampal CA3 cells. Nature. 1991;351:742–744. doi: 10.1038/351742a0. [DOI] [PubMed] [Google Scholar]
- 61.Wisden W, Morris BJ, Hunt SP. In situ hybridization with synthetic DNA probes. In: Chad J, Wheal H, editors. Molecular neurobiology—a practical approach, Vol. 2. IRL Press; Oxford: 1991. pp. 205–225. [Google Scholar]
- 62.Wo ZG, Oswald RE. Transmembrane topology of two kainate receptor subunits revealed by N-glycosylation. Proc Natl Acad Sci USA. 1994;91:7154–7158. doi: 10.1073/pnas.91.15.7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woodgett JR, Gould KL, Hunter T. Substrate specificity of protein kinase C. Use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur J Biochem. 1986;161:177–184. doi: 10.1111/j.1432-1033.1986.tb10139.x. [DOI] [PubMed] [Google Scholar]
- 64.Yamazaki M, Araki K, Shibata A, Mishina M. Molecular cloning of a cDNA encoding a novel member of the mouse glutamate receptor channel family. Biochem Biophys Res Commun. 1992;183:886–892. doi: 10.1016/0006-291x(92)90566-4. [DOI] [PubMed] [Google Scholar]
- 65.Yarowsky PJ, Carpenter DO. Aspartate: distinct receptors on Aplysia neurons. Science. 1976;192:807–809. doi: 10.1126/science.4895. [DOI] [PubMed] [Google Scholar]



